Abstract
Acivicin, a modified amino acid natural product, is a glutamine analog. Thus, it might interfere with metabolism by hindering glutamine transport, formation, or usage in processes such as transamidation and translation. This molecule prevented the growth of Escherichia coli in minimal medium unless the medium was supplemented with a purine or histidine, suggesting that the HisHF enzyme, a glutamine amidotransferase, was the target of acivicin action. This enzyme, purified from E. coli, was inhibited by low concentrations of acivicin. Acivicin inhibition was overcome by the presence of three distinct genetic regions when harbored on multicopy plasmids. Comprehensive transcript profiling using DNA microarrays indicated that histidine biosynthesis was the predominant process blocked by acivicin. The response to acivicin, however, was quite complex, suggesting that acivicin inhibition resonated through more than a single cellular process.
Interconnections among biochemical pathways remain an understudied question in modern biology. Currently, this problem is being addressed in several different bacterial systems. For example, Frodyma et al. (28) and Tsang et al. (77) investigated the metabolic integration (23) of vitamin synthetic pathways which are thought to have a rather low flux since cofactors are used in catalytic rather than structural quantities. We have chosen to explore metabolic integration by focusing on the action of an inhibitor, acivicin (Fig. 1), since it is believed to interact with glutamine amidotransferases (61, 62, 74, 84) and its antibacterial activity toward Bacillus subtilis is antagonized by histidine and purine nucleosides (35). These enzymes extract ammonia from glutamine prior to attaching it to an organic backbone.
FIG. 1.
Structures of some amino acids. Only R groups are depicted.
In Escherichia coli there are at least 12 distinct glutamine amidotransferases (58, 83) involved in biosynthesis, underscoring the importance of ammonia assimilation by processes in addition to transamination. Five are involved in amino acid biosynthesis: anthranilate synthase (EC 4.1.3.27, TrpE), asparagine synthase (EC 6.3.5.4, AsnB), carbamoyl phosphate synthetase (EC 6.3.5.5, CarAB [used in arginine as well as pyrimidine nucleotide production]), glutamate synthase (EC 1.4.1.13, GltBD), and imidazole glycerol phosphate (IGP) synthase (HisHF). Along with CarAB, another four (EC 6.3.4.2, PyrG, CTP synthetase; EC 2.4.2.14, PurF, glutamine 5-phospho-d-ribosyl-α-1-pyrophosphate [PRPP] amidotransferase; EC 6.3.5.3, PurL, 5'-phosphoribosyl-N-formyl glycinamide [FGAM] synthetase; and EC 6.3.4.1, GuaA, GMP synthetase) are enzymes of nucleotide biosynthesis. Two, PabAB (4-amino-4-deoxychorismate synthase, a component of the folate pathway) and NadE (EC 6.3.5.1, NAD synthetase), function in cofactor synthesis, while one, GlmS (EC 2.6.1.13), is involved in production of the cell wall precursor N-acetylglucosamine phosphate. Thus, antagonism of this enzyme family might be most revealing.
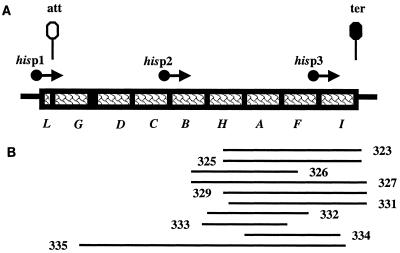
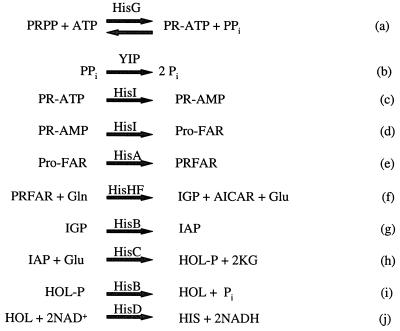
One glutamine amidotransferase, IGP synthase, is encoded by hisH and hisF; these two genes are components of the histidine operon, hisGDCBHAFI (Fig. 2A) (88). This amidotransferase occupies a central position in the eight-enzyme pathway from PRPP and ATP to histidine (Fig. 3). If this reaction or the immediately preceding HisA (pro-phosphoribosyl formimino-5-aminoimidazole-4-carboxamide ribonucleotide [PROFAR] isomerase)-catalyzed reaction is blocked, ATP is still condensed with PRPP and undergoes subsequent modification, including opening of its six-membered ring. Such blockages drain the purine nucleotide pools, effectively causing the metabolic economy to grind to a halt due to a lack of “currency,” presumably in the form of adenylates. Normally the amidotransferase reaction of the histidine biosynthetic pathway liberates 5-aminoimidazole-4-carboxamido-1-β-d-ribofuranosyl 5′-monophosphate (AICAR) as a by-product. The latter molecule, a purine biosynthetic intermediate, is salvaged in a process that leads to the resynthesis of ATP. This combined histidine-purine cycle is hence critical for cellular function, as demonstrated by the studies of Hartman et al. (36), Shedlovsky and Magasanik (70, 71), Johnston and Roth (44), and Taylor et al. (29, 42, 72, 73). Moreover, overproduction of HisHF has other deleterious consequences for cell division (3, 27, 57) independent of the above-mentioned adenylate drain. Thus, the HisHF enzyme is an attractive site for the study of metabolic integration.
FIG. 2.
(A) The histidine operon. his genes are indicated by boxes. Promoters are indicated by filled dots with arrows denoting direction of transcriptions. Sites of transcriptional termination are denoted by lollipops. (B) Plasmids that complement his point mutants, denoted by lines.
FIG. 3.
Histidine biosynthesis. Also shown is the reaction (b) catalyzed by yeast inorganic pyrophosphatase that drives reaction a to the right in a coupled in vitro system.
Due to the arrangement of the his genes within an operon (Fig. 2A) (88), it is difficult to eliminate function of an individual gene due to the polar nature of many his mutations. Furthermore, draining of adenylates by such mutants might provide a strong selective pressure for true reversion or pseudo-reversion. Hence, the ability to transiently compromise HisHF or HisA activity by the addition of a specific inhibitor is desirable. We demonstrate that acivicin has such HisHF-directed antagonism. The nutrients that prevent its inhibitory action, its specificity, and the consequences of its administration are investigated by the genetic, biochemical, and enzymological analyses of E. coli reported here.
MATERIALS AND METHODS
Abbreviations and nomenclature.
Standard bacterial nomenclature (8) is used. Biosynthetic intermediates are abbreviated as follows: PRFAR, N1-[(5′-phosphoribulosyl) formimino]-5-aminoimidazole-4-carboxamide ribonucleotide; IAP, imidazole acetol phosphate; HOL-P, l-histidinol phosphate; HOL, l-histidinol; and 2-KG, 2-ketoglutarate. Polypeptide nomenclature includes HisG (ATP phosphoribosyl transferase), the HisI (the bifunctional phosphoribosyl-ATP pyrophosphorohydrolase/phosphoribosyl-AMP cyclohydrolase), the HisH (glutamine amidotransferase) domain, the HisF (cyclase) domain, HisB (the bifunctional IGP dehydratase/HOL-P phosphatase), HisC (HOL-P aminotransferase), HisD (histidinol dehydrogenase), and YIP (yeast inorganic pyrophosphatase).
Chemicals and biochemical reagents.
Acivicin, glutamine, PRPP, and yeast inorganic pyrophosphatase were purchased from Sigma (St. Louis, Mo.). Purified E. coli HisHF enzyme (0.4 mg/ml, 7 U/mg) was a gift from V. J. Davisson, Purdue University.
Strains and plasmids.
Plasmids are described in Table 1. E. coli strains FB1 (ΔhisGDCBHAFI750) (12) and FB1/phisAGIE-tac were obtained from V. J. Davisson. The set of his operon point mutants was obtained from P. E. Hartman and has been described previously (30, 31). Salmonella enterica serovar Typhimurium Tn10 mutations were backcrossed into the wild type, selecting for tetracycline resistance as described elsewhere (20).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype | Source (reference) |
|---|---|---|
| E. coli | ||
| CS1562 | F− λ−tolC::mini-Tn10 supE42 rph-1 | C. Schnaitman (5) |
| DPD1675 | ilvB2101 ara thi Δ(proAB-lac) tolC::mini-Tn 10 | Lab strain (80) |
| DPD1692 | lac Kanr | Lab strain (17) |
| DPD1718 | lac KanrlacZ::recAφluxCDABE | Lab strain (17) |
| FB1 | ΔhisGDCBHAFI750 | V. J. Davisson (12) |
| hisG2243 | hisG2243 | PEH, JHUa (30) |
| hisG3857 | hisG3857 | PEH, JHU (30) |
| hisD921 | hisD921 | PEH, JHU (30) |
| hisC901 | hisC901 | PEH, JHU (30) |
| hisC904 | hisC904 | PEH, JHU (30) |
| hisB463 | hisB463 | PEH, JHU (30) |
| hisH4744 | F−hisH4744 nadB29 thi mtl xyl ara lac | PEH, JHU (31) |
| hisA323 | hisA323 | PEH, JHU (30) |
| hisF860 | hisF860 | PEH, JHU (30) |
| hisF891 | hisF891 | PEH, JHU (30) |
| hisI903 | hisI903 | PEH, JHU (30) |
| MG1655 | F− λ−rph-1 | D. Berg, Washington University (6) |
| RFM443 | rpsL galK2 lacΔ74 | Rolf Menzel (56) |
| S. enterica serovar Typhimurium | ||
| LT2 | S. enterica serovar Typhimurium + | K. Rudd, Miami |
| TT7542 | S. enterica serovar Typhimurium relA21::Tn10 | K. Rudd |
| TV101 | S. enterica serovar Typhimurium rfa | Lab strain (81) |
| LT2 relA | S. enterica serovar Typhimurium relA21::Tn10 | This study, P22 (TT7542) × LT2 → Tetr |
| Plasmids | ||
| pBR322 | Cloning vector | 68 |
| pDEW326 | pUC18 + his′BHAF′ | This work |
| pDEW327 | pUC18 + his′BHAFI | This work |
| pDEW335 | pUC18 + his′GDCBHAFI′ | This work |
| phisAGIE-tac | V. J. Davisson (21) | |
| pUC18 | Cloning vector | 68 |
| PVV101 | pBR322 + trpEDC′ | C. Yanofsky, Stanford University |
PEH, JHU, Philip E. Hartman, Johns Hopkins University.
Inhibition assays.
Disk diffusion was performed as described for sulfometuron methyl (47, 50), a modification of a previously described scheme (75). An alternative bioluminescent technique was also used (26). Briefly, an insertion of a recA promoter-Photorhabdus luminescens luxCDABE fusion within lacZ was crossed into strain DPD1692, selecting for kanamycin resistance. This strain, DPD1718, produces a high, baseline bioluminescence that is induced by DNA- damaging agents (82) and dampened by a wide range of metabolic inhibitors (11). Details of the construction have been described elsewhere (25). Both techniques are amenable to auxanography, a means to determine the pathway blocked by either mutation (20) or inhibitor action (47) through the supplementation with pools of nutrients. This method was used to determine those nutrients that allow metabolic function, be it growth or bioluminescence, in the presence of the inhibitor.
The ability of plasmids to alter the response to acivicin was also assayed using a bioluminescence-based protocol. Transformants (59) of strain DPD1718 harboring either pUC18 or pDEW327 were obtained by selecting for resistance to ampicillin (100 μg/ml) on Luria-Bertani plates (20). Single-colony isolates were inoculated into minimal E medium supplemented with thiamine, 0.4% glucose, and 100 μg of ampicillin per ml and incubated overnight at 37°C. Cultures were diluted into a modification of this medium that contained 50 instead of 100 μg of ampicillin per ml and shaken until they reached the exponential phase of growth. They were then exposed to acivicin in microtiter plates, and the response was monitored as a function of time using a standard method as published (79) except that the microtiter plates were incubated in a luminometer chamber maintained at 37°C. The resultant kinetic curves (data not shown) indicated that acivicin blocked luminescence shortly after its administration. This blockage was transient if the medium was supplemented appropriately or if the genetic constitution titrated out inhibitory effects. For simplicity, an endpoint, a modified response ratio (7, 79) obtained at 400 min, was calculated from the difference in luminescence obtained after treatment with a given concentration of acivicin for this time period divided by the difference in luminescence obtained with an untreated sample.
Genetic titration and complementation.
The method used has been described in general for E. coli (15). The specific plasmid libraries containing random segments of the E. coli W3110 genome have been described (26) and used (87; Z. Xue, D. R. Smulski, D. Delduco, S.-Y. Soon-Yong Choi, M. H. Jia, and R. A. LaRossa, unpublished data) elsewhere. The MIC of acivicin for strain DPD1675 was 1 μg/ml on E (20) minimal agar medium supplemented with 0.2% glucose, thiamine, and proline. Selection was for those few transformants (59) of pBR322- and pUC18-based genomic libraries of E. coli that would support growth on the above-described medium supplemented with ampicillin (100 μg/ml) and acivicin (3 μg/ml). Plasmid DNA was isolated from resistant clones, backcrossed by transformation (59) into strain DPD1675 to ascertain that resistance was a plasmid-specified phenotype, and sequenced as described previously (26, 87; Xue et al., unpublished).
Similarly, hisA and hisH mutants were transformed with the same libraries with selection for ampicillin-resistant prototrophs. In a like manner, the his-complementing plasmids were backcrossed as well as being transformed into a broad set of his point mutants. The extent of chromosomal DNA carried on each plasmid was determined by complementation of auxotrophs and sequencing the vector-chromosome junctions.
Substrate preparation.
PRFAR was synthesized using E. coli strain FB1/phisAGIE-tac based on published protocols of Davisson et al. (21) and Deras (22), with some modifications. The reaction scheme is shown in Fig. 3. PRPP (270 μmol) was reacted with excess ATP (400 μmol) in the presence of 72 U of inorganic pyrophosphatase and 9.6 mg of FB1/phisAGIE-tac extract in 86 mM potassium phosphate (pH 7.5)–28 mM MgCl2–3.5 mM EDTA. After the reaction flask was shaken at 30°C for 1 h, an additional 4.8 mg of FB1/phisAGIE-tac extract was added, and the reaction mixture was incubated for another 2 h. PRFAR was purified by applying the reaction mixture to a Q-Sepharose column (2.5 by 14 cm; Pharmacia) equilibrated with 60 mM NH4HCO3, and eluting with an NH4HCO3 gradient (60 to 300 mM over 300 ml). The fractionated eluant was analyzed by UV-visible light spectroscopy, and fractions in which A290/A260 was >1.0 were pooled and dried by lyophilization. The lyophilized product was redissolved in 0.1 M tetraethylammonium acetate (pH 7.0) and further purified by reverse-phase high-pressure liquid chromatography on a C18 column (Supelco LC18 column; preparative scale; 2.5 by 25 cm). PRFAR was eluted isocratically by 0.1 M tetraethylammonium acetate, monitored by optical density at 300 nm. Peak fractions containing PRFAR were pooled, lyophilized, and stored at −80°C. Purified PRFAR was analyzed by UV-visible light spectroscopy and gave characteristic absorbance peaks at A220 and A300.
HisHF enzyme assay.
The standard assay for E. coli HisHF (55) was performed as described elsewhere (45). The assay basis is the initial rate of substrate PRFAR disappearance as monitored by A300. Briefly 100 μM PRFAR is mixed with 5 mM glutamine in 50 mM Tris-HCl (pH 8.0), and the reaction is initiated by addition of purified HisHF at 0.02 U/ml (62 nM). The mixture was incubated at 25°C for 2 to 5 min. One unit of activity is defined as formation of 1 μmol of product per minute under the specified reaction conditions.
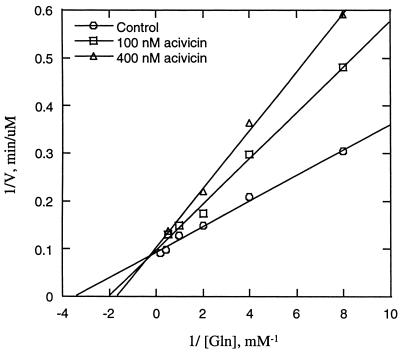
Ki determination.
To determine the inhibition constant (Ki) for acivicin, the HisHF assay was performed in the presence of 400 or 625 nM acivicin at glutamine concentrations of 0.125, 0.25, 0.5, 1, and 2 mM. The Ki for acivicin was determined from the reciprocal plot of 1/V versus 1/[glutamine].
HisHF inactivation by acivicin.
HisHF (1.0 μM) was preincubated at 25°C in 100 μl of 50 mM Tris-HCl (pH 8.0) with (i) 10 μM acivicin, (ii) 100 μM PRFAR, (iii) 10 μM acivicin and 100 μM PRFAR, and (iv) buffer only. An aliquot (12 μl) was removed at 0, 10, 20, 40, and 60 min and diluted into a reaction mixture (288 μl) containing 100 μM PRFAR, 5 mM glutamine, and 50 mM Tris-HCl (pH 8.0), and the initial rate of the reaction was measured as described above.
Gene expression profiling.
The basic dual-label, fluorescence-based method has been described in detail elsewhere (86). These experiments differed from those described previously in that the genes were spotted at a higher density, i.e., 9,000 spots/per slide, using a generation III DNA spotter (Molecular Dynamics, Sunnyvale, Calif.) such that an entire genome was spotted in duplicate on a single slide, negating the need for slide-to-slide correction. Genes were categorized into the functional groups of Riley and Labedan (67) as has been used in other transcript profiling exercises (85, 86).
RESULTS AND DISCUSSION
Nutritional supplementation.
Acivicin inhibited the growth of many E. coli K-12 and S. enterica serovar Typhimurium strains when grown on solidified minimal, but not rich, media. The presence of histidine or purines (guanine plus adenine), but not tryptophan, glutamate, or glucosamine-6-phosphate, significantly lessened the inhibition as monitored by disk diffusion assays (data not shown) with E. coli strain CS1562 (69) and S. enterica serovar Typhimurium strain TV101. Similar results were obtained when nutrient pools were used in auxanography (20) with a bioluminescent tester strain, DPD1718 (data not shown). The mixture of adenosine, histidine, phenylalanine, glutamine, and thymine was completely effective at preventing inhibition by acivicin, while supplementation with histidine, lysine, and the three branched chain amino acids was almost as effective an antidote. Other pools were incapable of preventing the inhibitory action. Histidine alone was not quite as effective an antidote as either mixture. This, together with the structural similarity between acivicin and glutamine, suggested that the target of acivicin in E. coli might be the HisHF enzyme.
These results were somewhat surprising since acivicin has been suggested to target other enzymes, notably glutamine amidotransferases, including GMP synthetase (41, 54), CTP synthetase (54), γ-glutamyl transpeptidase (38), formylglycineamidine ribonucleotide synthetase (24), and carbamoyl phosphate synthase (4), in mammalian and protozoan systems. Kis as low as 2 (4) to 5 (24) μM and as high as 420 μM (38) have been reported for these interactions.
Acivicin is a competitive inhibitor of glutamine for HisHF.
HisHF catalyzes the reaction of PRFAR and glutamine to form of IGP, AICAR, and glutamate (88). The kcat of E. coli HisHF is 8.5 s−1, the Km for glutamine is 240 μM, and the Km for PRFAR is 1.5 μM (45). We assayed the activity of HisHF in the presence of 100 or 400 nM acivicin, with glutamine concentrations ranging from 0.125 to 2 mM, while keeping PRFAR at 100 μM, a vast excess. A reciprocal plot of 1/V versus 1/[glutamine] was generated (Fig. 4), resulting in an estimate of 290 μM for the glutamine Km. This value was consistent with another determination noted above. Moreover, the Ki of acivicin was determined to be 140 nM. This indicates that acivicin is at least an order of magnitude more inhibitory in vitro toward HisHF than those enzymes that have been tested by others. Thus, acivicin was a potent inhibitor of HisHF in vitro.
FIG. 4.
Inhibition of E. coli IGP synthase (HisHF) by acivicin.
In a second experiment, HisHF activity was measured with excess (5 mM) glutamine and various concentrations of PRFAR (5, 12.5, 25, and 50 μM) in the presence or absence of 400 nM acivicin. No difference in the initial HisHF reaction rate was observed in the presence of the inhibitor (data not shown). This indicated that acivicin was a competitive inhibitor of glutamine, but not PRFAR, binding to HisHF.
The inactivation of HisHF by acivicin is accelerated by PRFAR.
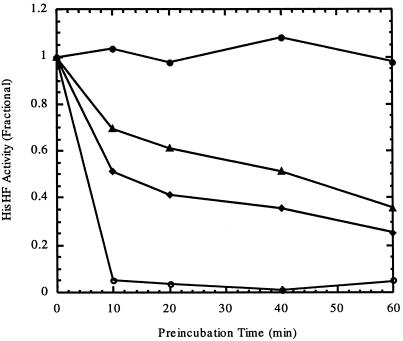
A possible mechanism of acivicin action is that it binds competitively to the glutamine binding site on HisH and inactivates the enzyme by covalent modification of an active site cysteine residue essential for glutamine amidotransferase activity. HisH and HisF of E. coli are isolated as a single heterodimer (45). It thus is possible that both the glutamine amidotransferase activity of HisH and the cyclase activity of HisF are carried out at one active site shared between the two polypeptides and that the binding of glutamine and PRFAR is cooperative. A second possibility is that separate substrate binding sites exist on the HisH and HisF polypeptides. After the glutamine amidotransferase reaction is carried out on the HisH domain, NH3 might be transferred to the PRFAR binding site on the HisF domain. To test this hypothesis, acivicin was used to probe the active site of HisHF together with PRFAR. HisHF was preincubated with 10 μM acivicin alone (1:10 ratio of enzyme to inhibitor), 100 μM PRFAR alone (1:100 ratio of enzyme to substrate), or both 10 μM acivicin and 100 μM PRFAR. The preincubation time varied from 0 to 60 min. At each time point, an aliquot of the mixture was removed and assayed for remaining activity. The fraction of residual activity was plotted versus preincubation time (Fig. 5). In the presence of either acivicin or PRFAR alone, there was a time-dependent loss of HisHF activity. This indicates that both acivicin and PRFAR were irreversible inhibitors of the reaction when incubated with HisHF alone. It is rather interesting that PRFAR inhibited the reaction irreversibly when it was added to the enzyme before the addition of glutamine. Even more interesting, when both acivicin and PRFAR were preincubated with HisHF, the enzyme activity at the first time point (10 min) decreased dramatically to almost the background level. We have not yet investigated if inactivation is more than additive. In addition, binding of PRFAR to the active site may promote covalent bond formation between glutamine and the catalytic cysteinyl residue of the HisH domain. These results were consistent with the proposed catalytic mechanism (66) and our hypothesis that the HisH domain and HisF domain share one active site that carries out both the glutamine amidotransferase and cyclase steps of the reaction. Moreover, they raise the possibility that the dominant factor contributing to the growth inhibition of a variety of microbial and eukaryotic cells could be the inactivation rates of various glutamine amidotransferases by acivicin as well as each enzyme's Ki for the small molecule.
FIG. 5.
Inactivation of E. coli IGP synthase (HisHF) by acivicin (triangles), PRFAR (diamonds), and acivicin plus PRFAR (open circles). The untreated enzyme is indicated by closed circles.
Reconstruction experiments.
Several salient features of his operon expression must be noted to aid in the interpretation of the following experiments. Three promoters (12) (Fig. 2A) are present in the operon although the primary promoter (hisp1) is more than 10-fold stronger than hisp2, which in turn is far stronger than hisp3. Furthermore, transcription from hisp1 occludes usage of hisp2 (88). Initiation at hisp1 is stimulated by the product of RelA, ppGpp (89), produced in response to starvation for any amino acid (13). Transcriptional read-through past att (43), the attenuator site, occurs when the in vivo level of histidyl-tRNAhis is low (88). Thus, the operon is subject to both global and specific regulatory circuitry.
Plasmids capable of complementing hisA and hisH mutants were found by selecting for prototrophic recombinants after transformation with genomic libraries. The vector junctions with the chromosomal inserts were determined by sequencing. These results are summarized in Fig. 2B.
The following experiments were performed with the sequenced plasmids (Fig. 2B) to corroborate the presumption that HisHF was the primary target of acivicin within E. coli. Two micrograms of acivicin created a zone of inhibition with a diameter of 38 ± 3 mm (n = 2) on the control strain. RFM443/pBR322. Strain RFM443/pUC18 yielded a zone of 34 ± 0 mm (n = 2). Strain RFM443/pDEW335 (his′GDCBHAFI′) had a greater tolerance; its zone of inhibition was 19 ± 1 mm. RFM443/pDEW327 (his′BHAFI′) was somewhat tolerant, having a zone of 27 ± 0 mm (n = 2) while RFM443/pDEW326 (his′BHAF′) was sensitive, displaying a diameter of 42 ± 2 mm (n = 2). pDEW335 (his′GDCBHAFI′) contains the nonoccluded hisp2 promoter, while the other two his plasmids lack this internal promoter. Further, pDEW326 (his′BHAF′) cannot specify an increased content of IGP synthase since only HisH and HisA polypeptides can be elevated. Thus, elevated expression of HisH, HisA, and HisF was sufficient for development of an acivicin-tolerant phenotype. Amplification of the trp control region, trpE and trpD expressing the two subunits of another glutamine amidotransferase, anthranilate synthase, did not result in tolerance; strain RFM443/pVV101 (trpEDC′) displayed a diameter of 39 ± 4 mm (n = 2). This result suggests that a glutamine amidotransferase cannot protect cells by simply acting as a macromolecular sponge (49) absorbing acivicin. Thus, amplification of the genes specifying the histidine biosynthetic glutamine amidotransferase, HisHF, conferred tolerance to the inhibitory agent.
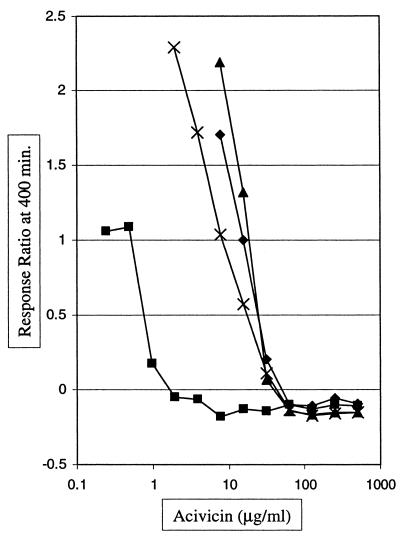
This result was further confirmed with a bioluminescence experiment (Fig. 6). A plasmid expressing only HisH, HisA, and HisF (pDEW327) and a control vector (pUC18) were placed in a bioluminescent E. coli strain, DPD1718. The bioluminescence of each recombinant was titrated with acivicin. The response ratio obtained 400 min after the administration of acivicin was plotted as a function of inhibitor concentration. As can be seen from Fig. 6, much more acivicin was needed to inhibit bioluminescence from the strain in which hisHAF was amplified, again suggesting that HisHF was the primary target of acivicin. The protection afforded by hisHAF amplification was similarly supplied by supplementation with l-histidine (Fig. 6; compare the curve of the control strain treated with l-histidine to that of the strain carrying the hisHAF amplification in the absence of l-histidine). Evidence for l-histidine supplementation enhancing the protective effect of hisHAF amplification was not obtained (Fig. 6); such effects strongly indicate that HisHF or HisA was the in vivo target of acivicin.
FIG. 6.
Titration of a bioluminescent response by acivicin. The bioluminescent host strain DPD1718 was transformed with either pUC18 or pDEW327. Squares, the strain contained pUC18 and the medium was not supplemented with histidine; diamonds, the strain contained pUC18 and the medium was supplemented with histidine; crosses, the strain contained pDEW327 (his′BHAFI) and the medium was not supplemented with histidine; triangles, the strain contained pDEW327 (his′BHAFI) and the medium was supplemented with histidine.
Genetic titration.
Libraries of random fragments of the E. coli genome in either pUC18 or pBR322, harbored in strain DPD1675, served as a source of genetic variation. Clones resistant to acivicin were selected, plasmids conferring the resistance phenotype were sequenced, and the precise locations of the resistance elements were thus determined. These resistance elements mapped to two regions distinct from his.
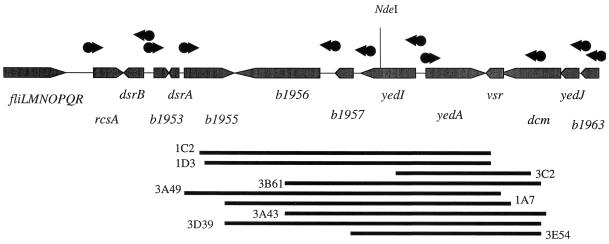
Nine plasmids mapped to one of these regions (9). Although each encompassed several genes in this region (Fig. 7), only a single gene, yedA, was present in each plasmid. The function of this gene has not yet been described, although homology to the PecM protein of Erwinia chrysanthemi (E value of 7e-19) has been noted by computational searches (1, 2). yedA is predicted to encode an integral membrane protein with nine regions spanning the lipid bilayer. It is tempting to speculate that YedA is a component of an acivicin export system.
FIG. 7.
yedA region of the E. coli chromosome. Lines below the map show the extents of various plasmids that confer acivicin resistance. Directions of transcription are indicated by arrowheads on symbols denoting genes; dots indicate promoters.
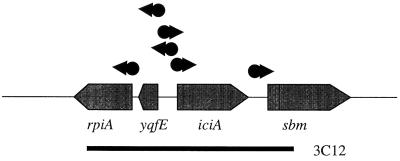
One other plasmid, mapping to a different region and containing intact sequences of iciA and yqfE, a divergently transcribed pair of genes, conferred resistance to acivicin (Fig. 8). Neither iciA nor yqfE alone resulted in an acivicin-resistant phenotype (data not shown). The function of yqfE is unknown, while those of IciA have been defined by in vivo and in vitro studies. IciA is a DNA binding protein that is a member of the LysR family of transcriptional regulators (76). It binds to a set of three double-stranded 13-mers within oriC, the origin of chromosomal replication. Such binding apparently antagonizes the creation of a bubble in the double helix; that bubble, stabilized by the specific binding to the same 13-mers in one single strand, is a prerequisite for the initiation of DNA synthesis (40). IciA is a pleiotropic regulator stimulating expression of nrd (33), dnaA (53), and perhaps other genes. Moreover, IciA in the presence of arginine inhibits its own synthesis (14). iciA transcription is also positively regulated by PhoB when phosphate is depleted (34). Further, IciA is a substrate for proteolytic degradation by the htrA-encoded protease Do (91), providing a mechanism in addition to its synthesis early in the cell cycle (93) for its periodic pattern of accumulation. Hence, amplification of a complex sensing system may confer resistance to acivicin. The relationship between yedA and the iciA region has not yet been addressed. Studies combining chromosomal mutations in one region and amplification of the second region on a multicopy plasmid may be informative. Thus, amplification of three distinct genetic regions (two, yedA and iciA-yqfE, defined by genetic titration and one, hisHAF, proven by reconstruction) gave rise to acivicin resistance.
FIG. 8.
iciA region of the E. coli chromosome, denoted as described for Fig. 7.
Gene expression profiling.
The growth of E. coli strain MG1655 in minimal medium with glucose as a carbon source was inhibited about 80% by 0.5 μg of acivicin per ml; such inhibition was completely prevented by coexposure to histidine. Histidine also reversed a greater acivicin challenge (2 μg/ml, 90% inhibition); the culture exposed to both the antagonist and the antidote grew at about 90% of the uninhibited rate (data not shown). Results from a representative DNA microarray experiment, in which E. coli MG1655 was challenged with acivicin at 2 μg/ml in minimal medium for 60 min, are presented in Tables 2 and 3. This treatment lowered the growth rate by about 85%. The structural similarity among acivicin, glutamine, and asparagine is illustrated in Fig. 1. This similarity was reflected in the gene expression profile described below.
TABLE 2.
Transcripts induced by acivicin treatment
| Gene | Location | Fold induction |
|---|---|---|
| ackA | b2296 | 2.0 |
| acrA | b0463 | 2.4 |
| acrR | b0464 | 2.6 |
| ahpC | b0605 | 3.4 |
| aldH | b1300 | 4.4 |
| amyA | b1927 | 2.3 |
| appA | b0980 | 4.3 |
| araA | b0062 | 5.5 |
| argA | b2818 | 3.6 |
| argB | b3959 | 5.7 |
| argC | b3958 | 8.5 |
| argD | b3359 | 2.8 |
| argE | b3957 | 6.4 |
| argG | b3172 | 7.7 |
| argH | b3960 | 2.2 |
| argR | b3237 | 4.0 |
| aroG | b0754 | 2.2 |
| aspC | b0928 | 7.5 |
| betA | b0311 | 2.4 |
| betB | b0312 | 2.4 |
| betI | b0313 | 4.3 |
| betT | b0314 | 8.0 |
| bfr | b3336 | 6.4 |
| blc | b4149 | 2.3 |
| cdsA | b0175 | 2.2 |
| chaA | b1216 | 2.4 |
| clpB | b2592 | 8.6 |
| cls | b1249 | 3.5 |
| csgA | b1042 | 3.2 |
| cspE | b0623 | 2.8 |
| cydA | b0733 | 2.8 |
| cynX | b0341 | 4.2 |
| dapB | b0031 | 5.2 |
| dfp | b3639 | 2.0 |
| eaeH | b0297 | 2.7 |
| entE | b0594 | 2.7 |
| fabB | b2323 | 2.9 |
| flgI | b1080 | 2.2 |
| frdB | b4153 | 2.7 |
| frdC | b4152 | 2.0 |
| frr | b0172 | 3.4 |
| ftn | b1905 | 10.6 |
| fucU | b2804 | 4.5 |
| glgS | b3049 | 3.3 |
| glpD | b3426 | 2.2 |
| grpE | b2614 | 2.5 |
| hemN | b3867 | 2.1 |
| hepA | b0059 | 4.8 |
| hisA | b2024 | 6.2 |
| hisB | b2022 | 14.3 |
| hisC | b2021 | 16.4 |
| hisD | b2020 | 11.8 |
| hisF | b2025 | 9.6 |
| hisG | b2019 | 12.1 |
| hisH | b2023 | 10.4 |
| hisI | b2026 | 13.6 |
| hisJ | b2309 | 2.8 |
| hisL | b2018 | 15.7 |
| hslS | b3686 | 34.4 |
| hslT | b3687 | 22.3 |
| htpX | b1829 | 2.6 |
| hyaF | b0977 | 2.0 |
| ilvI | b0077 | 2.4 |
| insB_1 | b0264 | 2.1 |
| intA | b2622 | 2.5 |
| kbl | b3617 | 2.1 |
| lacI | b0345 | 2.2 |
| lpdA | b0116 | 2.2 |
| melA | b4119 | 2.1 |
| melR | b4118 | 2.0 |
| menC | b2261 | 2.1 |
| metA | b4013 | 5.6 |
| metB | b3939 | 2.1 |
| metJ | b3938 | 4.7 |
| metK | b2942 | 4.3 |
| metR | b3828 | 2.8 |
| mobB | b3856 | 2.2 |
| mrdA | b0635 | 2.3 |
| mrp | b2113 | 3.4 |
| mscL | b3291 | 3.8 |
| msrA | b4219 | 2.7 |
| mtr | b3161 | 20.6 |
| murE | b0085 | 2.4 |
| nadC | b0109 | 4.2 |
| nagB | b0678 | 2.6 |
| narK | b1223 | 7.5 |
| nfo | b2159 | 3.4 |
| nfrA | b0568 | 2.2 |
| nrdD | b4238 | 7.8 |
| nrdG | b4237 | 5.6 |
| nrdH | b2673 | 3.0 |
| nrdI | b2674 | 2.2 |
| osmE | b1739 | 2.6 |
| osmY | b4376 | 3.8 |
| otsA | b1896 | 4.4 |
| otsB | b1897 | 3.4 |
| panB | b0134 | 4.4 |
| pckA | b3403 | 2.5 |
| phnB | b4107 | 7.8 |
| phnG | b4101 | 2.2 |
| proB | b0242 | 2.7 |
| pta | b2297 | 2.3 |
| ptsG | b1101 | 2.3 |
| purA | b4177 | 7.7 |
| qor | b4051 | 2.4 |
| recN | b2616 | 3.2 |
| ribE | b1662 | 2.1 |
| rna | b0611 | 4.8 |
| rob | b4396 | 2.8 |
| rpoH | b3461 | 3.2 |
| sdaA | b1814 | 3.0 |
| slp | b3506 | 3.0 |
| slyA | b1642 | 2.6 |
| slyD | b3349 | 2.1 |
| sohA | b3129 | 2.4 |
| soxS | b4062 | 2.8 |
| sseA | b2521 | 2.1 |
| sugE | b4148 | 2.5 |
| tdh | b3616 | 3.7 |
| thdF | b3706 | 2.3 |
| thrC | b0004 | 2.6 |
| thrL | b0001 | 6.6 |
| tolR | b0738 | 6.6 |
| torD | b0998 | 2.7 |
| treF | b3519 | 2.4 |
| trg | b1421 | 8.2 |
| tus | b1610 | 2.3 |
| ugpB | b3453 | 2.0 |
| umuD | b1183 | 2.2 |
| uspA | b3495 | 4.2 |
| uxaB | b1521 | 7.7 |
| xseB | b0422 | 3.2 |
| xylE | b4031 | 2.4 |
| yfiB | b2605 | 2.3 |
| ygjG | b3073 | 2.1 |
| yhaH | b3103 | 2.5 |
| yihP | b3877 | 2.2 |
| b0058 | b0058 | 4.1 |
| b0105 | b0105 | 2.8 |
| b0119 | b0119 | 2.6 |
| b0163 | b0163 | 2.1 |
| b0233 | b0233 | 2.2 |
| b0286 | b0286 | 2.2 |
| b0288 | b0288 | 2.0 |
| b0305 | b0305 | 2.2 |
| b0358 | b0358 | 2.2 |
| b0458 | b0458 | 3.5 |
| b0485 | b0485 | 2.6 |
| b0572 | b0572 | 3.5 |
| b0573 | b0573 | 2.6 |
| b0581 | b0581 | 2.7 |
| b0600 | b0600 | 2.1 |
| b0607 | b0607 | 6.8 |
| b0643 | b0643 | 5.3 |
| b0662 | b0662 | 2.2 |
| b0707 | b0707 | 3.7 |
| b0753 | b0753 | 2.5 |
| b0786 | b0786 | 2.4 |
| b0789 | b0789 | 2.5 |
| b0790 | b0790 | 2.7 |
| b0800 | b0800 | 2.7 |
| b0806 | b0806 | 2.2 |
| b0836 | b0836 | 2.9 |
| b0865 | b0865 | 7.0 |
| b0897 | b0897 | 3.1 |
| b0964 | b0964 | 2.1 |
| b0966 | b0966 | 3.8 |
| b1003 | b1003 | 2.5 |
| b1045 | b1045 | 2.6 |
| b1050 | b1050 | 4.7 |
| b1060 | b1060 | 2.5 |
| b1103 | b1103 | 3.2 |
| b1104 | b1104 | 3.1 |
| b1105 | b1105 | 3.0 |
| b1107 | b1107 | 2.5 |
| b1108 | b1108 | 6.1 |
| b1111 | b1111 | 4.2 |
| b1112 | b1112 | 3.2 |
| b1128 | b1128 | 2.2 |
| b1145 | b1145 | 2.0 |
| b1168 | b1168 | 2.1 |
| b1178 | b1178 | 2.0 |
| b1195 | b1195 | 2.2 |
| b1205 | b1205 | 5.3 |
| b1256 | b1256 | 3.5 |
| b1257 | b1257 | 3.8 |
| b1273 | b1273 | 2.0 |
| b1285 | b1285 | 3.1 |
| b1321 | b1321 | 2.7 |
| b1333 | b1333 | 3.0 |
| b1376 | b1376 | 7.0 |
| b1378 | b1378 | 2.0 |
| b1414 | b1414 | 2.2 |
| b1446 | b1446 | 3.1 |
| b1454 | b1454 | 2.3 |
| b1586 | b1586 | 6.6 |
| b1598 | b1598 | 2.1 |
| b1667 | b1667 | 3.7 |
| b1678 | b1678 | 2.0 |
| b1725 | b1725 | 2.1 |
| b1778 | b1778 | 2.5 |
| b1783 | b1783 | 2.7 |
| b1816 | b1816 | 2.1 |
| b1869 | b1869 | 2.0 |
| b1870 | b1870 | 2.2 |
| b1871 | b1871 | 2.3 |
| b1953 | b1953 | 4.7 |
| b1955 | b1955 | 3.7 |
| b2007 | b2007 | 2.3 |
| b2080 | b2080 | 7.2 |
| b2098 | b2098 | 2.4 |
| b2112 | b2112 | 2.5 |
| b2122 | b2122 | 2.5 |
| b2127 | b2127 | 4.9 |
| b2135 | b2135 | 2.1 |
| b2299 | b2299 | 3.2 |
| b2300 | b2300 | 3.2 |
| b2302 | b2302 | 3.6 |
| b2303 | b2303 | 2.8 |
| b2442 | b2442 | 2.2 |
| b2529 | b2529 | 2.1 |
| b2530 | b2530 | 2.7 |
| b2597 | b2597 | 23.6 |
| b2664 | b2664 | 3.2 |
| b2665 | b2665 | 4.7 |
| b2856 | b2856 | 2.6 |
| b2886 | b2886 | 2.1 |
| b2889 | b2889 | 2.1 |
| b2900 | b2900 | 2.0 |
| b2922 | b2922 | 2.3 |
| b2941 | b2941 | 5.0 |
| b2959 | b2959 | 3.5 |
| b2960 | b2960 | 2.6 |
| b3010 | b3010 | 2.1 |
| b3011 | b3011 | 2.1 |
| b3021 | b3021 | 2.2 |
| b3022 | b3022 | 3.1 |
| b3024 | b3024 | 3.1 |
| b3029 | b3029 | 2.1 |
| b3068 | b3068 | 4.5 |
| b3097 | b3097 | 2.6 |
| b3098 | b3098 | 2.9 |
| b3099 | b3099 | 2.2 |
| b3160 | b3160 | 2.5 |
| b3190 | b3190 | 2.8 |
| b3203 | b3203 | 2.2 |
| b3263 | b3263 | 2.9 |
| b3292 | b3292 | 2.3 |
| b3293 | b3293 | 2.5 |
| b3399 | b3399 | 2.5 |
| b3400 | b3400 | 3.4 |
| b3401 | b3401 | 2.0 |
| b3446 | b3446 | 2.7 |
| b3448 | b3448 | 4.3 |
| b3472 | b3472 | 2.5 |
| b3494 | b3494 | 3.7 |
| b3515 | b3515 | 2.7 |
| b3516 | b3516 | 2.0 |
| b3522 | b3522 | 3.5 |
| b3548 | b3548 | 3.8 |
| b3555 | b3555 | 2.3 |
| b3574 | b3574 | 2.2 |
| b3581 | b3581 | 2.8 |
| b3596 | b3596 | 4.0 |
| b3655 | b3655 | 2.6 |
| b3698 | b3698 | 2.6 |
| b3818 | b3818 | 3.1 |
| b3827 | b3827 | 2.0 |
| b3861 | b3861 | 2.1 |
| b3875 | b3875 | 2.4 |
| b3923 | b3923 | 3.7 |
| b3928 | b3928 | 2.2 |
| b3937 | b3937 | 2.7 |
| b3995 | b3995 | 2.0 |
| b4030 | b4030 | 6.7 |
| b4126 | b4126 | 2.7 |
| b4127 | b4127 | 2.3 |
| b4135 | b4135 | 3.4 |
| b4178 | b4178 | 3.8 |
| b4189 | b4189 | 2.6 |
| b4199 | b4199 | 2.8 |
| b4206 | b4206 | 2.6 |
| b4234 | b4234 | 2.0 |
| b4255 | b4255 | 2.5 |
| b4311 | b4311 | 2.0 |
| b4325 | b4325 | 2.6 |
| b4326 | b4326 | 9.4 |
TABLE 3.
Transcripts repressed by acivicin treatment
| Gene | Location | Fold induction |
|---|---|---|
| accC | b3256 | 0.45 |
| aceA | b4015 | 0.37 |
| aceK | b4016 | 0.48 |
| add | b1623 | 0.21 |
| ampE | b0111 | 0.45 |
| apaG | b0050 | 0.39 |
| appY | b0564 | 0.35 |
| apt | b0469 | 0.31 |
| aroA | b0908 | 0.20 |
| aroF | b2601 | 0.46 |
| aroH | b1704 | 0.45 |
| asd | b3433 | 0.41 |
| asnS | b0930 | 0.19 |
| asr | b1597 | 0.50 |
| atpA | b3734 | 0.44 |
| atpC | b3731 | 0.50 |
| atpD | b3732 | 0.45 |
| atpF | b3736 | 0.47 |
| avtA | b3572 | 0.48 |
| bioD | b0778 | 0.44 |
| btuR | b1270 | 0.28 |
| cfa | b1661 | 0.22 |
| cirA | b2155 | 0.03 |
| cspA | b3556 | 0.12 |
| cspC | b1823 | 0.14 |
| cybB | b1418 | 0.47 |
| cyoC | b0430 | 0.42 |
| cysA | b2422 | 0.27 |
| cysC | b2750 | 0.43 |
| cysD | b2752 | 0.48 |
| cysK | b2414 | 0.35 |
| cysM | b2421 | 0.29 |
| cysN | b2751 | 0.28 |
| cysP | b2425 | 0.42 |
| dnaG | b3066 | 0.39 |
| dppA | b3544 | 0.48 |
| dsbB | b1185 | 0.42 |
| efp | b4147 | 0.33 |
| evgA | b2369 | 0.21 |
| exbB | b3006 | 0.11 |
| exbD | b3005 | 0.10 |
| fecA | b4291 | 0.27 |
| fecI | b4293 | 0.11 |
| fecR | b4292 | 0.16 |
| fepB | b0592 | 0.47 |
| fhuE | b1102 | 0.25 |
| fimA | b4314 | 0.42 |
| fis | b3261 | 0.13 |
| fixX | b0044 | 0.49 |
| flgD | b1075 | 0.32 |
| fmu | b3289 | 0.37 |
| gadA | b3517 | 0.31 |
| gadB | b1493 | 0.41 |
| gatA | b2094 | 0.13 |
| gatB | b2093 | 0.47 |
| gatC | b2092 | 0.27 |
| gatD | b2091 | 0.49 |
| gatY | b2096 | 0.36 |
| gatZ | b2095 | 0.10 |
| gdhA | b1761 | 0.27 |
| gidB | b3740 | 0.42 |
| glnA | b3870 | 0.15 |
| glnS | b0680 | 0.00 |
| glpE | b3425 | 0.49 |
| gltD | b3213 | 0.22 |
| gltF | b3214 | 0.44 |
| gltJ | b0654 | 0.42 |
| gpt | b0238 | 0.47 |
| gst | b1635 | 0.07 |
| guaA | b2507 | 0.38 |
| guaB | b2508 | 0.42 |
| gusC | b1615 | 0.13 |
| gyrA | b2231 | 0.47 |
| hdhA | b1619 | 0.41 |
| hsdS | b4348 | 0.41 |
| iclR | b4018 | 0.41 |
| ilvB | b3671 | 0.46 |
| ilvC | b3774 | 0.04 |
| ilvD | b3771 | 0.35 |
| ilvE | b3770 | 0.35 |
| ilvG | b3767 | 0.09 |
| ilvM | b3769 | 0.28 |
| infB | b3168 | 0.47 |
| insA_3 | b0275 | 0.33 |
| insB_2 | b0274 | 0.49 |
| ksgA | b0051 | 0.25 |
| lepB | b2568 | 0.47 |
| leuC | b0072 | 0.44 |
| leuD | b0071 | 0.24 |
| livG | b3455 | 0.36 |
| livJ | b3460 | 0.13 |
| livK | b3458 | 0.18 |
| lysC | b4024 | 0.41 |
| malF | b4033 | 0.50 |
| malI | b1620 | 0.42 |
| manX | b1817 | 0.43 |
| marR | b1530 | 0.36 |
| mepA | b2328 | 0.48 |
| metE | b3829 | 0.13 |
| mgtA | b4242 | 0.46 |
| minC | b1176 | 0.44 |
| moaA | b0781 | 0.26 |
| msbA | b0914 | 0.07 |
| mukE | b0923 | 0.16 |
| mutT | b0099 | 0.48 |
| nadE | b1740 | 0.49 |
| narG | b1224 | 0.37 |
| narL | b1221 | 0.49 |
| nhaB | b1186 | 0.31 |
| nohB | b0560 | 0.47 |
| nusA | b3169 | 0.28 |
| ompA | b0957 | 0.11 |
| pdxH | b1638 | 0.27 |
| pheA | b2599 | 0.25 |
| phoP | b1130 | 0.20 |
| pncB | b0931 | 0.45 |
| pnp | b3164 | 0.34 |
| ppa | b4226 | 0.36 |
| pqiA | b0950 | 0.19 |
| prc | b1830 | 0.33 |
| priB | b4201 | 0.18 |
| priC | b0467 | 0.46 |
| prlA | b3300 | 0.29 |
| proX | b2679 | 0.32 |
| pyrB | b4245 | 0.05 |
| pyrI | b4244 | 0.20 |
| rbsA | b3749 | 0.35 |
| rhsD | b0497 | 0.44 |
| ribB | b3041 | 0.43 |
| rimL | b1427 | 0.23 |
| rnpA | b3704 | 0.22 |
| rpiR | b4089 | 0.43 |
| rplA | b3984 | 0.15 |
| rplB | b3317 | 0.13 |
| rplC | b3320 | 0.09 |
| rplD | b3319 | 0.11 |
| rplE | b3308 | 0.25 |
| rplF | b3305 | 0.29 |
| rplI | b4203 | 0.37 |
| rplJ | b3985 | 0.18 |
| rplK | b3983 | 0.25 |
| rplL | b3986 | 0.17 |
| rplM | b3231 | 0.16 |
| rplN | b3310 | 0.30 |
| rplO | b3301 | 0.37 |
| rplP | b3313 | 0.15 |
| rplR | b3304 | 0.41 |
| rplS | b2606 | 0.15 |
| rplU | b3186 | 0.48 |
| rplV | b3315 | 0.12 |
| rplW | b3318 | 0.11 |
| rplX | b3309 | 0.29 |
| rplY | b2185 | 0.14 |
| rpmA | b3185 | 0.43 |
| rpmB | b3637 | 0.22 |
| rpmD | b3302 | 0.46 |
| rpmG | b3636 | 0.25 |
| rpmH | b3703 | 0.38 |
| rpmJ | b3299 | 0.46 |
| rpoA | b3295 | 0.41 |
| rpsC | b3314 | 0.13 |
| rpsD | b3296 | 0.41 |
| rpsE | b3303 | 0.47 |
| rpsF | b4200 | 0.18 |
| rpsG | b3341 | 0.44 |
| rpsH | b3306 | 0.27 |
| rpsI | b3230 | 0.13 |
| rpsJ | b3321 | 0.10 |
| rpsK | b3297 | 0.39 |
| rpsM | b3298 | 0.39 |
| rpsN | b3307 | 0.26 |
| rpsP | b2609 | 0.17 |
| rpsQ | b3311 | 0.24 |
| rpsR | b4202 | 0.15 |
| rpsS | b3316 | 0.15 |
| rpsT | b0023 | 0.40 |
| rpsU | b3065 | 0.18 |
| secG | b3175 | 0.36 |
| sfsA | b0146 | 0.50 |
| sodA | b3908 | 0.45 |
| speD | b0120 | 0.46 |
| suhB | b2533 | 0.25 |
| surA | b0053 | 0.31 |
| thrA | b0002 | 0.32 |
| trmD | b2607 | 0.09 |
| trpL | b1265 | 0.46 |
| tyrB | b4054 | 0.45 |
| tyrS | b1637 | 0.17 |
| uidA | b1617 | 0.13 |
| uidB | b1616 | 0.05 |
| umuC | b1184 | 0.50 |
| upp | b2498 | 0.15 |
| vacJ | b2346 | 0.43 |
| xseA | b2509 | 0.42 |
| ygiC | b3038 | 0.35 |
| yjjR | b4366 | 0.08 |
| yjjS | b4367 | 0.12 |
| yjjT | b4371 | 0.40 |
| zwf | b1852 | 0.48 |
Indications that acivicin serves as an imposter of certain natural amino acids.
Transcription of glnA, encoding glutamine synthetase, was lowered more than sixfold. Expression of asnS, coding for asparaginyl-tRNA synthetase, was decreased more than fivefold; unfortunately, the spotted glnS PCR product was of poor quality (Y. Wei and R. LaRossa, unpublished data), precluding insight into its transcriptional response to acivicin.
Evidence that HisHF is the major target in vivo.
If the HisHF enzyme is inhibited by acivicin in vivo, then transcription should initiate frequently at the his promoter (75, 89), pass through the leader/attenuator (43), and traverse the structural genes. This expectation was indeed realized, as the eight structural genes, hisGDCBHAFI, and the leader hisL were up-regulated 6- to 16-fold by administration of 2 μg of acivicin per ml. Ranking of open reading frames (ORFs) by fold induction with acivicin placed his operon genes in the 4th (hisC), 5th (hisL), 6th (hisB), 7th (hisI), 8th (hisG), 9th (hisD), 11th (hisH), 12th (hisF), and 28th (hisA) positions.
Metabolic mayhem.
The term “metabolic mayhem” has been applied to the action of sulfonylurea herbicides in S. enterica serovar Typhimurium, which causes the cell to (i) signal methionine sufficiency (10) in the face of methionine limitation (51) and (ii) skew the ratios of 2-ketoacids (52) and acyl coenzyme A's (78), two classes of central precursor metabolites. Together, about 60% of the organic content of E. coli is derived from these two sets of central building blocks (48). A similar case may be evident when HisHF activity was limiting. The ATP pool was compromised (29, 42). The cell, however, did not respond by elevating the F0-F1 ATP synthase-specifying transcripts; rather, the atp operon was mildly repressed (Table 3), indicating that the capacity to convert ADP to ATP was not enhanced. Most purine biosynthetic transcripts were not affected appreciably by the acivicin administration, suggesting that the PurR regulon (92) was indifferent to this treatment. Evidence for modulating expression of two purine-related operons, however, was found. The bicistronic guaBA operon was down-regulated about twofold (Table 3), while the purA transcript was elevated more than sevenfold (Table 2). The latter result contradicted the thought (37) that the PurR-independent regulation of purA is posttranscriptional. Both the purA and guaBA operons are regulated by multiple regulatory circuits (92). guaBA is responsive to PurR (92), cyclic AMP receptor protein (39), and DnaA (92), while purA is controlled by both PurR and an adenine-dependent mechanism (92). These transcriptional data suggested that flux from IMP to AMP was being encouraged at the expense of forming GMP from the common intermediate IMP. Thus, the apparent inconsistencies in the transcriptional regulation of the purine regulon might be indicative of a baroque regulatory mechanism designed to maintain balance between the GTP and ATP pools.
Acivicin triggers the stringent response.
The just-mentioned induction and repression of gene expression suggest that the in vivo levels of histidyl-tRNA, glutaminyl-tRNA, and/or asparaginyl-tRNA may be lowered by acivicin treatment. Any such a drop would trigger the stringent response. This response has two basic elements, conservation of amino acid reserves by shutting off synthesis of ribosomes and other translational machinery (13) and a redirection of resources toward increasing amino acid biosynthesis (75, 89). Aspects of each are apparent in the gene expression profile of acivicin-treated cells. As expected for a treatment resulting in amino acid starvation, expression of the translational apparatus was decreased; 49 distinct ORFs encoding proteins involved in translation were down-regulated by a factor of 2 or more (Table 3).
Evidence for elevation of amino acid biosynthetic capacity was also found (Table 2). Seven arg genes were induced three- to ninefold by this treatment. Also highly induced were biosynthetic genes corresponding to the aspartate-derived family of amino acids. The gene, aspC, specifying the major transaminase responsible for aspartate formation was elevated eightfold. The leader transcript of the thr operon was elevated sevenfold, while expression of five met ORFs was enhanced two- to sixfold. Acivicin exposure also induced the lysine synthesis-involved dapB mRNA by a factor of 5. This subdivision of induced transcripts by metabolic origin of amino acids is unexpected. It suggests, moreover, that the global response to amino acid starvation may be more complex than suggested by current dogma or that acivicin's targets include factors other than HisHF.
In contrast, transcription of only a single amino acid transport gene, mtr, was elevated (Table 2). This elevation, by a factor of 21, was dramatic; mtr was the third most highly induced gene. Amino acid transport had been suggested to be under stringent control (13), based primarily on studies of branched chain amino acid transport (63). That suggestion may need to be reevaluated in light of these findings. The reported pleiotropic effects of the stringent response are quite broad (13). The transcriptional responses elicited by other amino acid antagonists as well as the dependence of these responses on relA will further define this regulon.
Other stress responses triggered by the acivicin challenge.
The two most highly induced genes were hslS and hslT, heat shock loci (65), whose transcripts were elevated 20- to 30-fold (Table 2). Other stress-responsive transcripts (Table 2) were highly induced, including clpB (8.6-fold), appA (4.3-fold), uspA (4.2-fold), ahpC (3.4-fold), rpoH (3.2-fold), slp (3-fold), rob (2.8-fold), cspE (2.8-fold), soxS (2.8-fold), htpX (2.6-fold), osmE (2.6-fold), sugE (2.5-fold), grpE (2.5-fold), sohA (2.4-fold), and slyD (2.1-fold). Also within this group of induced mRNA species were transcripts involved in osmotolerance, including betT (8-fold), otsA (4.4-fold), betI (4.3-fold), osmY (3.8-fold), otsB (2.7-fold), osmE (2.6-fold), treF (2.4-fold), betA (2.4-fold), and betB (2.4-fold). Expression of some genes involved in DNA and RNA metabolism was also heightened; elevated levels of hepA, rna, nfo, recN, and xseB transcripts were observed.
Expression of genes involved in iron metabolism was also elevated by acivicin treatment. Induced genes included ftn (11-fold), bfr (6.4-fold), and entE (2.7-fold). Transcripts of cydA, frdB, frdC, nrdD, nrdG, nrdH, nrdI, qor, and torD, involved in respiratory activity, were also elevated. Cause and effect are difficult to separate. The implied increase in iron metabolism could elevate the superoxide content, as suggested by the increased soxS mRNA titer. Such puzzles, as well as that involving the interplay between acivicin and respiration, await further study.
Unanticipated repression by acivicin.
Many changes were observed that were not predictable. Strikingly, the level of several amino acid biosynthetic transcripts was not up-regulated but rather reduced by histidine starvation (Table 3), including transcripts for genes specifying components of the aromatic pathways (aroA, aroF, aroH, pheA, trpL, and tyrB), the pyruvate family (avt, ilvB, ilvC, ilvD, ilvE, ilvG, ilvM, leuC, and leuD), sulfur amino acids (cysA, cysC, cysD, cysK, cysM, cysN, and metE), and the aspartate family (lysC). Greater than fivefold repression was observed for aroA, ilvC, ilvG, and metE. Each of these genes is distinctive; aroA (16) and ilvG (47) specify enzymes targeted by commercially important herbicides, while ilvC and metE encode enzymes that must be highly expressed (86) due to their poor performance as catalysts (32, 60, 90). Interestingly, expression of another most highly expressed, pyrimidine biosynthetic operon (86) was down-regulated by the acivicin challenge; pyrBI (specifying aspartate transcarbamylase) transcripts were greatly reduced. Surprisingly, genes implicated in uptake of amino acids or their precursors were often down-regulated. Included in this category were cysP, livJK, and proX. Certain carbon utilization transcripts were down-regulated. Repression of six gat and three uid genes was observed.
Like the data concerning expression of amino acid permease systems in response to acivicin treatment discussed earlier, the unexpected and strong repression of many highly expressed biosynthetic genes by acivicin treatment suggests that the response to amino acid starvation is not a uniform induction of appropriate defenses. Rather, the cellular logic appears to be more selective; the cost of synthesizing a poor catalyst like IlvC or MetE may outweigh the benefits associated with their action. Thus, we are surprised that the decision to correct the perceived imbalance or to wait for the insult to pass may be made on a gene-by-gene basis.
Unanticipated elevation of gene expression by acivicin administration.
Expression of three genes, ack (78), pta (78), and mrp (D. R. Smulski and R. A. LaRossa, unpublished data), whose inactivation leads to a sulfometuron methyl-sensitive phenotype in S. enterica serovar Typhimurium LT2 (52), was elevated after acivicin treatment. Other transcripts increased by this treatment were specified by the tdh-kbl operon. The corresponding gene products specified by this operon degrade threonine to pyruvate and ammonia through glycine and serine. Thus, the overall pathway involves tdh, kbl, gcvTHP, lpdA, glyA, and sdaA or metC (64). The lpdA and sdaA mRNA titers, as well as the tdh-kbl transcript levels, were elevated by acivicin treatment, suggesting increased flux through this pathway.
Summation.
Acivicin is a natural product produced by streptomycetes. Its ecological role may be to defend the home turf of the producing species, encouraging other microbes to emigrate toward less hostile environments. Such use in antibacterial warfare may be explained by theories concerning the evolution of translation (18, 19) and biosynthetic pathways. Hence, adoption of acivicin for use in cancer therapy represents its introduction into a novel niche, one occupied by cells lacking its intended target, HisHF. Thus, studies of the action of acivicin with other glutamine amidotransferases could represent more general aspects of the binding of glutamine to the amidotransferase enzyme family. In contrast, acivicin interacted much more avidly with HisHF from E. coli. This specific interaction is not limited to the E. coli enzyme; inhibition of the homologous eukaryotic enzyme, His7 from yeast, has been found (M. McCluskey and L. Huang, unpublished data). The relative contributions of acivicin's competition with glutamine and its inactivation of glutamine amidotransferases to the observed in vivo effects are worthy of further study.
That acivicin was targeted toward HisHF in vivo was demonstrated in several ways. Nutritional reversal of acivicin action by inclusion of histidine in the test medium was most suggestive. Amplification of the hisHAF portion of the histidine operon resulted in a resistance phenotype, also supporting the presumption that HisHF was the in vivo target. Finally, the gene expression profiling demonstrated that the cell was limited for histidine. Such starvation can both drain adenylates and limit histidyl-tRNA formation. In analogy to the detailed physiological and genetic studies of sulfometuron methyl action (48, 52), separation of these two consequences can be accomplished by determining the acivicin-induced change in the transcriptional profile of a feedback-insensitive hisG mutant grown in the presence of histidine. Given these results, it was somewhat surprising that a hisHAF-containing plasmid was not obtained from the multicopy libraries when acivicin resistance was used as a genetic selection. The large (>7-kbp) size of the operon may have contributed to this failure to obtain the expected result. Alternatively, the attenuation mechanism, unlike that of repression, may not be titrated out by the presence of the his operon on a multicopy plasmid.
Thus, several sorts of experiments were brought to bear on the interaction of E. coli and acivicin. Collectively, they supported the conclusion that acivicin inhibited HisHF while suggesting an intricate interaction between cell and inhibitor. Much of the gene expression profile could be understood by reference to the vast, preceding study of E. coli physiology. Without combining such knowledge with the realization that acivicin is a mimic of the natural amino acid l-glutamine, interpretation would be more difficult. Since inhibitor binding does not have to coincide with enzyme active sites, such mimicry may not always be as obvious as in the case of acivicin. Thus, we suggest that a multifaceted approach to inhibitor action remains the most likely path to definition of macromolecular targets. Especially informative may be the selection of missense mutants having a resistance phenotype; this approach has defined both inhibitor targets and the means by which these compounds are imported into the cell (46).
In addition, the gene expression profile also suggested several novel regulatory circuits. Each of these is worthy of further study. Two examples are noted. Transcripts of more than 150 genes of unknown function were elevated by exposure to acivicin; the dependence of these changes on various global regulatory mechanisms can be readily evaluated. The massive and apparently selective loss of highly expressed transcripts upon treatment is also provocative. Moreover, this global view suggested that the mRNA population is dramatically remodeled in response to acivicin. This unanticipated remodeling indicates that gene expression profiling will become a most important means for uncovering the pleiotropic responses to inhibitor action. Thus, comprehensive transcript profiling is an important tool for the biological detective; its findings, however, must be vigorously pursued by other methodologies if their authenticity is to be established.
ACKNOWLEDGMENTS
R.A.L. thanks Phil Hartman (Biology Department, Johns Hopkins University) for introducing him to the joys and complexities of gene action and histidine biosynthesis in 1972. We also extend thanks to Tom Cebula (Division of Molecular Biology, Food and Drug Administration) and a reviewer for their comments on this work and to our many colleagues who donated strains and other biological materials.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anton D N. Positive selection of mutants with cell envelope defects in a Salmonella typhimurium strain hypersensitive to the products of hisH and hisF. J Bacteriol. 1979;137:1271–1281. doi: 10.1128/jb.137.3.1271-1281.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki T, Oya H. Inactivation of Crithidia fasciculata carbamoyl phosphate synthase II by the antitumor drug acivicin. Mol Biochem Parasitol. 1987;23:173–181. doi: 10.1016/0166-6851(87)90153-8. [DOI] [PubMed] [Google Scholar]
- 5.Austin E A, Graves J F, Hite L A, Parker C T, Schnaitman C A. Genetic analysis of liposaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J Bacteriol. 1990;172:5312–5325. doi: 10.1128/jb.172.9.5312-5325.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachmann B J. Derivatives and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 7.Belkin S. A panel of stress-responsive luminous bacteria for monitoring wastewater toxicity. Methods Mol Biol. 1998;102:247–258. doi: 10.1385/0-89603-520-4:247. [DOI] [PubMed] [Google Scholar]
- 8.Berlyn M K, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 9.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 10.Bogosian G, Violand B N, Dorward-King E J, Workman W E, Jung P E, Kane J F. Biosynthesis and incorporation into protein of norleucine in Escherichia coli. J Biol Chem. 1989;264:531–539. [PubMed] [Google Scholar]
- 11.Bulich A A. A practical and reliable method for monitoring the toxicity of aquatic samples. Process Biochem. 1982;17:45–47. [Google Scholar]
- 12.Carlomagno M A, Chiariotti L, Alifano P, Napo A G, Bruni C B. Structure and function of the Salmonella typhimurium and Escherichia coli K-12 histidine operons. J Mol Biol. 1988;203:583–606. doi: 10.1016/0022-2836(88)90194-5. [DOI] [PubMed] [Google Scholar]
- 13.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 14.Celis R T. Repression and activation of arginine transport genes in Escherichia coli K12 by the ArgP protein. J Mol Biol. 1999;294:1087–1095. doi: 10.1006/jmbi.1999.3308. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee P K, Sternberg N L. A general genetic approach in Escherichia coli for determining the mechanism(s) of action of tumoricidal agents: application to DMP840, a tumoricidal agent. Proc Natl Acad Sci USA. 1995;92:8950–8954. doi: 10.1073/pnas.92.19.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comai L, Sen L C, Stalker D M. An altered aroA gene product confers resistance to the herbicide glyphosate. Science. 1983;221:370–371. doi: 10.1126/science.221.4608.370. [DOI] [PubMed] [Google Scholar]
- 17.Davidov Y, Rozen R, Smulski D R, Van Dyk T K, Vollmer A C, Elsemore D A, LaRossa R A, Belkin S. Improved bacterial SOS promoter::lux fusions for genotoxicity detection. Mutat Res. 2000;46:97–107. doi: 10.1016/s1383-5718(99)00233-8. [DOI] [PubMed] [Google Scholar]
- 18.Davies J. What are antibiotics? Archaic functions for modern activities. Mol Microbiol. 1990;4:1227–1232. doi: 10.1111/j.1365-2958.1990.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 19.Davies J, von Ahsen U, Wank H, Schroeder R. Evolution of secondary metabolite production: potential roles for antibiotics as prebiotic effectors of catalytic RNA reactions. Ciba Found Symp. 1992;171:24–32. doi: 10.1002/9780470514344.ch3. [DOI] [PubMed] [Google Scholar]
- 20.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 21.Davisson V J, Deras I L, Hamilton S E, More L L. A plasmid-based approach for the synthesis of a histidine biosynthesis intermediate. J Org Chem. 1994;59:137–143. [Google Scholar]
- 22.Deras I L. Ph.D. thesis. West Lafayette, Ind: Purdue University; 1996. [Google Scholar]
- 23.Downs D M, Escalante-Semerena J C. Impact of genomics and genetics on the elucidation of bacterial metabolism. Methods. 2000;20:47–54. doi: 10.1006/meth.1999.0904. [DOI] [PubMed] [Google Scholar]
- 24.Elliott W L, Weber G. In vivo inactivation of formylglycinamidine ribonucleotide synthetase in rat hepatoma. Biochem Pharmacol. 1985;34:243–248. doi: 10.1016/0006-2952(85)90131-5. [DOI] [PubMed] [Google Scholar]
- 25.Elsemore D A. Insertion of promoter region::luxCDABE fusions into the Escherichia coli chromosome. Methods Mol Biol. 1998;102:97–104. doi: 10.1385/0-89603-520-4:97. [DOI] [PubMed] [Google Scholar]
- 26.Elsemore, D. A., R. A. LaRossa, and T. K. Van Dyk. February 1998. A method for identifying the site of action of xenobiotic chemicals. WO98/38336 patent PCT/US98/03684.
- 27.Flores A, Casadesus J. Suppression of the pleiotropic effects of HisH and HisF overproduction identifies four novel loci on the Salmonella typyhimurium chromosome: osmY, sfiW, sfiC, and sfiY. J Bacteriol. 1995;177:4841–4850. doi: 10.1128/jb.177.17.4841-4850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frodyma M, Rubio A, Downs D M. Reduced flux through the purine biosynthetic pathway results in an increased requirement for coenzyme A in thiamine synthesis in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:236–240. doi: 10.1128/jb.182.1.236-240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galloway R J, Taylor B L. Histidine starvation and adenosine 5′-triphosphate depletion in the chemotaxis of Salmonella typhimurium. J Bacteriol. 1980;142:1068–1075. doi: 10.1128/jb.144.3.1068-1075.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrick-Silversmith L, Hartman P E. Histidine-requiring mutants of Escherichia coli K12. Genetics. 1970;66:231–244. doi: 10.1093/genetics/66.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldschmidt E P, Cater M S, Matney T S, Butler M A, Greene A. Genetic analysis of the histidine operon in Escherichia coli K12. Genetics. 1970;66:219–229. doi: 10.1093/genetics/66.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greene R C. Biosynthesis of methionine. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 542–560. [Google Scholar]
- 33.Han J S, Kwon H S, Yim J B, Hwang D S. Effect of IciA protein on the expression of the nrd gene encoding ribonucleoside diphosphate reductase in E. coli. Mol Gen Genet. 1998;259:610–614. doi: 10.1007/s004380050854. [DOI] [PubMed] [Google Scholar]
- 34.Han J S, Park J Y, Lee Y S, Thony B, Hwang D S. PhoB-dependent transcriptional activation of the iciA gene during starvation for phosphate in Escherichia coli. Mol Gen Genet. 1999;262:448–452. doi: 10.1007/s004380051104. [DOI] [PubMed] [Google Scholar]
- 35.Hanka L J, Martin D, Neil G L. A new antitumor antimetabolite (αS, 5S)-α-amino-3-chloro-4,5-dihydro-5-isoxazoleacetic acid (NSC-163501): antimicrobial reversal studies and preliminary evidence against L1210 mouse leukemia in vivo. Cancer Chemother Rep. 1973;57:141–148. [PubMed] [Google Scholar]
- 36.Hartman P E, Loper J C, Serman D. Fine structure mapping by complete transduction between histidine-requiring Salmonella mutants. J Gen Microbiol. 1960;22:323–353. doi: 10.1099/00221287-22-2-323. [DOI] [PubMed] [Google Scholar]
- 37.He B, Zalkin H. Regulation of Escherichia coli purA by purine repressor, one component of a dual control mechanism. J Bacteriol. 1994;176:1009–1013. doi: 10.1128/jb.176.4.1009-1013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussein A D, Walter R D. Purification and characterization of gamma-glutamyl transpeptidase from Ascaris suum. Mol Biochem Parasitol. 1996;77:41–47. doi: 10.1016/0166-6851(96)02573-x. [DOI] [PubMed] [Google Scholar]
- 39.Hutchings M I, Drabble W T. Regulation of the divergent guaBA and xseA promoters of Escherichia coli by the cyclic AMP receptor protein. FEMS Microbiol Lett. 2000;187:115–122. doi: 10.1111/j.1574-6968.2000.tb09146.x. [DOI] [PubMed] [Google Scholar]
- 40.Hwang D S, Kornberg A. Opposed action of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J Biol Chem. 1992;267:23087–23091. [PubMed] [Google Scholar]
- 41.Jayaram H N, Ardalan B, Deas M, Johnson R K. Mechanism of resistance of a variant of P388 leukemia to l-(alpha S,5S)-alpha-amino-3-chloro-4,5-dihydro-5-isoxazoleacetic acid (acivicin) Cancer Res. 1985;45:207–212. [PubMed] [Google Scholar]
- 42.Johnson M S, Taylor B L. Comparison of methods for the specific depletion of ATP in Salmonella typhimurium. Appl Environ Microbiol. 1993;59:3509–3512. doi: 10.1128/aem.59.10.3509-3512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnston H M, Roth J R. DNA sequence changes of mutations altering attenuator control of histidine operon of Salmonella typhimurium. J Mol Biol. 1981;145:735–756. doi: 10.1016/0022-2836(81)90312-0. [DOI] [PubMed] [Google Scholar]
- 44.Johnston H M, Roth J R. Histidine mutants requiring adenine: selection of mutants with reduced hisG expression in Salmonella typhimurium. Genetics. 1979;92:1–15. doi: 10.1093/genetics/92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klem T J, Davisson V J. Imidazole glycerol phosphate synthase: the glutamine amidotransferase in histidine biosynthesis. Biochemistry. 1993;32:5177–5186. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- 46.LaRossa R A. Mutant selections linking physiology, inhibitors, and genotypes. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2527–2587. [Google Scholar]
- 47.LaRossa R A, Schloss J V. The sulfonylurea herbicide sulfometuron methyl is an extremely potent and selective inhibitor of acetolactate synthase in Salmonella typhimurium. J Biol Chem. 1984;259:8753–8757. [PubMed] [Google Scholar]
- 48.LaRossa R A, Van Dyk T K. Metabolic mayhem caused by 2-ketoacid imbalances. Bioessays. 1987;7:125–130. doi: 10.1002/bies.950070308. [DOI] [PubMed] [Google Scholar]
- 49.LaRossa R A, Van Dyk T K. Physiological roles of the dnaK and groE stress proteins: catalysts of protein folding or macromolecular sponges? Mol Microbiol. 1991;5:529–534. doi: 10.1111/j.1365-2958.1991.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 50.LaRossa R A, Van Dyk T K. Utilization of sulfometuron methyl, an acetolactate synthase inhibitor, in molecular biological and metabolic studies of plants and microbes. Methods Enzymol. 1988;166:97–107. doi: 10.1016/s0076-6879(88)66015-0. [DOI] [PubMed] [Google Scholar]
- 51.LaRossa R A, Van Dyk T K, Smulski D R. A need for metabolic insulation: lessons from sulfonylurea genetics. In: Barak Z, Chipman D M, Schloss J V, editors. Biosynthesis of branched chain amino acids. New York, N.Y: VCH and Balaban Publishers; 1990. pp. 109–121. [Google Scholar]
- 52.LaRossa R A, Van Dyk T K, Smulski D R. Toxic accumulation of α-ketobutyrate caused by inhibition of the branched-chain amino acid biosynthetic enzyme acetolactate synthase in Salmonella typhimurium. J Bacteriol. 1987;169:1372–1378. doi: 10.1128/jb.169.4.1372-1378.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee Y S, Kim H, Hwang D S. Transcriptional activation of the dnaA gene, encoding the initiator for oriC replication by IciA protein, an inhibitor of in vitro origin replication in Escherichia coli. Mol Microbiol. 1996;19:389–396. doi: 10.1046/j.1365-2958.1996.485902.x. [DOI] [PubMed] [Google Scholar]
- 54.Lyons S D, Sant M E, Christopherson R I. Cytotoxic mechanisms of glutamine antagonists in mouse L1210 leukemia. J Biol Chem. 1990;265:11377–11381. [PubMed] [Google Scholar]
- 55.Martin R G, Berberich M A, Ames B N, Davis W W, Goldberger R F, Yourno J D. Enzymes and intermediates of histidine biosynthesis in Salmonella typhimurium. Methods Enzymol. 1971;17B:3–44. [Google Scholar]
- 56.Menzel R. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for β-galactosidases activity. Anal Biochem. 1989;181:40–50. doi: 10.1016/0003-2697(89)90391-6. [DOI] [PubMed] [Google Scholar]
- 57.Murray M L, Hartman P E. Overproduction of hisH and hisF gene products leads to inhibition of cell division in Salmonella. Can J Microbiol. 1971;18:671–681. doi: 10.1139/m72-105. [DOI] [PubMed] [Google Scholar]
- 58.Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 59.Nishimura A, Morita M, Nishimura Y, Sugino Y. A rapid and highly efficient method for preparation of competent Escherichia coli cells. Nucleic Acids Res. 1990;18:6169. doi: 10.1093/nar/18.20.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petersen J G, Holmberg S. The ILV5 gene of Saccharomyces cerevisiae is highly expressed. Nucleic Acids Res. 1986;14:9631–9651. doi: 10.1093/nar/14.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poster D S, Bruno S, Penta J, Neil G L, McGovern J P. Acivicin: an antitumor antibiotic. Cancer Clin Trials. 1981;4:327–330. [PubMed] [Google Scholar]
- 62.Prajda N. Enzyme targets of antiglutamine agents in cancer chemotherapy. Adv Enzyme Regul. 1985;24:207–223. doi: 10.1016/0065-2571(85)90077-9. [DOI] [PubMed] [Google Scholar]
- 63.Quay S C, Oxender D L. The relA locus specifies a positive effector of branched-chain amino acid transport. J Bacteriol. 1979;137:1059–1062. doi: 10.1128/jb.137.2.1059-1062.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reitzer L J. Sources of nitrogen and their utilization. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 380–390. [Google Scholar]
- 65.Richmond C S, Glaser J D, Mau R, Jin H, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rieder G, Merrick M J, Castorph H, Kleiner D. Function of hisF and hisH gene products in histidine biosynthesis. J Biol Chem. 1994;269:14386–14390. [PubMed] [Google Scholar]
- 67.Riley M, Labedan B. Escherichia coli gene products: physiological functions and common ancestories. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2118–2202. [Google Scholar]
- 68.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 69.Schnaitman C. Improved strains for target-based chemical screening. ASM News. 1991;57:612. [Google Scholar]
- 70.Shedlovsky A E, Magasanik B. A defect in histidine biosynthesis causing an adenine deficiency. J Biol Chem. 1962;237:3725–3730. [PubMed] [Google Scholar]
- 71.Shedlovsky A E, Magasanik B. The enzymatic basis of an adenine-histidine relationship in Escherichia coli. J Biol Chem. 1962;237:3731–3736. [PubMed] [Google Scholar]
- 72.Shioi J, Galloway R J, Niwano M, Chinnock R E, Taylor B L. Requirement of ATP in bacterial chemotaxis. J Biol Chem. 1982;257:7969–7957. [PubMed] [Google Scholar]
- 73.Smith J M, Roswell E H, Shioi J, Taylor B L. Identification of a site of ATP requirement for signal processing in bacterial chemotaxis. J Bacteriol. 1988;170:2698–2704. doi: 10.1128/jb.170.6.2698-2704.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith T K, Ikeda Y, Fujii J, Taniguchi N, Meister A. Different sites of acivicin binding and inactivation of gamma-glutamyl transpeptidases. Proc Natl Acad Sci USA. 1985;92:2360–2364. doi: 10.1073/pnas.92.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stephens J C, Artz S W, Ames B N. Guanosine 5′-diphosphate 3′-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino acid deficiency. Proc Natl Acad Sci USA. 1975;72:4389–4393. doi: 10.1073/pnas.72.11.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thony B, Hwang D S, Fradkin L, Kornberg A. iciA, an Escherichia coli gene encoding a specific inhibitor of chromosomal initiation of replication in vitro. Proc Natl Acad Sci USA. 1991;88:4066–4070. doi: 10.1073/pnas.88.10.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsang A W, Horswill A R, Escalante-Semerena J C. Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J Bacteriol. 1998;180:6511–6518. doi: 10.1128/jb.180.24.6511-6518.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Dyk T K, LaRossa R A. Involvement of ack-pta operon products in α-ketobutyrate metabolism by Salmonella typhimurium. Mol Gen Genet. 1987;207:435–440. doi: 10.1007/BF00331612. [DOI] [PubMed] [Google Scholar]
- 79.Van Dyk T K, Majarian W R, Konstantinov K B, Young R M, Dhurjati P S, LaRossa R A. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Dyk T K, Rosson R A. Photorhabdus luminescens luxCDABE promoter probe vectors. Methods Mol Biol. 1998;102:85–95. doi: 10.1385/0-89603-520-4:85. [DOI] [PubMed] [Google Scholar]
- 81.Van Dyk T K, Smulski D R, Chang Y-Y. Pleiotropic effects of poxA regulatory mutations of Escherichia coli and Salmonella typhimurium, mutations conferring sulfometuron methyl and α-ketobutyrate hypersensitivity. J Bacteriol. 1987;169:4540–4546. doi: 10.1128/jb.169.10.4540-4546.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vollmer A C, Belkin S, Smulski D R, Van Dyk T K, LaRossa R A. Detection of DNA damage by use of Escherichia coli carrying recA′::lux, uvrA′::lux, or alkA′::lux reporter plasmids. Appl Environ Microbiol. 1997;63:2566–2571. doi: 10.1128/aem.63.7.2566-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Webb E C. Enzyme nomenclature. Orlando, Fla: Academic Press; 1984. [Google Scholar]
- 84.Weber G, Natsumeda Y, Lui M S, Faderan M A, Liepnieks J J, Elliott W. Control of enzymic programs and nucleotide pattern in cancer by acivicin and tiazofurin. Adv Enzyme Regul. 1984;22:69–93. doi: 10.1016/0065-2571(84)90009-8. [DOI] [PubMed] [Google Scholar]
- 85.Wei Y, Lee J-M, LaRossa R A. The global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:2265–2272. doi: 10.1128/JB.183.7.2265-2272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei Y, Lee J-M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. High-density miroarrray mediated gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei Y, Vollmer A C, LaRossa R A. In vivo titration of mitomycin C action by four Escherichia coli genomic regions on multicopy plasmids. J Bacteriol. 2001;183:2259–2264. doi: 10.1128/JB.183.7.2259-2264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winkler M E. Biosynthesis of histidine. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 485–505. [Google Scholar]
- 89.Winkler M E, Roth D J, Hartman P E. Promoter- and attenuator-related metabolic regulation of the Salmonella typhimurium histidine operon. J Bacteriol. 1979;133:993–1000. doi: 10.1128/jb.133.2.830-843.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wittenbach V A, Rayner D R, Schloss J M. Pressure points in the biosynthetic pathway for branched-chain amino acids. In: Singh B K, Flores H E, Shannon J C, editors. Biosynthesis and molecular regulation of amino acids in plants. Rockville, Md: American Society of Plant Physiologists; 1992. pp. 69–88. [Google Scholar]
- 91.Yoo S J, Seol J H, Woo S K, Suh S W, Hwang D S, Ha D B, Chung C H. Hydrolysis of the IciA protein, an inhibitor of DNA replication initiation, by protease Do in Escherichia coli. FEBS Lett. 1993;327:17–20. doi: 10.1016/0014-5793(93)81029-y. [DOI] [PubMed] [Google Scholar]
- 92.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 561–579. [Google Scholar]
- 93.Zhou P, Bogan J A, Welch K, Pickett S R, Wang H J, Zaritsky A, Helmstetter C E. Gene transcription and chromosome replication in Escherichia coli. J Bacteriol. 1997;179:163–169. doi: 10.1128/jb.179.1.163-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]