Abstract
We have previously identified a locus, fsr, a homologue of staphylococcal agr loci, which positively regulates the expression of gelatinase and serine protease (encoded by gelE and sprE, respectively) in Enterococcus faecalis OG1RF. The expression of the three genes in the fsr locus, fsrA, fsrB, and fsrC, appears to be autoregulated, and we have shown that mutants with insertion disruptions in each of these three genes were significantly attenuated in a mouse peritonitis model compared to the parent strain. In the present study, we showed that fsrB and fsrC are highly expressed in the postexponential growth phase and that their expression is cell density dependent. Reverse transcriptase PCR using primers covering the intergenic regions in the fsr/gelE loci confirmed that fsrB and fsrC, as well as gelE and sprE, are cotranscribed. We also showed, using a nonpolar fsrB deletion mutant, that fsrB, the homologue of agrB of staphylococci with unknown function, is required for the regulatory function of fsr. Primer extension and analysis of transcriptional fusions indicated the presence of promoters immediately upstream of fsrA, of fsrB, and of gelE and that the fsrB and gelE promoters are fsr dependent, while the fsrA promoter is an fsr-independent weak constitutive promoter. Two conserved 7-bp direct repeats were found immediately upstream of the fsrB and gelE promoters, similar to the repeats found upstream of P2 and P3 promoters of the agr locus; deletions and mutations in the repeated sequences completely abolished the fsrB and gelE promoter activities, suggesting that the repeats are important for the regulatory function in the fsrB and gelE promoter regions.
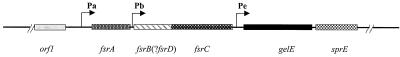
Enterococci are one of the leading causes of nosocomial infections, including urinary tract infections, bloodstream infections, wound infections, and endocarditis (12). In a previous study, we identified a locus, fsr (Enterococcus faecalis regulator), which contains three agr-like genes (22), immediately upstream of gelE (Fig. 1), which encodes a gelatinase (28), in E. faecalis strain OG1RF. These three agr-like genes, fsrA, fsrB, and fsrC, which show homology to agrA, agrB, and agrC, respectively, in the agr (accessory gene regulator) locus of Staphylococcus aureus, appeared to be autoregulated and to regulate the expression of gelE and sprE, a gene encoding a serine protease, which is downstream of gelE and which appeared to be cotranscribed with gelE (22). Mutants with insertion disruptions in these fsr genes showed significant delays in lethality in a mouse peritonitis model compared to wild-type OG1RF (22).
FIG. 1.
Open reading frames in fsr/gelE loci. Line, chromosome; boxes, genes and open reading frames; ?fsrD, possible agrD homologue at the 3′ end of fsrB; arrows, promoters indicating directions of transcription. Pa, fsrA promoter; Pb, fsrB promoter; Pe, gelE promoter.
S. aureus agr/hld loci temporally control the expression of various virulence factors by positively regulating the expression of secreted proteins such as alpha-toxin, β-toxin, δ-toxin, enterotoxin B, toxic shock syndrome toxin 1, and a serine protease and by negatively regulating the expression of surface proteins such as protein A, coagulase, and fibronectin-binding protein in the postexponential growth phase (3, 7, 8, 10, 23). The agr locus in S. aureus consists of four genes, agrA, agrB, agrC, and agrD, which all appear to be required for the Agr function (15, 16, 19, 23). The expression of agr genes is autoregulated, and the expression of agrB, agrC, and agrD is driven by agr-dependent promoter P2, while another agr-dependent promoter, P3, in the opposite direction from P2, regulates the expression of the P3 transcript (referred to as RNAIII), which is the real effector of the Agr response (15, 16). The expression of agrA appears to be driven by a weak constitutive promoter upstream of agrA (19). The agr genes encode a quorum-sensing system in which an autoinducing peptide encoded by agrD, possibly processed and/or secreted by AgrB, functions as an autoinducer to activate the expression of the agr genes and RNAIII (5, 6). AgrC and AgrA, which are the sensor transducer and the response regulator of typical bacterial two-component systems, respectively, are thought to sense cell density through the autoinducing peptide and subsequently regulate the expression of virulence properties (5, 15, 16). Based on the cross-activation and cross-inhibition by autoinducing peptides, S. aureus strains can be divided into at least three different groups, in which the pheromones from the strains in one group cross-activate the agr expression of other strains in that group but inhibit the agr expression of strains in the other groups (5).
In this work, we investigated whether fsrB of E. faecalis is required for fsr functions by deletion mutagenesis. We also characterized the fsr/gelE loci by reverse transcriptase PCR (RT-PCR), Northern blot analysis, primer extension, and gene fusion analyses. Our results suggest that fsrB is required for fsr functions, that the expression of fsrA, fsrB, fsrC, gelE, and sprE is driven by three different promoters, and that the expression of fsrB and fsrC is cell density dependent. Our data also suggest that two 7-bp direct repeats upstream of fsrB and gelE promoters are important for the regulation of fsrB and gelE expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Most of the bacterial strains and plasmids used in this study are listed in Table 1. E. faecalis OG1RF has been described previously (13); an additional eight gelatinase-positive (Gel+) E. faecalis strains from different clinical sources and geographical areas shown by pulsed-field gel electrophoresis to represent distinct strains were also used. Escherichia coli DH5α was used as the host strain for routine cloning. Shuttle vector pTCV-lac (20), which contains a promoterless lacZ, was used for detection of promoter activity in E. faecalis. Luria-Bertani broth and agar were used for E. coli culture, and brain heart infusion (BHI; Difco Laboratories, Detroit, Mich.) was used for E. faecalis culture unless otherwise stated. The concentrations of antibiotics used for selection were as follows: ampicillin, 50 μg/ml (E. coli); erythromycin, 250 (E. coli) and 10 μg/ml (E. faecalis); kanamycin (KAN), 50 (E. coli) and 2,000 μg/ml (E. faecalis).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant properties | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | E. coli host strain for pBluescript SK(−) | Stratagene |
| DH5α(pTCV-lac) | E. coli DH5α containing plasmid pTCV-lac | 20 |
| TX4577 | E. coli DH5α containing plasmid pTEX4577 | 27 |
| E. faecalis | ||
| OG1RF | Gel+ serine protease positive (Spr+) Rifr Fusr | 13 |
| TX5240 | OG1RF fsrA mutant with pTEX4577 insertion in fsrA; Gel− Spr− Kanr | 22 |
| TX5241 | OG1RF fsrB mutant with pTEX4577 insertion in fsrB; Gel− Spr− Kanr | 22 |
| TX5242 | OG1RF fsrC mutant with pTEX4577 insertion in fsrC; Gel− Spr− Kanr | 22 |
| TX5266 | OG1RF fsrB deletion mutant, deletion from bp 79 to 684 of fsrB; Gel− Spr− | This study |
| Plasmids | ||
| pBluescript SK(−) | Cloning vector; Ampr | Stratagene |
| pTEX4577 | Suicide vector in E. faecalis derived from pBluescript SK(−); Kanr | 27 |
| pTCV-lac | Shuttle vector containing promoterless lacZ; Kanr Eryr | 20 |
| pTEX5267 | pTEX4577 containing fsrB flanking regions (917-bp 5′ region: bp −839 to +78, amplified using BDF1 and BDR1 primers; 1,065-bp 3′ region: 45 bp before stop codon to 1,020 bp after stop codon, amplified using DBF2 and DBR2 primers), used for construction of fsrB deletion mutant; Kanr | This study |
| pTEX5268 | fsrA promoter cloned upstream of lacZ in pTCV-lac, from bp −406 to −6 (401 bp, amplified using APRF1 and APRR1 primers) relative to fsrA start codon; Kanr Eryr | This study |
| pTEX5269 | fsrB promoter cloned upstream of lacZ in pTCV-lac, from bp −110 to −8 (103 bp, amplified using BPRF1 and BPRR1 primers) relative to fsrB start codon; Kanr Eryr | This study |
| pTEX5270 | gelE promoter cloned upstream of lacZ in pTCV-lac, from bp −218 to −16 (203 bp, amplified using EPRF1 and EPRR1 primers) relative to gelE start codon; Kanr Eryr | This study |
| pTEX5298 | fsrB promoter region (bp −90 to −8 relative to fsrB start codon, amplified using BPRF1 and BMP1 primers) cloned upstream of lacZ in pTCV-lac; Kanr Eryr | This study |
| pTEX5299 | fsrB promoter region (bp −72 to −8 relative to fsrB start codon, amplified using BPRF1 and BMP2 primers) cloned upstream of lacZ in pTCV-lac; Kanr Eryr | This study |
| pTEX5300 | fsrB promoter region (bp −85 to −8 relative to fsrB start codon, amplified using BPRF1 and BMP4 primers) cloned upstream of lacZ in pTCV-lac; Kanr Eryr | This study |
| pTEX5301 | fsrB promoter region (bp −91 to −8 relative to fsrB start codon in which bp −70 to −65 were altered from AAGGAA to TTCCTT, amplified using BPRF1 and BMP5 primers) cloned upstream of lacZ in pTCV-lac; Kanr Eryr | This study |
| pTEX5302 | fsrB promoter region (bp −72 to −8 relative to fsrB start codon) with putative gelE promoter regulatory region (bp −188 to −170 relative to gelE start codon), amplified using BPRF1 and BMP6 primers, cloned upstream of lacZ in pTCV-lac; Kanr Eryr | This study |
| pTEX5303 | gelE promoter region (bp −188 to −16 relative to gelE start codon, amplified using EPRF1 and EMP1 primers) cloned upstream of lacZ in pTCV-lac; Kanr Eryr | This study |
| pTEX5304 | gelE promoter region (bp −170 to −16 relative to gelE start codon, amplified using EPRF1 and EMP2 primers) cloned upstream of lacZ in pTCV-lac; Kanr Eryr | This study |
| pTEX5305 | gelE promoter region (bp −188 to −16 relative to gelE start codon in which bp −167 to −161 were altered from AAGGAA to TTCCTT, amplified using EPRF1 and EMP5 primers) cloned upstream of lacZ in pTCV-lac; Kanr Eryr | This study |
| pTEX5306 | gelE promoter region (bp −170 to −16 relative to gelE start codon) with putative fsrB promoter regulatory region (bp −91 to −73 relative to fsrB start codon), amplified using EPRF1 and BMP6 primers, cloned upstream of lacZ in pTCV-lac; Kanr Eryr | This study |
DNA techniques.
Routine isolation of plasmid DNA from E. coli was performed as previously described (2). Large-scale preparation of plasmid DNA was carried out using the Midi kit or Maxi kit (Qiagen, Valencia, Calif.). Transformation of E. faecalis was accomplished by the method described previously by Li et al. (9) using a Gene Pulser (Bio-Rad, Hercules, Calif.). Genomic DNA from E. faecalis was prepared according to the method described by Wilson (29). PCR amplification of DNA was performed on a DNA thermal cycler (Perkin-Elmer Corp., Norwalk, Conn.) using synthetic oligonucleotide primers and Taq DNA polymerase from Life Technologies (Gaithersburg, Md.). The primers used in the PCR amplification are listed in Table 2.
TABLE 2.
Primers
| Primer | Length (mer) | Sequence (5′–3′)a, location, and relevant properties | Reference or source |
|---|---|---|---|
| Used in fsrB deletion mutagenesis | |||
| BDF1 | 31 | AAA GAG CTC GAA AGG GAT GAG TGA ACA AAT G, from bp −839 to −818 to start codon of fsrB, outer primer used for amplification of fsrB 5′ region | This study |
| BDR1 | 31 | CCG AAT TCC TTT GTC CAT TGT GTT TTT CCT G, from bp 78 to 56 in fsrB, inner primer used for amplification of fsrB 5′ region | This study |
| BDF2 | 31 | CCG AAT TCT GGA TGG GAC AAA CTG AAA AAC A, from bp 54 to 23 before fsrB stop codon, inner primer used for amplification of fsrB 3′ region | This study |
| BDR2 | 31 | CCG GTA CCT AGC CAA CAA ACG AAT CAC AAC C, from bp 1020 to 999 after fsrB stop codon, outer primer used for amplification of fsrB 3′ region | This study |
| Used for cotranscription study | |||
| fsrARTF1 | 22 | ACA ATA CTT CTA ACG CTT TTG C, from bp 70 to 49 after fsrA start codon | This study |
| orf1RTR1 | 20 | TAG TGG CGT GAC CTA CAT TG, from bp 28 to 9 before orf1 stop codon | This study |
| fsrBRTF1 | 21 | GAT CTT CCT GAT CCA TAT CCA, from bp 52 to 32 after fsrB start codon | This study |
| fsrARTR1 | 23 | CAA GGC ACT ATT TCT TAC TTA GG, from bp 22 before fsrA stop codon to bp 1 after fsrA stop codon | This study |
| fsrCRTF1 | 21 | ACT CGT AAA AAG ACA AAA ACG, from bp 71 to 51 after fsrC start codon | This study |
| fsrBRTR1 | 20 | TGA TGG TGT GGG AAC TAA GC, from bp 103 to 84 before fsrB stop codon | This study |
| gelERTF1 | 21 | AGT ATT GCT TTA TCC TCC CTA, from bp 51 to 71 before gelE start codon | This study |
| fsrCRTR1 | 21 | CTA AAA GTA ATC CAG AAG AGC, from bp 164 to 144 before fsrC stop codon | This study |
| sprERTF1 | 22 | ATC AGA AAC AAA AAA CCA GCA C, from bp 74 to 53 after sprE start codon | This study |
| gelERTR1 | 19 | CTC GTG ATG CGA TGC TTG C, from bp 148 to 130 before gelE stop codon | This study |
| Used for primer extension study | |||
| APE1 | 22 | ACA ATA CTT CTA ACG CTT TTG C, from bp 70 to 49 after fsrA start codon | This study |
| BPE2 | 21 | TAA GCC AGA AAA ATA AAC GGT, from bp 108 to 88 after fsrB start codon | This study |
| EPE2 | 21 | CCA ACA AAG ATG CCT GTA CCT, from bp 50 to 30 after gelE start codon | This study |
| Used for study of promoter activity | |||
| APRF1 | 32 | CGG GAT CCT CTC AAG AAG ATA TAA AAT TAC AC, from bp −6 to −29 to fsrA start codon, used for amplification of fsrA promoter | This study |
| APRR1 | 28 | CGG AAT TCG CGA CTC ATT CAA CAG GAA G, from bp −406 to −387 to fsrA start codon, used for amplification of fsrA promoter | This study |
| BPRF1 | 29 | CGG GAT CCT CCT CTT CAA GTA TTG CAC TA, from bp −8 to −28 to fsrB start codon, used for amplification of fsrB promoter | This study |
| BPRR1 | 30 | CGG AAT TCC AAG GCA CTA TTT CTT ACT TAG, from bp −110 to −89 to fsrB start codon, used for amplification of fsrB promoter | This study |
| EPRF1 | 30 | CGG GAT CCT TCC CCA GTT TCC TTT TAT TTC, from bp −16 to −37 to gelE start codon, used for amplification of gelE promoter | This study |
| EPRR1 | 29 | CGG AAT TCG CTA TGG TAT TGA GTT ATG AG, from bp −218 to −198 to gelE start codon, used for amplification of gelE promoter | This study |
| BMP1 | 28 | CGG AAT TCA GGG AGG GAT AAT GAC TAA T, from bp −90 to −71 to fsrB start codon | This study |
| BMP2 | 32 | CGG AAT TCA TTA AGG AAT TAT CTA TCT ATT AG, from bp −72 to −49 to fsrB start codon | This study |
| BMP4 | 57 | CGG AAT TCG GGA TAA TGA CTA ATT AAG GAA TTA TCT ATC TAT TAG TCG CTA TAT TCG, from bp −85 to −37 to fsrB start codon | This study |
| BMP5 | 63 | CGG AAT TCT AGG GAG GGA TAA TGA CTA ATA TTC CTT TTA TCT ATC TAT TAG TCG CTA TAT TCG, from bp −91 to −37 to fsrB start codon, bp −70 to −64 sequence TAA GGA A was changed to ATT CCT T | This study |
| BMP6 | 62 | CGG AAT TCA GGG AAA AAT GTC GGC TGA TTA AGG AAT TAT CTA TCT ATT AGT CGC TAT ATT CG, from bp −188 to −170 to gelE start codon and from bp −72 to −37 to fsrB start codon | This study |
| EMP1 | 27 | CGG AAT TCA GGG AAA AAT GTC GGC TGA, from bp −188 to −170 to gelE start codon | This study |
| EMP2 | 30 | CGG AAT TCA TTA AGG AAT TTA GAT AGT GCC, from bp −170 to −149 to gelE start codon | This study |
| EMP5 | 55 | CGG AAT TCA GGG AAA AAT GTC GGC TGA TTT TCC TTT TTA GAT AGT GCC GGT TAG G, from bp −188 to −142 to gelE start codon, bp −167 to −162 sequence AAG GAA was changed to TTC CTT | This study |
| EMP6 | 56 | CGG AAT TCT AGG GAG GGA TAA TGA CTA ATT AAG GAA TTT AGA TAG TGC CGG TTA GG, from bp −91 to −73 to fsrB start codon and bp −170 to −142 to gelE start codon | This study |
| Vlac1 | 23 | GTT GAA TAA CAC TTA TTC CTA TC, flanking pTCV-lac cloning sites, used for sequencing inserts in pTCV-lac | 20 |
| Vlac2 | 21 | CTT CCA CAG TAG TTC ACC ACC, flanking pTCV-lac cloning sites, used for sequencing inserts in pTCV-lac | 20 |
| Others | |||
| lytF1 | 20 | ACA CCA ACC ACA GAA ACT AC, from bp 226 to 245 in E. faecalis autolysin gene | This study |
| lytR1 | 20 | GGC AAT AAA TTC TGA AGG AC, from bp 555 to 536 in E. faecalis autolysin gene | This study |
| gelEF1 | 21 | TGG TTG TGA TTC GTT TGT TGG, from bp −508 to −561 to gelE start codon | This study |
| GBR2 | 22 | TGA CCA GAA CAG ATT CAC TTG G, from bp 9 to 30 before gelE stop codon | This study |
Linker sequences are underlined.
DNA sequencing and sequence analysis.
Automated sequencing was used to determine nucleotide sequence by the dideoxy chain termination method (21, 26). PCR sequencing was carried out using the Taq DyeDeoxy terminator cycle sequencing kit (ABI, Foster City, Calif.), and the reactions were analyzed by an ABI model 373A DNA sequencer. DNA inserts in pBluescript SK(−) or in plasmid pTEX4577 (27) were sequenced using T3 and T7 primers. Other primers used for sequencing are listed in Table 2. To determine whether there is sequence variation in 3′ ends of fsrB, which shows homology to agrD of S. aureus, the 3′ ends of fsrB from eight different E. faecalis strains were amplified by PCR using fsrBF1 and fsrCRTF1 primers (Table 2). The PCR products were purified by a DNA cleanup kit (Promega, Madison, Wis.) and sequenced using the fsrCRTF1 primer.
DNA sequence analysis was accomplished using the Genetics Computer Group sequence analysis package, version 7.2 (University of Wisconsin, Madison). For the DNA and protein homology search, BLAST sequence comparison programs were applied using GenEMBL and/or SWISS-PROT databases. BESTFIT or GAP was used for comparing two DNA or peptide sequences, and PILEUP was used for multiple sequence alignment.
Deletion mutagenesis.
Primers used for fsrB deletion mutagenesis are listed in Table 2. To make a deletion in fsrB, 5′ and 3′ flanking regions of fsrB were amplified by PCR, ligated together by linkers (EcoRI) designed in the two inner primers, and inserted into a mutagenesis vector (27), pTEX4577, using two linkers (with SacI and KpnI recognition sites) designed in the two outer primers. The resulting construct, pTEX5267, was then transformed into OG1RF by electroporation as previously described (9), and single-crossover mutants were selected on BHI-KAN agar plates. The single-crossover mutants were expected to be still gelatinase positive because of the duplication of the flanking regions and putative promoters immediately upstream of fsrB. Since this single-crossover event creates duplicated fragments of the regions flanking the target gene, subsequent recombination between these duplicated fragments would lead to the loss of the mutagenesis vector and one copy of the duplicated flanking sequences and give rise to a KAN-sensitive wild type or deletion mutant. To identify fsrB deletion mutants, we first plated the cultures of the single-crossover fsrB mutant grown overnight without KAN onto Todd-Hewitt agar containing 3% gelatin to score for the loss of gelatinase activity because we predicted that the deletion of fsrB would abolish the expression of gelE. Colonies that were gelatinase production negative were then scored for the loss of KAN resistance and were further confirmed as deletion mutants by PCR using two primers flanking fsrB (primers BDF1 and BDR2) and by sequencing the PCR product. Pulsed-field gel electrophoresis (13) was used to verify that the deletion mutants were not contaminants.
Detection of gelatinase and serine protease activities.
The production of gelatinase and serine protease in E. faecalis strains was detected by methods previously described by using Todd-Hewitt agar (Difco Laboratories) containing 3% gelatin and zymogram gels containing 0.05% casein (Novex, San Diego, Calif.) and by using 20-fold-concentrated supernatants from overnight cultures (22).
Northern blot analysis and RT-PCR.
Isolation of total RNA from E. faecalis, Northern blot analysis, and RT-PCR were carried out as previously described by Qin et al. (22). Radioactive DNA probes for Northern blot analysis were prepared using the random-primer DNA labeling system from Life Technologies according to the protocol supplied. Primers used for RT-PCR are listed in Table 2.
Time course of fsr and gelE expression.
To study the time course of fsr and gelE gene expression in wild-type OG1RF and fsr mutants, overnight cultures of OG1RF and the fsrC insertion mutant (TX5242) were diluted 1:40 in BHI and incubated at 37°C. Total RNA was isolated from cells harvested at different time points. Northern blotting and hybridization using fsrC and gelE probes were utilized to determine the expression levels of fsr and gelE genes.
Cell density-dependent fsr expression.
Northern blot analysis was used to determine the expression levels of fsr and gelE genes at different cell concentrations. Cells of OG1RF and the fsrC gene disruption mutant (TX5242) from the cultures at early exponential (2 h after inoculation) and postexponential (4 h after inoculation) phases were harvested by centrifugation, resuspended in BHI to the desired cell concentrations, which were determined by measuring the optical density at 600 nm (OD600; 1 OD600 unit for an E. faecalis culture in BHI = 1.25 × 109 CFU/ml), and then incubated at 37°C for 45 min before isolation of total RNA for Northern blot analysis using fsrB and gelE genes as probes.
Determination of cotranscription in fsr/gelE loci.
Our previous Northern blot analysis suggested that some genes in the fsr/gelE loci are cotranscribed (22). To verify these results, RT-PCR using the primers flanking the intergenic region between two adjacent genes was applied to determine the cotranscription of the genes in the fsr/gelE loci. The primers used in the study of cotranscription are listed in Table 2.
Primer extension analysis and manual DNA sequencing.
Primer extension was used to map the 5′ end of the transcripts and to locate putative promoters in fsr/gelE loci. Primers complementary to sense DNA were designed from the sequences downstream of the start codons of desired genes (Table 2). Primer extension was carried out as previously described (25) with slight modification. In brief, primers were end labeled with 32P using T4 polynucleotide kinase (Life Technologies) and [γ-32P]ATP. For annealing RNA with the primer, 10 to 30 μg of total RNA was mixed with 2 × 106 cpm of 32P-labeled primer and 3 M sodium acetate (pH 4.8) was added to a final concentration of 0.3 M, followed by precipitation with ethanol. The pellet containing the RNA and primer was then dissolved in 30 μl of S1 solution (80% deionized formamide, 40 mM PIPES [piperazine-N,N′-bis{2-ethanesulfonic acid}] buffer [pH 6.4], 1 mM EDTA, 0.4 M NaCl) and heated for 10 min at 80°C. The mixture was then incubated at 37°C overnight to anneal the primer to the RNA template. The next day, RNA and primer were precipitated by ethanol.
For primer extension, primer extension mixture was prepared by mixing 4 μl of 5× First Strand buffer (Life Technologies; 250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2), 2 μl of 50 mM MgCl2, 2 μl of 10 mM deoxynucleoside triphosphates, 1 μl of 20 mM dithiothreitol, 0.5 μl of RNasin (RNase inhibitor; 40 U/μl), 1 μl of Superscript reverse transcriptase (200 U; Life Technologies), and 9.5 μl of diethyl pyrocarbonate-treated water (to a final volume of 20 μl). The hybridized RNA-primer pellet was then dissolved in the primer extension mixture, heated for 5 min at 70°C, and incubated at 37°C for 60 min. The RNA in the reaction mixture was removed by adding 1 μl of 0.5 M EDTA and 0.5 μl of 2-mg/ml DNase-free RNase and incubating for 30 min at 37°C. The cDNA in the reaction mixture was then precipitated with ethanol and resuspended in 4 μl of formamide loading buffer (0.95 ml of deionized formamide, 0.1 ml of 5× Tris-borate-EDTA buffer [25]) and 4 μl of 2× stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanole FF). The primer extension product was analyzed on 5% acrylamide DNA sequencing gel along with DNA sequencing reactions using the same primer according to the method previously described (25).
A manual DNA sequencing reaction was carried out with a T7 Sequenase 2.0 DNA sequencing kit from Amersham Life Science (Cleveland, Ohio) according to the protocol from the supplier using plasmid pAM-S1 (28) as the DNA template.
Construction of transcriptional fusion and β-galactosidase activity assay.
In order to confirm the putative promoters identified from primer extension and to determine the strength and the regulation of these promoters, sequences of putative promoters were amplified by PCR and cloned into transcriptional fusion vector pTCV-lac (20) using the EcoRI and BamHI restriction sites to generate transcriptional fusion to the promoterless lacZ reporter gene in the vector. pTCV-lac is a 12-kb broad-host-range shuttle vector containing erythromycin and KAN resistance genes which can be expressed in both gram-positive and -negative organisms and a promoterless β-galactosidase-encoding lacZ gene with a gram-positive ribosome binding site. The orientation and sequences of the cloned promoter fragments were confirmed by restriction analysis and DNA sequencing analysis using Vlac1 and Vlac2 primers (Table 2). The primers used in the amplification of wild-type promoters and promoters with mutations are listed in Table 2.
β-Galactosidase activities of E. faecalis strains containing pTCV-lac or fusion constructs were detected using the method previously described by Poyart and Trieu-Cuot (20). The β-galactosidase activities were calculated using the following formula: units of activity = 1,000 × (OD420 − 1.60 × OD550)/(t × v × OD600) (OD420 and OD550 are the densities measured from the reaction; OD600 is the cell density of the culture measured before the β-galactosidase activity assay; t is the time of reaction in minutes; v is the volume of culture used in the assay in milliliters; the light scattering correction factor 1.60 was used instead of 1.75, which is used for E. coli, because for E. faecalis OD420 [light scattering] ≈ 1.60 × OD550).
SDS-PAGE gel and 2-D gel electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as previously described (14). For analysis of proteins in supernatants of E. faecalis cultures, supernatants were concentrated 20-fold and dialyzed against 10 mM Tris-HCl (pH 7.4). Two-dimensional (2-D) gel electrophoresis was performed using the method previously described (17). Surface proteins from OG1RF and fsr gelE mutants were prepared by the method previously described (30) and 50 or 100 μg of surface proteins was used for each 2-D gel electrophoresis. The proteins in the 2-D gels were visualized by silver staining as previously described (4).
RESULTS
Time course of fsr and gelE expression.
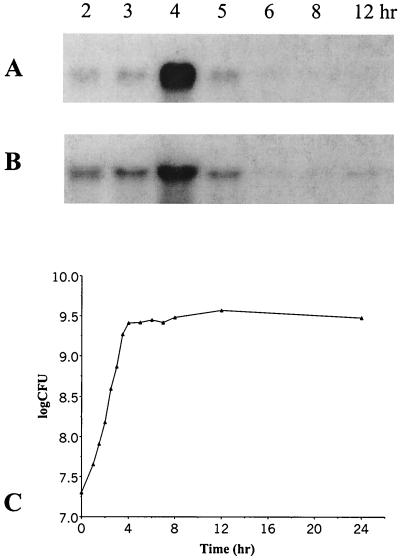
The expression of fsr (using fsrC as the examined gene) and gelE in wild-type OG1RF and an fsrC gene disruption mutant (TX5242; Table 1) (22) during growth was determined by Northern blot analysis. The expression of fsrC in OG1RF was at low but detectable levels from 2 to 3 h after inoculation (early and middle exponential phases), peaked at 4 h (postexponential phase), and diminished to undetectable levels after 12 h (Fig. 2A and C). A similar expression pattern of gelE was observed in OG1RF (Fig. 2B and C). These results indicate that fsrC and gelE are highly expressed in postexponential phase in wild-type OG1RF. No fsrC or gelE expression was detected in different phases during growth in the fsrC disruption mutant (data not shown).
FIG. 2.
Northern blot analysis of the time course of fsrC and gelE expression. (A) Northern blot analysis during growth using an fsrC probe. (B) Northern blot analysis during growth using a gelE probe. Lanes: total RNA from E. faecalis OG1RF cells from 2-, 3-, 4-, 5-, 6-, 8-, and 12-h cultures. RNA isolation, Northern blotting, and hybridization were performed as described in Materials and Methods. (C) Growth curve of OG1RF. Bacterial cells were cultured as described in Materials and Methods. Cell concentrations (CFU per milliliter) were determined by serial dilutions and plating on BHI agar in triplicate.
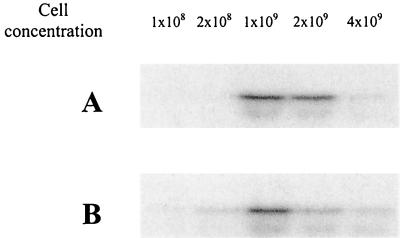
To investigate whether the expression of fsr genes is cell density dependent as it is for their homologues in S. aureus (6), the expression levels of fsrC were analyzed at different cell concentrations by Northern blotting. The levels of fsrC mRNA increased as the cell concentrations increased from 5 × 107 to 1 × 109 CFU/ml and began to drop after the cell concentration exceeded 1 × 109 CFU/ml, regardless of the growth phase of the bacterial cells used in the experiment (Fig. 3), suggesting that the fsr gene expression is cell density dependent.
FIG. 3.
Northern blot analysis of fsrC expression at different cell densities. OG1RF cells from early exponential (2-h) and postexponential (4-h) phases were harvested by centrifugation and resuspended in BHI to desired concentrations and incubated at 37°C for 45 min before RNA was isolated for Northern blot analysis using the fsrC probe. (A) OG1RF cells from 2-h culture. (B) OG1RF cells from 4-h culture. The cell concentration (CFU per milliliter) used in each lane is indicated.
Determination of gene cotranscription by RT-PCR.
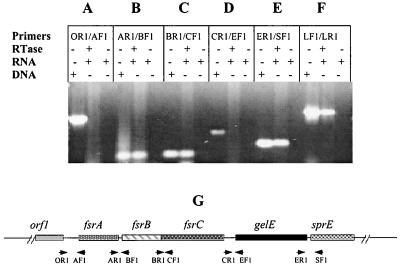
Our previous Northern blot results suggested that fsrB and fsrC, as well as gelE and sprE, are cotranscribed (22). To further confirm this finding, we applied RT-PCR using the primer pairs that cover the intergenic regions of fsr/gelE loci. RT-PCR using primer pairs fsrBRTF1 and fsrARTR1 and fsrCRTF1 and fsrBRTR1 encompassing fsrA, fsrB, and fsrC intergenic regions showed amplified bands with the predicted sizes (Fig. 4B and C), indicating that fsrB and fsrC are cotranscribed and that the transcript from fsrA reads through to fsrB. Positive amplification was also observed using primer pair sprERTF1 and gelERTR1, which covers the noncoding region between gelE and sprE (Fig. 4E), indicating the cotranscription of gelE and sprE. No signal was detected using the primer pairs (orf1RTR1 and fsrARTF1 and fsrCRTR1 and gelERTF1) between orf1 and fsrA and fsrC and gelE (Fig. 4A and D), suggesting that orf1 is not on the same transcript as fsrA and that transcription of gelE is initiated from its own promoter.
FIG. 4.
RT-PCR analysis of gene cotranscription in fsr/gelE loci. Every three lanes divided by a solid line represents one set of experiments using the same primers. In each set, the first lane shows PCR using chromosomal DNA as the template, which serves as a positive control for PCR, the second lane shows an RT-PCR with (+) RNA and reverse transcriptase (RTase) but without (−) chromosomal DNA; the third lane shows an RT-PCR with RNA but without RTase and chromosomal DNA, which serves as a control to determine the contamination of DNA in RNA samples. (A to E) RT-PCR analysis using primers covering the intergenic regions between orf1 and fsrA, fsrA and fsrB, fsrB and fsrC, fsrC and gelE, and gelE and sprE, respectively. (F) RT-PCR using two internal primers (lytF1 and lytR1) of E. faecalis autolysin, a positive control for RT-PCR. Primers: OR1, orfRTR1; AF1, fsrARTF1; AR1, fsrARTR1; BF1, fsrBRTF1; BR1, fsrBRTR1; CF1, fsrCRTF1; CR1, fsrCRTR1; EF1, gelERTF1; ER1, gelERTR1; SF1, sprERTF1; LF1, lytF1; LR1, lytR1. (G) Diagram illustrating the positions of the primers used in the RT-PCR analysis. Solid line, chromosomal DNA; boxes, genes; arrows, primers.
Construction and analysis of fsrB deletion mutant.
In our previous study of fsr gene expression in fsr insertion mutants, we found that an insertion in fsrB abolished the expression of fsrC, which is downstream of fsrB, and that expression and production of gelE and sprE were undetectable in an fsrB insertion mutant while both were readily detected in wild-type OG1RF (22). It was not clear whether the effect of the insertion in fsrB on the regulatory functions of fsr was due to the polar effect on fsrC or the loss of fsrB function. To determine whether fsrB is required for the regulatory function of the fsr locus, an fsrB deletion mutant was generated using suicide vector pTEX4577, containing the flanking regions of fsrB (pTEX5267). A KAN-resistant single-crossover fsrB mutant was obtained after electroporating pTEX5267 into OG1RF. Two putative deletion mutants were then obtained after growing the single-crossover mutant without KAN and screening about 5,000 colonies for the loss of gelatinase activity and KAN resistance. PCR analysis of the putative deletion mutants using two primers (BDF1 and BDR2) of the flanking regions of fsrB gave a shorter product than that resulting from analysis of wild-type OG1RF as expected, and sequencing the PCR product confirmed that these KAN-sensitive clones had the expected deletion in fsrB from bp 79 to 684 (data not shown). One of the fsrB deletion mutants was designated TX5266.
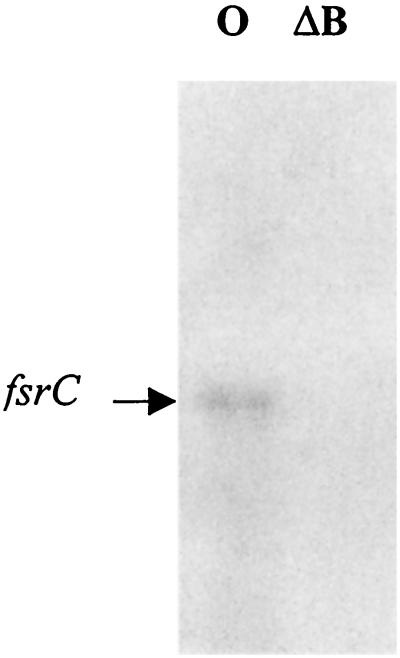
Gelatinase activity was not detected with TX5266 even after 72 h of incubation at 37°C, and serine protease activity was also not detectable in this fsrB deletion mutant on a casein zymogram gel, while both activities were readily detected for wild-type OG1RF (data not shown). Northern blot analysis using an internal fragment of the fsrC gene as a probe did not show any detectable signal for the fsrB deletion mutant, while OG1RF showed a 2.2-kb band (Fig. 5), suggesting that fsrB is also necessary for the regulatory functions of the fsr locus.
FIG. 5.
Northern blot analysis of fsrB deletion mutant (TX5266). RNA from postexponential-phase cultures of OG1RF and the fsrB deletion mutant (TX5266) was isolated as described in Materials and Methods. Northern blots were probed with the fsrC probe. Arrow, band that hybridized with the fsrC probe. Lanes: O, OG1RF; ΔB, fsrB deletion mutant TX5266 (deleted from bp 79 to 684 in fsrB).
Mapping 5′ ends of fsr and gelE transcripts by primer extension.
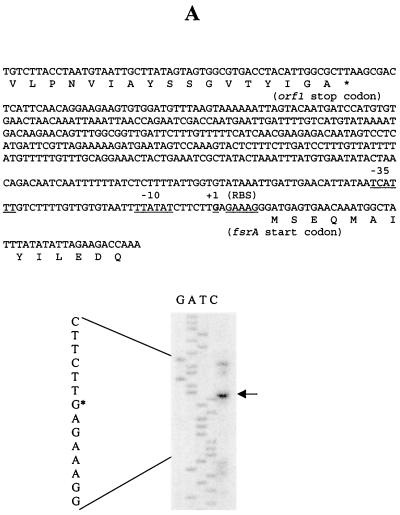
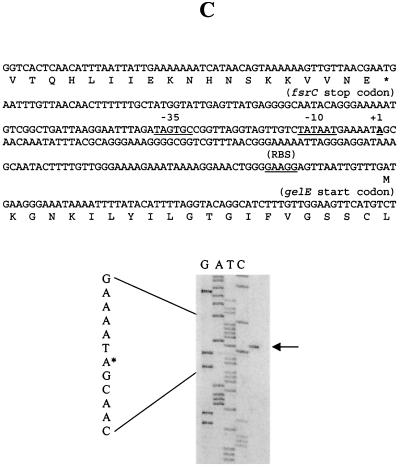
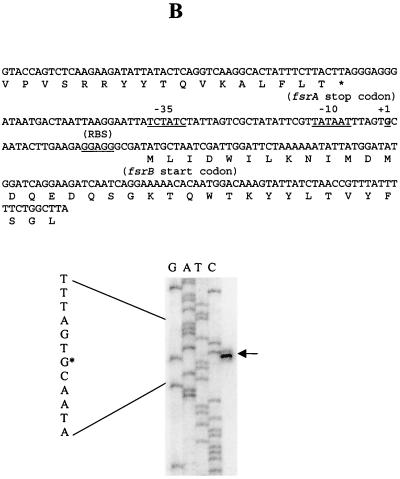
With primers complementary to the sense strands downstream of the start codons of fsrA, fsrB, and gelE, the 5′ ends of fsrA, fsrB, and gelE transcripts were mapped to 9, 23, and 119 bp upstream of fsrA, fsrB, and gelE start codons, respectively, and potential −10 and −35 sequences were identified immediately upstream of the 5′ ends of these transcripts (Fig. 6).
FIG. 6.
Determination of the 5′ ends of the fsrA, fsrB, and gelE transcripts by primer extension. Primer extension was carried out as described in Materials and Methods. The primer extension products were run alongside sequencing reactions obtained with the same primers. Arrows, primer extension products in the sequencing gels; asterisks, locations of the 5′ ends of the sequences (left of each gel). The sequences encompassing the 5′ ends of the transcripts are shown on the top of each gel; putative −35, −10, and ribosome binding site (RBS) sequences are underlined, and the transcription start sites (+1) are in boldface and underlined. Primer extensions using primers APE1 (A), BPE2 (B), and EPE2 (C) are shown.
Study of fsrA, fsrB, and gelE promoter activities in wild-type and fsr mutant strains.
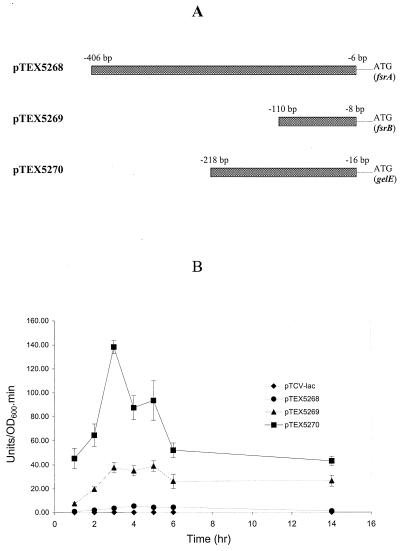
To confirm activity of the fsrA, fsrB, and gelE promoter sequences identified above and to investigate the regulatory functions in these promoter regions, plasmid pTEX5268 (containing the putative fsrA promoter), pTEX5269 (containing the putative fsrB promoter), and pTEX5270 (containing the putative gelE promoter) were constructed. Plasmid pTEX5268 was generated by cloning a 401-bp PCR fragment from the intergenic region between orf1 and fsrA (from −406 to −6 bp relative to the fsrA start codon) upstream of the promoterless lacZ reporter gene in shuttle vector pTCV-lac (20) (Fig. 7A). Similarly, pTEX5269 and pTEX5270 were constructed by fusing 103- and 203-bp fragments covering the intergenic regions between fsrA and fsrB (from bp −110 to −8 relative to the fsrB start codon) and between fsrC and gelE (from bp −218 to −16 relative to the gelE start codon) with the promoterless lacZ in pTCV-lac (Fig. 7A). These three constructs, containing putative fsrA, fsrB, and gelE promoters, were introduced into wild-type OG1RF, and the specific β-galactosidase activities in different growth phases were analyzed. The β-galactosidase activities of fsrB and gelE promoter constructs peaked after 3 to 5 h of growth, while fsrA promoter activity remained relatively low and constant (Fig. 7B). The gelE promoter (pTEX5270) had the strongest promoter activity compared to fsrA and fsrB promoters. Based on the β-galactosidase activities, the maximum activity of the gelE promoter during growth (at 3 h) was about 3 and 26 times greater than that of fsrB (at 5 h) and fsrA (at 4 h), respectively.
FIG. 7.
(A) Constructs of fsrA, fsrB, and gelE promoter lacZ fusions. DNA fragments (boxes) containing putative fsrA, fsrB, and gelE promoters were cloned in front of a promoterless lacZ in shuttle vector pTCV-lac, resulting in plasmids pTEX5268 (putative fsrA promoter in pTCV-lac), pTEX5269 (putative fsrB promoter in pTCV-lac), and pTEX5270 (putative gelE promoter in pTCV-lac), respectively. The distances of each end of the fragments from fsrA, fsrB, and gelE start codons are indicated. (B) Determination of fsrA, fsrB, and gelE promoter activities of lacZ fusion constructs in OG1RF. The β-galactosidase activity assay was performed as described in Materials and Methods. β-Galactosidase activity was represented as units per unit of optical density at 600 nm (OD600) of cells per minute. Error bars, standard deviations.
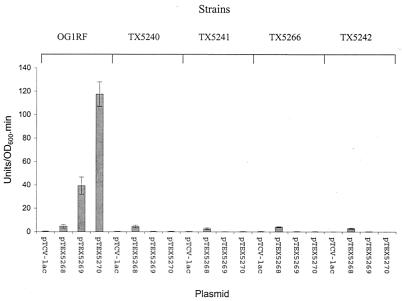
When the fsrA, fsrB, and gelE promoter fusion constructs were introduced into the fsr mutants, only the fsrA promoter showed promoter activities in these mutants (Fig. 8), and the fsrA promoter activity in each mutant was comparable to its activity in wild-type OG1RF. Neither the fsrB nor the gelE promoters were active in any fsr mutants (Fig. 8), suggesting that the fsrB and the gelE promoters are fsr dependent, while the fsrA promoter is an fsr-independent constitutive promoter.
FIG. 8.
fsrA, fsrB, and gelE promoter activities in fsr mutants. E. faecalis cells containing different plasmid constructs grown for 4 h were used for the β-galactosidase activity assay. Error bars, standard deviations. Strains were as follows: TX5240 (fsrA gene disruption mutant) (20), TX5241 (fsrB gene disruption mutant) (20), TX5242 (fsrC gene disruption mutant) (20), TX5266 (fsrB deletion mutant). Plasmids in the different strains were as follows: pTCV-lac (vector without insert as negative control), pTEX5268 (fsrA promoter in pTCV-lac), pTEX5269 (fsrB promoter in pTCV-lac), and pTEX5270 (gelE promoter in pTCV-lac).
Identification of regulatory sequences in fsrB and gelE promoter regions.
Since the promoter activity assay mentioned above and our previous Northern blot analysis indicated that the expression of both fsrB and gelE is regulated by the fsr locus, we analyzed the sequences of the fsrB, gelE, and fsrA promoter regions for possible sequence homology. Sequence alignment revealed two conserved segments immediately upstream of the −35 regions of the fsrB and gelE promoters (Fig. 9). Within these two homologous regions are two 7-bp imperfect repeats, which are separated by 14 bp (Fig. 9), suggesting possible regulatory sequences in these regions. The fsrA promoter region did not show significant sequence similarity to either the fsrB or the gelE promoter regions.
FIG. 9.
Schematic map of fsrB and gelE promoter regions in different lacZ fusion constructs and their β-galactosidase activities. (A) Different gelE promoter constructs, pTEX5270, pTEX5303, pTEX5304, pTEX5305, and pTEX5306, and their β-galactosidase activities in OG1RF. (B) Different fsrB promoter constructs, pTEX5269, pTEX5298, pTEX5299, pTEX5300, pTEX5301, and pTEX5302, and their β-galactosidase activities in OG1RF. The sequences for pTEX5270 and pTEX5269 (top lines of panels A and B) only show the regions containing the gelE and fsrB promoter and the repeated sequences; the sequences further upstream are not shown. All the gelE promoter constructs have the same sequences downstream of the promoter to bp −16 from gelE start codon (dots, sequence not shown), and all the fsrB promoter constructs have the same sequences downstream of the promoter to bp −8 from the fsrB start codon. The position labeled at the end of each line is relative to the gelE or fsrB start codon. Solid lines (A and B), sequences identical to that of the top sequences (changes in the promoter sequences are shown for constructs pTEX5305, pTEX5306, pTEX5301, and pTEX5302). Repeated sequences upstream of gelE and fsrB promoters are underlined; putative transcription start sites (+1) are in boldface and boxed; −10 and −35 sequences are also underlined. The β-galactosidase activity of each construct is indicated at the right.
In order to investigate the importance of these conserved repeated sequences upstream of fsrB and gelE promoters, different lacZ fusion constructs with deletions or mutations in the repeated sequences of fsrB and gelE promoters were made in shuttle vector pTCV-lac (20) (Fig. 9) and assayed for their β-galactosidase activities in wild-type OG1RF. Constructs pTEX5298 (containing bp −90 to −8 relative to the fsrB start codon) and pTEX5303 (containing bp −188 to −16 relative to the gelE start codon) (Fig. 9) containing the fsrB and gelE promoters with just an additional 34-bp sequence immediately upstream of the −35 regions, which include both the repeated sequences, exhibited promoter activities similar to those exhibited by pTEX5269 (containing bp −110 to −8 relative to the fsrB start codon; Fig. 7A) and pTEX5270 (containing bp −218 to −16 relative to the gelE start codon; 7A), respectively (Fig. 9). This suggested that the fsrB and gelE promoters with the upstream repeated sequences are sufficient to maintain their promoter activities. However, deletions of the most-upstream repeats (constructs pTEX5299, pTEX5300, and pTEX5304) or mutations in the closer repeats (constructs pTEX5301 and pTEX5305) upstream of the fsrB and gelE promoters completely abolished the fsrB and gelE promoter activities (Fig. 9), suggesting that these repeats are important for the regulation of fsrB and gelE expression. In addition, when the fsrB promoter without its own repeated region was fused to the repeated region of the gelE promoter (construct pTEX5302), it was not only active but also showed about twofold-higher activity than the fsrB promoter with its original repeated sequences (Fig. 9). Similarly, exchange of fsrB repeated sequences for those of the gelE promoter (construct pTEX5306) also generated a functional gelE promoter (Fig. 9), suggesting that these repeated sequences upstream of fsrB and gelE promoters have a similar regulatory function. None of these fusion constructs exhibited promoter activity in an fsrC gene disruption mutant (data not shown), indicating that the regulatory function of the repeated sequences is mediated by fsr.
Sequences of 3′ end of fsrB from different clinical isolates.
To investigate whether there is strain variation in the 3′ end sequence of fsrB, a region whose last 50-amino-acid sequence shows homology to that of AgrD of S. aureus, the 3′ end of fsrB was amplified by PCR from eight distinct Gel+ strains from different clinical sources and geographical areas and sequenced. DNA sequence analysis of the last 150-bp sequences at the 3′ end of fsrB revealed that five strains had identical sequences while three had only single silent-base-pair changes compared to OG1RF (data not shown), indicating that the 3′ ends of fsrB are conserved among different gelatinase-producing E. faecalis strains. The 150-bp 3′ end sequence of fsrB from OG1RF is also identical to this region of E. faecalis strain V583 in the TIGR E. faecalis genome database.
DISCUSSION
In S. aureus, the four agr genes (agrA, agrB, agrC, and agrD) are all required for the regulatory functions of the agr locus (15, 16). We have previously shown by Northern blotting that insertion disruption of the three agr homologues in E. faecalis, fsrA, fsrB, and fsrC, abolished the expression of fsr genes and of gelE and sprE (22). However, it was not clear whether inactivation of fsr regulatory functions by insertion in fsrB was caused by the loss of fsrB functions or by its polar effect on fsrC, which appeared to be cotranscribed with fsrB. We have shown here that a nonpolar deletion in fsrB in E. faecalis which maintained the fsrB promoter also led to the elimination of fsrC expression and production of gelatinase and serine protease, as demonstrated by Northern blot analysis and zymogram gel analysis, confirming that fsrB (the agrB homologue) is required for the fsr regulatory functions. As corroboration of our previous results (22), it appears that all three fsr genes are required for fsr functions.
It has been reported that the agr/hld loci in S. aureus consist of three promoters, the P1 promoter, which controls the expression of agrA (19), and two divergent but nonoverlapping promoters, P2 and P3, which control the transcription of the P2 transcript (encoding agrB, agrD, and agrC) and the P3 transcript (encoding RNAIII), respectively (15, 16). The P1 promoter is a weak constitutive promoter (19), while P2 and P3 are agr-dependent promoters (15). Our primer extension results suggest that a separate promoter is located immediately upstream of fsrA, of fsrB, and of gelE. Gene fusion analysis of these putative promoters with a promoterless lacZ in OG1RF and fsr mutants indicated that, like the P1 promoter in S. aureus, the fsrA promoter is a weak and constitutive fsr-independent promoter and that fsrB and gelE promoters are more-active promoters than the fsrA promoter and that their expression is fsr dependent, as the P2 and P3 promoters are agr dependent (15). Our RT-PCR experiments using the primers covering the intergenic regions in fsr/gelE loci further indicated that fsrA, fsrB, and fsrC and gelE and sprE are cotranscribed. Analysis of DNA sequences in fsr/gelE loci revealed one possible rho-independent transcription terminator (24) in the intergenic region between fsrC and gelE genes (218 bp upstream of gelE), which contains two 13-bp inverted repeats, suggesting that transcription from fsr genes would not read through to gelE and sprE. From the promoter fusion analysis results, together with the results of the RT-PCR and previous Northern blot analysis (22), we can conclude that the transcription of fsrA starts from the fsrA promoter and may read through to fsrB and possibly to fsrC, that the expression of fsrB and fsrC is mainly under the control of the fsrB promoter, and that the expression of gelE and sprE is regulated by the gelE promoter.
In the agr/hld loci in S. aureus, Morfeldt et al. have previously identified two 7-bp interrupted repeats (separated by 14 bp) upstream of the P3 promoter that are required for the agr-dependent expression of the P3 transcript (RNAIII) (11). Similar repeats were found upstream of P2, and the repeats upstream of P2 compete with those of P3 for binding to the SarA protein (11). Similar repeated sequences upstream of the sapA promoter, which is required for activation of transcription of sapA, the gene encoding a bacteriocin named sakacin A, has also been found in Lactobacillus sake Lb706 (1). By sequence alignment of the intergenic regions between fsrA and fsrB and between fsrC and gelE, which contain fsrB and gelE promoters, respectively, we found two conserved 7-bp repeats, which are also separated by 14 bp, immediately upstream of the fsrB and gelE promoters. Cloned fsrB and gelE promoter regions containing their repeated sequences (pTEX5298 and pTEX5303) displayed promoter activities similar to those of cloned fsrB and gelE promoter regions containing almost the entire intergenic sequences (pTEX5269 and pTEX5270, which had additional 20- and 30-bp sequences upstream of the repeats compared to pTEX5298 and pTEX5303), suggesting that the promoters with the repeated sequences are sufficient to perform the promoter activities. Moreover, deletion or changes of these repeated sequences upstream of fsrB and gelE completely abolished the promoter activities of the fsrB and gelE promoters, further indicating that these repeated sequences are required for the fsr-dependent regulation of fsrB and gelE promoters, a mechanism that appears to be similar to that of the Agr system in S. aureus (11).
In this study, we also examined the possibility that fsr genes regulate the expression of some surface proteins and other secreted proteins besides gelatinase and serine protease. The results of SDS-PAGE analysis of proteins in supernatants from fsr mutants and OG1RF showed that the fsr mutants had more protein bands in their supernatants than wild-type OG1RF even though a 29-kDa band and a 34-kDa band, presumably the serine protease and gelatinase, were clearly no longer present in the supernatants from the mutants (data not shown). The increase in the number of protein bands in the fsr mutants may be due the lack of the two proteases in the supernatants so that proteins released from dead cells or other sources were not degraded by these enzymes, since the supernatant from a gelE insertion mutant (TX5128) (22, 27), which did not produce either of these proteases, also showed a pattern of protein bands similar to those from fsr mutants. Analysis of surface protein profiles of fsr gene disruption mutants and an fsrB deletion mutant compared to that of the parental strain, OG1RF, using 2-D gel electrophoresis showed that protein patterns of fsr mutants were similar to that of OG1RF and that all the fsr mutants had identical protein patterns on 2-D gels (unpublished preliminary data). However, the intensities of at least three spots with the sizes of 44, 33, and 18 kDa on the 2-D gel of fsr mutants were greater than the intensities of those of OG1RF, suggesting increases in the production of these proteins in the fsr mutant strains. The pattern of surface proteins from a gelE insertion mutant (TX5128) (22, 27) was similar to that of surface proteins from OG1RF. Whether fsr genes in E. faecalis regulate the expression of surface proteins, like their homologues in S. aureus, could be further addressed by isolating and partially sequencing these proteins and subsequent studying the expression of the genes encoding these proteins.
Ji et al. have previously shown that the regulatory functions of the Agr system in S. aureus are mediated by a secreted oligopeptide pheromone, encoded by agrD, which functions as a cell density signal (6). We have not yet identified an AgrD-like peptide in E. faecalis even though we identified an agrD homologue in the last 150 bp of the 3′ end of fsrB by sequence analysis. Nakayama et al. have reported in abstract form the isolation of an 11-amino-acid pheromone, whose sequence matches that of 220 to 230 amino acids of the C terminus of FsrB, from the supernatant of an E. faecalis strain and demonstrated that the isolated pheromone could induce the expression of gelatinase in a pheromone concentration-dependent manner (J. Nakayama, Y. Cao, A. D. L. Akkermans, W. M. deVos, and H. Nagasawa, Abstr. 1st Int. ASM Conf. Enterococci, abstr. 21, 2000). The sequence of the pheromone was identical to that of a segment in the C terminus of FsrB. In our present study, we found that the expression of fsr genes is cell density dependent, as it is for their homologues in S. aureus, and that the expression of fsr genes peaked at a cell concentration of 109 CFU/ml, a concentration that is about the concentration of cells of E. faecalis at postexponential phase, consistent with the time course of fsr expression during growth. These results suggest that the regulation of fsr gene expression is similar to that of agr in S. aureus, which is mediated by a quorum-sensing system encoded by agr and which is most active in postexponential phase (5, 6, 15).
Strains of S. aureus can be classified into at least three different groups based on the cross-activation and inhibition by the autoinducing peptides of the Agr systems (5). For strains studied to date, the autoinducing peptides from strains in the same group are identical and can induce the expression of agr genes in the strains of the same group but inhibit agr expression in strains from other groups (5). The sequence similarity among the precursors of the autoinducing peptides (encoded by agrD) from strains in different groups is very limited (5). As noted above, the last 50 amino acids at the C terminus of FsrB, which show 28% identity and 47% similarity to those of AgrD of S. aureus, appear to be the AgrD equivalent in E. faecalis. To test whether E. faecalis strains that contain the fsr locus can also be divided into different groups based on their fsr sequences, we partially sequenced the 3′ ends of fsrB from eight distinct strains and compared the last 150-bp sequences of fsrB from these eight Gel+ strains with those from strains OG1RF and V583. From the sequence analysis results, we only detected single-base-pair changes with no difference in the deduced amino acid sequences among these 10 strains, suggesting that, unlike the agr genes in S. aureus, the fsr genes in E. faecalis are conserved among strains.
In conclusion, the fsrB gene as well as fsrA and fsrC are all required for the regulatory functions of the fsr locus. The expression of fsrA is under the control of a weak and constitutive promoter, the fsrA promoter, and the transcription of fsrB and fsrC as well as that of gelE and sprE are regulated by two fsr-dependent promoters, the fsrB and gelE promoters, respectively. Two directly repeated sequences immediately upstream of the fsrB and gelE promoters are necessary for the activation of fsrB and gelE promoters in an fsr-dependent manner. The expression of fsr genes in E. faecalis OG1RF is cell density dependent and is most active in the postexponential phase. While these aspects of regulation are similar to those for the agr locus of S. aureus, unlike agr, which is present in all S. aureus strains studied (5, 18), these fsr genes are found in only some E. faecalis strains (22) (but in 100% of gelatinase-producing strains). Analysis of the 3′ end of fsrB indicates that this locus is much more conserved than agr.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI33516 and AI47923 from the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, to B. E. Murray.
We thank Steve Norris and Jerry Howell of the Department of Pathology of University of Texas Medical School at Houston for their help with 2-D gel electrophoresis.
REFERENCES
- 1.Axelsson L, Holck A. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Bacteriol. 1995;177:2125–2137. doi: 10.1128/jb.177.8.2125-2137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung A L, Eberhardt K, Heinrichs J H. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun. 1997;65:2243–2249. doi: 10.1128/iai.65.6.2243-2249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guevara J J, Johnston D A, Ramagali L S, Martin B A, Capetillo S, Rodriguez L V. Quantitative aspects of silver deposition in proteins resolved in complex polyacrylamide gels. Electrophoresis. 1982;3:197–205. [Google Scholar]
- 5.Ji G, Beavis R, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 6.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornblum J, Kreiswirth B N, Projan S J, Ross H, Novick R P. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, Skurray R A, editors. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 373–402. [Google Scholar]
- 8.Lebeau C, Vandenesch F, Greenland T, Novick R P, Etienne J. Coagulase expression in Staphylococcus aureus is positively and negatively modulated by an agr-dependent mechanism. J Bacteriol. 1994;176:5534–5536. doi: 10.1128/jb.176.17.5534-5536.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Weinstock G M, Murray B E. Generation of auxotrophic mutants of Enterococcus faecalis. J Bacteriol. 1995;177:6866–6873. doi: 10.1128/jb.177.23.6866-6873.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindberg M, Jonsson K, Muller H, Jonsson H, Signas C, Hook M, Raja R, Raucci G, Anantharamaiah G M. Fibronectin-binding proteins in Staphylococcus aureus. In: Novick R P, Skurray R A, editors. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 343–356. [Google Scholar]
- 11.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 12.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray B E, Singh K V, Ross R P, Heath J D, Dunny G M, Weinstock G M. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nallapareddy S R, Qin X, Weinstock G M, Hook M, Murray B E. The Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun. 2000;68:5218–5224. doi: 10.1128/iai.68.9.5218-5224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 16.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Farrell P Z, Goodman H M, O'Farrell P H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 18.Otto M, Sussmuth R, Vuong C, Jung G, Gotz F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999;450:257–262. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- 19.Peng H L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poyart C, Trieu-Cuot P. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to β-galactosidase in gram-positive bacteria. FEMS Microbiol Lett. 1997;156:193–198. doi: 10.1111/j.1574-6968.1997.tb12726.x. [DOI] [PubMed] [Google Scholar]
- 21.Prober J M, Trainor G L, Dam R J, Hobbs F W, Robertson C W, Zagursky R J, Cocuzza A J, Jensen M A, Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987;238:336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- 22.Qin X, Singh K V, Weinstock G M, Murray B E. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun. 2000;68:2579–2586. doi: 10.1128/iai.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick R P. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh K V, Qin X, Weinstock G M, Murray B E. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J Infect Dis. 1998;178:1416–1420. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 28.Su Y A, Sulavik M C, He P, Makinen K K, Makinen P L, Fiedler S, Wirth R, Clewell D B. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect Immun. 1991;59:415–420. doi: 10.1128/iai.59.1.415-420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, David D M, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Brooklyn, N.Y: Green Publishing Associates; 1994. pp. 2.4.1–2.4.2. [Google Scholar]
- 30.Xu Y, Jiang L, Murray B E, Weinstock G M. Enterococcus faecalis antigens in human infections. Infect Immun. 1997;65:4207–4215. doi: 10.1128/iai.65.10.4207-4215.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]