Abstract
d-Galactan I is an O-antigenic polymer with the repeat unit structure [→3)-β-d-Galf-(1→3)-α-d-Galp-(1→], that is found in the lipopolysaccharide of Klebsiella pneumoniae O1 and other gram-negative bacteria. A genetic locus containing six genes is responsible for the synthesis and assembly of d-galactan I via an ATP-binding cassette (ABC) transporter-dependent pathway. The galactosyltransferase activities that are required for the processive polymerization of d-galactan I were identified by using in vitro reactions. The activities were determined with endogenous lipid acceptors in membrane preparations from Escherichia coli K-12 expressing individual enzymes (or combinations of enzymes) or in membranes reconstituted with specific lipid acceptors. The d-galactan I polymer is built on a lipid acceptor, undecaprenyl pyrophosphoryl-GlcpNAc, a product of the WecA enzyme that participates in the biosynthesis of enterobacterial common antigen and O-antigenic polysaccharide (O-PS) biosynthesis pathways. This intermediate is directed into d-galactan I biosynthesis by the bifunctional wbbO gene product, which sequentially adds one Galp and one Galf residue from the corresponding UDP-sugars to form a lipid-linked trisaccharide. The two galactosyltransferase activities of WbbO are separable by limiting the UDP-Galf precursor. Galactosyltransferase activity in membranes reconstituted with exogenous lipid-linked trisaccharide acceptor and the known structure of d-galactan I indicate that WbbM catalyzes the subsequent transfer of a single Galp residue to form a lipid-linked tetrasaccharide. Chain extension of the d-galactan I polymer requires WbbM for Galp transferase, together with Galf transferase activity provided by WbbO. Comparison of the biosynthetic pathways for d-galactan I and the polymannose E. coli O9a antigen reveals some interesting features that may reflect a common theme in ABC transporter-dependent O-PS assembly systems.
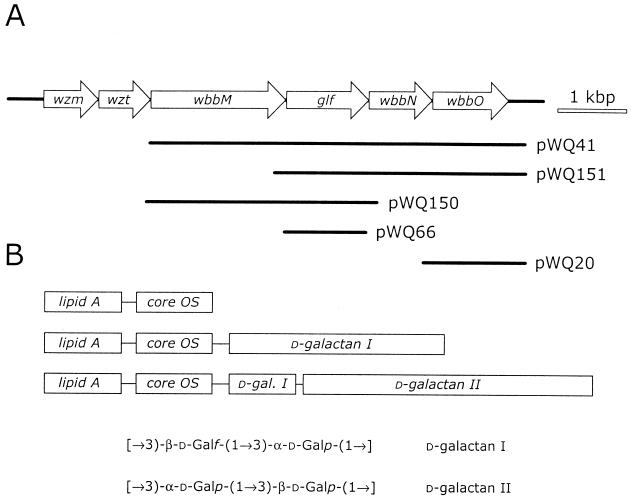
Lipopolysaccharide (LPS) is the major component of the outer leaflet of the gram-negative bacterial outer membrane. In members of the family Enterobacteriaceae, LPS consists of three structural domains: the hydrophobic lipid A, the core oligosaccharide, and the O-antigenic polysaccharide (O-PS). The O-PS structures are hypervariable. In the klebsiellae, there are 11 O-PS structures, but structural similarities lead to some serological cross-reactivities, so the actual number of unique O-serotypes is smaller (reviewed in references 12 and 50). The clinically prevalent O1 antigen contains two structurally distinct O-PS domains composed of the repeat units [→3)-β-d-Galf-(1→3)-α-d-Galp-(1→] (d-galactan I) and [→3)-α-d-Galp-(1→3)-β-d-Galp-(1→] (d-galactan II).
Genetic (5, 8) and chemical (26, 27, 55) analyses indicate that d-galactan I chains are linked directly to the lipid A core structure. d-Galactan II is confined to the distal end of some of the available d-galactan I chains (Fig. 1B). d-Galactan II provides the epitope or epitopes that define the O1 antigen (55). The presence of d-galactan II is required for the resistance of the bacteria to complement-mediated killing in the host, and therefore Klebsiella pneumoniae mutants that produce only d-galactan I are serum sensitive (33).
FIG. 1.
Organization of the genetic locus for d-galactan I biosynthesis and structural features of the LPS in K. pneumoniae O1. (A) Genetic organization of the his-linked O-PS biosynthesis locus from K. pneumoniae O1. wzm and wzt encode components of an ABC transporter for export of the polysaccharide. The wbbM, glf, and wbbO genes encode functions involved in polymerization of the polysaccharide, whereas the role of the wbbN gene product remains unclear. The functions ascribed to each gene (where known) are described in the text. The fragments contained in each of the plasmids used in this study (Table 1) are indicated. (B) Schematic representation of the organization of the LPS of K. pneumoniae O1 (for details, see the text). The O antigen biosynthesis locus shown in panel A is sufficient for synthesis of d-galactan I. Additional and currently unknown genes are required for d-galactan II biosynthesis.
There are two major pathways for the biosynthesis of LPS O-PS. These are designated Wzy dependent and ATP-binding cassette (ABC) transporter dependent (reviewed in references 52 and 53). Both pathways are widely distributed among different genera and are involved in the synthesis of diverse O-PS structures. A third pathway (synthase dependent) is currently confined to a single O-PS (17). In the Wzy-dependent mechanism, undecaprenyl pyrophosphoryl (und-PP)-linked oligosaccharide repeat units are assembled at the cytoplasmic face of the inner membrane. These intermediates are then exported to the periplasmic face, where they provide the direct substrates for polymerization. Export and polymerization require the activity of members of the Wzx and Wzy protein families, respectively. In the ABC transporter-dependent mechanism, the polymer is built by processive transfer of glycosyl residues to an initiating und-PP-linked lipid intermediate at the cytoplasmic face of the inner membrane. After the lipid-linked polymer is synthesized at the cytoplasmic face of the inner membrane, it is exported to the periplasmic face by an ABC transporter (5, 45, 58). The various O-PS biosynthesis pathways are believed to converge with the presence of und-PP-linked polymer at the periplasmic face of the inner membrane, at which point the nascent O-PS is ligated to preformed lipid A core. The completed LPS molecule is then translocated to the outer leaflet of the outer membrane by a process that is still undetermined.
In members of the family Enterobacteriaceae, initiation of ABC transporter-dependent O-PS synthesis requires und-PP-GlcNAc (7, 44), formed by the action of the UDP-GlcNAc::undecaprenylphosphate GlcNAc-1-phosphate transferase encoded by the wecA (formerly rfe) gene. The wecA gene is found in virtually all strains of Enterobacteriaceae (29) and is located in the enterobacterial common antigen (ECA) biosynthesis locus (34, 37). Its activity was originally characterized in the initiation of ECA biosynthesis (34). However, WecA is a versatile initiating transferase that also participates in some Wzy-dependent O-PS synthesis pathways (1) and in the synthase-dependent pathway (16, 17). Thus, both the initial and terminal steps in O-PS assembly may be conserved, with the pathways differing in the cellular location and mechanism of the polymerization process.
Genes at the his-linked O-PS biosynthesis locus in K. pneumoniae O1 are required for the expression of d-galactan I (5, 8, 55), but the locations and identities of genes required for d-galactan II biosynthesis remain unknown. The his-linked locus contains six genes (Fig. 1) whose products form an ABC transporter-dependent O-PS assembly system. The first two genes in the locus, wzm and wzt (formerly rfbAB), encode the transmembrane and ATP-binding components, respectively, of the ABC-2 transporter (5). The remaining four genes (wbbM, glf, wbbN, and wbbO) are proposed to be involved in the synthesis of d-galactan I. The glf gene product is a UDP-galactopyranosye mutase, which catalyzes the reversible interconversion of uridine 5′-diphospho-α-d-galactopyranose (UDP-Galp) and uridine 5′-diphospho-α-d-galactofuranose (UDP-Galf) (28, 30, 36); these two sugar nucleotides provide the precursors for d-galactan I synthesis. The precise roles played by WbbM and WbbN are unknown, but some information is available for WbbO. When WbbO is expressed in E. coli K-12, the lipid A core of the host LPS is modified by the addition of a trisaccharide, β-d-Galf-(1→3)-α-d-Galp-(1→3)-β-d-GlcpNAc (7). The addition of the GlcpNAc residue results from WecA activity (10), and the lipid A-core-linked trisaccharide is missing in a wecA mutant (7). These data are consistent with WbbO serving as a bifunctional galactosyltransferase that transfers two galactosyl residues to und-PP-GlcNAc, but conclusions are complicated by the potential involvement of endogenous transferase activities in the E. coli background. Mutations in the O-PS biosynthesis locus in E. coli K-12 prevent formation of its native Wzy-dependent O-PS (31, 57), but the remaining activities can contribute to LPS synthesis in the presence of other (heterologous) gene products (25, 56, 57). For example, the analysis of WbbO activity in the E. coli in vivo system is dependent on the relaxed specificity of the E. coli K-12 Wzx protein (10), which exports und-PP-linked oligosaccharides across the inner membrane to provide substrate for the Wzy polymerase. Although Wzx can export hybrid und-PP-linked oligosaccharides, it is not able to export completed d-galactan I (7). Consequently, there is no WbbO-mediated LPS modification in an E. coli background without any Wzx activity. It is therefore possible that the extent of the modification of lipid A core in E. coli K-12 expressing WbbO reflects a size restriction imposed by Wzx selectivity. WbbO could potentially transfer additional residues to form und-PP-linked intermediates of various sizes, with the only molecules detected being those that are exported and ligated to lipid A core. Thus, the identities and precise activities of galactosyltransferases required for processive synthesis of d-galactan I are unknown.
Although the genetic loci for a number of O-PS biosynthesis systems have been sequenced and analyzed recently, relatively few of the glycosyltransferase enzymes have been defined biochemically. In most cases, assignment to an ABC transporter-dependent pathway is confined to identification of the genes encoding the ABC transporter (wzm and wzt). The processive polymerization process itself has only been studied in detail in synthesis of the polymannose E. coli O9a antigen (see reference 23 and references therein). O9a polymer synthesis is initiated with und-PP-GlcNAc formed by the action of WecA. The WbdC mannosyltransferase then adds one residue in a reaction that is confined to polymer initiation. Two multifunctional mannosyltransferases (WbdA and WbdB) then act in an alternating fashion to generate the repeating unit domain.
Our objective in analyzing the d-galactan I system was to determine whether there are common patterns of enzyme activities in the ABC transporter-dependent pathways. Here we report in vitro experiments that demonstrate that WecA, WbbO, and WbbM are sufficient for d-galactan I synthesis. The WbbO galactosyltransferase is shown to be bifunctional, with one activity that is confined to synthesis of an acceptor for polymerization of the repeat unit domain and another activity that, together with WbbM, is required throughout polymerization. Although there are some differences in the precise roles of proteins in the d-galactan I and O9a syntheses, the pathways do share some interesting features that may reflect a common theme in this type of O-PS assembly system.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown at 37°C in Luria broth (LB) (35) or on LB agar. When required, antibiotics were added to the following final concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 34 μg ml−1; kanamycin, 30 μg ml−1; and tetracycline, 20 μg ml−1. For general cloning, plasmid constructs were made according to standard procedures with the vector pBC KS(+) (Stratagene). Alternatively, the pBAD vectors (11) were used to provide arabinose-inducible expression. Transformation was performed by electroporation with a Bio-Rad Gene Pulser under conditions described elsewhere (4).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype, phenotype, or description | Reference or source |
|---|---|---|
| E. coli strain CWG288 | K-12 lacZ trp Δ(sbcB-rfb) upp rel rpsL galE::Tn10 [λDE3] | 28 |
| Plasmids | ||
| pMAV11 | pACYC184-derivative carrying wecA from E. coli K-12; Cmr | 57;;1 |
| pJK2363 | pGEM4 carrying rfpB from S. dysenteriae type 1: Apr | 24;;1 |
| pWQ20 | pTrc99A derivative carrying wbbO; Apr | 7;;1 |
| pWQ41 | pBC KS(+) derivative carrying wbbM-glf-wbbN-wbbO on a 5.3-kb PstI fragment; Cmr | R. Köplin, unpublished data |
| pWQ66 | pET30a(+)- derivative containing glf; Kmr | 28;;1 |
| pWQ150 | pBAD18-Km derivative containing wbbM-glf cloned on a 3.5-kb EcoRI fragment; Kmr | This study |
| pWQ151 | pBAD24 derivative containing glf-wbbN-wbbO cloned on a 3.8-kb EcoRV fragment; Apr | This study |
Enzyme preparation.
Membrane preparations provided the source of galactosyltransferase activity and were made with minor modifications of the method originally described by Osborn et al. (38). Briefly, a 5-ml overnight culture was diluted in 100 ml of LB, and the culture was incubated with shaking the until late exponential phase (optical density at 600 nm [OD600] of ∼1.5). The bacteria were collected by centrifugation and washed once with 100 ml of cold saline (0.9% [wt/vol] NaCl) and then with 10 ml of cold buffer A (50 mM Tris acetate [pH 8.5], 1 mM EDTA). The cells were then resuspended in 10 ml of cold buffer A and sonicated five times for 30 s with intermittent cooling on ice. Cell debris was removed by centrifugation at 4,000 × g for 10 min. The supernatant was then centrifuged at 111,000 × g for 20 min at 4°C, and the resulting membrane pellet was resuspended in 0.3 ml of buffer A and stored at −70°C.
In vitro glycosyltransferase activity.
Glycosyltransferase activity was measured by the incorporation of radioactivity from UDP-[14C]α-d-galactopyranose (UDP-Galp; [U=14C]Galp; 278 mCi mmol−1; NEN) or UDP-[14C]α-d-N-acetylglucosamine (UDP-GlcNAc; 10.2 mCi mmol−1; ICN) into chloroform-methanol-extractable lipid intermediates. The in vitro reaction mixture contained approximately 800 μg of protein and 72 pmol (∼ 45,000 cpm) of radiolabeled NDP-sugar substrate in 0.1 ml of buffer B (50 mM Tris acetate [pH 8.5], 10 mM MgCl2, 1 mM EDTA). UDP-galactopyranose mutase (Glf) was added to some reactions. The Glf-containing enzyme extract was prepared as a cell-free lysate of CWG288(pWQ66), as described elsewhere (28). The 2 μl of extract added to the standard reaction mixture contained approximately 2 μg of Glf enzyme. Galactosyltransferase reactions were performed at room temperature and were terminated by extraction with either chloroform-methanol (C:M) or aqueous phenol. For C:M extraction, 1.3 ml of C:M (3:2) was added. After vigorous mixing, the insoluble debris was removed by low-speed centrifugation in a benchtop centrifuge. The organic phase containing lipid intermediates was transferred to a fresh tube, 150 μl of 40 mM MgCl2 was added to the extract, and the suspension was mixed. The upper aqueous phase was removed, and the organic phase was washed twice with pure solvent upper phase (PSUP) (38). The hot aqueous phenol extraction is a scaled down modification of the procedure established by Westphal and Jann (51). Prior to phenol extraction, unincorporated substrate was removed by washing the membranes three times in 0.6 ml of 40 mM MgCl2. The membranes were resuspended in 0.3 ml of 40 mM MgCl2, and an equal volume of 90% (wt/vol) phenol was added. The extraction was performed at 65°C for 30 min with frequent mixing. After cooling, the phases were separated by centrifugation in a benchtop centrifuge, and the upper aqueous fraction was collected. The phenol phase was reextracted with an equal volume of 40 mM MgCl2, and the aqueous phases were pooled. Radioactivity in the C:M or aqueous phenol extracts was measured with EcoLite scintillation fluid (ICN) in a Tricarb 2000 liquid scintillation counter (Canberra Packard).
Preparation of the butanol-soluble lipid intermediate acceptors and membrane reconstitution.
An overnight culture (10 ml) of E. coli CWG288 or CWG288 containing the appropriate plasmid was diluted into 200 ml of fresh LB containing appropriate antibiotics, and incubation was continued until the culture reached the mid-exponential phase (OD600 of ∼0.6). Where appropriate, isopropyl-β-d-thiogalactopyranoside (IPTG) was added (5 mM final concentration), and the incubation was continued for a further 3 h. The bacterial cells were harvested by centrifugation, and the membrane fraction was prepared as described above. Bulk lipid acceptor was prepared from a reaction mixture containing the membranes, 40 μM UDP-Galp, 40 μM UDP-GlcNAc, and 20 mM dithiothreitol in 10 ml of buffer B. The reaction mixture was incubated for 30 min at room temperature. The lipid intermediates were extracted twice with 1.5 volume of butan-1-ol, and the pooled upper butanol phases were washed once with an equal volume of 40 mM MgCl2. The butanol extract was concentrated with a stream of air to ∼0.3 ml and washed once with an equal volume of 40 mM MgCl2. A 0.1-ml aliquot of the extract was then used to prepare an aqueous suspension of lipid intermediates, and membrane reconstitution was performed by multiple freeze-thaw cycles, according to the methods described by Jann et al. (14). Prior to routine (0.1 ml) galactosyltransferase assays, 5 μl of lipid extract was added to the membranes for reconstitution.
Thin-layer chromatography (TLC) separation of lipid intermediates.
Dried C:M extracts were dissolved in 150 μl of C:M (3:2 ratio), and approximately 2,000 cpm (5 to 20 μl) of radioactive material was applied to a Silica Gel 60 plate (EM Science, Gibbstown, N.J.). The plate was developed twice with a solvent consisting of chloroform-methanol-water (65:25:4) (3) and exposed to Kodak BioMax film.
Tricine-SDS-PAGE and Western blot analysis.
Prior to loading, membrane preparations were boiled for 10 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (13) and then treated with proteinase K (final concentration of 1 μg ml−1) at 60°C for 1 h. All samples were separated on commercially available 10 to 20% Tricine gels (Novex) according to the manufacturer's instructions and electrophoretically transferred to nitrocellulose membranes (49). d-Galactan I-specific polyclonal antibody was prepared in rabbits immunized with formalin-killed K. pneumoniae CWK37, a derivative of serotype O1 lacking d-galactan II (7). Cross-reactive antibodies were removed by repeated absorption with E. coli K-12 cells. Alkaline phosphatase-conjugated antirabbit antibody (Cedarlane Laboratories, Hornby, Ontario, Canada) was used as the secondary antibody, and detection was performed with nitroblue tetrazolium chloride–5-bromo-4-chloro-3-indolylphosphate.
RESULTS
WbbO is a bifunctional galactosyltransferase.
Previous results showed that wbbO expression in E. coli K-12 resulted in modification of the lipid A core of the host LPS by the addition of a trisaccharide, β-d-Galf-(1→3)-α-d-Galp-(1→3)-β-d-GlcpNAc (7). The reaction was dependent on WecA activity and functional Wzx supplied by the host. The structural data were consistent with WbbO serving as a bifunctional galactosyltransferase that transfers two galactosyl residues to und-PP-GlcNAc, but interpretation of the data was complicated by the potential involvement of endogenous activities in the E. coli background. Therefore, analysis was performed in vitro to unequivocally determine the activity of WbbO.
Membranes prepared from E. coli CWG288 harboring the appropriate plasmids were used as a source of enzyme. This strain carries a galE::Tn10 mutation that eliminates UDP-glucose-4-epimerase and thus prevents the interconversion of UDP-Galp and UDP-Glcp. As a result, UDP-Galp substrate is directed specifically to d-galactan I synthesis and redirection of radiolabel from UDP-Galp into other glycoconjugates via UDP-Glcp is prevented. E. coli CWG288 also has a chromosomal Δsbc-rfb deletion in the O-PS biosynthesis region that eliminates the glf (UDP-Galf synthesis) and wzx genes that form part of the E. coli K-12 O-antigen biosynthesis (rfb) cluster (46, 57). In the absence of plasmid-encoded Wzx or ABC transporter activity, any polymers or oligosaccharides of d-galactan I formed in CWG288 membranes are retained as biosynthetic (lipid-linked) intermediates, rather than being exported and ligated to LPS lipid A core (7).
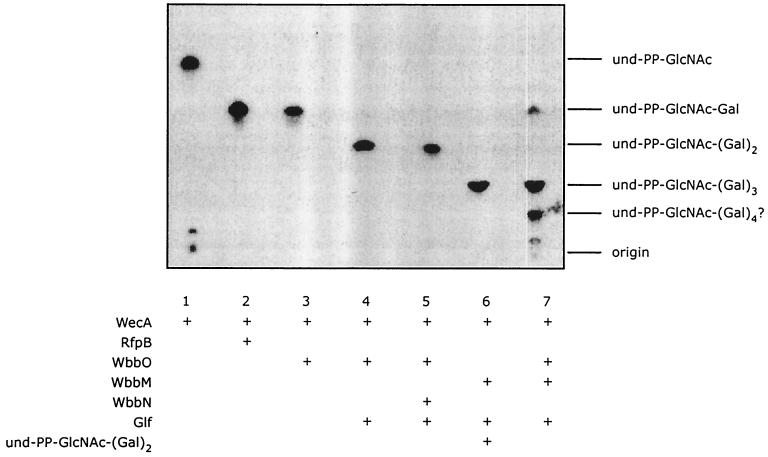
The lipid intermediates synthesized by membrane preparations were labeled with a radiolabeled NDP-sugar substrate, extracted in C:M, and separated by TLC. The resulting autoradiogram is shown in Fig. 2. Two standards were routinely used. The first consisted of und-PP-[14C]GlcNAc, synthesized by WecA. When E. coli membranes were incubated with UDP-[14C]GlcNAc to generate und-PP-GlcNAc via WecA from the chromosomal copy, the amounts of incorporation were small (data not shown). To increase und-PP-GlcNAc synthesis, E. coli overexpressing WecA (from pMAV11) was used as a source of enzyme. The majority of the resulting radioactive material (und-PP-GlcNAc) migrated as a single component in TLC (Fig. 2, lane 1). Variable trace amounts of larger material were detected near the origin. This larger material likely reflects extended ECA intermediates, as would be predicted in an enzyme preparation that would contain small residual amounts of the other ECA precursors. The second standard was und-PP-GlcNAc-Galp, formed by the sequential action of WecA and the monospecific galactosyltransferase (RfpB) from Shigella dysenteriae (9, 24). In the biosynthesis of the S. dysenteriae O1 antigen, RfpB transfers an α-(1→3)-linked Galp residue to und-PP-GlcNAc. In the absence of any added plasmid, E. coli CWG288 membranes show no incorporation of radioactivity from UDP-[14C]Galp into C:M-extractable lipids (Table 2). When RfpB is expressed from pJK2363, a significant amount of radioactive lipid intermediate is made with the slower migration consistent with the predicted product, und-PP-GlcNAc-Galp (Fig. 2, lane 2). As expected, the radioactive incorporation was eliminated when pJK2363 was expressed in a wecA mutant (data not shown).
FIG. 2.
TLC separation of lipid-linked intermediates synthesized and extracted in C:M (3:2) from membrane preparations of E. coli CWG288 expressing various glycosyltransferases. The relevant enzymes and exogenous lipid in each reaction are identified below the appropriate lane, and the predicted products are shown to the right. The reaction in lane 1 shows radioactivity incorporated from UDP[14C]GlcNAc, whereas all other samples were extracted from membranes incubated with UDP[14C]Galp. The following plasmids were used: lane 1, pMAV11 (wecA); lane 2, pJK2363 (rfpB); lane 3, pWQ20 (wbbO); lane 4, pWQ20 with added Glf extract; lane 5, pWQ151 (glf wbbN wbbO); lane 6, pWQ150 (wbbM glf) plus exogenous und-PP-GlcNAc-Galp-Galf; lane 7, pWQ20 plus pWQ150.
TABLE 2.
Incorporation of radioactivity from UDP-[14C]galactose into lipid-linked oligo- and polysaccharides
| Plasmid | Enzymea | [14C]Gal incorporation (pmol)
|
C:M (3:2)/hot water-phenol extract incorporation ratio | |
|---|---|---|---|---|
| C:M (3:2) extract | Hot water-phenol extract | |||
| None | <0.1 | |||
| pWQ20 | WbbO + Glf extract | 12.3 | 13.9 | 1:1.1 |
| pWQ41 | WbbM, Glf, WbbN, WbbO | 1.5 | 16.5 | 1:11 |
| pWQ20 + pWQ150 | WbbM, Glf, WbbO | 1.2 | 18.6 | 1:15.5 |
Membranes were prepared from E. coli CWG288, and incorporation was determined under standard reaction conditions.
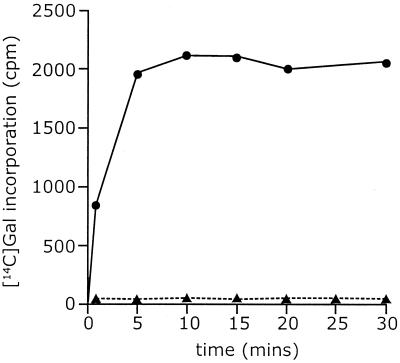
Membranes from cells expressing WbbO rapidly incorporated radioactivity from UDP-[14C]Galp into C:M-extractable material in a time-dependent manner (Fig. 3). The reaction was irreversible (data not shown). On TLC, the extracted material was separated into a single component that comigrated with und-PP-GlcNAc-Galp (Fig. 2, lane 3). No incorporation was detected when the membranes were prepared from a wecA mutant host strain expressing WbbO (data not shown). The in vitro results, together with the structure of WbbO-modified LPS (7), indicated that identical products were obtained with either RfpB or WbbO incubated with UDP-Galp. However, in these membranes, UDP-Galf is limiting, so the membranes were augmented with an aliquot of cytoplasmic extract from E. coli CWG288(pWQ66) overexpressing Glf (28). The equilibrium of the Glf-mediated interconversion is such that only about 5% of UDP-Galp is converted into UDP-Galf by the UDP-Galp mutase in the absence of any transferase to draw off the UDP-Galf product (28, 30). Supplementation of the WbbO-containing membranes with Glf-containing CWG288 lysate resulted in almost quantitative recovery of a higher-molecular-weight lipid intermediate with a migration consistent with a und-PP-linked trisaccharide (Fig. 2, lane 4). Addition of lysate from CWG288 lacking Glf had no effect (data not shown). The “second' galactosyltransferase activity is not the result of an unexpected endogenous activity in the membranes utilizing und-PP-GlcNAc-Galp as an acceptor, since E. coli CWG288 membranes prepared from cells expressing RfpB cannot form the und-PP-linked trisaccharide in the presence of Glf (data not shown).
FIG. 3.
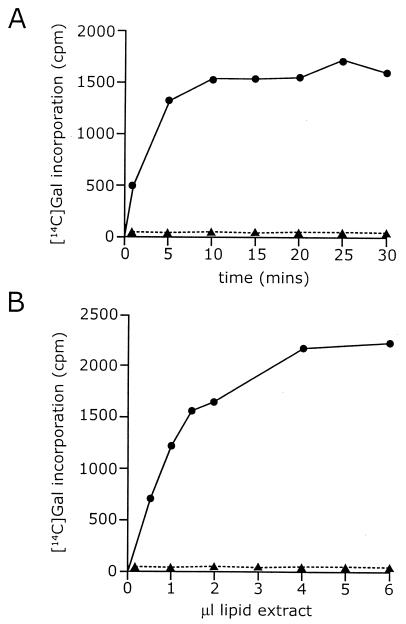
Galactosyltransferase activity of WbbO. The figure shows a time course of incorporation of radioactivity from UDP-[14C]Galp into lipid-linked intermediates catalyzed by membranes of E. coli CWG288(pWQ20) (solid circles). The control (solid triangles) shows membranes from E. coli CWG288 containing no plasmid.
Collectively, these data and previous structural studies support the conclusion that WbbO is a novel bifunctional galactosyltransferase that uses und-PP-GlcNAc as its acceptor to form und-PP-GlcNAc-Galp-Galf. Furthermore, the two activities of the WbbO enzyme could be distinguished by limitation of UDP-Galf so that the enzyme must function by a mechanism involving sequential galactosyl transfer rather than by a process in which both residues are transferred simultaneously to the acceptor.
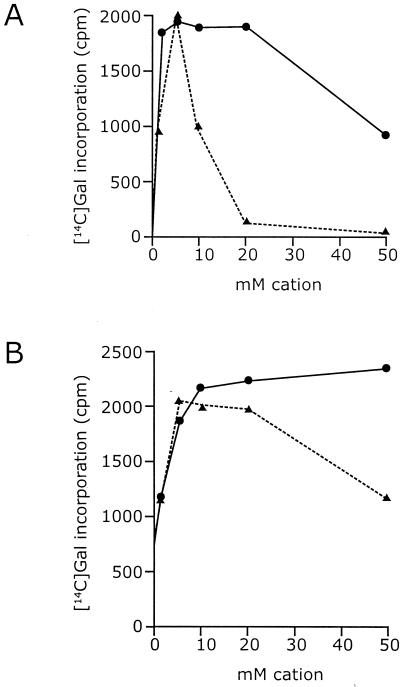
The WbbO-mediated galactosyl transfer reactions show activity over a broad pH range, with an optimum of pH 8.5. WbbO requires divalent cations, showing optimal activity in the presence of 5 to 20 mM Mg2+ (Fig. 4A). This requirement can be replaced by Mn2+. However, the higher concentrations of the two cations had different effects on activity. Concentrations of Mg2+ above 20 mM result in a 30 to 50% decrease in activity of WbbO, whereas the same concentration of Mn2+ results in >90% inhibition.
FIG. 4.
Cation dependence of WbbO and WbbM activity. Standard reaction mixtures were prepared in buffer B with MgCl2 omitted. MgCl2 (solid line) or MnCl2 (dashed line) was added to the concentrations shown, and the reaction mixtures were incubated for 30 min. Panel A shows incorporation of radioactivity from UDP=[14C]Galp into C:M extract by membranes containing WbbO. Panel B shows WbbM-mediated incorporation in membranes reconstituted with exogenous und-PP-GlcNAc-Galp-Galf.
The wbbM gene product is a galactopyranosyltransferase.
The next step in assembly, transfer of a Galp residue, is predicted by the known structure of d-galactan I. To identify the galactosyltransferase responsible, membranes containing WecA, WbbO, and Glf activity, together with either WbbM or WbbN, were examined for their ability to synthesize a lipid intermediate larger than the product formed by WecA, WbbO, and Glf alone. Membranes prepared from CWG288(pWQ151) expressing glf-wbbN-wbbO were only capable of synthesizing the same intermediate as membranes with WbbO and Glf (Fig. 2, lane 5), indicating the inability of WbbN to extend the endogenous lipid-linked trisaccharide intermediate. In contrast, membranes containing WbbO, Glf, and WbbM generated larger products (Fig. 2, lane 7).
To unequivocally establish WbbM as the enzyme responsible for catalyzing the next step in the biosynthesis of d-galactan I, plasmid pWQ150 was constructed, which put wbbM and glf under the control of the arabinose-inducible pBAD promoter. Membrane preparations from E. coli CWG288(pWQ150) were unable to incorporate radioactivity from UDP-[14C]Galp into C:M extract (data not shown). However, when the membrane preparations were reconstituted with an exogenous acceptor comprising the butanol extract from CWG288 membranes containing WbbO and Glf (i.e., und-PP-GlcNAc-Galp-Galf), radioactivity was rapidly incorporated from UDP-[14C]Galp into C:M extract (Fig. 5A). The incorporation of radioactivity was dependent on the amount of acceptor lipid used in the reconstitution (Fig. 5B). In contrast, no activity was detected when the membranes were reconstituted with butanol extracts from the host strain (E. coli CWG288) (Fig. 5A) or with the RfpB reaction product (und-PP-GlcNAc-Galp) (data not shown). The WbbM reaction product was extracted from the reconstituted membranes with C:M and separated by TLC (Fig. 2, lane 6). The product showed a size increase consistent with an increase of one galactosyl residue. Taking into consideration the known structure of d-galactan I, these data suggest that WbbM transfers a Galp residue to generate the lipid-linked tetrasaccharide, und-PP-GlcNAc-Galp-Galf-Galp.
FIG. 5.
Galactosyltransferase activity of WbbM in reconstituted membranes. Panel A shows a time course of incorporation of radioactivity from UDP-[14C]Galp into lipid-linked intermediates catalyzed by membranes of E. coli CWG288(pWQ150). The membranes were tested with (solid line) and without (dashed line) reconstitution with exogenous und-PP-GlcNAc-Galp-Galf. Panel B shows the dependence of incorporation on the amount of exogenous acceptor (solid line). The control (dashed line) shows the absence of activity in membranes reconstituted with lipid extracted from CWG288.
Like WbbO activity, the WbbM reaction in reconstituted membranes was active over a broad pH range, with an optimum of pH 8.5. The transfer of [14C]Galp from precursor to und-PP-trisaccharide also showed a requirement for divalent cations (Fig. 4B). Close to 95% of maximal activity resulted from addition of 10 mM Mg2+, with no inhibition at higher concentrations. Mn2+ could substitute for Mg2+, but concentrations above 20 mM were inhibitory. Levels of incorporation increased up to 50 mM Mg2+. In the case of WbbM, the effects of divalent cations could reflect their involvement in effective lipid reconstitution rather than enzymatic activity per se, and there is no obvious way to resolve these alternatives.
Polymer extension requires only WbbM and WbbO.
Plasmid pWQ41 contains the wbbM-glf-wbbN-wbbO genes necessary for the synthesis of d-galactan I (7), and it was expected that E. coli CWG288(pWQ41) would provide a source of lipid intermediates with a higher degree of polymerization. Although these membranes incorporated large amounts of radioactivity from UDP-[14C]Galp, relatively little radioactivity could be extracted by using C:M (Table 2). Solvents such as C:M (2:1) have been used to extract und-PP-linked intermediates as large as octasaccharides (38). The inability to detect incorporation of radioactivity into the C:M extracts was interpreted as reflecting rapid elongation of the lipid-linked polymer by the processive galactosyltransferases. In attempts to trap shorter intermediates, the duration of incubation was reduced and the incubation temperature was reduced. Neither approach was effective. Therefore, to isolate a full range of d-galactan I oligosaccharides and polymers, membranes were extracted with hot aqueous phenol. This treatment cleaves lipid intermediates due to the labile linkage between phosphate and the unsaturated α-isoprene of undecaprenol, releasing lipid-free phosphoryated glycans and oligosaccharides into the aqueous phase (21). The d-galactan I structure is stable under these conditions, because the same extraction is used in the isolation of K. pneumoniae O1 LPS for structural studies (55). Thus, the difference in incorporation in samples extracted in phenol-water compared to that in C:M reflects products with higher degrees of polymerization. E. coli CWG288(pWQ41) released a significant amount of radioactivity into the phenol-water extract, with an 11.1-fold increase over the incorporation into C:M (Table 2). In contrast, equivalent levels of incorporation were detected in the C:M and aqueous phenol extracts from membranes expressing wbbO with Glf-containing extract, as would be expected for und-PP-linked trisaccharides (Table 2).
To confirm the identity of the polymeric material in membranes from CWG288(pWQ41), membranes were solubilized in SDS, separated by Tricine-SDS-PAGE, and examined by Western immunoblotting with polyclonal serum specific for d-galactan I (Fig. 6). These membranes contained high-molecular-weight d-galactan I. Note that LB contains sufficient galactose to overcome the galE defect in CWG288 and allow synthesis of d-galactan I to occur during growth in strains harboring pWQ41.
FIG. 6.
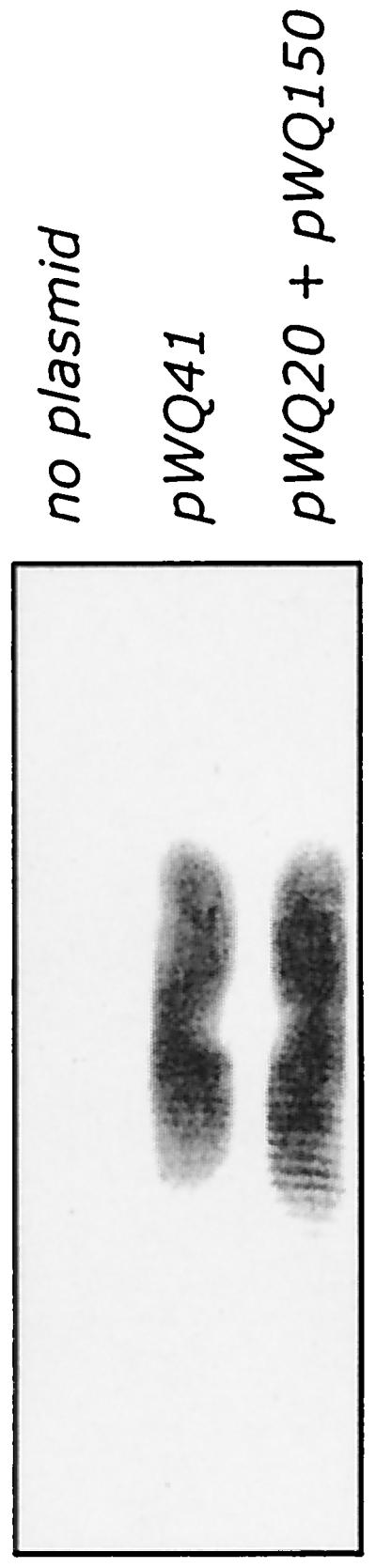
Lipid-linked d-galactan I polymer formed in membranes from E. coli CWG288 expressing combinations of galactosyltransferases. The figure shows a Western immunoblot reacted with d-galactan I-specific antibodies. d-Galactan I was synthesized in membranes from cells containing pWQ41 (wbbM glf wbbN wbbO) and pWQ20 (wbbO) plus pWQ150 (wbbM glf).
The known structure of d-galactan I dictates that the step following WbbO in the assembly pathway involves a galactofuranosyltransferase, and we initially expected that WbbN would perform that function. However, constructs containing WbbN were unable to incorporate radiolabeled galactose or extend und-PP-GlcNAc-Galp-Galf-Galp acceptor reconstituted into membrane preparations (data not shown). This result could potentially reflect an inability to reconstitute the system with larger lipid intermediates. However, a larger lipid radiolabeled intermediate was detected in the C:M extract from membranes prepared from E. coli CWG288 containing both plasmids pWQ20 (wbbO) and pWQ150 (wbbM-glf). The lipid extract from these membranes contained compounds with TLC migrations consistent with und-PP-GlcNAc-Galp and und-PP-GlcNAc-Galp-Galf-Galp as well as a larger compound (Fig. 2, lane 7). Extraction with hot phenol-water revealed that these membranes were also fully capable of generating higher-molecular-weight intermediates (Table 2). When membranes from this strain were subjected to Tricine-SDS-PAGE and Western immunoblotting with polyclonal antisera specific for d-galactan I (Fig. 6), high-molecular-weight d-galactan polymer was detected in these preparations. No d-galactan I was detected on membranes from CWG288 containing no plasmid (Fig. 6) or in membranes from the strain transformed with pWQ150, pWQ151, or pWQ20 (data not shown) alone. The WbbO and WbbM galactosyltransferases are therefore sufficient for the synthesis of d-galactan I.
DISCUSSION
d-Galactan I and structures based on d-galactan I are found in a variety of Klebsiella O serotypes. The structures of the glycan backbones of the O1 and O8 antigens are identical, but they differ in the presence of O-acetyl groups at either the 2 or 6 position of Galf residues in d-galactan I in serotype O8 (19). The O1 and O8 antigens cross-react due to the shared d-galactan II antigen. The serotype O2a antigen is comprised only of d-galactan I, while serotype O2a, 2c has d-galactan I together with another structurally (and serologically) distinct domain (54). The LPS from serotype reference strains for O9 and O2a, 2e, 2 h contains d-galactan I substituted for nonstoichiometrically with O-acetyl groups and an α-(1→2)-linked Galp side group attached to the backbone Galp residues: these serotypes only differ in the extent of substitution (18, 32). Finally, serotype O2a, 2f, 2g contains d-galactan I in which the d-Galp residues are stoichiometrically substituted for with an α-(1→4)-linked Galp side group (18). d-Galactan I is also found in O-PS from other gram-negative bacteria. For example, Serratia plymuthica produces d-galactan I and d-galactan II (2). d-Galactan I is also found in Serratia marcescens O16 and O20 (39, 40) and Pasteurella hemolytica serotypes 4 and T10 (41, 43). S. marcescens O24 forms a glycan structure identical to that of the Klebsiella O2a, 2f, 2g antigen (39).
Biosynthetic data are not available for all of the d-galactan I-based structures, but some relevant genetic observations have been made. The d-galactan I-biosynthesis loci in Klebsiella serotypes form at least three clonal groups based on the extent of nucleotide sequence similarity, as is evident in hybridization studies (19, 20). However, the genetic organization and gene products of the O1 and O8 loci are conserved (20). Gene probes derived from the K. pneumoniae O1 d-galactan I biosynthesis locus also hybridize at low stringency to DNA from both S. marcescens O16 and O20. The cluster from O16 has been sequenced and has the same genetic organization as K. pneumoniae O1 (48). The corresponding S. marcescens gene products are highly conserved and in some cases are functionally interchangeable with their K. pneumoniae counterparts. From the data available, it is reasonable to assume that the biosynthesis of d-galactan I follows the same pathway in different gram-negative bacteria. Interestingly the prototype ABC transporter-dependent O-PS assembly system involves the E. coli O9a polymannose antigen. The same O-PS structure is found in K. pneumoniae serotype O3, and the E. coli O9a antigen is thought to have arisen through a recombination event involving transfer of O-PS biosynthesis genes from Klebsiella O3 to E. coli (47).
WbbO is a novel bifunctional galactosyltransferase capable of transferring α-linked Galp as well as β-linked Galf residues to the und-PP-GlcNAc acceptor. Several independent lines of evidence lead to the conclusion that the resulting compound contains the trisaccharide β-d-Galf-(1→3)-α-d-Galp-(1→3)-β-d-GlcpNAc linked to und-PP. These data include the in vitro dependence of the first galactosyl transfer on the well-characterized WecA reaction product, the in vitro dependence of the second galactosyl transfer reaction on UDP-galactopyranose mutase (Glf) activity, and the previously determined composition and linkages of the modified LPS in E. coli K-12 expressing WbbO (7). The WbbO enzyme has been assigned as a retaining glycosyltransferase belonging to family 4 (6) based on features of the amino acid sequence as described by P. M. Coutinho and B. Henrissat (http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html). As might be expected from their shared activities, RfpB belongs to the same family. The mechanism involved in the bifunctionality of WbbO is unknown. The RfpB and WbbO proteins are similar in size and share considerable similarity over 122 residues at the C terminus (37.7% identity, 52.4% similarity) (7). Whether the divergent N-terminal domain of WbbO contains the β-galactofuranosyltransferase activity that distinguishes WbbO from RfpB is unclear. This region shows no informative similarity to other glycosyltransferases or to any other known proteins in the databases. It is also conceivable that the enzyme has overlapping sites for Galp and Galf transferase activities. The fact that the two steps of WbbO activity can be uncoupled may provide a means of identifying the active site in the future.
Since the α-galactopyranosyltransferase activity of WbbO transfers residues to a GlcNAc acceptor, it is not surprising that it is confined to the initiation stages of d-galactan I biosynthesis. In this respect, the Galp transferase activity of WbbO is analogous to WbdC activity in assembly of the O9a antigen of E. coli (23). WbdC adds a single mannose residue to serve as a specific “adapter” between the versatile und-PP-GlcNAc initial acceptor and the repeat unit domain subsequently formed by WbdB and WbdA. However, unlike WbdC, the role of the WbbO protein is not confined to synthesis of the “adapter” for d-galactan I biosynthesis. Its β-galactofuranosyltransferase activity also contributes, together with WbbM, to processive polymerization of the repeat unit structure of d-galactan I. In both contexts, the WbbO protein must transfer a β-d-Galf residue to a Galp acceptor to form the β-d-Galf-(1→3)-α-d-Galp linkage. In the polymerization of the O9a antigen, WbdB and WbdC appear to transfer more than one mannosyl residue to the growing chain (23), and such activities provide a convenient mechanism by which the repeat unit structure with alternating pairs of α-(1→2) and α-(1→3) linkages can be maintained.
WbbM was assigned to glycosyltransferase family 8 (http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html) based on amino acid sequence features, and its role as a retaining α-galactosyltransferase is now established. One interesting feature of WbbM is its size. With a size of 73,286 Da predicted by sequence data and confirmed by expression experiments (5), WbbM is bigger than most other monofunctional bacterial galactosyltransferases. Interestingly the WbdA protein involved in O9a antigen synthesis is also much larger than the other related mannosyltransferases. While the size of WbdA must take into account the fact that it appears to have two distinct domains, each containing a motif conserved among several bacterial α-mannosyltransferases (22), its size also reflect an involvement in other roles. For example it might coordinate the alternately acting enzymes to maintain what is a very rapid and efficient processive polymerization process. As shown here, the rate of transfer of galactosyl residues to the und-PP-GlcNAc acceptor in the presence of all of the required enzymes is sufficiently fast that it is difficult to identify any shorter lipid intermediates extractable in C:M.
The role of the wbbN gene product in d-galactan I biosynthesis remains unclear at this time. While the current data indicate that this gene is expendable from the perspective of polymerization, the observations do not necessarily preclude a glycosyltransferase function. In fact, database searches (data not shown) indicate some local similarities shared by WbbN and some known and predicted β-glycosyltransferases, including ExoO, a β-glucosyltransferase involved in succinoglycan biosynthesis in Sinorhizobium meliloti (42). It is conceivable that WbbN participates in some other way in the assembly of d-galactan I-containing LPS. One possibility would involve termination of the elongation process. To date it is still unknown how the chain length of O-PS is controlled in the ABC transporter-dependent pathway (reviewed in reference 53). In the E. coli O8 antigen (the synthesis of which is similar to that of O9a), polymannose chains terminate in 3-O-methylmannose, and this has been proposed to reflect a discrete termination process (15). The biosynthetic source of the modified mannosyl residue is unknown. To date, we have found no evidence for any modified glycose units terminating d-galactan I (M. B. Perry and C. Whitfield, unpublished data), and the lipid-linked d-galactan I synthesized in membranes shows no obvious differences in chain length in the presence or absence of WbbN (Fig. 6). Interestingly, it is not possible to generate plasmid constructs with a wbbN deletion in the d-galactan I biosynthesis cluster. In contrast, deletion of either wbbO or wbbM can be readily achieved (R. Köplin and C. Whitfield, unpublished data). While this information is suggestive of an important role for WbbN in the assembly of d-galactan I in vivo, it does not provide any further clues as to its function. The locus for the biosynthesis of the E. coli O9a antigen also contains one gene of unknown function (wbdD) located in the vicinity of the known mannosyltransferase genes (wbdA, wbdB, and wbdC). The WbdD and WbbN proteins share no sequence similarity and may have different roles. Determination of their precise function(s) in O-PS assembly awaits further study.
ACKNOWLEDGMENTS
J. Klena, P. Rick, and M. Valvano generously provided bacterial strains and plasmids. We acknowledge the contributions of R. Köplin, who made pWQ41, and P. Amor, who prepared d-galactan I antiserum. We thank B. Clarke for critically reviewing the manuscript.
Financial support for this work was provided by grants to C.W. and A.J.C. from the Natural Sciences and Engineering Research Council of Canada. C.W. is a Canadian Institutes of Health Research (CIHR) Senior Investigator.
REFERENCES
- 1.Alexander D C, Valvano M A. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J Bacteriol. 1994;176:7079–7084. doi: 10.1128/jb.176.22.7079-7084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auken H M, Oxley D, Wilkinson S G. Structural and serological characterisation of an O-specific polysaccharide from Serratia plymuthica. FEMS Microbiol Lett. 1993;111:295–300. doi: 10.1111/j.1574-6968.1993.tb06401.x. [DOI] [PubMed] [Google Scholar]
- 3.Behrens N H, Leloir L F. Dolichol monophosphate glucose: an intermediate in glucose transfer in liver. Proc Natl Acad Sci USA. 1970;66:153–159. doi: 10.1073/pnas.66.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binotto J, MacLachlan P R, Sanderson P R. Electrotransformation of Salmonella typhimurium LT2. Can J Microbiol. 1991;37:474–477. doi: 10.1139/m91-078. [DOI] [PubMed] [Google Scholar]
- 5.Bronner D, Clarke B R, Whitfield C. Identification of an ATP-binding cassette transport system required for translocation of lipopolysaccharide O-antigen side chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol Microbiol. 1994;14:505–519. doi: 10.1111/j.1365-2958.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 6.Campbell J A, Davies G J, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke B R, Bronner D, Keenleyside W J, Severn W B, Richards J C, Whitfield C. Role of Rfe and RfbF in the initiation of biosynthesis of d-galactan I, the lipopolysaccharide O antigen from Klebsiella pneumoniae serotype O1. J Bacteriol. 1995;177:5411–5418. doi: 10.1128/jb.177.19.5411-5418.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke B R, Whitfield C. Molecular cloning of the rfb region of Klebsiella pneumoniae serotype O1:K20: the rfb gene cluster is responsible for synthesis of the d-galactan I O polysaccharide. J Bacteriol. 1992;174:4614–4621. doi: 10.1128/jb.174.14.4614-4621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fält I C, Schweda E K H, Weintraub H, Sturm S, Timmis K N, Lindberg A A. Expression of the Shigella dysenteriae type-1 lipopolysaccharide repeating unit in Escherichia coli K-12/Shigella dysenteriae type-1 hybrids. Eur J Biochem. 1993;213:573–581. doi: 10.1111/j.1432-1033.1993.tb17796.x. [DOI] [PubMed] [Google Scholar]
- 10.Feldman F M, Marolda C L, Monteiro M A, Perry M B, Parodi A J, Valvano M A. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J Biol Chem. 1999;274:35129–35138. doi: 10.1074/jbc.274.49.35129. [DOI] [PubMed] [Google Scholar]
- 11.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen D S, Mestre F, Albertí S, Hernández-Allés S, Álvarez D, Doménech-Sánchez A, Gil J, Merino S, Tomás J M, Benedi V J. Klebsiella pneumoniae lipopolysaccharide O typing: revision of prototype strains and O-group distribution among clinical isolates from different sources and countries. J Clin Microbiol. 1999;37:56–62. doi: 10.1128/jcm.37.1.56-62.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jann K, Goldemann G, Weisgerber C, Wolf-Ullisch C, Kanegasaki S. Biosynthesis of the O9 antigen of Escherichia coli. Initial reaction and overall reaction. Eur J Biochem. 1982;127:157–164. doi: 10.1111/j.1432-1033.1982.tb06850.x. [DOI] [PubMed] [Google Scholar]
- 15.Jansson P-E, Lönngren J, Widmalm G. Structural studies of the O-antigen polysaccharides of Klebsiella O5 and Escherichia coli O8. Carbohydr Res. 1985;145:59–66. doi: 10.1016/s0008-6215(00)90412-9. [DOI] [PubMed] [Google Scholar]
- 16.Keenleyside W J, Perry M B, MacLean L L, Poppe C, Whitfield C. A plasmid-encoded rfbO:54 gene cluster is required for biosynthesis of the O:54 antigen in Salmonella enterica serovar Borreze. Mol Microbiol. 1994;11:437–448. doi: 10.1111/j.1365-2958.1994.tb00325.x. [DOI] [PubMed] [Google Scholar]
- 17.Keenleyside W J, Whitfield C. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J Biol Chem. 1996;271:28581–28592. doi: 10.1074/jbc.271.45.28581. [DOI] [PubMed] [Google Scholar]
- 18.Kelly R F, MacLean L L, Perry M B, Whitfield C. Structures of the O-antigens of Klebsiella serotypes O2(2a, 2e), O2(2a, 2e, 2h), and O2(2a, 2f, 2g), members of a family of related D-galactan O-antigens in Klebsiella spp. J Endotoxin Res. 1995;2:131–140. [Google Scholar]
- 19.Kelly R F, Severn W B, Richards J C, Perry M B, MacLean L L, Tomás J M, Merino S, Whitfield C. Structural variation in the O-specific polysaccharides of Klebsiella pneumoniae serotype O1 and O8 lipopolysaccharide: evidence for clonal diversity in rfb genes. Mol Microbiol. 1993;10:615–625. doi: 10.1111/j.1365-2958.1993.tb00933.x. [DOI] [PubMed] [Google Scholar]
- 20.Kelly R F, Whitfield C. Clonally diverse rfb gene clusters are involved in expression of a family of related d-galactan O antigens in Klebsiella species. J Bacteriol. 1996;178:5205–5214. doi: 10.1128/jb.178.17.5205-5214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent J L, Osborn M J. Properties of the O-specific hapten formed in vivo by mutant strains of Salmonella typhimurium. Biochemistry. 1968;7:4396–4408. doi: 10.1021/bi00852a036. [DOI] [PubMed] [Google Scholar]
- 22.Kido N, Sugiyama T, Yokochi T, Kobayashi H, Okawa Y. Synthesis of Escherichia coli O9a polysaccharide requires the participation of two domains of WbdA, a mannosyltransferase encoded within the wb* gene cluster. Mol Microbiol. 1998;27:1213–1221. doi: 10.1046/j.1365-2958.1998.00765.x. [DOI] [PubMed] [Google Scholar]
- 23.Kido N, Torgov V I, Sugiyama T, Uchiya K, Sugihara H, Komatsu T, Kato N, Jann K. Expression of the O9 polysaccharide of Escherichia coli: sequencing of the E. coli O9 rfb gene cluster, characterization of mannosyl transferases, and evidence for an ATP-binding cassette transport system. J Bacteriol. 1995;177:2178–2187. doi: 10.1128/jb.177.8.2178-2187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klena J D, Ashford II R S, Schnaitman C A. Role of Escherichia coli K-12 rfa genes and the rfp gene of Shigella dysenteriae 1 in generation of lipopolysaccharide core heterogeneity and attachment of O antigen. J Bacteriol. 1992;174:7297–7307. doi: 10.1128/jb.174.22.7297-7307.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kogan G, Haraguchi G, Hull S I, Hull R A, Shashkov A, Jann B, Jann K. Structural analysis of O4-reactive polysaccharides from recombinant Escherichia coli. Changes in the O-specific polysaccharide induced by cloning of the rfb genes. Eur J Biochem. 1993;214:259–265. doi: 10.1111/j.1432-1033.1993.tb17919.x. [DOI] [PubMed] [Google Scholar]
- 26.Kol O, Wieruszeski J-M, Strecker G, Fournet B, Zalisz R, Smets P. Structure of the O-specific polysaccharide chain of Klebsiella pneumoniae O1:K2 (NCTC 5055) lipopolysaccharide. A complementary elucidation. Carbohydr Res. 1992;236:339–344. doi: 10.1016/0008-6215(92)85028-x. [DOI] [PubMed] [Google Scholar]
- 27.Kol O, Wieruszeski J-M, Strecker G, Montreuil J, Fournet B. Structure of the O-specific polysaccharide chain from Klebsiella pneumoniae O1:K2 (NCTC 5055) lipopolysaccharide. Carbohydr Res. 1991;217:117–125. doi: 10.1016/0008-6215(91)84122-u. [DOI] [PubMed] [Google Scholar]
- 28.Köplin R, Brisson J R, Whitfield C. UDP-galactofuranose precursor required for formation of the lipopolysaccharide O antigen of Klebsiella pneumoniae serotype O1 is synthesized by the product of the rfbDKPO1 gene. J Biol Chem. 1997;272:4121–4128. doi: 10.1074/jbc.272.7.4121. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn H-M, Meier-Dieter U, Mayer H. ECA, the enterobacterial common antigen. FEMS Microbiol Rev. 1988;54:195–222. doi: 10.1111/j.1574-6968.1988.tb02743.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee R, Monsey D, Weston A, Duncan K, Rithner C, McNeil M. Enzymatic synthesis of UDP-galactofuranose and an assay for UDP-galactopyranose mutase based on high-performance liquid chromatography. Anal Biochem. 1996;242:1–7. doi: 10.1006/abio.1996.0419. [DOI] [PubMed] [Google Scholar]
- 31.Liu D, Reeves P R. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- 32.MacLean L L, Whitfield C, Perry M B. Characterization of the polysaccharide antigen of Klebsiella pneumoniae O:9 lipopolysaccharide. Carbohydr Res. 1993;239:325–328. doi: 10.1016/0008-6215(93)84231-t. [DOI] [PubMed] [Google Scholar]
- 33.McCallum K L, Schoenhals G, Laakso D, Clarke B R, Whitfield C. A high molecular weight fraction of smooth lipopolysaccharide in Klebsiella serotype O1:K20 contains a unique O-antigen epitope and determines resistance to nonspecific serum killing. Infect Immun. 1989;57:3816–3822. doi: 10.1128/iai.57.12.3816-3822.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick P D. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- 35.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 36.Nassau P M, Martin S L, Brown R E, Weston A, Monsey D, McNeil M R, Duncan K. Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J Bacteriol. 1996;178:1047–1052. doi: 10.1128/jb.178.4.1047-1052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohta M, Ina K, Kusuzaki K, Kido N, Arakawa Y, Kato N. Cloning and expression of the rfe-rff gene cluster of Escherichia coli. Mol Microbiol. 1991;5:1853–1862. doi: 10.1111/j.1365-2958.1991.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 38.Osborn J M, Cynkin M A, Gilbert J M, Muller L, Singh M. Synthesis of bacterial O-antigens. Methods Enzymol. 1972;28:583–601. [Google Scholar]
- 39.Oxley D, Wilkinson S G. Structure of the O-specific galactan from the lipopolysaccharide of the reference strain for Serratia marcescens serogroup O24. Carbohydr Res. 1989;195:117–122. doi: 10.1016/0008-6215(89)85094-3. [DOI] [PubMed] [Google Scholar]
- 40.Oxley D, Wilkinson S G. Structures of neutral glycans isolated from the lipopolysaccharides of reference strains for Serratia marcescens serogroups O16 and O20. Carbohydr Res. 1989;193:241–248. doi: 10.1016/0008-6215(89)85122-5. [DOI] [PubMed] [Google Scholar]
- 41.Perry B M, Babiuk L A. Structure of the polysaccharide chain of Pasteurella haemolytica (serotype 4) lipopolysaccharide. Can J Biochem Cell Biol. 1984;46:1163–1165. [Google Scholar]
- 42.Reuber T L, Walker G C. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 43.Richards J C, Leitch R A. Elucidation of the structure of Pasteurella haemolytica serotype T10 lipopolysaccharide O-antigen by NMR spectroscopy. Carbohydr Res. 1989;186:275–286. doi: 10.1016/0008-6215(89)84041-8. [DOI] [PubMed] [Google Scholar]
- 44.Rick P D, Hubbard G L, Barr K. Role of the rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J Bacteriol. 1994;176:2877–2884. doi: 10.1128/jb.176.10.2877-2884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rocchetta H L, Lam J S. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J Bacteriol. 1997;179:4713–4724. doi: 10.1128/jb.179.15.4713-4724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P R. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugiyama T, Kido N, Kato Y, Koide N, Yoshida T, Yokochi T. Generation of Escherichia coli O9a serotype, a subtype of E. coli O9, by transfer of the wb* gene cluster of Klebsiella O3 into E. coli via recombination. J Bacteriol. 1998;180:2775–2778. doi: 10.1128/jb.180.10.2775-2778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szabo M, Bronner D, Whitfield C. Relationships between the rfb gene clusters required for biosynthesis of identical d-galactose-containing O antigens in Klebsiella pneumoniae serotype O1 and Serratia marcescens serotype O20. J Bacteriol. 1995;177:1544–1553. doi: 10.1128/jb.177.6.1544-1553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towbin T M, Staehelin T, Gordon G. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trautmann M, Ruhnke M, Rukavina T, Held T K, Cross A S, Marre R, Whitfield C. O antigen seroepidemiology of Klebsiella clinical isolates and implications for immunoprophylaxis of Klebsiella infections. Clin Diagn Lab Immunol. 1997;4:550–555. doi: 10.1128/cdli.4.5.550-555.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westphal O, Jann K. Bacterial lipopolysaccharide extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 52.Whitfield C. Biosynthesis of lipopolysaccharide O-antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 53.Whitfield C, Amor P A, Köplin R. Modulation of surface architecture of Gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol Microbiol. 1997;23:629–638. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]
- 54.Whitfield C, Perry M B, MacLean L L, Yu S-H. Structural analysis of the O-antigen side chain polysaccharides in the lipopolysaccharides of Klebsiella serotypes O2(2a), O2(2a, 2b), and O2(2a, 2c) J Bacteriol. 1992;174:4913–4919. doi: 10.1128/jb.174.15.4913-4919.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitfield C, Richards J C, Perry M B, Clarke B R, MacLean L L. Expression of two structurally distinct d-galactan O antigens in the lipopolysaccharide of Klebsiella pneumoniae serotype O1. J Bacteriol. 1991;173:1420–1431. doi: 10.1128/jb.173.4.1420-1431.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao Z, Liu H, Valvano M A. Acetylation of O-specific lipopolysaccharides from Shigella flexneri 3a and 2a occurs in Escherichia coli K-12 carrying cloned S. flexneri 3a and 2a rfb genes. J Bacteriol. 1992;174:7500–7508. doi: 10.1128/jb.174.23.7500-7508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao Z, Valvano M A. Genetic analysis of the O-specific lipopolysaccharide biosynthesis region (rfb) of Escherichia coli K-12 W3110: identification of genes that confer group 6 specificity to Shigella flexneri serotypes Y and 4a. J Bacteriol. 1994;176:4133–4143. doi: 10.1128/jb.176.13.4133-4143.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L, Al-Hendy A, Toivanen P, Skurnik M. Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: similarities to the dTDP-L-rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol Microbiol. 1993;9:309–321. doi: 10.1111/j.1365-2958.1993.tb01692.x. [DOI] [PubMed] [Google Scholar]