Abstract
Recent technological advances in the field of artificial intelligence hold promise in addressing medical challenges in breast cancer care, such as early diagnosis, cancer subtype determination and molecular profiling, prediction of lymph node metastases, and prognostication of treatment response and probability of recurrence. Radiomics is a quantitative approach to medical imaging, which aims to enhance the existing data available to clinicians by means of advanced mathematical analysis using artificial intelligence. Various published studies from different fields in imaging have highlighted the potential of radiomics to enhance clinical decision making. In this review, we describe the evolution of AI in breast imaging and its frontiers, focusing on handcrafted and deep learning radiomics. We present a typical workflow of a radiomics analysis and a practical “how-to” guide. Finally, we summarize the methodology and implementation of radiomics in breast cancer, based on the most recent scientific literature to help researchers and clinicians gain fundamental knowledge of this emerging technology. Alongside this, we discuss the current limitations of radiomics and challenges of integration into clinical practice with conceptual consistency, data curation, technical reproducibility, adequate accuracy, and clinical translation. The incorporation of radiomics with clinical, histopathological, and genomic information will enable physicians to move forward to a higher level of personalized management of patients with breast cancer.
Keywords: breast cancer, radiomics, medicinal imaging, personalized medicine, quantitative biomarkers, artificial intelligence
1. Introduction
Summary: this review summarizes the state of the art of radiomics in breast imaging and provide clinicians and researchers with the basis for a practical approach to this emerging field.
Key points:
- The current limitations in the diagnosis of breast cancer lie in not detecting valuable prognostic and predictive information inherent in the heterogeneity of this disease;
- Thanks to the advancement in AI technologies, radiomics can extract qualitative and quantitative information from images that can support clinicians in the management of patient with breast cancer;
- Radiomics showed valuable applications in breast imaging distinguishing between malignant and benign lesions, assessing the tumour subtype and its grade, molecular expressions, and predict response to therapy and the risk of recurrence.
Breast cancer (BC) is the most diagnosed tumour (excluding skin cancers), with an increasing incidence, and is the second leading cause of death from malignancy among females worldwide [1]. Though concerns regarding early detection and accurate diagnosis have been raised, continued efforts are required towards application of precision medicine in BC.
Currently, the diagnosis of early invasive BC relies on radiological evaluation; essentially, based on mammography (with or without contrast) (Figure 1), breast ultrasound (US) (Figure 2) and contrast-enhanced magnetic resonance imaging (MRI) (Figure 3), supplemented by pathological confirmation of malignancy on radiologically obtained tissue samples [2,3]. However, such diagnostic approach has major limitations. Firstly, the sensitivity and positive predictive value are suboptimal [4]; secondly, biopsy is invasive and uncomfortable for women; thirdly, there is a long turnaround time for test results [5]. Moreover, BC is a heterogeneous disease with a significant chance that some of its features, including significant characteristics, remain undetected, meaning that valuable prognostic and predictive information can be missed. Due to such heterogeneity and dynamic tumour biology, indications for re-biopsy are increasing [6]. This is pertinent in the current era of personalised medicine, which relies on early diagnosis of disease, in a patient with specific characteristics, and the subsequent individually tailored treatments, aiming to deliver the right treatment to the right patient at the right time. Accordingly, the ultimate goals of the modern breast imaging are to detect BC as early as possible, then to classify the lesion and predict its clinical course and its biological aggressiveness to optimise treatment in a specific patient [7,8].
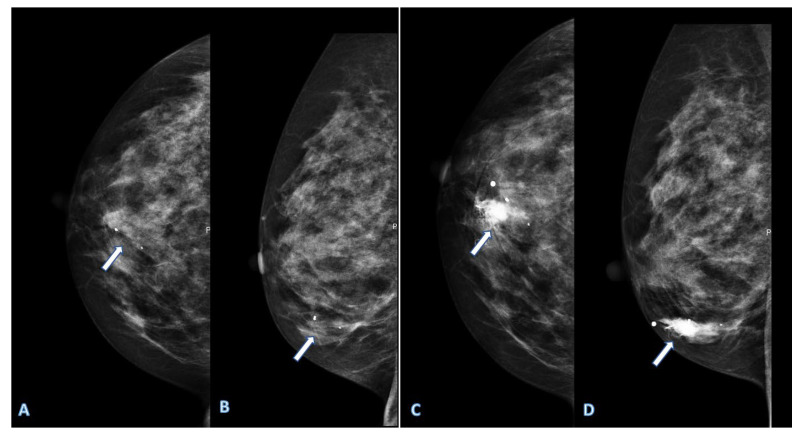
Figure 1.
Digital mammography of a right breast shows some calcifications in an area (arrows) with increased radiopacity in cranio-caudal (A) and medio-lateral (B) mammograms. Contrast enhanced mammography shows an increased enhancement in the same area (C,D), which is suspicious for breast cancer.
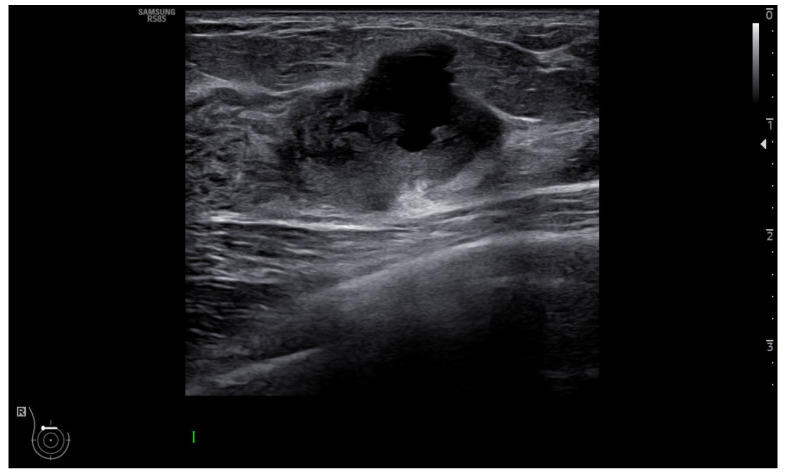
Figure 2.
Ultrasound shows a heterogeneous hypoechoic mass lesion with small lobulations located in the upper quadrant of the left breast, highly suspicious for breast cancer.
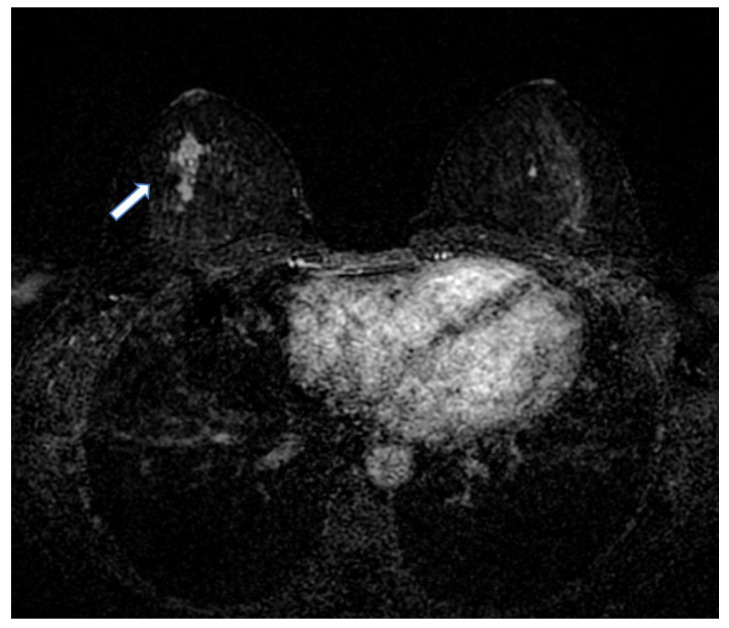
Figure 3.
Breast MRI shows an enhancement (arrow) in the right breast suspicious for breast cancer.
Continued exponential growth of medical imaging has led to an advancement in radiomics, which provides countless quantitative biomarkers extracted from modern diagnostic images, including detailed tumoral characterization of BC [9]. In particular, recent technological advances in the field of artificial intelligence (AI) applied to image analysis, via software based on machine learning (ML) and deep learning (DL), hold promise in addressing medical challenges in cancer detection, treatment assessment, prediction of treatment response and monitoring of disease progression [9,10,11,12,13,14,15,16,17,18]. In the current breast imaging practice, evaluation of BC is largely qualitative, including subjective evaluations such as tumour morphology, enhancement curves, and anatomic relationship to the surrounding tissues. However, to achieve the goal of personalised medicine, a quantitative evaluation is also required [19,20]. Therefore, radiomics is an emerging field of extreme interest, dealing with quantitative evaluation of images and extraction of designated features [21,22,23]. Data derived from a radiomics investigation, such as intensity, shape, texture, and wavelength [22,24,25,26,27], can be input to ML or DL algorithms, providing information to differentiate malignant and benign tumours, assess cancer genetics, predict treatment response, and contribute to more robust models that combine multidisciplinary information [11,16,22,28,29,30].
This review discusses the state of the art of radiomics, in both research and clinical applications, and its role in achieving personalised management in patients with BC. This paper will also try to demystify radiomics for clinicians by illustrating its limitations and challenges, as well as the opportunities it provides as a decision-support tool in cancer management.
2. Strengths and Limitations of Current Breast Imaging Techniques
Breast cancer screening with mammography has significantly lowered the breast cancer mortality rate; however, its sensitivity is limited in dense breasts. In this situation, ultrasound is a helpful imaging modality to examine the glandular and ductal components, investigate the axillary lymph nodes, and avoid the use of ionizing radiation. For these reasons, ultrasound is the primary diagnostic imaging technique in women under the age of 40.
Dynamic contrast-enhanced (DCE) MRI and contrast-enhanced spectral mammography (CESM) offer the advantage of adding a functional evaluation to the morphological data through the administration of contrast medium that exploits neo-angiogenesis occurring in tumoral lesions [31,32,33,34]. Nowadays, MRI is accepted as the most sensitive imaging technique for detecting and staging breast cancer and is of great help in the study of particularly complex breasts, or of patients with an elevated hereditary–familial risk of developing breast cancer [35]. With a similar performance to MRI, CESM can also visualize microcalcifications and possibly reduce the rate of false-positive findings and benign biopsies [32,36].
Radiomics features of BC can be extracted from MRI [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53], US [54], PET/CT [55,56], CEM and Mx and tomosynthesis images [51,57]. The most-used imaging biomarkers in breast cancer are those derived from perfusion and diffusion imaging. The perfusion imaging is both an MRI and CEM technique based on intravenously administered contrast agents, that enables spatially resolved quantification of the hemodynamic status of tissue. The first assumption is that the contrast media administration induces time-dependent changes in tissue signal, which can be monitored by the dynamic acquisition of images before, during and after contrast media injection. The second postulation is that pathological tissues have different hemodynamic properties from normal tissues, that can be depicted by perfusion biomarkers [58]. Diffusion-weighted imaging (DWI) is an MRI technique that uses quantitative estimation of random motion of water molecules as a surrogate for tissue characterisation, and it can be successfully employed in oncology to detect pathological structural changes [27,28,58].
Image guided percutaneous breast lesion biopsy can be performed using mammography, US, or MRI visualisation. The choice of the biopsy method is generally dictated by the imaging characteristics of the lesion, patient factors, and modality availability.
Compared to surgical biopsy, percutaneous imaging-guided biopsy is less invasive, faster, more cosmetically satisfactory, and the recovery time is shorter [19,59,60]. In the case of vacuum-assisted biopsies, the procedure may also be curative as small lesions can be removed completely [61]. All these percutaneous procedures also allow the placement of a harmless lesion marker, which is useful for the recognition of the target lesion during surgery or after neoadjuvant therapy (NAT—chemotherapy performed before surgery). The NAT may lead to resolution of the tumour without detectable mass, and lesion markers may be helpful to confirm complete treatment response [16,18,19].
Although percutaneous image-guided procedures are generally safe and highly accurate, there is still the risk of infrequent (<1 in 1000) complications, which include pain, bleeding, post procedural infections, and, rarely, pneumothorax and pseudoaneurysms [62]. In addition to relatively easily managed complications of haematomas and infections, some complications such as an arterial pseudoaneurysm or a pneumothorax, may be more difficult to manage. Although controversial, neoplastic seeding may occur, quoted in the literature in up to 1–2 cases per 1000 US-guided biopsies [63]. Moreover, as percutaneous biopsies only sample a part of the lesion, the examination may not be conclusive. In fact, in 3–9% of cases the malignant potential of lesions is uncertain, and up to 33% of these manifest eventually as clinically significant malignancy [63].
3. What Radiomics Is and How it Works in Breast Imaging Workflow
Radiomics assumes that radiological images contain more information than is visible to human eyes. Thus, radiomics is a translational field of research aiming to find associations between qualitative and quantitative information extracted from medical imaging and clinical data, to support evidence-based clinical decision making [9]. The extraction of quantitative features from radiological images allows the creation of high-dimensional data with clinical data. This is followed by data mining to extract valuable information for decision support models.
Radiomic workflow involves the following steps:
Images acquisition;
Images segmentation;
Features extraction;
Features selection;
Model construction.
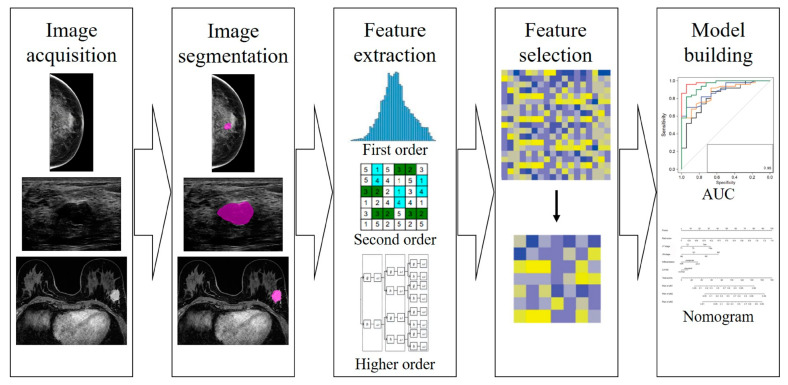
Such steps are shown in Figure 4.
Figure 4.
Different steps of radiomics workflow in breast imaging.
In breast imaging, the pixel value of MRI, US, and mammography, does not express physical properties of the tissue, such as Hounsfield Unit in computed tomography, but it is dependent on acquisition parameters.
In addition, MRI signal may change even with the same reconstruction parameters for two consecutive acquisitions, while US acquisition is also operator dependent [64,65]. To overcome this limitation, it is a good practice to acquire all imaging data using the same device and imaging parameters to ensure the stability of the features and record the parameters meticulously in order to facilitate the reproducibility of the study.
When this approach is not feasible, harmonization is mandatory to ensure robustness of features and generalizability of the model obtained [66].
Image segmentation with delineation of Regions of Interest (ROI) is a crucial part of the radiomic workflow. ROIs limit the area of analysis, and their delineation can be obtained manually, in a semi-automatic or fully automatic way.
Manual segmentation may introduce observer-bias, as studies have shown that many radiomic features are not robust against intra- and inter-observer variations concerning ROI delineation [67]. Consequently, studies using manual image segmentation with manual correction should perform assessments of intra- and inter-observer reproducibility of the derived radiomic features and exclude non-reproducible features from further analyses [68].
Semi-automatic segmentation may potentially reduce these issues. It has been demonstrated to work well for relatively homogeneous lesions; however, inhomogeneous lesions with poorly defined boundaries require intensive user correction [65].
Fully automatic segmentation based on DL networks is rapidly emerging, and many different algorithms have already been trained for image segmentation tasks of various organs. Such algorithms need ad hoc training and quality control, with manually contoured images as reference [9,69]. Generalizability of trained algorithms, however, is a major drawback, given that applying those algorithms on a different dataset often results in complete failure [9,68].
Feature extraction is the calculation of mathematical expressions used to quantify characteristics of the grey levels within the ROIs. Since many ways and formulas exist to calculate those features, adherence to the Image Biomarker Standardization Initiative guidelines is recommended.
Features can be extracted either directly from the images or after applying different filters, and they are usually categorised into the following subgroups:
Shape features describe the shape of the traced ROI and its geometric properties such as volume, maximum diameter along different orthogonal directions, maximum surface, tumour compactness, and sphericity.
First-order statistics features describe the distribution of individual voxel values without concern for spatial relationships. These are histogram-based properties reporting the mean, median, maximum, minimum values of the voxel intensities on the image, as well as their skewness (asymmetry), kurtosis (flatness), uniformity, and randomness (entropy).
Second-order statistics features include the so-called textural features, which are obtained by calculating the statistical inter-relationships between neighbouring voxels. They provide a measure of the spatial arrangement of the voxel intensities and, hence, of intra-lesion heterogeneity.
Higher-order statistics features are obtained by statistical methods after applying filters or mathematical transforms to the images.
The subsequent step involves feature selection, in order to exclude all the features that are either not reproducible or not strongly related to the outcome.
Selection can be made through statistical methods or through ML methods. The first starts from all the features provided by the calculation tool and performs a preliminary analysis to select the most repeatable and reproducible parameters and to subsequently reduce them by correlation and redundancy analysis [70].
Alternatively, ML techniques, underlying the idea that computers may learn from past examples and detect hard-to-discern patterns from large and complex data sets, may lead to the selection of appropriate features [71].
Finally, the remaining, non-correlated and highly relevant features can be used as input to the model for the respective classification task (e.g., discriminate between malignant or benign lesion).
Models are usually built by splitting the dataset into training and test sets, and most robust models are usually validated using a totally external dataset, to ensure generalizability of the obtained results [72].
4. The Role of Artificial Intelligence and Big Data in Radiomics
Mammography was one of the first imaging modalities to incorporate AI techniques, beginning with traditional computer-aided detection (CAD) [73]. CAD systems for mammography have been available for over a decade, meaning that the application of more recent ML and DL techniques to mammography has an existing benchmark for comparison [73].
Since then, significant advances in imaging analysis and the development of high-throughput methods have facilitated the rapid and simultaneous extraction and correlation of multiple imaging parameters [74].
Artificial intelligence is often associated with radiomics, given that it can be exploited in different steps of the radiomic workflow. For instance, AI can perform the task of image segmentation, before radiomic features are extracted. However, this approach is not often used, because with DL, the steps of image segmentation, feature extraction and classification are usually performed as a unique task [75].
When handcrafted features are extracted, ML algorithms, such as random forests, neural networks, linear regression, logistic regression, least absolute shrinkage, and selection operator, can help in feature selection before building the model [76].
AI studies must pass through rigorous validation steps including defining the imaging data sets (training, validation, and test sets), defining the ‘ground truth’ reference standard, having a detailed description of the training approach and metrics of model performance, and having validation or testing of the algorithm with external data. Three independent data sets (training, validation, and test sets) are needed: first, the AI algorithms are trained on an initial set of images according to a reference standard; second, the final algorithm is validated on a separate set of images; third, an external set of images is used to report the final statistical results of the AI algorithm [77].
AI methods can relate imaging-based characteristics to clinical, histopathology, or genomic data, contributing to precision medicine [9].
Moreover, currently unknown correlations between observed phenotypes and genotypes may be discovered through the mappings between imaging data and genomic data, providing possibilities to improve early detection and better management of the disease [8,21,22,23].
5. Recent Radiomics’ Application in Breast Cancer Care
The recently developed AI algorithms on vast amounts of imaging data has led to satisfactory models for the application of radiomics in breast cancer care, and even the patients approve the introduction of AI in clinical practice although only as a support to radiologist, and not in substitution thereof [78]. Indeed, radiomics already showed valuable applications in breast imaging practice: it may distinguish between malignant and benign lesions, assess the tumour subtype and its grade, assess the molecular expressions, and predict response to therapy and the risk of recurrence [5,9,16,17,18,20]. With the ability to infer the molecular profile of the tumour, a specific mutation or genotype, or even defining treatment possibilities and prognosis in BC patients, radiomics data may substitute physical breast biopsies in the near future [9,17,20].
Although radiomics and radio-genomics have great potentialities and offer some promising applications for personalised medicine [21,22], independent validation datasets are still needed to confirm the diagnostic and prognostic value of such technologies. They still need time before playing a significant practical role in cancer research and even more time to reach clinical practice. This is essentially due to the limitations of the available big data, which often lacks complete characterisation of the patients, poor integration of individual datasets, and a widespread misperception about their use and sharing [79].
The following are the updated state of the art of radiomics’ application in breast cancer care.
5.1. Radiomics as a Virtual Biopsy in Breast Cancer Diagnosis and Classification
The early detection and characterization of BC is crucial to improve outcomes in women because small non-metastatic disease can be effectively treated with curative intent [2,80,81]. The diagnosis of breast cancer currently relies on radiological and clinical evaluation, confirmed by histopathological examination. However, such an approach has limitations of suboptimal sensitivity, long turnaround time for test results, the invasiveness of the procedure, and the risk that some features of target lesions may remain undetected, requiring repeat biopsy.
Radiomics, through the extraction of quantitative peculiar features of BC from imaging data, may identify diagnostic information of breast cancer, potentially reducing the need for invasive biopsies, and facilitating an approach that is as personalised as possible for each patient. From the perspective of truly personalised management of breast cancer, based on early diagnosis and individually tailored treatments, radiomics is rising as a means to obtain information from diagnosis to molecular profiling, and treatment response assessment indeed, without the need of a physically biopsied tissue sample.
Zhou et al. [51] used 99 texture and histogram parameters from 133 patients who underwent DCE-MRI to differentiate between benign and malignant BC with 91% of accuracy. Xie et al. [82] analysed radiomics features extracted from 134 BC with similar accuracy, comparing triple negative breast cancer (TNBC) to the non-TNBC at breast MRI.
In 2018, a retrospective study [83] analysed unenhanced DWI-based radiomics to determine the malignant nature of suspicious breast lesions detected on screening mammography, decreasing the false-positive results in lesions classified as BI-RADS 4 or 5 at screening mammography while retaining sensitivity greater than 98%.
Li et al. [84] analysed the radiomic features from mammography in 182 patients (106 malignant and 76 benign), showing that the performance of the combined lesion and parenchyma classifier in the differentiation of malignant and benign findings was better than that which only used the lesion features.
In 2019, a sub-study of a multi-centre and prospective study leaded by Tagliafico et al. [85] applied a radiomics approach to tomosynthesis for the first time to differentiate normal from malignant breast tissue in patients with dense breasts in a small number of 40 patients, showing encouraging results.
Luo et al. [86] used radiomics features extracted from breast US of 315 patients to discriminate benign from malignant lesions.
In 2017, Fan et al. [43] analysed a combined model of DCE-MRI-based radiomics features and clinical information to predict molecular subtype of BC, with 0.87 of AUC value. Similar promising results were reported in the 2019 by Xie et al. [87] with texture features extracted from unenhanced breast MRI, and the 2020 by Demircioglu et al. [88], on a population of 98 women, showed the usability of a simplified and rapid approach to tumour for MRI-based decoding and phenotyping of BC.
Despite such encouraging results, radiomics technology is still not ready to substitute tissue biopsy in the near feature, and even then, they will require the aid of other parameters to be correctly interpreted and acted upon [9,20].
5.2. Prediction of Response to Neoadjuvant Chemotherapy
In the last decade, NAT has been increasingly used to treat operable BC, and it is associated with a favourable treatment response in 30% of women with aggressive BC, and decreases the rate of recurrence by up to 50% [2].
The achievement of pathological complete response (pCR) is a powerful prognostic factor for long-term outcome, and it is considered as the only currently validated biomarker of survival. However, it can only be assessed at surgery so far [89,90]. Therefore, radiomics may allow a non-invasive and earlier detection of treatment resistant lesions, to avoid the unnecessary toxicity of chemotherapy, and delayed access to other, potentially effective, therapies.
Several recent studies proposed prediction models of pCR to NAT in BC based on MRI [16,20,89,91,92,93], and valuating the pCR prediction by the extraction of radiomics features from pre-NAT breast MRI, obtaining statistically significant results [9,16,18,50,94,95,96,97].
In 2020, Choudhery et al. [97] used morphological and three-dimensional textural features to predict the molecular subtype and the pCR in 259 BC patients treated with NAT, showing significant association with pCR and residual cancer burden in BC. In 2017, Braman et al. [96] evaluated radiomic features based of both peri- and intra-tumoral regions on pre-treatment DCE-MRI to predict the pCR to NAT in 117 BC patients, demonstrating that peri-tumoral radiomics contributed to the successful prediction of the pCR of BC patients, yielding a maximum AUC of 0.74 within the testing set.
Previously, other authors [43,50,94,95,98] showed that quantitative analyses of radiomic features from pre-treatment breast DCE-MRI data in BC patients could be used as valuable image markers that are associated with pCR to NAT.
In the above-mentioned studies, DCE had been used more frequently than DWI to extract radiomics feature as it can provide the tumour’s kinetic characteristics of the contrast agent by producing pharmacokinetic maps.
In a multicentre study, Liu et al. [91] utilized multiple MRI sequences, including DWI, to predict pCR to NAT in BC patients. A total of 586 patients were enrolled, and a radiomic score was calculated using 13,950 features. Quantitative analyses extracted from MRI provide a promising tool for predicting tumour response in patients with advanced BC and show the potential and practical value in the clinic.
In 2021, our team performed a retrospective mono-centric study with the aim to assess radiomics with MRI for the early prediction of pCR in 83 BC patients undergoing NAT, investigating the correlation between pre-NAT radiomics with DCE-MRI features and disease-free survival (DFS), and the correlation between post-NAT (residual BC tissue) radiomics features and DFS [18]. Using 136 representative radiomics features selected through cluster analysis from the 1037 extracted features, a radiomic score was calculated to predict the response to NAT, with AUC of 0.64. After combining the clinical, biological and radiomics models, the AUC was 0.83, showing that MRI-based radiomic features slightly improved the pre-treatment prediction of pCR to NAT, in addiction to biological characteristics. The identification of the non-pCR patients in high-risk subgroups (defined by radiomics features), if confirmed on larger cohorts, could be helpful to identify such patients, to avoid unnecessary treatment.
5.3. Radiomics for Predicting Lymph Node Metastasis
Axillary lymph node (ALN) status is among the most important breast cancer prognostic factors [99,100]. Radiomics showed encouraging results in predicting the presence of ALN metastasis. Accordingly, several studies aimed to develop and validate radiomic nomograms as new tools based on radiomic signatures and clinical-pathologic risk factors to stratify patients more precisely in risk categories [101,102,103,104].
Radiomic nomograms proved to be reliable either when radiomic signatures were extracted from mammography (AUC up to 0.88) [101], CEM (AUC up to 0.79) [104] and MRI (AUC 0.89) [103]. In the latter, Yu et al. also assessed preoperative identification of ALN metastases and individual DFS in patients with early-stage BC [103].
More recently, the same team [105] implemented the approach with a multi-omics signature incorporating MRI multi-sequence key radiomic features of ALN and tumour regions with clinicopathologic characteristics and molecular subtype.
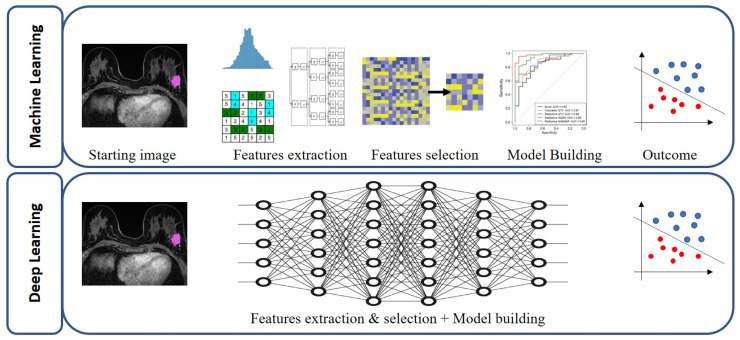
Recurring limitations of the above-mentioned studies were reliance on the retrospective design of the studies themselves, the heterogeneity of imaging parameters, and the absence of standardization in the extraction and developing of radiomic features, highlighting the common issues of radiomics studies. Cattell et al. [106] aimed to compare the generalizability of conventional radiomics vs. deep learning based features (using DCE-MRI) in an independent test set with dissimilar resolution, while developing a prediction model for preoperative prediction of SLN metastases. Interestingly, the features based on DL outperformed the conventional radiomic model accuracy, particularly in the independent testing set of dissimilar resolution, indicating that such features can ultimately result in a more generalizable model. Figure 5 shows the difference between ML and DL approach, in which the steps of feature extraction, selection and classification are performed as a unique task.
Figure 5.
Difference between machine learning (ML) approach and deep learning (DL) approach, in which the steps of feature extraction, selection and classification are performed as a unique task.
5.4. Radiomics for Predicting Breast Cancer Recurrence
Recurrence is the principal cause of breast cancer-related death [107]. Radiomics may play a role in the prediction of the risk of BC recurrence with the double potential benefits to reduce overtreatment in the low-risk patients and to reduce the undertreatment in the high-risk patients with BC.
Even for the prediction of BC recurrence, the most used imaging modality in the field of radiomics is the breast MRI. Three recent studies [53,108,109] found a statistically significant correlation between the risk of recurrence and radiomic features extracted from pre-treatment breast DCE-MRI in patients with invasive BC. In 2018, Park et al. [53] evaluated 294 MRI and drawn ROIs on entire tumour volume from each of the four distinct dynamic-series resulting in a radiomic signature which was significantly associated with worse DFS. In 2020, Chitalia et al. [108] analysed the MRI of 95 BC patients, extracting radiomics features from the entire tumour volume using only the first and second post- contrast images: 22 radiomics features were selected to create a tumour heterogeneity index that identified phenotypes of low, medium, and high intra-tumour heterogeneity with statistically significant differences in 10-year recurrence-free survival across phenotypes. In 2019, Mazurowski et al. [109] evaluated radiomics features of the MRI of 892 BC patients and found tumour size, textural and volumetric measures related to the enhancement had the strongest association with DFS in the univariate analysis. Most of these variables were also independently prognostic of outcomes in a multivariate analysis, after controlling for clinical and pathologic variables. Moreover, the authors also evaluated the prognostic value of each of the selected variables for specific clinically relevant subsets of patients (e.g., patients with ER/PR positive tumours, who underwent NAT or received hormonal therapy), and this increases their applicability in clinical settings.
A recent retrospective study [110] demonstrated that combining MRI radiomic features of BCs with MRI radiomic features of normal parenchyma from the contralateral breast, helps in the prediction of BC recurrence, suggesting that the underlying breast environment may contribute to recurrence more than the cancer biology of the tumour alone.
In 2020, Koh et al. found an association between MRI radiomics features and systemic recurrence in patients with TNBC. They selected 32 features on the second phase of DCE-MRI of patients with TNBC before any treatment. Their radiomics model, obtained by combining the Rad score with clinical and pathologic data, better predicted systemic recurrence than that of the clinical model alone. However, when they tried to validate the results with a different MRI scanner, the external validation did not show the radiomics model to be superior to the clinical model.
In addition to MRI, even mammography and US were used to extract radiomics features for prediction of histological findings.
In 2021 Xiong et al. [111] developed the first radiomics signature based on US to predict DFS in women with invasive BC. Their radiomics nomogram proved to be superior to the clinicopathological nomogram in terms of clinical usefulness. However, a limit of this US radiomic signature was that it was not reliable for DFS prediction when different sonographic platforms were used.
In a multicentric study, Yu et al. [112] developed and validated a recurrence risk model based on radiomics features from US for TNBC patients. The radiomics model, incorporating a radiomics signature and three prognostic variables, had a better diagnostic performance than that of a radiomics signature and clinicopathological model when used alone.
In the same year, Dasgupta et al. [113] conducted a prospective study to investigate the role of pre-treatment quantitative US radiomics in predicting recurrence for patients with locally advanced BC. They obtained 95 radiomics features including spectral features, texture, and texture-derivatives ones that have been demonstrated to be related to tissue microstructural elastic properties. These parameters were selected using the support vector machine-based model that demonstrated an accuracy of 82% to identify BC patients developing recurrence.
Concerning mammography, in 2021, Mao et al. [114] developed a mammography-based radiomics model for predicting the risk of BC: the multivariate logistic regression model including radiomics signature and clinical risk factors (tumour grade and HER 2) showed good performance yielding AUC of 0.92 in the training set.
Overall, these studies have shown that that adding radiomics to the standard radiological workflow would improve the prognostic value of breast imaging.
6. Future Trends and Modern Perspectives
Precise identification of cancer subtype and biomarkers of tumour immune biology play an increasingly important role in prognostication and treatment selection. The oncological drive towards multifocal biopsy of heterogenous lesions, multisite biopsy in metastatic disease, and resampling following therapy calls for non-invasive alternatives [115]. Fusion of data collated in The Cancer Imaging Archives (TCIA) and The Cancer Genome Atlas (TCGA), with increasingly capable radiomics workflows and the emergence of radiogenomics stands poised to provide the requisite solutions [116,117,118,119]. Non-invasive biomarkers for predicting response to specific treatments are being validated, and radiomics signatures are en route to support clinical practice. Innovation will be explosive, and radiologists are the essential partners in testing the translational applications of these technologies [20].
7. Conclusions
Data from the above-mentioned studies show the advancement in AI technologies in healthcare and support the potential utility of radiomics analysis in determining breast cancer biomarker from diagnostic imaging. However, standardization and transparency across all stages of the radiomics workflow and refinement of AI algorithms are required for improving reproducibility of radiomics analysis.
The assessment of clinical relevance and impact prior to study commencement, increased level of evidence using studies with large enough datasets and external validation, and its combination with established methods will help to move the field towards clinical implementation. As radiomics applications in breast cancer care include diagnosis, prognostication, and prediction of treatment response, a multidisciplinary collaboration among radiologists, data scientists and imaging scientists is demanded.
Multi-centre prospective oncological and radiological translational research using real-world heterogeneous datasets may develop radiomics techniques as the paradigm shifts towards minimally invasive techniques in the scenario of personalised medicine.
Glossary
| Diffusion Weighted Imaging (DWI) | MRI technique based on the random Brownian motion measure of water molecules within a voxel of tissue. |
| Machine learning | A branch of artificial intelligence involving use and development of computer systems that are able to learn and adapt without following explicit instructions, by using algorithms and statistical models to analyse and draw inferences from patterns in data. |
| Radiomic features | Characteristics of a tissue or a lesion that can be extracted in form of data. |
| ROI | acronym of Region of Interest, the portion of an image over which radiomics features are extracted. It can be delineated manually, in semi-automatic of fully automatic way. |
| Radiomic signature | Computational model which aims to address either unmet clinical needs, mostly in the field of oncologic imaging, as biomarkers, for example, or to compare radiomics performance with that of radiologists. |
Author Contributions
Conception and design: F.P., P.D.M. and A.R. (Anna Rapino); administrative support: E.C. and G.C.; provision of study materials or patients: all authors; collection and assembly of data: all authors; data analysis and interpretation: all authors; manuscript writing: all authors; final approval of manuscript: all authors. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
All authors declare that they have no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.World Cancer Research Fund International Breast Cancer Statistics. 2023. [(accessed on 28 December 2022)]. Available online: https://www.wcrf.org/cancer-trends/breast-cancer-statistics/
- 2.Tirada N., Aujero M., Khorjekar G., Richards S., Chopra J., Dromi S., Ioffe O. Breast Cancer Tissue Markers, Genomic Profiling, and Other Prognostic Factors: A Primer for Radiologists. Radiographics. 2018;38:1902–1920. doi: 10.1148/rg.2018180047. [DOI] [PubMed] [Google Scholar]
- 3.Gradishar W.J., Anderson B.O., Balassanian R., Blair S.L., Burstein H.J., Cyr A., Elias A.D., Farrar W.B., Forero A., Giordano S.H., et al. Breast Cancer, Version 4.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018;16:310–320. doi: 10.6004/jnccn.2018.0012. [DOI] [PubMed] [Google Scholar]
- 4.Torre L.A., Siegel R.L., Ward E.M., Jemal A. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol. Biomark. Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 5.Tagliafico A.S., Piana M., Schenone D., Lai R., Massone A.M., Houssami N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast. 2019;49:74–80. doi: 10.1016/j.breast.2019.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Luca F., Rotunno G., Salvianti F., Galardi F., Pestrin M., Gabellini S., Simi L., Mancini I., Vannucchi A.M., Pazzagli M., et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget. 2016;7:26107–26119. doi: 10.18632/oncotarget.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotili A., Trimboli R.M., Penco S., Pesapane F., Tantrige P., Cassano E., Sardanelli F. Double reading of diffusion-weighted magnetic resonance imaging for breast cancer detection. Breast Cancer Res. Treat. 2020;180:111–120. doi: 10.1007/s10549-019-05519-y. [DOI] [PubMed] [Google Scholar]
- 8.Pesapane F., Rotili A., Penco S., Nicosia L., Cassano E. Digital Twins in Radiology. J. Clin. Med. 2022;11:6553. doi: 10.3390/jcm11216553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pesapane F., Rotili A., Agazzi G.M., Botta F., Raimondi S., Penco S., Dominelli V., Cremonesi M., Jereczek-Fossa B., Carrafiello G., et al. Recent Radiomics Advancements in Breast Cancer: Lessons and Pitfalls for the Next Future. Curr. Oncol. 2021;28:2351–2372. doi: 10.3390/curroncol28040217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Codari M., Schiaffino S., Sardanelli F., Trimboli R.M. Artificial Intelligence for Breast MRI in 2008-2018: A Systematic Mapping Review. AJR Am. J. Roentgenol. 2019;212:280–292. doi: 10.2214/AJR.18.20389. [DOI] [PubMed] [Google Scholar]
- 11.Pesapane F., Codari M., Sardanelli F. Artificial intelligence in medical imaging: Threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur. Radiol. Exp. 2018;2:35. doi: 10.1186/s41747-018-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker A.S., Mueller M., Stoffel E., Marcon M., Ghafoor S., Boss A. Classification of breast cancer in ultrasound imaging using a generic deep learning analysis software: A pilot study. Br. J. Radiol. 2018;91:20170576. doi: 10.1259/bjr.20170576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker A.S., Marcon M., Ghafoor S., Wurnig M.C., Frauenfelder T., Boss A. Deep Learning in Mammography: Diagnostic Accuracy of a Multipurpose Image Analysis Software in the Detection of Breast Cancer. Investig. Radiol. 2017;52:434–440. doi: 10.1097/RLI.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhary K., Poirion O.B., Lu L., Garmire L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2018;24:1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abajian A., Murali N., Savic L.J., Laage-Gaupp F.M., Nezami N., Duncan J.S., Schlachter T., Lin M., Geschwind J.-F., Chapiro J. Predicting Treatment Response to Intra-arterial Therapies for Hepatocellular Carcinoma with the Use of Supervised Machine Learning-An Artificial Intelligence Concept. J. Vasc. Interv. Radiol. 2018;29:850–857.e1. doi: 10.1016/j.jvir.2018.01.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pesapane F., Agazzi G.M., Rotili A., Ferrari F., Cardillo A., Penco S., Dominelli V., D’Ecclesiis O., Vignati S., Raimondi S., et al. Prediction of the Pathological Response to Neoadjuvant Chemotherapy in Breast Cancer Patients With MRI-Radiomics: A Systematic Review and Meta-analysis. Curr. Probl. Cancer. 2022;46:100883. doi: 10.1016/j.currproblcancer.2022.100883. [DOI] [PubMed] [Google Scholar]
- 17.Nicosia L., Bozzini A.C., Ballerini D., Palma S., Pesapane F., Raimondi S., Gaeta A., Bellerba F., Origgi D., De Marco P., et al. Radiomic Features Applied to Contrast Enhancement Spectral Mammography: Possibility to Predict Breast Cancer Molecular Subtypes in a Non-Invasive Manner. Int. J. Mol. Sci. 2022;23:15322. doi: 10.3390/ijms232315322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pesapane F., Rotili A., Botta F., Raimondi S., Bianchini L., Corso F., Ferrari F., Penco S., Nicosia L., Bozzini A., et al. Radiomics of MRI for the Prediction of the Pathological Response to Neoadjuvant Chemotherapy in Breast Cancer Patients: A Single Referral Centre Analysis. Cancers. 2021;13:4271. doi: 10.3390/cancers13174271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi E.M.C., Invento A., Pesapane F., Pagan E., Bagnardi V., Fusco N., Venetis K., Dominelli V., Trentin C., Cassano E., et al. Diagnostic performance of image-guided vacuum-assisted breast biopsy after neoadjuvant therapy for breast cancer: Prospective pilot study. Br. J. Surg. 2023;110:217–224. doi: 10.1093/bjs/znac391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pesapane F., Suter M.B., Rotili A., Penco S., Nigro O., Cremonesi M., Bellomi M., Jereczek-Fossa B.A., Pinotti G., Cassano E. Will traditional biopsy be substituted by radiomics and liquid biopsy for breast cancer diagnosis and characterisation? Med. Oncol. 2020;37:29. doi: 10.1007/s12032-020-01353-1. [DOI] [PubMed] [Google Scholar]
- 21.Lambin P., Leijenaar R.T.H., Deist T.M., Peerlings J., de Jong E.E.C., van Timmeren J., Sanduleanu S., Larue R.T.H.M., Even A.J.G., Jochems A., et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 22.Yip S.S.F., Parmar C., Kim J., Huynh E., Mak R.H., Aerts H. Impact of experimental design on PET radiomics in predicting somatic mutation status. Eur. J. Radiol. 2017;97:8–15. doi: 10.1016/j.ejrad.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Parekh V.S., Jacobs M.A. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer. 2017;3:1–9. doi: 10.1038/s41523-017-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davnall F., Yip C.S., Ljungqvist G., Selmi M., Ng F., Sanghera B., Ganeshan B., Miles K.A., Cook G.J., Goh V. Assessment of tumor heterogeneity: An emerging imaging tool for clinical practice? Insights Imaging. 2012;3:573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aerts H.J., Velazquez E.R., Leijenaar R.T., Parmar C., Grossmann P., Carvalho S., Bussink J., Monshouwer R., Haibe-Kains B., Rietveld D., et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahmim A., Salimpour Y., Jain S., Blinder S.A., Klyuzhin I.S., Smith G.S., Mari Z., Sossi V. Application of texture analysis to DAT SPECT imaging: Relationship to clinical assessments. Neuroimage Clin. 2016;12:e1–e9. doi: 10.1016/j.nicl.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pesapane F., Patella F., Fumarola E.M., Panella S., Ierardi A.M., Pompili G.G., Franceschelli G., Angileri S.A., Biasina A.M., Carrafiello G. Intravoxel Incoherent Motion (IVIM) Diffusion Weighted Imaging (DWI) in the Periferic Prostate Cancer Detection and Stratification. Med. Oncol. 2017;34:35. doi: 10.1007/s12032-017-0892-7. [DOI] [PubMed] [Google Scholar]
- 28.Patella F., Franceschelli G., Petrillo M., Sansone M., Fusco R., Pesapane F., Pompili G., Ierardi A.M., Saibene A.M., Moneghini L., et al. A multiparametric analysis combining DCE-MRI- and IVIM -derived parameters to improve differentiation of parotid tumors: A pilot study. Future Oncol. 2018;14:2893–2903. doi: 10.2217/fon-2017-0655. [DOI] [PubMed] [Google Scholar]
- 29.King A.D., Chow K.K., Yu K.H., Mo F.K., Yeung D.K., Yuan J., Bhatia K.S., Vlantis A., Ahuja A.T. Head and neck squamous cell carcinoma: Diagnostic performance of diffusion-weighted MR imaging for the prediction of treatment response. Radiology. 2013;266:531–538. doi: 10.1148/radiol.12120167. [DOI] [PubMed] [Google Scholar]
- 30.Peng S.L., Chen C.F., Liu H.L., Lui C.C., Huang Y.J., Lee T.H., Chang C.-C., Wang F.-N. Analysis of parametric histogram from dynamic contrast-enhanced MRI: Application in evaluating brain tumor response to radiotherapy. NMR Biomed. 2013;26:443–450. doi: 10.1002/nbm.2882. [DOI] [PubMed] [Google Scholar]
- 31.Nicosia L., Bozzini A.C., Palma S., Montesano M., Signorelli G., Pesapane F., Latronico A., Bagnardi V., Frassoni S., Sangalli C., et al. Contrast-Enhanced Spectral Mammography and tumor size assessment: A valuable tool for appropriate surgical management of breast lesions. Radiol. Med. 2022;127:1228–1234. doi: 10.1007/s11547-022-01561-x. [DOI] [PubMed] [Google Scholar]
- 32.Suter M.B., Pesapane F., Agazzi G.M., Gagliardi T., Nigro O., Bozzini A., Priolo F., Penco S., Cassano E., Chini C., et al. Diagnostic accuracy of contrast-enhanced spectral mammography for breast lesions: A systematic review and meta-analysis. Breast. 2020;53:8–17. doi: 10.1016/j.breast.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penco S., Rotili A., Pesapane F., Trentin C., Dominelli V., Faggian A., Farina M., Marinucci I., Bozzini A., Pizzamiglio M., et al. MRI-guided vacuum-assisted breast biopsy: Experience of a single tertiary referral cancer centre and prospects for the future. Med. Oncol. 2020;37:36. doi: 10.1007/s12032-020-01358-w. [DOI] [PubMed] [Google Scholar]
- 34.Bakker M.F., de Lange S.V., Pijnappel R.M., Mann R.M., Peeters P.H.M., Monninkhof E.M., Emaus M.J., Loo C.E., Bisschops R.H., Lobbes M.B., et al. Supplemental MRI Screening for Women with Extremely Dense Breast Tissue. N. Engl. J. Med. 2019;381:2091–2102. doi: 10.1056/NEJMoa1903986. [DOI] [PubMed] [Google Scholar]
- 35.Kuhl C., Weigel S., Schrading S., Arand B., Bieling H., Konig R., Tombach B., Leutner C., Rieber-Brambs A., Nordhoff D., et al. Prospective multicenter cohort study to refine management recommendations for women at elevated familial risk of breast cancer: The EVA trial. J. Clin. Oncol. 2010;28:1450–1457. doi: 10.1200/JCO.2009.23.0839. [DOI] [PubMed] [Google Scholar]
- 36.Nicosia L., Bozzini A.C., Palma S., Montesano M., Pesapane F., Ferrari F., Dominelli V., Rotili A., Meneghetti L., Frassoni S., et al. A Score to Predict the Malignancy of a Breast Lesion Based on Different Contrast Enhancement Patterns in Contrast-Enhanced Spectral Mammography. Cancers. 2022;14:4337. doi: 10.3390/cancers14174337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parikh J., Selmi M., Charles-Edwards G., Glendenning J., Ganeshan B., Verma H., Mansi J., Harries M., Tutt A., Goh V. Changes in primary breast cancer heterogeneity may augment midtreatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology. 2014;272:100–112. doi: 10.1148/radiol.14130569. [DOI] [PubMed] [Google Scholar]
- 38.Pickles M.D., Lowry M., Gibbs P. Pretreatment Prognostic Value of Dynamic Contrast-Enhanced Magnetic Resonance Imaging Vascular, Texture, Shape, and Size Parameters Compared With Traditional Survival Indicators Obtained From Locally Advanced Breast Cancer Patients. Investig. Radiol. 2016;51:177–185. doi: 10.1097/RLI.0000000000000222. [DOI] [PubMed] [Google Scholar]
- 39.Kim J.H., Ko E.S., Lim Y., Lee K.S., Han B.K., Ko E.Y., Hahn S.Y., Nam S.J. Breast Cancer Heterogeneity: MR Imaging Texture Analysis and Survival Outcomes. Radiology. 2017;282:665–675. doi: 10.1148/radiol.2016160261. [DOI] [PubMed] [Google Scholar]
- 40.Li H., Zhu Y., Burnside E.S., Huang E., Drukker K., Hoadley K.A., Fan C., Conzen S.D., Zuley M., Net J.M., et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2:16012. doi: 10.1038/npjbcancer.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holli K., Laaperi A.L., Harrison L., Luukkaala T., Toivonen T., Ryymin P., Dastidar P., Soimakallio S., Eskola H. Characterization of breast cancer types by texture analysis of magnetic resonance images. Acad. Radiol. 2010;17:135–141. doi: 10.1016/j.acra.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Kato F., Oyama-Manabe N., Li R., Cui Y., Tha K.K., Yamashita H., Kudo K., Shirato H. Identifying Triple-Negative Breast Cancer Using Background Parenchymal Enhancement Heterogeneity on Dynamic Contrast-Enhanced MRI: A Pilot Radiomics Study. PLoS ONE. 2015;10:e0143308. doi: 10.1371/journal.pone.0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan M., Li H., Wang S., Zheng B., Zhang J., Li L. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS ONE. 2017;12:e0171683. doi: 10.1371/journal.pone.0171683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y., Li H., Guo W., Drukker K., Lan L., Giger M.L., Ji Y. Deciphering Genomic Underpinnings of Quantitative MRI-based Radiomic Phenotypes of Invasive Breast Carcinoma. Sci. Rep. 2015;5:17787. doi: 10.1038/srep17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.James D., Clymer B.D., Schmalbrock P. Texture detection of simulated microcalcification susceptibility effects in magnetic resonance imaging of breasts. J. Magn. Reson. Imaging. 2001;13:876–881. doi: 10.1002/jmri.1125. [DOI] [PubMed] [Google Scholar]
- 46.Dong Y., Feng Q., Yang W., Lu Z., Deng C., Zhang L., Lian Z., Liu J., Luo X., Pei S., et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur. Radiol. 2018;28:582–591. doi: 10.1007/s00330-017-5005-7. [DOI] [PubMed] [Google Scholar]
- 47.Bickelhaupt S., Paech D., Kickingereder P., Steudle F., Lederer W., Daniel H., Götz M., Gählert N., Tichy D., Wiesenfarth M., et al. Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography. J. Magn. Reson. Imaging. 2017;46:604–616. doi: 10.1002/jmri.25606. [DOI] [PubMed] [Google Scholar]
- 48.Li H., Zhu Y., Burnside E.S., Drukker K., Hoadley K.A., Fan C., Conzen S.D., Whitman G.J., Sutton E.J., Net J.M., et al. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology. 2016;281:382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obeid J.P., Stoyanova R., Kwon D., Patel M., Padgett K., Slingerland J., Takita C., Alperin N., Yepes M., Zeidan Y.H. Multiparametric evaluation of preoperative MRI in early stage breast cancer: Prognostic impact of peri-tumoral fat. Clin. Transl. Oncol. 2017;19:211–218. doi: 10.1007/s12094-016-1526-9. [DOI] [PubMed] [Google Scholar]
- 50.Fan M., Wu G., Cheng H., Zhang J., Shao G., Li L. Radiomic analysis of DCE-MRI for prediction of response to neoadjuvant chemotherapy in breast cancer patients. Eur. J. Radiol. 2017;94:140–147. doi: 10.1016/j.ejrad.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 51.Zhou J., Zhang Y., Chang K.T., Lee K.E., Wang O., Li J., Lin Y., Pan Z., Chang P., Chow D., et al. Diagnosis of Benign and Malignant Breast Lesions on DCE-MRI by Using Radiomics and Deep Learning With Consideration of Peritumor Tissue. J. Magn. Reson. Imaging. 2019;51:798–809. doi: 10.1002/jmri.26981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J., Sun D., Chen L., Fang Z., Song W., Guo D., Ni T., Liu C., Feng L., Xia Y., et al. Radiomics Analysis of Dynamic Contrast-Enhanced Magnetic Resonance Imaging for the Prediction of Sentinel Lymph Node Metastasis in Breast Cancer. Front. Oncol. 2019;9:980. doi: 10.3389/fonc.2019.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park H., Lim Y., Ko E.S., Cho H.H., Lee J.E., Han B.K., Choi J.S., Park K.W. Radiomics Signature on Magnetic Resonance Imaging: Association with Disease-Free Survival in Patients with Invasive Breast Cancer. Clin. Cancer Res. 2018;24:4705–4714. doi: 10.1158/1078-0432.CCR-17-3783. [DOI] [PubMed] [Google Scholar]
- 54.Guo Y., Hu Y., Qiao M., Wang Y., Yu J., Li J., Chang C. Radiomics Analysis on Ultrasound for Prediction of Biologic Behavior in Breast Invasive Ductal Carcinoma. Clin. Breast Cancer. 2018;18:e335–e344. doi: 10.1016/j.clbc.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Antunovic L., Gallivanone F., Sollini M., Sagona A., Invento A., Manfrinato G., Kirienko M., Tinterri C., Chiti A., Castiglioni I. [(18)F]FDG PET/CT features for the molecular characterization of primary breast tumors. Eur. J. Nucl. Med. Mol. Imaging. 2017;44:1945–1954. doi: 10.1007/s00259-017-3770-9. [DOI] [PubMed] [Google Scholar]
- 56.Ou X., Zhang J., Wang J., Pang F., Wang Y., Wei X., Ma X. Radiomics based on (18) F-FDG PET/CT could differentiate breast carcinoma from breast lymphoma using machine-learning approach: A preliminary study. Cancer Med. 2019;9:496–506. doi: 10.1002/cam4.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakai A., Onishi Y., Matsui M., Adachi H., Teramoto A., Saito K., Fujita H. A method for the automated classification of benign and malignant masses on digital breast tomosynthesis images using machine learning and radiomic features. Radiol. Phys. Technol. 2019;13:27–36. doi: 10.1007/s12194-019-00543-5. [DOI] [PubMed] [Google Scholar]
- 58.Le Bihan D. Intravoxel incoherent motion perfusion MR imaging: A wake-up call. Radiology. 2008;249:748–752. doi: 10.1148/radiol.2493081301. [DOI] [PubMed] [Google Scholar]
- 59.Penco S., Rizzo S., Bozzini A.C., Latronico A., Menna S., Cassano E., Bellomi M. Stereotactic vacuum-assisted breast biopsy is not a therapeutic procedure even when all mammographically found calcifications are removed: Analysis of 4,086 procedures. AJR Am. J. Roentgenol. 2010;195:1255–1260. doi: 10.2214/AJR.10.4208. [DOI] [PubMed] [Google Scholar]
- 60.Abbate F., Bacigalupo L., Latronico A., Trentin C., Penco S., Menna S., Viale G., Cassano E., Bellomi M. Ultrasound-guided vacuum assisted breast biopsy in the assessment of C3 breast lesions by ultrasound-guided fine needle aspiration cytology: Results and costs in comparison with surgery. Breast. 2009;18:73–77. doi: 10.1016/j.breast.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Thomassin-Naggara I., Lalonde L., David J., Darai E., Uzan S., Trop I. A plea for the biopsy marker: How, why and why not clipping after breast biopsy? Breast Cancer Res. Treat. 2012;132:881–893. doi: 10.1007/s10549-011-1847-x. [DOI] [PubMed] [Google Scholar]
- 62.Liberman L. Percutaneous image-guided core breast biopsy. Radiol. Clin. N. Am. 2002;40:483–500. doi: 10.1016/S0033-8389(01)00011-2. [DOI] [PubMed] [Google Scholar]
- 63.Bick U., Trimboli R.M., Athanasiou A., Balleyguier C., Baltzer P.A.T., Bernathova M., Borbély K., Brkljacic B., Carbonaro L.A., Clauser P., et al. Image-guided breast biopsy and localisation: Recommendations for information to women and referring physicians by the European Society of Breast Imaging. Insights Imaging. 2020;11:12. doi: 10.1186/s13244-019-0803-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madabhushi A., Udupa J.K. New methods of MR image intensity standardization via generalized scale. Med. Phys. 2006;33:3426–3434. doi: 10.1118/1.2335487. [DOI] [PubMed] [Google Scholar]
- 65.Rizzo S., Botta F., Raimondi S., Origgi D., Fanciullo C., Morganti A.G., Bellomi M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018;2:36. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saltybaeva N., Tanadini-Lang S., Vuong D., Burgermeister S., Mayinger M., Bink A., Andratschke N., Guckenberger M., Bogowicz M. Robustness of radiomic features in magnetic resonance imaging for patients with glioblastoma: Multi-center study. Phys. Imaging Radiat. Oncol. 2022;22:131–136. doi: 10.1016/j.phro.2022.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baessler B., Weiss K., Pinto Dos Santos D. Robustness and Reproducibility of Radiomics in Magnetic Resonance Imaging: A Phantom Study. Investig. Radiol. 2019;54:221–228. doi: 10.1097/RLI.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 68.van Timmeren J.E., Cester D., Tanadini-Lang S., Alkadhi H., Baessler B. Radiomics in medical imaging-“how-to” guide and critical reflection. Insights Imaging. 2020;11:91. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zanca F., Brusasco C., Pesapane F., Kwade Z., Beckers R., Avanzo M. Regulatory Aspects of the Use of Artificial Intelligence Medical Software. Semin. Radiat. Oncol. 2022;32:432–441. doi: 10.1016/j.semradonc.2022.06.012. [DOI] [PubMed] [Google Scholar]
- 70.Ollers M., Bosmans G., van Baardwijk A., Dekker A., Lambin P., Teule J., Thimister W., Rhamy A., De Ruysscher D. The integration of PET-CT scans from different hospitals into radiotherapy treatment planning. Radiother. Oncol. 2008;87:142–146. doi: 10.1016/j.radonc.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 71.Suzuki K. Overview of deep learning in medical imaging. Radiol. Phys. Technol. 2017;10:257–273. doi: 10.1007/s12194-017-0406-5. [DOI] [PubMed] [Google Scholar]
- 72.Collins G.S., Reitsma J.B., Altman D.G., Moons K.G. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD statement. Ann. Intern. Med. 2015;162:55–63. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 73.Katzen J., Dodelzon K. A review of computer aided detection in mammography. Clin. Imaging. 2018;52:305–309. doi: 10.1016/j.clinimag.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 74.Bitencourt A., Daimiel Naranjo I., Lo Gullo R., Rossi Saccarelli C., Pinker K. AI-enhanced breast imaging: Where are we and where are we heading? Eur. J. Radiol. 2021;142:109882. doi: 10.1016/j.ejrad.2021.109882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheth D., Giger M.L. Artificial intelligence in the interpretation of breast cancer on MRI. J. Magn. Reson. Imaging. 2020;51:1310–1324. doi: 10.1002/jmri.26878. [DOI] [PubMed] [Google Scholar]
- 76.Eschrich S., Yang I., Bloom G., Kwong K.Y., Boulware D., Cantor A., Coppola D., Kruhøffer M., Aaltonen L., Orntoft T.F., et al. Molecular staging for survival prediction of colorectal cancer patients. J. Clin. Oncol. 2005;23:3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 77.Mongan J., Moy L., Kahn C.E., Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020;2:e200029. doi: 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pesapane F., Rotili A., Valconi E., Agazzi G.M., Montesano M., Penco S., Nicosia L., Bozzini A., Meneghetti L., Latronico A., et al. Women’s perceptions and attitudes to the use of AI in breast cancer screening: A survey in a cancer referral centre. Br. J. Radiol. 2023;96:20220569. doi: 10.1259/bjr.20220569. [DOI] [PubMed] [Google Scholar]
- 79.Pesapane F., Volonte C., Codari M., Sardanelli F. Artificial intelligence as a medical device in radiology: Ethical and regulatory issues in Europe and the United States. Insights Imaging. 2018;9:745–753. doi: 10.1007/s13244-018-0645-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.American Cancer Society Breast Cancer Survival Rates by Stage. 2019. [(accessed on 9 January 2023)]. Available online: http://www.cancer.org/cancer/breastcancer/detailedguide/breast-cancer-survival-by-stage.
- 81.Bevers T.B., Helvie M., Bonaccio E., Calhoun K.E., Daly M.B., Farrar W.B., Garber J.E., Gray R., Greenberg C.C., Greenup R., et al. Breast Cancer Screening and Diagnosis, Version 3.2018, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018;16:1362–1389. doi: 10.6004/jnccn.2018.0083. [DOI] [PubMed] [Google Scholar]
- 82.Xie T., Wang Z., Zhao Q., Bai Q., Zhou X., Gu Y., Peng W. Machine Learning-Based Analysis of MR Multiparametric Radiomics for the Subtype Classification of Breast Cancer. Front. Oncol. 2019;9:505. doi: 10.3389/fonc.2019.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bickelhaupt S., Jaeger P.F., Laun F.B., Lederer W., Daniel H., Kuder T.A., Wuesthof L., Paech D., Bonekamp D., Radbruch A., et al. Radiomics Based on Adapted Diffusion Kurtosis Imaging Helps to Clarify Most Mammographic Findings Suspicious for Cancer. Radiology. 2018;287:761–770. doi: 10.1148/radiol.2017170273. [DOI] [PubMed] [Google Scholar]
- 84.Li H., Mendel K.R., Lan L., Sheth D., Giger M.L. Digital Mammography in Breast Cancer: Additive Value of Radiomics of Breast Parenchyma. Radiology. 2019;291:15–20. doi: 10.1148/radiol.2019181113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tagliafico A.S., Valdora F., Mariscotti G., Durando M., Nori J., La Forgia D., Rosenberg I., Caumo F., Gandolfo N., Houssami N., et al. An exploratory radiomics analysis on digital breast tomosynthesis in women with mammographically negative dense breasts. Breast. 2018;40:92–96. doi: 10.1016/j.breast.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 86.Luo W.Q., Huang Q.X., Huang X.W., Hu H.T., Zeng F.Q., Wang W. Predicting Breast Cancer in Breast Imaging Reporting and Data System (BI-RADS) Ultrasound Category 4 or 5 Lesions: A Nomogram Combining Radiomics and BI-RADS. Sci. Rep. 2019;9:11921. doi: 10.1038/s41598-019-48488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie T., Zhao Q., Fu C., Bai Q., Zhou X., Li L., Gu Y., Peng W. Differentiation of triple-negative breast cancer from other subtypes through whole-tumor histogram analysis on multiparametric MR imaging. Eur. Radiol. 2019;29:2535–2544. doi: 10.1007/s00330-018-5804-5. [DOI] [PubMed] [Google Scholar]
- 88.Demircioglu A., Grueneisen J., Ingenwerth M., Hoffmann O., Pinker-Domenig K., Morris E., Haubold J., Forsting M., Nensa F., Umutlu L. A rapid volume of interest-based approach of radiomics analysis of breast MRI for tumor decoding and phenotyping of breast cancer. PLoS ONE. 2020;15:e0234871. doi: 10.1371/journal.pone.0234871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tan W., Yang M., Yang H., Zhou F., Shen W. Predicting the response to neoadjuvant therapy for early-stage breast cancer: Tumor-, blood-, and imaging-related biomarkers. Cancer Manag. Res. 2018;10:4333–4347. doi: 10.2147/CMAR.S174435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tyagi N.K., Dhesy-Thind S. Clinical practice guidelines in breast cancer. Curr. Oncol. 2018;25((Suppl. S1)):S151–S160. doi: 10.3747/co.25.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Z., Li Z., Qu J., Zhang R., Zhou X., Li L., Sun K., Tang Z., Jiang H., Li H., et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin. Cancer Res. 2019;25:3538–3547. doi: 10.1158/1078-0432.CCR-18-3190. [DOI] [PubMed] [Google Scholar]
- 92.Xiong Q., Zhou X., Liu Z., Lei C., Yang C., Yang M., Zhang L., Zhu T., Zhuang X., Liang C., et al. Multiparametric MRI-based radiomics analysis for prediction of breast cancers insensitive to neoadjuvant chemotherapy. Clin. Transl. Oncol. 2019;22:50–59. doi: 10.1007/s12094-019-02109-8. [DOI] [PubMed] [Google Scholar]
- 93.Drukker K., Li H., Antropova N., Edwards A., Papaioannou J., Giger M.L. Most-enhancing tumor volume by MRI radiomics predicts recurrence-free survival "early on" in neoadjuvant treatment of breast cancer. Cancer Imaging. 2018;18:12. doi: 10.1186/s40644-018-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cain E.H., Saha A., Harowicz M.R., Marks J.R., Marcom P.K., Mazurowski M.A. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: A study using an independent validation set. Breast Cancer Res. Treat. 2018;173:455–463. doi: 10.1007/s10549-018-4990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Teruel J.R., Heldahl M.G., Goa P.E., Pickles M., Lundgren S., Bathen T.F., Gibbs P. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed. 2014;27:887–896. doi: 10.1002/nbm.3132. [DOI] [PubMed] [Google Scholar]
- 96.Braman N.M., Etesami M., Prasanna P., Dubchuk C., Gilmore H., Tiwari P., Plecha D., Madabhushi A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19:57. doi: 10.1186/s13058-017-0846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choudhery S., Gomez-Cardona D., Favazza C.P., Hoskin T.L., Haddad T.C., Goetz M.P., Boughey J.C. MRI Radiomics for Assessment of Molecular Subtype, Pathological Complete Response, and Residual Cancer Burden in Breast Cancer Patients Treated With Neoadjuvant Chemotherapy. Acad. Radiol. 2020;29:S145–S154. doi: 10.1016/j.acra.2020.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahmed A., Gibbs P., Pickles M., Turnbull L. Texture analysis in assessment and prediction of chemotherapy response in breast cancer. J. Magn. Reson. Imaging. 2013;38:89–101. doi: 10.1002/jmri.23971. [DOI] [PubMed] [Google Scholar]
- 99.Galimberti V., Cole B.F., Viale G., Veronesi P., Vicini E., Intra M., Mazzarol G., Massarut S., Zgajnar J., Taffurelli M., et al. Axillary dissection versus no axillary dissection in patients with breast cancer and sentinel-node micrometastases (IBCSG 23-01): 10-year follow-up of a randomised, controlled phase 3 trial. Lancet Oncol. 2018;19:1385–1393. doi: 10.1016/S1470-2045(18)30380-2. [DOI] [PubMed] [Google Scholar]
- 100.Hieken T.J., Trull B.C., Boughey J.C., Jones K.N., Reynolds C.A., Shah S.S., Glazebrook K.N. Preoperative axillary imaging with percutaneous lymph node biopsy is valuable in the contemporary management of patients with breast cancer. Surgery. 2013;154:831–838. doi: 10.1016/j.surg.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 101.Tan H., Wu Y., Bao F., Zhou J., Wan J., Tian J., Lin Y., Wang M. Mammography-based radiomics nomogram: A potential biomarker to predict axillary lymph node metastasis in breast cancer. Br. J. Radiol. 2020;93:20191019. doi: 10.1259/bjr.20191019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang J., Wang T., Yang L., Wang Y., Li H., Zhou X., Zhao W., Ren J., Li X., Tian J., et al. Preoperative Prediction of Axillary Lymph Node Metastasis in Breast Cancer Using Mammography-Based Radiomics Method. Sci. Rep. 2019;9:4429. doi: 10.1038/s41598-019-40831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu Y., Tan Y., Xie C., Hu Q., Ouyang J., Chen Y., Gu Y., Li A., Lu N., He Z., et al. Development and Validation of a Preoperative Magnetic Resonance Imaging Radiomics-Based Signature to Predict Axillary Lymph Node Metastasis and Disease-Free Survival in Patients With Early-Stage Breast Cancer. JAMA Netw. Open. 2020;3:e2028086. doi: 10.1001/jamanetworkopen.2020.28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mao N., Yin P., Li Q., Wang Q., Liu M., Ma H., Dong J., Che K., Wang Z., Duan S., et al. Radiomics nomogram of contrast-enhanced spectral mammography for prediction of axillary lymph node metastasis in breast cancer: A multicenter study. Eur. Radiol. 2020;30:6732–6739. doi: 10.1007/s00330-020-07016-z. [DOI] [PubMed] [Google Scholar]
- 105.Yu Y., He Z., Ouyang J., Tan Y., Chen Y., Gu Y., Mao L., Ren W., Wang J., Lin L., et al. Magnetic resonance imaging radiomics predicts preoperative axillary lymph node metastasis to support surgical decisions and is associated with tumor microenvironment in invasive breast cancer: A machine learning, multicenter study. EbioMedicine. 2021;69:103460. doi: 10.1016/j.ebiom.2021.103460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cattell R., Ying J., Lei L., Ding J., Chen S., Serrano Sosa M., Huang C. Preoperative prediction of lymph node metastasis using deep learning-based features. Vis. Comput. Ind. Biomed. Art. 2022;5:8. doi: 10.1186/s42492-022-00104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fisher B., Jeong J.H., Anderson S., Bryant J., Fisher E.R., Wolmark N. Twenty-five-year follow-up of a randomized trial comparing radical mastectomy, total mastectomy, and total mastectomy followed by irradiation. N. Engl. J. Med. 2002;347:567–575. doi: 10.1056/NEJMoa020128. [DOI] [PubMed] [Google Scholar]
- 108.Chitalia R.D., Rowland J., McDonald E.S., Pantalone L., Cohen E.A., Gastounioti A., Feldman M., Schnall M., Conant E., Kontos D. Imaging Phenotypes of Breast Cancer Heterogeneity in Preoperative Breast Dynamic Contrast Enhanced Magnetic Resonance Imaging (DCE-MRI) Scans Predict 10-Year Recurrence. Clin. Cancer Res. 2020;26:862–869. doi: 10.1158/1078-0432.CCR-18-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mazurowski M.A., Saha A., Harowicz M.R., Cain E.H., Marks J.R., Marcom P.K. Association of distant recurrence-free survival with algorithmically extracted MRI characteristics in breast cancer. J. Magn. Reson. Imaging. 2019;49:e231–e240. doi: 10.1002/jmri.26648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park G.E., Kim S.H., Lee E.B., Nam Y., Sung W. Ipsilateral Recurrence of DCIS in Relation to Radiomics Features on Contrast Enhanced Breast MRI. Tomography. 2022;8:596–606. doi: 10.3390/tomography8020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiong L., Chen H., Tang X., Chen B., Jiang X., Liu L., Feng Y., Liu L., Li L. Ultrasound-Based Radiomics Analysis for Predicting Disease-Free Survival of Invasive Breast Cancer. Front. Oncol. 2021;11:621993. doi: 10.3389/fonc.2021.621993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu F., Hang J., Deng J., Yang B., Wang J., Ye X., Liu Y. Radiomics features on ultrasound imaging for the prediction of disease-free survival in triple negative breast cancer: A multi-institutional study. Br. J. Radiol. 2021;94:20210188. doi: 10.1259/bjr.20210188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dasgupta A., Bhardwaj D., DiCenzo D., Fatima K., Osapoetra L.O., Quiaoit K., Saifuddin M., Brade S., Trudeau M., Gandhi S., et al. Radiomics in predicting recurrence for patients with locally advanced breast cancer using quantitative ultrasound. Oncotarget. 2021;12:2437–2448. doi: 10.18632/oncotarget.28139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mao N., Yin P., Zhang H., Zhang K., Song X., Xing D., Chu T. Mammography-based radiomics for predicting the risk of breast cancer recurrence: A multicenter study. Br. J. Radiol. 2021;94:20210348. doi: 10.1259/bjr.20210348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tselikas L., Sun R., Ammari S., Dercle L., Yevich S., Hollebecque A., Ngo-Camus M., Nicotra C., Deutsch E., Deschamps F., et al. Role of image-guided biopsy and radiomics in the age of precision medicine. Chin. Clin. Oncol. 2019;8:57. doi: 10.21037/cco.2019.12.02. [DOI] [PubMed] [Google Scholar]
- 116.Cancer Genome Atlas Research Network. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.The Cancer Genome Atlas. TCGA. [(accessed on 8 January 2023)]; Available online: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.
- 118.The Cancer Imaging Archive. TCIA. [(accessed on 8 January 2019)]. Available online: http://www.cancerimagingarchive.net.
- 119.Sun R., Limkin E.J., Vakalopoulou M., Dercle L., Champiat S., Han S.R., Verlingue L., Brandao D., Lancia A., Ammari S., et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.