Significance
Despite extensive investigations on structure–function relationships of membrane G protein-coupled receptors (GPCR), the effects of membrane components, such as cholesterol and its derivatives, on oligomerization preferences remains to be clarified. Here, using double electron–electron resonance (DEER) spectroscopy to measure distances between nitroxide spin labels, we show that the β1-adrenergic receptor (β1AR) monomer–dimer equilibrium in dodecyl-β-D-maltoside micelles is modulated by sterols. Global fitting of DEER echo curves for spin-labeled β1AR upon titration with sodium cholate and cholesteryl hemissucinate demonstrates that saturation of micelles with the former induces receptor dimerization, while specific binding of the latter to β1AR inhibits dimerization and stabilizes the monomeric form. These results illustrate how quantitative analysis of DEER data can contribute to studies of GPCR oligomerization.
Keywords: G protein-coupled receptor, dimerization, cholesterol, spin-labeling, electron paramagnetic resonance
Abstract
G protein-coupled receptors (GPCR) activate numerous intracellular signaling pathways. The oligomerization properties of GPCRs, and hence their cellular functions, may be modulated by various components within the cell membrane (such as the presence of cholesterol). Modulation may occur directly via specific interaction with the GPCR or indirectly by affecting the physical properties of the membrane. Here, we use pulsed Q-band double electron–electron resonance (DEER) spectroscopy to probe distances between R1 nitroxide spin labels attached to Cys163 and Cys344 of the β1-adrenergic receptor (β1AR) in n-dodecyl-β-D-maltoside micelles upon titration with two soluble cholesterol analogs, cholesteryl hemisuccinate (CHS) and sodium cholate. The former, like cholesterol, inserts itself into the lipid membrane, parallel to the phospholipid chains; the latter is aligned parallel to the surface of membranes. Global quantitative analysis of DEER echo curves upon titration of spin-labeled β1AR with CHS and sodium cholate reveal the following: CHS binds specifically to the β1AR monomer at a site close to the Cys163-R1 spin label with an equilibrium dissociation constant ~1.4 ± 0.4 mM. While no direct binding of sodium cholate to the β1AR receptor was observed by DEER, sodium cholate induces specific β1AR dimerization ( ~35 ± 6 mM and a Hill coefficient n ~ 2.5 ± 0.4) with intersubunit contacts between transmembrane helices 1 and 2 and helix 8. Analysis of the DEER data obtained upon the addition of CHS to the β1AR dimer in the presence of excess cholate results in dimer dissociation with species occupancies as predicted from the individual KD values.
G protein-coupled receptors (GPCR) are part of a ubiquitous group of seven-transmembrane domain cell surface receptors in eukaryotes, comprising over 800 members encoded in the human genome and constituting about 15% of all membrane proteins (1). Despite extensive research and significant advances in GPCR pharmacology (2–4) and structural biology (5–8), many gaps remain in regard to the conformational and oligomerization properties of GPCRs.
The activity of GPCRs, like other membrane proteins, is dependent on the composition of the lipid environment, for example, the presence of cholesterol (9) which constitutes up to 25 to 35% of mammalian cell membranes depending on tissue type. The steroid core of cholesterol is more rigid and shorter in length than the phospholipids chains, and hence, accumulation of cholesterol in lipid bilayers affects the physical properties of the membrane (10, 11). The focus of the current work, the β1-adrenergic receptor (β1AR), is a member of the adrenoreceptor family of class A GPCRs that stimulates the sympathetic nervous system. β1AR is mostly expressed in cardiac tissue and regulates the heart rate and strength of cardiac contraction. In the case of β1AR, cholesterol acts as a negative allosteric modulator by restricting conformational flexibility and inhibiting the transition from inactive to active states of the receptor (12). The mechanism, however, whereby cholesterol exerts its effect on β1AR is still poorly understood. Modulation of GPCR properties by components of the membrane can occur in potentially two ways: a direct mechanism involving specific interaction with the GPCR that affects ligand binding and disrupts the signaling cascade and an indirect, membrane-mediated mechanism in which the physical properties of the membrane bilayer are altered that subsequently impact the activity of the GPCR (13).
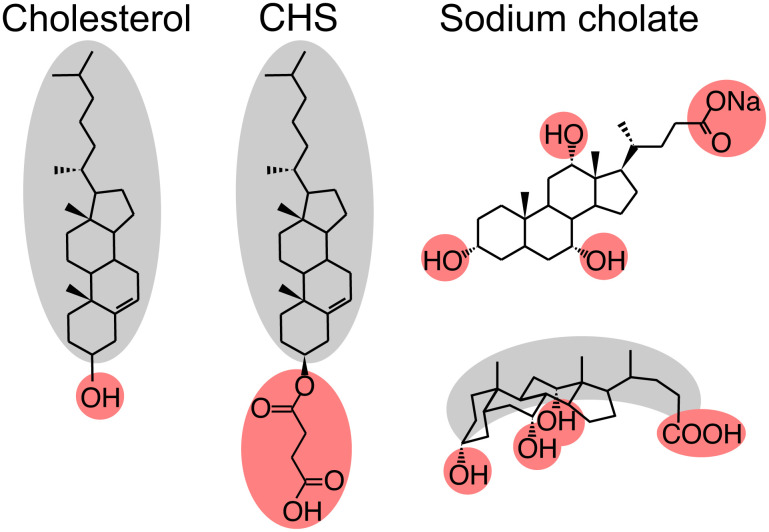
Here, we use Q-band pulsed electron paramagnetic resonance (EPR) spectroscopy-based double electron–electron resonance (DEER) (14, 15) to examine the effect of two different sterol analogs of cholesterol on the structural and oligomeric preferences of β1AR in n-dodecyl-β-D-maltoside (DDM) micelles by probing long-range distances between a pair of R1 nitroxide spin labels (16). The two sterols are cholesteryl hemisuccinate (CHS), a soluble analog of cholesterol, and sodium cholate, a bile salt derivative of cholesterol (Fig. 1). Both sterols can affect the fluidity, thickness, water penetration, and intrinsic curvature of the cell membrane (10, 17). CHS, like cholesterol, penetrates the membrane, with the hydrophobic steroid core parallel to the phospholipid chains and the hydrophilic tail on the membrane surface, increasing the thickness and decreasing the fluidity of the membrane, thereby changing the overall geometry of the lipid environment (Fig. 1). Cholate, on the other hand, is oriented parallel to the surface of the lipid bilayer: the convex hydrophobic surface formed by the rigid steroid core is partially inserted into the membrane, while the hydrophilic concave surface, comprising three hydroxyl groups, aligns with the phospholipid head groups on the surface of the membrane (Fig. 1) (18–21). As a result, cholate reduces the thickness and increases the fluidity of the membrane and at very high concentrations can even dissolve membranes.
Fig. 1.
Chemical structure of cholesterol, CHS, and sodium cholate. Cholesterol and CHS comprise hydrophobic (gray) and hydrophilic (pink) regions that allow cholesterol and CHS to penetrate the lipid membrane, parallel to the phospholipid chains. Sodium cholate, on the other hand, has distinct, opposing polar (pink) and hydrophobic (gray) surfaces, such that sodium cholate aligns parallel to the membrane surface. The chemical structure and steric representation of sodium cholate are displayed on the top and bottom, respectively (18).
DEER probes pairwise distances between nitroxide spin labels ranging from 15 to 80 Å in the absence of deuteration (15, 22) and up to 160 Å with deuteration (23–25). DEER is especially powerful for studying heterogeneous systems such as GPCRs as the DEER-derived P(r) pairwise distance distributions can serve to characterize multiple conformations and their relative populations (16, 26–28). DEER has also been used to structurally characterize the oligomeric structure of native rhodopsin (29) and Anabaena sensory rhodopsin (30). Global analysis of the complete set of DEER echo curves (31) obtained for C163-R1/C344-R1 spin-labeled β1AR upon titration with the two sterols enables us to quantify the binding equilibria and shows that while CHS binds to a specific site on β1AR in close proximity to Cys163, directly stabilizing the monomeric form, cholate promotes β1AR dimerization, in all likelihood by interacting with and altering the properties of DDM micelles.
Results and Discussion
Site-Specific Nitroxide Spin-Labeling and Assignment of Peaks in the DEER-Derived P(r) Distribution for β1AR in the Sterol-Free State.
We chose to reconstitute β1AR in DDM as this detergent has been extensively used as a membrane mimetic in structural studies on GPCRs. DDM has a low critical micelle concentration ≈ 0.17 mM and a relatively small size (70 to 72 kDa); pure DDM micelles adopt an oblate ellipsoid shape with polar and equatorial axes of 21 and 34 Å, respectively; and insertion of sterols occurs in the flatter regions of DDM micelles increasing their hydrodynamic radius and inducing pseudo-bicelle geometry (32).
To investigate the effects of CHS and cholate on the structural preferences of β1AR by DEER, we made use of the naturally occurring cysteines present in a construct of thermostabilized turkey β1AR optimized for expression in Escherichia coli (33) (see Experimental Procedures for details of purification and reconstitution, and SI Appendix, Figs. S1 A and B). Of the 10 naturally occurring cysteines, four are involved in disulfide bridges located in the extracellular loop, two (Cys302 and Cys124) are buried within the interior of the transmembrane domain and are not accessible for spin-labeling, and the remaining four (Cys85, Cys133, C163, and C344) are potentially accessible for spin-labeling (34). DEER data collected on a series of single point mutations (C85A, C85V, C133V, C163L, C344A, and C344S; SI Appendix, Table S1) indicated that only Cys163 and C344 were labeled with the R1 nitroxide: the DEER echo curves and DEER-derived P(r) distributions for the spin-labeled C85A, C85V, and C133V mutants are identical to those of wild type (SI Appendix, Fig. S2), while no significant dipole–dipole oscillations above background are observable in the DEER echo curves for the spin-labeled C163L, C344A, and C344S mutants (SI Appendix, Fig. S3). Both Cys163 and Cys344 are located on the cytoplasmic side of β1AR, at the C-terminal ends of transmembrane helices TM4 and TM7, respectively (Fig. 2). The side chain of Cys163 is in close proximity to various detergent molecules seen in several β1AR crystal structures (35, 36), while Cys344 is positioned C-terminal to the highly conserved NPxxY motif found in GPCRs and is an ideal reporter of global conformational changes.
Fig. 2.
Q-band DEER measurements on R1-nitroxide spin-labeled β1AR (C163-R1/C344-R1) in DDM micelles. (A) Sterol-free, (B) CHS-bound, and (C) cholate/DDM micelles. The experimental DEER echo curves (Left) are shown in blue, and the corresponding best-fit curves obtained by Tikhonov regularization using the program DeerLab (37) are in red; the dashed black line represents the background. The P(r) distributions (Middle) derived from the experimental DEER data using validated Tikhonov regularization are shown in blue (with the light blue shading representing the upper and lower error estimates); the P(r) distributions calculated from the molecular coordinates (31) of the β1AR crystal structure [Protein Data Bank (PDB) 2Y00 (35)] using the calcPr helper function in Xplor-NIH (38) are displayed in red. The Right panels display the corresponding structures. The locations of the R1 nitroxide spin labels at Cys344-R1 and Cys163-R1 are shown as red spheres, and the CHS and HEGA-10 (2CV) molecules are shown as sticks. β1AR in the 2Y00 structure was crystallized with two CHS molecules: CHS1 is in close proximity to Cys163-R1 and is highlighted in lilac, while CHS2 is shown in blue. The cholate-induced dimer of β1AR was modeled using the monomer coordinates from the 2Y00 structure (35) placed in the dimer orientation observed in the 4GPO crystal structure (39). Note that in the P(r) distributions shown for the cholate-induced dimer in (C) (Middle), there is a small peak at ~23 Å present in both the structure-based simulated (red trace) and experimental DEER-derived (blue trace) P(r) distributions. The 23 Å peak in the experimental P(r) distribution obtained by Tikhonov regularization is likely due to overfitting since it is not consistently present over the course of the cholate titration (Fig. 3 and SI Appendix, Fig. S13) and the upper and lower confidence limits are large in relation to the peak height (see SI Appendix, Fig. S5 for a more detailed discussion). In the case of the simulated P(r) distribution, the 23 Å peak arises from one of the possible three positions of the C163-R1 nitroxide (Rightmost panel in C) (SI Appendix, Table S2).
The P(r) distribution derived from the DEER echo curve for sterol-free spin-labeled β1AR (C163-R1/C344-R1), obtained by validated Tikhonov regularization (37), displays three main peaks at 31, 34, and 39 Å (Fig. 2A). Simulations using an R1 nitroxide conformational library (31, 40) in Xplor-NIH (38) indicates that these arise from three distinct positions (a, b, and c) of the C163-R1 side chain trapped during freeze quenching giving rise to three pairwise intramolecular distances between C344-R1 and C163-R1 (Fig. 2A and SI Appendix, Table S2 and Fig. S4A). The presence of the detergent molecule located in the vicinity of C163-R1 in the β1AR crystal structure (35, 36) is essential for good qualitative agreement between the experimentally-derived and simulated P(r) distributions, underlying the importance of interactions between detergent molecules such as DDM and the surface of β1AR.
Sterol Modulation of β1AR Structural Preferences.
Cholesterol constitutes a major component of eukaryotic cell membranes and regulates GPCR function, dynamics, and oligomerization preferences (9). Due to its poor solubility, cholesterol is often replaced by CHS in structural and biophysical studies. Both cholesterol and CHS are frequently cocrystallized with GPCRs in X-ray and cryoelectron microscopy structures. In the case of β2AR, a close homolog to β1AR, cholesterol was found to bind to a specific site, denoted as the cholesterol consensus motif (CCM), formed by a cleft between TM2–4 (41). The same site was also found to be occupied by cholesterol or CHS in several β1AR structures (35, 42, 43). C163-R1 is located in close proximity to the CCM and therefore represents an ideal reporter to study the binding of CHS to β1AR by DEER. Upon addition of CHS to spin-labeled β1AR (C163-R1/C344-R1) the peak at 31 Å in the P(r) distribution of sterol-free β1AR disappears while that at 34 Å undergoes a corresponding increase in intensity (Fig. 2B). The P(r) distributions (31, 38, 40) calculated from the β1AR crystal structure (2Y00) (35) with (Fig. 2B) and without (Fig. 2A) CHS in binding site 1 indicate that CHS sterically occludes the occupancy of C163-R1 in site a, leading to the displacement of C163-R1 from position a to position b (SI Appendix, Table S2 and Fig. S4B).
In contrast to CHS, the addition of sodium cholate to spin-labeled β1AR (C163-R1/C344-R1) does not alter the positions of the three main peaks at 31, 34, and 39 Å characteristic of the DEER-derived P(r) distribution of sterol-free β1AR; however, a minor peak at 26 Å in the P(r) distribution of sterol-free β1AR (Fig. 2A) increases in intensity, as does the peak at 31 Å (Fig. 2C). The 4GPO crystal structure of β1AR (39) is oligomeric and displays two dimer interfaces: dimer interface 1 formed by helices TM1, TM2 and H8 and dimer interface 2 formed by helices TM4 and TM5 (SI Appendix, Fig. S5A). Simulation of the P(r) distributions (31, 38, 40) for the two intermolecular interfaces indicates that the dimer observed by DEER corresponds to dimer interface 1: the 26 Å peak arises from two intersubunit distances between C344-R1 on one subunit, and C344’-R1 and C163’-R1a of the other subunit; further, in addition to the intramolecular C344-R1–C163-R1a distance, the peak at 31 Å also contains a contribution from the intersubunit C344-R1–C163’-R1b,c distances (SI Appendix, Table S2 and Fig. S5B). Noncovalent dimerization of β1AR in the presence of cholate and the absence of dimer when CHS is bound was confirmed by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) under conditions where the β1AR/DDM complex was not fully denatured upon treatment with SDS (see SI Appendix, Fig. S1 legend for a full discussion). Although unusual for nonmembrane proteins, noncovalent dimerization under reducing conditions has been previously observed by SDS-PAGE for another GPCR membrane protein, namely the CCR3 chemokine receptor (44).
Quantitative Global Analysis of CHS and Cholate DEER Titration Data.
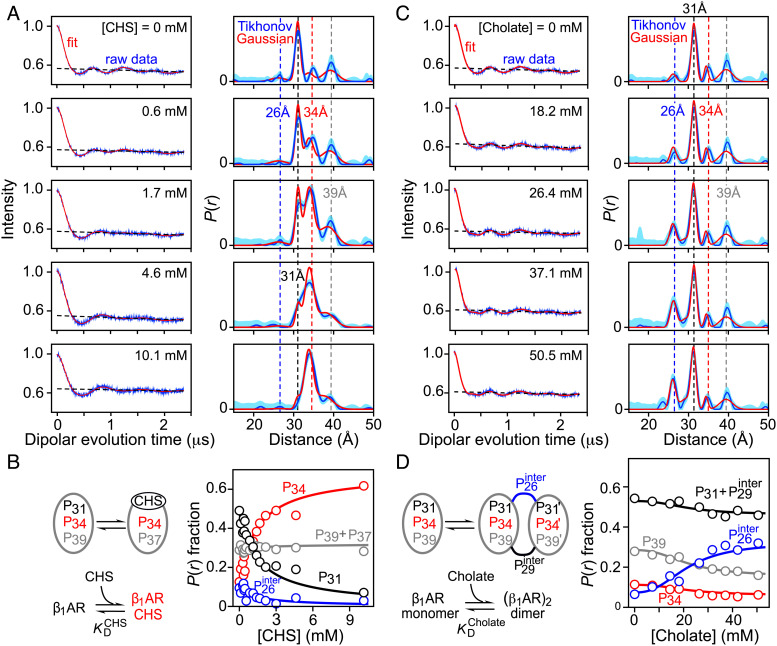
Fig. 3 A and C show a subset of DEER echo curves and corresponding P(r) distributions for spin-labeled β1AR (C163-R1/C344-R1) upon titration with CHS and cholate, respectively (the complete titration datasets are shown in SI Appendix, Figs. S7–S13, respectively). As noted above, the P(r) distribution for sterol free β1AR has three major peaks at 31, 34 and 39 Å, and a peak at 26 Å corresponding to a minor (<10%) population of dimer. Titration with either CHS or cholate changes the relative intensities (and therefore the underlying populations) of these peaks. Importantly, the P(r) distributions obtained by model-free validated Tikhonov regularization (DeerLab) (37) and deep neural network analysis [DeerNet (45, 46)], and by model-dependent Gaussian fitting [DD/GLADDvu (47, 48)] are fully consistent with one another (Fig. 3 and SI Appendix, Fig. S6). The accuracy of the peak position and width of any component in the P(r) distribution derived from a four-pulse DEER experiment depends upon the maximum length of the acquired dipolar evolution time, tmax: the upper limits for the determination of accurate peak position and width (in Å) are given by the empirical relationships ~50(tmax/2)1/3 and ~40(tmax/2)1/3 (with tmax in microseconds), respectively (15). For a distance of 40 Å, this means that tmax should be at least 2 μs; in our experiments we used tmax = 2.5 μs. Note that use of a longer tmax value was precluded owing to phase memory (transverse) relaxation (49), caused by flip-flop transitions of protons in close proximity to the nitroxide unpaired electron (50, 51), resulting in rapid decay of the DEER echo curves as the samples were not deuterated (25, 52, 53).
Fig. 3.
Quantitative analysis of Q-band DEER data obtained for nitroxide spin-labeled β1AR (C163-R1/C344-R1) upon titration with CHS and cholate. (A) DEER echo curves recorded for ~60 μM β1AR (C163-R1/C344-R1) in DDM as a function of CHS concentration (Left panels: raw experimental data, blue; best-fit curves from global Gaussian modeling, red; background, black dashed lines) and corresponding P(r) distributions (Right) obtained either by validated Tikhonov regularization [blue, with upper and lower error estimates shown by the light blue shading (37)] or global Gaussian modeling with five Gaussians (31) in which peak positions and widths are treated as global parameters and the fractional population of the bound state is determined by the equilibrium dissociation constant (red). (The complete titration dataset and analysis is provided in SI Appendix, Figs. S7–S10.) (B) Simple binding scheme of CHS to β1AR (Left) and fractional populations of the peaks in the P(r) distribution as a function of CHS concentration. The circles are the fractional populations obtained from the global four-Gaussian fits to the DEER data in which the peak positions and widths are treated as global parameters; the continuous lines are obtained from the global fit to the raw DEER data using a five-Gaussian model that, in addition, includes the concentration dependence of the fraction CHS-bound species (pbound) determined by the simple binding isotherm given by [(CHS)/ ]/[1 + (CHS)/ ], where is the equilibrium dissociation constant. The decrease in the peak intensity at ~26 Å (shown in blue) is due to the disappearance of residual dimer (~9% in sterol-free β1AR) and occurs concomitantly with the binding of CHS. Note that the total area under the P(r) distribution is always normalized to 1 at every point in the titration. The reduced χ2 and global parameters of the fits (peak positions and widths, and ) are provided in Table 1. (C) DEER echo curves recorded for ~60 µM β1AR (C163-R1/C344-R1) in DDM as a function of cholate concentration (Left panels: raw experimental data, blue; best-fit curves from global Gaussian modeling, red; background, black dashed line) and corresponding P(r) distributions (Right) obtained either by validated Tikhonov regularization (blue, with upper and lower error estimates shown by the light blue shading) or global Gaussian modeling with five Gaussians in which peak positions and widths are treated as global parameters and the fractional population of dimer is determined by a cooperative dimerization model (red). (D) β1AR dimerization upon binding of cholate to DDM micelles (Left) and fractional populations of the peaks in the P(r) distribution as a function of cholate concentration. The circles are the fractional populations obtained from the global four-Gaussian fits to the DEER data in which the peak positions and widths are global parameters; the continuous lines are obtained from the global fit to the raw DEER data using a five-Gaussian model that, in addition, includes the concentration dependence of the fraction of dimeric species (pdimer) determined by the Hill equation given by /{ + }, where is the equilibrium dissociation constant, n the Hill coefficient, and the fraction dimer in the absence of cholate. Note that the total area under the P(r) distribution is always normalized to 1 at every point in the titration. (The complete cholate titration dataset and analysis is provided in SI Appendix, Figs. S11–S13). The reduced χ2 and global parameters of the fits (peak positions and widths, and and the Hill coefficient n) are provided in Table 2. Note that the fractional contribution of the peaks arising from intramolecular distances decreases over the course of the cholate titration; this is because the total area under the P(r) distribution is normalized to 1 at every point in the titration. A renormalized plot is shown in SI Appendix, Fig. S14, where the contribution from the intramolecular peaks is normalized to 1 and remains constant throughout the titration (since there are two intramolecular distances in the dimer, C163-R1–C344-R1 and C163’-R1–C344’-R1, corresponding to 1 in monomer units), and that from intersubunits peaks is normalized to 1.5 (since there are three intermolecular distances in the dimer, C344-R1–C344’-R1, C344-R1–C163’-R1, and C163-R1–C344’-R1, corresponding to 1.5 in monomer units).
The use of Gaussian fitting for quantitative global analysis of DEER titration data in terms of fractional occupancies of the peaks in the P(r) distribution is critical for three reasons (31, 40): a) Gaussian fitting enables one to directly quantify the area under each P(r) peak; b) the peak positions and widths can be restrained to be invariant over the course of the titration; and c) Gaussian fitting is beneficial when dealing with a broad peak, such as the peak at 39 Å, that could potentially represent a heterogeneous ensemble of states. Initially, the DEER echo curves for both the CHS and cholate titration data were fit globally to a four-Gaussian model to describe the P(r) distribution with the peak positions at ~26, ~31, ~34, and ~39 Å and corresponding widths treated as global parameters. The results for the CHS and cholate titration data are summarized in Tables 1 and 2. The overall normalized χ2 values for these global four-Gaussian fits are only minimally larger than the mean normalized χ2 obtained by fitting each DEER echo curve individually using validated Tikhonov regularization (37), as expected since the global Gaussian fit is more constrained. Moreover, the distribution of residuals between observed and calculated DEER echo curves is essentially random for both the individual Tikhonov regularization fits (SI Appendix, Figs. S7 and S11 for CHS and cholate, respectively) and the global four-Gaussian fits (SI Appendix, Figs. S8 and S12 for CHS and cholate, respectively). Plots of the fractional occupancies of the peaks in the P(r) distributions derived from the global four-Gaussian fits as a function of CHS and cholate concentration are shown in Fig. 3 B and D, respectively. It is important to note that the total area under the calculated P(r) distributions is invariant and normalized to 1 at each point in the titrations (i.e., the appearance of new peaks in the P(r) distributions is not reflected in an increase in area under the curves).
Table 1.
Normalized χ2 and values of optimized parameters for the global fits to the complete set of Q-band DEER echo curves recorded on β1AR (C163-R1/C344-R1) upon titration with CHS using Gaussian models
| DeerLab individual fits using Tikhonov regularization* | Global fits to complete CHS titration dataset† | ||
|---|---|---|---|
| Four-Gaussian fit‡ | Five-Gausssian fit with equilibrium binding model§ | ||
| Normalized χ2 | 0.94 ± 0.08 | 1.10 | 1.13 |
| (mM) | - | - | 1.4 ± 0.4 |
| Gaussian 1 (P26) | |||
| Mean (Å) | 26.2 ± 0.1 | 26.1 ± 0.2 | 26.2 ± 0.2 |
| Width (Å) | - | 1.8 ± 0.2 | 1.8 ± 0.2 |
| Gaussian 2 (P31) | |||
| Mean (Å) | 31.2 ± 0.1 | 31.1 ± 0.1 | 31.2 ± 0.1 |
| Width (Å) | - | 0.6 ± 0.1 | 0.6 ± 0.1 |
| Gaussian 3 (P34) | |||
| Mean (Å) | 34.6 ± 0.4 | 34.1 ± 0.1 | 33.9 ± 0.1 |
| Width (Å) | - | 1.4 ± 0.1 | 1.1 ± 0.1 |
| Gaussian 4 (P39) | |||
| Mean (Å) | 39.7 ± 0.5 | 39.4 ± 0.1 | 39.4 ± 0.1 |
| Width (Å) | - | 1.7 ± 0.1 | 1.9 ± 0.1 |
| Gaussian 4’ (P37) | |||
| Mean (Å) | - | - | 37.4 ± 0.1 |
| Width (Å) | - | - | 2.2 ± 0.1 |
*Averages for model-free fits to the individual DEER echo curves for the CHS titration using DeerLab with validated Tikhonov regularization (37).
†Gaussian global fits based on the program DD/GLADDvu (47, 48), incorporated into an inhouse Python program (31) to the complete set of Q-band DEER echo curves recorded over a series of CHS concentrations (0 to 10.1 mM). In both the four- and five-Gaussian global fits, the mean distances and widths are constrained to be equal for all CHS titrations points.
‡In the four-Gaussian global fit, only the mean distances and corresponding widths are constrained to be invariant.
§In the five-Gaussian global fit, the dependence of the fractional populations of the distance peaks on CHS concentration, as specified by a simple equilibrium binding model with an equilibrium dissociation constant (Fig. 2) included as an optimized parameter. Gaussian 1 (P26) corresponds to intersubunit distances (see Table 2, footnote §) arising from a small population of dimer (<10%) in the sterol-free state that is absent in the CHS-bound. Gaussians 2 (P31), 3 (P34), and 4 (P39) correspond to intramolecular distances between the Cys344-R1 nitroxide and three distinct positions (a, b, and c) of the Cys163-R1 nitroxide in the free state; in the CHS-bound state, the Cys163-R1 nitroxide occupies two distinct positions represented by Gaussians 3 (P34) and 4’ (P37).
Table 2.
Normalized χ2 and values of optimized parameters for the global fits to the complete set of Q-band DEER echo curves recorded on β1AR (C163-R1/C344-R1) upon titration with cholate using Gaussian models
| DeerLab individual fits using Tikhonov regularization* | Global fits to complete cholate titration dataset† | ||
|---|---|---|---|
| Four-Gaussian fit‡ | Five-Gausssian fit with Hill binding model§ | ||
| Normalized χ2 | 0.93 ± 0.09 | 1.14 | 1.18 |
| (mM) | - | - | 35 ± 6 |
| Hill coefficient n | - | - | 2.5 ± 0.4 |
| - | - | 0.09 ± 0.01 | |
| Gaussian 1 (P26) | |||
| Mean (Å) | 26.1 ± 0.3 | 26.1 ± 0.1 | 25.9 ± 0.1 |
| Width (Å) | - | 1.2 ± 0.1 | 0.8 ± 0.1 |
| Gaussian 2 (P31) | |||
| Mean (Å) | 31.1 ± 0.1 | 31.0 ± 0.1 | 31.1 ± 0.1 |
| Width (Å) | - | 0.6 ± 0.1 | 0.7 ± 0.1 |
| Gaussian 2’ (P29) | |||
| Mean (Å) | - | - | 29.3 ± 0.3 |
| Width (Å) | - | - | 1.4 ± 0.2 |
| Gaussian 3 (P34) | |||
| Mean (Å) | 34.9 ± 0.1 | 33.9 ± 0.3 | 34.2 ± 0.1 |
| Width (Å) | - | 0.4 ± 0.3 | 0.5 ± 0.4 |
| Gaussian 4 (P39) | |||
| Mean (Å) | 39.7 ± 0.1 | 39.4 ± 0.1 | 39.4 ± 0.1 |
| Width (Å) | - | 1.6 ± 0.1 | 1.6 ± 0.1 |
*Averages for model-free fits to the individual DEER echo curves for the CHS titration using DeerLab with validated Tikhonov regularization (37).
†Gaussian global fits based on the program DD/GLADDvu (47, 48), incorporated into an inhouse Python program (31), to the complete set of Q-band DEER echo curves recorded over a series of cholate concentrations (0 to 50 mM). In both the four- and five-Gaussian global fits, the mean distances and widths are constrained to be equal for all CHS titrations points.
‡In the four-Gaussian global fit, only the mean distances and corresponding widths are constrained to be invariant.
§In the five-Gaussian global fit, the dependence of the fractional populations of the distance peaks in the P(r) distribution on cholate concentration, as specified by a Hill binding model for which the equilibrium dissociation constant , the Hill coefficient n, and the fraction dimer present in the absence of cholate, , are optimized in the fit. The Cys163-R1 nitroxide label occupies three distinct positions, a, b, and c. Gaussian 1 comprises two intersubunit distances: C344-R1–C344’-R1 and C344-R1–C163’-R1a; Gaussians 2 (P31), 3 (P34), and 4 (P39) comprises intramolecular distances between C344-R1 and the three positions of C163-R1; and Gaussian 2’ (P29) comprises intersubunit distances between C344-R1 and C163’-R1a/C163’-R1b.
We then proceeded to fit the CHS and cholate DEER titration data incorporating an equilibrium binding and dimer association model, respectively, that also incorporates the concentration dependence of the CHS-bound and dimeric β1AR species, using essentially the same approach described previously to globally fit time-resolved DEER EPR data for the binding of a substrate to calmodulin (31). The binding of CHS to β1AR was represented by a simple equilibrium binding isotherm where the fraction-bound species, pbound, is given by (CHS)/[ + (CHS)], where is the equilibrium dissociation constant (Fig. 3B). Dimerization of β1AR upon addition of cholate was described by the empirical Hill equation where the fraction dimer, pdimer, is given by /{ + n}, where is the equilibrium dissociation constant, n is the Hill coefficient, and is the fraction dimer in the absence of cholate (Fig. 3D). For both titration datasets, the number of Gaussians had to be extended to five to ensure a random distribution of residuals between experimental and calculated DEER echo curves comparable to that obtained from individual fits using Tikhonov regularization (Fig. 4 and SI Appendix, Figs. S9 and S13A for the CHS and cholate data, respectively). In the case of CHS, the peak at ~39 Å in the P(r) distribution is represented by two peaks, one at 39.4 ± 0.1 Å corresponding to the sterol-free state and the other at 37.4 ± 0.1 Å, corresponding to the CHS-bound state (Table 1). For cholate, the peak at ~31 Å in the P(r) distribution is represented by two peaks, one at 31.1 ± 0.1 Å corresponding to the intramolecular C344-R1–C163-R1a distance and the other at 29.3 ± 0.3 Å corresponding to the intersubunit C344-R1–C163’-R1a,b distances (Table 2). The disappearance of the small peak at ~26 Å (shown in blue in Fig. 2B) over the course of the CHS titration is due to the dissociation of residual dimer (~9%), present in sterol-free β1AR and occurs concomitantly with the binding of CHS (i.e., since CHS is not a reducing agent, residual dimer in sterol-free β1AR cannot be attributed to any intermolecular covalent disulfide-linked species). The normalized χ2 values for the global five-Gaussian fits, incorporating the concentration dependence of the species, to the CHS and cholate titration DEER data are close to 1 and essentially the same as those obtained from the global four-Gaussian fits where only peaks positions and widths are restrained (Tables 1 and 2). The optimized values of the equilibrium dissociation constants and are 1.4 ± 0.4 and 35 ± 6 mM, respectively, the value of the Hill coefficient n is 2.5 ± 0.4, and the fraction dimer, , in the absence of ligand is 0.09 ± 0.01. Plots of the calculated P(r) distributions from the global five-Gaussian fits over the course of the titration compared to the P(r) distributions obtained from the individual Tikhonov regularization fits are shown in Fig. 3A and SI Appendix, Fig. S10 for the CHS titration, and in Fig. 3B and SI Appendix, Fig. S13B for the cholate titration. Finally, a comparison of the fractional peak occupancies as a function of CHS and cholate concentration obtained with the global four- (circles) and five- (continuous lines) Gaussian fits are shown in Fig. 3 B and D, respectively.
Fig. 4.
Comparison of the residuals as a function of dipolar evolution time for the global Gaussian fits to the Q-band DEER titration data for CHS and cholate binding to spin-labeled β1AR (C163-R1/C344-R1)-DDM micelles versus the individual fits obtained by Tikhonov regularization. (A) CHS and (B) cholate titrations. The residuals for the global Gaussian fits (incorporating the concentration dependence of the CHS-bound and dimeric species) are shown in red, while those for the individual Tikhonov regularization fits are in blue. The concentration of β1AR is ~60 μM. The complete datasets are provided in SI Appendix, Figs. S7–S9 for the CHS titration, and SI Appendix, Figs. S11–S13 for the cholate titration. The reduced χ2 values for the fits to the CHS and cholate DEER titration data are provided in Tables 1 and 2, respectively.
CHS Dissociates the Cholate-Induced β1AR Dimer.
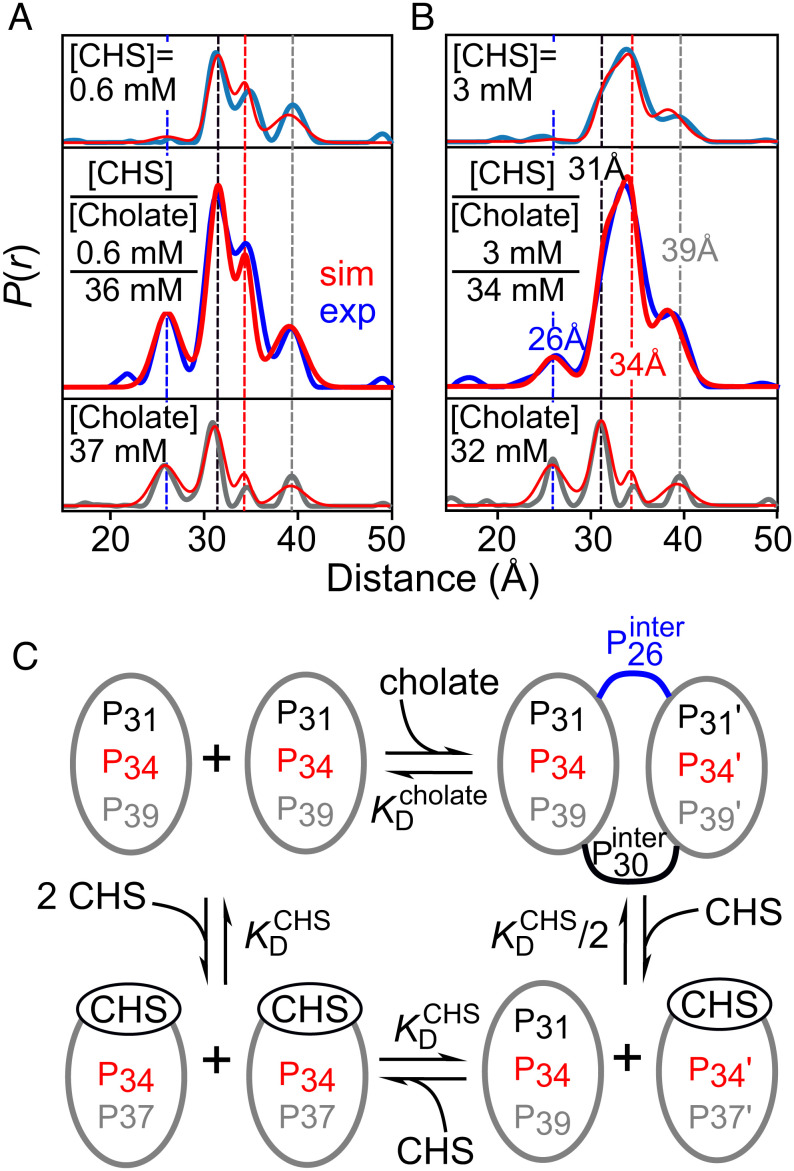
The effect of addition of CHS on cholate-induced β1AR dimerization was investigated by recording DEER data on ~50 μM spin-labeled β1AR (C163-R1/C344-R1) in the presence of 34 to 36 mM cholate and 0.6 or 3 mM CHS. The resulting DEER-derived P(r) distributions are shown in Fig. 5 A and B (Middle), and the corresponding DEER echo curves are shown in SI Appendix, Fig. S15. In the presence of CHS, the intensity of the 26 Å peak attributable to the cholate-induced dimeric form is reduced and the peak at ~34 Å arising from the CHS-bound monomeric state increases. The equilibria depicting the processes involved are shown in Fig. 5C. In the scheme shown in Fig. 5C, binding of CHS to only a single subunit of the dimer is required to dissociate the dimer into monomers.
Fig. 5.
CHS dissociates the cholate-induced dimer of nitroxide spin-labeled β1AR (C163-R1/C344-R1). P(r) distance distributions obtained for spin-labeled β1AR (~50 μM) in DDM with (A) 0.6 mM and (B) 3 mM CHS in the presence of 34 to 36 mM cholate are shown in the Middle panels. For comparison the Top and Bottom panels show the P(r) distributions obtained in the presence of either CHS (Top) or cholate (Bottom). (C) Depiction of the equilibria involved in cholate-induced dimerization upon binding of cholate to DDM micelles and dimer dissociation by specific binding of CHS to β1AR. In panels A and B, the light blue (Top), blue (Middle, denoted as “exp”), and gray (Bottom) P(r) distributions are obtained from individual fits to the experimental DEER echo curves using Tikhonov regularization (37). The P(r) distributions depicted by the thin red lines in the Top and Bottom panels are obtained from global fits to the DEER titration data using the global five-Gaussian fits (31) incorporating the respective equilibria for CHS and cholate binding, respectively (see Fig. 3 and Tables 1 and 2). The P(r) distributions depicted by the thick red lines (denoted as “sim” in the Middle panels) are obtained from a population-weighted linear combination of the species in panel C with the fractional populations calculated using the values (Tables 1 and 2) of the equilibrium dissociation constants, and in the case of cholate-induced dimerization the Hill coefficient as well, determined from the global fits to the DEER titration data shown in Fig. 3. The small peaks below ~26 Å seen in some of the experimental P(r) distributions obtained by Tikhonov regularization arise from overfitting artefacts as their upper and lower confidence limits are large in relation to the corresponding peak heights.
Comparison of the P(r) distributions obtained by individual fits using validated Tikhonov regularization with those calculated from a population-weighted average of the P(r) distributions for the fully saturated CHS and cholate bound states obtained from the global five-Gaussian fits (Fig. 3 and Tables 1 and 2), where the fractional populations are calculated using the scheme in Fig. 5C with the equilibrium dissociation constants and Hill coefficient given in Tables 1 and 2, display excellent qualitative agreement. Thus, binding of CHS to β1AR, even in the presence of close to saturating amounts of cholate, dissociates the dimer, and CHS and cholate do not compete for the same binding site. Further, the fact that the fractional populations of monomer and dimer in the presence of variable concentrations of CHS and cholate are determined by the experimental equilibrium dissociation constants obtained from the individual CHS and cholate titration data (Fig. 3 and Tables 1 and 2 and SI Appendix, Figs. S10 and S13) indicates that cholate-induced β1AR dimerization is fully reversible and cannot be attributed to the formation of any intermolecular disulfide-linked species.
Concluding Remarks.
In summary, using DEER we have shown that the soluble cholesterol analog CHS specifically binds to spin-labeled β1AR (C163-R1/C344-R1) solubilized in DDM micelles. Binding occurs in close proximity to Cys163-R1, thereby altering the distribution of the Cys163-R1 spin-label through steric occlusion (Fig. 3A). The CHS titration data can be described by a simple binding isotherm with an equilibrium dissociation constant of ~1.4 mM (Fig. 3B). Sodium cholate, on the other hand, promotes dimerization of spin-labeled β1AR (C163-R1/C344-R1) in a cooperative manner with an equilibrium dissociation constant of ~35 mM and a Hill coefficient of ~2.5 (Fig. 3 C and D). Interaction with cholate results in the appearance of new intersubunit distances involving C344-R1 and C163-R1 without perturbing the intramolecular pairwise distances between the same spin labels. According to the 4GPO structure of β1AR (39), two dimer interfaces are possible with intersubunit contacts between TM1, TM2, and H8 in dimer interface 1 and between TM4 and TM5 in dimer interface 2. Dimer interface 1, which was previously observed in the crystal structure of β2AR (54), is fully consistent with our DEER data on spin-labeled β1AR (SI Appendix, Fig. S5C). Further, the observation that a distinct parallel orientation of the two subunits in the spin-labeled β1AR dimer, corresponding to dimer interface 1 (Fig. 2C), is observed by DEER excludes any random dimerization that might be caused by hydrophobic mismatch (55).
Although direct interaction of cholate with spin-labeled β1AR (C163-R1/C344-R1) was not observed by DEER, we cannot ascertain whether the effect of cholate arises from direct binding of cholate to β1AR at a site distant from the two spin labels (and therefore distinct from the CHS binding site monitored by DEER) or, more likely, due to superficial penetration of cholate on the surface of DDM micelles, thereby altering their physical properties. Further, CHS binding to β1AR has an inhibitory effect on receptor dimerization. Thus, CHS not only dissociates the small fraction (~9%) of residual dimer present in the sterol-free β1AR sample (Fig. 3A) but also dissociates the dimer in the presence of close to saturating concentrations of cholate (Fig. 5).
One of the functions of cholesterol appears to relate to its role in controlling oligomerization of GPCRs. Thus, the crystal structure of β2AR is a symmetric dimer with two cholesterol molecules located at dimer interface 1 and an additional two cholesterol molecules bound to the CCM (54). This suggests that cholesterol may promote dimerization by stabilizing the β2AR dimer. In contrast to β2AR, however, β1AR was found to form parallel oligomers under sterol-free conditions (39), indicating that dimerization can occur independently of cholesterol binding. This is in agreement with our findings: namely that residual β1AR dimer (~9%) was found in the absence of any added cholesterol analog, and no direct evidence for specific cholate binding to β1AR was observed for cholate-induced dimerization. The detection of a small fraction of β1AR dimeric species in the sterol-free state and the complete conversion of monomer to dimer in the presence of cholate is intriguing given that the role of dimerization in class A GPCRs remains under debate (56). For both β1AR and its β2AR homolog, constitutive dimers have been detected in vivo using bioluminescence energy transfer (BRET) (57, 58). Due to the highly heterogeneous composition of membranes in living cells, however, various environmental factors may influence GPCR oligomerization, and independent studies of the impact of individual components within the cell membrane are very limited.
To the best of our knowledge, dimerization of β1AR upon addition of cholate and its inhibition by specific binding of CHS are unique findings that further contribute to an understanding of GPCR oligomerization and, hence, the mechanism of GPCR signal transduction.
Experimental Procedures
β1AR Expression, Purification and Spin Labeling.
Thermostabilized turkey β1AR was expressed and purified in E. coli Rosetta (DE3)-competent cells (Sigma-Aldrich) as described previously (33). Unless otherwise stated, all purification steps were performed at 4 °C. Purification comprised the following steps: solubilization of β1AR with 1.5% DDM, 0.7% CHAPS [(3-((3-cholamidoproly) dimethylammonio)-1-propanesulfonate)], and 0.15% CHS; Ni-nitriloacetic acid (Qiagen) affinity chromatography using a 50% Ni agarose resin (ThermoFisher); cleavage of thioredoxin 1 (TrxA) and maltose-binding protein fusion tags with human rhinovirus 3C protease (ThermoFisher); separation of β1AR by chromatography using a Sepharose Fast Flow cation exchange (Cytiva) column; further purification with receptor specific alprenolol agarose affinity chromatography (Cube Biotech); and finally, size exclusion chromatography using a Superose 6 Increase 10/300 GL (Sigma Aldrich) column in 20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM ethylene diamine tetraacetic acid (EDTA), and 2 mM DDM. The fractions from the final column were analyzed by SDS-PAGE; the molecular mass of purified β1AR was confirmed by matrix-assisted laser desorption/ionization mass spectrometry (SI Appendix, Fig. S2 A and B). Finally, purified β1AR in DDM was concentrated to ~60 μM using a centrifugal concentrator with a molecular weight cutoff of 50,000 (VivaSpin20, Sartorius) and stored at 4 °C.
R1 nitroxide labeling was carried out with S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL; Toronto Research Chemicals). A 10-fold molar excess of MTSL was added to β1AR (~60 μM) in DDM, incubated for 2 h at room temperature and 12 h at 4 °C. Excess spin label was removed by successive washes (in 20 mM Tris, pH 7.5, 100 mM NaCl and 1 mM EDTA) using centrifugal concentrator devices (59) and further concentrated. The final concentration of the stock solution of spin-labeled β1AR in DDM was ~114 μM. Flash freezing of R1 nitroxide spin-labeled β1AR samples (final concentration ~60 μM) in buffer containing 30% (vol/vol) deuterated glycerol was carried out by directly placing the EPR tubes (1 mm inner diameter/1.6 mm outer diameter; VitroCom) containing 15 μL of sample into liquid nitrogen.
Assignment of the DEER-derived probability distance P(r) distributions to specific cysteine pairs was carried by recording DEER data for a series of single point mutants: C85A, C85V, C133V, C163L, C344A, and C344S (SI Appendix, Figs. S3 and S4 and Table S1). Mutagenesis was carried out by GenScript and the quality of the plasmids was additionally verified by plasmid sequencing (Plasmidsaurus).
CHS and Cholate Titrations.
For the titrations with CHS and cholate (Figs. 2 and 3), appropriate amounts of concentrated ligand stock solutions (20 mM CHS in a 1:10 w/w CHS:DDM solution from ThermoFisher comprising 10 mg/mL CHS and 100 mg/mL DDM; or 200 mM sodium cholate in buffer containing 20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM EDTA) were added to the 114 μM stock solution of spin-labeled β1AR (C163-R1/C344-R1) in DDM, supplemented with 30% v/v (final concentration) deuterated glycerol. The final concentration of spin-labeled β1AR used in the individual CHS and cholate DEER titration experiments was ~60 μM. Samples were incubated for 15 min at room temperature and kept at 4 °C prior to liquid nitrogen flash freezing in the EPR tubes. For the experiments where both CHS and cholate were added (Fig. 5), appropriate amounts of cholate and CHS from the stock ligand solutions were added in quick succession to the stock solution of spin-labeled β1AR in DDM (final concentration of ~50 μM), incubated for 15 min at room temperature, and kept at 4 °C prior to flash freezing in liquid nitrogen.
Q-Band DEER.
Pulsed EPR data were collected at Q-band (33.8 GHz) at a temperature of 50 K on a Bruker E-580 spectrometer equipped with a 300-W traveling-wave tube amplifier, a model ER5107D2 resonator, and a cryo-free cooling unit, as described previously (60). Four-pulse DEER experiments were acquired using a conventional four-pulse sequence (14). The observer and electron-electron double resonance (ELDOR) pump pulses were separated by about 80 MHz, with the observer π/2 and π pulses set to 12 and 24 ns, respectively, and the ELDOR π pulse set to 10 ns. The pump frequency was centered at the Q-band nitroxide spectrum located at +40 MHz from the center of the resonator frequency. The τ1 value of 400 ns for the first echo-period time was incremented eight times in 16 ns steps to average 2H modulation; the position of the ELDOR pump pulse was incremented in steps of Δt = 4 ns. The bandwidth of the overcoupled resonator was 120 MHz. All DEER echo curves were acquired for tmax = 2.5 μs with the total length (T = 2τ2) of the dipolar evolution time set to 6 μs; data collection was not extended to the full τ2 (3 μs) range because of a “2 + 1” echo perturbation of the DEER echo curves at a time of about τ1 from the final observed π pulse.
P(r) Distributions and Analysis.
P(r) pairwise distance distributions were obtained from the DEER echo curves either by model-free validated Tikhonov regularization using the program DeerLab (37) (for example, Fig. 2A) or by a sum of Gaussians to directly fit the experimental DEER data (including background correction with a best-fit exponential decay) using the program DD/GLADDvu (47, 48) (Fig. 2 A and B). Validated Tikhonov regularization was performed with bootstrap analysis for uncertainty quantification via the bootan function in the DeerLab library, with the number of bootstrap samples evaluated set to 1,000 (37). For global fitting and analysis of the titration series with CHS and sodium cholate, we imported the fitting routines from the program DD/GLADDvu (47, 48) into a home-written Python program and included terms that allowed us to treat peak positions and widths as global optimized parameters (31, 40). Minimization was carried out using the least-squares Levenberg–Marquard algorithm. In addition, global fits to all DEER data for a given titration series with either CHS or cholate were also carried out incorporating the appropriate equations describing CHS-binding or cholate-induced dimerization equilibria, respectively, to calculate the ligand concentration dependence of the species (Fig. 2), in a manner similar to that described previously to fit time-resolved DEER data to a transient kinetic scheme (31). The raw DEER echo curves in digital format and the Python scripts used to globally fit the data have been deposited in Figshare (DOI:10.6084/m9.figshare.21810408) (61).
Calculation of P(r) Distributions from Molecular Coordinates.
P(r) distance distributions between R1 nitroxide spin labels were calculated from molecular coordinates as described previously (31, 40) using a conformational library for the nitroxide spin labels in the program Xplor-NIH)(38)) via a modified version of the calcPr helper program. The populations within the library of R1 residue side-chain conformations consist of an intrinsic component modulated by overlap with nearby heavy atoms, such that the population of a particular conformation goes to zero when it closely approaches a heavy atom (including heteroatoms). While the nitroxide spin-label conformational library was derived for soluble proteins, the results of the calculations on β1AR are still extremely helpful in terms of understanding the origins of experimental DEER-derived P(r) distributions, and moreover good qualitative agreement is obtained between the experimental and simulated P(r) distributions (Fig. 2 A–C and SI Appendix, Fig. S5).
The β1AR structure [PDB code 2Y00 (35)] used for the P(r) calculations was cocrystalized with two CHS molecules (CHS1 and CHS2) and several Hega-10 (denoted as 2CV in Fig. 2C and in the PDB coordinates) detergent molecules. CHS1 is located in close proximity to the R1 nitroxide spin label at Cys163-R1, while CHS2 is located in a region with no spin label reporters in close proximity. Therefore, only the CHS1 binding site was considered for the purpose of the P(r) calculations. For sterol-free β1AR, both CHS molecules were removed, while, for CHS–β1AR complex, the CHS ligands were kept in the positions identified by the 2Y00 X-ray structure (35). The Hega-10 detergent molecule 2CV located on the surface of β1AR close to the Cys163-R1 spin label was found to have a significant impact on the calculated P(r) distance distributions. Removing the 2CV detergent molecule from the calculations led to significant changes in the calculated P(r) distribution, which did not agree with our experimental data. This mismatch clearly demonstrates the importance of occupying this position with a detergent molecule, and therefore the 2CV molecule was kept for all further calculations. The sterol-free β1AR calculations revealed four possible positions of the Cys344-R1 spin label and five for Cys163-R1 (SI Appendix, Fig. S6, Top). The best qualitative match with the experimental data was obtained by removing three Cys344-R1 and two Cys163-R1 nitroxide tag conformations, indicating that these positions are not significantly populated under our experimental conditions.
To obtain the best qualitative agreement with experimental DEER data, several modifications were performed for the calculations on the CHS–β1AR complex. a) To populate the R1 nitroxide spin label at the Cys163-R1b position, as observed experimentally, we restricted the occupancy at the Cys163-R1c position. The need for this modification suggests the presence of an additional detergent molecule(s) on the surface of the β1AR that prevents movement of the Cys163-R1 spin label. b) The CHS1 molecule was shifted from the TM4 helix toward the TM2 helix allowing more space for the Cys163-R1 spin label (SI Appendix, Fig. S4 and Table S2). Interestingly, β2AR was crystallized with two cholesterol molecules in the CCM located in the cleft formed by the TM2–4 helices (41), which corresponds to CHS1 binding site of β1AR. The optimized CHS position for the CHS–β1AR complex corresponds to the center of the cholesterol dimer in the β2AR complex with cholesterol (41, 54).
Calculations for the β1AR dimer were performed using the monomeric β1AR crystal structure [PDB code: 2Y00 (35)] and the parallel orientations of dimeric conformations observed in the 4GPO β1AR crystal structure (39). Separate calculations for the two possible dimer interfaces revealed that dimer interface 1, which includes the TM1, TM2, and H8 helices, displays short distances around 26 Å (SI Appendix, Fig. S5 and Table S2). Dimer interface 2 formed by intersubunit contacts between the TM4 and TM5 helices does not have any intersubunit distances between R1 spin labels below 38 Å, indicating that the 26 Å distance, which is observed experimentally, is characteristic of dimer interface 1.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Drs. Stefan Grzesiek and Marco Rogowski for providing the β1AR plasmid, Drs. James Baber and Dan Garrett for technical assistance, and Drs. John Louis and Rodolfo Ghirlando for useful discussions. This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH (G.M.C., Grant DK-029023).
Author contributions
N.K., T.S., and G.M.C. designed research; N.K., T.S., and C.D.S. performed research; N.K. contributed new reagents/analytic tools; N.K., T.S., and G.M.C. analyzed data; and N.K., T.S., and G.M.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: H.M.A.-H., Columbia University; and D.J.W., University of Maryland.
Data, Materials, and Software Availability
All other data are included in the article and/or SI Appendix. The raw DEER echo curves in digital format and the Python scripts used in global fitting have been deposited on figshare (DOI:10.6084/m9.figshare.21810408) (61).
Supporting Information
References
- 1.Shimada I., Ueda T., Kofuku Y., Eddy M. T., Wuthrich K., GPCR drug discovery: Integrating solution NMR data with crystal and cryo-EM structures. Nat. Rev. Drug Discov. 18, 59–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolb P., et al. , Structure-based discovery of β2-adrenergic receptor ligands. Proc. Natl. Acad. Sci. U.S.A. 106, 6843–6848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoichet B. K., Kobilka B. K., Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol. Sci. 33, 268–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wisler J. W., Rockman H. A., Lefkowitz R. J., Biased G Protein-coupled receptor signaling: Changing the paradigm of drug discovery. Circulation 137, 2315–2317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbaum D. M., Rasmussen S. G., Kobilka B. K., The structure and function of G-protein-coupled receptors. Nature 459, 356–363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manglik A., Kobilka B., The role of protein dynamics in GPCR function: Insights from the β2AR and rhodopsin. Curr. Opin. Cell Biol. 27, 136–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilger D., Masureel M., Kobilka B. K., Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 25, 4–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wingler L. M., Lefkowitz R. J., Conformational basis of G protein-coupled receptor signaling versatility. Trends Cell Biol. 30, 736–747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar P., Chattopadhyay A., Cholesterol in gpcr structures: Prevalence and relevance. J. Membr. Biol. 255, 99–106 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Rosenhouse-Dantsker A., Bukiya A. N., Eds. Cholesterol Modulation of Protein Function in "Advances in Experimental Medicine and Biology", (Springer, 2019), vol. 1115. [Google Scholar]
- 11.Soubias O., Sodt A. J., Teague W. E., Hines K. G., Gawrisch K., Physiological changes in bilayer thickness induced by cholesterol control GPCR rhodopsin function. Biophys. J., in press 10.1016/j.bpj.2022.11.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abiko L. A., et al. , Filling of a water-free void explains the allosteric regulation of the β1-adrenergic receptor by cholesterol. Nat. Chem. 14, 1133–1141 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Jakubik J., El-Fakahany E. E., Allosteric modulation of GPCRs of Class A by cholesterol. Int. J. Mol. Sci. 22, 1953 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pannier M., Veit S., Godt A., Jeschke G., Spiess H. W., Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 142, 331–340 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Jeschke G., DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 63, 419–446 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Hubbell W. L., Cafiso D. S., Altenbach C., Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 7, 735–739 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Serdiuk T., et al. , A cholesterol analog stabilizes the human β2-adrenergic receptor nonlinearly with temperature. Sci. Signal 15, eabi7031 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garidel P., Hildebrand A., Knauf K., Blume A., Membranolytic activity of bile salts: Influence of biological membrane properties and composition. Molecules 12, 2292–2326 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jojart B., et al. , Mixed micelles of sodium cholate and sodium dodecylsulphate 1:1 binary mixture at different temperatures–experimental and theoretical investigations. PLoS One 9, e102114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coreta-Gomes F. M., et al. , Interaction of bile salts with model membranes mimicking the gastrointestinal epithelium: A study by isothermal titration calorimetry. Langmuir 31, 9097–9104 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Neves M. C., et al. , Interaction of bile salts with lipid bilayers: An atomistic molecular dynamics study. Front. Physiol. 10, 393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duss O., Yulikov M., Allain F. H. T., Jeschke G., Combining NMR and EPR to determine structures of large rnas and protein-rna complexes in solution. Methods Enzymol. 558, 279–331 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Ward R., et al. , EPR distance measurements in deuterated proteins. J. Magn. Reson. 207, 164–167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Mkami H., Norman D. G., EPR distance measurements in deuterated proteins. Methods Enzymol. 564, 125–152 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Schmidt T., Wälti M. A., Baber J. L., Hustedt E. J., Clore G. M., Long distance measurements up to 160 A in the GroEL tetradecamer using Q-band DEER EPR spectroscopy. Angew. Chem. Int. Ed. Engl. 55, 15905–15909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerch M. T., et al. , Viewing rare conformations of the β2 adrenergic receptor with pressure-resolved DEER spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 31824–31831 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elgeti M., Hubbell W. L., DEER analysis of GPCR conformational heterogeneity. Biomolecules 11, 778 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichenwallner J., Liu B., Balo A. R., Ou W. L., Ernst O. P., Electron paramagnetic resonance spectroscopy on G-protein-coupled receptors: Adopting strategies from related model systems. Curr. Opin. Struct. Biol. 69, 177–186 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Zhao D. Y., et al. , Cryo-EM structure of the native rhodopsin dimer in nanodiscs. J. Biol. Chem. 294, 14215–14230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milikisiyants S., et al. , Oligomeric structure of Anabaena sensory rhodopsin in a lipid bilayer environment by combining solid-state NMR and long-range DEER constraints. J. Mol. Biol. 429, 1903–1920 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Schmidt T., et al. , Time-resolved DEER EPR and solid-state NMR afford kinetic and structural elucidation of substrate binding to Ca2+-ligated calmodulin. Proc. Natl. Acad. Sci. U.S.A. 119, e2122308119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson A. A., et al. , GPCR stabilization using the bicelle-like architecture of mixed sterol-detergent micelles. Methods 55, 310–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abiko L. A., Rogowski M., Gautier A., Schertler G., Grzesiek S., Efficient production of a functional G protein-coupled receptor in Escherichia coli for structural studies. J. Biomol. NMR 75, 25–38 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frei J. N., et al. , Conformational plasticity of ligand-bound and ternary GPCR complexes studied by 19F NMR of the β1-adrenergic receptor. Nat. Commun. 11, 669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warne T., et al. , The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature 469, 241–244 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warne T., et al. , Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454, 486–491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fabregas Ibanez L., Jeschke G., Stoll S., DeerLab: A comprehensive software package for analyzing dipolar electron paramagnetic resonance spectroscopy data. Magn. Reson. (Gott) 1, 209–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwieters C. D., Bermejo G. A., Clore G. M., Xplor-NIH for molecular structure determination from NMR and other data sources. Protein Sci. 27, 26–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J., Chen S., Zhang J. J., Huang X. Y., Crystal structure of oligomeric β1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat. Struct. Mol. Biol. 20, 419–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt T., Wang D., Jeon J., Schwieters C. D., Clore G. M., Quantitative agreement between conformational substates of holo calcium-loaded calmodulin detected by double electron-electron resonance EPR and predicted by molecular dynamics simulations. J. Am. Chem. Soc. 144, 12043–12051 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanson M. A., et al. , A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure 16, 897–905 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christopher J. A., et al. , Biophysical fragment screening of the β1-adrenergic receptor: Identification of high affinity arylpiperazine leads using structure-based drug design. J. Med. Chem. 56, 3446–3455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu X., et al. , Binding pathway determines norepinephrine selectivity for the human β1AR over β2AR. Cell Res. 31, 569–579 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang M., et al. , Milligram production and biological activity characterization of the human chemokine receptor CCR3. PLoS One 8, e65500 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Worswick S. G., Spencer J. A., Jeschke G., Kuprov I., Deep neural network processing of DEER data. Sci. Adv. 4, eaat5218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keeley J., et al. , Neural networks in pulsed dipolar spectroscopy: A practical guide. J. Magn. Reson. 338, 107186 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Brandon S., Beth A. H., Hustedt E. J., The global analysis of DEER data. J. Magn. Reson. 218, 93–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hustedt E. J., Stein R. A., McHaourab H. S., Protein functional dynamics from the rigorous global analysis of DEER data: Conditions, components, and conformations. J. Gen. Physiol. 153, e201711954 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huber M., et al. , Phase memory relaxation times of spin labels in human carbonic anhydrase II: Pulsed EPR to determine spin label location. Biophys. Chem. 94, 245–256 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Canarie E. R., Jahn S. M., Stoll S., Quantitative structure-based prediction of electron spin decoherence in organic radicals. J. Phys. Chem. Lett. 11, 3396–3400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt T., Clore G. M., T(m) filtering by 1H-methyl labeling in a deuterated protein for pulsed double electron-electron resonance EPR. Chem. Commun. (Camb) 56, 10890–10893 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El Mkami H., Ward R., Bowman A., Owen-Hughes T., Norman D. G., The spatial effect of protein deuteration on nitroxide spin-label relaxation: Implications for EPR distance measurement. J. Magn. Reson. 248, 36–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baber J. L., Louis J. M., Clore G. M., Dependence of distance distributions derived from double electron-electron resonance pulsed EPR spectroscopy on pulse-sequence time. Angew. Chem. Int. Ed. Engl. 54, 5336–5339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cherezov V., et al. , High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milovanovic D., et al. , Hydrophobic mismatch sorts SNARE proteins into distinct membrane domains. Nat. Commun. 6, 5984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo H., et al. , Methods used to study the oligomeric structure of G-protein-coupled receptors. Biosci. Rep. 37, BSR20160547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Angers S., et al. , Detection of β2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proc. Natl. Acad. Sci. U.S.A. 97, 3684–3689 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mercier J. F., Salahpour A., Angers S., Breit A., Bouvier M., Quantitative assessment of β1- and β2-adrenergic receptor homo- and heterodimerization by bioluminescence resonance energy transfer. J. Biol. Chem. 277, 44925–44931 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Van Eps N., et al. , Gi- and Gs-coupled GPCRs show different modes of G-protein binding. Proc. Natl. Acad. Sci. U.S.A. 115, 2383–2388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt T., Schwieters C. D., Clore G. M., Spatial domain organization in the HIV-1 reverse transcriptase p66 homodimer precursor probed by double electron-electron resonance EPR. Proc. Natl. Acad. Sci. U.S.A. 116, 17809–17816 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt T., Kubatova N., Clore G. M., Digital data deposition and Python global fitting scripts for “Quantitative analysis of sterol-modulated monomer-dimer equilibrium of the β1-adrenergic receptor by DEER spectroscopy”. Chemphyschem., 17, 2987–2991 (2023), 10.6084/m9.figshare.21810408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All other data are included in the article and/or SI Appendix. The raw DEER echo curves in digital format and the Python scripts used in global fitting have been deposited on figshare (DOI:10.6084/m9.figshare.21810408) (61).