Significance
This comprehensive cross-sectional study demonstrates widespread infection of WTD with SARS-CoV-2 across the State of New York. We showed cocirculation of three major SARS-CoV-2 variants of concern (VOCs; Alpha, Delta, and Gamma) in this species, long after their last detection in humans. Interestingly, the viral sequences recovered from WTD were highly divergent from SARS-CoV-2 sequences recovered from humans, suggesting rapid adaptation of the virus in WTD. The impact of these mutations on the transmissibility of the virus between WTD and from WTD to humans remains to be determined. Together, our findings indicate that WTD—the most abundant large mammal in North America—may serve as a reservoir for variant SARS-CoV-2 strains that no longer circulate in the human population.
Keywords: one health, white-tailed deer, wildlife reservoir, spillover, SARS-CoV-2
Abstract
The spillover of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from humans to white-tailed deer (WTD) and its ability to transmit from deer to deer raised concerns about the role of WTD in the epidemiology and ecology of the virus. Here, we present a comprehensive cross-sectional study assessing the prevalence, genetic diversity, and evolution of SARS-CoV-2 in WTD in the State of New York (NY). A total of 5,462 retropharyngeal lymph node samples collected from free-ranging hunter-harvested WTD during the hunting seasons of 2020 (Season 1, September to December 2020, n = 2,700) and 2021 (Season 2, September to December 2021, n = 2,762) were tested by SARS-CoV-2 real-time RT–PCR (rRT-PCR). SARS-CoV-2 RNA was detected in 17 samples (0.6%) from Season 1 and in 583 samples (21.1%) from Season 2. Hotspots of infection were identified in multiple confined geographic areas of NY. Sequence analysis of SARS-CoV-2 genomes from 164 samples demonstrated the presence of multiple SARS-CoV-2 lineages and the cocirculation of three major variants of concern (VOCs) (Alpha, Gamma, and Delta) in WTD. Our analysis suggests the occurrence of multiple spillover events (human to deer) of the Alpha and Delta lineages with subsequent deer-to-deer transmission and adaptation of the viruses. Detection of Alpha and Gamma variants in WTD long after their broad circulation in humans in NY suggests that WTD may serve as a wildlife reservoir for VOCs no longer circulating in humans. Thus, implementation of continuous surveillance programs to monitor SARS-CoV-2 dynamics in WTD is warranted, and measures to minimize virus transmission between humans and animals are urgently needed.
The coronavirus disease 2019 (COVID-19) was declared a pandemic in March 2020 and as of January 2023 has incurred over 659 million human cases and more than 6.6 million deaths globally (1). COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a new zoonotic virus for which most of the first known human infections were linked to the Huanan Seafood Wholesale Market in Wuhan, China, where several live wild animal species were sold (2). SARS-CoV-2 is a single-stranded RNA virus within the Sarbecovirus subgenus, Betacoronavirus genus, of the family Coronaviridae (3). Analysis of the genome sequence of SARS-CoV-2 revealed high similarity to coronaviruses circulating in bats in China, suggesting that bats are the most likely source of the ancestral virus that originated SARS-CoV-2 (4). While the closest bat coronaviruses (RaTG13, RmYN02, RpYN06, and PrC31) are phylogenetically related to SARS-CoV-2, they present several mutations across the genome that distinguish them from SARS-CoV-2, indicating that direct transmission of the virus from bats to humans was unlikely (4, 5). These observations point to the involvement of a yet unidentified animal species that served as an intermediate host and enabled spillover of the virus into humans (6).
Comparative sequence analyses of the main cellular receptor for SARS-CoV-2, the angiotensin-converting enzyme 2 (ACE2), from more than 400 animal species suggested a broad host range for the virus (7). Notably, the ACE2 of three species of deer, including reindeer (Rangifer tarandus), Père David’s deer (Elaphurus davidianus), and white-tailed deer (WTD) (Odocoileus virginianus), share a high similarity to the human ACE2 and were predicted to allow binding and entry of SARS-CoV-2 into deer cells (7). We and others have confirmed these in silico predictions and demonstrated that WTD are highly susceptible to SARS-CoV-2 infection (8, 9). Most importantly, intranasal inoculation of SARS-CoV-2 in WTD resulted in virus replication and shedding, which led to efficient deer-to-deer transmission (8–10). These findings placed WTD—a species broadly distributed in North America with an estimated population size of 30 million animals (11)—at the center of investigations, which demonstrated exposure of free-ranging WTD to SARS-CoV-2 and highlighted their potential to serve as a reservoir for SARS-CoV-2 in North America. Notably, results from the first studies revealed prevalence rates varying from ~30 to 40%, with animals testing positive for SARS-CoV-2 antibodies and/or viral RNA (12–16). Importantly, detection of SARS-CoV-2 RNA in respiratory secretions and tissues from several of the sampled animals suggested recent infection with the virus (12–14). Additionally, analysis of SARS-CoV-2 sequences recovered from WTD revealed multiple introductions from humans and suggested onward transmission of the virus in free-ranging deer populations (12–14). The pathways of human-to-deer transmission of SARS-CoV-2, however, are not yet understood. These findings highlight the need to establish surveillance programs that will allow continuous monitoring of the circulation, distribution, and evolution of SARS-CoV-2 in WTD populations.
In this study, we conducted a comprehensive cross-sectional investigation to assess the prevalence, genetic diversity, and evolution of SARS-CoV-2 in WTD in the State of New York (NY). All samples included in our study (n = 5,462) consisted of retropharyngeal lymph nodes (RPLNs)—one of the major sites of SARS-CoV-2 replication in WTD (10)—that were collected as part of the New York State’s Chronic Wasting Disease (CWD) Surveillance Program during two hunting/CWD testing seasons (Season 1, 2020, and Season 2, 2021).
Results
Prevalence of SARS-CoV-2 Infection in WTD in New York.
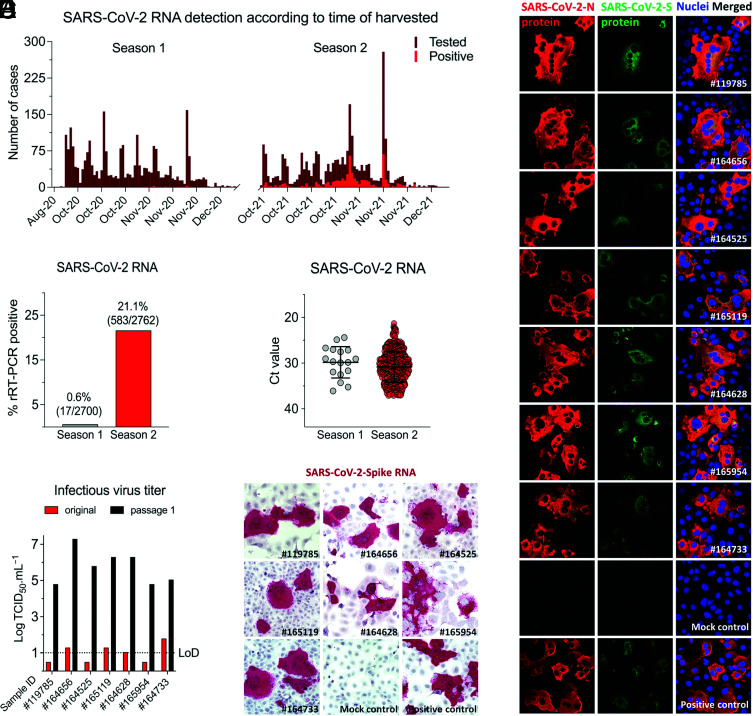
A total of 5,462 RPLN samples collected as part of the New York State’s CWD Surveillance Program from wild hunter-harvested WTD during the hunting seasons of 2020 (Season 1: September 2020 through December 2020) and 2021 (Season 2: July 2021 through December 2021) were tested for SARS-CoV-2 by real-time RT–PCR. In Season 1, positive samples were detected in October and November 2020, whereas in Season 2, positive cases were detected between October and December 2021, with peak positivity being detected between November 1 and 15, 2021 (Fig. 1A). SARS-CoV-2 RNA was detected in 17 of 2,700 (0.6%) samples from Season 1 and in 583 of 2,762 (21.1%) samples collected in Season 2 (Fig. 1B). Viral RNA loads, based on rRT–PCR cycle threshold (Ct) values, varied slightly between the seasons (mean Ct values for Season 1 was 29.8 ± 3.4 [mean ± SD, range from 24 to 37], whereas for Season 2, mean Ct was 30.8 ± 3.4 [range from 21 to 37]) (Fig. 1C); however, no statistical differences were observed in Ct values between the two seasons (P > 0.2). All RPLN samples with Ct values lower than 30 (11 samples from Season 1 and 205 samples from Season 2) were subjected to virus isolation in Vero E6 TMPRRSS2 cells (8, 10, 17). Infectious virus was recovered from seven samples collected in Season 2, and active virus replication was confirmed by virus titrations in the original sample and on passage 1, and by in situ hybridization (ISH) and immunofluorescence (IFA) on passage 2 in Vero cells (Fig. 1 D–F). Together, these results showed a significant increase (35-fold, chi-square = 585.5, P < 0.0001) in the prevalence of SARS-CoV-2 infection between the 2020 and 2021 seasons and demonstrate a broad circulation of the virus in WTD in NY.
Fig. 1.
Prevalence of SARS-CoV-2 infection in WTD in NY. A total of 5,462 retropharyngeal lymph node (RPLN) samples were collected from free-ranging hunter-harvested WTD during the hunting Season 1 (September to December 2020, n = 2,700) and Season 2 (September to December 2021, n = 2,762) in the State of New York (NY). (A) RPLN tested for the presence of SARS-CoV-2 viral RNA using rRT–PCR. The number of samples tested and SARS-CoV-2–positive samples in Season 1 and Season 2 is shown in the graphic. (B) Distribution of positive samples per season. Number of SARS-CoV-2 RNA-positive RPLN from WTD. (C) SARS-CoV-2 RNA detection by rRT–PCR presented as cycle threshold (Ct) values. All the Ct values for positive samples from season 1 (n = 17) and season 2 (n = 583) are shown. (D) Infectious SARS-CoV-2 recovered from seven samples (sample #IDs depicted in the x axis). (E) Virus isolation was confirmed by in situ hybridization (ISH) assay labeling viral RNA and (F) immunofluorescence assay using a monoclonal antibody anti-SARS-CoV-2 nucleoprotein (N) (red) and a polyclonal antibody anti-SARS-CoV-2 Spike protein (green). Nuclear counterstain was performed with DAPI (blue). (Magnification, 40×).
Hotspots of SARS-CoV-2 Infection among WTD in New York.
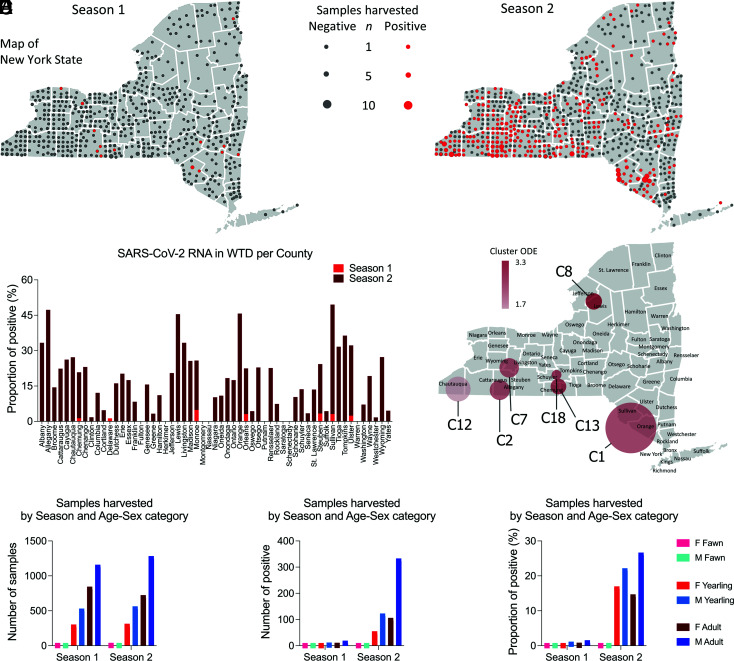
The samples tested in our study were collected statewide from WTD harvested in 57 of 62 counties in New York. The geographic distribution of the samples from Season 1 and Season 2 and those that tested positive for SARS-CoV-2 is presented in Fig. 2A. In Season 1, 17 positive samples were detected in ten of the 62 counties in the state including Orleans (n = 1) and Monroe County (n = 2) in the Finger Lakes region, Steuben (n = 4) and Chemung County (n = 1) in the Southern Tier, Delaware (n = 1), Sullivan (n = 3), and Ulster (n = 2) in the Hudson Valley region, Columbia (n = 1) and Greene County (n = 1) in the Capital Region, and Clinton County (n = 1) in the North Country (Fig. 2A). In Season 2, the increased prevalence of SARS-CoV-2 infection in WTD was accompanied by a marked expansion in the geographic distribution of the virus with positive cases being detected in 48 counties (Fig. 2 A and B). The proportion of positive samples detected in each county is presented in Fig. 2B. Positive SARS-CoV-2 cases were detected in nine of the ten geographic regions of the state (Western New York, Finger Lakes, Southern Tier, Central New York, North Country, Mohawk Valley, Capital Region, Mid-Hudson, and Long Island) (Fig. 2A). No samples were collected in New York City. The counties with the highest number of cases were Allegany (n = 72), Cattaraugus (n = 32), and Steuben (n = 31) in the Southern Tier region, Chautauqua (n = 62) in the Western New York region, and Orange (n = 32) and Sullivan (n = 44) in the Hudson Valley region. Together, these counties accounted for 47% of the cases detected across the state (n = 273). A summary of the samples tested and SARS-CoV-2 positivity detected by county is presented in SI Appendix, Table S1.
Fig. 2.
Demographics of WTD population sampled and tested for SARS-CoV-2. Retropharyngeal lymph node (RPLN) samples were collected from free-ranging hunter-harvested WTD during the hunting Season 1 (September to December 2020, n = 2,700) and Season 2 (September to December 2021, n = 2,762) in the State of New York (NY). (A) Sampling distribution and positivity across NY. SARS-CoV-2 RNA was detected in 17 samples (0.6%) from Season 1 and 583 samples (21.1%) from Season 2. (B) Proportion of SARS-CoV-2–positive samples in Season 1 and Season 2 per county of NY. (C) High-risk areas or hotspots of SARS-CoV-2 infection in WTD in NY. Spatial cluster analysis performed with the samples from Season 2 (n = 2,762). Nineteen spatial clusters (C1 to C19) containing from 3 to 57 positive samples collected within a radius of 10.6 to 55 km from each other were identified. Seven hotspots with high-risk for SARS-CoV-2 infection in WTD (relative risk [RR] > 1.76) are highlighted in the map. (D) Total number of collected samples based on sex and age of WTD during Season 1 and Season 2. Animals were distributed in three age groups as follows: <1.5 y old (fawns), ≥1.5 and <2.5 y old (yearlings), and ≥2.5 y old (adults). (E) Number of SARS-CoV-2–positive samples based on sex and age of WTD during Season 1 and Season 2. (F) Proportion of positive samples based on sex and age of WTD during Season 1 and Season 2.
To identify high-risk areas or hotspots of SARS-CoV-2 infection in WTD in NY, spatial cluster analysis was performed with the samples from Season 2. This analysis included all 2,762 samples tested in Season 2 and relied on a likelihood ratio test and explored the maximum number of positive SARS-CoV-2 cases that were detected in a given geographic location. Nineteen spatial clusters (C1 to C19) containing 3 to 57 positive samples were identified (SI Appendix, Table S2). These SARS-CoV-2–positive WTD clusters comprised samples collected within a radius of 10.6 to 80 km from each other (SI Appendix, Table S2). Among the 19 clusters identified, seven hotspots with high risk for SARS-CoV-2 infection in WTD (relative risk [RR] > 1.76) were detected (Fig. 2C and SI Appendix, Table S2), most of them (C2, C7, C13, and C18) in the Southern Tier region of NY. One cluster (C12, RR = 1.76) was formed by samples collected in Chautauqua in the Western New York Region and a small cluster (C8, RR = 3.38) in the North Country region of the state (Fig. 2C). From the Southern Tier clusters, C2 (RR = 2.7) involved two counties (Allegany and Cattaraugus); C13 (RR = 2.85), C7 (RR = 2.63), and C18 (RR = 2.74) comprised one county each. The larger cluster identified in our analysis, C1 at Hudson Valley (n = 89 cases), involved three counties (Sullivan, Orange, and Ulster). These results revealed several hotspots of SARS-CoV-2 infection in WTD in NY, most of which overlap with geographic areas with the highest deer population densities and deer harvest rates in the state.
Male WTD Are at a Higher Risk of Infection with SARS-CoV-2.
The distribution of positive SARS-CoV-2 cases based on sex and age of the animals sampled in our study was also investigated. Sex and age of all WTD harvested from Season 1 and Season 2 were determined, and all animals were distributed into three age groups as follows: <1.5 y old (fawns), ≥1.5 and <2.5 y old (yearlings), and ≥2.5 y old (adults). Overall, the number of males in each age group sampled during Season 1 and Season 2 was slightly higher than the number of females (Fig. 2D), with the number of males that tested positive for SARS-CoV-2 in all age groups being 2 to 3 times higher than the number of females in the corresponding age groups (Fig. 2E). As shown in Fig. 2F, the proportion of males that tested positive in all age groups was also markedly higher than the proportion of positive females detected. Logistic regression analysis confirmed that males were more likely to test positive for SARS-CoV-2 than females (OR = 1.952, 95% CI = 1.591 to 2.407), with adult males being more likely to become infected with SARS-CoV-2 than younger yearling males (OR = 1.906, 95% CI = 1.55 to 2.35) (SI Appendix, Table S3). These results suggest that adult male WTD are at a higher risk of infection with SARS-CoV-2.
Cocirculation of Multiple SARS-CoV-2 Variants of Concern (VOCs) in WTD.
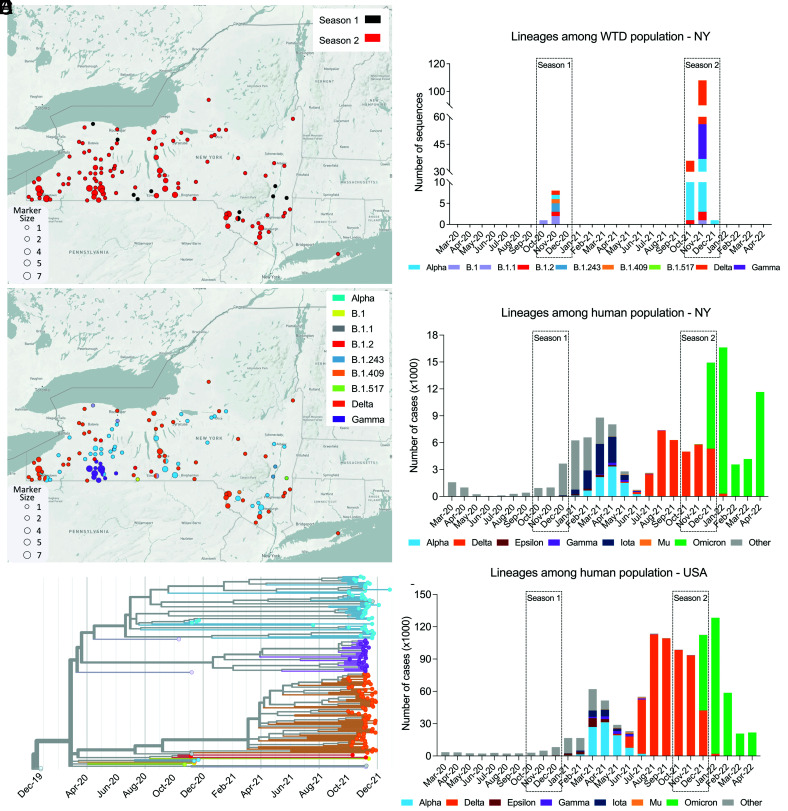
To assess the genetic makeup and diversity of SARS-CoV-2 in WTD, we performed whole genome sequencing on 216 samples (including 11 samples from Season 1 and 205 samples from Season 2) that tested positive for SARS-CoV-2 and presented a rRT–PCR Ct value <30. Consensus genome sequences of 216 samples were assembled and subjected to SARS-CoV-2 lineage classification using Pangolin version 4.0.6 (18). Of the 216 samples sequenced, 164 genomes passed the Pangolin QC thresholds for minimum length and maximum N content (≤30%) and were used for downstream analyses. A total of nine SARS-CoV-2 genomes were recovered from samples from Season 1 and 155 from samples from Season 2. The geographic distribution of the samples from which complete or near-complete SARS-CoV-2 genome sequences were obtained according to the season is presented in Fig. 3A. Nine lineages including three major VOCs (Alpha [B.1.1.7], Gamma [B.1.1.28, P.1], and Delta [B.1.617.2]) were detected in WTD samples in NY. The SARS-CoV-2 sequences recovered in Season 1 were classified as B.1, B.1.1, B.1.2, B1.243, B.1.409, B.1.507, and Alpha, whereas in Season 2, B.1.1, B.1.2, B.1.517, Alpha, Gamma, and Delta lineages were detected. The geographic distribution of these lineages across the state is presented in Fig. 3B. Most of the samples sequenced in our study were classified within one of the three major VOCs identified as Alpha, Gamma, or Delta. Interestingly, Alpha and Delta variants were detected in several geographic regions throughout the state, while the samples classified as Gamma (n = 27) formed a localized cluster in the Southern Tier County of Allegany (Fig. 3B). Notably, there was one sample detected in Season 1 (161392; GISAID EPI_ISL_13610582 and GenBank OP006342) that was classified by Pangolin as B.1.617.2 (Delta), but the lineage for this sample was unassigned by Scorpio (18). Analysis of the mutation profile of the sample revealed the presence of only 8 of 20 B.1.617.2 lineage-defining mutations (ORF1b, P314L; S, T19R, T478K, and D614G; ORF7a, T120I; and N D63G, R203M, and D377Y); thus, a lineage was not assigned to this sample.
Fig. 3.
Distribution of SARS-CoV-2 lineages across NY and phylogenetic relatedness of viral genomes. (A) Map of the State of New York (NY) and geographic distribution of the 164 samples from which complete or near-complete SARS-CoV-2 genome sequences were obtained and according to the season is presented (9 and 155 genomes from Season 1 and Season 2, respectively). (B) Map of NY and geographic distribution of the SARS-CoV-2 lineages detected in free-ranging WTD in this study. (C) Phylogenetic tree showing 164 SARS-CoV-2 genomes obtained from WTD in NY across time. (D) The distribution of SARS-CoV-2 lineages among the WTD population in NY during Season 1 and Season 2. (E) Monthly distribution of SARS-CoV-2 human cases (March 2020 to April 2022) from NY and (F) the United States. The colored stacked bars represent number of each SARS-CoV-2 lineage.
Phylogenetic analysis performed with the SARS-CoV-2 sequences revealed three large clusters corresponding to three SARS-CoV-2 VOCs (Alpha, Gamma, and Delta) identified in the sampled WTD population (Fig. 3C). Together, these results demonstrate the concomitant circulation of Alpha, Gamma, and Delta VOCs in WTD in NY in 2021.
Given that most of the samples that were identified as Alpha, Gamma, or Delta variants in WTD in our study were detected between October and December 2021 (Season 2), we analyzed the circulation of these VOCs in humans. Our analyses involved a large number of human SARS-CoV-2 cases per lineage based on available sequencing information for the State of New York and the United States over a time frame (March 2020 to April 2022) overlapping the WTD sampling that was performed in this study. These data clearly show an overlap in the detection of the Delta variant in humans and in WTD in NY and across the United States (Fig. 3 D, E, and F). Notably, while the Alpha and Gamma variants were circulating in WTD in NY in November and December 2021 (Fig. 3D), detection of these VOCs in humans peaked between April and June 2021, with only sporadic detections after August (Fig. 3E). Three Alpha sequences were reported in NY in September 2021 based on data available at GISAID, with a single last detection occurring in February 2022, while the two last Gamma sequences were detected in NY in October 2021. All these latest Alpha and Gamma detections in humans in the state occurred in New York City, which was not included in the WTD sampling in this study. Analysis of the SARS-CoV-2 sequences recovered from humans in NYC or rural NY State (86,142 sequences from NYC and 34,686 sequences from rural NY) revealed a very similar scenario and distribution of these variants over time (SI Appendix, Fig. S1). Importantly, the last detection of Alpha and Gamma sequences in rural NY in humans preceded their last detection in NYC. Interestingly, the Alpha variant was detected in multiple geographic locations in WTD in NY in a time that there was no evidence that the virus was broadly circulating in humans in the state. The time lapse between peak detection of Alpha and Gamma variants in humans and in WTD suggests that these variants may have become established in WTD in NY.
Phylogenomic Analysis of SARS-CoV-2 Variants Detected in WTD.
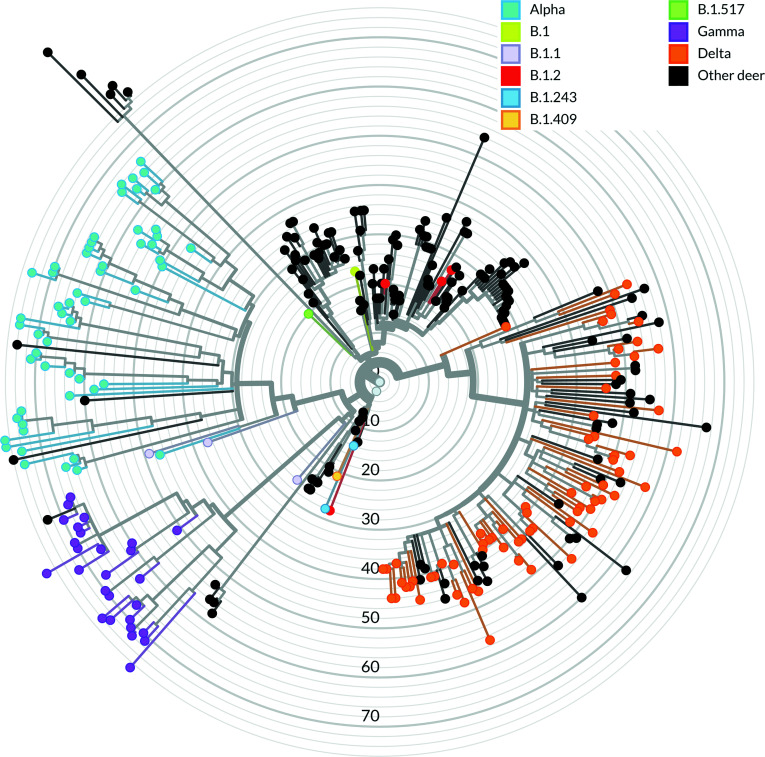
The phylogenetic relationship of the WTD SARS-CoV-2 samples characterized in our study (n = 164) was compared with other sequences derived from WTD (n = 159) available in the EpiCoV database in Global Initiative on Sharing All Influenza Data (GISAID). As of August 2022, most available SARS-CoV-2 sequences in GISAID were classified as B.1-like or Delta lineages, whereas most sequences obtained in this study were classified as Delta variant lineage, followed by Alpha and Gamma lineages, respectively. Within each of the major lineages identified in this study (Alpha, Gamma, and Delta), samples obtained from close geographic locations (e.g., same county or neighboring counties) formed clusters of closely related SARS-CoV-2 sequences (Fig. 4). Interestingly, the closest phylogenetic relationship was observed among the Gamma sequences, with all sequences obtained forming a monophyletic cluster (Fig. 4). Pairwise nucleotide analysis demonstrated that the Gamma lineage sequences share up to 99.99% similarity. All Gamma sequences were obtained from WTD harvested 6 to 25 km from each other, within Allegany County. Consistent with the broader geographic distribution throughout state, the Alpha and Delta lineage sequences formed multiple smaller clusters, with the Delta sequences from NY being interspersed with Delta variant clusters detected in WTD in other states of the United States (Fig. 4).
Fig. 4.
Phylogenomic analysis of SARS-CoV-2 derived from WTD. Phylogeny of 164 SARS-CoV-2 sequences derived from WTD in the State of New York during Season 1 and Season 2 in the context of 159 SARS-CoV-2 sequences derived from WTD available in GISAID. The node colors (sky blue: Alpha; purple: Gamma; and orange: Delta) represent SARS-CoV-2 VOCs derived from WTD sequences from this study, and the black nodes represent the Pan-US SARS-CoV-2 sequences derived from WTD.
Evolution and Mutation Profile of SARS-CoV-2 in WTD.
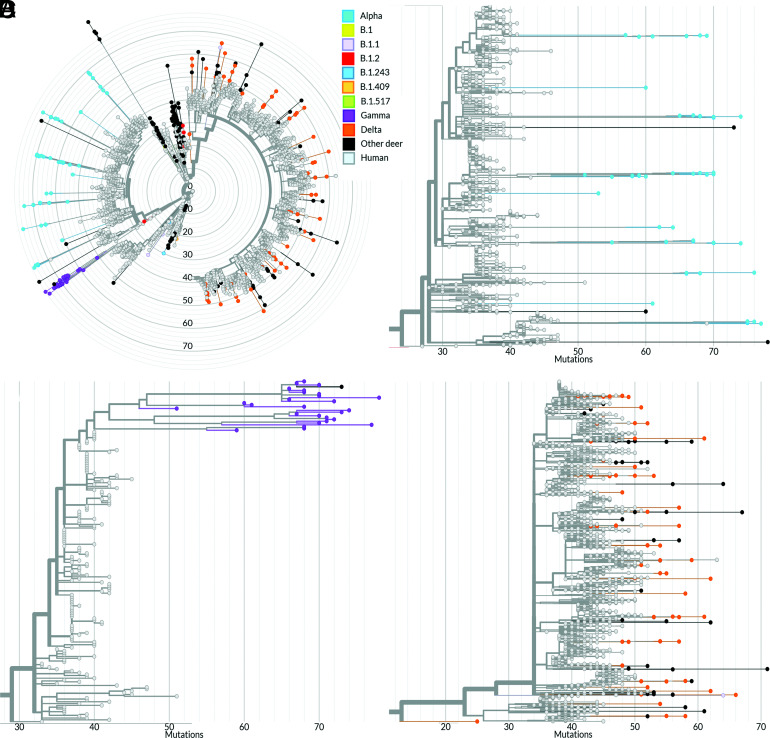
The genetic relationship of the WTD SARS-CoV-2 sequences with human SARS-CoV-2 sequences available in GISAID was also investigated. A phylogenetic analysis including all 164 sequences characterized in this study plus 159 WTD SARS-CoV-2 sequences and 3,837 human SARS-CoV-2 genomes available in GISAID was performed (Fig. 5A). The human sequences included in our analysis were obtained between March 5, 2020, and January 30, 2022. No direct links between SARS-CoV-2 sequences detected in WTD and in humans in NY were observed. Interestingly, WTD sequences from the Alpha (Fig. 5B) and Delta lineages (Fig. 5D) from NY formed 11 and 27 divergent phylogenetic branches, respectively, that were interspersed among the human sequences. All Gamma WTD sequences detected here formed a monophyletic group highly divergent from the human-derived Gamma lineage sequences (Fig. 5 A and C).
Fig. 5.
Phylogenomic analysis of SARS-CoV-2 derived from humans and WTD. Divergence between WTD and human-derived SARS-CoV-2 sequences. The node colors (sky blue: Alpha; purple: Gamma; and orange: Delta) represent SARS-CoV-2 VOCs derived from WTD from this study; black nodes represent the Pan-US SARS-CoV-2 sequences derived from WTD, and gray nodes represent SARS-CoV-2 sequences derived from human. The X axis shows the number of mutations compared with the reference sequence Wuhan-1 (GenBank accession number MN908947.3). (A) Phylogenetic tree comprising 164 WTD-derived sequences and 3,837 human-derived sequences. Magnification of the Alpha (B), Gamma (C), and Delta (D) clades.
Phylogenetic analysis of SARS-CoV-2 detected in WTD revealed a high genetic diversity and marked evolution of the viruses detected in this animal species when compared with human-derived SARS-CoV-2 sequences. The WTD SARS-CoV-2 Alpha and Gamma lineage sequences presented numerous mutations (~50 to 80) in comparison with the reference sequence Wuhan-1 (GenBank accession number MN908947.3) (Fig. 5 B and C). Analysis of the WTD SARS-CoV-2 Delta lineage sequences revealed similar genetic divergence from the human Delta sequences; however, the number of mutations accumulated in sequences of this lineage in WTD was lower (range, 40 to 65) than the observed mutations in the Alpha and Gamma sequences (Fig. 5D). The substitution rate in human SARS-CoV-2 sequences was estimated at 24.4 nucleotide substitutions per year, whereas in WTD, the substitution rates were estimated at 35.3, 36.3, and 26.9 substitutions per year for the Alpha, Gamma, and Delta sequences, respectively (SI Appendix, Fig. S2).
Potential host-adaptive mutations were observed in the Alpha, Gamma, and Delta lineages detected in WTD (SI Appendix, Tables S4–S7). These are nondefining mutations for their respective lineage or sublineage and present low frequency among human-derived SARS-CoV-2 sequences (global frequency < 0.5%). Mutations in the spike (S) gene are of particular interest due to their potential to result in the evasion of immune responses. Among the mutations observed in our dataset, S:Q564L was detected in 11 of 27 (40.7%) Gamma sequences. This mutation is present in only 167 sequences from the complete GISAID database and in only one sequence of 68,540 Gamma sequences (as of June 6, 2022). Another mutation in the S gene, S1252F, was detected in nine Alpha (16.4%) and one B.1 lineage sequences from close geographic locations in NY. Among the putative host-adaptive mutations observed in the S gene, four occurred in the receptor-binding domain (RBD): P384S and V445A in one Alpha sequence each and T323I and Y449H in one Gamma sequence each. No mutations in the S RBD were detected in Delta sequences. Only the RBD V445A mutation, detected in three geographically related Alpha lineage sequences, occurred in more than one sample (6.3%) (SI Appendix, Table S4). Other potential host-adaptive mutations were observed in WTD from different regions of NY or in samples from NY and other states (SI Appendix, Table S4). For example, the S mutation W258C was detected in eight Alpha variant sequences (27.8%) from WTD from different locations of NY, while S mutation L1203F was detected in four Alpha and three Delta sequences from NY and one B.1.2 from Iowa (IA). Mutations outside the S gene were also observed (SI Appendix, Table S5). The ORF1a mutation L4111F is a strong candidate for host-specific adaptation. It was detected in fourteen sequences from NY, including nine Gamma (33.3%) and six Delta sequences, and in five B.1 sequences from WTD from Canada and one Alpha sequence obtained from WTD in Pennsylvania.
Homoplasy analysis was performed to identify additional sites with potential strong host-adaptive mutations of the SARS-CoV-2 VOCs in WTD (SI Appendix, Table S6). The analysis identified three mutations (ORF1a:T3150I, S:P25S, and S:T29I) with a consistency index <0.5 that presented a low frequency in human-derived sequences. Other mutations were also detected in WTD from other states and in different lineages (SI Appendix, Fig. S3). The mutation in ORF1a:L1853F was detected in one B.1 and five Alpha sequences from NY, three B.1.311 and ten B.1.2 sequences from IA, and one Delta sequence from Maine (ME) and Canada (SI Appendix, Fig. S3A). The ORF1a:L4111F mutation was detected in nine Gamma and one Delta sequences in NY, one Alpha sequence from Pennsylvania (PA), and five B.1 sequences from Canada (SI Appendix, Fig. S3B). This mutation is also among mutations above 30% frequency in its respective lineage (SI Appendix, Table S5). The S mutation T29I was detected in six Alpha lineage sequences from NY State, one B.1.2 from IA, and one Delta sequence from Kansas (KS) (SI Appendix, Fig. S3C). Other homoplastic mutations were also detected here mostly on ORF1a and represented by the substitution of leucine or serine to phenylalanine (L or S to F) (SI Appendix, Table S6).
Dispersal of SARS-CoV-2 Variants Within Hotspots of Infection in WTD in New York.
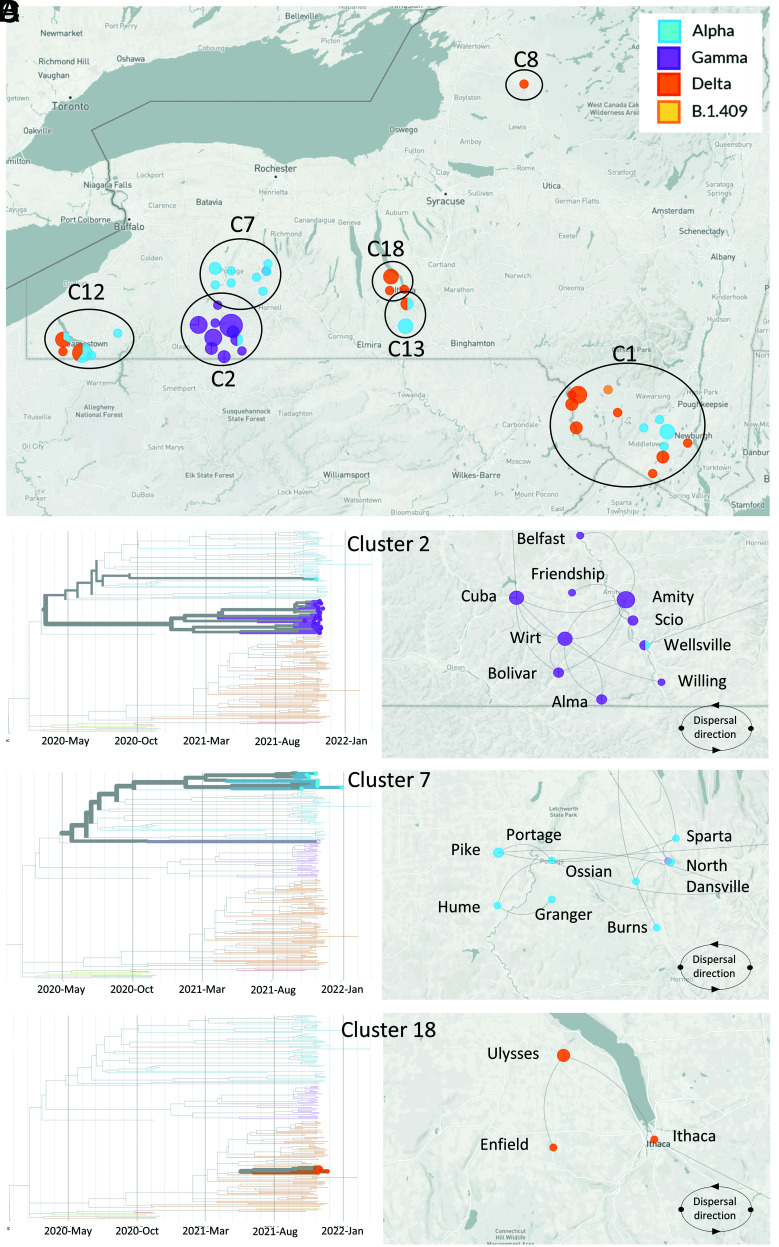
The geographic dispersal dynamics of SARS-CoV-2 in high-risk clusters identified using spatial analysis were investigated. The SARS-CoV-2 sequences obtained from seven high-risk clusters (C1, C2, C7, C8, C12, C13, and C18) were included in our phylogeographic reconstructions (Fig. 6A). C7 (n = 10) and C18 (n = 5) comprised a single SARS-CoV-2 variant, which was identified as Alpha and Delta, respectively. C1 (n = 10) contained sequences identified as Alpha (n = 4), Delta (n = 4), and B.1.409 (n = 1) variants. Cluster C2 contained mostly Gamma (n = 27) and one Alpha sequence, while C12 comprised Alpha (n = 7) and Delta variant (n = 6) sequences, and C13 comprised Alpha (n = 4), Delta (n = 2), and a single B.1.1 (n = 1) variant sequence.
Fig. 6.
Dispersal of SARS-CoV-2 within hotspots of infection in WTD in NY. (A) Map of the State of New York (NY) and sampling location and lineage classification of sequences obtained within the seven high-risk clusters. Clusters are identified as C1, C2, C7, C8, C12, C13, and C18. Phylogenetic reconstruction and dispersal analysis of (B) C2, (C) C7, and (D) C18. Directions of dispersal lines are counterclockwise.
For the phylogeographic dispersal analysis, we focused on C2, C7, and C18 as representative clusters of the three major SARS-CoV-2 VOCs identified in our study, Gamma, Alpha, and Delta, respectively. The phylogenetic relationship and dispersal pathways were inferred based on whole genome sequences and the town and date of sample collection. This approach allowed spatial reconstruction of the dispersal history of the viral lineage(s) within each identified cluster. C2 was geographically restricted to Allegany County and comprised all the Gamma variant sequences detected in our study. These sequences formed a monophyletic branch of closely related sequences (Fig. 6B). Phylogeographic dispersal analysis of the Gamma sublineages detected in this cluster revealed an intricate dispersal pathway with clear connections between the sequences. Analysis of the directionality of dispersion of the Gamma sequences in C2 points to the samples detected in the town of Cuba as the central focus from which the virus may have spread to Amity, Wirt, and Bolivar and then to other herds in surrounding towns in the area, including Friendship, Willing, and Alma (Fig. 6B).
The Alpha variant C7 involved the counties of Livingston, Wyoming, and Allegany. A total of 10 Alpha variant sequences were recovered from samples within this cluster, which were collected in nine different towns in the region (Dansville, North Dansville, Ossian, Portage, and Sparta in Livingston County; Burns, Hume, and Granger in Allegany County; and Pike in Wyoming County). These sequences formed a large branch in the phylogenetic tree that formed two smaller branches of closely related sequences (Fig. 6C). Dispersal analysis of the Alpha sequences detected in this cluster revealed two potential dispersal pathway(s) with links between the detected sequences. It appears that the sample detected in Portage may be the central foci in C7 from where the virus may have spread to eastern (Sparta, North Dansville, and Ossian) and western locations (Pike, Hume, and Granger) (Fig. 6C).
The Delta variant C18 represents the smallest (n = 5) of the clusters in which we analyzed the spatial dispersal of SARS-CoV-2. This cluster was restricted to Tompkins County, and samples were detected in the towns of Ulysses, Ithaca, and Enfield. The Delta sequences detected in C18 formed a unique branch in the phylogenetic tree with highly related sequences (Fig. 6D). Phylogeographic dispersal analysis of the Delta sequences detected in this cluster revealed a clear connection between the sequences with the samples detected in the town of Ulysses appearing to be the central foci of the cluster, which further dispersed into nearby Enfield and Ithaca (Fig. 6D). The phylogenetic relationship of the SARS-CoV-2 sequences detected in WTD and their divergence from contemporary samples recovered from humans demonstrates the circulation and indicates onward transmission of Alpha, Gamma, and Delta SARS-CoV-2 VOCs in WTD in NY.
Discussion
WTD are highly susceptible to SARS-CoV-2 infection and efficiently transmit the virus through direct or indirect contact to other susceptible animals as demonstrated by our research group and collaborators from the National Animal Disease Center (8, 10). Follow-up testing for SARS-CoV-2 in captive and free-ranging WTD in different regions of the United States and Canada has shown that a significant proportion of WTD may have been exposed or infected with SARS-CoV-2 in North America (12–16).
In the present study, we used samples from a well-established surveillance program for CWD in wild deer in the State of New York (19) to conduct a comprehensive cross-sectional investigation and assess the prevalence of SARS-CoV-2 infection in WTD over two deer hunting seasons (2020 and 2021) during the COVID-19 pandemic. During these seasons, 253,990 and 211,269 WTD were harvested by hunters in NY, respectively, with ~1% of the animals being sampled and tested for CWD (2,700 and 2,762, respectively) and subsequently included in our study. Of the samples tested for SARS-CoV-2, 17 (0.6%) were positive in 2020 and 583 (21.1%) in 2021. The sampling strategy used by the NYS Department of Environmental Conservation emphasizes locations and age classes of deer that are at a greater risk for CWD infection using weighted surveillance and a qualitative risk assessment. The sampling calculations are determined by three primary criteria: i) deer sex and age classes, with emphasis given to older bucks and does; ii) proximity to geographic risk factors, such as captive cervid facilities, taxidermy and deer processing centers, and proximity with CWD-endemic states (like PA); and finally, iii) the deer population size, with areas with larger deer counts (estimated based on the number of deer harvested by square kilometer) receiving more intense surveillance. This sampling strategy allowed us to evaluate the infection rate and dynamics of SARS-CoV-2 infection using samples that are representative of the adult WTD population distributed across the State of New York (~1M WTD). Our results showed a striking 35-fold increase in SARS-CoV-2 prevalence and consequently a broader geographic distribution of the infection between 2020 (10/62 positive counties, Season 1) and 2021 (57/62 positive counties, Season 2). The low positivity rate in Season 1 could be due to the fact that most samples in our study were collected prior to the peak SARS-CoV-2 cases in humans in NY, which occurred between December 2020 and February 2021. Importantly, previous studies reporting higher SARS-CoV-2 prevalence in WTD in Ohio and Iowa detected most positive samples after the peak human cases in each state (12, 14), whereas a study in Pennsylvania reported prevalence of 16.3% (20 of 163 positive samples), with most positive samples being detected between October and November 2021 (13). Differences in prevalence between the different studies could be caused by inherent differences in the target WTD populations (free ranging vs. captive), sample type and sampling strategy (active vs. opportunistic), differences in the ecology of the virus and the hosts (humans and WTD), differences in geographic locations, and differences in human and WTD population densities, which could all affect the nature, duration, and extent of human–deer interactions and consequently virus spillover from humans to deer.
Spatial clustering analysis based on positive SARS-CoV-2 detections revealed several hotspots or areas with a high concentration of cases in WTD in NYS. Notably, when the viral sequence information was compared with the spatial clusters, several clusters of closely related SARS-CoV-2 variants overlapped with these defined geographic hotspots, indicating local circulation and transmission of the virus within the deer populations in these areas. These findings corroborate observations of early studies, which found evidence of potential transmission of SARS-CoV-2 among wild WTD in the states of Ohio and Iowa in the United States or in the province of Ontario in Canada (12, 14). Interestingly, the broad geographic distribution of the Alpha and Delta SARS-CoV-2 variants in our study and the identification of several clusters of closely related viruses within confined geographic locations highlight the occurrence of multiple viral spillover events from humans to WTD between 2020 and 2021, which was followed by deer-to-deer transmission of the virus.
Evidence of the circulation of Alpha and Gamma VOCs in WTD in NY ~4 to 5 mo after their last reported detection in humans was one of the most intriguing findings of our study. When we analyzed human SARS-CoV-2 sequences detected in rural NY—where the WTD samples included in our study were collected—we observed that the last detection of the Alpha and Gamma VOCs in humans in rural NY occurred in August 2021. Although the possibility of encrypted circulation of these VOCs in humans in rural NY and their potential spillover into WTD cannot be formally excluded, the time lapse between their last detection in humans in these areas and their detection in WTD together with their marked evolution from human SARS-CoV-2 sequences suggests that these variants may have become established in WTD in NY. Detection of the Alpha VOC in WTD in multiple counties across a broad geographic range of the state with no detections described in humans in these areas further supports this hypothesis. Future surveillance in WTD populations will be critical to answer this question definitively and to evaluate whether variant displacement—similar to what was observed in humans—also happens in this host reservoir with a specific VOC becoming the dominant virus circulating in WTD populations.
Transmission of the virus between WTD can occur through direct contact or indirectly, likely through respiratory droplets and fomites (8–10). While assessing SARS-CoV-2 transmission pathways within WTD populations, animal age and sex must be considered as direct or indirect contact between animals can be increased by distinct social behaviors of different age groups and sex. For example, maternal groups consisting of females and fawns are relatively isolated during the spring and summer, while males typically have large home ranges and have increased movement and contact with other deer, especially during the breeding season (15, 20). Interestingly, results from our study, in which sample collection overlaps with the breeding season (September to December), indicate that adult male deer are at a significantly higher risk of becoming infected with SARS-CoV-2 than other sex and age classes. This observation is consistent with previous studies that have shown higher prevalence of antibodies or SARS-CoV-2 RNA in adult male WTD (14–16) and highlights the potential epidemiological role of males in the maintenance and transmission of the virus within wild WTD populations. Although the pathways for transmission of SARS-CoV-2 from humans to WTD remain largely unknown, human activities such as feeding wildlife or targeted baiting of hunting prey (e.g., WTD) could provide the opportunity for human-to-WTD transmission of the virus. Animal feed and feed ingredients are known to promote and enhance survival of several animal viruses, including coronaviruses (21), and thus, wildlife feeding practices should be investigated as a risk factor and avoided due to their potential to enable spillover of SARS-CoV-2 from humans to WTD and perhaps other susceptible wildlife species (e.g., deer mice, mink, raccoon, and red fox; Fig. 7) (22–26).
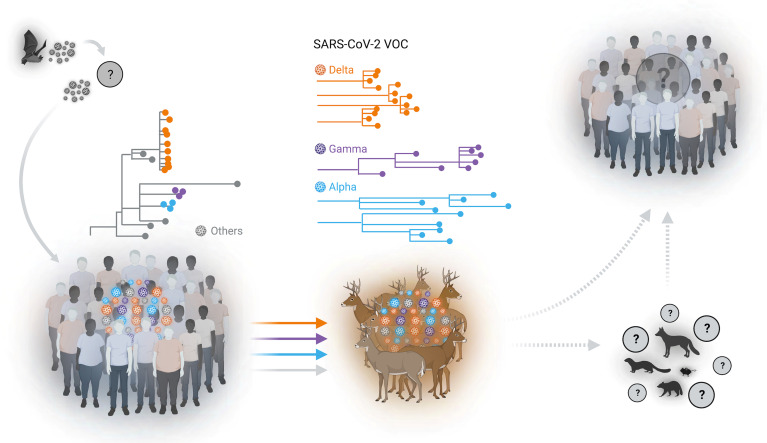
Fig. 7.
Spillover of SARS-CoV-2 from humans to WTD. Multiple spillover events of SARS-CoV-2 from humans to wild WTD population were identified in several geographic areas in the State of New York (NY). Sequence analysis of SARS-CoV-2 genomes demonstrated the circulation of classical SARS-CoV-2 lineages and the cocirculation of three major VOCs (Alpha, Gamma, and Delta) in WTD. Our analysis suggests the occurrence of multiple spillover events (human to deer) of SARS-CoV-2 lineages with subsequent deer-to-deer transmission and emergence of genetically diverse viruses in this species. The potential for spillback of variant SARS-CoV-2 to humans or yet spillover from WTD to other susceptible wildlife species (e.g., mink, deer mice, raccoon, and red fox) remains unknown.
In our study, we have not found direct sequence links between the viruses circulating in WTD in NY with any of the available SARS-CoV-2 sequences recovered from humans. In fact, the viral sequences obtained from WTD are highly divergent from available human-derived SARS-CoV-2 sequences, with some strains accumulating over 70 nucleotide mutations across the genome. High genetic divergence from human SARS-CoV-2 sequences was observed among sequences from all three major variants (Alpha, Gamma, and Delta) identified in our study. Notably, Alpha and Gamma sequences presented the highest divergence to human sequences, which aligns with the fact that these variants were likely introduced earlier and circulated longer in WTD populations than Delta variant viruses. The impact of such mutations on the virus ability to transmit between WTD or from WTD to humans is still unknown and should be investigated in the future as it may shed light on mechanisms that affect the risk of deer-to-human transmission of the virus. The peak detection of Alpha and Gamma variants in humans in NY occurred between March and June 2021, with only sporadic detections occurring after August, while the Delta variant was the dominant variant circulating in humans in NY during the time of sample collection from WTD included in our study. These findings indicate independent evolution of SARS-CoV-2 VOCs in WTD and demonstrate parallel circulation of viral variants (Alpha and Gamma) that had been replaced by the Delta variant in humans, thus highlighting the potential that WTD may serve as a wildlife reservoir for nearly extinct SARS-CoV-2 variants. Thus, it is extremely important to continue to monitor WTD populations in the areas sampled in our study to assess the dynamics of the identified VOCs and of other deer-associated variants that might evolve and emerge in this host and potentially spillback into humans.
To date, only one report described the detection of a single WTD-like SARS-CoV-2 variant recovered from humans in Canada, which suggests potential deer-to-human transmission of the virus (27). Deer hunting is a widespread practice in North America that is regulated by the states and used as a method of population control for this highly prolific animal species. In 2020, for example, ~6.5 million WTD were harvested in the United States and Canada (28). This practice provides opportunities for close contact between a potentially infected reservoir and humans along with wildlife rehabilitation, captive cervid ownership, and zoological collections. In the present study, we recovered infectious virus from seven RPLN samples that tested positive for SARS-CoV-2 RNA, corroborating findings from previous studies that detected infectious virus in samples collected from wild WTD (12, 14). Although the proportion of live virus–positive samples detected was low, the findings are important in that they demonstrate that there is a risk of contact with infectious SARS-CoV-2 during handling and processing of WTD carcasses, which could lead to spillback via deer-to-human transmission of the virus. Our previous experimental data, assessing the infectious and transmission dynamics of SARS-CoV-2 in WTD, demonstrated a short window (~5 d) of infectiousness, a period in which experimentally inoculated animals excrete the virus in oral and nasal secretions and carry infectious/replicating virus in respiratory and lymphoid tissues and are able to transmit the virus to contact animals (10). The short viral replication window in tissues may have contributed to the low detection of infectious virus in the samples tested in our study. Additional factors such as time from sample collection to testing and suboptimal sample storage conditions during transportation to the laboratory may also have negatively affected the success of virus isolation in our study.
The high number of mutations observed in SARS-CoV-2 samples recovered from WTD in relation to human samples indicates that the virus has been circulating and evolving within the deer population. Some of the observed mutations could be a response to host adaptation. The mutation ORF1a:L4111F, for example, is observed in WTD-derived samples from multiple geographic regions and is present at a low frequency (<0.5%) among all human sequences available in the GISAID database. This suggests that this mutation emerged independently in deer after spillover from humans. The absence or very low frequency of fixed mutations in the S gene suggests that host-specific adaptation was not necessary for human-to-deer spillover, reinforcing the notion of SARS-CoV-2 as a “generalist” virus (29). The presence of homoplasious and high-frequency mutations in ORF1a and ORF1b emphasizes the importance of also characterizing non-Spike protein mutations as they may also play a role in virus host range and species specificity. A few mutations detected in the S gene lie near amino acid residues that define the Delta, Alpha, and Gamma VOCs (SI Appendix, Table S7). The mutation S:R683W detected in Delta WTD sequences, for example, is located close to the S:P681R site, which enhances cleavage of the S into the S1 and S2 subunits (30). Two other substitutions detected in Alpha WTD samples, S:D568N and S:I569T, lie in close proximity to the S:A570D mutation, which may play an important role modulating the conformation of S RBD, and was shown to contribute to Alpha VOC infectivity in humans (31). It is possible that these S substitutions might increase affinity of S protein for the WTD ACE2 receptor ortholog (32); however, additional experiments are needed to determine their actual biological function and assess whether they contribute to SARS-CoV-2 infection, replication, and transmission between WTD and from WTD to humans.
The strengths of our study include i) the large number of samples collected across much of the territory of the State of New York and surveyed over two consecutive deer hunting seasons during the COVID-19 pandemic; ii) the isolation of infectious SARS-CoV-2 viruses from tissues of free-ranging animals demonstrating the potential infectiousness and risk of human exposure; and iii) identification and detection of SARS-CoV-2 variants in WTD several months after their last detection in humans. The limitations of the study include i) the low rate of isolation of SARS-CoV-2, which could be a result of suboptimal conditions of transportation of the samples to the laboratory, which may have decreased virus viability; iii) the samples included were collected cross-sectionally for CWD testing at single time points from the animals sampled and not over time; iii) the fact that encrypted circulation of the Alpha and Gamma VOCs in humans cannot be formally ruled out; and iv) the fact that serum samples were not available for parallel testing as detection of antibodies combined with SARS-CoV-2 RNA could reveal higher rates of exposure to the virus in the target WTD populations.
The evidence generated in this study demonstrates widespread dissemination of SARS-CoV-2 in wild WTD populations across the State of New York and further indicates the cocirculation of three major VOCs in this new wildlife host. Notably, circulation of SARS-CoV-2 in WTD results in emergence of genetically diverse viruses in this species. These observations highlight the need to establish continuous surveillance programs to monitor the circulation, distribution, and evolution of SARS-CoV-2 in WTD populations and to establish measures to minimize additional virus introductions in animals that may lead to spillback of deer-adapted SARS-CoV-2 variants to humans.
Materials and Methods
Sample Collection.
RPLNs from WTD collected for the CWD Surveillance Program in New York State (NYS) were used in this study to investigate the presence of SARS-CoV-2. Samples were collected and submitted to the Animal Health Diagnostic Center (AHDC) at the Cornell University for ELISA testing. A total of 5,462 RPLNs collected during the hunting seasons of 2020 (Season 1, September to December 2020, n = 2,700) and 2021 (Season 2, September to December 2021, n = 2,762) were included in this study. All samples were stored at −80 °C upon arrival at the AHDC until tested for SARS-CoV-2. The WTD sex, age, date of harvest, and location to the level of town were collected with the sample to use for analysis.
Nucleic Acid Extraction and rRT–PCR.
RPLN tissues (200 mg) were minced with a sterile scalpel, resuspended (1/10 w/v) in PBS, and homogenized using magnetic beads. For the RNA extraction, 200 µL of the homogenate was used as input using the MagMax Core Extraction Kit (Thermo Fisher) and the automated KingFisher Flex nucleic acid extractor (Thermo Fisher) following the manufacturer’s recommendations. The rRT–PCR was performed using the EZ-SARS-CoV-2 real-time RT–PCR assay (Tetracore Inc.), which detects both genomic and subgenomic viral RNAs for increased diagnostic sensitivity. The analytic sensitivity of the EZ-SARS-CoV-2 rRT–PCR assay has been estimated at 1.75 SARS-CoV-2 N RNA copies per reaction in clinical samples (33). An internal inhibition control was included in all reactions. Positive and negative amplification controls were run side by side with test samples.
Virus Isolation.
For virus isolation, RPLNs, which tested positive for SARS-CoV-2 by rRT–PCR with Ct values lower than 30, were subjected to virus isolation under biosafety level 3 conditions at the Cornell University. Vero E6/TMPRSS2 (JCRB Cell Bank, JCRB1819) were cultured in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), L-glutamine (2 mM), penicillin (100 U.mL−1), streptomycin (100 μg.mL−1), and Geneticin (1 mg.mL−1). Twenty-four–well plates were seeded with ~75,000 Vero E6/TMPRSS2 cells per well 24 h prior to sample inoculation. Cells were rinsed with phosphate-buffered saline (PBS) (Corning) and inoculated with 200 μL of the homogenate, and inoculum adsorbed for 1 h at 37 °C with 5% CO2. Mock-inoculated cells were used as negative controls. After adsorption, cells were rinsed with PBS once, and replacement cell culture media were added, and cells were incubated at 37 °C with 5% CO2 and monitored daily for cytopathic effect (CPE) for 3 d. SARS-CoV-2 infection in CPE-positive cultures was confirmed with an immunofluorescence assay (IFA) as described previously (8). Positive samples were harvested and reinoculated into Vero E6/TMPRSS2 cell cultures in chamber slides (Thermo Scientific Nunc Lab-Tek II Chamber Slide System; Fisher Scientific). At 24 h after inoculation, cells were fixed and stained with a monoclonal anti-SARS-CoV-2 antibody against N (mouse mAb anti-SARS-CoV-2 nucleoprotein clone B61G11) (8) and a polyclonal antibody anti-S protein (rabbit pAb anti-SARS-CoV-2 Spike S2; Sino Biological). DAPI staining was used to counterstain cell nuclei, and images were obtained using a confocal microscope. Additionally, ISH was also performed in Vero E6/TMPRSS2 cells at 24 h after infection following procedures previously established in our laboratory (34). Cell cultures with no CPE were frozen, thawed, and subjected to two additional blind passages/inoculations in Vero E6/TMPRSS2 cell cultures. At the end of the third passage, the cell cultures were subjected to IFA as previously described by us (8, 10).
Titrations in the VI-positive original samples and on passage 1 of the virus isolates in cell culture were performed by limiting dilution in Vero E6/TMPRRSS2 cells as previously described by us (8, 10). Viral titers were determined using the Spearman and Karber method and expressed as median tissue culture infectious dose 50 per ml (TCID50.mL−1).
Data Acquisition.
All WTD-derived SARS-CoV-2 genomes (n = 159) were retrieved from GISAID (accessed May 1, 2022). All human-derived SARS-CoV-2 genomes (n = 120,828) from the State of New York, United States, were retrieved, and a local BLAST database was built to obtain 3,837 sequences with the highest nucleotide similarities to WTD-derived SARS-CoV-2 genomes in the current study.
SARS-CoV-2 Genome Sequencing.
Total RNA was reverse-transcribed using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) as described in the protocol available at https://doi.org/10.17504/protocols.io.br54m88w. The cDNA from each sample was used four times for parallel generation of tiled amplicons with ARTIC V3 and ARTIC V4.1 primers (IDT) and library preparation using a modified ARTIC network’s nCoV-2019 sequencing protocol v2 (35). The minor modifications include the use of Q5 High-Fidelity 2X Master Mix and the use of 4 µL cDNA in a 25 µL PCR. Additionally, the annealing and denaturation temperatures were dropped to 64 °C and 95 °C, respectively. Purified amplicons were diluted 1:10 and quantified before used as an input for two sequencing library preparations (V3 and V4.1 primers) that were loaded onto R9.4 flow cells in a MinION Mk1B (Oxford Nanopore Technologies, ONT).
Genomic Analysis.
Raw FAST5 reads were basecalled and demultiplexed using Guppy v5.0.16 in a high accuracy mode. The output FASTQ files were processed through the ARTIC bioinformatics pipeline using the applicable primer scheme (V3 or V4.1) to generate consensus sequences (https://github.com/artic-network/artic-ncov2019). Lineage classification was performed using Pangolin version 4.0.6 (18). Only sequences passing Pangolin QC thresholds for minimum length and maximum N content were used for downstream analyses.
Interactive Phylogenomic and Phylogeographic Analyses.
For phylogenetic analysis of WTD-derived SARS-CoV-2 genomes, two curated datasets were created. The first dataset consisted of all 164 SARS-CoV-2 from the current study and 159 deer-derived SARS-CoV-2 genomes downloaded from the GISAID database. The second dataset consisted of 3,837 human-derived SARS-CoV-2 genomes and 323 deer-derived SARS-CoV-2 genomes from the present study and GISAID database (36). Phylogenetic analysis of both datasets was performed by using procedures implemented in Nextstrain (36). Briefly, multiple sequence alignment was performed using Nextalign; maximum likelihood tree was inferred using IQ-TREE through Augur tool kit and data visualization through Auspice. Maps in figures 3A and 3B were generated using Microreact (37).
Mutation Analysis.
Sequences from the present study were grouped according to its VOC classification, and nucleotide sequence alignments were performed individually for each group of VOCs using MAFFT v7.453 (38). Variations in relation to reference genome (GenBank accession number MN908947.3) were identified in Geneious Prime 2019 software. Lineage-defining nonsynonymous mutations were obtained from the Lineage Comparison tool of Outbreak web interface (39) and excluded from the dataset. The remaining mutations were used to screen for possible host-adaptive mutations. The phylogenetic tree with WTD-derived sequences was screened for homoplasies using HomoplasyFinder to strengthen findings of putative host-adaptive mutations (40). Nucleotide positions with a consistency index <0.5 were considered for further analysis.
Timescale Distribution of Human-Derived SARS-CoV-2.
To place the deer-derived SARS-CoV-2 VOCs from the current study in the context of VOCs circulating in human population, the epidemiological metadata (VOCs and date of collection) of a total of 120,828 (NY) and 250,000 (United States) human-derived SARS-CoV-2 genome sequences from March 2020 to April 2022 were retrieved from GISAID. The SARS-CoV-2 sequences from NY were further separated into sequences from NYC (86,142) and rural NY (34,686). Stacked bar graphs showing monthly cases of different VOCs in human population of NY and United States or NYC and rural NY were created (37) using GraphPad Prism 9 (GraphPad Software).
SaTScan Spatial Analysis.
The space-time discrete Bernoulli model within SaTScan software version 10.0.2 (42) was used to identify statistically significant clusters of SARS-CoV-2 infection in the NY WTD population. For each sample, the geographic location (latitude and longitude) was determined based on the centroid of town using the US Census Bureau County TIGER/Line Shapefiles for County Subdivisions, which are equivalent to towns, cities, and other similar administrative divisions. To detect high rates of SARS-CoV-2 in deer clusters, the Monte Carlo hypothesis test method and 999 replications were applied to detect a high prevalence rate of SARS-CoV-2 (42). The spatial clusters were visualized in QGIS (geographic information system) mapping software version 3.16.16 (43).
Relative Risk (RR) and Logistic Regression Analysis.
The measure of RR was used to identify locations with a greater risk of having SARS-CoV-2 in deer following the equation RR=(c/e)/((C-e)/(C-e)), where c represents the total number of observed cases in the town, e represents the total number of expected cases in a town, and C is the total number of observed cases in New York State. If the RR was larger than 1, the location had a greater risk of having a positive deer. A logistic regression analysis conducted on deer outside the clusters was used to determine whether the difference between deer grouped by age, sex, and season is true in the comparison of age, sex, and season for SARS-CoV-2 infection.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
The work was supported in part by the USDA National Institute of Food and Agriculture (award no. 2021-68014-33635) and by the USDA Animal and Plant Health Inspection Service (agreement no. AP20VSD&B000C020). We thank the Cornell Biosafety team for the support. We also thank the Animal Health Diagnostic Center at the Cornell University for the use of extraction and real-time PCR equipment. The NYS Department of Environmental Conservation was instrumental in tissue collection and cooperation in data sharing for this study. Tetracore Inc. generously provided the real-time PCR reagents used to test the samples in this study. We also gratefully acknowledge all data contributors, i.e., the authors and their originating laboratories responsible for obtaining the specimens and their submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based.
Author contributions
M.M., K.L.S., and D.G.D. designed research; L.C.C., M.M., S.L.B., N.A.H., L.M.C., S.A., and D.G.D. performed research; L.C.C., S.L.B., L.M.C., M.R.R.E., and K.L.S. contributed new reagents/analytic tools; L.C.C., M.M., S.L.B., N.A.H., S.A., M.R.R.E., K.L.S., and D.G.D. analyzed data; and L.C.C., M.M., S.L.B., and D.G.D. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All SARS-CoV-2 consensus genomes are deposited in GISAID (https://www.gisaid.org/; accession numbers are available in SI Appendix, Table S8), and raw reads have been submitted to NCBI’s Short Read Archive (BioProject Number PRJNA872140).
Supporting Information
References
- 1.WHO, “COVID-19 weekly epidemiological update” (WHO; Tech. Rep. 81, 2021). [Google Scholar]
- 2.Huang C., et al. , Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya A. E., et al. , The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544 (2020), 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau S. K. P., et al. , Possible bat origin of severe acute respiratory syndrome Coronavirus 2. Emerg. Infect. Dis. 26, 1542–1547 (2020), 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami S., et al. , Detection and characterization of bat sarbecovirus phylogenetically related to SARS-CoV-2, Japan. Emerg. Infect. Dis. 26, 3025–3029 (2020), 10.3201/eid2612.203386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F., The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020), 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damas J., et al. , Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.2010146117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer M. V., et al. , Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. J. Virol. 95, e00083-21 (2021), 10.1128/jvi.00083-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cool K., et al. , Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. Emerg. Microbes Infect. 11, 95–112 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martins M., et al. , From Deer-to-Deer: SARS-CoV-2 is efficiently transmitted and presents broad tissue tropism and replication sites in white-tailed deer. PLOS Pathog. 18, e1010197 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanberry B. B., Addressing regional relationships between white-tailed deer densities and land classes. Ecol. Evol. 11, 13570–13578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuchipudi S. V., et al. , Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer. Proc. Natl. Acad. Sci. U.S.A. 119, e2121644119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marques A. D., et al. , Multiple introductions of SARS-CoV-2 alpha and delta variants into white-tailed deer in Pennsylvania. mBio 13, e0210122 (2022), 10.1128/mbio.02101-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale V. L., et al. , SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 602, 481–486 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palermo P. M., Orbegozo J., Watts D. M., Morrill J. C., SARS-CoV-2 neutralizing antibodies in white-tailed deer from Texas. Vector Borne Zoonotic Dis. 22, 62–64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandler J. C., et al. , SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc. Natl. Acad. Sci. U.S.A. 118, e2114828118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuyama S., et al. , Enhanced isolation of SARS-CoV-2 by TMPRSS2- expressing cells. Proc. Natl. Acad. Sci. U.S.A. 117, 7001–7003 (2020), 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambaut A., et al. , A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5, 1403–1407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DEC, "Chronic wasting disease" (New York State's Department of Environmental Conservation; Tech. Rep. 2021, 2022). [Google Scholar]
- 20.O’Brien D. J., et al. , Epidemiology of mycobacterium bovis in free-ranging white-tailed deer, Michigan, USA, 1995–2000. Prev. Vet. Med. 54, 47–63 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Dee S. A., et al. , Survival of viral pathogens in animal feed ingredients under transboundary shipping models. PLoS One 13, e0194509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francisco R., et al. , Experimental Susceptibility of North American Raccoons (Procyon lotor) and Striped Skunks (Mephitis mephitis) to SARS-CoV-2. Frontiers in Veterinary Science, 8 (2022), 10.3389/fvets.2021.715307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shriner S. A., et al. , SARS-CoV-2 exposure in escaped mink, Utah, USA. Emerg. Infect. Dis. 27, 988–990 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin B. D., et al. , SARS-CoV-2 infection and transmission in the North American deer mouse. Nat. Commun. 12, 3612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosco-Lauth A. M., et al. , Survey of peridomestic mammal susceptibility to SARS-CoV-2 infection. Emerg. Infect. Dis. 27, 2073–2080 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porter S. M., Hartwig A. E., Bielefeldt-Ohmann H., Bosco-Lauth A. M., Root J. J., Susceptibility of wild canids to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Emerg. Infect. Dis. 28, 1852–1855 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickering B., et al. , Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission.. Nature Microbiology, 7, 2011–2024. (2022), 10.1038/s41564-022-01268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Deer Association, “Deer report” (National Deer Association Tech. Rep. 2021, 2021), p. 68. [Google Scholar]
- 29.MacLean O. A., et al. , Natural selection in the evolution of SARS-CoV-2 in bats created a generalist virus and highly capable human pathogen. PLoS Biol. 19, e3001115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., et al. , Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 39, 110829 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang T. J., et al. , Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function. Nat. Struct. Mol. Biol. 28, 731–739 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Harvey W. T., et al. , SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laverack M., et al. , Clinical evaluation of a multiplex real-time RT-PCR assay for detection of SARS-CoV-2 in individual and pooled upper respiratory tract samples. Arch. Virol. 166, 2551–2561 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carvallo F. R., Severe SARS-CoV-2 infection in a cat with hypertrophic cardiomyopathy. Viruses 13, 1510 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quick J., nCoV-2019 sequencing protocol v2 (GunIt) (Version 2, 2021). 10.17504/protocols.io.bdp7i5rn Accessed 29 June 2022. [DOI]
- 36.Hadfield J., et al. , Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Argimón S., et al. , Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2, e000093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013), 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsueng G., et al. , Outbreak.info research library: A standardized, searchable platform to discover and explore COVID-19 resources. Serv. Biol. bioRxiv [Preprint] (2022), 10.1101/2022.01.20.477133 Accessed 6 June 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crispell J., Balaz D., Gordon S. V., Homoplasyfinder: A simple tool to identify homoplasies on a phylogeny. Microb. Genom. 5, e000245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulldorff M., Information Management Services, Inc. SaTScanTM (Version 10.0.2, 2022) www.satscan.org. Accessed 29 June 2022.
- 42.Hedman H. D., et al. , Spatial analysis of chronic wasting disease in free-ranging white-tailed deer (Odocoileus virginianus) in Illinois, 2008–2019. Transbound. Emerg. Dis. 68, 2376–2383 (2021). [DOI] [PubMed] [Google Scholar]
- 43.QGIS.Org, QGIS geographic information system version 3.16.16. https://www.qgis.org. Accessed 29 June 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All SARS-CoV-2 consensus genomes are deposited in GISAID (https://www.gisaid.org/; accession numbers are available in SI Appendix, Table S8), and raw reads have been submitted to NCBI’s Short Read Archive (BioProject Number PRJNA872140).