Abstract
A yeast two-hybrid screen searching for chromosomally encoded proteins that interact with the Agrobacterium tumefaciens VirB8 protein was carried out. This screen identified an interaction candidate homologous to the partial sequence of a gene that had previously been identified in a transposon screen as a potential regulator of virG expression, chvD. In this report, the cloning of the entire chvD gene is described and the gene is sequenced and characterized. Insertion of a promoterless lacZ gene into the chvD locus greatly attenuated virulence and vir gene expression. Compared to that of the wild-type strain, growth of the chvD mutant was reduced in rich, but not minimal, medium. Expression of chvD, as monitored by expression of β-galactosidase activity from the chvD-lacZ fusion, occurred in both rich and minimal media as well as under conditions that induce virulence gene expression. The ChvD protein is highly homologous to a family of ATP-binding cassette transporters involved in antibiotic export from bacteria and has two complete Walker box motifs. Molecular genetic analysis demonstrated that disruption of either Walker A box, singly, does not inactivate this protein's effect on virulence but that mutations in both Walker A boxes renders it incapable of complementing a chvD mutant strain. Constitutive expression of virG in the chvD mutant strain restored virulence, supporting the hypothesis that ChvD controls virulence through effects on virG expression.
Agrobacterium tumefaciens is a gram-negative bacterium capable of transferring DNA from the tumor-inducing (Ti) plasmid into the nucleus of a higher plant cell, where it can be integrated into the chromosomal genome and expressed (for a review, see reference 19). The processes that are involved in this transfer have been extensively studied and include (i) activation of virulence gene expression by plant-derived xenobiotics such as phenolic compounds and monosaccharides, (ii) production and processing of proteins and protein-DNA complexes that will be transferred into the plant cell, and (iii) construction of a complex in the bacterial membrane system that is used to transport these substrates.
Activation of virulence gene expression in Agrobacterium is under complex control, and the system has not yet been completely characterized. The central regulatory scheme is comprised of the VirA and VirG proteins, which are highly homologous to two-component systems found in a wide variety of prokaryotes (8, 41). VirA is a membrane-bound histidine kinase that is critical for signal input, and VirG is the response regulator that, after phosphorylation by VirA, can efficiently bind to the vir gene promoters and activate gene expression. Multiple signals are involved in this regulatory pathway. In most strains, phenolic compounds are absolutely required for the induction of vir gene expression, though the site of signal recognition—e.g., VirA or some other protein—and the mechanism of signal transduction have not been elucidated (9, 26, 33). Monosaccharides such as arabinose or glucose lower the concentrations of phenolic compounds necessary for activation of vir gene expression (3, 36). This occurs as a result of the activities of ChvE, a chromosomally encoded sugar-binding protein that interacts with VirA (10, 37). In addition to phenolic compounds and monosaccharides, several other environmental cues are known to affect virulence gene expression through the VirA-VirG system. Low pH stimulates vir gen expression by at least two mechanisms: one that is VirA dependent and one that activates the P2 promoter of virG independently of VirA (11, 27, 43). Low PO4 has been shown to upregulate expression of virG through effects on the P1 promoter of virG (40). Finally, an elevated temperature downregulates vir gene induction, probably because of the temperature sensitivity of VirA (30). In none of these cases are the mechanisms of signal recognition or transduction understood.
The VirB proteins comprise a membrane-bound complex that is necessary for T-DNA transport to the plant cell and are homologous to the type IV transporter systems used by many bacteria to export either proteins or DNA-protein complexes (13, 42). Because VirB8 localizes to specific sites in the membrane system (24), we undertook a search for proteins that may serve as VirB8 anchors by using the periplasmic domain of VirB8 (VirB8′) as bait in a Saccharomyces cerevisiae two-hybrid screen of the Agrobacterium genome. This screen yielded two proteins that are strong interactors with VirB8. Genetic analysis demonstrated that one of these is involved in virulence but that the other is not. The gene affecting virulence, chvD, has been previously identified in a screen searching for mutations that affect the capacity of the virG promoter to be activated at low pH and low PO4 (43). This gene, originally discovered in a transposon mutagenesis screen of A. tumefaciens chromosomes, was not isolated. In this study, the intact chvD gene was cloned and sequenced and its expression, role in vir gene expression, and effect on growth in rich medium were characterized.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Table 1 lists the plasmids, bacterial strains, and yeast strains used in this study. Plasmids were introduced into Escherichia coli and A. tumefaciens by electroporation.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1/λpir | RK2 tra regulon, pir negative, host for pir-dependent plasmid | 22 |
| DH5α | λ−φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1 | GIBCO BRL |
| A. tumefaciens | ||
| A348 | C58 chromosomal background carrying pTiA6 | 18 |
| AB20 | A348 chvD gene was disrupted by insertion of the pZL101 suicide plasmid, Kanr | This study |
| AB21 | A348 “21-5” gene was disrupted by insertion of the pZL102 suicide plasmid, Kanr | This study |
| AB22 | A348 chvD gene was disrupted by insertion of the pZL103 suicide plasmid, Kanr | This study |
| S. cerevisiae EGY 48 | MATα trp1 his3 ura3 lexAops-LEU2 | 20 |
| Plasmids | ||
| pBSII | ColE1 Ampr, cloning vector | Stratagene |
| pJB20 | Broad-host-range plasmid, derivative of pUCD2; IncW Ampr Tetr | 5 |
| pJB93 | A 528-bp virB8 EcoRI fragment without a transmembrane domain in pJK202, HIS3 Ampr | This study |
| pJG4-5 (pV0) | Cloning prey vector for the construction of activator fusion, TRP1 Ampr | 20 |
| pJK202 | Cloning bait vector for the construction of LexA fusions, HIS3 Ampr | 20 |
| pMutA-G665D | IncRi derivative plasmid carrying virA (G665D) and wild-type virG, Ampr | 29 |
| pSH18-34 | Contains Gal1-LexAop-lacZ, a reporter gene in yeast, URA3 Ampr | 20 |
| pV1 | Modified version of pJG4-5 with EcoRI site carrying +1 reading frame, TRP1 Ampr | This study |
| pV2 | Modified version of pJG4-5 with EcoRI site carrying +2 reading frame, TRP1 Ampr | This study |
| pV01-1 | 835-bp Tsp509I fragment of A348 DNA containing a portion of chvD in pJG4-5, TRP1 Ampr | This study |
| pV21-5 | 468-bp Tsp509I fragment of A348 DNA in pV2, TRP1 Ampr | This study |
| pVK112 | R6K suicide vector, lacZY for transcriptional fusions, Kanr | 22 |
| pYW12 | pJB20 derivative, with lac promoter followed by the multiple cloning site from pUC19; Ampr | 39 |
| pYW15c | Broad-host-range expression vector, PN25-MCS-STOP, IncW and pBR322ori, Ampr | 39 |
| pYW48 | pYW15 derivative plasmid with PN25∷virG and wild-type virA, IncW Ampr | 39 |
| pZL35 | 2.7-kb EcoRV fragment containing the chvD gene from pZL78 cloned into pBSII, Ampr | This study |
| pZL39 | 2.7-kb EcoRV chvD-containing fragment from pZL78 cloned into pJB20 ScaI site, Tetr | This study |
| pZL78 | 7.8-kb HindIII fragment including chvD open reading frame cloned into pBSII, Ampr | This study |
| pZL81 | 2.7-kb PCR fragment containing chvD open reading frame and promoter cloned into pYW12 KpnI site; Ampr | This study |
| pZL82 | pZL81 derivative with mutation in Walker A1 ATP-binding site of chvD; Ampr | This study |
| pZL83 | pZL81 derivative with mutation in Walker A2 ATP-binding site of chvD; Ampr | This study |
| pZL84 | pZL81 derivative with mutations in both Walker A ATP-binding sites of chvD; Carr | This study |
| pZL101 | 449-bp chvD (nucleotides 335–784) PCR fragment from pV01-1 cloned into EcoRI site of pVK112 in the same orientation as lacZ; Kanr | This study |
| pZL102 | 398-bp v21-5 (nucleotides 28–426) PCR fragment from pV21-5 cloned into EcoRI site of pVK112; Kanr | This study |
| pZL103 | 449-bp chvD (nucleotides 335–784) PCR fragment from pV01-1 cloned into EcoRI site of pVK112 in the orientation opposite to that of lacZ; Kanr | This study |
DNA manipulations.
Standard methods were used for plasmid isolation, PCR amplification, restriction digestion, and DNA gel electrophoresis and blotting. Genomic DNA was extracted from A. tumefaciens strain A348 by the method described in reference 12. For Southern hybridization, probe DNA was labeled with digoxigenin-11-UTP (Boehringer Mannheim) during PCR amplification and signal was detected using the chemiluminescent CPD-Star detection system according to the recommended procedure of the manufacturer (Boehringer Mannheim).
vir gene induction, growth, and virulence assays.
The bacterial growth media and growth conditions as well as procedures for vir gene induction have been described previously (9). To determine the difference in growth activities among wild-type A348, AB20, and its derivatives, the strains were grown in liquid AB minimal medium (43) with antibiotic at 25°C overnight. Equal amounts of cells (based on their optical densities at 600 nm [OD600s]) were then transferred to L broth, and the OD600 of each culture was determined every 4 h. Virulence assays were performed according to the method of Banta et al. (4). Briefly, overnight cultures (adjusted to an OD600 of 0.5) of Agrobacterium were cocultivated with Nicotiana tabacum cv. Havana 425 leaf squares on hormone-free MS medium (32) supplemented with 300 μM acetosyringone (AS). After 2 days, the leaf squares were transferred to hormone-free MS medium containing vancomycin (200 μg/ml) and timentin (200 μg/ml) and cultured at 25°C in the dark. After 10 days, the tumors on each leaf piece were counted and photographed.
Immunoblotting.
Equal numbers of cells grown in AB induction medium (43) with or without AS (Sigma) were collected, resuspended in sample buffer, and boiled for 8 min. Ten microliters of sample was loaded, and proteins were separated by sodium dodecyl sulfate–10 to 12% polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane by electrotransfer, and probed with antibodies directed against VirA (39), VirE2 (7), and VirB8 (15) by chemiluminescent-detection procedures described previously (5).
Yeast two-hybrid vector construction, genomic libraries, and library screening.
The pJG4-5 prey vector was mutagenized so that the EcoRI cloning site for fusion protein production existed in altered reading frames. This was accomplished by cloning double-stranded oligonucleotides, with an altered EcoRI cloning site that carried a different reading frame, into pJG4-5 digested with EcoRI and XhoI. For vector 1 (pV1), the mutagenic oligonucleotide consisted of oligonucleotides 1a (5′AATTGGGAATTCGGCCGAC3′) and 1b (5′TCGAGTCGGCCGAATTCCC3′); for vector 2 (pV2), it consisted of oligonucleotides 2a (5′AATTGAATTCGGCCGAC3′) and 2b (5′TCGAGTCGGCCGAATTC3′). Each modified vector was confirmed by DNA sequence analysis. Genomic libraries in these prey vectors were constructed with A348 genomic DNA partially digested with Tsp509I, a 4-bp cutter that yields the same 5′ overhangs as EcoRI. The digested sample was size selected by purifying the 500- to 3,000-bp fragments from a preparative agarose gel. This DNA was cloned into the EcoRI sites of the three pJG4-5-derived prey vectors and transformed into E. coli DH5α with 39,200 colonies recovered from the library in pJG4-5, 40,400 colonies from pV1, and 21,300 colonies from pV2. Plasmid pJB93, a bait plasmid, was constructed by amplifying a 528-bp PCR fragment encoding amino acid residues 56 to 231 of virB8 on plasmid pJW239 (6) with primers JB7-8 (5′CGAATTCCCCGTTGAGCAGGCTTCTGCCC3′) and JB8-8 (5′CGAATTCATGGTGCGCCCTGGCCTAC3′). This fragment, lacking the N-terminal transmembrane domain of VirB8 (16), was cloned into the EcoRI site of plasmid pJK202, yielding pJB93, and is referred to as VirB8′. For the two-hybrid library screen, the prey plasmids were isolated with the three libraries and transformed into S. cerevisiae strain EGY48 that contains pSH18-34, a reporter plasmid carrying the Gal1-lexop-lacZ gene cluster, and the VirB8′ bait plasmid pJB93. The pJG4-5 empty vector was also transformed into EGY48(pJB93, pSH18-34) as a control. The basic protocol and all strains and plasmids used in the assay are as described by Golemis et al. (20). Positive clones in the library screens were identified as only those colonies passing both the β-galactosidase production and the leucine prototrophy screens. After sequence analysis, two clones were chosen for further characterization. pV01-1 was chosen because it had homology to a previously identified virulence gene, chvD (43), and pV21-5 was chosen because it had an approximately 15-kDa peptide fused to the activator domain of the prey vector.
Insertion mutagenesis of chvD and v21-5.
The suicide plasmid pVK112 carries a unique EcoRI site followed by a promoterless lacZ reporter gene as well as a kanamycin selection marker (22). It also contains the vegetative origin of R6K but lacks the plasmid's pir gene and can therefore replicate only in strains that contain pir on a separate replicon, such as E. coli S17-1/λpir, but not in Agrobacterium. pZL101 contains a 449-bp internal chvD PCR fragment derived from pV01-1 cloned such that the chvD fragment is in the same orientation as the promoterless lacZ gene, while pZL103 carries the same fragment cloned into pVK112 in the opposite orientation. pZL102 contains a 398-bp internal v21-5 PCR fragment cloned in the same orientation as the lacZ gene of the pVK112. Homologous recombination of these plasmids in the respective Agrobacterium chromosomal locus creates an insertion mutation at the gene along with a lacZ transcriptional fusion (22). pZL101 and pZL102 were electroporated into A348, and the cells were plated on kanamycin (100 μg/ml) plus X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The blue colonies are the candidate mutants in which the chromosomal chvD gene or v21-5 gene was disrupted by insertion of pZL101 or pZL102, yielding strain AB20 or AB21, respectively. In the case of pZL103, white, kanamycin-resistant colonies were obtained and yielded strain AB22. The mutants were confirmed by PCR with one primer annealing to the lacZ gene (AlacZ, 5′CCGCCACATATCCTGATCTT3′) and another primer being external to the 5′ terminus of the chvD or v21-5 fragments cloned into pVK112 (for chvD, 5′CCGCTACAACGAATTGATGA3′, and for v21-5, 5′TTCGTTCTCCCGGGTACTGC3′). The integration events were also checked by plasmid rescue in which the genomic DNA of each putative mutant was digested with restriction enzymes (BstEII, NotI, and XmnI) that do not cut within the plasmids. These fragments were circularized with T4 DNA ligase and electroporated into S17-1/λpir. Primers external and internal to the chvD gene were used on these rescued plasmids to determine the precise insertion sites by sequence. Finally, the genomic DNAs of these two mutants were digested with BamHI and XmnI, Southern blotted, and probed with PCR fragments from chvD, v21-5, and neo (from pVIK112). The predicted extra hybridization band or larger band was seen from the mutant but not from wild-type A348, confirming the insertion.
Cloning of the full-length chvD gene.
BamHI, ClaI, EcoRV, HindIII, and SmaI digests of A348 genomic DNA were electrophoresed on an agarose gel and transferred by capillary blotting to a Hybond-N membrane (Amersham). A 450-bp chvD probe was obtained by incorporation of alkali-labile digoxigenin-11-dUTP (Boehringer Mannheim) during PCR amplification from pV01-1 with the primers ChvD1 (5′CCGACGAAACGGCGGATGA3′) and AchvD-1 (5′CCTTCTGGCGCGAGGCGTC3′). The hybridization was carried out under stringent conditions as recommended by the manufacturer, and detection was performed with a chemiluminescent CPD-Star detection system purchased from Boehringer Mannheim. A single 7.8-kb hybridization band from the HindIII digest was detected. A preparative HindIII digest was electrophoresed, and the DNA in the 7.0- to 8.5-kb range was isolated and cloned into pBluescript II (pBSII) and then transformed to DH5α. Approximately 2,000 transformants were screened by colony hybridization using the chvD probe, and a single colony containing the 7.8-kb HindIII fragment was identified. The plasmid carrying this fragment was designated pZL78.
Sequence analysis of chvD.
A 2.7-kb EcoRV fragment encompassing the whole chvD gene was subcloned from pZL78 into pBSII, yielding pZL35, and into the broad-host-range vector pJB20, yielding pZL39. pZL78 and pZL35 were sequenced using T3 and T7 primers and primers from known chvD sequences (AChvD4, 5′CCTCATCCGCCGTTTCGTCG3′; ChvD3, 5′GTTATGACGAGCTGGTGGAAG3′). Additional new primers were designed on the basis of determined chvD sequence results and used to obtain additional sequences upstream and downstream of the chvD gene. The DNA sequences were compiled and analyzed through BLAST searches (2).
Construction of mutations in the NTP-binding sites of chvD.
Mutations in the chvD Walker A ATP-binding sites were generated by overlap extension PCR mutagenesis (31). PCR was carried out with Pfu DNA polymerase (Stratagene) and primers designated as follows: chvD35, 5′CGGGGTACCCCGAAAACATCCTGCCAGAGC3′; chvd35-stop, 5′CGGGGTACCCCGACAAGAGGATCAGCGCGT3′; walkA1, 5′ACGGCGCCGGTGAATCGACCGTTCT3′; AwalkA1, 5′AGAACGGTCGATTCACCGGCGCCGT3′; walkA2, 5′ACGGTGCAGGTGAGACCACACTGTT3′; and AwalkA2, 5′AACAGTGTGGTCTCACCTGCACCGT3′. Primer chvD-35 and primer chvD35-stop contain 9 nucleotides for constructing a KpnI site (underlined). A 2.5-kb PCR fragment was amplified from pZL35 and cloned into pYW12, yielding pZL81. Primers walkA1 and AwalkA1 contain a mutation in the first ATP-binding site, and primers walkA2 and AwalkA2 contain a mutation in their second ATP-binding sites, which were underlined in the primers. The first Walker A box ATP-binding site (Gly-Pro-Asn-Gly-Ala-Gly-Lys-Ser) between amino acid residues 39 and 46 was mutated by the primers walkA1 and AwalkA1. The Lys-45 codon was changed to Glu with four primers and two rounds of PCR. The first-round PCR had two reactions and used pZL35 as the template; one reaction was with primers chvD-35 and Awalk1, and the other was with primers chvD-stop and walkA1. These two PCR products were run on a 1% agarose gel, isolated, and then combined in a new PCR mixture with primers chvD-35 and chvD-stop. The amplified fragment that had the mutation in the Walker A ATP-binding site was digested by KpnI and cloned into pYW12 to make pZL82. The Lys-45 change to Glu was confirmed by sequencing. In the same way, pZL81 was mutagenized at the second Walker A ATP-binding site between amino acid residues 350 and 357 (Gly-Pro-Asn-Gly-Ala-Gly-Lys-Thr) with primers walkA2 and AwalkA2, resulting in Lys-356 being changed to Glu in plasmid pZL83. pZL84, containing chvD with mutations at each of these two ATP-binding sites, was constructed with walkA2 and AwalkA2 and pZL82 as the template. The absence of random mutations was verified by sequence analysis.
Nucleotide sequence accession number.
The GenBank accession number of the 2.7-kb fragment containing chvD and the surrounding sequence is AY027490.
RESULTS
Construction and screening of an Agrobacterium genomic library in the yeast two-hybrid system.
VirB8, encoded by pTiA6, is a membrane-bound protein with a large C-terminal periplasmic domain. This region of the protein, referred to here as VirB8′, includes amino acids 164 to 692 of the VirB8 protein. Earlier studies using the yeast two-hybrid system (reference 17 and our unpublished observations) demonstrated interactions between VirB8′ and itself, VirB9, and VirB10. Given its apparently critical role in the construction of the VirB complex (24), we sought to determine whether VirB8′ also interacted with other Agrobacterium proteins. For these studies we constructed genomic libraries using total DNA from A. tumefaciens strain A348, which carries the two Agrobacterium chromosomes, the Ti plasmid, and the so-called cryptic plasmid pAtC58 (18). A partial Tsp509I digest was size selected (0.5 to 3 kb) and cloned into three different prey vectors (see Materials and Methods) that contain an EcoRI cloning site in each open reading frame. Screening of these libraries with VirB8′ in the bait yielded 10 to 15 colonies per vector that were positive for β-galactosidase activity and were leucine prototrophic, as is expected for interactors. Western analysis demonstrated that five of these interactors had significant fusions (>5 kDa) to the activator domain of the prey vector and were characterized further. The plasmids carrying positive interactors were isolated, transformed into E. coli, and used for sequence analysis.
Characterization of positive interactors.
Sequence analysis of the positive interactors showed that three had identical 835-bp inserts in pV0 (= pJG4-5) and that two had identical 478-bp inserts in pV2. BLAST searches showed that the inserts pV01-1, pV01-4, and pV01-5 had 100% homology with the partial sequence available for chromosomal virulence gene D (chvD) in A. tumefaciens but that the inserts in pV21-2 and pV21-5 had no homologies in the database. As described in Materials and Methods, internal fragments of approximately 400 bp from either pV01-1 or pV21-5 were cloned into pVIK112, a plasmid that carries a promoterless lacZ gene and cannot replicate in Agrobacterium (22), yielding pZL101 or pZL102, respectively. The resultant plasmids must recombine into either the chromosome or resident plasmids in order to confer antibiotic resistance on the cell. Recombination of this plasmid into the appropriate target site was confirmed (see Materials and Methods), and the strains were tested for virulence and their capacity to express β-galactosidase. With AB21 (insert from pV21-5), the lacZ gene was expressed in both rich (L broth) and minimal AB medium, indicating insertion into a functional operon, but there was no effect on virulence.
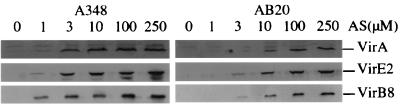
The insertion of pZL101 into the chvD locus resulted in a strain (AB20) that expressed β-galactosidase under all conditions tested, and phenolic compounds that induce the vir genes did not affect this expression either positively or negatively (data not shown). A second insertion at the chvD locus, with pZL103, inserted lacZ in the opposite orientation. The strain carrying this insertion, AB22, did not exhibit β-galactosidase activity under any of the conditions tested. Both AB20 and AB22 were, as expected, attenuated in virulence when they were tested on either Kalanchoe or tobacco (see below). A severe reduction in the capacity of AB20 to induce the expression of virulence genes in response to AS was observed (Fig. 1). Finally, both mutant strains exhibited a pronounced reduction in growth rate compared to that of the wild type in L broth but not minimal AB medium (data not shown; see below).
FIG. 1.
vir gene expression in A. tumefaciens strains A348 (wild type) and AB20 (chvD mutant). Bacterial cells were induced with the indicated concentrations of AS, and immunoblots were probed with antibodies directed against the indicated Vir proteins.
Cloning and sequencing of chvD.
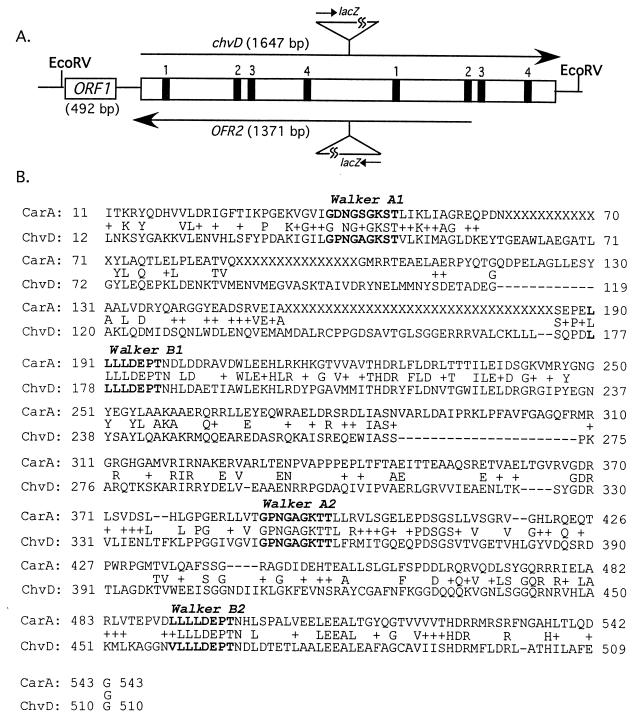
Because the mutations in AB22 and AB20, as well as the original chvD mutation in strain A348 chvD1 (43), were generated via insertional mutagenesis, the possibility existed that one or more downstream open reading frames of a potential operon are responsible for the observed phenotypes. To address this issue, the entire chvD gene was cloned and sequenced as described in Materials and Methods. Briefly, Southern blots of chromosomal DNA, digested with a variety of restriction enzymes and probed with the 449-bp fragment of chvD, revealed a 7.8-kb HindIII fragment that carried at least part of the chvD gene. This fragment was cloned into pBSII, and a 2.7-kb EcoRV fragment of this plasmid was shown to carry the chvD-homologous sequences. The 2.7-kb EcoRV fragment was subcloned into pBSII and sequenced. Analysis of these data, as well as some additional upstream sequences from the 7.8-kb HindIII fragment, demonstrated that the chvD open reading frame is 1,647 bp (549 amino acids). BLAST searches demonstrated that the ChvD protein is homologous to a variety of ATP-binding cassette (ABC) transporters, particularly members of a group of bacterial ABC transporters involved in antibiotic export (Fig. 2). For example, it shares 31% identity and 48% similarity with the carbomycin resistance (carA) gene of Streptomyces thermotolerans (35) and 28% identity and 45% similarity with the pristinamysin resistance (vgaB) gene of Staphylococcus aureus (1). Finally, the ChvD protein contains two complete sets of ABC consensus sequences but no obvious hydrophobic domains, which would be expected if it were inserted into the membrane.
FIG. 2.
(A) Map of ChvD indicating sites of Walker A boxes (box 1), the LSGG signatures (box 2), Walker B boxes (box 3), and the conserved histidine found in most ABC transporters (box 4). The orientation of the lacZ gene from the inserted plasmids pZL101 (upper) and pZL103 (lower) are also shown. (B) BLAST comparison of the amino acid sequences of ChvD and CarA, where the middle line indicates identical (letters) and conserved (+) amino acids.
Two additional potential open reading frames have been identified on the 2.7-kb EcoRV fragment carrying chvD. First, 5′ to the chvD translation start site is a 492-bp open reading frame (ORF1) with 245 bp separating these two coding regions (Fig. 2). No significant hydrophobic domains are predicted for this protein, and BLAST searches did not reveal significant homologies to any known proteins. Second, the antisense strand of chvD encodes a 497-amino acid open reading frame (ORF2), if we assume that GTG (at position 2082 of the 2.7-kb EcoRV fragment) is the start codon (Fig. 2). The predicted protein has two potential membrane-spanning domains, based on prediction programs of Czero et al. (14) and Hoffman and Stoffel (21), but BLAST searches did not reveal significant homology to known proteins.
Complementation of the chvD mutant strain AB20.
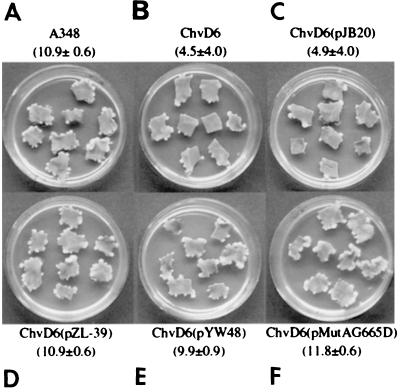
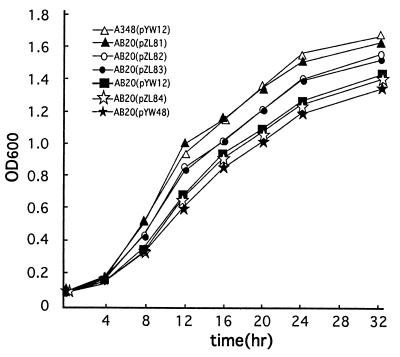
Two strategies were used to test the role of chvD in virulence and vir gene expression. The first was to clone the 2.7-kb EcoRV fragment described above into the broad-host-range IncW vector pJB20, yielding pZL39, and to determine whether it could complement the chvD mutation in strain AB20. Strain AB20 or AB20(pJB20) induced significantly fewer tumors than the wild-type strain, A348 (Fig. 3A to C). In contrast, AB20(pZL39), containing the wild-type chvD gene, exhibited completely restored virulence (Fig. 3D). These results demonstrate that no open reading frame downstream of chvD is involved in the attenuated virulence phenotype. A second complementation strategy was used to determine whether the chvD-mediated effect on virulence and vir gene expression lies upstream or downstream of the VirA-VirG signaling cascade. In this case, pYW48, containing virG constitutively expressed from the PN25 promoter of the T5 coliphage (39) or pMutA-G665D, containing a constitutively active form of VirA (29), was electroporated into AB20, and the resultant strain was tested in a tobacco leaf explant tumor assay. Full virulence was restored in both cases (Fig. 3E and F), demonstrating that the effects of chvD activity are not necessary if VirG is constitutively expressed or activated, consistent with a model in which chvD is involved in controlling virG expression (43). In terms of growth in L broth, AB20(pZL81), carrying the wild-type chvD gene, exhibited restored growth compared to that of AB20 with the vector pYW12 while the AB20(pYW48) strain, which contains constitutively expressed virG and complements the virulence phenotype, continued to exhibit the reduced growth phenotype (Fig. 4). Similarly, pMutA-G665D did not restore the capacity of AB20 to grow optimally in L broth (data not shown).
FIG. 3.
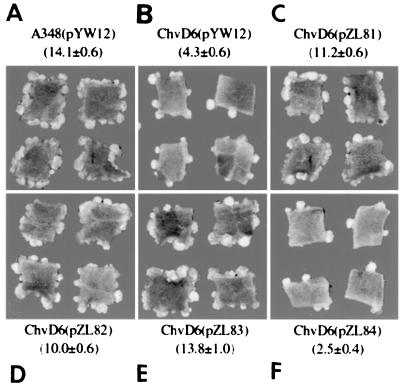
Tobacco leaf virulence assay. Complementation of chvD mutant strain AB20. (A) A348 (wild type); (B) AB20; (C) AB20(pJB20); (D) AB20(pZL39); (E) AB20(pYW48); (F) AB20(pMutA-G665D). The mean numbers of tumors/explant ± standard errors (n = 14) for the experiments are shown.
FIG. 4.
Growth in L broth of wild-type A348(pYW12) and the AB20 mutant complemented with the following plasmids: pYW12 (vector), pYW48 (constitutively expressed virG), pZL81 (wild-type chvD), pZL82 (chvD Walker A1 mutation), pZL83 (chvD Walker A2 mutation), and pZL84 (chvD Walker A1 and A2 mutations). Mean OD600s (n = 3) ± standard errors (error bars not visible) are shown.
Mutagenesis of chvD ATP-binding sites.
An important question is whether the ATPase activity of ChvD is necessary for the effects on virulence. Therefore, the Walker A1 and Walker A2 sites of chvD (Fig. 2) were mutagenized either singly or together and tested for effects on growth in rich medium, virulence, and vir gene expression. The capacity for the AB20 strain to grow in L broth was partially disrupted when ChvD contained the Walker A box mutations (Fig. 4). However, mutation in either Walker A box site individually (pZL82 and pZL83) resulted in chvD genes that could still fully complement the AB20 mutant strain for virulence and vir gene expression. The strains induced as many tumors (Fig. 5) and induced virulence gene expression (Fig. 6) as well as the wild-type strain A348. When the chvD gene carried both mutations (pZL84), the resultant gene could not complement the AB20 strain in any of these three assays (Fig. 4 to 6).
FIG. 5.
Tobacco leaf virulence assay of strain AB20 complemented with chvD containing mutations at the Walker A ATP binding sites. (A) Wild-type A348(pYW12); (B) AB20(pYW12); (C) AB20(pZL81); (D) AB20(pZL82); (E) AB20(pZL83); (F) AB20(pZL84). The mean numbers of tumors/explant ± standard errors (n = 14) for all experiments are shown.
FIG. 6.
vir gene expression in Walker A box ATP-binding site mutants. Shown are the results of an immunoblot analysis of VirB8 expression in strains induced with either 1 or 3 μM AS. A lanes, wild-type A348(pYW12); B lanes, AB20(pYW12); C lanes, AB20(pZL81); D lanes, AB20(pZL82); E lanes, AB20(pZL83); F lanes, AB20(pZL84).
DISCUSSION
Phenolic compounds, sugars, and low pH and low PO4 concentrations are all involved in maximizing expression of the virulence genes on the Ti plasmid of A. tumefaciens. This complex set of environmental signals is integrated by the VirA-VirG two-component system, with input arriving from both known and as yet unidentified proteins (41, 8, 9). The experiments presented here identify chvD as encoding an ABC transporter homologue and demonstrate that it is a critical member of the virulence regulatory pathway, most likely affecting the capacity of virG to be expressed.
ChvD is most closely related to a class of bacterial ABC transporters that are involved in antibiotic resistance, including, for example, carA of Streptomyces thermotolerans (35) and vgaB of Staphylococcus aureus (1). These have been classified in the ABC-A2 subset of ABC transporters (38). The ATP-binding proteins of these transporters are characterized by having two complete sets of Walker A and B boxes and ABC signature motifs. They have no membrane-spanning domains but, rather, interact with membrane-spanning proteins that are the apparent pore involved in the export process. As in the case of ChvD, the membrane-spanning proteins of this subfamily of ABC transporters are often not part of the same operon as the ATP-binding protein (38). Preliminary studies have not yet identified any antibiotics that are more lethal to mutant strain AB20 than to the wild-type strain. However, strains carrying a disruption in the chvD gene grow poorly in comparison to the wild type in rich medium but equivalent to the wild type in minimal medium. This phenotype is consistent with the hypothesis that this ABC transporter system has an activity that can export potentially inhibitory compounds, within the L broth, that gain entry into the cell. Supporting evidence for this concept is that mutation at the conserved lysine in either Walker A box of ChvD results in genes that can partially restore the mutant growth phenotype. Usually, however, such mutations in the ATP-binding site of ABC transporter systems completely disrupt their transport activities reflecting the fact that the biochemical mechanism of almost all ABC transporter ATP-binding proteins requires two functional ATP-binding sites for transport activity (34). Biochemical characterization will be required to determine whether the single point mutations in ChvD completely disrupt its ATPase activity or, alternatively, whether the activity of the mutant ChvD reflects dimerization to provide sufficient ATP-binding sites for partial activity.
The relationship between the transport activity of ChvD and virulence is uncertain. Disruption of two other Agrobacterium chromosomal virulence proteins, ChvI and ChvG, also results in poor growth in rich medium (12, 28) and lowered virulence. The relationship between this apparent two-component control system and ChvD is not known. Of significant interest is that, despite their effects on growth, mutations at the conserved lysine of either Walker A box in the ChvD protein did not disrupt this protein's effects on virulence or vir gene expression. Assuming that the growth phenotype reflects ChvD-mediated transport activity, these results suggest that the role of ChvD in regulation of vir gene expression and virulence is not dependent on a fully functional transport activity. Such dual functions have been observed, for example, in ChvE, which is the periplasmic sugar-binding protein of an ABC transporter sugar uptake system in A. tumefaciens but also affects, independently of sugar uptake, vir gene expression (23). Similarly, the ecsA (ATP-binding protein) and ecsB (membrane-spanning protein) genes of Bacillus subtilis have been proposed to work coordinately to regulate transcription independently of transport activities (25). However, when both Walker boxes of ChvD carried mutations at the conserved lysine, vir gene expression, virulence, and growth in L broth were completely disrupted. Perhaps some level of transport activity is necessary for ChvD activity in terms of virulence. However, because antibody directed against ChvD is not yet available, the possibility that the two Walker A box mutations within the protein render it unstable has not been addressed.
Complementation analysis demonstrated that a 2.7-kb fragment encoding the ChvD protein is capable of restoring optimal vir gene expression, virulence, and growth in L broth of the chvD mutant to wild-type levels. This result demonstrates that the chvD gene is not followed by a downstream open reading frame involved in controlling these phenotypes. It is unlikely that ORF2, encoded by the opposite stand of the chvD coding region, is responsible for any of the phenotypes investigated here. First, the insertion of a transcriptional lacZ fusion in this orientation (strain AB22) did not result in β-galactosidase activity, indicating that, under the conditions tested, this strand is not transcribed. Second, while mutation at the Walker A1 site of ChvD (K→E) resulted in a mutation in ORF2 (P→L), the mutation at the Walker A2 site of ChvD (K→E) was silent in Orf2 (L→L). The strain carrying this Walker A2 mutation [AB20(pZL83)] had a mutant phenotype with regard to growth in L broth (Fig. 4), thus demonstrating that Orf2 must not be involved in this phenotype. Third, the AB20 strain carrying a version of ChvD with both Walker box mutations (pZL84) was not complemented in either growth or virulence assays, in contrast to the strains with individual mutations (pZL82 and pZL83), which exhibited partially restored growth in L broth and wild-type virulence. Since ORF2 in pZL82 is identical to ORF 2 in pZL84, these results demonstrate that the observed phenotypes are the result of ChvD activities or disruptions thereof.
The original chvD mutant strain, A348 chvD1, was isolated based on the reduced ability of a virG::lacZ fusion to be expressed in the presence of low PO4 concentrations (43). To further clarify the position of ChvD activity in the pathways regulating vir gene expression and the potential role of its transport function in virulence, two different plasmids carrying a mutant form of virG or virA were tested for their capacity to complement the chvD mutant strain AB20. The first of these, pYW48, carries wild-type virG expressed from the constitutive PN25 promoter of the T5 coliphage (39) along with wild-type virA, while the second, pMutA-G665D, carries wild-type virG expressed from its own promoter and a constitutively active form of VirA (29). In both cases, virulence was restored whereas the strains continued to exhibit the mutant phenotype in terms of the capacity for growth in L broth. Thus, ChvD-mediated transport activity does not affect regulatory processes downstream of virG expression. Rather, these results are consistent with the previously published data demonstrating that a transposon insertion into chvD downregulates the activity of the virG promoter (43).
The modified pJG4-5 prey vectors described here allow insertion of sequences into the EcoRI site of the activation protein fusion in all three open reading frames. This finding is particularly useful in the construction of genomic libraries from any prokaryotic organism. Using libraries of Agrobacterium genomic DNA in such vectors resulted in the isolation of fusion proteins that interacted with the periplasmic domain of VirB8, a result that led to the ultimate isolation and characterization of the chvD gene. At this point, however, the biological relevance of this interaction is not clear. First, the domain of VirB8 used in the screen of the yeast two-hybrid Agrobacterium genomic library is periplasmic (16) whereas the predicted location of ChvD is cytoplasmic. Second, strains carrying deletions of VirB8 exhibit wild-type expression from a plasmid carrying a virB promoter driving lacZ (A. Yin and A. N. Binns, unpublished data), indicating that VirB8-ChvD interaction is not required for ChvD's stimulating activity. This does not eliminate the possibility, however, that degradation or breakdown products of VirB8 in the cytoplasm may yield peptides that interact with ChvD and repress its activity, thereby reducing vir gene expression.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM47369 and NSF grant MCB98-17149. C.A.M. is supported by NIH training grant 5-T32-GM08216-14.
We thank David Lynn, Yulei Wang, and Jutta Bohne for discussion and plasmids.
REFERENCES
- 1.Allignet J, El Sohl N. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogamin A and related compounds. Gene. 1997;202:133–138. doi: 10.1016/s0378-1119(97)00464-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ankenbauer R G, Nester E W. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J Bacteriol. 1990;172:6442–6446. doi: 10.1128/jb.172.11.6442-6446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banta L M, Joerger R D, Howitz V R, Campbell A M, Binns A N. Glu-255 outside the predicted ChvE binding site in VirA is crucial for sugar enhancement of acetosyringone perception by Agrobacterium tumefaciens. J Bacteriol. 1994;176:3242–3249. doi: 10.1128/jb.176.11.3242-3249.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaupré C F, Bohne J, Dale E M, Binns A N. Interactions between VirB9 and VirB10 proteins involed in movement of DNA from Agrobacterium tumefaciens to plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger B R, Christie P J. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993;175:1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binns A N, Beaupré C F, Dale E M. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J Bacteriol. 1995;177:4890–4899. doi: 10.1128/jb.177.17.4890-4899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binns A N, Howitz V R. The genetic and chemical basis of recognition in the Agrobacterium: plant interaction. Curr Top Microbiol Immunol. 1994;192:119–138. doi: 10.1007/978-3-642-78624-2_6. [DOI] [PubMed] [Google Scholar]
- 9.Campbell A M, Tok J, Zhang J, Wang Y, Stein M, Lynn D G, Binns A N. Xenognosin sensing in virulence: is there a phenol receptor in Agrobacterium tumefaciens? Chem Biol. 2000;7:65–76. doi: 10.1016/s1074-5521(00)00065-x. [DOI] [PubMed] [Google Scholar]
- 10.Cangelosi G A, Ankenbauer R G, Nester E W. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci USA. 1990;87:6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C H, Winans S W. Resection and mutagenesis of the acid pH-inducible P2 promoter of the Agrobacterium tumefaciens virG gene. J Bacteriol. 1996;178:4717–4720. doi: 10.1128/jb.178.15.4717-4720.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles T C, Nester E W. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1993;175:6614–6625. doi: 10.1128/jb.175.20.6614-6625.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czero M, Wallin E, Simon E, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic proteins: the dense alignment surface method. Prot Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 15.Dale E M, Binns A N, Ward J E., Jr Construction and characterization of Tn5virB, a transposon that generates nonpolar mutations, and its use to define virB8 as an essential virulence gene in Agrobacterium tumefaciens. J Bacteriol. 1993;175:887–891. doi: 10.1128/jb.175.3.887-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das A, Xie Y-H. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol Microbiol. 1998;27:405–414. doi: 10.1046/j.1365-2958.1998.00688.x. [DOI] [PubMed] [Google Scholar]
- 17.Das A, Xie Y H. The Agrobacterium tumefaciens T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol. 2000;182:758–763. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garfinkel D J, Simpson R B, Ream L W, White F F, Gordon M P, Nester E W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981;27:143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 19.Gelvin S B. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 20.Golemis E, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. In: Ausubel F M, et al., editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1999. pp. 20.1.1–20.21.40. [Google Scholar]
- 21.Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 22.Kalogeraki V S, Winans S C. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/s0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- 23.Kemmer J M, Liang X Y, Nester E W. The Agrobacterium tumefaciens virulence gene chvE is a part of a putative ABC-type sugar transport operon. J Bacteriol. 1997;179:2452–2458. doi: 10.1128/jb.179.7.2452-2458.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar R B, Xie Y H, Das A. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol Microbiol. 2000;36:608–617. doi: 10.1046/j.1365-2958.2000.01876.x. [DOI] [PubMed] [Google Scholar]
- 25.Leskelä S, Wahlström E, Hyyryläinen H-L, Jacobs M, Pavla A, Sarvas M, Kontinen V P. Ecs, an ABC transporter of Bacillus subtilis: dual signal transduction functions affecting expression of secreted proteins as well as their secretion. Mol Microbiol. 1999;31:533–543. doi: 10.1046/j.1365-2958.1999.01194.x. [DOI] [PubMed] [Google Scholar]
- 26.Lohrke S M, Nechaev S, Yang H, Severinov K, Jin S J. Transcriptional activation of Agrobacterium tumefaciens virulence gene promoters in Escherichia coli requires the A. tumefaciens RpoA gene, encoding the alpha subunit of RNA polymerase. J Bacteriol. 1999;181:4533–4539. doi: 10.1128/jb.181.15.4533-4539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantis N J, Winans S C. The Agrobacterium tumefaciens vir gene transcriptional activator virG is transcriptionally induced by acid pH and other stress stimuli. J Bacteriol. 1992;174:1189–1196. doi: 10.1128/jb.174.4.1189-1196.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantis N J, Winans S C. The chromosomal response regulatory gene chvI of Agrobacterium tumefaciens complements an Escherichia coli phoB mutation and is required for virulence. J Bacteriol. 1993;175:6626–6636. doi: 10.1128/jb.175.20.6626-6636.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean B G, Greene E A, Zambryski P C. Mutants of Agrobacterium VirA that activate vir gene expression in the absence of the inducer acetosyringone. J Biol Chem. 1994;269:2645–2651. [PubMed] [Google Scholar]
- 30.Melchers L S, Regensburg-Tuïnk T J G, Bourret R B, Sedee N J A, Schilperoort R A, Hooykaas P J J. Membrane topology and functional analysis of the sensory protein VirA of Agrobacterium tumefaciens. EMBO J. 1989;8:1919–1925. doi: 10.1002/j.1460-2075.1989.tb03595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikarlian I, Sergreant A. Modification of the overlap extension method for extensive mutagenesis on the same template. Methods Mol Biol. 1996;57:193–202. doi: 10.1385/0-89603-332-5:193. [DOI] [PubMed] [Google Scholar]
- 32.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–496. [Google Scholar]
- 33.Peng W T, Lee Y W, Nester E W. The phenolic recognition profiles of Agrobacterium tumefaciens VirA protein are broadened by a high level of sugar binding protein ChvE. J Bacteriol. 1998;180:5632–5638. doi: 10.1128/jb.180.21.5632-5638.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 35.Schoner B E, Geistlich M, Rosteck P, Rao R N, Seno E, Reynolds P, Cox K, Burgett S, Hershberger C L. Sequence similarity between macrolide resistance determinants and ATP binding transport proteins. Gene. 1992;115:93–96. doi: 10.1016/0378-1119(92)90545-z. [DOI] [PubMed] [Google Scholar]
- 36.Shimoda N, Toyoda-Yamamoto A, Nagamine J, Usami S, Katayama M, Sakagami Y, Machida Y. Control of expression of Agrobacterium vir genes by synergistic actions of phenolic signal molecules and monosaccharides. Proc Natl Acad Sci USA. 1990;87:6684–6688. doi: 10.1073/pnas.87.17.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimoda N, Toyoda-Yamamoto A, Shinsuke S, Machida Y. Genetic evidence for an interaction between the VirA sensor protein and the ChvE sugar-binding protein of Agrobacterium. J Biol Chem. 1993;268:26552–26558. [PubMed] [Google Scholar]
- 38.Suarin W, Hofnung M, Dassa E. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol. 1999;48:22–41. doi: 10.1007/pl00006442. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Mukhopadhyay A, Howitz V R, Binns A N, Lynn D G. Construction of an efficient expression system for Agrobacterium tumefaciens based on the coliphage T5 promoter. Gene. 2000;242:105–112. doi: 10.1016/s0378-1119(99)00541-7. [DOI] [PubMed] [Google Scholar]
- 40.Winans S C. Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J Bacteriol. 1990;172:2433–2438. doi: 10.1128/jb.172.5.2433-2438.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winans S C. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winans S C, Burns D L, Christie P J. Adaptation of the conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winans S C, Kerstetter R A, Nester E W. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J Bacteriol. 1988;170:4047–4054. doi: 10.1128/jb.170.9.4047-4054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]