Significance
Sea turtle populations have undergone recent global declines. We analyzed de novo assembled genomes for both extant sea turtle families through the Vertebrate Genomes Project to inform their conservation and evolutionary biology. These highly conserved genomes were differentiated by localized gene-rich regions of divergence, particularly within microchromosomes, suggesting that these genomic elements play key functional roles in the evolution of sea turtles and possibly other vertebrates. We further demonstrate that dissimilar evolutionary histories impact standing genomic diversity and genetic load, and are critical to consider when using these metrics to assess adaptive potential and extinction risk. Our results also demonstrate how reference genome quality impacts inferences of comparative and conservation genomics analyses that need to be considered in their application.
Keywords: marine turtle, gene evolution, conservation genomics, genetic diversity, demographic history
Abstract
Sea turtles represent an ancient lineage of marine vertebrates that evolved from terrestrial ancestors over 100 Mya. The genomic basis of the unique physiological and ecological traits enabling these species to thrive in diverse marine habitats remains largely unknown. Additionally, many populations have drastically declined due to anthropogenic activities over the past two centuries, and their recovery is a high global conservation priority. We generated and analyzed high-quality reference genomes for the leatherback (Dermochelys coriacea) and green (Chelonia mydas) turtles, representing the two extant sea turtle families. These genomes are highly syntenic and homologous, but localized regions of noncollinearity were associated with higher copy numbers of immune, zinc-finger, and olfactory receptor (OR) genes in green turtles, with ORs related to waterborne odorants greatly expanded in green turtles. Our findings suggest that divergent evolution of these key gene families may underlie immunological and sensory adaptations assisting navigation, occupancy of neritic versus pelagic environments, and diet specialization. Reduced collinearity was especially prevalent in microchromosomes, with greater gene content, heterozygosity, and genetic distances between species, supporting their critical role in vertebrate evolutionary adaptation. Finally, diversity and demographic histories starkly contrasted between species, indicating that leatherback turtles have had a low yet stable effective population size, exhibit extremely low diversity compared with other reptiles, and harbor a higher genetic load compared with green turtles, reinforcing concern over their persistence under future climate scenarios. These genomes provide invaluable resources for advancing our understanding of evolution and conservation best practices in an imperiled vertebrate lineage.
Sea turtles recolonized marine environments over 100 Mya (1, 2) and are now one of the most widely distributed vertebrate groups on the planet (3). Leatherback turtles (Dermochelys coriacea) represent the only remaining species of the family Dermochelyidae, which diverged from the Cheloniidae (hard-shelled sea turtles) about 60 Mya (4). Unique morphological (Fig. 1A) and physiological traits allow leatherback turtles to exploit cool, highly productive pelagic habitats (5, 6), while green turtles (Chelonia mydas) and other hard-shelled species largely inhabit warmer nearshore habitats following an early pelagic life stage. Most previous research in this group has focused on organismal and ecological adaptations (7), but the genomic basis of traits that differentiate or unite these species is not well understood.
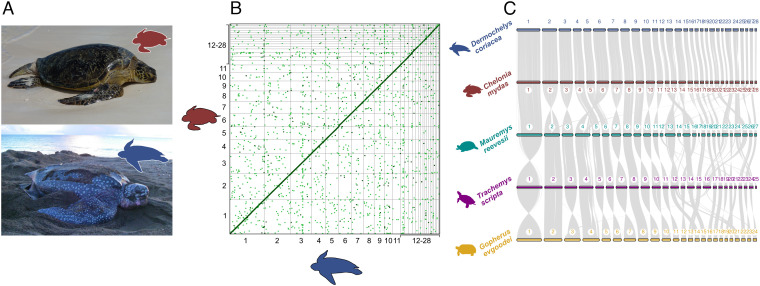
Fig. 1.
(A) Green turtle (C. mydas); photo credit: NOAA NMFS PIFSC under USFWS Permit #TE-72088A-3, and leatherback turtle (D. coriacea); photo credit: Ricardo Tapilatu. (B) Dot plot showing regions with an identity greater than 0.5 across the entire genomes of green (red) and leatherback (blue) turtles. (C) Gene synteny and collinearity among leatherback turtle (blue), green turtle (red), Chinese pond turtle (Mauremys reevesii; green), pond slider turtle (Trachemys scripta; purple) and Goode's thornscrub tortoise (Gopherus evgoodei; yellow). Each bar represents chromosomes with respective numbers, and gray lines represent homolog gene connections.
Anthropogenic pressures have caused substantial population declines in sea turtles, with contemporary populations representing mere fractions of their historical abundances (8, 9). Although sea turtles spend most of their life in the ocean, they also exhibit long-distance migrations to natal rookeries for terrestrial reproduction (7, 10, 11). Consequently, they are threatened by human activities in both terrestrial and marine environments, including direct harvest of meat and eggs (12), fisheries bycatch (13), coastal development (14, 15), pollution (16), disease (17), and climate change (18, 19), which is exacerbated by their temperature-dependent mechanism of sex determination (TSD) altering population dynamics (20, 21). The IUCN lists most sea turtle species as vulnerable or endangered, and while decades of conservation efforts have fueled positive trends for some populations (22), others continue to decline (23). In particular, leatherback turtles have undergone extensive declines (>95% in some populations) over the last century (24–27), including the extirpation of the Malaysian nesting population (28). Leatherback turtle recovery is impeded by relatively low hatching success compared with other sea turtle species (29). In contrast, many green turtle populations have recently increased following conservation actions (22), but their continued recovery remains threatened by anthropogenic activities and high incidence of the neoplastic disease fibropapillomatosis (FP), a viral-mediated tumor disease that disproportionately impacts this species (30).
Genomic data have been instrumental in advancing understanding of species’ evolutionary histories and ecological adaptations (31–33), and providing critical information for conservation management (34–37). However, this research has been hampered in taxa where genomic resources remain limited. In particular, the lack of high-quality reference genomes, which are essential for accurate comparative evolutionary analyses (38, 39) and robust estimates of a range of metrics to inform conservation biology such as inbreeding, hybridization, disease susceptibility, genetic load, and adaptation (36, 40, 41), impede this work in threatened species. A draft genome for the green turtle was assembled almost a decade ago (42), and provided important insights into turtle evolution. However, errors, gaps, misassemblies, and fragmentation in draft genomes can lead to spurious inferences, potentially masking signals of interest (38, 43) and impeding effective management strategies (41). Well-annotated, chromosomal-level reference genomes can resolve these issues, improving our understanding of the genomic underpinnings of ecological and evolutionary adaptations (39, 44). For example, high-quality genomes with accurate annotations have enabled examination of gene changes associated with recolonization of the marine environment by terrestrial vertebrates, including the loss of olfactory receptor (OR) gene families (32, 45). Comparative genomic analyses have also demonstrated adaptive diversity in genes underlying reptilian immunity (46), and high-quality genomes have provided key insights into mammalian disease susceptibility (33, 47, 48). Equivalent investigations are critical for sea turtles, with diseases such as FP adversely impacting populations across the globe (30), information on immune genes is needed for devising effective conservation strategies (49).
We assembled chromosome-level reference genomes for leatherback and green turtles as part of the Vertebrate Genomes Project (VGP), and leveraged these resources to address questions centered around evolutionary history and conservation. Specifically, we provide insights into the genomic underpinnings of phenotypic traits that separate and unite these two species by examining genome synteny and regions of divergence. Given the contrasting recent population trends of these two species, we additionally used whole genome resequencing data of individuals representative of global populations to compare key conservation-relevant metrics, including patterns of diversity and deleterious variants, and reconstructed demographic histories to inform assessments of future vulnerability. These genomes represent two of the most contiguous reptilian genomes assembled to date, and our results provide a foundation for further hypothesis-driven investigations into the evolutionary adaptation and conservation of this imperiled vertebrate lineage.
Results
Genome Quality.
Reference genomes for the leatherback and green turtles were generated using four genomic technologies following the VGP pipeline v1.6 (39), with minor modifications (see Methods). A total of 100% of the leatherback and 99.8% of the green turtle assembled sequences were placeable within chromosomes. The assembled genomes were near full-length (~2.1 GB), with annotations of all 28 known chromosomes for both species, composed of 11 macrochromosomes (>50 Mb) and 17 microchromosomes (<50 Mb) (SI Appendix, Table S1 and Fig. S1). These genomes are among the highest quality genomes assembled for nonavian reptiles to date in terms of both contiguity and completeness (Dataset S1), with the leatherback turtle assembly representing the first reptile genome where all scaffolds were assigned to chromosomes. Scaffold N50s were high for both genomes (SI Appendix, Table S1). We annotated 18,775 protein-coding genes in the leatherback and 19,752 in the green turtle genomes (see below for analysis of these gene differences). For the leatherback and green turtles, 96.9% and 97.5% of these genes were supported at >95% of their length from experimental evidence and/or high-quality protein models from related species (see Methods). The numbers of protein-coding genes are within the range of other reptiles (Dataset S1) and include 97.7% and 98.2% complete BUSCO copies for leatherback and green turtles based on Sauropsida models (50), which are similar to or higher than all other assembled reptilian genomes to date (SI Appendix, Fig. S2).
Genome Architecture.
Despite diverging over 60 Mya (4), leatherback and green turtles show extremely high genome synteny and collinearity (Fig. 1 B and C and SI Appendix, Figs. S6 and S7), with Progressive CACTUS revealing 95% sequence identity across the length of the genomes (SI Appendix, Table S3). After multiple rounds of manual curation to correct artifacts of misassemblies, few large structural rearrangements between the two species remained, including inversions of up to 7 Mb on chromosomes 12, 13, 24, and 28 (SI Appendix, Fig. S6). The high collinearity between species included near-complete end-to-end contiguous synteny for nine of 28 chromosomes (SI Appendix, Fig. S6). The remaining 19 chromosomes exhibited at least one small region of reduced collinearity (RRC) between the species, with RRCs representing a total of ~83.4 Mb (~3.9%) and ~110.5 Mb (~5.2%) of the leatherback and green turtle genome lengths, respectively. Eight chromosomes exhibited small RRCs (0.1 to 3 Mb), and 11 contained RRCs that were between 3 and 18 Mb in length (Fig. 2 A–D and Dataset S3). Analyses of coding regions revealed a similar pattern of strong collinearity between the two species (Fig. 1C and SI Appendix, Fig. S6), particularly within the macrochromosomes, which contain more than 80% of the total length of the genomes. The two genomes also displayed similar percentages of repetitive elements (REs), which were almost exclusively transposable elements (TEs) and unclassified repeats (SI Appendix, Fig. S8). The landscape of TE superfamily composition over evolutionary time was also similar between the two species, with the exception of REs with low Kimura values (<5%), which appeared at a higher frequency in the leatherback turtle genome (see SI Appendix, section I for full analyses).
Fig. 2.
(A–D) Dotplots (identity values; dark green = 1 to 0.75, green = 0.75 to 0.5, orange = 0.5 to 0.25 and yellow = 0.25 to 0) showing four of the regions with reduced collinearity (RRC) identified within chromosomes and associated with higher copy numbers of immune system (IS), ORs, or zinc finger domain genes in the green turtle relative to leatherback turtle (see also SI Appendix, Fig. S6 and Tables S3–S5 and Dataset S3 for full details of all RRCs). RRC positions are marked with gray squares on the dot plots (Left; with leatherback turtle on the X-axes and green turtle on the Y-axes) and gene collinearity maps (Right) for each chromosome highlighting the connections among specific gene families in different colors. (E) Gene family evolution of ORs class I (red) and class II (black) for amniote phylogeny. Gene numbers are presented on the nodes and gain/loss along each branch are presented below branches. Small scale bar represents substitutions/site, and big scale bar represents divergence times (MA). The blue dashed line shows the estimated divergence between the two sea turtle families. (F) Number of unique and shared orthogroups and single- and multicopy genes between the two sea turtles (coding genes including genes with rearrangement). The boxes outlined in black denote shared orthogroups, with the higher multicopy in the green turtle due to greater gene copies within orthogroups. (G) Comparison of gene counts between both species per multigenic orthogroup, depicting only those orthogroups where both species have different numbers of genes and a minimum number of five genes for one of the species. Bubbles above the diagonal represent higher counts for the green turtle and below for the leatherback turtle. The size of the bubbles represents the number of orthogroups with the same gene count combination.
Gene Families and Gene Functional Analysis.
Gene function analysis of localized RRCs revealed that most contained genes with higher copy numbers in the green turtle compared with the leatherback (Fig. 2 A–D and Dataset S3). Nineteen chromosomes had RRCs with higher gene copy numbers in the green turtle, and of these, ten contained genes associated with immune system, olfactory reception, and/or zinc-finger protein-coding genes. Many of the same gene families were also detected as high-diversity exonic regions via separate, independent analyses (SI Appendix, section I), reinforcing their importance in the divergent evolution of these species. In addition to localized RRCs, higher gene copy numbers in the green turtle occurred in many gene orthologous groups (orthogroups) across the entire genome, and generally in variable multicopy genes (Fig. 2 F and G). Copy number variation accounted for most of the nearly one thousand more genes annotated in the green turtle genome relative to the leatherback (Fig. 2 F and G and SI Appendix, Table S1). We detected no evidence of collapsed multicopy genes in the leatherback turtle assembly across multiple analyses (see Methods and SI Appendix, Table S4), supporting this as a biological signal rather than technical artifact of the assemblies.
Olfactory receptors (ORs) represented the largest orthogroups in both genomes, and differences in copy numbers were connected to many of the identified RRCs. All OR class I genes were clustered at the beginning of chromosome 1, and the green turtle had higher copy numbers in this region (Fig. 2 A–D). This area also contained a cluster of OR class I genes in at least three additional testudinid species (SI Appendix, Fig. S10), and is the only divergent region across the very large chromosome 1 in the turtles analyzed. In contrast, OR class II genes were spread across several chromosomes in both sea turtle species, with higher copy numbers again in the green turtle found within RRCs (Fig. 2 B–D). The instability and rapid evolution of OR gene numbers in turtles is further illustrated in the expansion-contraction analysis of orthogroups (Fig. 2E and Dataset S6 A–D), which showed that OR class I genes underwent a modest contraction in the ancestral sea turtle lineage, followed by an expansion in the green turtle but a further contraction in the leatherback turtle. Similar trends were detected for OR class II genes, but with a greater magnitude of contraction in the ancestral sea turtle lineage followed by a further contraction for the leatherback turtle and only a small expansion for the green turtle (Fig. 2E).
Another important RRC (RRC14) encompassed the major histocompatibility complex (MHC), which plays a critical role in vertebrate immunity and is particularly relevant to sea turtle conservation due to the threat of FP and other diseases (32). In addition to the MHC region, this RRC includes several copies of OR class II genes, zinc-finger protein-coding genes and other genes involved with immunity, such as butyrophilin subfamily members and killer cell lectin-like receptors (Fig. 2D and Dataset S3). Invariably, the green turtle carried higher numbers of all the multicopy genes present in RRC14. RRCs on other chromosomes similarly showed increased levels of zinc-finger protein genes in the green turtle, including the RRCs labeled 6A, 11A, 14A, and 28 (Dataset S3). In particular, zinc-finger protein genes were highly prevalent on chromosomes 14 and 28 in both sea turtles, representing more than 50% of all the protein domains present on these chromosomes (SI Appendix, Fig. S11). Finally, all but three genes with known roles in TSD in reptiles (Dataset S7) were located as single-copy genes within both sea turtle genomes, with homologous copies located in the same region of the chromosomes in both species (see SI Appendix, section I for full analyses).
Macro and Microchromosomes.
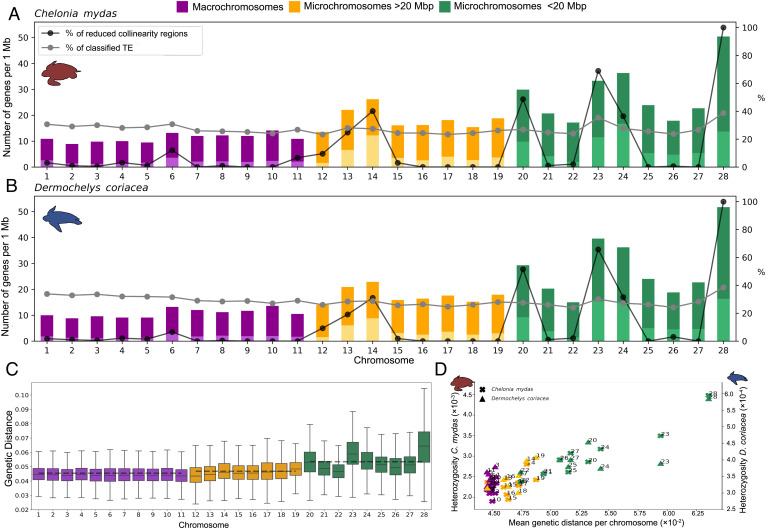
Microchromosomes contained significantly higher proportions of genes than macrochromosomes (Fig. 3 A and B; green turtle: F(2, 25) = 16.46, P < 0.01; leatherback turtle: F(2, 25) = 16.35, P < 0.01), and gene content was strongly positively correlated with GC content (SI Appendix, Fig. S13; green turtle R2 = 0.81, P < 0.01; leatherback turtle R2 = 0.87, P < 0.01). These patterns were particularly apparent in small (<20 Mb) microchromosomes, where GC content reached 50%, compared with the 43 to 44% genome-wide averages. Within chromosome groups, larger proportions of multicopy genes were generally associated with higher total gene counts (green turtle: R2 0.84, P < 0.01; leatherback turtle: R2 = 0.92, P < 0.01), and chromosomes with the highest multicopy genes numbers had increased proportions of RRCs (Fig. 3 A and B; green turtle: R2 = 0.69, P < 0.01; leatherback turtle: R2 = 0.81, P < 0.01).
Fig. 3.
Number of genes, genetic distance between species, and heterozygosity within species in macrochromosomes, small (<20 Mb), and intermediate (>20 Mb) microchromosomes. (A) Relation between the number of genes, percentage of RRCs, and classified TE per chromosome for the green and (B) leatherback turtles. Dark colors indicate the total number of genes and light colors indicate the number of multicopy genes. (C) Average genetic distance between green and leatherback turtles per chromosome. (D) Relation between genetic distance and heterozygosity per chromosome for each species.
Mean genetic distances for single-copy regions between the two sea turtles were also higher in small microchromosomes (0.053) compared with both intermediate (>20 Mb) microchromosomes (0.047), and macrochromosomes (0.045) (Fig. 3C; F(2, 25) = 21.98, P < 0.01). However, examination of intermediate microchromosome and macrochromosome RRCs revealed elevated genetic distances in these regions that approached the values observed in small microchromosomes (SI Appendix, Table S5). Genetic distances were also significantly positively correlated with heterozygosity (green turtle: R2 = 0.97, P < 0.01; leatherback turtle R2 = 0.97, P < 0.01), which was significantly higher in small microchromosomes for both species (Fig. 3D; green turtle: F(2, 25) = 15.72, P < 0.01; leatherback turtle: F(2, 25) = 5.09, P < 0.05).
Genome Diversity.
Genome-wide nucleotide diversity was almost a magnitude of order lower in leatherback compared with green turtles (mean repeat masked π = 2.86 × 10−4 and 2.46 × 10−3, respectively; t(5.52) = 36.9, P < 0.001; Fig. 4A and SI Appendix, Figs. S15–S17 and Table S7). Despite having largely similar gene content identified in the annotation, this strong pattern was also observed in coding regions (Fig. 4A; t(5.52) = 37.7, P < 0.001), such that leatherback turtles possess much less standing functional variation, possibly impacting their adaptive capacity to future novel environmental conditions. The strikingly low genomic diversity of leatherback turtles is also less than almost all other reptile species examined (SI Appendix, Fig. S19; but see ref. 51), including Chelonoidis abingdonii, where low diversity has been considered a contributing factor to their extinction (52). Contrastingly, genomic diversity of the green turtle fell in the midrange for reptiles, as well as for mammals examined using similar methods (53, 54). Finally, within both species, heterozygosity was lower in coding regions (mean π = 2.77 × 10−4 and 2.18 × 1−3 for leatherback and green turtles; Fig. 4A) relative to noncoding regions (mean π = 3.18 × 10−4 and 2.64 × 10−3; leatherbacks: [t(4) = −8.9, P < 0.01] and greens: [t(5) = −30.9, P < 0.01]), as expected from selection pressures driving higher sequence conservation in these functional genomic regions.
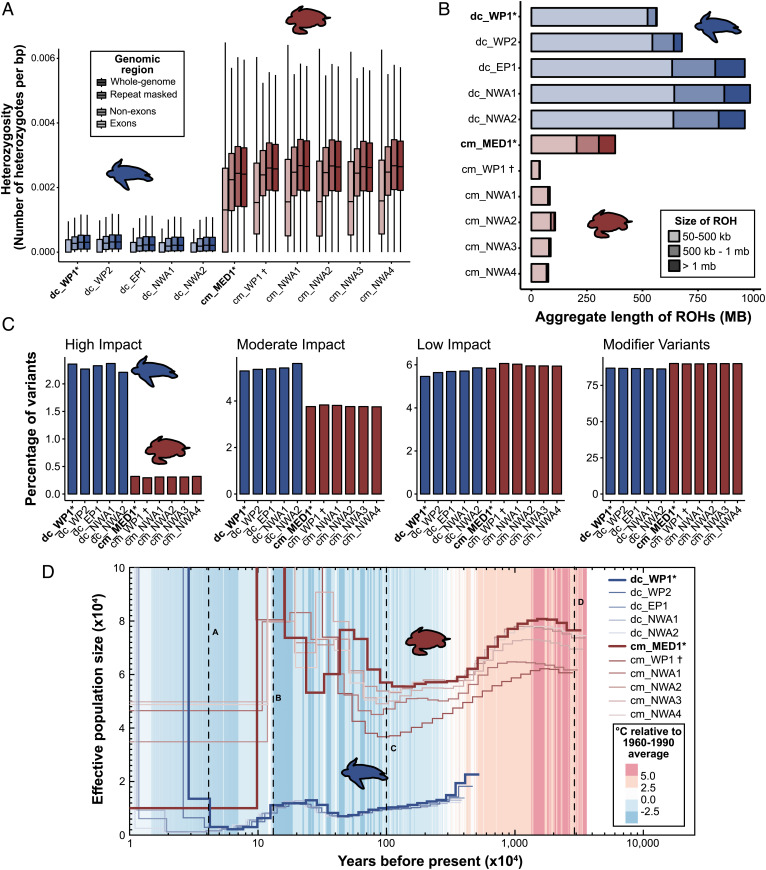
Fig. 4.
Data are presented for the leatherback (blue) and green (red) turtle genomes, including reference individuals for both species (*), and the individual used to generate the draft genome (†; Wang et al. 2013). (A) estimates of heterozygosity for the entire genome, repeat-masked genome, exon and nonexon regions, with outliers removed. (B) accumulated lengths of runs of homozygosity (ROH). (C) predicted impacts of variants from within coding regions. (D) Pairwise sequential Markovian coalescent plot (PSMC) of demographic history of both species overlayed with temperature. Letters indicating portions of the PSMC curves (A–D) are geological events referred to in the main text and SI Appendix, section I.
Runs of Homozygosity (ROH).
In addition to lower genome-wide heterozygosity, leatherbacks had a greater total number of ROHs (>50 kb) than green turtles (mean NROH = 4,510 and 829, respectively), as well as a greater total aggregate length of the genome in ROH (range =26.1 to 45.5% in leatherback turtles; 1.8 to 17.7% in green turtles). The mean length of ROHs was also significantly higher in leatherback (LROH = 183.9 kb) compared with green turtles (LROH = 154.9 kb) (t(7429.4) = −8.85, P < 0.01). Length distribution breakdown showed that leatherbacks have a higher aggregate length of all categories of ROHs relative to the green turtles (Fig. 4B and SI Appendix, Fig. S22). Short ROHs (50 to 500 kb) had the highest total aggregate length in leatherbacks, with a mean aggregate length of 597 Mb (Fig. 4B), suggesting long-term low population sizes in the leatherback turtle.
Within species, overall ROH distributions were generally similar between samples representative of different populations for leatherback turtles, although individuals from the Northwest Atlantic and East Pacific populations displayed slightly higher total aggregate lengths of ROHs than those from the West Pacific population, primarily due to greater aggregate lengths of medium and long ROHs (Fig. 4B). Among green turtles, the aggregate length of ROHs in all categories were generally small and similar across individuals, with the clear exception of the genome reference sample that originated from the Mediterranean population. This individual displayed higher numbers and lengths of long ROHs (>1 Mb) compared with all other green turtles (n = 50 compared with <5, and aggregate length = 74 Mb compared with <4 Mb), suggesting higher levels of recent inbreeding relative to the other green turtle populations represented in our dataset. Comparative analyses mapping this individual to the two previous green turtle assemblies failed to detect these long ROHs (SI Appendix, Fig. S23), demonstrating the importance of highly contiguous reference genomes for detecting biologically important patterns using this conservation-relevant metric.
Genetic Load.
Coding region variants with predicted high (e.g., stop-codon gain or loss) or moderate impacts were significantly more common in leatherback compared with green turtles (Fig. 4C; high-impact variants: t(4.18) = −65.7, P < 0.001; moderate impact variants: t(4.51) = −29.5, P < 0.001). Conversely, low-impact and modifier (i.e., variants predicted to cause negligible impacts) variants were significantly more common in green turtles (Fig. 4C; low-impact variants: t(5.88) = 4.0, P < 0.01; modifier variants: t(5.33) = 31.8, P < 0.001). The missense-to-silent mutation ratio was also higher in leatherbacks than green turtles (t(7.19) = −72.3, P < 0.001; mean = 0.99 and 0.70), further suggesting that genetic load is higher in the leatherback turtles. Within species, there was limited variation between individuals for all variant categories (Fig. 4C).
Demographic History.
Pairwise Sequential Markovian Coalescence (PSMC) analyses indicated different historical effective population sizes (Ne) between the two sea turtle species (Fig. 4D). Ne for all leatherback turtle populations represented in our dataset have been relatively small and sustained over time, ranging in size from approximately 2,000 to 21,000 over the last 10 My, up until the Last Glacial Maximum (LGM) and at the lower end of this range for most of the last 5 My. This pattern is consistent between all individuals examined, with similar timings and magnitudes of Ne fluctuations until recent history (Fig. 4D). In contrast, green turtles have experienced wider variation and a higher overall Ne in general, fluctuating between approximately 50,000 and 125,000, until the late Pleistocene, with estimates varying by population (Figs. 4D and SI Appendix, Fig. S24). While Ne for leatherback turtles is relatively low, it modestly increased prior to the Eemian warm period (Fig. 4D [B]), followed by a subsequent decrease during this period until the LGM (Fig. 4D [A]) when all populations exhibit sharp spikes in Ne possibly due to interocean gene flow following warming after the LGM. In contrast, green turtles generally displayed three distinct peaks in Ne (Fig. 4D), associated with ocean connectivity changes following the closure of the Tethys Sea [D], during the Pleistocene period [C], and prior to the Eemian warming period (Fig. 4D [B]). While the patterns of Ne are broadly similar within green turtles, the timing and magnitude of these fluctuations varied between populations (SI Appendix, Fig. S24).
Discussion
Divergence in Localized RRCs and Microchromosomes amidst High Global Genome Synteny.
The ancestral lineage leading to leatherback and green turtles diverged over 60 Mya (4), giving rise to species that are adapted to dissimilar habitats, diets, and modes of life. Despite high overall levels of genome synteny between the species, RRCs and small microchromosomes were associated with higher concentrations of multicopy gene families, as well as heightened nucleotide diversity and genetic distances between species, suggesting that these genomic elements may be important sources of variation underlying phenotypic differentiation. Higher heterozygosity despite richer gene content in the microchromosomes suggests that these regions accumulate variation and may have a high adaptation value. Though our results here do not demonstrate direct causality, we have identified candidate regions and gene families that can be targeted in further studies quantifying evidence for positive selection and their roles in sea turtle adaptation and speciation.
The high global stability of macro- and microchromosomes between sea turtle families aligns with recent work showing similar patterns across reptiles, including birds, emphasizing the importance of microchromosomes in vertebrate evolution (55). Higher evolutionary rates in microchromosomes have been documented in intraspecific (56) and interspecific (57) avian studies, so it is possible that the characteristics of microchromosomes and RRCs we observed are not unique to sea turtles, but rather, are prevalent among vertebrates. The mechanisms driving these patterns are not well-understood, but could be related to higher recombination rates in micro- compared with macrochromosomes (58) that result in higher nucleotide diversity and lower haplotype sharing. Once generated, balancing selection may play a role in maintaining variation in these gene-dense regions, but more work is needed across taxa to determine the broad support for these hypotheses. The prevalence of localized genomic differentiation and underlying mechanisms among other closely or more distantly related vertebrate groups has yet to be widely evaluated due to a lack of equivalent quality genomic resources, but this is rapidly changing. Our detailed analyses of RRCs, microchromosomes, and their associated genes were only possible due to the high-quality of the assembled genomes because these analyses can be sensitive to genome fragmentation and misassemblies (39). For example, the RRCs and many microchromosomes could not be detected using the draft green turtle genome due to fragmentation and sequence gaps (SI Appendix, Figs. S3 and S4). As chromosomal-level genomes across all vertebrate lineages become available, our work provides a roadmap for identifying genomic regions harboring contrasting expansion/contractions of gene families and diversity levels. For taxa with highly conserved genomes like sea turtles, analyses of RRCs and microchromosomes are likely important to understand their divergent evolutionary histories and the phenotypic connections of the genes within them.
Contrasting Sensory and Immune gene Evolution between Sea Turtle Families.
Sea turtles have complex sensory systems and can detect both volatile and water-soluble odorants, which are imperative for migration, reproduction, and identification of prey, conspecifics, and predators (59–63). However, leatherback and green turtles occupy dissimilar ecological niches, depending on different sensory cues. While leatherback turtles almost exclusively inhabit the pelagic environment posthatching, performing large horizontal and vertical migrations to seek out patches of gelatinous prey (64), green turtles recruit to neritic coastal and estuarine habitats as juveniles, and can have highly variable diets (65, 66). Sea turtle nasal cavity morphology also differs between species, with leatherback turtle cavities relatively short, wide, and more voluminous than chelonids (67–69), suggesting reduced requirements for olfactory reception. OR genes encode proteins used to detect olfactory cues, with the number of genes correlated with the number of detectable odorants (70), and linked to the chemical complexity of the inhabited environment (71). The two major groups of ORs in amniotic vertebrates are separated by their affinities with hydrophilic molecules (class I) or hydrophobic molecules (class II) (72). Class I OR genes may be particularly important in aquatic adaptation (32), and expansions of class I ORs in testudines, including green turtles, have been previously reported. However, the accuracy of these estimates for complex gene families using short-read assemblies has been uncertain because they may be prone to misassembly (32, 42, 73). We detected an additional 93 class I OR genes in our green turtle genome compared with those reported in the draft green turtle genome (42), suggesting they can be erroneously collapsed in short-read assemblies. Our reconstruction of both classes of OR gene evolution throughout the sea turtle lineage revealed that after ancestral contractions, gene copy evolution diverged in opposite directions between the two sea turtles. The greater loss of class II compared with class I OR genes in the ancestral sea turtle lineage likely reflects relaxed selection for detection of airborne odorants, as has been observed in other lineages that recolonized marine environments (74). However, as sea turtles continue to use terrestrial habitats for reproduction, they may need to retain some of these capabilities, which could explain why the observed contraction was weaker than those in exclusively marine species (e.g., the vaquita Phocaena sinus; Fig. 2E).
The strong class I OR expansion in the green turtle may be related to its distribution in complex neritic habitats and variable diet, requiring detection of a high diversity of waterborne odorants, while the continued loss of ORs in the leatherback turtle could be a consequence of its more specialized diet and the lower chemosensory-complexity of pelagic habitats. Although leatherback turtles can detect chemical cues from their prey, sensory experiments have indicated that visual cues are more important for food recognition in this species (75, 76). Additionally, while the precise mechanisms underpinning philopatry in sea turtles remain unclear, green turtles are thought to use olfactory cues to reach specific natal nesting beaches following long-distance navigation guided by magnetoreception (61, 63). In contrast, leatherback turtles exhibit more ‘straying’ from natal rookeries than other species, and such relaxed philopatry may be related to reduced reliance on olfactory cues to hone in on specific beaches.
Diversity within the highly complex MHC region is a key component in the vertebrate immune response to pathogens, with greater gene copy numbers and heterozygosity linked to lower disease susceptibility (77). While both sea turtle species contained most of the core MHC-related genes, the green turtle had more copies of genes involved in adaptive and innate immunity. Pathogen prevalence and persistence is often greater in neritic habitats than open ocean habitats (78), so green turtles may be exposed to higher pathogen loads and diversity than leatherback turtles (79). However, reptilian immune systems are understudied compared with other vertebrates, with few studies of MHC genes conducted in turtles (80). Thus, it is not yet understood how immune gene diversity translates into disease susceptibility or ecological adaptation in sea turtles, which is critical for their conservation as FP continues to threaten population recoveries around the globe (30). Although this viral-mediated tumor disease occurs in all sea turtle species, prevalence and recovery greatly vary between and within species, making it plausible that harboring certain genes, copy numbers, or specific alleles may play important roles in disease dynamics. Despite decades of research on this disease (30) only one study on the immunogenomic factors governing FP susceptibility or resilience has been conducted (81), in part due to difficulty in accurately quantifying hypervariable and complex MHC loci with short-read sequencing technologies (82). Our reference genomes now enable studies to accurately interrogate these complex gene families to advance our fundamental understanding of immune gene evolution in testudines.
Differential Genomic Diversity and Demographic Histories.
Genomic diversity is a critical metric for evaluating extinction risk and adaptive potential to environmental perturbation (83–85), with heterozygosity positively correlated with individual fitness (see reviews by refs. 86 and 87). Understanding the causes and consequences of genomic diversity is imperative for leatherback turtles in particular, where contemporary populations have sharply declined due to human activities (25). The exceptionally low genomic diversity observed in leatherback turtles broadly aligns with previous estimates (88, 89), but our PSMC and ROH results indicate that this is likely a consequence of long-term low effective population sizes and historical bottleneck events, rather than losses during recent population declines. This is consistent with mitochondrial analyses suggesting that contemporary populations radiated from a small number of matriarchal lineages within a single refugium following the Pleistocene (89). In contrast, higher heterozygosity, limited ROHs, and larger, more variable historical Ne in green turtles likely reflects radiation from many refugia and frequent admixing of populations (90).
Regardless of the causes of current genomic diversity levels, the amount of standing variation can have important implications for species’ future persistence (91), especially given the adaptive capacity likely required to keep pace with rapid anthropogenic global change. Although genome-wide diversity estimation does not require high-quality reference genomes, they enable deeper examination of diversity patterns relevant to conservation. The use of our reference genomes demonstrated that diversity is very low within coding regions of leatherback turtle genomes, indicating limited standing functional variation that may have implications for their adaptive potential to novel conditions. Additionally, leatherback turtles exhibited a higher genetic load compared with green turtles, and this signal was consistent across all samples, regardless of population. Leatherback turtles have substantially lower hatching success compared with other sea turtle species (29), potentially related to the heightened genetic load and low heterozygosity (92, 93), and may combine with other factors to slow population recoveries despite conservation efforts. However, other species with low diversity have rebounded following population declines and/or appear to have purged deleterious alleles through long-term low population sizes (94–96), thereby limiting the impacts on viability (54, 94, 97). Although our results of greater genetic load despite long-term low Ne suggest this is not the scenario for leatherback turtles, further in-depth research on these topics enabled by the presented reference genomes will clarify these relationships for leatherback and other sea turtle species to guide conservation recommendations.
Although patterns of diversity, genetic load, and demographic histories were generally consistent within species, ROH analyses revealed a striking exception of the green turtle reference individual from the Mediterranean. This isolated population has undergone severe decline over the last century due to human exploitation (98), and our results indicate that consequent inbreeding is likely occurring, which may impact recovery. The specific individual was from the Israel green turtle rookery, estimated to have only 10 to 20 nesting females in the last decade (99, 100), but it is unclear whether Israel is demographically isolated from other rookeries in the region (100, 101). Further research is needed to understand whether inbreeding is a concern only for this nesting aggregation, or the Mediterranean population more broadly. These findings also highlight the utility of ROH metrics even in animals with longer generation times, and the importance of using highly contiguous genomes for accurate ROH assessment to inform conservation.
While it is widely documented that environmental changes can strongly impact species’ abundances and distributions (102–104), following an initial decrease associated with declining temperatures, Ne of leatherback turtles remained relatively constant throughout substantial temperature fluctuations in the Pleistocene. As ectotherms, reptiles are sensitive to climatic thermal fluctuations; however, leatherback turtles exhibit unique physiological adaptations that produce regional endothermy and facilitate exploitation of cold-water habitats (6), potentially leading them to being less susceptible to periods of cooler temperatures. The long-term lower Ne of leatherback turtles may be associated with this species’ greater mass and trophic position (105). In contrast, wide fluctuations for green turtles appear correlated with climatic events, beginning with the closure of the Tethys Sea, which altered ocean connectivity and represented a period of increasing temperatures that may have opened more suitable habitat. As temperatures subsequently decreased, Ne also decreased; however, temperature fluctuations during the Pleistocene were associated with additional increases in Ne. While warmer temperatures presumably allowed for larger population sizes of green turtles, large spikes in Ne around the Eemian warming, particularly for the Mediterranean individual, are likely associated with admixing of previously isolated populations due to warm-water corridors allowing movement between populations and ocean basins (106). While our overall estimates and trends for both species were broadly concordant with previous studies (89, 107, 108), a recent study using multiple sequentially Markovian coalescent (MSMC2) analyses found steep declines in Ne for green turtles >100,000 y before present (108), which was not detected in our PSMC analyses. Since this decline was also not detected in a prior study using PSMC on the draft green turtle genome (107), and demographic inferences are generally robust to genome quality (109, 110), this is likely a consequence of the different methods, with MSMC analyses inferring larger Ne for more ancient time scales (109).
Enabling Future Research and Conservation Applications.
In addition to the insights reported here, the reference genomes for both extant sea turtle families provide invaluable resources to enable a wide breadth of previously unattainable fundamental and applied research. Combined with other forthcoming genomes (39), comparative genomics analyses can reveal the genomic basis for long-standing traits of interest such as adaptation to saltwater, diving capacity, and long-distance natal homing. Studies leveraging these reference genomes alongside whole-genome sequencing of archival samples can assess how genomic erosion, inbreeding, and mutational load are linked to population size, trajectories, and conservation measures in global sea turtle populations. For instance, the fact that leatherback turtles have persisted with low diversity and Ne for extended periods offers hope for their recovery, but given that some populations have now been reduced to only a few hundred individuals (111), research quantifying purging of deleterious alleles, inbreeding depression, and adaptive capacity within populations is urgently needed (112). We emphasize that high-quality reference genomes are not required for all research goals, and combined with other recent studies (109, 110, 113), our findings provide clear guidance on when they may, or may not, be necessary to generate accurate results to inform conservation. For example, genome-wide diversity estimates are typically robust to assembly quality, but detection of long ROHs can be strongly affected. As ROH metrics are increasingly used to guide species management plans (114–116), it is important for researchers to understand how genome quality may impact their analyses and inferences. Additionally, many conservation applications that may not explicitly require whole-genome data can also directly benefit from the utility of these reference genomes, including the development of amplicon panels and molecular assays to investigate TSD mechanisms and adaptive capacity under climate change, and assessing linkages between immune genes and disease risk. Finally, with global distributions and long-distance migratory connectivity, sea turtle conservation requires international collaboration that has been previously hampered by difficulty comparing datasets between laboratories. Existing anonymous markers (e.g., microsatellites and restriction-site based SNP markers) can now be anchored to these genomes, and new ones can be optimized for conservation-focused questions and shared across the global research community, facilitating large-scale syntheses and equitable capacity building for genomics research. While ongoing anthropogenic impacts continue to threaten the viability of sea turtles to persist, combined with the critical work of reducing major threats such as fisheries bycatch and habitat loss, these genomes will enable research that make critical contributions to recovering imperiled populations.
Methods
Reference Sample Collections, Genome Assembly, and Annotation.
Ultra-high molecular weight DNA was isolated from blood collections, and biopsies of internal organs for RNA were collected opportunistically from recently deceased or euthanized animals. Raw data were deposited into the VGP Genome Ark and NCBI Short-Read Archive (SRA; see Data Accessibility Statement). We assembled both genomes using four genomic technologies following the VGP pipeline v1.6 (39) with minor modifications. Short- and long-read transcriptome data (RNA-Seq and Iso-Seq) were generated from tissues known for their high transcript diversity in each species to enable accurate, species-specific annotations. These data, plus homology-based mapping from other species, were used to annotate the genomes using the standardized NCBI pipeline (117). We performed annotation as previously described (39, 118), using the same RNA-Seq, Iso-Seq, and protein input evidence for the prediction of genes in both species. Full details for all methods are provided in SI Appendix, section I.
Genome Quality Analysis.
We used the pipeline assembly-stats from https://github.com/sanger-pathogens/assembly-stats to estimate scaffold N50, size distributions, and assembly size. BUSCO analysis (115) and QV value estimations (116) were conducted to assess the overall completeness, duplication, and relative quality of the assemblies. We used D-GENIES (118) with default parameters to conduct dot plot mapping of the entire genomes and each individual chromosome to evaluate the synteny between leatherback and green turtle genomes, and Haibao Tang JCVI utility libraries following the MCScan pipeline (119) to verify their contiguity. Incongruences in gene synteny blocks were manually investigated using Artemis Comparison Tool (120), identifying possible regions that could be caused by artifacts during assembly, and correcting these. The final curated assemblies were analyzed using the genome evaluation pipeline (https://git.imp.fu-berlin.de/cmazzoni/GEP) to obtain all final QC plots and summary statistics.
Identification and Analysis of RRCs.
Using dot plots, 20 Mb windows were visually screened to identify regions of reduced collinearity (RRCs; SI Appendix, Fig. S5). Several genomic features (e.g., GC content, repeat elements) were compared between RRCs and equisized regions directly up- and down-stream to determine whether these were influencing collinearity (Dataset S5). Interproscan (119) was used to identify the functions of genes found within RRCs, and overall GO-term proportions for each chromosome were estimated using PANTHER (120); SI Appendix, Fig. S25). The two sea turtle genomes were aligned using Progressive Cactus (121, 122) to examine whether RRCs presented patterns of sequence divergence and/or gene duplication between the species.
Gene Families and Gene Functional Analysis.
To estimate the timing of OR gene family evolution in sea turtles, we used computational analysis of gene family evolution (CAFEv5; (123). CAFE uses phylogenomics and gene family sizes to identify expansions and contractions. We used a dataset containing 8 species of turtle, 4 nonturtle reptiles, 3 mammals, and 1 amphibian using OrthoFinder (124, 125). OR orthogroups were grouped based on subfamily (class I and class II; see ref. 73), and an ultrametric phylogeny was generated by gathering 1:1 orthologs. We then aligned OrthoFinder amino acid sequences for each orthogroup and generated a phylogenetic tree. See SI Appendix, section I for searches of other specific genes.
Genetic Diversity and Demographic History.
The halSnps pipeline (126) was used to estimate genetic distance between species by computing interspecific single variants based on alignments obtained with Progressive Cactus (121, 122). Genetic distances were calculated for 10,000-bp windows across the genome, where each window included only single alignments in the Cactus output. Differences in genetic distance, gene content, GC content, and heterozygosity between macro-, intermediate micro-, and small microchromosomes were tested using one-way ANOVAs for each species. Regression analyses were used to test for correlations between these measures across chromosomes.
For genome diversity, ROH, demographic history, and genetic load analyses, we included whole-genome resequencing data for additional individuals representing multiple global populations in each species (SI Appendix, Table S6 and section I). We calculated genome-wide heterozygosity using a method adapted from (127) using 100-kb nonoverlapping windows. Heterozygosity was calculated for the entire genome, repeat-masked genome, exons, and nonexons. Statistical comparisons between species were made using t tests. We subsequently applied the heterozygosity pipeline to generate genome-wide heterozygosity for additional reptilian species sourced from NCBI SRA, where species-specific reference genomes were available (SI Appendix, section I).
ROHs were identified by generating a SNP-list using the analysis of next generation sequencing data [ANGSD; (128)] pipeline. ANGSD was parameterized to output files configured for use as input for the PLINK ROH analysis (129). ROHs were then further characterized into size classes approximately based on (130).
Estimates of deleterious allele accumulation were conducted using snpEff (131). We estimated the impacts of variants (SNPs and INDELs) from coding regions using the species-specific genome annotations generated for both species. gVCFs were generated for each individual followed by joint-genotyping using GATK (132), allowing the reference individuals to include homozygous alleles found in other individuals. Combined VCFs were separated for each individual and filtered using based on depth of coverage (⅓×–2× mean coverage). snpEff predicts variant impacts and bins them into ‘high-’, ‘moderate’, or ‘low-’ impact categories, and outputs a list of genes that have predicted variant effects. We ran the snpEff analysis on all individuals for both species, and compared the percentages of each variant type between species using t tests.
PSMC (133) analyses of demographic history were employed for all individuals for both species. We used SAMtools (134) and BCFtools (135) to call genotypes with base and mapping quality filters of >Q30, before filtering for insert size (50 to 5,000bp) and allele balance (AB), and retaining only biallelic sites with an AB of <0.25 and >0.75. We then ran PSMC analysis using the first 10 scaffolds (84% of total genome length). We scaled our outputs using a generation time of 30 y (SI Appendix, section I), and a mutation rate of 1.2 × 10−8 (107).
Data Accessibility Statement.
Genome assemblies have been deposited on NCBI GenBank. The NCBI GenBank accession numbers for the leatherback turtle assembly (rDerCor1) are GCF_009764565.3 and GCA_009762595.2 for the annotated primary and original alternate haplotypes in BioProject PRJNA561993, and for the green turtle assembly (rCheMyd1) are GCF_015237465.2 and GCA_015220195.2 for primary and alternate haplotypes respectively in BioProject PRJNA561941. The raw data used for assemblies are available on the Vertebrate Genome Ark (https://vgp.github.io/genomeark/). The leatherback turtle data generated for the purpose of assembly annotation was deposited in the SRA under accession numbers SRX8787564-SRX8787566 (RNA-Seq) and SRX6360706-SRX6360708 (ISO-Seq). Green turtle data generated for annotation were deposited in SRA under accessions SRX10863130-SRX10863133 (RNA-Seq) and as SRX11164043-SRX11164046 (ISO-Seq). The NovaSeq 6000 DNA-Seq data for the green turtle resequencing, including raw reads, are deposited in NCBI (https://www.ncbi.nlm.nih.gov/) under BioProject ID: PRJNA449022. All scripts used for downstream analyses following genome assembly and annotation have been deposited on GitHub under repository https://github.com/bpbentley/sea_turtle_genomes.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Acknowledgments
We thank the St. Croix Sea Turtle Program, USFWS, NOAA-SWFSC California in-water leatherback research team, and the New England Aquarium for assistance with leatherback sample collection, and the Israel National Sea Turtle Rescue Centre, NOAA PIFSC-MTBAP, and Thierry Work (USGS) for assistance with green turtle sample collection. We thank Estefany Argueta and Jamie Adkins for assistance with literature searches and library preparations, and Phillip Morin, Andrew Foote, Anna Brüniche-Olsen, Annabel Beichman, Morgan McCarthy, David Adelson, and Yuanyuan Cheng for invaluable discussions and comments on the manuscript. Green turtle sequencing was performed by the Long Read Team of the DRESDEN-concept Genome Center, DFG NGS Competence Center, part of the Center for Molecular and Cellular Bioengineering (CMCB), Technische Universität Dresden and MPI-CBG. For green turtle resequenced samples, we thank Jessica Farrell, Whitney Crowder, Brooke Burkhalter, Nancy Condron, and the veterinary and rehabilitation staff and volunteers of the University of Florida’s (UF) Sea Turtle Hospital at Whitney Laboratories, staff of the South Carolina Aquarium, UF’s Interdisciplinary Center for Biotechnology Research Core for sequencing services, and to the Florida FWC and South Carolina DNR for assistance with permitting. We thank Erin LaCasella for help procuring leatherback samples for resequencing from the NOAA-SWFSC Marine Mammal and Sea Turtle Research Collection. This work was completed in part with resources provided by the University of Massachusetts' Green High Performance Computing Cluster (GHPCC). Funding was provided by the University of Massachusetts Amherst, NSF-IOS (grant #1904439 to L.M.K.), NOAA-Fisheries, National Research Council postdoctoral fellowship program to L.M.K., VGP, Rockefeller University, to E.D.J., HHMI to E.D.J., the Sanger Institute, Max-Planck-Gesellschaft, and grant contributions from Tom Gilbert, Paul Flicek, R.W.M., Karen A. Bjorndal, Alan B. Bolten, Ed Braun, N.J.G., T.M.-B., and A.F.S. We acknowledge CONICYT-DAAD for scholarship support to T.C.-V., the São Paulo Research Foundation to E.K.S.R.–FAPESP (grant #2020/10372-6). BeGenDiv is partially funded by the German Federal Ministry of Education and Research (BMbF, Förderkennzeichen 033W034A). The work of F.T.-N. and P.M. was supported by the Intramural Research Program of the National Library of Medicine, NIH. The work of M.P. was partially funded through the Federal Ministry of Education and Research (grant 01IS18026C). H.P. was supported by a Formació de Personal Investigador fellowship from Generalitat de Catalunya (FI_B100131). M.K. was supported by “la Caixa” Foundation (ID 100010434; code LCF/BQ/PR19/11700002), the Vienna Science and Technology Fund (WWTF), and the City of Vienna (VRG20-001). Funding for green turtle resequencing was provided by a Welsh Government Sêr Cymru II and the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 663830-BU115 and the Sea Turtle Conservancy, Florida Sea Turtle Grants Program (17-033R).
Author contributions
B.P.B., E.W.M., E.D.J., C.J.M., and L.M.K. designed research; B.P.B., T.C.-V., E.K.S.R., H.P., L.S.A., A. Alexander, S.M.B., P.M., M.K., M.P., J.M., B.H., M.U.-S., G.F., K.H., W.C., A.T., Y.S., S.P., J.W., K.Y., J.R.P., K.S., S.R.B., Y.L., H.B.S., P.S., B.T.H., R.W.M., D.W.M., A.F.S., D.J.D., N.J.G., S.W., F.T.-N., M.F.N., T.M.-B., A. Antunes, Y.T., P.H.D., O.F., C.J.M., and L.M.K. performed research; B.P.B., T.C.-V., E.K.S.R., H.P., L.S.A., A. Alexander, P.M., M.K., M.P., M.U.-S., G.F., K.H., W.C., A.T., Y.S., S.P., J.W., A.F.S., A.S., F.T.-N., T.M.-B., A. Antunes, O.F., C.J.M., and L.M.K. analyzed data; S.M.B. and Y.T. collected samples; K.Y. provided whole genome resequencing data for additional individuals that were added to the analyses.; Y.L. provided samples; D.J.D. provided whole genome resequencing data for additional individuals that were added to the analyses; and B.P.B., T.C.-V., E.K.S.R., H.P., L.S.A., A. Alexander, M.K., G.F., K.H., J.R.P., E.V.T., H.B.S., P.S., R.W.M., N.J.G., A.F.S., S.W., F.T.-N., T.M.-B., A. Antunes, P.H.D., E.W.M., E.D.J., C.J.M., and L.M.K. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Blair P. Bentley, Email: bbentley@umass.edu.
Camila J. Mazzoni, Email: mazzoni@izw-berlin.de.
Lisa M. Komoroske, Email: lkomoroske@umass.edu.
Data, Materials, and Software Availability
All genomic data and scripts data have been deposited in VGP GenomeArk (136, 137) Github (138).
Supporting Information
References
- 1.Hirayama R., Oldest known sea turtle. Nature 392, 705–708 (1998). [Google Scholar]
- 2.Shaffer H. B., McCartney-Melstad E., Near T. J., Mount G. G., Spinks P. Q., Phylogenomic analyses of 539 highly informative loci dates a fully resolved time tree for the major clades of living turtles (Testudines). Mol. Phylogenet. Evol. 115, 7–15 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Pike D. A., Climate influences the global distribution of sea turtle nesting: Sea turtle nesting distributions. Glob. Ecol. Biogeogr. 22, 555–566 (2013). [Google Scholar]
- 4.Thomson R. C., Spinks P. Q., Shaffer H. B., A global phylogeny of turtles reveals a burst of climate-associated diversification on continental margins. Proc. Natl. Acad. Sci. U.S.A. 118, e2012215118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davenport J., Temperature and the life-history strategies of sea turtles. J. Therm. Biol. 22, 479–488 (1997). [Google Scholar]
- 6.Frair W., Ackman R. G., Mrosovsky N., Body temperature of Dermochelys coriacea: Warm turtle from cold water. Science 177, 791–793 (1972). [DOI] [PubMed] [Google Scholar]
- 7.Pritchard P. C. H., “Evolution, phylogeny, and current status” in The Biology of Sea Turtles, Lutz P. L., Musick J. A., Eds. (CRC Press, 1996), pp. 1–28. [Google Scholar]
- 8.McClenachan L., Jackson J. B. C., Newman M. J. H., Conservation implications of historic sea turtle nesting beach loss. Front. Ecol. Environ. 4, 290–296 (2006). [Google Scholar]
- 9.Jackson J. B., et al. , Historical overfishing and the recent collapse of coastal ecosystems. Science 293, 629–637 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Grassman M. A., Owens D. W., McVey J. P., R. M. M., Olfactory-based orientation in artificially imprinted sea turtles. Science 224, 83–84 (1984). [DOI] [PubMed] [Google Scholar]
- 11.Lohmann K. J., Lohmann C. M. F., There and back again: Natal homing by magnetic navigation in sea turtles and salmon. J. Exp. Biol. 222, jeb184077 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Tomillo P. S., Saba V. S., Piedra R., Paladino F. V., Spotila J. R., Effects of illegal harvest of eggs on the population decline of leatherback turtles in Las Baulas Marine National Park, Costa Rica. Conserv. Biol. 22, 1216–1224 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Fossette S., et al. , Pan-atlantic analysis of the overlap of a highly migratory species, the leatherback turtle, with pelagic longline fisheries. Proc. Biol. Sci. 281, 20133065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaska Y., et al. , Natural and anthropogenic factors affecting the nest-site selection of Loggerhead Turtles, Caretta caretta, on Dalaman-Sarıgerme beach in South-west Turkey: (Reptilia: Cheloniidae). Zool. Middle East 50, 47–58 (2010). [Google Scholar]
- 15.Von Holle B., et al. , Effects of future sea level rise on coastal habitat. J. Wildl. Manage. 83, 694–704 (2019). [Google Scholar]
- 16.Mrosovsky N., Ryan G. D., James M. C., Leatherback turtles: The menace of plastic. Mar. Pollut. Bull. 58, 287–289 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Chaloupka M., Balazs G. H., Work T. M., Rise and fall over 26 years of a marine epizootic in Hawaiian green sea turtles. J. Wildl. Dis. 45, 1138–1142 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Hawkes L. A., Broderick A. C., Godfrey M. H., Godley B. J., Climate change and marine turtles. Endanger. Species Res. 7, 137–154 (2009). [Google Scholar]
- 19.Wallace B. P., et al. , Global conservation priorities for marine turtles. PLoS One 6, e24510 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yntema C. L., Mrosovsky N., Incubation temperature and sex ratio in hatchling loggerhead turtles: a preliminary report. Mar. Turtle Newsl. 11, 9–10 (1979). [Google Scholar]
- 21.Jensen M. P., et al. , Environmental warming and feminization of one of the largest sea turtle populations in the world. Curr. Biol. 28, 154–159.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Mazaris A. D., Schofield G., Gkazinou C., Almpanidou V., Hays G. C., Global sea turtle conservation successes. Sci Adv 3, e1600730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.IUCN, The IUCN red list of threatened species (2021). (April 16, 2021).
- 24.Martínez L. S., et al. , Conservation and biology of the leatherback turtle in the Mexican Pacific. Chelonian Conserv. Biol. 6, 70–78 (2007). [Google Scholar]
- 25.Santidrián Tomillo P., et al. , Reassessment of the leatherback turtle (Dermochelys coriacea) nesting population at Parque Nacional Marino Las Baulas, Costa Rica: Effects of conservation efforts. Chelonian Conserv. Biol. 6, 54–62 (2007). [Google Scholar]
- 26.Spotila J. R., Reina R. D., Steyermark A. C., Plotkin P. T., Paladino F. V., Pacific leatherback turtles face extinction. Nature 405, 529–530 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Laúd OPO Network, Enhanced, coordinated conservation efforts required to avoid extinction of critically endangered Eastern Pacific leatherback turtles. Sci. Rep. 10, 4772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan E., Liew H., Decline of the leatherback population in Terengganu, Malaysia, 1956–1995. Chelonian Conserv. Biol. 2, 196–203 (1996). [Google Scholar]
- 29.Eckert K. L., Wallace B. P., Frazier J. G., Eckert S. A., Pritchard P. C. H., Synopsis of the Biological Data on the Leatherback Sea Turtle, Dermochelys coriacea (U.S. Department of Interior, Fish and Wildlife Service, 2012). [Google Scholar]
- 30.Jones K., Ariel E., Burgess G., Read M., A review of fibropapillomatosis in Green turtles (Chelonia mydas). Vet. J. 212, 48–57 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Zhang G., et al. , Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan I., et al. , Olfactory receptor subgenomes linked with broad ecological adaptations in Sauropsida. Mol. Biol. Evol. 32, 2832–2843 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Jebb D., et al. , Six reference-quality genomes reveal evolution of bat adaptations. Nature 583, 578–584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahon B. J., Teeling E. C., Höglund J., How and why should we implement genomics into conservation? Evol. Appl. 7, 999–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Supple M. A., Shapiro B., Conservation of biodiversity in the genomics era. Genome Biol. 19, 131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandies P., Peel E., Hogg C. J., Belov K., The value of reference genomes in the conservation of threatened species. Genes 10, 864 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hohenlohe P. A., Funk W. C., Rajora O. P., Population genomics for wildlife conservation and management. Mol. Ecol. 30, 62–82 (2020), 10.1111/mec.15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Goodsell J., Norgren R. B. Jr., Limitations of the rhesus macaque draft genome assembly and annotation. BMC Genomics 13, 206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhie A., et al. , Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuentes-Pardo A. P., Ruzzante D. E., Whole-genome sequencing approaches for conservation biology: Advantages, limitations and practical recommendations. Mol. Ecol. 26, 5369–5406 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Formenti G., et al. , The era of reference genomes in conservation genomics. Trends Ecol. Evol. 37, 197–202 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Wang Z., et al. , The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Genet. 45, 701–706 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whibley A., Kelley J. L., Narum S. R., The changing face of genome assemblies: Guidance on achieving high-quality reference genomes. Mol. Ecol. Resour. 21, 641–652 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Peona V., et al. , Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird-of-paradise. Mol. Ecol. Resour. 21, 263–286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan Y., et al. , Comparative genomics provides insights into the aquatic adaptations of mammals. Proc. Natl. Acad. Sci. U.S.A. 118, e2106080118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gemmell N. J., et al. , The tuatara genome reveals ancient features of amniote evolution. Nature 584, 403–409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson R. N., et al. , Adaptation and conservation insights from the koala genome. Nat. Genet. 50, 1102–1111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hubert J.-N., Zerjal T., Hospital F., Cancer- and behavior-related genes are targeted by selection in the Tasmanian devil (Sarcophilus harrisii). PLoS One 13, e0201838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmerman L. M., The reptilian perspective on vertebrate immunity: 10 years of progress. J. Exp. Biol. 223, jeb214171 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Seppey M., Manni M., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Wan Q.-H., et al. , Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 23, 1091–1105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quesada V., et al. , Giant tortoise genomes provide insights into longevity and age-related disease. Nat Ecol Evol 3, 87–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morin P. A., et al. , Reference genome and demographic history of the most endangered marine mammal, the vaquita. Mol. Ecol. Resour. 21, 1008–1020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson J. A., et al. , Genomic flatlining in the endangered island fox. Curr. Biol. 26, 1183–1189 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Waters P. D., et al. , Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc. Natl. Acad. Sci. U.S.A. 118, e2112494118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Megens H.-J., et al. , Comparison of linkage disequilibrium and haplotype diversity on macro- and microchromosomes in chicken. BMC Genet. 10, 86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Axelsson E., Webster M. T., Smith N. G. C., Burt D. W., Ellegren H., Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 15, 120–125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodionov A. V., Micro vs. macro: Structural-functional organization of avian micro- and macrochromosomes. Genetika 32, 597–608 (1996). [PubMed] [Google Scholar]
- 59.Endres C. S., Lohmann K. J., Detection of coastal mud odors by loggerhead sea turtles: A possible mechanism for sensing nearby land. Mar. Biol. 160, 2951–2956 (2013). [Google Scholar]
- 60.Endres C. S., Putman N. F., Lohmann K. J., Perception of airborne odors by loggerhead sea turtles. J. Exp. Biol. 212, 3823–3827 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Manton M., Karr A., Ehrenfeld D. W., Chemoreception in the migratory sea turtle, Chelonia mydas. Biol. Bull. 143, 184–195 (1972). [Google Scholar]
- 62.Kitayama C., et al. , Behavioral effects of scents from male mature Rathke glands on juvenile green sea turtles (Chelonia mydas). J. Vet. Med. Sci. 82, 1312–1315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Endres C. S., et al. , Multi-modal homing in sea turtles: Modeling dual use of geomagnetic and chemical cues in island-finding. Front. Behav. Neurosci. 10, 19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dodge K. L., Logan J. M., Lutcavage M. E., Foraging ecology of leatherback sea turtles in the Western North Atlantic determined through multi-tissue stable isotope analyses. Mar. Biol. 158, 2813–2824 (2011). [Google Scholar]
- 65.Arthur K. E., Boyle M. C., Limpus C. J., Ontogenetic changes in diet and habitat use in green sea turtle (Chelonia mydas) life history. Mar. Ecol. Prog. Ser. 362, 303–311 (2008). [Google Scholar]
- 66.Seminoff J. A., et al. , Large-scale patterns of green turtle trophic ecology in the eastern Pacific Ocean. Ecosphere 12, e03479 (2021). [Google Scholar]
- 67.Kitayama C., et al. , Morphological features of the nasal cavities of hawksbill, olive ridley, and black sea turtles: Comparative studies with green, loggerhead and leatherback sea turtles. PLoS One 16, e0250873 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kondoh D., Kitayama C., Kawai Y. K., The nasal cavity in sea turtles: Adaptation to olfaction and seawater flow. Cell Tissue Res. 383, 347–352 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Yamaguchi Y., et al. , Computed tomographic analysis of internal structures within the nasal cavities of green, loggerhead and leatherback sea turtles. Anat. Rec. 304, 584–590 (2021). [DOI] [PubMed] [Google Scholar]
- 70.Niimura Y., Nei M., Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J. Hum. Genet. 51, 505–517 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yohe L. R., Fabbri M., Hanson M., Bhullar B.-A.S., Olfactory receptor gene evolution is unusually rapid across Tetrapoda and outpaces chemosensory phenotypic change. Curr. Zool. 66, 505–514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saito H., Chi Q., Zhuang H., Matsunami H., Mainland J. D., Odor coding by a mammalian receptor repertoire. Sci. Signal. 2, ra9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vandewege M. W., et al. , Contrasting patterns of evolutionary diversification in the olfactory repertoires of reptile and bird genomes. Genome Biol. Evol. 8, 470–480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu A., et al. , Convergent degeneration of olfactory receptor gene repertoires in marine mammals. BMC Genomics 20, 977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Constantino M. A., Salmon M., Role of chemical and visual cues in food recognition by leatherback posthatchlings (Dermochelys coriacea L). Zoology 106, 173–181 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Warraich N., Wyneken J., Blume N., Feeding behavior and visual field differences in loggerhead and leatherback sea turtles may explain differences in longline fisheries interactions. Endanger. Species Res. 41, 67–77 (2020). [Google Scholar]
- 77.Siddle H. V., Marzec J., Cheng Y., Jones M., Belov K., MHC gene copy number variation in Tasmanian devils: Implications for the spread of a contagious cancer. Proc. Biol. Sci. 277, 2001–2006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Escobar L. E., et al. , A global map of suitability for coastal Vibrio cholerae under current and future climate conditions. Acta Trop. 149, 202–211 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Zhang L., et al. , Massive expansion and functional divergence of innate immune genes in a protostome. Sci. Rep. 5, 8693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elbers J. P., Taylor S. S., Others, Major histocompatibility complex polymorphism in reptile conservation. Herpetol. Conserv. Biol. 11, 1–12 (2016). [Google Scholar]
- 81.Martin K. R., Mansfield K. L., Savage A. E., Adaptive evolution of major histocompatibility complex class I immune genes and disease associations in coastal juvenile sea turtles. R Soc. Open Sci. 9, 211190 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vekemans X., et al. , Whole-genome sequencing and genome regions of special interest: Lessons from major histocompatibility complex, sex determination, and plant self-incompatibility. Mol. Ecol. 30, 6072–6086 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moritz C., Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst. Biol. 51, 238–254 (2002). [DOI] [PubMed] [Google Scholar]
- 84.Fernandez-Fournier P., Lewthwaite J. M. M., Mooers A. Ø., Do we need to identify adaptive genetic variation when prioritizing populations for conservation? Conserv. Genet. 22, 205–216 (2021). [Google Scholar]
- 85.Borrell Y. J., et al. , Heterozygosity-fitness correlations in the gilthead sea bream Sparus aurata using microsatellite loci from unknown and gene-rich genomic locations. J. Fish Biol. 79, 1111–1129 (2011). [DOI] [PubMed] [Google Scholar]
- 86.DeWoody J. A., Harder A. M., Mathur S., Willoughby J. R., The long-standing significance of genetic diversity in conservation. Mol. Ecol. 30, 4147–4154 (2021). [DOI] [PubMed] [Google Scholar]
- 87.Willi Y., et al. , Conservation genetics as a management tool: The five best-supported paradigms to assist the management of threatened species. Proc. Natl. Acad. Sci. U.S.A. 119, e2105076119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Komoroske L. M., et al. , A versatile rapture (RAD-Capture) platform for genotyping marine turtles. Mol. Ecol. Resour. 19, 497–511 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Dutton P. H., Bowen B. W., Owens D. W., Barragan A., Davis S. K., Global phylogeography of the leatherback turtle (Dermochelys coriacea). J. Zool. 248, 397–409 (1999). [Google Scholar]
- 90.Jensen M. P., et al. , The evolutionary history and global phylogeography of the green turtle (Chelonia mydas). J. Biogeogr. 46, 860–870 (2019). [Google Scholar]
- 91.Kardos M., et al. , The crucial role of genome-wide genetic variation in conservation. Proc. Natl. Acad. Sci. U.S.A. 118, e2104642118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dobrynin P., et al. , Genomic legacy of the African cheetah, Acinonyx jubatus. Genome Biol. 16, 277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mattila A. L. K., et al. , High genetic load in an old isolated butterfly population. Proc. Natl. Acad. Sci. U.S.A. 109, E2496–E2505 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robinson J. A., Brown C., Kim B. Y., Lohmueller K. E., Wayne R. K., Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr. Biol. 28, 3487–3494.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dussex N., et al. , Population genomics of the critically endangered kākāpō. Cell Genomics 1, 100002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kyriazis C. C., Wayne R. K., Lohmueller K. E., Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evol. Lett. 5, 33–47 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dewoody Y. D., Dewoody J. A., On the estimation of genome-wide heterozygosity using molecular markers. J. Hered. 96, 85–88 (2005). [DOI] [PubMed] [Google Scholar]
- 98.Casale P., et al. , Mediterranean sea turtles: Current knowledge and priorities for conservation and research. Endanger. Species Res. 36, 229–267 (2018). [Google Scholar]
- 99.Tikochinski Y., et al. , Mitochondrial DNA STR analysis as a tool for studying the green sea turtle (Chelonia mydas) populations: The Mediterranean Sea case study. Mar. Genomics 6, 17–24 (2012). [DOI] [PubMed] [Google Scholar]
- 100.Tikochinski Y., et al. , Mitochondrial DNA short tandem repeats unveil hidden population structuring and migration routes of an endangered marine turtle. Aquat. Conserv. 28, 788–797 (2018), 10.1002/aqc.2908. [DOI] [Google Scholar]
- 101.Karaman S., et al. , Population genetic diversity of green turtles, Chelonia mydas, in the Mediterranean revisited. Mar. Biol. 169, 77 (2022). [Google Scholar]
- 102.Hewitt G. M., Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 183–95 (2004), discussion 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seersholm F. V., et al. , Rapid range shifts and megafaunal extinctions associated with late Pleistocene climate change. Nat. Commun. 11, 2770 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen I.-C., Hill J. K., Ohlemüller R., Roy D. B., Thomas C. D., Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (2011). [DOI] [PubMed] [Google Scholar]
- 105.Brüniche-Olsen A., Kellner K. F., Belant J. L., DeWoody J. A., Life-history traits and habitat availability shape genomic diversity in birds: Implications for conservation. Proc. Biol. Sci. 288, 20211441 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van der Zee J. P., et al. , The population genomic structure of green turtles (Chelonia mydas) suggests a warm-water corridor for tropical marine fauna between the Atlantic and Indian oceans during the last interglacial. Heredity 127, 510–521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fitak R. R., Johnsen S., Green sea turtle (Chelonia mydas) population history indicates important demographic changes near the mid-Pleistocene transition. Mar. Biol. 165, 110 (2018). [Google Scholar]
- 108.Vilaça S. T., et al. , Divergence and hybridization in sea turtles: Inferences from genome data show evidence of ancient gene flow between species. Mol. Ecol. 30, 6178–6192 (2021), 10.1111/mec.16113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patton A. H., et al. , Contemporary demographic reconstruction methods are robust to genome assembly quality: A case study in tasmanian devils. Mol. Biol. Evol. 36, 2906–2921 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nadachowska-Brzyska K., Burri R., Smeds L., Ellegren H., PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers. Mol. Ecol. 25, 1058–1072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.United States National Marine Fisheries Service, U.S. Fish and Wildlife Service, “Endangered species act status review of the leatherback turtle (Dermochelys coriacea)” (United States National Marine Fisheries Service, 2020). [Google Scholar]
- 112.Khan A., et al. , Genomic evidence for inbreeding depression and purging of deleterious genetic variation in Indian tigers. Proc. Natl. Acad. Sci. U.S.A. 118, e2023018118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Prasad A., Lorenzen E. D., Westbury M. V., Evaluating the role of reference-genome phylogenetic distance on evolutionary inference. Mol. Ecol. Resour. 22, 45–55 (2022). [DOI] [PubMed] [Google Scholar]
- 114.Brüniche-Olsen A., Kellner K. F., Anderson C. J., DeWoody J. A., Runs of homozygosity have utility in mammalian conservation and evolutionary studies. Conserv. Genet. 19, 1295–1307 (2018). [Google Scholar]
- 115.Foote A. D., et al. , Runs of homozygosity in killer whale genomes provide a global record of demographic histories. Mol. Ecol. 30, 6162–6177 (2021), 10.1111/mec.16137. [DOI] [PubMed] [Google Scholar]
- 116.Robinson J. A., et al. , The critically endangered vaquita is not doomed to extinction by inbreeding depression. Science 376, 635–639 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O’Leary N. A., et al. , Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pruitt K. D., et al. , RefSeq: An update on mammalian reference sequences. Nucleic Acids Res. 42, D756–D763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blum M., et al. , The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mi H., et al. , PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49, D394–D403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Paten B., et al. , Cactus graphs for genome comparisons. J. Comput. Biol. 18, 469–481 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Armstrong J., et al. , Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587, 246–251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mendes F. K., Vanderpool D., Fulton B., Hahn M. W., CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics, 10.1093/bioinformatics/btaa1022 (2020). [DOI] [PubMed]
- 124.Emms D. M., Kelly S., OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Emms D. M., Kelly S., OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hickey G., Paten B., Earl D., Zerbino D., Haussler D., HAL: A hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics 29, 1341–1342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Robinson J. A., et al. , Genomic signatures of extensive inbreeding in Isle Royale wolves, a population on the threshold of extinction. Sci Adv 5, eaau0757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Purcell S., et al. , PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ceballos F. C., Joshi P. K., Clark D. W., Ramsay M., Wilson J. F., Runs of homozygosity: Windows into population history and trait architecture. Nat. Rev. Genet. 19, 220–234 (2018). [DOI] [PubMed] [Google Scholar]
- 131.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McKenna A., et al. , The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li H., et al. , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vertebrate Genomes Project, Reference genomes and associated read data for the green turtle (Chelonia mydas). GenomeArk. https://genomeark.github.io/vgp-curated-assembly/Chelonia_mydas/. Deposited 28 May 2021.
- 137.Vertebrate Genomes Project, Reference genomes and associated read data for the leatherback turtle (Dermochelys coriacea). GenomeArk. https://genomeark.github.io/vgp-curated-assembly/Dermochelys_coriacea/. Deposited 28 May 2021.
- 138.Bentley B., et al. , Scripts associated with genome analyses of green and leatherback turtles. Github. https://github.com/bpbentley/sea_turtle_genomes. Deposited 24 June 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Data Availability Statement
Genome assemblies have been deposited on NCBI GenBank. The NCBI GenBank accession numbers for the leatherback turtle assembly (rDerCor1) are GCF_009764565.3 and GCA_009762595.2 for the annotated primary and original alternate haplotypes in BioProject PRJNA561993, and for the green turtle assembly (rCheMyd1) are GCF_015237465.2 and GCA_015220195.2 for primary and alternate haplotypes respectively in BioProject PRJNA561941. The raw data used for assemblies are available on the Vertebrate Genome Ark (https://vgp.github.io/genomeark/). The leatherback turtle data generated for the purpose of assembly annotation was deposited in the SRA under accession numbers SRX8787564-SRX8787566 (RNA-Seq) and SRX6360706-SRX6360708 (ISO-Seq). Green turtle data generated for annotation were deposited in SRA under accessions SRX10863130-SRX10863133 (RNA-Seq) and as SRX11164043-SRX11164046 (ISO-Seq). The NovaSeq 6000 DNA-Seq data for the green turtle resequencing, including raw reads, are deposited in NCBI (https://www.ncbi.nlm.nih.gov/) under BioProject ID: PRJNA449022. All scripts used for downstream analyses following genome assembly and annotation have been deposited on GitHub under repository https://github.com/bpbentley/sea_turtle_genomes.
All genomic data and scripts data have been deposited in VGP GenomeArk (136, 137) Github (138).