Significance
Intercellular proteases, including ClpXP, degrade damaged or unneeded proteins. Peptide tags allow specific protein substrates to be recognized by the ClpX unfoldase/translocase component of ClpXP and by an adaptor, SspB, which tethers itself to ClpX and enhances degradation of the tagged protein by ClpXP. Our cryo-EM structure of ClpXP bound to SspB and a tagged substrate shows that SspB and ClpX simultaneously contact the degradation tag and reveal changes in the structure of ClpX and its interaction with ClpP. These structural changes appear to be a prelude to an initial ClpX translocation step that pulls the substrate away from SspB and initiates degradation by allowing substrate unfolding and further translocation of the unfolded substrate into the proteolytic chamber of ClpP.
Keywords: AAA+ proteases, tethering adaptors, cryo-EM structure, adaptor-mediated proteolysis
Abstract
Energy-dependent protein degradation by the AAA+ ClpXP protease helps maintain protein homeostasis in bacteria and eukaryotic organelles of bacterial origin. In Escherichia coli and many other proteobacteria, the SspB adaptor assists ClpXP in degrading ssrA-tagged polypeptides produced as a consequence of tmRNA-mediated ribosome rescue. By tethering these incomplete ssrA-tagged proteins to ClpXP, SspB facilitates their efficient degradation at low substrate concentrations. How this process occurs structurally is unknown. Here, we present a cryo-EM structure of the SspB adaptor bound to a GFP-ssrA substrate and to ClpXP. This structure provides evidence for simultaneous contacts of SspB and ClpX with the ssrA tag within the tethering complex, allowing direct substrate handoff concomitant with the initiation of substrate translocation. Furthermore, our structure reveals that binding of the substrate·adaptor complex induces unexpected conformational changes within the spiral structure of the AAA+ ClpX hexamer and its interaction with the ClpP tetradecamer.
Proteolytic adaptors alter the repertoire of substrates degraded by individual proteases (1). For example, in Escherichia coli, the SspB adaptor changes the proteolytic fate of ssrA-tagged proteins, which are produced when protein biosynthesis on a ribosome stalls and the tmRNA system completes translation by attaching an ssrA tag to the C terminus of the partial protein (2, 3). Translation can stall at many different codons in any protein-coding segment of the E. coli genome, and thus, ssrA-tagged proteins are highly diverse in terms of sequence, size, and structure (4). Importantly, the ssrA tag serves as a degron for the cytoplasmic AAA+ ClpXP and ClpAP proteases (5). Thus, any ssrA-tagged protein in the E. coli cytoplasm can potentially be degraded, ridding the cell of truncated, useless, and potentially dangerous protein fragments and permitting the amino acids in these aberrant molecules to be recycled.
Each subunit of the SspB homodimer consists of a native domain, which binds to the first seven residues of the ssrA tag (AANDENYALAA), and a disordered C-terminal region, ending with a short segment that binds to the N domain of ClpX (6–10). These SspB-mediated interactions tether ssrA-tagged proteins to ClpXP and stimulate their degradation by decreasing KM and increasing Vmax (2, 10). Concurrently, SspB redirects degradation by blocking recognition and proteolysis of ssrA-tagged proteins by the ClpAP protease (11).
ClpXP consists of one or two hexamers of ClpX and the double-ring ClpP tetradecamer (12). The ClpX ring is assembled by packing of six AAA+ modules of the hexamer in a shallow spiral around an axial channel that serves as a conduit to the degradation chamber of ClpP (13–15). ClpXΔN·ClpP, which lacks the SspB-binding N-terminal domain of ClpX, is active in degradation of some protein substrates (16, 17), including ssrA-tagged proteins, and has been used for most prior cryo-EM studies (13–15). Early biochemical experiments established that positively charged “RKH” loops that reside at the top of the ClpXP axial channel aid in initial ssrA tag recruitment (18). Following recruitment, the C-terminal ALAA residues of the ssrA tag bind in the upper portion of a closed ClpX channel, as shown in a “recognition-complex” structure (13). Subsequent channel opening is then needed to allow tag translocation and eventual substrate unfolding. Following this multistep ssrA-tag recognition, ClpX or ClpXΔN uses translocation steps powered by ATP hydrolysis to unfold any native structure present in the substrate and to spool the unfolded polypeptide through the axial channel and into ClpP for degradation (12, 19).
The structural manner in which SspB interacts with ClpXP and stimulates substrate degradation is poorly understood. For example, when ClpX-tethered SspB releases the ssrA-tagged substrate relative to tag recruitment, tag recognition, and pore opening of the ClpX channel is unclear. Moreover, the molecular mechanisms by which adaptors stimulate degradation by AAA+ proteases are understudied, with only the minimal requirements having been established (20, 21).
Here, we present a cryo-EM structure of a ClpXP·GFP-ssrA·SspB complex (hereafter protease·substrate·adaptor complex) that demonstrates that SspB directly positions the C-terminal residues of the ssrA tag in the upper part of a closed axial channel of ClpX, almost exactly as in an adaptor-free recognition complex (13). These simultaneous contacts between SspB, ClpX, and the ssrA tag allow direct hand-off of the substrate from the adaptor to the protease. Surprisingly, we also find that binding of the substrate·adaptor causes a substantial alteration in the spiral arrangement of ClpX subunits and how they interact with ClpP. We discuss the implications of this latter finding for the mechanism by which ClpXP and other AAA+ proteases engage protein substrates as a prelude to ATP-fueled unfolding and eventual degradation.
Results and Discussion
Structure of the Protease·Substrate·Adaptor Complex.
We combined E. coli ClpX, ClpP, SspB, and Aequorea victoria green-fluorescent protein with an appended E. coli ssrA tag (GFP-ssrA) in the presence of ATPγS and determined a cryo-EM structure of the complex (SI Appendix, Figs. S1–S3 and Table S1). ATPγS supports assembly of ClpXP and binding to SspB and GFP-ssrA but is hydrolyzed too slowly to support GFP-ssrA unfolding and degradation (22, 23).
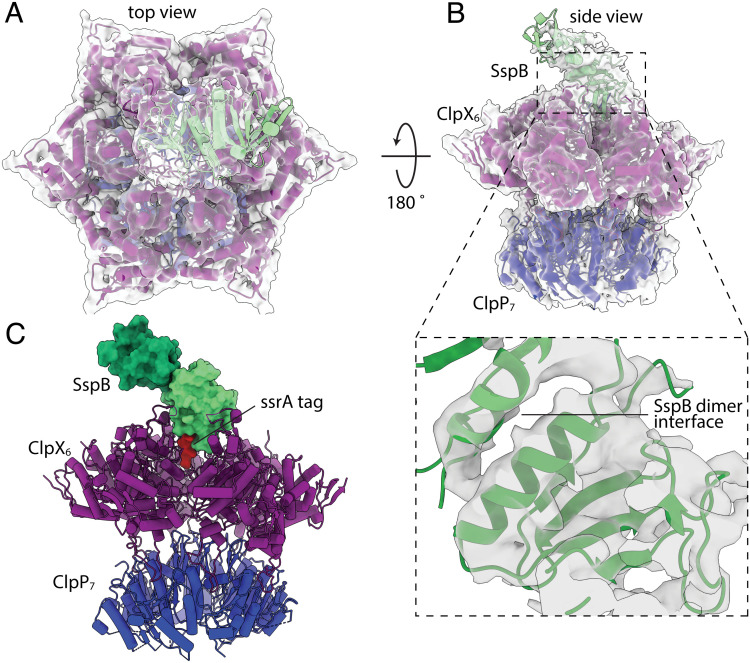
In our cryo-EM structure, we observed density for the large and small AAA+ domains of ClpX, for the ssrA tag, for both heptameric rings of ClpP, and for the two native domains of SspB, although density for the one proximal to ClpX was better resolved (Fig. 1). By contrast, the ClpX N domains, the C-terminal residues of SspB, and the native barrel of GFP were not visible in the density map, presumably as a consequence of these structures occupying multiple orientations given that the ClpX N domains are flexibly joined to the AAA+ modules and a short unstructured region separates native GFP from the ssrA tag. ATPγS or ADP was bound at the interfaces between the large and small AAA+ domains of all six ClpX subunits.
Fig. 1.
Protease·substrate·adaptor cryo-EM density map and atomic model. Overlay of density map (gray) and model (cartoon representation) of the protease·substrate·adaptor structure in top (A) and side (B) views. The Inset in panel B depicts the map and model for the proximal SspB subunit and dimer interface. (C) Atomic model of the complex. The native domains of the proximal and distal SspB subunits are shown in surface representation and colored light and dark green, respectively. The ssrA tag is shown in surface representation and colored red. ClpX is shown in cartoon representation and colored purple. The heptameric ring of ClpP nearest ClpX is shown in cartoon representation and colored blue.
Protease and Adaptor Simultaneously Contact Substrate during Delivery.
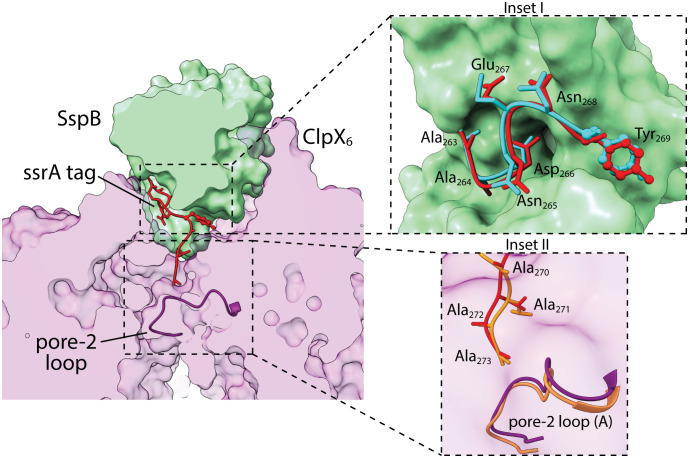
In our protease·substrate·adaptor structure, the SspB subunit proximal to ClpX positioned the ssrA tag to make bridging interactions with the top portion of a closed axial channel of ClpX (Fig. 2). This bridging geometry in the protease·substrate·adaptor delivery complex would allow subsequent opening of the axial channel and complete transfer of the substrate to the protease. For example, an ATP-fueled ClpX translocation step could open the channel by pulling the ssrA tag deeper into the pore. Such a motion could simultaneously release the substrate from the SspB adaptor, which because of its bulk could not enter the channel. Subsequent translocation steps would then result in substrate unfolding and degradation.
Fig. 2.
SspB·ssrA and ClpX·ssrA contacts. Substrate delivery complex clipped in plane to highlight contacts between the proximal subunit of SspB (green surface representation), the ssrA tag (red stick or ball-and-stick representation), and ClpX (light purple; surface representation). The pore-2 loop (dark purple backbone representation) of chain A of ClpX blocks the axial channel and contacts the C terminus of the ssrA tag. Inset I highlights SspB contacts with the ssrA tag. The conformation of an ssrA tag from a crystal structure (pdb 1OX9) bound to SspB (cyan stick or ball-and-stick representation) is also shown. Inset II depicts contacts between ClpX and the C-terminal residues of the ssrA tag overlaid with models of the same tag residues and pore-2 loop (orange) from the recognition complex (pdb 6WRF).
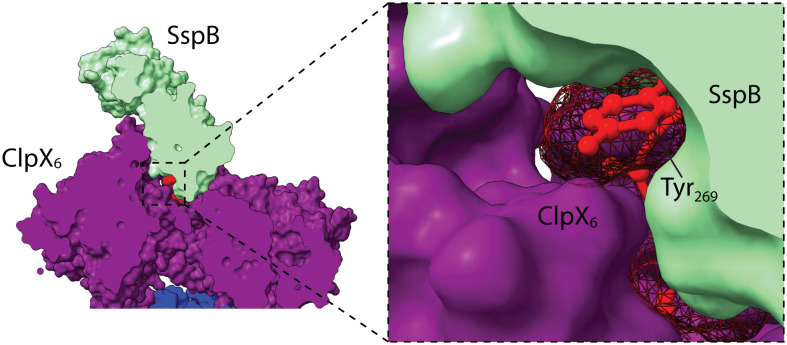
The seven N-terminal residues of the ssrA tag in our cryo-EM structure had essentially the same conformation and contacted SspB in the same manner observed previously in an ssrA·SspB crystal structure (6) (Fig. 2). Moreover, the four C-terminal residues of the tag were positioned similarly in our current structure and in the recognition-complex structure (13), further supporting a bridging followed by a handoff mechanism. In both the new structure and recognition complex, the pore-2 loop of ClpX subunit A closed the axial channel and contacted the C terminus of the ssrA tag (Fig. 2). Mutation of the pore-2 loop weakens ClpXP affinity for a cross-linked ssrA-SspB complex more than 60-fold (24), and thus, the contacts between the pore-2 loop and the ssrA tag in our structure make substantial contributions to the thermodynamic stability of the protease·substrate·adaptor complex. SspB and ClpX also contacted opposite faces of the aromatic ring of the single tyrosine in the ssrA tag in the structure (Fig. 3), emphasizing the very tight packing in this region. This close packing may result in minor steric or electrostatic clashes and could explain why inserting several residues in this region of the tag strengthens ssrA·SspB binding to ClpX (25).
Fig. 3.
SspB–ssrA–ClpX interface. SspB and ClpX contact opposite sides of the aromatic ring of the tyrosine in the ssrA tag, which is shown in red ball-and-stick representation with a mesh surface.
RKH Loops Contact SspB but Provide Little Stabilization.
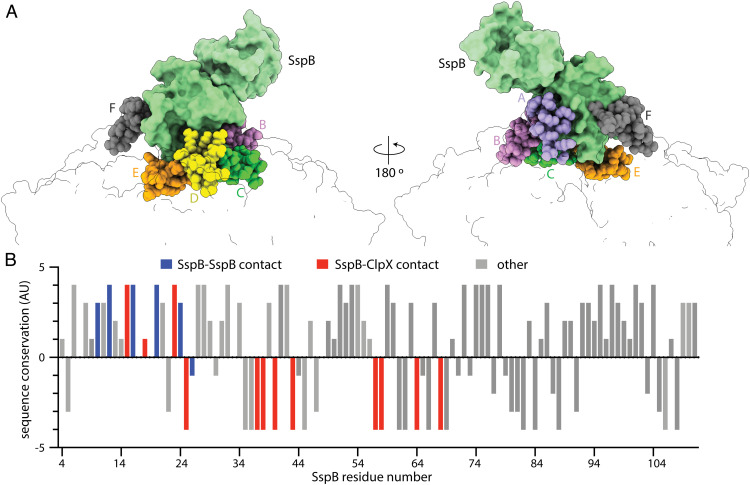
ClpX has the axial pore-1 and pore-2 loops common to all proteolytic AAA+ enzymes but also contains family-specific RKH loops, which surround the entrance to the translocation channel and play roles in substrate and adaptor binding and specificity (12, 13, 15, 24). In our structure, multiple ClpX RKH loops contacted the proximal SspB subunit (Fig. 4), as anticipated by cross-linking results (24).
Fig. 4.
SspB contacts with ClpX RKH loops are poorly conserved. (A) The folded domain of SspB proximal to ClpX (green surface representation) is contacted by six RKH loops (sphere representation colored by subunit identity). (B) Conservation score of SspB residues calculated by Consurf (https://consurf.tau.ac.il/consurf_index.php) plotted against sequence position. Positive values indicate higher sequence conservation. Residues classified and bars colored based on SspB residue contacts. Gray bars (other) note residues lacking ClpX or SspB interdimer contacts. For this analysis, SspB residues within 5 Å from interacting molecules were considered as contacts.
Interestingly, however, our covariation analysis using RaptorX (26) or GREMLIN (27) showed no significant mutual-sequence information between ClpX and SspB. Moreover, only the dimer interface and ssrA-binding groove of SspB showed high levels of sequence conservation, whereas most residues contacted by ClpX in our structure were poorly conserved (Fig. 4). These findings and the very low affinity of tail-less SspB dimers for ClpX (28) suggest that the interactions between the RKH loops and SspB are roughly neutral with respect to free energy, with favorable interactions canceled by unfavorable interactions or entropic costs. By this model, SspB binding is stabilized mainly by its tethering to the N domains of ClpX and by the bridging of the ssrA-tag between SspB and ClpX. Relatively weak binding of substrate-free SspB to ClpX could be biologically important in avoiding competition between substrate-free SspB adaptors and the degradation of substrates without ssrA tags that need unrestricted access to the ClpX channel.
Conformational Changes in ClpXP Are Induced by Substrate·SspB Binding.
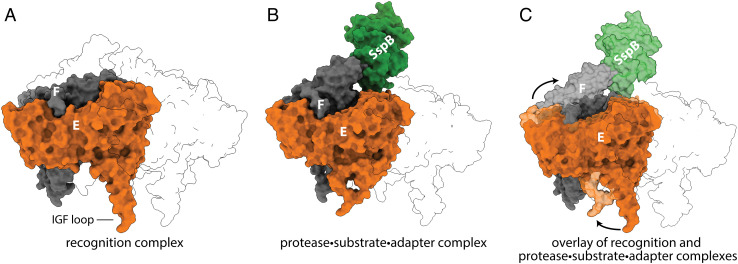
In our protease·substrate·adaptor complex, the large AAA+ domains of ClpX subunits A, B, C, D, and E formed a shallow spiral, as observed in previous ClpXP structures (13–15). Notably, however, the large AAA+ domain of subunit F moved out of the spiral and up toward SspB, as a consequence of a rotation relative to its small AAA+ domain (Fig. 5). This movement of subunit F was accompanied by smaller movements of subunit E and a minor shift in subunit A. Movies S1 and S2 show morphs between the protease·substrate·adaptor complex and recognition complex viewed from different perspectives.
Fig. 5.
Structural changes in ClpX caused by the binding of GFP-ssrA·SspB. (A) ClpX subunits F (gray) and E (orange) are shown in the recognition complex (pdb 6WRF) with other subunits in transparent outline. (B) Positions of subunits F and E and SspB (green) in the protease·substrate·adaptor structure. (C) Overlay of the recognition complex in darker shades with the protease·substrate·adaptor complex in lighter shades highlights the upward movement of subunit F and leftward movement of subunit E.
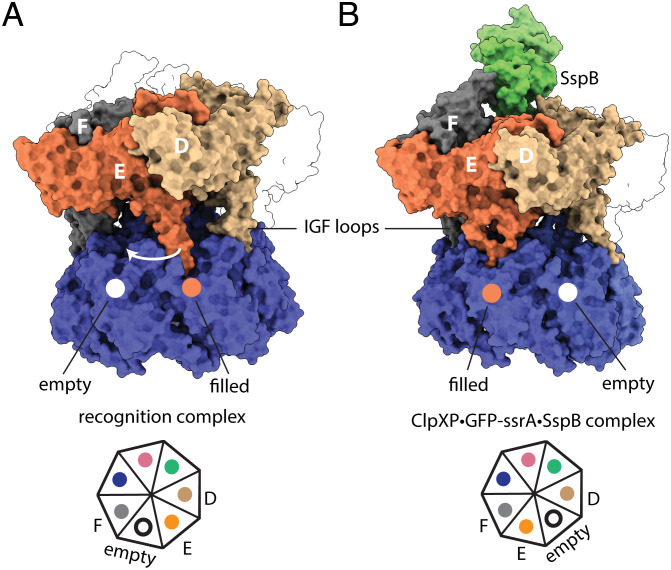
These movements of ClpX subunits in the protease·substrate·adaptor complex resulted in changes in the contacts between ClpX and ClpP. The six IGF loops of a ClpX hexamer fit into six of the seven docking clefts in a heptameric ring of ClpP, leaving one cleft empty. In prior structures of E. coli ClpXP (13, 14), this empty cleft was located between the IGF loops of ClpX subunits E and F. In our protease·substrate·adaptor structure, by contrast, the empty cleft was located between the IGF loops of subunits D and E (Fig. 6).
Fig. 6.
Substrate·adaptor mediated movement of IGF loops. (A) The empty ClpP cleft is located between the IGF loops of subunits E and F in the recognition complex (pdb 6WRF). (B) In the protease·substrate·adaptor complex, the empty cleft was located between ClpX subunits E and D.
Does the ClpX conformation observed in our protease·substrate·adaptor structure have a specialized function? Biophysical experiments indicate that ClpXP adopts a kinetic state distinct from a translocation complex state when it tries to unfold a substrate (29). Thus, the ClpX conformation in our protease·substrate·adaptor complex might correspond to a state capable of applying an efficient unfolding force. Such a conformation could help ClpX to resist the equal-and-opposite force imposed by a distorted substrate during a denaturation attempt and thus improve the chances of successful unfolding. If this surmise is correct, then similar conformations of the ClpX hexamer should be observed in SspB-free but substrate-engaged structures, where “substrate-engaged” is defined as a state in which one additional translocation step would result in substrate denaturation.
Summary.
Our protease·substrate·adaptor structure provides a glimpse of a critical step in SspB delivery of ssrA-tagged substrates for ClpXP degradation and reveals a unique conformation of the ClpX hexamer. It also provides an explanation for the ability of SspB to lower KM for ClpXP degradation of ssrA-tagged substrates, namely that ClpX·ssrA contacts are augmented by interactions between ssrA·SspB and SspB·ClpX to increase affinity.
Materials and Methods
Protein Expression Purification.
E. coli ClpP, E. coli ClpX (containing a neutral K408E mutation), and E. coli SspB were expressed and purified as reported (30). A. victoria GFP-ssrA was expressed and purified as described (31).
Cryo-EM Sample Preparation and Analysis.
For cryo-EM sample preparation, ClpX6 (5.4 μM), ClpP14 (1.8 μM), GFP-ssrA (10 μM), and SspB2 (10 μM) were incubated with ATPγS (5 mM) in 20 mM HEPES (pH 7.5), 20 mM KCl, 5 mM MgCl2, 10% glycerol, and 0.032% Nonidet P40. These protein ratios ensured that ClpP was bound by a single ClpX hexamer with sufficient SspB and GFP-ssrA to saturate ClpX. The sample was plunge-frozen on 300-mesh Quantifoil copper grids, which had been glow-discharged for 60 s in an easiGlow glow discharger (Pelco) at 15 mA and blotted using a Vitrobot Mk IV instrument (Thermo Fisher Scientific) for 3 s with a blot force of 10 (25 °C; 100% relative humidity). For the ClpXP·GFP-ssrA·SspB structure, 9,520 movies were collected at a magnification of 105,000 X and detected in superresolution mode on a Gatan K3 detector for an effective pixel size of 0.435 Å (superresolution mode) with EPU (Thermo Fisher Scientific) on a Titan Krios G3i (Thermo Fisher Scientific), operating at an acceleration voltage of 300 kV. Movies were collected as 30 frames with a total exposure on specimen of 75.98 e−/Å2. Defocus ranged from −1.0 to −2.5 μm.
Frames in each movie were binned (twofold), aligned, gain-corrected, and dose-weighted using MotionCor2 (32) to generate micrographs. The contrast transfer function (CTF) was estimated using CTFFIND4 (33). RELION 3.1 (34) was used for 2D/3D classification and refinement (SI Appendix, Figs. S1 and S2). After several rounds of 2D classification, particles were reextracted and combined using the join star tool in RELION, and the resulting 391,668 particles were selected for 3D reconstruction.
First, particles were subjected to a 3-class 3D classification with pose estimation using a 40-Å low-pass filtered ClpP map (EMDB: EMD-20434) as the initial model. No masks were used at this stage. The best-resolved class contained 236,728 particles. These particles underwent autorefinement without symmetry (C1), and postprocessing yielded a ClpXP map at 3.8 Å resolution. Three rounds of CTF-refinement and particle polishing followed by an additional round of 3D autorefinement improved the overall resolution of this map to 3.2 Å.
Next, a mask encompassing SspB, ClpX, and the cis-ring of ClpP was generated, and particle subtraction was applied to the particle stacks to focus on this region of the map. RELION reconstruct was applied to this subtracted particle stack to generate an initial model for a 3-class 3D classification with pose estimation. A final round of 3D autorefinement on the best-resolved 3D class (236,728 particles) and postprocessing produced a map that focused on SspB·ClpX·ClpPcis-ring at a resolution of 3.7 Å. This map and associated half maps were then rescaled using a calibrated pixel size (0.416 Å) that was determined by analyzing the real space correlation between publicly available apo-ferritin density maps and maps of apo-ferritin determined using this microscope under an identical imaging condition. The final rescaled maps were used to estimate local and global resolution, as well as directional resolution (35) (SI Appendix, Fig. S3).
To build atomic models, ClpX, ClpP, and SspB structures (pdb codes 6PP8, 6PPE, and 1OX9) were docked into EM maps using ChimeraX-1.3 (36). ClpX domains were rigid-body-refined using Coot (37), and real-space refinement was performed using Phenix 1.14 (38). The ssrA degron was modeled and refined using Coot and Phenix. All model building relied on the rescaled but unsharpened maps noted above.
Supplementary Material
Appendix 01 (PDF)
Morph between the protease·substrate·adaptor complex (PDB 8ET3) and recognition complex (PDB 6WRF) as viewed from subunit E of ClpX (orange cartoon representation). As SspB (green surface representation) docks with ClpX (cartoon representation and surface outline) in the protease·substrate·adaptor complex, ClpX subunit F (gray cartoon representation) moves up to contact SspB and the IGF loop of ClpX subunit E moves from one binding cleft of ClpP to the adjacent, formerly empty, cleft.
Morph between the recognition complex (PDB 6WRF) and protease·substrate·adaptor complex (PDB 8ET3) as viewed from subunit F of ClpX.
Acknowledgments
This study was supported by NIH grants R01-GM144542 and R35-GM141517 and NSF-CAREER grant 2046778. Samples were prepared at the Automated Cryogenic Electron Microscopy Facility in MIT.nano and screened on a Talos Arctica microscope, which was a gift from the Arnold and Mabel Beckman Foundation. We thank Ed Brignole and Sora Kim for their advice and assistance.
Author contributions
A.G., X.F., T.A.B., J.H.D., and R.T.S. designed research; A.G., X.F., J.H.D., and R.T.S. performed research; A.G., X.F., J.H.D., and R.T.S. analyzed data; T.A.B., J.H.D., and R.T.S. supervised research; and A.G., J.H.D., and R.T.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: D.N.B., University of Massachusetts Medical School; and H.K.S., Korea University.
Contributor Information
Joseph H. Davis, Email: jhdavis@mit.edu.
Robert T. Sauer, Email: bobsauer@mit.edu.
Data, Materials, and Software Availability
The structure has been deposited in the Protein Data Base (PDB entry 8ET3), and the cryo-EM density map has been deposited in the Electron Microscopy Data Base (EMBD entry 28585). The electron micrographs have been deposited in the Electron Microscopy Public Image Archive (EMPIAR entry 11349).
Supporting Information
References
- 1.Mahmoud S. A., Chien P., Regulated proteolysis in bacteria. Annu. Rev. Biochem. 87, 677–696 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levchenko I., Seidel M., Sauer R. T., Baker T. A., A specificity-enhancing factor for the ClpXP degradation machine. Science 289, 2354–2356 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Keiler K. C., Waller P. R., Sauer R. T., Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Moore S. D., Sauer R. T., The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76, 101–124 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Gottesman S., Roche E., Zhou Y., Sauer R. T., The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12, 1338–1347 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song H. K., Eck M. J., Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine. Mol. Cell 12, 75–86 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Levchenko I., Grant R. A., Wah D. A., Sauer R. T., Baker T. A., Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag. Mol. Cell 12, 365–372 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Wah D. A., et al. , Flexible linkers leash the substrate binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol. Cell 12, 355–63 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Bolon D. N., Grant R. A., Baker T. A., Sauer R. T., Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol. Cell 16, 343–350 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Chien P., Grant R. A., Sauer R. T., Baker T. A., Structure and substrate specificity of an SspB ortholog: Design implications for AAA+ adaptors. Structure 5, 1296–1305 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Flynn J. M., et al. , Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U.S.A. 98, 10584–10589 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker T. A., Sauer R. T., ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1823, 15–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fei X., Bell T. A., Barkow S. R., Baker T. A., Sauer R. T., Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates. Elife 9, e61496 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fei X., et al. , Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate. Elife 9, e52774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ripstein Z. A., Vahidi S., Houry W. A., Rubinstein J. L., Kay L. E., A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery. Elife 9, e52158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh S. K., et al. , Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J. Biol. Chem. 276, 29420–29429 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Martin A., Baker T. A., Sauer R. T., Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature 437, 1115–1120 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Farrell C. M., Baker T. A., Sauer R. T., Altered specificity of a AAA+ protease. Mol. Cell 25, 161–166 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauer R. T., Fei X., Bell T. A., Baker T. A., Structure and function of ClpXP, a AAA+ proteolytic machine powered by probabilistic ATP hydrolysis. Crit. Rev. Biochem. Mol. Biol. 57, 188–204 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis J. H., Baker T. A., Sauer R. T., Engineering synthetic adaptors and substrates for controlled ClpXP degradation. J. Biol. Chem. 284, 21848–21855 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis J. H., Baker T. A., Sauer R. T., Small-molecule control of protein degradation using split adaptors. ACS Chem. Biol. 6, 1205–1213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wah D. A., Levchenko I., Baker T. A., Sauer R. T., Characterization of a specificity factor for an AAA+ ATPase: Assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chem. Biol. 9, 1237–1245 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Martin A., Baker T. A., Sauer R. T., Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat. Struct. Mol. Biol. 15, 139–145 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Martin A., Baker T. A., Sauer R. T., Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell 29, 441–450 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hersch G. L., Baker T. A., Sauer R. T., SspB delivery of substrates for ClpXP proteolysis probed by the design of improved degradation tags. Proc. Natl. Acad. Sci. USA 101, 12136–12141 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J., Wang S., Wang Z., Xu J., Protein contact prediction by integrating joint evolutionary coupling analysis and supervised learning. Bioinformatics 31, 3506–3513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ovchinnikov S., Kamisetty H., Baker D., Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife 3, e02030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolon D. N., Wah D. A., Hersch G. L., Baker T. A., Sauer R. T., Bivalent tethering of SspB to ClpXP is required for efficient substrate delivery: A protein-design study. Mol. Cell 13, 443–449 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Saunders R. A., Stinson B. M., Baker T. A., Sauer R. T., Multistep substrate binding and engagement by the AAA+ ClpXP protease. Proc. Natl. Acad. Sci. U.S.A. 117, 28005–28013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levchenko I., Grant R. A., Flynn J. M., Sauer R. T., Baker T. A., Versatile modes of peptide recognition by the AAA+ adaptor protein SspB. Nat. Struct. Mol. Biol. 12, 520–525 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Kim Y. I., Burton R. E., Burton B. M., Sauer R. T., Baker T. A., Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell 5, 639–648 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan Y. Z., et al. , Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. D60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. D66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Morph between the protease·substrate·adaptor complex (PDB 8ET3) and recognition complex (PDB 6WRF) as viewed from subunit E of ClpX (orange cartoon representation). As SspB (green surface representation) docks with ClpX (cartoon representation and surface outline) in the protease·substrate·adaptor complex, ClpX subunit F (gray cartoon representation) moves up to contact SspB and the IGF loop of ClpX subunit E moves from one binding cleft of ClpP to the adjacent, formerly empty, cleft.
Morph between the recognition complex (PDB 6WRF) and protease·substrate·adaptor complex (PDB 8ET3) as viewed from subunit F of ClpX.
Data Availability Statement
The structure has been deposited in the Protein Data Base (PDB entry 8ET3), and the cryo-EM density map has been deposited in the Electron Microscopy Data Base (EMBD entry 28585). The electron micrographs have been deposited in the Electron Microscopy Public Image Archive (EMPIAR entry 11349).