Significance
There is considerable genetic diversity in the TP53 gene, with hundreds of missense single nucleotide polymorphisms (SNPs) that may alter p53 function. While cancer-causing TP53 mutations are well characterized, many of the germline missense variants remain unclassified because their impact on p53 function and cancer risk is unknown. Our characterization of shared gain of function (GOF) activities and elucidation of a common gene expression signature amongst a subset of these hypomorphic variants has the potential to allow us to better inform affected individuals of their associated cancer risk and therapeutic outcomes.
Keywords: p53, NF-kB, gene signature, mutant p53, cancer
Abstract
Missense mutations that inactivate p53 occur commonly in cancer, and germline mutations in TP53 cause Li Fraumeni syndrome, which is associated with early-onset cancer. In addition, there are over two hundred germline missense variants of p53 that remain uncharacterized. In some cases, these germline variants have been shown to encode lesser-functioning, or hypomorphic, p53 protein, and these alleles are associated with increased cancer risk in humans and mouse models. However, most hypomorphic p53 variants remain un- or mis-classified in clinical genetics databases. There thus exists a significant need to better understand the behavior of p53 hypomorphs and to develop a functional assay that can distinguish hypomorphs from wild-type p53 or benign variants. We report the surprising finding that two different African-centric genetic hypomorphs of p53 that occur in distinct functional domains of the protein share common activities. Specifically, the Pro47Ser variant, located in the transactivation domain, and the Tyr107His variant, located in the DNA binding domain, both share increased propensity to misfold into a conformation specific for mutant, misfolded p53. Additionally, cells and tissues containing these hypomorphic variants show increased NF-κB activity. We identify a common gene expression signature from unstressed lymphocyte cell lines that is shared between multiple germline hypomorphic variants of TP53, and which successfully distinguishes wild-type p53 and a benign variant from lesser-functioning hypomorphic p53 variants. Our findings will allow us to better understand the contribution of p53 hypomorphs to disease risk and should help better inform cancer risk in the carriers of p53 variants.
The p53 tumor suppressor gene plays a central role in cancer, as best illustrated by the fact that TP53 is mutated in over 50% of human tumors (1, 2). Germline mutations in TP53 are present in families with Li Fraumeni syndrome (LFS), who develop multiple cancers of the brain, breast, bone, and adrenal cortex as early as the first decade of life (3). p53 also is a central player in the response of cells to genotoxic stress, and mutations in TP53 are associated with cancer therapy resistance and poor prognosis (4, 5). Consistent with the latter premise, numerous studies have shown that missense mutant forms of p53 protein can possess oncogenic functions: these are referred to as “gain of function (GOF)”. Prominent GOF activities by mutant p53 include the activation of SREBP1/2 (6), growth factor receptor recycling (7, 8), and activation of NF-κB signaling (9). It is important to note that despite widespread evidence for mutant p53 GOF, some tumor types do not show evidence for GOF (10). Typically, GOF activities of mutant p53 play roles in late-stage tumors, and GOF mutants are often associated with increased metastatic potential of tumors (11–15).
Unlike most tumor suppressor genes and oncogenes, there is significant germline coding region variation in TP53, with an estimated 200 germline missense variants in different ethnic groups. In some cases, these missense germline variants have been shown to impair but not completely inactivate p53 function; we refer to such variants as “hypomorphs.” We previously described one hypomorphic variant of TP53, Pro47Ser (P47S, rs1800371), which occurs in 1 to 2% of African-descent individuals and is associated with increased cancer incidence in a mouse model (16) and in humans (17). We showed that P47S protein is selectively defective in the ability to regulate p53 target genes associated with ferroptosis or iron–lipid–mediated cell death. This ferroptotic defect is correlated with increased production of glutathione and coenzyme A in P47S cells (18, 19). This ferroptotic defect leads to iron accumulation in P47S mice; this led to our discovery that P47S is significantly associated with a disorder called Iron Overload in African Americans (20). Interestingly, the increase in glutathione in P47S cells leads to increased activation of mTOR due to increased Rheb-mTOR association; this leads to improved fitness and muscle recovery in P47S mice, suggesting that this allele may have been selected for at one time (21).
We recently began studying another p53 variant that is specific to African-descent populations, Tyr107His (Y107H, rs368771578). Like P47S, Y107H has been described as “partially functional,” or hypomorphic (22). We show here that the P47S and Y107H hypomorphic variants, despite their localization in different domains of p53, both show increased propensity to misfold compared to wildtype (WT) p53. Specifically, we show that these variants adopt a protein conformation commonly associated with tumor-derived mutant forms of p53. Mutant p53 is known to enhance NF-κB activity (9), and we show that these two hypomorphs are both associated with increased NF-κB activity in unstressed human cells and mouse tissues. We then use machine-learning approaches on RNA sequencing data from unstressed lymphoblastoid cells and identify a 143-gene signature that accurately distinguishes p53 hypomorphs from WT p53 and even a benign variant. These findings suggest that we may be able to broadly screen individuals with genetic variants of p53 and better inform them of their cancer risk.
Results
Altered p53 Target Gene Expression in Lymphoblastoid Cell Lines (LCLs) from P47S and Y107H Individuals.
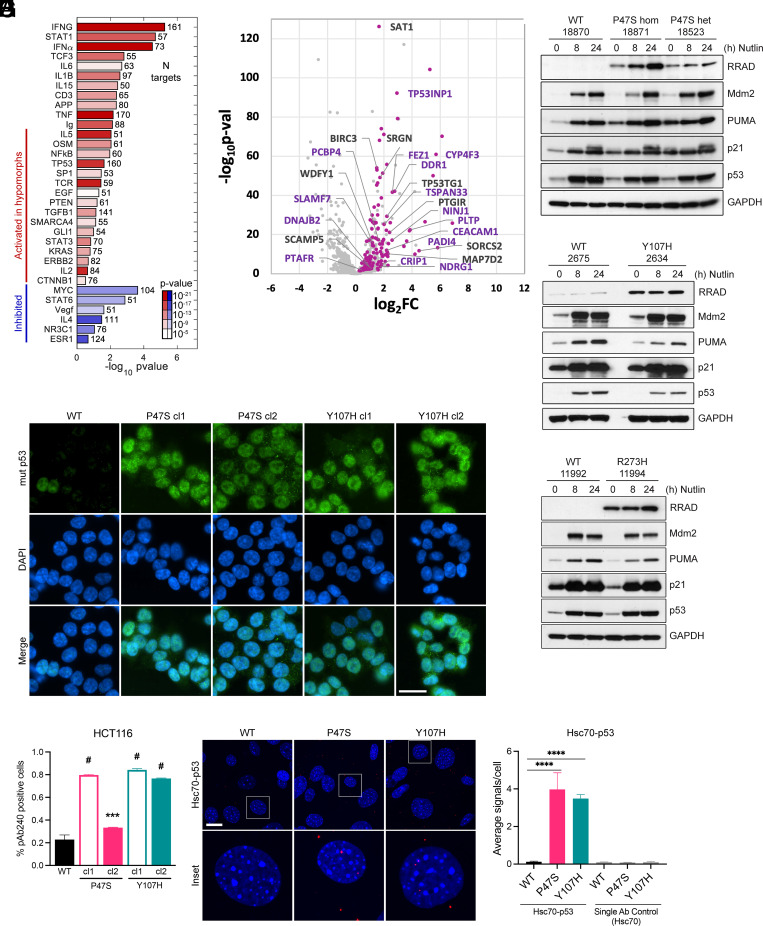
We sought to assess the p53 transcriptional response in LCLs isolated from individuals containing two different germline genetic variants of TP53: P47S (Pro47Ser, rs1800371) and Y107H (Tyr107His, rs368771578). Toward this goal, we conducted RNA sequencing (RNA Seq) and western blot analysis following Nutlin-3a treatment in P47S and Y107H L CL lines, compared to age- and sex-matched WT controls from the same region of Africa (Y107H) or the same family (P47S). Because these hypomorphic variants are located in distinct functional domains of p53 (P47S in transactivation domain 2 and Y107H in the DNA binding domain of p53), we were surprised to find three things in common between them. First, we found the evidence for an enhanced NF-κB gene signature in hypomorph cells relative to controls, even under unstressed conditions (Fig. 1A). Second, we identified a small subset of p53 target genes that showed impaired transactivation in both hypomorph lines following Nutlin-3a treatment compared to controls (Fig. 1B); this occurred despite equal induction of p53 and the p53-induced targets MDM2, p21/CDKN1A, and PUMA in P47S, Y107H, and WT cells (Fig. 1 C and D). Third, we discovered one p53 target, RRAD, that was markedly increased at the protein level in P47S and Y107H cells, both at baseline and following Nutlin-3a treatment, compared to controls. RRAD was highest in homozygous P47S LCLs, suggesting that this is not a dominant negative effect on WT p53, and also was increased in P47S and Y107H heterozygous LCLs relative to controls (Fig. 1 C and D). RRAD was also higher in LCLs from another heterozygous p53 hypomorph, G334R (23), compared to a WT sibling control (SI Appendix, Fig. S1A), and in heterozygous LCLs with a Li Fraumeni mutant, R273H (Fig. 1E). The latter finding led us to perform quantitative RT-PCR analysis of WT, Y107H, and R273H LCLs treated with Nutlin-3a; this analysis confirmed that induction of canonical p53 targets like MDM2 and CDKN1A was indistinguishable in WT and Y107H LCLs but was significantly impaired in LCLs containing the Li Fraumeni mutant R273H (SI Appendix, Fig. S1 B and C). Overall, our analyses indicate that the P47S and Y107H hypomorphs are defective in the transactivation of only a small subset of p53 target genes and thus can be classified as hypomorphic. They also support that there is a correlation between the RRAD level and the presence of inactive (Li Fraumeni mutant) as well as hypomorphic p53 (P47S and Y107H).
Fig. 1.
p53 hypomorphs adopt a mutant conformation and express high levels of RRAD. (A) IPA analysis showing significance and effect on regulators with their targets significantly enriched among the genes differentially expressed in p53 hypomorphs P47S and Y107H. The bars represent activation Z-score of the regulators predicted by IPA based on direction of target changes, color represents significance P value, and the number after the bar represents the number of targets affected. (B) Log2 fold change plotted against –Log10(P-value) for genes that respond to Nutlin-3a treatment after 24 h in WT. Highlighted pink are the genes that are activated to a greater extent in WT than in Y107H in response to treatment and labelled with names are the genes that behave similarly in P47S. The purple labels correspond to known p53 target genes. (C–E) LCL cell lines expressing hypomorphic p53 either (C) P47S—homozygous and heterozygous, (D) Y107H heterozygous, or (E) R273H heterozygous compared to regionally/family matched WT p53 LCL cell lines were treated with 10 μM Nutlin-3a for the indicated time points and harvested. Whole cell lysates were subjected to western blot analysis for the indicated antibodies. (F) WT, P47S, and Y107H HCT116 cells were analyzed for mutant p53 (pAb240) conformation by immunofluorescence analysis. (Scale bar, 25 μm.) (G) Quantification of the fraction of cells staining for mutant conformation (pAb240). Fractions indicated are such that 1.0 = 100%. Data are presented as mean ± SD. n = 25 random fields of view (>250 cells) from each of 2 independent experiments. ***P < 0.001; #P < 0.0001 by one-way ANOVA followed by Dunnett’s multiple comparisons test. (H) Primary MEFs generated from WT, P47S, and Y107H mice were used to assess mutant p53 conformation by p53-Hsc70 interaction using PLA. Representative images are shown for each condition (Scale bar, 20 μm.) White boxes denote the enlarged insets. (I) Quantification of the average number of PLA signals per cell. Values are expressed as mean ± SD. n = 6 random fields of view (>100 cells) from each of two independent experiments. ****P < 0.0001 by one-way ANOVA followed by Dunnett’s multiple comparisons test.
p53 Hypomorphs Adopt a Misfolded, Mutant Conformation.
RRAD is regulated by both p53 and NF-κB (24). Mutant forms of p53, including the R273H mutant, bind and enhance the NF-κB activity (9). This led us to predict that the P47S and Y107H variants might both misfold into the common, denatured conformation that is characteristic of mutant p53 and specifically recognized by the monoclonal antibody pAb240 (25, 26). We further reasoned that P47S and Y107H LCLs might show increased NF-κB activity. To test the first hypothesis, we used CRISPR-Cas9 knock-in technology to generate two clones each of the P47S and Y107H hypomorphs, in the background of HCT116 cells. The creation and validation of these lines are described in the Materials and Methods; equivalent induction of p53 and target genes is shown in SI Appendix, Fig. S1D. We also used CRISPR/Cas9 to generate the Y107H knock-in mouse using C57Bl/6 embryos; these Y107H mice were backcrossed to a C57Bl/6 background for five generations, and the entire p53 coding region was sequenced to ensure the absence of second-site mutations. Mouse embryonic fibroblasts (MEFs) from these mice show equivalent induction of p53 following genotoxic stress compared to WT (SI Appendix, Fig. S1E). Immunofluorescent staining of HCT116 clones with the pAb240 mutant conformation-specific antibody detected a significantly increased percentage of mutant p53 conformation-positive cells in P47S and Y107H knock-in HCT116 clones compared to the parental WT p53 cell line; there were approximately 20% pAb240 positive cells in WT, and 35 to 80% pAb-240 positive cells in hypomorph clones (Fig. 1 F and G). Staining with the pan-p53 antibody was indistinguishable (SI Appendix, Fig. S1F). As an orthogonal approach, we analyzed primary MEFs isolated from mice containing germline versions of WT p53, P47S, or Y107H, all generated in the Humanized p53 knock-in background (16). In these WT, P47S, and Y107H MEFs, we used proximity ligation analysis (PLA) to assess the interaction of p53 with Hsc70, as this interaction is specific for misfolded, mutant p53 (27, 28). PLA of the Hsc70–p53 interaction in early passage primary MEFs revealed significantly increased association of Hsc70 with P47S and Y107H compared to WT p53 (Fig. 1 H and I; see SI Appendix, Fig. S1E for the negative control). These data indicate that the P47S and Y107H hypomorphs both have increased propensity to misfold into a mutant conformation. This is similar to what we showed previously for the G334R hypomorph of p53 (23).
We next sought to determine whether the level of RRAD might correlate with the level of misfolded p53 in the cell. First, we noted that there was increased RRAD in homozygous P47S LCLs compared to heterozygous P47S (Fig. 1C). Second, we found that the level of RRAD was significantly greater in cells containing the P47S variant compared to R273H; the latter is generally believed to be predominantly folded into a WT conformation (SI Appendix, Fig. S1F). Next, we sought to confirm our RNA Seq findings that a subset of p53 target genes were transactivated less in P47S and Y107H LCLs; western blot analysis confirmed that the p53 target proteins PADI4 and CEACAM-1 exhibit decreased induction in P47S and Y107H LCLs relative to matched WT controls (SI Appendix, Fig. S1 G and H). In contrast, the p53 targets LCN15, MDM2, PUMA, and p21/CDKN1A were induced to equivalent levels in hypomorph and WT LCLs (SI Appendix, Fig. S1 G and H). The combined data indicate that increased RRAD levels and decreased ability to transactivate a subset of target genes is shared between the P47S and Y107H hypomorphic variants of p53.
P47S and Y107H Cells Exhibit Elevated NF-κB Signaling.
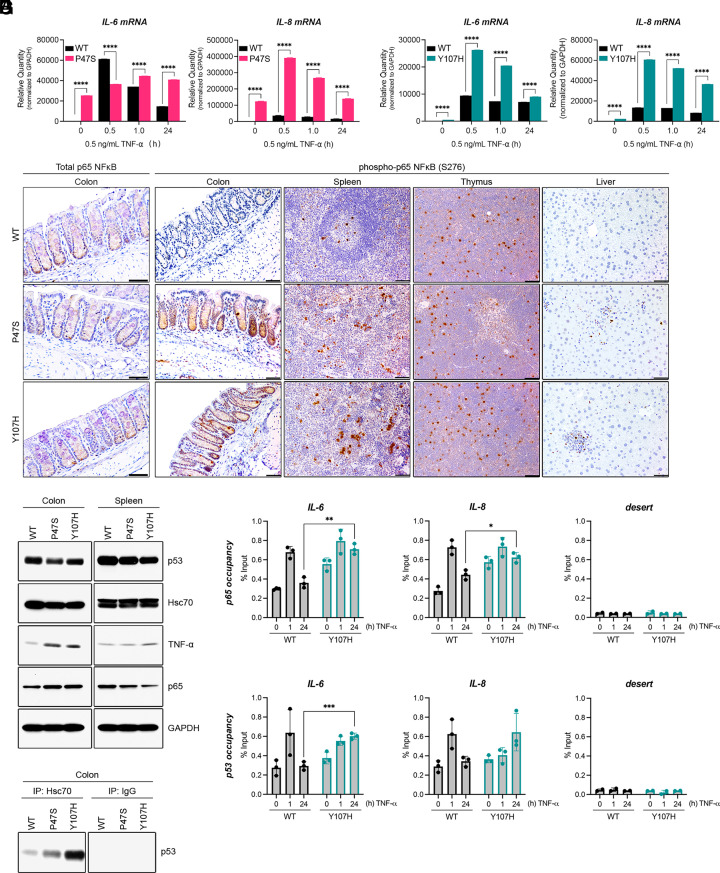
Tumor-derived mutant forms of p53 can bind and stabilize the p65 subunit of NF-κB on chromatin, leading to increased steady-state level of NF-κB target genes in tumors with mutant p53 (9, 29, 30). We hypothesized that the increased mutant conformation of p53 in P47S and Y107H cells might lead to increased NF-κB activity. To address this, we treated LCLs heterozygous for P47S and Y107H with TNF-α and analyzed the induction of IL-6 and IL-8, two well-known NF-κB target genes by quantitative real-time polymerase chain reaction (q-RT-PCR). In both cases, we compared this induction to matched WT LCL lines. This analysis revealed that both P47S and Y107H LCLs exhibit hyperactive NF-κB signaling compared to matched WT controls, both at baseline and in response to stimulation with TNF-α (Fig. 2 A and B). Expression of NF-κB target genes remained elevated for 24 h in p53 hypomorph cells relative to WT controls, similar to the previously reported “GOF” activity of mutant p53 (9). In contrast, we failed to see increased transcriptional response to another stimulus, epidermal growth factor (EGF), in p53 hypomorph cells relative to control (SI Appendix, Fig. S2 A and B).
Fig. 2.
p53 hypomorphs exhibit elevated NF-κB signaling. (A and B) LCL cell lines expressing WT or hypomorphic p53, either (A) P47S or (B) Y107H were treated with 0.5 ng/mL TNF-α for the indicated periods. RNA was extracted and analyzed by Q-RT-PCR with primers specific for IL-6 and IL-8 mRNA. Values were normalized for GAPDH mRNA in the same sample. Values are expressed as mean ± SD. Data are representative of n = 6 technical replicates from two independent experiments. ****P < 0.0001 by one-way ANOVA followed by Tukey’s multiple comparisons test. (C) Representative immunohistochemical images of colon, spleen, thymus, and liver sections obtained from 10-wk-old male mice of the indicated genotypes. Tissue sections were stained for total p65 (colon) or for phospho-p65 NF-κB (Ser276). (Scale bar, 50 μm.) (D) Hsc70 immunoprecipitation in colon tissue harvested from 10-wk-old male mice of the indicated genotypes. Immunoprecipitation of Hsc70 or IgG negative control was followed by immunoblotting for p53. (E) Whole cell lysates prepared from the colon and spleen of 10-wk-old male mice of the indicated genotypes were analyzed by western blotting with the indicated antibodies. (F and G) WT and Y107H LCLs were treated with 0.5 ng/mL TNF-α for 0, 1, or 24 h and subjected to ChIP with (9) p65 NF-κB or (9) p53 antibodies, followed by Q-RT-PCR analysis with primers specific to the NF-κB site of the IL-6 and IL-8 promoters or a gene desert (negative control). Values are presented as percentage of input ± SD. The data represent three technical replicates and are representative of two biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001 as determined by two-tailed Student’s t test.
To corroborate our findings, we next sought to probe NF-κB signaling in tissues from p53 hypomorph mice. We used immunohistochemistry to analyze the level of total and phospho-p65 (S276) in the colon, spleen, thymus, and liver of mice containing the P47S and Y107H variants, compared to WT p53. We found that there was equivalent staining for, and level of, total p65 in all tissues of all three genotypes of mice (Fig. 2C, Left and SI Appendix, Fig. S2 C and D) but markedly elevated phospho-p65 staining in the colons and spleens of P47S and Y107H hypomorph mice relative to WT controls (Fig. 2C). In contrast, there were no differences in phospho-p65 staining in the thymus or livers of these mice compared to controls, suggesting that these findings were cell type-specific. We next monitored the Hsc70–p53 interaction in the colon and thymus and found increased Hsc70 association with P47S and Y107H in the colon, consistent with the premise that increased misfolded p53 tracks with increased NF-κB activity (Fig. 2 D and E). We saw equivalent Hsc70–p53 interaction in the thymus of WT, P47S, and Y107H mice (SI Appendix, Fig. S2C). These data indicate that increased NF-κB activation occurs in a subset of tissues from hypomorph mice and is correlated with the level of Hsc70–p53 interaction.
One of the most consistent phenotypes conferred by mutant p53 is increased invasiveness (14). We, therefore, next explored the propensity for invasion in MEFs and HCT116 cells containing P47S, Y107H, or WT p53. To do this, we performed invasion assays with cells that were untreated or that were treated with TNF-α to stimulate migration. We found that MEFs from P47S and Y107H mice exhibited significantly increased migration and invasion compared to WT controls, both when untreated and in the presence of TNF-α (SI Appendix, Fig. S2 F and G). Similar results were obtained in P47S and Y107H HCT116 clones, compared to WT (SI Appendix, Fig. S2E). In sum, multiple clones of human and mouse cells containing the P47S and Y107H hypomorphs showed increased invasive potential, both at steady state and following stimulation with TNF-α.
Increased p65 Occupancy on Chromatin in Y107H LCLs.
Mutant p53 binds to and prolongs NF-κB occupancy on chromatin at the promoters of NF-κB target genes (9). We therefore next performed chromatin immunoprecipitation (ChIP) for p65 and p53 at the promoters of NF-κB target genes in WT and Y107H hypomorph LCLs. This analysis revealed increased occupancy of p65 at the promoters of IL-6 and IL-8 in Y107H cells, compared to WT, both at steady state and following treatment with TNF-α (Fig. 2F). Notably, we also found evidence for p53 occupancy at the NF-κB sites in these promoters (Fig. 2G). Primers for a promoter desert and IgG controls were clear of signal, and control CDKN1A primers showed comparable signals in WT and Y107H LCLs (Fig. 2 F and G and SI Appendix, Fig. S2 F–H).
Altered Biochemical Properties of the Y107H Hypomorph Can Be Reversed by the p53 Refolding Agent Arsenic Trioxide (ATO).
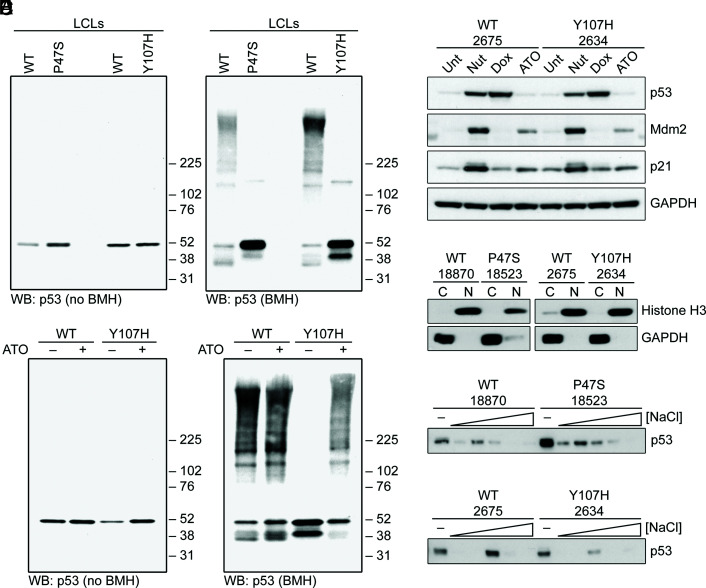
We next sought to analyze the ability of hypomorphic p53 to participate in protein–protein interactions, including the ability to form tetramers and higher-order oligomers. To do this, we employed the cross-linking agent bismaleimidohexane (BMH), which we have previously shown can detect species consistent with p53 tetramers and octamers, along with higher-order protein complexes (18). As expected, untreated LCLs from P47S and Y107H showed similar steady-state levels of p53 compared to matched WT cells (Fig. 3A). Following BMH cross-linking, there were marked differences in the pattern of cross-linked species in hypomorph cells compared to WT p53: the majority of WT p53 existed in higher molecular weight complexes, consistent with p53 tetramers and octamers, while the P47S and Y107H proteins appear to exist predominantly as monomers, along with minor species consistent with the size of p53 dimers (Fig. 3A). Interestingly, the higher molecular weight complexes could be restored for the Y107H hypomorph by incubation with ATO, which is reported to refold mutant p53 into a WT conformation (31). ATO had no effect on WT p53 protein complex formation (Fig. 3B) nor was this concentration of ATO sufficient to stabilize p53 (Fig. 3C). To explore the ability of hypomorphs to bind to chromatin, we next analyzed WT, P47S, and Y107H LCLs for the presence of p53 in nuclear, salt-extracted fractions. We did not see significant differences in the percent of WT, P47S, or Y107H p53 in the salt-extractable, presumably chromatin-bound, state (Fig. 3 D–F). The combined data support the premise that p53 hypomorphs do not show overt differences in chromatin binding compared to WT p53, but they show reduced propensity to engage in higher-order tetramers and oligomers in unstressed cells.
Fig. 3.
Altered biochemical properties of hypomorphic p53 proteins. (A) WT, P47S, and Y107H LCLs were treated with or without 1 mM BMH for 30 min followed by immunoblotting for p53. (B) WT, P47S, and Y107H LCLs were treated with vehicle or 1 μg/mL ATO for 16 h and then fixed with or without 1 mM BMH for 30 min followed by immunoblotting for p53. (C) WT or Y107H LCLs were untreated or treated with 10 μM Nutlin-3a (Nut), 1 μM doxorubicin (Dox), or 1 μg/mL ATO for 24 h. Whole-cell lysates were analyzed by western blotting with the indicated antibodies. (D) Cytosolic and nuclear fractions isolated from WT, P47S, and Y107H LCLs were analyzed by western blotting with the indicated antibodies. Histone H3 and GAPDH served as nuclear and cytosolic controls, respectively, to confirm efficiency of subcellular fractionation. (E and F) Chromatin-bound proteins were extracted from nuclear pellets isolated from WT, (E) P47S, and (F) Y107H LCLs using sequentially increasing salt concentrations (0 to 500 mM NaCl) and analyzed by western blotting for p53.
Identification of a Gene Expression Signature for Hypomorphic p53.
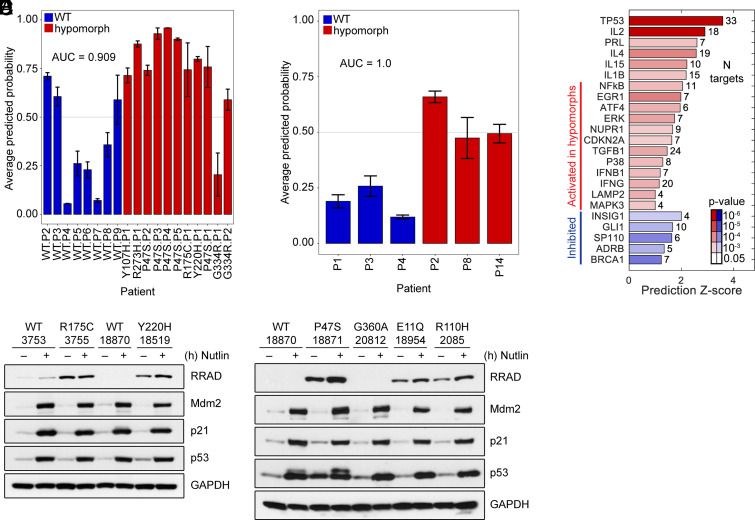
Our finding of increased NF-κB and altered gene expression in unstressed LCLs from P47S and Y107H individuals prompted us to test the possibility that these and other hypomorphs might share a common gene signature. Toward this goal, we compared RNA Seq data from RNA isolated from eighteen samples. This included eight different LCL lines with WT p53 (of varied ethnicity, sex, and age) along with ten different hypomorph cell lines: Y107H; P47S (five independent lines); R175C; Y220H; G334R; and R273H (Li Fraumeni mutant). With the exception of one P47S line, all LCLs were heterozygous. All lines were analyzed in unstressed conditions in biological triplicates. Gene expression data were used to train a support vector machine classifier model that identified a 143-gene signature with the potential to distinguish between WT p53 and a hypomorph (Fig. 4A and SI Appendix, Fig. S3A). This 143-gene signature was then assessed against a blinded independent test set of six new samples: of this independent set, two were previously unanalyzed WT samples, two were previously unanalyzed hypomorphs (P47S), and two samples were of unknown functional status (G360A and E11Q). This model accurately predicted all four WT and P47S hypomorph variants. It classified E11Q as a hypomorph and G360A as WT, suggesting that the latter is a benign variant (Fig. 4B and SI Appendix, Fig. S3B). Ingenuity pathways analysis (IPA) of these genes indicated that significant regulators in the hypomorph gene signature were, not surprisingly, p53 and NF-κB (Fig. 4C). Importantly, we next made an effort to confirm the accuracy of our gene signature predictions in the validation set by using the steady-state level of RRAD as a surrogate for hypomorphic p53. Our RRAD western blots confirmed the conclusions of our gene signature prediction that E11Q is likely a hypomorph (high level of RRAD), while G360A is likely a benign variant (undetectable RRAD, Fig. 4 D and E).
Fig. 4.
p53 hypomorphs share a predictive gene signature. (A) Classification scores for training set samples obtained through leave one out cross-validation based on total 143 unique genes. Probability of 0.5 is used as the classification threshold to calculate accuracy. (B) Classification scores for independent test set. (C) IPA analysis showing enrichment and effect on regulators with targets significantly enriched among the 143 genes from the classifier. (D) Western blot analysis of WT, R175C, and Y220H LCLs treated with 10 μM Nutlin-3a for 24 h. (E) Western blot analysis of WT, P47S, G360A, E11Q, and R110H LCLS treated with 10 μM Nutlin-3a for 24 h.
Discussion
Information from three large studies is presently used in order to classify somatic and germline missense variants of p53 as inactivating (pathogenic) versus benign, or passenger, mutations (22, 32, 33). There are some noted strengths of these studies, which were all conducted in an unbiased manner. First, they use multiple read-outs of p53 function, including transcription, etoposide-induced cell death, and the ability to function in a dominant negative manner. And second, they have reasonably high throughput and have been used to analyze hundreds to thousands of missense and other p53 variants. There are caveats to these studies, however. One study employs a yeast-based assay wherein reporter genes from only eight p53 targets are queried; in some cases, these eight reporters are inconsistent with each other (22). Two of these studies are performed in cancer cell lines where p53 variants are overexpressed to supra-physiological levels (32, 33); in one study, this is followed by 12 d of exposure to etoposide (32). Although these studies have been highly valuable in the categorization of fully inactivating and Li Fraumeni mutants of p53, in almost all cases, they fail to accurately categorize more subtle hypomorphic mutants. In support of the latter premise, we find that transient transfection and overexpression of mutant p53, or p53 hypomorphs, do not lead to increased p-p65 or to increased levels of RRAD (SI Appendix, Fig. S3C). Importantly, hypomorphic or lesser-functioning mutants of p53 are estimated to occur in up to 26% of tumors, so this issue is clinically quite important (34). One consequence of the failure of current assays to capture hypomorphs is that despite clear evidence of increased cancer risk in mice and humans (16, 17), the P47S hypomorph is presently classified as benign (35). Similarly, the G334R variant is found in families with multiple generations of cancer including cousins with pediatric adrenocortical carcinoma (a hallmark of p53 mutation) (23); however, this variant is presently classified as a variant of uncertain significance. As such, individuals with these variants remain underinformed about their potential for cancer risk and ineligible for advanced cancer screening by MRI that is offered to Li Fraumeni carriers. Recently, some groups have attempted to address this issue, by establishing the functionality of p53 variants in nontransformed cell lines containing endogenous p53 variants following exposure to genotoxic stress (36, 37). However, such studies have been low throughput, and they require a genotoxic stress. Moreover, their data interpretation may be confounded by the existence of other variants in the p53 pathway that influence the transcriptional response, such as SNP309 in MDM2 (38). Our identification of a gene signature for a p53 hypomorph from unstressed cells has the potential to markedly impact the categorization of these variants and to better inform carriers of cancer risk and the optimal choices for cancer therapy.
There are some caveats to our study. First, the increased RRAD protein in our hypomorph LCLs first led us to test p53 misfolding and NF-κB activation. However, we subsequently discovered that part of the overexpression of RRAD protein in hypomorph LCLs occurs at the post-transcriptional level. Specifically, the RNA level of RRAD is comparable in Nutlin-treated WT, P47S, and Y107H LCLs, but the level of RRAD protein is significantly higher in P47S and Y107H LCLs (Fig. S3 D–G); the mechanism underlying this post-transcriptional regulation remains to be determined. Additionally, we see clear evidence for p53 misfolding in our HCT116 hypomorph tumor lines, whereas we do not see increased RRAD level in these hypomorph cells relative to WT. Therefore, at present, the association of overexpressed RRAD with hypomorphic p53 appears to be restricted to nontransformed LCLs. We also note that our gene signature is created from immortalized LCLs; ideally, this signature should be adapted for analysis in primary peripheral blood mononuclear cells, and this is our next goal. Finally, two questions remain unanswered from this study. The first concerns whether the increased NF-κB activity in p53 hypomorphic cells and mice contributes to the increased cancer risk evident in these mice and humans. Our preliminary studies suggest that this may not be the case. Specifically, we exposed WT, P47S, and Y107H mice to three cycles of dextran sulfate sodium (DSS) to induce colitis; while we found an increased hyperplasia score and two examples of precancerous lesions in P47S mice, this was not the case in Y107H mice, which were indistinguishable in response to DSS from WT mice (SI Appendix, Fig. S4 A–G). The second question concerns our finding that P47S and Y107H proteins show impaired oligomerization in unstressed cells. Mutant-conformation p53 is reported to show impaired ability to oligomerize compared to WT protein (39), and other p53 hypomorphs occur in the oligomerization domain (including the R337H and G334R alleles); however, we were surprised by the extent of the oligomerization defect in these LCLs. We are continuing to explore the contribution of this defect to the altered p53 function in hypomorphic cell lines and mice and to the increased cancer risk in P47S and Y107H mice.
Materials and Methods
All experimental procedures involving mice were conducted in accordance with protocols approved by The Institutional Animal Care & Use Committee at The Wistar Institute and conformed to the guidelines outlined in the NIH Guide for the Care and Use of Laboratory Animals (40). Materials and experimental procedures for cell culture, animal models, generation, and validation of CRISPR knock-in HCT116 cells and mice, RNA isolation, qRT-PCR, immunoblotting, immunoprecipitation, tissue staining and immunohistochemistry, proximity ligation analyses, BMH cross-linking, invasion assays, nuclear fractionation, chromatin immunoprecipitation, RNA Seq analysis and gene signature, DSS colitis study, and statistical analysis are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We acknowledge the Histotechnology Facility, Imaging Facility, and Genomics Facility at the Wistar Institute, especially Fred Keeney. We acknowledge the Molecular Pathology and Imaging Core at the University of Pennsylvania, funded by the Center for Molecular Studies in Digestive and Liver Diseases (NIH-P30-DK050306). We thank the Genome Engineering and iPSC Center (GEiC) at Washington University in St. Louis for gRNA validation and cell line engineering services. We sincerely thank Noam Auslander (Wistar Institute) for helpful discussions. We are grateful to David Lu (summer intern from Cornell University) for assistance with data analysis and Janice Reynaga (University of Pennsylvania) for assistance with some of the experiments. The research support for this study was provided by NIH grants CA102184 (PI Murphy) and CA238611 (PI Murphy). J.C.L. received support from T32 CA009171-43 and the Wistar Accelerator Postdoctoral Award; A.I. was supported in part from T32 GM008216. T.B. was supported through R00 CA241367.
Author contributions
J.C.L., J.I.-J.L., D.L.G., and M.E.M. designed research; J.C.L., J.I.-J.L., A.I., T.K., N.L.C., N.A.K., H.D., and A.V.K. performed research; T.B. contributed new reagents/analytic tools; J.C.L., J.I.-J.L., A.I., T.K., D.G., A.V.K., and M.E.M. analyzed data; and J.C.L. and M.E.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
[RNA Seq] data have been deposited in NCBI Gene Expression Omnibus (GSE209837) (41). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Levine A. J., Spontaneous and inherited TP53 genetic alterations. Oncogene 40, 5975–5983 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine A. J., p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 20, 471–480 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Guha T., Malkin D., Inherited TP53 mutations and the Li-Fraumeni syndrome. Cold Spring Harb. Perspect. Med. 7, a026187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowe S. W., Ruley H. E., Jacks T., Housman D. E., p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74, 957–967 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Lowe S. W., et al. , p53 status and the efficacy of cancer therapy in vivo. Science 266, 807–810 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Freed-Pastor W. A., et al. , Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller P. A., et al. , Mutant p53 drives invasion by promoting integrin recycling. Cell 139, 1327–1341 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Muller P. A., et al. , Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene 32, 1252–1265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooks T., et al. , Mutant p53 prolongs NF-kappaB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 23, 634–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boettcher S., et al. , A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 365, 599–604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang G. A., et al. , Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–872 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Olive K. P., et al. , Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119, 847–860 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Bargonetti J., Prives C., Gain-of-function mutant p53: History and speculation. J. Mol. Cell Biol. 11, 605–609 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller P. A., Vousden K. H., Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25, 304–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oren M., Rotter V., Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2, a001107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jennis M., et al. , An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes. Dev. 30, 918–930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy M. E., et al. , A functionally significant SNP in TP53 and breast cancer risk in African-American women. NPJ Breast Cancer 3, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leu J. I., Murphy M. E., George D. L., Mechanistic basis for impaired ferroptosis in cells expressing the African-centric S47 variant of p53. Proc. Natl. Acad. Sci. U.S.A. 116, 8390–8396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leu J. I., Murphy M. E., George D. L., Functional interplay among thiol-based redox signaling, metabolism, and ferroptosis unveiled by a genetic variant of TP53. Proc. Natl. Acad. Sci. U.S.A. 117, 26804–26811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh K. S., et al. , African-centric TP53 variant increases iron accumulation and bacterial pathogenesis but improves response to malaria toxin. Nat. Commun. 11, 473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gnanapradeepan K., et al. , Increased mTOR activity and metabolic efficiency in mouse and human cells containing the African-centric tumor-predisposing p53 variant Pro47Ser. Elife 9, e55994 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato S., et al. , Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. U.S.A. 100, 8424–8429 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers J., et al. , A rare TP53 mutation predominant in Ashkenazi Jews confers risk of multiple cancers. Cancer Res. 80, 3732–3744 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szoltysek K., et al. , RRAD, IL4I1, CDKN1A, and SERPINE1 genes are potentially co-regulated by NF-kappaB and p53 transcription factors in cells exposed to high doses of ionizing radiation. BMC Genomics 19, 813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gannon J. V., Greaves R., Iggo R., Lane D. P., Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 9, 1595–1602 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y. M., Bradbury D., Russell N., Expression of different conformations of p53 in the blast cells of acute myeloblastic leukaemia is related to in vitro growth characteristics. Br. J. Cancer 68, 851–855 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinds P. W., Finlay C. A., Frey A. B., Levine A. J., Immunological evidence for the association of p53 with a heat shock protein, hsc70, in p53-plus-ras-transformed cell lines. Mol. Cell Biol. 7, 2863–2869 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinhasi-Kimhi O., Michalovitz D., Ben-Zeev A., Oren M., Specific interaction between the p53 cellular tumour antigen and major heat shock proteins. Nature 320, 182–184 (1986). [DOI] [PubMed] [Google Scholar]
- 29.Di Minin G., et al. , Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol. Cell 56, 617–629 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Weisz L., et al. , Mutant p53 enhances nuclear factor kappaB activation by tumor necrosis factor alpha in cancer cells. Cancer Res. 67, 2396–2401 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Chen S., et al. , Arsenic trioxide rescues structural p53 mutations through a cryptic allosteric site. Cancer Cell 39, 225–239.e228 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Giacomelli A. O., et al. , Mutational processes shape the landscape of TP53 mutations in human cancer. Nat. Genet. 50, 1381–1387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotler E., et al. , A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol. Cell 71, 178–190.e178 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Klimovich B., et al. , p53 partial loss-of-function mutations sensitize to chemotherapy. Oncogene 41, 1011–1023 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doffe F., et al. , Identification and functional characterization of new missense SNPs in the coding region of the TP53 gene. Cell Death Differ. 28, 1477–1492 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raad S., et al. , Blood functional assay for rapid clinical interpretation of germline TP53 variants. J. Med. Genet. 58, 796–805 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerdoumi Y., et al. , Germline TP53 mutations result into a constitutive defect of p53 DNA binding and transcriptional response to DNA damage. Hum. Mol. Genet. 26, 2591–2602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bond G. L., et al. , A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119, 591–602 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Walerych D., et al. , Wild-type p53 oligomerizes more efficiently than p53 hot-spot mutants and overcomes mutant p53 gain-of-function via a “dominant-positive” mechanism. Oncotarget 9, 32063–32080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.National Research Council, Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 41.Leung J. C., et al. , Common activities and predictive gene signature identified for genetic hypomorphs of TP533. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2098375. Deposited 26 July 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
[RNA Seq] data have been deposited in NCBI Gene Expression Omnibus (GSE209837) (41). All study data are included in the article and/or SI Appendix.