Abstract
Reverse genetics is used to evaluate the roles in vivo of allosteric regulation of Escherichia coli glycerol kinase by the glucose-specific phosphocarrier of the phosphoenolpyruvate:glycose phosphotransferase system, IIAGlc (formerly known as IIIglc), and by fructose 1,6-bisphosphate. Roles have been postulated for these allosteric effectors in glucose control of both glycerol utilization and expression of the glpK gene. Genetics methods based on homologous recombination are used to place glpK alleles with known specific mutations into the chromosomal context of the glpK gene in three different genetic backgrounds. The alleles encode glycerol kinases with normal catalytic properties and specific alterations of allosteric regulatory properties, as determined by in vitro characterization of the purified enzymes. The E. coli strains with these alleles display the glycerol kinase regulatory phenotypes that are expected on the basis of the in vitro characterizations. Strains with different glpR alleles are used to assess the relationships between allosteric regulation of glycerol kinase and specific repression in glucose control of the expression of the glpK gene. Results of these studies show that glucose control of glycerol utilization and glycerol kinase expression is not affected by the loss of IIAGlc inhibition of glycerol kinase. In contrast, fructose 1,6-bisphosphate inhibition of glycerol kinase is the dominant allosteric control mechanism, and glucose is unable to control glycerol utilization in its absence. Specific repression is not required for glucose control of glycerol utilization, and the relative roles of various mechanisms for glucose control (catabolite repression, specific repression, and inducer exclusion) are different for glycerol utilization than for lactose utilization.
In Escherichia coli, glucose controls utilization of several other carbon sources, including lactose, melibiose, maltose, and glycerol (14, 27, 29, 30, 32). Effects of glucose on the expression of genes needed for metabolism of other sugars, e.g., lactose, formed the foundation for much of the initial understanding of molecular genetic control mechanisms. Glucose effects were found to involve both positive and negative control aspects. At the level of transcriptional control, these two opposing aspects for expression of the lac operon are mediated by the cyclic AMP (cAMP)-cAMP receptor protein complex (for catabolite repression) and by the lac repressor (for specific repression), respectively. The specific repression is relieved by binding of an inducer. Subsequent studies have revealed that glucose acts to modulate the level of cAMP and the level of the inducer. These controls are exerted by two different forms of IIAGlc, the glucose-specific phosphocarrier of the phosphoenolpyruvate:glycose phosphotransferase system (PTS). The form of IIAGlc that is phosphorylated at an active-site histidine residue participates in the increase of cAMP by activation of adenylate cyclase, and the form of IIAGlc that is unphosphorylated binds to lactose permease and prevents lactose uptake. Because the latter process prevents uptake of the inducer, this mechanism is termed inducer exclusion. IIAGlc-dependent PTS-mediated inducer exclusion is an important regulatory concept that unifies several aspects of genetic, allosteric, and metabolic controls. The finding of both positive and negative control mechanisms raises the issue of their relative roles in glucose control. In the case of the lac operon, recent studies show that specific repression coupled to inducer exclusion is the dominant mechanism for glucose control of lactose utilization (6, 11, 36). In lacI strains, glucose control is abolished, which is seen as loss of the repression of β-galactosidase and elimination of the plateau during diauxic growth on glucose-lactose (11). A similar phenotype is seen for strain PPA586, an MG1655 derivative with lacY(S209I), in which the lactose permease is thought to be resistant to IIAGlc inhibition (6, 8, 33).
Glycerol kinase (EC 2.7.1.30; ATP-glycerol 3-phosphotransferase) is a component of a regulatory network in E. coli by which glucose and other carbon sources control the utilization of glycerol and the gene expression that is needed for glycerol metabolism (14, 27, 29, 32). The proteins involved in glycerol metabolism are encoded by the elements of the glp regulon, which displays a complex genetic structure (3, 5, 37, 39). It contains five operons, which are located at three different chromosomal loci. Glucose modulation of glycerol utilization involves both regulation of transcription and posttranslational control of glycerol kinase catalytic activity. Control of transcription of the regulon elements is analogous to the lac operon and involves both positive control by cAMP-cAMP receptor protein and negative control by a specific repressor that is encoded by the glpR gene. DNA-binding sites for the specific repressor in the glpFKX operon have been identified both in the 5′ upstream region and internally within the glpK coding sequence (37). The inducer for expression of the glp elements is sn-glycerol 3-phosphate (15), which is both the product of the reaction that is catalyzed by glycerol kinase during glycerol catabolism and an important metabolite in phospholipid biosynthesis under all growth conditions. The catalytic activity of glycerol kinase is controlled posttranslationally by the allosteric inhibitors fructose 1,6-bisphosphate (FBP) and IIAGlc (19, 26, 42). These allosteric effectors display V-system regulation (19, 23) that allows efficient control that is not dependent on changes in the concentrations of the substrates. The unusual existence of two very different allosteric effectors that are involved in glucose control has been noted and postulated to be the basis for the extreme effectiveness of glucose in preventing glycerol consumption (14).
Several years ago, Zwaig and Lin identified the mutant E. coli strain 43 on the basis of its loss of glucose control of glycerol utilization (42). They showed that the glycerol kinase from strain 43 had lost sensitivity to inhibition by FBP; the role of IIAGlc was unknown at that time. We isolated the glpK22 allele from strain 43 and showed that it contains a mutation that results in a single amino acid substitution in glycerol kinase, G-304-S (21). The variant enzyme encoded by the glpK22 allele was purified and characterized. It was found to show greatly reduced sensitivity to FBP inhibition, in agreement with the earlier work, and to show weak activation by IIAGlc with greatly reduced apparent affinity for binding IIAGlc. Thus, this variant glycerol kinase has lost sensitivity to inhibition by both allosteric effectors. This finding raises the question of the relative roles of the regulation by each glycerol kinase allosteric effector in glucose inhibition of glycerol utilization. Results of a study with Salmonella suggest that both allosteric inhibitors may be required for glucose control in that organism (19).
In other investigations of the novel allosteric regulation of glycerol kinase, we identified, purified, and characterized in vitro variant enzymes that are insensitive to one of the allosteric inhibitors while retaining normal sensitivity to the other inhibitor as well as normal catalytic behavior. Glycerol kinase A-65-T is not sensitive to inhibition by FBP but displays normal regulation by IIAGlc and normal catalytic properties (16). Glycerol kinase T-477-N is not sensitive to inhibition by IIAGlc but displays normal regulation by FBP and normal catalytic properties (see below). These variants provide the basis for reverse genetic investigations of the quantitative contributions of each of the allosteric inhibitors to glucose inhibition of glycerol utilization and control of expression of the glp regulon in vivo.
Because of the complex genetic structure of the glp regulon and the many-faceted control network, it is necessary to place the glpK alleles encoding the variant enzymes into the normal chromosomal context. This report describes the construction and characterization of strains with chromosomal glpK alleles. Three different genetic backgrounds were utilized for the constructions. Initially, strain DG1 was used, because it is a ΔglpK (16) strain, to facilitate the constructions. Unexpectedly, however, strain DG1 and its derivatives were found to carry the glpR208 allele (9). This allele is functionally identical to the glpR2 allele, and these strains were used here as GlpR− strains. Strain MC4100 was used as a glpR+ strain because other investigators have performed extensive studies of the glp operator structures and transcriptional control in this strain and some of its derivatives (13, 38, 40). Strain MG1655 was used as a second glpR+ strain because it is a prototrophic strain and has been used as the background for extensive investigations of the mechanisms of glucose control of how other carbon sources are used (6, 7). The effects of the specific alterations in glycerol kinase allosteric regulatory properties on diauxic growth on glucose-glycerol, glycerol utilization, and levels of glycerol kinase specific activity are described. The results reveal that, in E. coli, FBP inhibition of glycerol kinase is quantitatively dominant in glucose control of glycerol utilization, which is independent of inducer exclusion mediated by specific repression.
MATERIALS AND METHODS
Materials.
Chemicals and enzymes were purchased from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise indicated. Purified IIAGlc was generously provided by Saul Roseman, Department of Biology, Johns Hopkins University, Baltimore, Md.
Strains and culture methods.
Biological materials that were used in these studies are listed in Table 1. All strains were grown in Luria-Bertani (LB) medium or minimal M9 (18) medium with carbohydrate added as indicated. MacConkey glycerol agar was prepared according to the instructions of the manufacturer (Difco). Antibiotics were added when indicated to obtain final concentrations of 75 μg/ml for ampicillin, 8 μg/ml for tetracycline, 200 μg/ml for streptomycin, and 40 μg/ml for kanamycin.
TABLE 1.
Strains and plasmids used in these studies
| Strain or plasmid | Genotype | Source or referencea |
|---|---|---|
| E. coli strain | ||
| DG1 | TB1 ΔglpK202 | 16 |
| HfrH | F+ | Ry Young |
| HfrPK3 | F+ayi-15 lacY1 leuB6 supE44 thi-1 thr-1 tonA21 | Ry Young |
| JM83 | F+ara glpR208 Δ(lac-proAB) rpsL(Strr) [φ80dlacΔ(lacZ)M15] thi | Ry Young; 17 |
| JW383 | thr-1 ara-14 eda-50 fhuA31 Δ(galK-attλ)99 hisG4(Oc) lacY1λ−leuB6 metF159(Am) mtl-1 rpsL136(Strr) thi-1 tsx-78 xylA5 zii-510::Tn10 | CGSC; 34 |
| Lin 43 | HfrC fadL701 glpK22 glpR2 phoA8 pit-10 relA1 sopT1 tonA22 T2r | CGSC; 42 |
| MC4100 | F−araD139 Δ(argF-lac)U169 deoC1 flbB5301 ptsF25 rbsR relA1 rpsL150(Strr) | CGSC |
| MG1655 | λ−rph-1 | CGSC |
| TB1 | JM83 glpR208 hsdR(rK− mK+) | 1 |
| ts1962 | F−lac-28 proA23 ptsI19(Ts) rpsL101(Strr) trp-30 | CGSC; 12 |
| AP100 | ptsI19(Ts) rpsL101(Strr) | ts1962 × HfrH |
| AP110 | AP100 ΔglpK202 zii-510::Tn10 | AP100 × P1.KH10 |
| KH1 | DG1 zii-510::Tn10 | DG1 × P1.JW383 |
| KH5 | HfrPK3 ΔglpK202 zii-510::Tn10 | Hfr PK3 × P1.KH1 |
| KH10 | KH1 pro+ | KH1 × HfrH |
| KH11 | DG1 glpK+pro+ | DG1 × Hfr PK3 |
| KH12 | KH10 glpK203 | KH10 × KH5/pA65T |
| KH15 | KH10 glpK204 | KH10 × KH5/pT477N |
| KH18 | MC4100 ΔglpK202 zii-510::Tn10 | MC4100 × P1.KH1 |
| KH24 | MC4100 glpK204 zii-510::Tn10 | MC4100 × P1.KH15 |
| KH34 | MC4100 glpK203 zii-510::Tn10 | MC4100 × P1.KH12 |
| KH37 | KH10 glpK22 | KH10 × KH5/pCH5 |
| KH38 | MC4100 glpK22 zii-510::Tn10 | MC4100 × P1.KH37 |
| KH51 | MG1655 ΔglpK202 zii-510::Tn10 | MG1655 × P1.KH1 |
| KH52 | MG1655 zii-510::Tn10 | MG1655 × P1.KH1 |
| KH58 | MG1655 glpK203 zii-510::Tn10 | MG1655 × P1.KH12 |
| KH59 | MG1655 glpK204 zii-510::Tn10 | MG1655 × P1.KH15 |
| KH61 | MG1655 glpK22 zii-510::Tn10 | MG1655 × P1.KH38 |
| Plasmids | ||
| pA65T | pHG165 + HindIII fragment glpK203 | 16 |
| pCH5 | pHG165 + HindIII fragment glpK22 | 21 |
| pCJ102 | pBR322 + HindIII fragment glpK | 23 |
| pCJ102/T477N | pBR322 + HindIII fragment glpK204 | This study |
| pHG165 | Cloning vector; ampr | 35 |
| pT477N | pHG165 + HindIII fragment glpK204 | This study |
CGSC, E. coli Genetic Stock Center, Yale University, New Haven, Conn.
Enzyme assays and protein determinations.
Glycerol kinase enzyme activity was measured by using the continuous ADP-coupled spectrophotometric assay at pH 7.0 and 25°C (24). Other additions are as indicated in the tables and figures. One unit of glycerol kinase catalyzes the formation of 1 μmol of ADP per min in this assay. For studies with purified glycerol kinase, the enzyme concentration was determined from the A280 (16) and varied from 0.1 to 0.5 μg/ml. Kinetic data were analyzed for initial-velocity or inhibition parameters as previously described (23). For studies with cellular extracts, the protein concentration was determined using the Bio-Rad assay with bovine serum albumin as a standard.
glpK alleles.
Alleles that encode the A-65-T (glpK203) and G-304-S (glpK22) glycerol kinases were derivatives of plasmid pHG165 (35) and have been described previously (16, 21). The allele for the T-477-N glycerol kinase (glpK204) was constructed by using the Kunkel method of site-directed mutagenesis, as described previously for the construction of other site-directed variants of glycerol kinase (23). The variant enzyme was purified to >95% homogeneity and characterized in vitro by using previously described protocols (23). The catalytic properties that are reported are the average of three experiments using enzyme purified from two independent isolates of the variant enzyme. No significant differences were found in the properties of the different isolates.
Integration of variant genes into the genome.
The glpK alleles were moved from the pHG165-derived plasmids into the chromosome of strain KH10 by conjugation. Strain KH5, transformed with a plasmid carrying the glpK allele, was mated with strain KH10 (18). Exconjugants with a functional glpK gene were selected on minimal-glycerol plates with streptomycin. Individual colonies were purified on selection medium several times. Purified colonies were screened on LB and LB-AMP plates to identify AMPs exconjugants to ensure loss of the plasmid. AMPs exconjugants were plated on MacConkey glycerol agar to screen for those able to ferment glycerol, i.e., those putatively bearing a chromosomal copy of a functional allele. Strains bearing mutations in glpK were identified by determining the glycerol kinase regulatory phenotype with enzyme activity and inhibition assays of cell extracts prepared as follows. AMPs colonies that fermented glycerol on MacConkey glycerol agar were grown overnight in 2 ml of LB, 1.5 ml of the culture was transferred to a microcentrifuge tube, and the cells were pelleted by microcentrifugation. The cell pellet was resuspended in the same volume of standard buffer (0.1 M triethanolamine HCl, 2 mM glycerol, 1 mM EDTA, 1 mM β-mercaptoethanol, adjusted to pH 7.0 at room temperature by using NaOH) and disrupted by sonication. Cellular debris was removed using centrifugation, and the supernatant was transferred to a new tube. Glycerol kinase activity and the total protein concentration of the extract were determined as described above. The reported specific activities were corrected for glycerol-independent ATPase activity in the cellular extracts by subtracting the rate obtained in assays of extracts from ΔglpK strains. The correction was 0.1 to 0.3 U/mg for different strains and growth conditions. Extracts were diluted at least 50-fold into the assays, thus reducing the glycerol concentration from the extract to a negligible level. Glycerol kinase activity in the solution was then dependent on addition of glycerol to the assay. The same reaction rate was observed for the respective ΔglpK strains and for the glpK+ strains assayed without added glycerol.
Strains derived by this procedure carry glpK alleles in the chromosome in a glpR208 background and have a tetracycline marker near the glpK gene. This marker was used for P1 transduction (18) with P1vir to move the glpK alleles into the MC4100 and MG1655 genetic backgrounds. Transductants were selected on LB-tetracycline plates and screened on MacConkey glycerol agar to confirm the presence of glycerol kinase activity. The glycerol kinase regulatory phenotypes of these strains were determined as described above after overnight growth in minimal-glycerol (0.2%) medium.
Diauxic growth curves.
Strains were incubated at 37°C overnight in 4 ml of M9 minimal-glucose (0.2%) medium. Three milliliters of the overnight culture was transferred to a sterile microcentrifuge tube, and the cells were collected by centrifugation. The cell pellet was resuspended in 1 ml of 1× M9 salts and washed. The washed cells were resuspended in 1 ml of 1× M9 salts, and the optical density at 600 nm (OD600) was determined. Aliquots of these cells were used to inoculate 80 ml of M9 medium containing either 2.5 mM glucose or 2.5 mM glucose plus 5 mM glycerol to an OD600 of ∼0.02. Growth at 37°C in a rotary shaker (250 rpm) was monitored by determining the OD600. Periodically, 5-ml aliquots were removed from the growing culture and placed on ice. Cells and medium were separated by centrifugation. The cells were resuspended in 1 ml of standard buffer and disrupted with sonication. After clarification by centrifugation, the resulting cellular extracts were assayed for glycerol kinase activity and protein concentration. The concentration of glycerol in the medium was determined by using end-point assays with glycerol kinase in the ADP-coupled assay. The concentration of glucose was determined at the Texas Veterinary Medical Diagnostic Laboratory using endpoint assays with hexokinase and glucose-6-phosphate dehydrogenase in a Hitachi 911 analyzer.
Sedimentation velocity experiments.
Glycerol kinases and IIAGlc.were exhaustively dialyzed in the same beaker against 0.1 M triethanolamine-HCl (pH 7.0), which also contained 2 mM glycerol, 1 mM β-mercaptoethanol, and 10 μM ZnCl2. Protein samples were diluted with filtered dialysate to 0.3 mg of glycerol kinase per ml (5 μM [subunits]) and to 0.54 mg of IIAGlc per ml (30 μM) when present. All samples were clarified by centrifugation prior to ultracentrifugation and then loaded into cells assembled with 12-mm-optical-pathlength double-sector Epon charcoal-filled centerpieces and sapphire windows. Approximately 0.2 ml of sample and 0.25 ml of buffer were loaded into the sample and reference channels, respectively. Samples were run in a Beckman model XL-A analytical ultracentrifuge at 35,000 rpm and 25°C for 90 min. Scans were performed at 280 nm and collected without pausing, allowing 4 min to elapse between scans. Because IIAGlc lacks tryptophans or tyrosines (31), it is transparent at the 280-nm wavelength. Each run was conducted either with all cells containing glycerol kinase or with all cells containing glycerol kinase and IIAGlc, giving three independent measurements. Data were analyzed using the SVEDBERG program (version 6.38) from J. Philo (25).
Rescue of growth in lactose medium.
Strains were grown to saturation at 30°C in 5 ml of M9 minimal-glucose (0.2%) medium with ampicillin for plasmid-harboring strains. Cells from 3 ml of the cultures were collected by centrifugation in a sterile tube, then washed and resuspended in 1× M9 medium, and the OD600 was determined. Aliquots of these cells were used to inoculate 25 ml of prewarmed M9 minimal-lactose (0.2%) medium with ampicillin when appropriate, at an OD600 of 0.025. Cells were incubated in a rotary shaker (250 rpm) at 42°C, and the OD600 was determined to monitor growth.
RESULTS AND DISCUSSION
Construction and characterization of allosteric regulatory variant glycerol kinase T-477-N.
Two of the allosteric variant glycerol kinases used in these studies, A-65-T and G-304-S, were described previously, and their catalytic and regulatory properties have been determined by in vitro studies with the purified enzymes (16, 21). The T-477-N glycerol kinase was constructed, purified, and characterized as described in Materials and Methods. The substituted amino acid position is in the IIAGlc binding site on glycerol kinase and is >25 Å from the active site and >65 Å from the FBP-binding site (10, 20). Examination of the structure of the glycerol kinase-IIAGlc complex suggests that the variant T-477-N enzyme will prevent IIAGlc binding because of increased side-chain volume and polarity; the methyl group of T-477 is in a nonpolar environment. Previous work has shown that the catalytic activity of glycerol kinase is reduced by substitutions to negatively charged amino acid side chains in this region (4), so the neutral polar amino acid asparagine was used for substitution of T-477. Initial-velocity studies of the substrate dependence of the catalytic properties of the T-477-N variant enzyme yielded the following kinetic parameters for this enzyme: Vmax, 13 ± 2 U/mg; KATP, 7 ± 2 μM; Kgol, 6 ± 3 μM; and KiATP, 53 ± 45 μM. The values of these parameters were not significantly different from those obtained for the normal enzyme: Vmax, 15 ± 1 U/mg; KATP, 9 ± 2 μM; Kgol, 5 ± 2 μM; KiATP, 54 ± 23 μM. The kinetic properties showed apparent negative cooperativity with respect to ATP that was similar to the response obtained for the normal enzyme, showing that this aspect of the regulatory behavior of the enzyme was not altered qualitatively by the substitution.
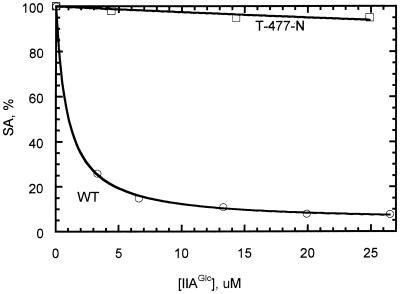
Effects of the substitution on allosteric regulation by FBP and IIAGlc were assessed by initial-velocity studies. In assays at pH 7.0, 25°C, and 0.4 μg of glycerol kinase per ml, FBP inhibition showed positive cooperativity and yielded the following parameters for the T-477-N glycerol kinase: Imax, 89 ± 1%; Hill coefficient (nH), 1.8 ± 0.1; and Kapp, 0.3 ± 0.02 mM. The values were not significantly different from the parameters obtained for the normal enzyme: Imax, 93 ± 1%; nH, 1.7 ± 0.1; Kapp, 0.26 ± 0.02 mM. Figure 1 shows IIAGlc inhibition for normal and T-477-N glycerol kinases. The highest concentration of IIAGlc that was used corresponded approximately to the total concentration of IIAGlc (phosphorylated and unphosphorylated) estimated to occur in vivo (27). Inhibition parameters of the normal glycerol kinase agreed with previous results (23). Because of the small extent of inhibition of the T-477-N glycerol kinase, the fitting algorithm could not estimate both Imax and Kapp; consequently, the value for Imax was fixed at the value obtained for normal glycerol kinase. The resultant fit to the data obtained for the T-477-N glycerol kinase showed that the substitution decreased the apparent affinity for IIAGlc binding by about 400-fold if the substitution did not affect the extent of inhibition. This result is consistent with the expected effect of increasing the size and polarity of the amino acid at this position, on the basis of the crystal structure of the complex of IIAGlc with the normal glycerol kinase (10).
FIG. 1.
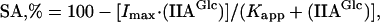 |
We utilized two methods to examine binding of IIAGlc to the T-477-N variant glycerol kinase. In the first method, sedimentation velocity measurements were performed as described in Materials and Methods to assess formation of a complex between IIAGlc and glycerol kinase. For the normal glycerol kinase, the apparent sedimentation coefficient was increased from 11 ± 0.06 S to 13.3 ± 0.06 S when 30 μM IIAGlc was added to the ultracentrifuge cell. This concentration of IIAGlc showed saturation behavior for inhibition of normal glycerol kinase in kinetics assays (Fig. 1), and the increased value of the sedimentation coefficient with IIAGlc indicated its binding to glycerol kinase. The sedimentation coefficient of the T-477-N variant glycerol kinase in the absence of IIAGlc was 10.6 ± 0.1 S and was 10.5 ± 0.06 S with 30 μM IIAGlc included in the cell. Thus, addition of IIAGlc did not alter the sedimentation coefficient of the T-477-N variant, indicating that IIAGlc does not bind to this variant under these conditions. We estimated that this result indicates a reduction of at least 200-fold for the IIAGlc binding affinity for the variant relative to the normal glycerol kinase. This result is consistent with that from the inhibition kinetics studies and supports the assumption that was made in fitting the inhibition data, namely that the substitution decreases the binding affinity but does not alter the extent of inhibition.
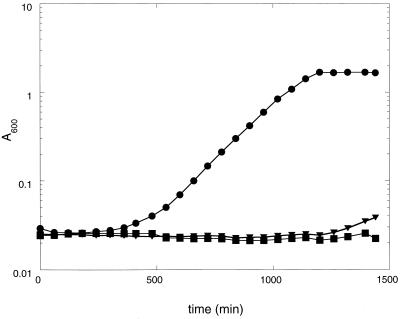
The second method provided a measure of IIAGlc binding to glycerol kinase in vivo. Strain AP110 did not grow on lactose at the restrictive temperature (42°C). This is believed to be a consequence of the ts variant enzyme I in this strain (12), resulting in inability of the PTS to phosphorylate IIAGlc to relieve its inhibition of the lactose permease. Figure 2 shows that growth of AP110 cells on lactose could be rescued by transformation with the plasmid pCJ102, which constitutively expresses the normal glycerol kinase (22). Growth of cells that harbor the plasmid vector pBR322 alone was not rescued. In contrast, growth of cells that express the T-477-N variant glycerol kinase from the same plasmid was not rescued significantly in the same time period. Analogous results were obtained on minimal-lactose plates on which just-visible colonies appeared after 5 days of growth at the restrictive temperature for AP110 cells in which the T-477-N variant glycerol kinase was expressed. Cells that contained the vector alone showed no growth after 5 days, and cells in which the normal glycerol kinase was overexpressed produced large colonies after 2 days. Addition of exogenous glycerol was not required for these rescue experiments, and the expression of the glycerol kinases did not affect discernibly the growth rate of the strains on minimal glucose (0.1%) plates at the permissive temperature (30°C). The absence of an effect on the rate of growth on glucose is consistent with simulations indicating that the amount of IIAGlc greatly exceeds the amount required for growth (28). Rescue of growth on lactose by overexpression of normal glycerol kinase is consistent with binding of the glycerol kinase to the unphosphorylated form of IIAGlc, thus titrating the inhibitor away from the lactose permease and allowing entry of lactose into the cell. Both of the glycerol kinases were greatly overexpressed constitutively from the pBR322 construct, and the specific activity in crude extracts was the same for the normal and T-477-N glycerol kinases: 10 U/mg for liquid cultures that were grown overnight at 42°C in LB. Thus, the lack of efficient rescue by the variant glycerol kinase was not due to differences in the level of expression or protein stability. The very high level of glycerol kinase may account for the binding of IIAGlc in the absence of added glycerol; alternatively, ATP or sn-glycerol 3-phosphate binding to the glycerol kinase may promote IIAGlc binding. The level of specific activity was >10-fold higher than the level observed for expression from the chromosome during growth on glycerol (see below); yet, no effective rescue was observed with the T-477-N variant glycerol kinase, leading to the expectation that the level of binding to IIAGlc was even lower at one-tenth the glycerol kinase concentration.
FIG. 2.
Glycerol kinase-dependent rescue of growth on lactose. Cells of strain AP110 that harbor plasmids with different glpK alleles were inoculated into minimal-lactose medium and incubated with shaking at 42°C. Growth was monitored by determining the OD600. ●, pCJ102 (normal glycerol kinase); ▪, pBR322 (vector); ▾, pCJ102-T477N (T-477-N glycerol kinase).
Results of the sedimentation velocity experiments and the in vivo rescue experiments indicated that the binding of IIAGlc to the T-477-N variant glycerol kinase is very weak relative to the normal glycerol kinase. Thus, the T-477-N substitution practically abolished binding and inhibition by IIAGlc at these concentrations, and the T-477-N glycerol kinase provided the desired altered allosteric regulatory phenotype of greatly reduced inhibition by IIAGlc with no significant change for the catalytic or other regulatory properties of the enzyme.
Construction and characterization of mutant strains with chromosomal alleles for allosteric regulatory variant glycerol kinases.
Strains with chromosomal glpK alleles that encode the allosteric regulatory variants of glycerol kinase, T-477-N, A-65-T, and G-304-S, were constructed in the three genetic backgrounds as described under Materials and Methods. The glycerol kinase regulatory phenotype of each strain was verified by assays of enzyme activity in cell extracts. Results of those assays are summarized in Table 2. In each case, the allosteric regulatory phenotype was the same as that observed for the respective purified glycerol kinase. The extract from the strain with the normal glycerol kinase displayed inhibition of glycerol kinase by both FBP and IIAGlc (K+ glycerol kinase) that, with the putative glycerol kinase A-65-T, showed inhibition by IIAGlc but was insensitive to FBP (Ki glycerol kinase); that, with the putative glycerol kinase T-477-N, showed inhibition by FBP but was resistant to IIAGlc inhibition (Kr glycerol kinase); and that with the putative glycerol kinase G-304-S was insensitive to FBP and resistant to IIAGlc inhibition (Ki,r glycerol kinase). The designations of the regulatory defect (Ki, Kr) follow earlier nomenclature (19). The same results were obtained for all three genetic backgrounds.
TABLE 2.
Glycerol kinase regulatory phenotypesa
| Strain | FBP inhibition | IIAGlc inhibition | GlpR/GlpK phenotypeb |
|---|---|---|---|
| KH11 | 0.4 | 0.4 | R−/K+ |
| KH12 | 1.0 | 0.5 | R−/Ki |
| KH15 | 0.4 | 1.0 | R−/Kr |
| KH37 | 0.9 | 1.1 | R−/Ki,r |
| MC4100 | 0.3 | 0.2 | R+,K+ |
| KH34 | 1.1 | 0.6 | R+/Ki |
| KH24 | 0.4 | 1.1 | R+/Kr |
| KH38 | 1.1 | 1.1 | R+/Ki,r |
| MG1655 | 0.5 | 0.3 | R+/K+ |
| KH58 | 1.1 | 0.4 | R+/Ki |
| KH59 | 0.5 | 1.0 | R+/Kr |
| KH61 | 1.0 | 1.0 | R+/Ki,r |
Extracts from overnight cultures of the indicated strains were prepared, and the specific activity of glycerol kinase was determined as described in Materials and Methods. Strains KH11, KH12, KH15, and KH37 were grown in LB, and the remaining strains were grown in minimal glycerol (0.2%) medium. For evaluation of the regulatory phenotype, FBP or IIAGlc was added to the assay to a final concentration of 5 mM or 3 μM, respectively. The inhibition is expressed as the ratio of the specific activity with added inhibitor to the specific activity without the inhibitor. Specific activities for the uninhibited enzymes are shown in Table 3.
Abbreviations: K+, normal glycerol kinase; Ki, FBP-insensitive glycerol kinase; Kr, IIAGlc-resistant glycerol kinase; Ki,r, FBP-insensitive and IIAGlc-resistant glycerol kinase; R−, glpR208; R+, glpR+.
Growth rates and glycerol kinase specific activity after overnight growth in minimal glucose, minimal glycerol, and LB media were determined for all of the strains. Differences were seen for growth rates of strains KH11, MC4100, and MG1655 in each of the different media (data not shown). However, in strains with variants of glycerol kinase, the growth rate was sensitive to the allosteric regulatory phenotype of glycerol kinase only for cells expressing Ki or Ki,r glycerol kinase, for which the growth rate in glycerol was increased in all genetic backgrounds. Strains with Ki or Ki,r glycerol kinase also showed a strong fermentation phenotype on MacConkey glycerol agar in all genetic backgrounds. This phenotype was displayed as a large red disk in the agar surrounding the colonies and was seen for both plasmid-borne and chromosomal copies of these alleles. The phenotype has segregated with the mutations following conjugation or transduction in all the strains that have been examined. Strains with Kr glycerol kinase showed a normal fermentation phenotype.
Table 3 shows the specific activity of glycerol kinase in extracts prepared after overnight growth in several media. In glucose-grown cells, the apparent levels of enzyme activity were not significantly different from the apparent activity obtained for the respective ΔglpK strains. This apparent activity was not dependent on addition of glycerol to the assay and thus reflected sources of ADP other than glycerol kinase. The basal level of glycerol kinase specific activity in glucose-grown cells was too low to be measured by using the ADP-coupled assay (<0.1 U/mg). Glycerol kinase enzyme activity was repressed by glucose in both glpR208 and glpR+ strains, in agreement with an earlier report (41).
TABLE 3.
Dependence of glycerol kinase specific activity on growth medium and genetic background
| Strain | GlpR/GlpK phenotype | Avg sp act ± SD (U/mg)a on growth medium (no. determinations)
|
||
|---|---|---|---|---|
| Minimal glucose | Minimal glycerol | LB medium | ||
| KH11 | R−/K+ | 0.1 ± 0.2 (4) | 0.8 ± 0.4 (6) | 2.1 ± 0.6 (9) |
| KH12 | R−/Ki | 0.1 ± 0.1 (4) | 0.2 ± 0.1 (6) | 2.1 ± 0.9 (7) |
| KH15 | R−/Kr | 0 ± 0.02 (4) | 0.7 ± 0.1 (6) | 1.9 ± 0.6 (6) |
| KH37 | R−/Ki,r | 0 ± 0.1 (4) | 0.2 ± 0.1 (6) | 1.2 ± 0.3 (7) |
| MC4100 | R+/K+ | 0 ± 0.1 (5) | 1.1 ± 0.7 (8) | 0.1 ± 0.2 (7) |
| KH34 | R+/Ki | 0 ± 0.1 (5) | 0.2 ± 0.1 (7) | 0 ± 0.02 (3) |
| KH24 | R+/Kr | 0 ± 0.1 (5) | 1.3 ± 0.7 (7) | 0 ± 0.2 (3) |
| KH38 | R+/Ki,r | 0 ± 0.1 (4) | 0.1 ± 0.1 (5) | 0 ± 0.2 (4) |
| MG1655 | R+/K+ | 0 ± 0.04 (6) | 1.0 ± 0.2 (7) | 0 ± 0.1 (5) |
| KH58 | R+/Ki | 0 ± 0.1 (6) | 0.2 ± 0.1 (3) | 0 ± 0.1 (3) |
| KH59 | R+/Kr | 0 ± 0.1 (5) | 0.6 ± 0.3 (4) | 0 ± 0.04 (3) |
| KH61 | R+/Ki,r | 0 ± 0.04 (5) | 0.2 ± 0.1 (6) | 0 ± 0.02 (3) |
Specific activities were determined as described in Materials and Methods. Numbers in parenthesis indicate the number of independent determinations, and the uncertainties are shown as the sample standard deviation. The values shown were corrected by subtracting the apparent specific activities obtained for the respective ΔglpK strains: KH18, 0.3 ± 0.1 U/mg; KH10 and KH51, 0.2 ± 0.1 U/mg. The apparent specific activities obtained with the ΔgkpK strains are not dependent on the addition of glycerol to the enzyme assay; i.e., they are not glycerol kinase activity but reflect other sources of ADP that are measured by the coupled assay.
The level of enzyme activity of glycerol kinase in cells that were grown on glycerol showed little dependence on the glycerol 3-phosphate repressor phenotype; the levels were the same for KH11, MC4100, and MG1655. However, the expression level did depend on the glycerol kinase regulatory phenotype. The level was reduced in strains with Ki or Ki,r glycerol kinases but not in strains with K+ or Kr glycerol kinases. Despite this lower level of glycerol kinase activity, the strains with Ki or Ki,r glycerol kinases grew more rapidly on glycerol and displayed the enhanced glycerol fermentation phenotype on MacConkey glycerol plates.
In LB, the glycerol kinase specific activity level showed little dependence on the glycerol kinase regulatory phenotype but was dependent on the glycerol 3-phosphate repressor phenotype. In the glpR208 strains, high levels of glycerol kinase specific activity were obtained. The levels of specific activity were considerably reduced for the glpR+ strains and were not significantly above the apparent level observed for the respective ΔglpK strains. The low levels of expression of the Ki or Ki,r glycerol kinases in minimal glycerol medium did not appear to be related to instability of the enzymes or inherent differences in expressibility because the same high level of specific activity was seen for the normal and variant glycerol kinases in the glpR208 genetic background during growth on LB.
The glpK22 allele was used in these experiments to verify that the effects of Ki,r glycerol kinase on glucose control were not changed by the different genetic backgrounds used here relative to the initial report of those effects (41). The properties of the strains which expressed this variant that were constructed in this work, including the reduced level of glycerol kinase specific activity following growth in minimal glycerol and the glpR allele dependence of the level of glycerol kinase specific activity during growth on LB, agree with those properties described in the earlier work. Diauxic growth curves (data not shown) also agreed with earlier work: a normal plateau for a glpR+ strain and elimination of the plateau in a glpR strain. Thus, the effects of the glpK22 allele on mechanisms of glucose control in strains used here were not distinguishable from the effects that were observed earlier in a different strain, and the present strains provided suitable genetic backgrounds for assessing the contribution of each of the glycerol kinase allosteric control mechanisms to regulation of glycerol metabolism.
Roles of glycerol kinase allosteric regulation in glucose control of glycerol utilization and glycerol kinase expression.
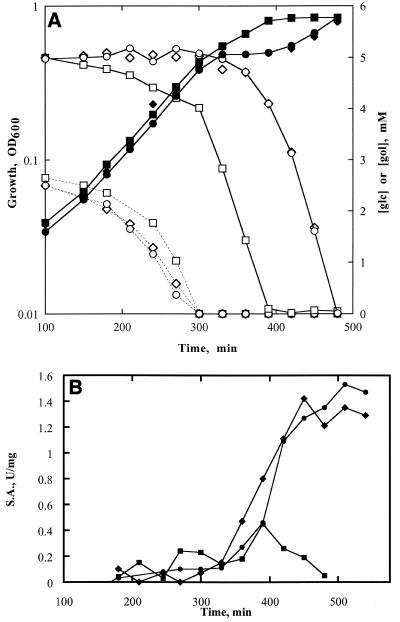
The fundamental observation of glucose control of utilization of other carbon sources is diauxic growth, which is exhibited as a biphasic growth curve. Figure 3A displays growth curves obtained in minimal medium with 2.5 mM glucose plus 5 mM glycerol for strain MC4100. This growth showed the expected two phases, which are separated by a plateau. Figure 3A also shows the basis for the diauxic growth. During the first phase, glucose is utilized while glycerol utilization is inhibited. During the second growth phase, which occurred after the glucose was consumed, glycerol was utilized. Figure 3B shows that the level of glycerol kinase specific activity was barely detectable during the first phase and was induced to higher levels for the phase of glycerol utilization. Thus, glucose prevented both utilization of glycerol and expression of glycerol kinase.
FIG. 3.
Roles of glycerol kinase allosteric regulation in diauxic growth, glucose-glycerol utilization, and expression of glycerol kinase in the MC4100 genetic background. Cultures were prepared as described in Materials and Methods. (A) Filled symbols show growth of strains 34 (R+/Ki) (▪), 24 (R+/Kr) (♦), and MC4100 (R+/K+) (●), which were measured as the change in OD600 following inoculation. Open symbols connected by dashed lines show the concentration of glucose, and open symbols connected by solid lines show the concentration of glycerol in the medium. (B) Specific activity (S.A.) of glycerol kinase. ●, MC4100 (R+/K+); ▪, strain 34 (R+/Ki); ♦, strain 24 (R+/Kr).
The consequences of the specific changes in the allosteric regulatory properties of glycerol kinase for glucose control of glycerol utilization in derivatives of strain MC4100 also are shown in Fig. 3A and B. The curves for strain 24 (R+/Kr) were practically identical to those obtained for strain MC4100 (R+/K+). Thus, loss of IIAGlc inhibition of glycerol kinase, i.e., inducer exclusion, did not affect the diauxic growth, glycerol utilization, or expression of glycerol kinase under these conditions. In contrast, the curves obtained for strain 34 (R+/Ki) differed greatly from those of strain MC4100. Loss of FBP inhibition of glycerol kinase affected all three aspects of glucose control: (i) the plateau in the growth curve was eliminated, (ii) glycerol was consumed throughout the growth and the rate of glucose utilization during the first phase was reduced, and (iii) glycerol kinase specific activity remained at a low level throughout the growth. Thus, FBP inhibition was the quantitatively dominant allosteric regulatory mechanism for control of glycerol kinase in vivo in E. coli. A dominant role for FBP inhibition was consistent with the observation that 16 of 18 glycerol-specific revertants of a ptsI strain expressed glycerol kinase that was no longer inhibited by FBP (2). For the other two genetic backgrounds, the effects of the altered glycerol kinase allosteric regulatory properties on the diauxic growth curves, carbon source utilization, and glycerol kinase specific activity were the same as those shown for the MC4100 genetic background (data not shown). Thus, the role of allosteric regulation of glycerol kinase was not dependent on the genetic background.
For each of the strains described here, growth rates and glycerol kinase specific activities in minimal glucose (2.5 mM) medium alone and the first phase in minimal glucose (2.5 mM) plus glycerol (5 mM) medium were indistinguishable (data not shown). Thus, growth on glucose was not affected by the different glycerol kinase regulatory phenotypes, suggesting that the PTS was not altered. The behaviors of strains MG1655 and KH52 with respect to growth rates, diauxic growth, and glycerol kinase specific activities were identical; thus the presence of the Tn10 element did not affect the properties of the strains under these conditions.
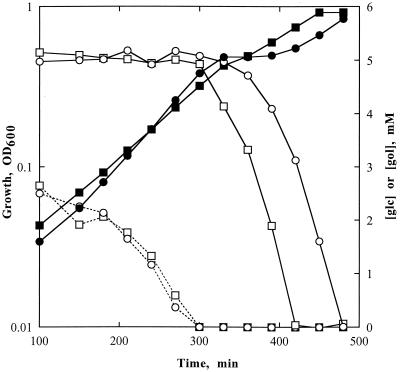
The unexpected lack of effect of loss of IIAGlc inhibition of glycerol kinase, i.e., inducer exclusion, on glucose control of glycerol utilization raises questions about the role of specific repression by the glycerol phosphate repressor. We showed previously that glucose represses expression of glycerol kinase in glpR strains, including strain KH11, during growth on glucose plus glycerol (9). Figure 4 compares diauxic growth curves and glycerol utilization for strains KH11 (R−/K+) and MC4100 (R+/K+). Diauxic growth and glycerol kinase specific activities in strains with the glpR2 or glpR208 allele are indistinguishable from those of ΔglpR strains, indicating that these alleles encode nonfunctional repressors (9). The results (Fig. 4) show that glucose control of glycerol utilization during the first phase of growth also was not dependent on a functional glycerol phosphate repressor.
FIG. 4.
Growth and glucose and glycerol utilization in the glpR208 and glpR+ genetic backgrounds. Cultures were prepared as described in Materials and Methods. The filled symbols show growth measured as the change in OD600 following inoculation. Open symbols connected by dashed lines show the concentration of glucose, and open symbols connected by solid lines show the concentration of glycerol in the medium. ○/●, MC4100 (R+/K+); □/▪, KH11 (R−/K+).
In all three genetic backgrounds, strains that expressed FBP-insensitive glycerol kinase showed loss of glucose control of glycerol utilization, diauxic growth, and glycerol kinase expression. The behavior of these strains during growth on glycerol was very similar to that of lacI (11) or lacY(S209I) (6, 8, 33) strains during growth on lactose. Allosteric inhibition of glycerol kinase by IIAGlc alone provided no discernible glucose control in the FBP-insensitive glycerol kinase strains described here. The low level of glycerol kinase in these strains might be expected to contribute to loss of IIAGlc inhibition by favoring dissociation of the glycerol kinase-IIAGlc complex. However, the initial identification of FBP-insensitive glycerol kinase was based on a screen for loss of glucose control of glycerol utilization, and the enzyme was expressed at higher levels from a plasmid-borne copy of the glpK203 allele lacking the upstream genetic control elements (16). Cells that expressed the normal glycerol kinase from the same plasmid background showed glucose control at the higher level of expression. Glucose control was abolished for the strain with FBP-insensitive glycerol kinase at even the higher level of expression, which suggests that the absence of effective IIAGlc control observed here was not due to the lower level of expression of the chromosomal copy of the glpK203 allele. The absence of effective IIAGlc control in strains with FBP-insensitive glycerol kinase could reflect synergy in the binding of FBP and IIAGlc, in which FBP enhances the binding of IIAGlc. However, results of isothermal titration calorimetry experiments are consistent with independent binding of these allosteric effectors to the normal glycerol kinase in vitro (I. Luque, D. W. Pettigrew, and E. Freire, unpublished data).
In contrast to the complete loss of glucose control of glycerol utilization that is associated with the FBP-insensitive glycerol kinase, changes in glucose control are not associated with the IIAGlc-resistant glycerol kinase under these conditions. All of the strains with the IIAGlc-resistant glycerol kinase show diauxic growth curves, inhibition of glycerol utilization, and repression of glycerol kinase specific activity that are not discernibly different from those responses in strains with the normal glycerol kinase. Glucose control of glycerol utilization and repression of glycerol kinase activity also are not dependent on a functional glycerol 3-phosphate repressor, i.e., specific repression. The lack of dependence of glucose control on IIAGlc inhibition of glycerol kinase is completely consistent with the lack of dependence on specific repression. Independence from specific repression is implicit in the earlier publications in which the glpK22 (42) and glpK203 (16) alleles were identified; in both cases, glpR strains were used, and the glpR glpK+ control strains showed normal glucose control. The consequences of this observation for PTS regulation of carbon source utilization appear to have been unrecognized previously. The lack of dependence of glucose control of glycerol utilization on specific repression suggests that cAMP-dependent catabolite repression may be the dominant mechanism of glucose repression of glycerol kinase activity levels during diauxic growth. Thus, the mechanisms by which glucose controls glycerol utilization by E. coli differ significantly from those by which it controls lactose utilization (6, 11, 36). The differences in the relative roles of catabolite repression, specific repression, and inducer exclusion in regulation of use of these two carbon sources may reflect the differences in genetic structure (operon versus regulon) and/or the differences with respect to the nature of the inducer (unusual disaccharide versus normal metabolite in lipid metabolism).
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (GM-49992 and GM-38759) and by the Texas Agricultural Experiment Station (grant H-6559). A.C.P. was supported in part by NIH Chemistry/Biology Interface Training grant T32-GM08523.
We thank Ry Young for strains and Donna Barker, Audra Boettcher, and Geneva Sampson for expert technical assistance.
REFERENCES
- 1.Baldwin T O, Berends T, Bunch T A, Holzman T F, Rausch S K, Shamansky L, Treat M L, Ziegler M M. Cloning of the luciferase structural genes from Vibrio harveyi and expression of bioluminescence in Escherichia coli. Biochemistry. 1984;23:3663–3667. doi: 10.1021/bi00311a014. [DOI] [PubMed] [Google Scholar]
- 2.Berman M, Lin E C C. Glycerol-specific revertants of a phosphoenolpyruvate phosphotransferase mutant: suppression by the desensitization of glycerol kinase to feedback inhibition. J Bacteriol. 1971;105:113–120. doi: 10.1128/jb.105.1.113-120.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donahue J L, Bownas J L, Niehaus W G, Larson T J. Purification and characterization of glpX-encoded fructose 1,6-bisphosphatase, a new enzyme of the glycerol 3-phosphate regulon of Escherichia coli. J Bacteriol. 2000;182:5624–5627. doi: 10.1128/jb.182.19.5624-5627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feese D M, Faber H R, Bystrom C E, Pettigrew D W, Remington S J. Glycerol kinase from Escherichia coli and an Ala65→Thr mutant—the crystal structures reveal conformational changes with implications for allosteric regulation. Structure. 1998;6:1407–1418. doi: 10.1016/s0969-2126(98)00140-3. [DOI] [PubMed] [Google Scholar]
- 5.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 6.Hogema B M, Arents J C, Bader R, Eijkemans K, Inada T, Aiba H, Postma P W. Inducer exclusion by glucose 6-phosphate in Escherichia coli. Mol Microbiol. 1998;28:755–765. doi: 10.1046/j.1365-2958.1998.00833.x. [DOI] [PubMed] [Google Scholar]
- 7.Hogema B M, Arents J C, Bader R, Eijkemans K, Yoshida H, Takahashi H, Alba H, Postma P W. Inducer exclusion in Escherichia coli by non-PTS substrates—the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol Microbiol. 1998;30:487–498. doi: 10.1046/j.1365-2958.1998.01053.x. [DOI] [PubMed] [Google Scholar]
- 8.Hogema B M, Arents J C, Bader R, Postma P W. Autoregulation of lactose uptake through the LacY permease by enzyme IIAGlc of the PTS in Escherichia coli K-12. Mol Microbiol. 1999;31:1825–1833. doi: 10.1046/j.1365-2958.1999.01319.x. [DOI] [PubMed] [Google Scholar]
- 9.Holtman C K, Thurlkill R, Pettigrew D W. Unexpected presence of defective glpR alleles in various strains of Escherichia coli. J Bacteriol. 2001;183:1459–1461. doi: 10.1128/JB.183.4.1459-1461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurley J H, Faber H R, Worthylake D, Meadow N D, Roseman S, Pettigrew D W, Remington S J. Structure of the regulatory complex of Escherichia coli IIIglc with glycerol kinase. Science. 1993;259:673–677. [PubMed] [Google Scholar]
- 11.Inada T, Kimata K, Aiba H. Mechanism responsible for glucose-lactose diauxie in Escherichia coli: challenge to the cAMP model. Genes Cells. 1996;1:293–301. doi: 10.1046/j.1365-2443.1996.24025.x. [DOI] [PubMed] [Google Scholar]
- 12.Jones-Mortimer M C, Kornberg H L. Genetic control of inducer exclusion by Escherichia coli. FEBS Lett. 1974;48:93–95. doi: 10.1016/0014-5793(74)81070-7. [DOI] [PubMed] [Google Scholar]
- 13.Larson T J, Cantwell J S, van Loo-Bhattacharya A T. Interaction at a distance between multiple operators controls the adjacent, divergently transcribed glpTQ-glpACB operons of Escherichia coli K-12. J Biol Chem. 1992;267:6114–6121. [PubMed] [Google Scholar]
- 14.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 307–342. [Google Scholar]
- 15.Lin E C C. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- 16.Liu W Z, Faber R, Feese M, Remington S J, Pettigrew D W. Escherichia coli glycerol kinase: role of a tetramer interface in regulation by fructose 1,6-bisphosphate and phosphotransferase system regulatory protein IIIglc. Biochemistry. 1994;33:10120–10126. doi: 10.1021/bi00199a040. [DOI] [PubMed] [Google Scholar]
- 17.Messing J. A multipurpose cloning system based on the single-stranded DNA bacteriophage M13. Recombinant DNA technical bulletin. NIH publication no. 79–99. Vol. 2. Bethesda, Md: National Institutes of Health; 1979. pp. 43–48. [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 19.Novotny J M, Frederickson W L, Waygood E B, Saier M H., Jr Allosteric regulation of glycerol kinase by enzyme IIIglc of the phosphotransferase system in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1985;162:810–816. doi: 10.1128/jb.162.2.810-816.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ormo M, Bystrom C E, Remington S J. Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate. Biochemistry. 1998;37:16565–16572. doi: 10.1021/bi981616s. [DOI] [PubMed] [Google Scholar]
- 21.Pettigrew D W, Liu W Z, Holmes C, Meadow N D, Roseman S. A single amino acid change in Escherichia coli glycerol kinase abolishes glucose control of glycerol utilization in vivo. J Bacteriol. 1996;178:2846–2852. doi: 10.1128/jb.178.10.2846-2852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettigrew D W, Ma D-P, Conrad C A, Johnson J R. Escherichia coli glycerol kinase. Cloning and sequencing of the glpK gene and the primary structure of the enzyme. J Biol Chem. 1988;263:135–139. [PubMed] [Google Scholar]
- 23.Pettigrew D W, Meadow N D, Roseman S, Remington S J. Cation-promoted association of Escherichia coli phosphocarrier protein IIAGlc with regulatory target protein glycerol kinase—substitutions of a zinc(II) ligand and implications for inducer exclusion. Biochemistry. 1998;37:4875–4883. doi: 10.1021/bi971634u. [DOI] [PubMed] [Google Scholar]
- 24.Pettigrew D W, Smith G B, Thomas K P, Dodds D C. Conserved active site aspartates and domain-domain interactions in regulatory properties of the sugar kinase superfamily. Arch Biochem Biophys. 1998;349:236–245. doi: 10.1006/abbi.1997.0444. [DOI] [PubMed] [Google Scholar]
- 25.Philo J S. An improved function for fitting sedimentation velocity data for low-molecular-weight solutes. Biophys J. 1997;72:435–444. doi: 10.1016/S0006-3495(97)78684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Postma P W, Epstein W, Schuitema A R J, Nelson S O. Interaction between IIIGlc of the phosphoenolpyruvate:sugar phosphotransferase system and glycerol kinase of Salmonella typhimurium. J Bacteriol. 1984;158:351–353. doi: 10.1128/jb.158.1.351-353.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohwer J M, Hofmeyr J-H S, Postma P W. Retro-regulation of the bacterial phosphotransferase system: a kinetic model. In: Larsson C, Påhlman I-L, Gustafsson L, editors. Biothermokinetics in the post genomic era. Proceedings of the 8th International Meeting on BioThermoKinetics. Göteborg, Sweden: Göteborg University; 1998. pp. 340–344. [Google Scholar]
- 29.Roseman S. The bacterial phosphoenolpyruvate:glycose phosphotransferase system. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms: cellular and molecular biology. Washington, D.C.: ASM Press; 1994. pp. 151–160. [Google Scholar]
- 30.Roseman S, Meadow N D. Signal transduction by the bacterial phosphotransferase system. Diauxie and the crr gene. J Biol Chem. 1990;265:2993–2996. [PubMed] [Google Scholar]
- 31.Saffen D W, Presper K A, Doering T L, Roseman S. Sugar transport by the bacterial phosphotransferase system. Molecular cloning and structural analysis of the Escherichia coli ptsH, ptsI, and crr genes. J Biol Chem. 1987;262:16241–16253. [PubMed] [Google Scholar]
- 32.Saier M H., Jr Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Microbiol Rev. 1989;53:109–120. doi: 10.1128/mr.53.1.109-120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saier M H, Jr, Straud H, Massman L S, Judice J J, Newman M J, Feucht B U. Permease-specific mutations in Salmonella typhimurium and Escherichia coli that release the glycerol, maltose, melibiose, and lactose transport systems from regulation by the phosphoenolpyruvate:sugar phosphotransferase system. J Bacteriol. 1978;133:1358–1367. doi: 10.1128/jb.133.3.1358-1367.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart G A B, Lubinsky-Mink S, Jackson C G, Cassel A, Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986;15:172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- 36.Stulke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 37.Weissenborn D L, Wittekindt N, Larson T J. Structure and regulation of the glpFK operon encoding glycerol diffusion facilitator and glycerol kinase of Escherichia coli K-12. J Biol Chem. 1992;267:6122–6131. [PubMed] [Google Scholar]
- 38.Yang B, Gerhardt S G, Larson T J. Action at a distance for glp repressor control of glpTQ transcription in Escherichia coli K-12. Mol Microbiol. 1997;24:511–521. doi: 10.1046/j.1365-2958.1997.3651733.x. [DOI] [PubMed] [Google Scholar]
- 39.Yang B, Larson T J. Multiple promoters are responsible for transcription of the glpEGR operon of Escherichia coli K-12. Biochim Biophys Acta. 1998;1396:114–126. doi: 10.1016/s0167-4781(97)00179-6. [DOI] [PubMed] [Google Scholar]
- 40.Zhao N, Oh W, Trybul D, Thrasher K S, Kingsbury T J, Larson T J. Characterization of the interaction of the glp repressor of Escherichia coli K-12 with single and tandem glp operator variants. J Bacteriol. 1994;176:2393–2397. doi: 10.1128/jb.176.8.2393-2397.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zwaig N, Kistler W S, Lin E C C. Glycerol kinase, the pacemaker for the dissimilation of glycerol in Escherichia coli. J Bacteriol. 1970;102:753–759. doi: 10.1128/jb.102.3.753-759.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zwaig N, Lin E C C. Feedback inhibition of glycerol kinase, a catabolic enzyme in Escherichia coli. Science. 1966;153:755–757. doi: 10.1126/science.153.3737.755. [DOI] [PubMed] [Google Scholar]