Significance
Influenza A viruses (IAVs) represent a major worldwide health threat. Wild aquatic birds are the natural reservoir of IAVs, which also infect other species including domestic poultry and mammals. Pigs are “mixing vessels” where IAVs from different strains can reassort, posing a continuous threat of zoonotic infections, and animal–human reassortant IAVs episodically generate severe pandemics. We demonstrate large-scale quantitative assessment of reassortment between two circulating human IAVs using high-throughput droplet microfluidics. Our method will enable i) a better understanding of the rules underlying reassortment and ii) scrutinization of reassortment between zoonotic animal viruses and human seasonal viruses, to identify those representing a threat to human health. This provides an additional predictive parameter to complement existing Pandemic Influenza Risk Assessment Tools.
Keywords: droplet microfluidics, single-cell RNA-seq, influenza, genetic reassortment, direct coupling analysis
Abstract
The segmented RNA genome of influenza A viruses (IAVs) enables viral evolution through genetic reassortment after multiple IAVs coinfect the same cell, leading to viruses harboring combinations of eight genomic segments from distinct parental viruses. Existing data indicate that reassortant genotypes are not equiprobable; however, the low throughput of available virology techniques does not allow quantitative analysis. Here, we have developed a high-throughput single-cell droplet microfluidic system allowing encapsulation of IAV-infected cells, each cell being infected by a single progeny virion resulting from a coinfection process. Customized barcoded primers for targeted viral RNA sequencing enabled the analysis of 18,422 viral genotypes resulting from coinfection with two circulating human H1N1pdm09 and H3N2 IAVs. Results were highly reproducible, confirmed that genetic reassortment is far from random, and allowed accurate quantification of reassortants including rare events. In total, 159 out of the 254 possible reassortant genotypes were observed but with widely varied prevalence (from 0.038 to 8.45%). In cells where eight segments were detected, all 112 possible pairwise combinations of segments were observed. The inclusion of data from single cells where less than eight segments were detected allowed analysis of pairwise cosegregation between segments with very high confidence. Direct coupling analysis accurately predicted the fraction of pairwise segments and full genotypes. Overall, our results indicate that a large proportion of reassortant genotypes can emerge upon coinfection and be detected over a wide range of frequencies, highlighting the power of our tool for systematic and exhaustive monitoring of the reassortment potential of IAVs.
Influenza A viruses (IAVs) are human and animal respiratory pathogens that represent a worldwide major public health threat. In humans, they recur every year as “seasonal influenza”, due to antigenic variation as a result of polymerase errors. In addition, the segmented nature of their genome, made up of eight negative-sense single-stranded viral RNAs (vRNAs), provides IAVs with further evolutionary potential: upon coinfection of a single cell by distinct viruses, progenies that harbor combinations of genomic segments derived from the different parental viruses can emerge from a process termed genetic reassortment.
Aquatic birds, poultry, swine, bats, and other mammals are hosts to a dynamic pool of IAVs (1) which pose ever-changing threats as a source of zoonotic infections, especially as pigs can act as a “mixing vessel” for swine, avian, and human IAVs to undergo genetic reassortment (2, 3). Genetic reassortment between cocirculating human IAVs increases genetic diversity during the evolution of seasonal viruses, and reassortment between highly divergent animal and human IAVs can lead to novel and potentially deadly viruses. Reassortment between human and avian IAVs led to the emergence of the 1957 and 1968 pandemic strains (4). The importance of IAV reassortment as a public health concern was highlighted in 2009 by the emergence of the H1N1pdm09 virus in humans, a complex reassortant swine virus combining segments derived from avian, swine, and human IAVs (5, 6) and more recently by the increased prevalence of avian H7N9 (7) and swine H1N1 multireassortant IAVs with increased human infectivity (8).
Genetic reassortment is intimately linked to the copackaging of the eight different IAV genomic segments via cis-acting packaging signals present on each vRNA. These signals, accountable for direct vRNA–vRNA interactions, display segment and subtype specificity and have been mapped throughout the entire length of the vRNAs (9–19). Hence, the current model for vRNA packaging, which also relies on structural (20) and genetic data (21–25), is that vRNA–vRNA interactions between adjacent packaging signals control the bundling of the eight viral segments and their copackaging into virions with a conserved “7 + 1” pattern, with one central segment surrounded by seven others (16, 26, 27). Direct contacts between segments have been observed by electron tomography (10, 28). In addition, the amino acid sequence of the viral nucleoprotein (NP) may control selective packaging jointly with packaging signals on the vRNAs (29).
It has previously been reported that IAV reassortment can occur at high frequency in vivo between phenotypically identical parental viruses in the absence of constraints imposed by segment mismatches, although frequencies vary between virus strains and host species (30–35). Even though the theoretical number of genotypes that can emerge from reassortment between two strains is 256 (28), the full panel has never been observed and certain genes tend to cosegregate (11, 36, 37), suggesting that genetic reassortment is biased (reviewed in refs. 12 and 38). Incompatibilities among heterologous vRNAs due to sequence differences in the packaging signals and/or incompatibilities among heterologous proteins can both strongly impact the outcome of mixed IAV infection (reviewed in refs. 12 and 38–40). Progress in characterizing cross-species incompatibilities has been made in recent years (38). However, the rules underlying reassortment remain largely unknown, primarily because characterization of reassortant viruses has required genotyping via RT-PCR followed by sequencing or, alternatively, by high-resolution melt analysis (30, 39, 41) of clonal isolates obtained upon plaque purification or limiting dilution. With these procedures, reassortment cannot be studied comprehensively; so far, a maximum of ~400 (35, 42) to ~700 (43) reassortant genomes have been analyzed at a time, and not all were performed under uniform experimental settings.
Droplet microfluidics systems (44), in which aqueous pL to nL droplets in inert fluorinated carrier oil are used as independent microreactors containing single cells (45), combined with droplet barcoding and next generation sequencing (NGS) technologies have enabled single-cell RNA sequencing (scRNA-seq) transcriptomic analysis on thousands of individual cells (46, 47). Commercial droplet microfluidic systems have been applied for virus and host transcriptomic analysis of IAV-infected cells (48–51). More recently, it was also shown that IAV can propagate within drops containing single cells, enabling analysis of viral populations produced from individual infected cells (52).
Here, we report a droplet microfluidics approach to systematically study IAV genetic reassortment on a large scale. Our approach adapts a proven droplet microfluidics systems for scRNA-seq (46) to perform high-throughput targeted sequencing of vRNAs. The experimental and bioinformatic pipelines allowed the analysis of 18,422 individual viral genomes resulting from coinfection of cultured cells with two distinct IAVs, among which 9,438 displayed reassortant genotypes. Results were highly reproducible across technical and biological replicates, confirmed that genetic reassortment is far from random, recalled some previously identified reassortant gene combinations and allowed quantification of reassortant genotypes including rare events.
Results
A High-Throughput Droplet-Based Single-Virus RNA Sequencing System Workflow to Study IAV Genetic Reassortment.
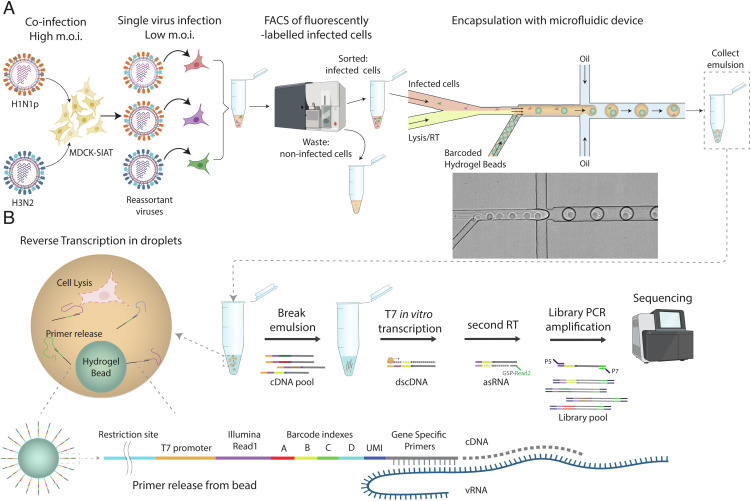
We have developed a procedure, termed µFlu, that combines molecular virology, droplet microfluidics, NGS, and bioinformatics to systematically study IAV genetic reassortment at an unprecedented scale (Fig. 1). The µFlu procedure was applied to two human seasonal strains of IAV, H1N1pdm09 (A/Bretagne/7608/2009) (termed H1N1p) and H3N2 (A/Centre/1003/2012) (termed H3N2). Coinfection of Madin-Darby Canine Kidney (MDCK)-SIAT (sialyltransferase) (53) “producer" cells with H1N1p (m.o.i = 10 pfu/cell) and H3N2 (m.o.i = 5 pfu/cell) was performed in triplicate (Co-INF1, Co-INF2, and Co-INF3). Flow cytometry of MDCK-SIAT cells separately infected with H1N1p and H3N2 and stained with anti-H1N1p-HA and anti-H3N2-M2 antibodies revealed that 30 to 38% of cells exposed to both strains were coinfected (SI Appendix, Fig. S1A and Table S1). The viral progeny from coinfections (SI Appendix, Table S1) were next used to infect fresh MDCK-SIAT “indicator" cells (m.o.i = 0.01 pfu/cell). For every viral stock, two independent low m.o.i. infections were performed generating six replicates, R1.1, R1.2, R1.3, R2.1, R2.2, and R2.3 (SI Appendix, Fig. S1E). Following infection, cells were fixed and fluorescently labeled with anti-HA and anti-M2 antibodies, and single infected cells (1.16 to 3.03% of total cells) were collected/enriched by fluorescence-activated cell sorting (FACS), using forward scatter and side scatter to remove cell debris and cell aggregates (SI Appendix, Fig. S1 B and C). As a control, cells were separately infected, in duplicate, at high m.o.i. with either parental virus, sorted by FACS, and mixed 1:1 generating “Premix” controls 1 and 2 (SI Appendix, Fig. S1 D and E). Additional controls were also performed, where cells were separately infected at high m.o.i. with the H1N1p and H3N2 viruses and either treated separately until the end of the µFlu procedure (No-MixCtrl) or encapsulated and reverse transcribed separately prior to mixing for the rest of the library preparation steps (Library-MixCtrl) (SI Appendix, Fig. S1E). Under such conditions, reassortment cannot occur since coencapsulation of cells infected by H1N1p and cells infected by H3N2 viruses is not possible.
Fig. 1.
Single-cell targeted RNA-seq using barcoded hydrogel beads to study genetic reassortment of IAVs. (A) MDCK-SIAT “producer” cells are coinfected at high m.o.i. with human seasonal H3N2 and H1N1p viruses and the progeny, which contains reassortant and parental viruses, is used to infect MDCK-SIAT “indicator” cells at low m.o.i. to achieve single virus infections. At 14 h post-infection, cells expressing viral antigens are sorted by FACS and then loaded onto a microfluidic device for coencapsulation of single cells together with single barcoded hydrogel beads (HgBs), lysis buffer, and RT reagents (SI Appendix, Fig. S18 and Movies S1 and S2). (B) In the droplets, primers are released from the beads by digestion with BclI restriction endonuclease prior to RT. After in-drop RT, the emulsion is broken and subsequent steps (second-strand synthesis, in vitro transcription by T7 RNA polymerase, a second RT reaction using GSP-Read2 primers and library PCR for Illumina sequencing) are performed in bulk. The design of barcoded RT primers linked to HgBs is schematized. UMI: Unique Molecular Identifier; GSP: Gene Specific Primer. P5 and P7: Illumina P5 and P7 primer sites.
A microfluidic system, adapted from refs. 46 and 54, was used to achieve encapsulation of 0.5 × 105 to 1 × 105 low m.o.i. infected “indicator" cells from each experiment in 120 pL droplets, together with lysis buffer, reverse transcription (RT) reagents, BclI restriction endonuclease, and a single hydrogel bead (HgB) (Fig. 1A). Each bead carried ~109 barcoded single-stranded DNA gene-specific primers (GSPs) for cDNA synthesis on vRNAs (Fig. 1B). The primers on each bead carried the same barcode (BC), while primers on different beads carried different BCs (46). The BCs were produced by split-and-pool ligation (55). After in-drop RT, emulsions were broken, and all subsequent steps of library preparation were performed in bulk (Fig. 1B). A linear amplification step using in vitro transcription by T7 RNA polymerase was used (46, 54) to reduce the imbalance between segments. Illumina libraries were prepared and sequenced on a NovaSeq 6000 S1 (300 cycles paired-end). All the cDNAs from a single cell, and hence a single virus, in principle carry the same droplet-specific BC allowing the individual viral genomes to be characterized by NGS. “Premix” controls were processed in the same way. Two NGS runs were performed: NGS1 to analyze R1.1, R1.2, and R1.3 together with the No-MixCtrl, the Library-MixCtrl and the Premix 1 controls, and NGS2 to analyze R2.1, R2.2, and R2.3 and Premix 2 (SI Appendix, Fig. S1E).
Bioinfomatic Analysis and Reproducibility of µFlu Reassortment Datasets.
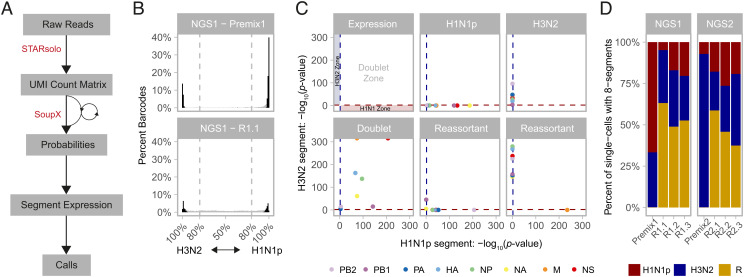
We developed a bioinformatic pipeline to accurately identify the viral progeny captured in the scRNA-seq data resulting from the µFlu experimental workflow (Fig. 2 A–C and SI Appendix, Fig. S2). Our results were robust to a variety of parameter choices (SI Appendix, Figs. S3 and S4), and we present herein those based on one such set (SI Appendix, Supplementary Methods and Supplementary Results). After removal of data from BCs with <125 unique molecular identifier (UMI) counts, we analyzed 54,918 BCs (corresponding to cell-containing droplets) across all experiments (SI Appendix, Tables S3 and S4 and Dataset S1). As we were interested in analyzing reassortant patterns, we wanted to be sure to exclude any droplets containing more than one infected cell (i.e., “doublets”). As such, we conservatively excluded any BCs harboring ≥1 vRNA from both H1N1p and H3N2 strains from further analysis (SI Appendix, Table S4 and Dataset S2). Doublet rates were high among experiments where coencapsulation of cells infected with differing viral strains was possible (combined six replicates of reassortment experiments: 37.70%; Premix1 + Premix2: 23.12%), while doublet rates were low in experiments where cells infected with the different strains were never encapsulated together (No-MixCtrl 2.39%, Library-MixCtrl 0.66%) (SI Appendix, Figs. S1E and S5A). This suggests that artifactual doublets associated with molecular biology steps (e.g., stray BCs and template switching) are relatively infrequent in this setting and that the high doublet rates in Premix and coinfection experiments are more likely attributable to coencapsulation of cell aggregates that form after FACS and/or changes in cell density due to sedimentation during injection. Across all replicates, following the exclusion of potential doublets, data from 37,492 single-cell-containing droplets remained.
Fig. 2.
Processing of scRNA-seq data. (A) Overview of the bioinformatic pipeline. FASTQs were aligned with STARsolo to a reference containing both the H1N1p and H3N2 segmented IAV genomes. (B) Histogram of the percentage of BCs as a function of the percentage of UMIs aligning to the H1N1p and H3N2 genomes for the NGS1—Premix (Top) and NGS1—R1.1 (Bottom) experiments; see SI Appendix, Fig. S3 for histograms of all experiments. Dashed lines distinguish BCs harboring >80% of their UMIs from a single strain that were used in the first iteration of the global contamination fraction (rho) estimation with SoupX, using UMIs from the minor strain as presumed contamination. Black bars represent BCs used in the second iteration of rho estimation, the empirically estimated rho, again using UMIs from the minor strain as presumed contamination (gray bars represent BCs that were not used). See SI Appendix, Supplementary Methods for details. (C) Segment expression and calling rules. We then used a statistical framework implemented in SoupX to test whether the UMI counts for each segment and BC were expressed at a level significantly higher than expected when accounting for potential contamination from the ambient pool. A schematic representation of the rules is shown in the "Expression" panel. FDR-adjusted P values for H1N1p (x-axis) and H3N2 (y-axis) segment expression are plotted for five representative BCs from NGS1-R1.1, corresponding to a pure H1N1p, pure H3N2, a doublet, and two reassortants. A FDR threshold of 1% (dotted lines) was used to define whether there was evidence of segment expression for each gene and strain. (D) Percentage of reassortants (R) and pure strains (H1N1p and H3N2) among single-cell-containing droplets with evidence for expression of all eight segments for the two premix controls and all six reassortment experiments.
Focusing on 4,952 single cells across all replicates for which we had evidence for all eight viral segments (SI Appendix, Table S4 and Dataset S2), we found that the average rate of reassortment across coinfection experiments was 51.08% ± 8.42% SD (Fig. 2D, yellow bars), while the false-positive reassortment rate was <1%, based on the control experiments (Fig. 2D and SI Appendix, Fig. S5B). Thus, less than 2% of observed reassortants are artifacts. The false-positive reassortants can be attributed to events happening after breaking the emulsion (e.g., stray BCs and template switching) as the false-positive reassortment rate was similar in Premix1 + Premix2 (where coencapsulation of cells infected by H1N1p and cells infected by H3N2 viruses in drops is possible) and in No-MixCtrl and Library-MixCtrl (where there is no coencapsulation of cells infected by H1N1p and cells infected by H3N2 viruses) (SI Appendix, Fig. S5B). Barnard’s test of independence (56) was performed by constructing a 2 × 2 table with the frequency counts of reassortant genomes and parental genomes between the Premix control experiments and individual replicates of NGS1 and NGS2 as categorical values. In all cases, the categorical values are “not independent” (P < 10−5), indicating that the reassortment rates in all six replicate experiments are different from the false-positive reassortment rates in the Premix control experiments (SI Appendix, Table S5). We observed similar patterns of reassortment across all replicates (SI Appendix, Fig. S6 A–C and E–G and Dataset S2), as demonstrated by the high Pearson correlation (57) for reassortant genotype frequencies (r = 0.77 to 0.88, P < 10−26), single segment frequencies (r = 0.64 to 0.92, P < 10−3) and pairwise segment combinations frequencies (r = 0.72-0.94, P < 10−18) between pairs of replicates (SI Appendix, Table S6). This justifies that data from R1.1, R1.2, and R1.3 can be merged (SI Appendix, Fig. S6D), as can data from R2.1, R2.2, and R2.3 (SI Appendix, Fig. S6H). Furthermore, the high Pearson correlation of reassortant genotype frequencies (r = 0.919, P < 10−64), single-segment frequencies (r = 0.957, P < 10−8), and pairwise segment frequencies (r = 0.955, P < 10−59) between these two merged data sets (SI Appendix, Table S6) justifies the creation of a “merged dataset” with data from R1.1, R1.2, R1.3, R2.1, R2.2, and R2.3, which contains 2,592 reassortant eight-segment genomes, and which is used from hereon (Dataset S2).
Reassortment between Two Circulating Seasonal H1N1 and H3N2 Strains Generates >60% of All Possible Gene Combinations, with a Limited Number of High-Frequency Gene Combinations.
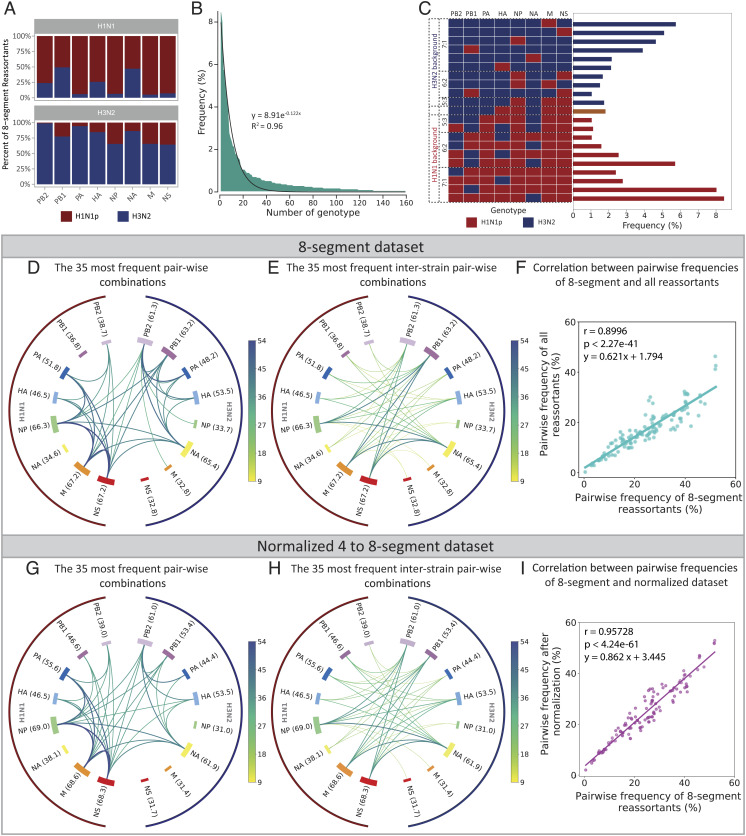
The frequencies of genotypes, single segments, and pairs of cosegregating segments, were analyzed among the 2,592 reassortant eight-segment genomes (Fig. 3). Barnard’s test of independence (56) was performed by constructing a 2 × 2 table with the mean intrastrain and interstrain pair-wise segment frequency counts between the experimental dataset and theoretical values of random reassortment. For random reassortment, intrastrain and interstrain pair-wise segment frequency would each be equal to 0.5. For the merged dataset (and all replicates), the categorical values are “not independent,” i.e., P < 10−5 (SI Appendix, Table S7), clearly indicating that reassortment is nonrandom. The cumulative counts of each segment found in a nonhomologous H1N1p or H3N2 genetic background (i.e., in genomes containing ≥5 segments from the opposite strain) are shown in Fig. 3A. In reassortant genomes where ≥ 5 segments were derived from H1N1p (top), the most frequent H3N2 segments were PB2, PB1, HA, and NA, while PA, NP, M, and NS segments were rarely present. Noticeably, this distribution was nonsymmetrical, as in genomes where ≥5 segments derived from H3N2 (bottom), the H1N1p segments PB1, NP, M, and NS were the most frequently incorporated, while the H1N1p segments PB2 and PA were rarely detected. A total of 159 different reassortant genotypes were observed out of the 254 theoretical possibilities (Dataset S2) but at very different frequencies, approximating to a negative exponential distribution (Fig. 3B): The 21 most frequent genotypes (Fig. 3C) represent only 13% of detected genotypes but make up 66% of all reassortant genomes (Dataset S2). Among reassortants with an H1N1p genetic background (red bars in Fig. 3C), the most frequent genotypes were 7:1 reassortants containing H3N2-NA or -PB1 or 6:2 reassortants containing H3N2-NA and -PB1, in agreement with previously published studies (13, 36). Conversely, the incorporation of single H1N1p-M, -NS, -NP, and -PB1 segments dominated in the H3N2 genetic background. Genomes wherein the number of segments from H1N1p and H3N2 are more imbalanced are more frequent, indicating a more frequent cosegregation of segments from the same strain (SI Appendix, Fig. S7).
Fig. 3.
Reassortant genotypes detected using the µFlu system. (A) Frequency of each segment in a nonhomologous H1N1p or H3N2 genetic background, i.e., in eight-segment genomes including ≥5 segments from the opposite strain (N = 1,260 and 1,123, respectively). Blue: H3N2 origin. Red: H1N1p origin. (B) Distribution of frequency (%) of all observed eight-segment reassortant genotypes (N = 159). The black line is a fit to a negative exponential curve. (C) Frequency of the 21 reassortant genotypes each representing >1% of all observed eight-segment reassortant genomes. (D and E) Chord plots of the 35 most frequent inter- and intrastrain pairwise segment combinations (D) and the 35 most frequent pairwise interstrain segment combinations (E) for the eight-segment reassortant genomes (N = 2,592). (F) Comparison between observed pairwise segment frequency of all reassortants (nonnormalized 2- to 8-segment dataset) and observed pairwise segment frequency of eight-segment reassortants. The Pearson’s correlation is r = 0.8996 (P < 10−40). (G and H) Chord plots of the 35 most frequent inter- and intra-strain pairwise segment combinations (G) and the 35 most frequent pairwise interstrain segment combinations (H) for 4- to 8-segment reassortant genomes (N = 9,324) after dropout normalization. Line thickness is proportional to the pairwise segment frequency (in percent), line color represents the pairwise segment frequency (in percent), and the size of the bar for each segment represents the single segment frequency (percent value mentioned in brackets). In all cases, the merged data set (combining NGS1 and NGS2) was used. (I) Comparison between pairwise segment frequency of the normalized 4- to 8-segment dataset and observed pairwise frequency of eight-segment reassortants. The Pearson’s correlation is r = 0.95728 (P < 10−60).
To investigate this, the frequencies of the pairwise combinations of segments within reassortant genomes were analyzed. For any pair of segments a and b, there are four possible combinations, aH1N1p-bH1N1p, aH3N2-bH3N2, aH1N1p-bH3N2, and aH3N2-bH1N1p, and across all segment pairs, the average frequency of these combinations among reassortant genomes was 32.5%, 30.2%, 18.7%, and 18.7%, respectively, confirming the tendency of segments from the same strain to cosegregate. In the reassortant viruses, certain segments are more likely to derive from one strain than the other: NP, M, and NS are more frequently from H1N1p (66%, 67%, and 67%, respectively), PB2, PB1, and NA are more frequently from H3N2 (61%, 63%, and 65%, respectively), while PA and HA derive roughly equally from both strains (52% and 47% from H1N1, respectively) (Fig. 3D and SI Appendix, Figs. S8 and S9 A–C and S10). Focusing on pairwise interstrain segment combinations only (Fig. 3E and SI Appendix, Figs. S9 A–C and S11), all possible pairwise interstrain combinations of segments were observed, with a wide range of frequencies from 0.5% (H1N1p-PB2:H3N2-PA) to >40% (40.7% for H1N1p-NP:H3N2-PB1 and 41% for H1N1p-M:H3N2-PB1) of all reassortant genotypes.
We sought to increase the statistical power of our analysis by including data from “dropout genomes,” with one to six missing (and nonidentified) segments, termed “dropout segments” (SI Appendix, Table S4). Missing segments can result from incomplete capture during RT. Primers were optimized to maximize the efficient and uniform amplification of all segments using all 16 H1N1p and H3N2 in vitro transcribed vRNAs. Next-generation sequencing results obtained with the final set of primers used in this study are shown in SI Appendix, Fig. S12A. Cellular expression levels of all 16 vRNAs, in the combined control experiments (No-MixCtrl, Library-MixCtrl, Premix1, and Premix2) (SI Appendix, Fig. S12B), all free of reassortment, globally reflect RT primer efficiency as measured on vRNA transcripts (SI Appendix, Fig. S12A). The dropout segment frequency varied depending on the segment nature and H1N1p/H3N2 background (SI Appendix, Fig. S12 C–E and Supplementary Methods), which may be due to differences in vRNA levels and/or differences in the efficiency of cDNA synthesis. Using data from the 9,438 reassortant genomes containing 2 to 8 segments (2,592 from the eight-segment dataset plus 6,846 from 2- to 7-segment dataset) slightly biased the data, although frequencies of cosegregated pairs of segments remained similar to data from eight-segment genomes (SI Appendix, Fig. S9 A–C compared to SI Appendix, Figs. S9 D–F and S10 and S11 compared to SI Appendix, Figs. S13 and S14). To correct for this bias, the data were normalized using the pairwise frequencies of the eight-segment reassortant genomes to take into account the different dropout frequencies for the different segments (SI Appendix, Supplementary Methods and Figs. S8 and S15). The normalized data from the 9,324 reassortant genomes (2,592 from the eight-segment dataset plus 6,732 from 4- to 7-segment dataset) is shown in Fig. 3 G and H (intra- plus inter-strain or interstrain combinations only) and closely match those obtained analyzing the eight-segment data (Fig. 3 D and E). For pairwise segment frequencies, the best fit by linear regression of the 2- to 8-segment nonnormalized reassortant dataset to data from eight-segment reassortants is y = 0.621x + 1.794 (r = 0.8996, P < 10−40) (Fig. 3F), whereas for the normalized 4- to 8-segment dataset, the best fit is y = 0.862x + 3.445 (r = 0.95728, P < 10−60) (Fig. 3I), indicating that normalization corrects for underestimation of pairwise reassortments and increases the correlation with the eight-segment data. Importantly, although we observed differences in the overall mean frequencies of the H1N1p and H3N2 single segments and of the H1N1p-H1N1p and H3N2-H3N2 pairwise segments between experiments, both in the eight-segment and dropout normalized datasets (SI Appendix, Fig. S12 F and G), the H1N1p-H3N2 mean pairwise frequencies did not appear to be sensitive to the background (SI Appendix, Fig. S12 H and I).
Modeling and Prediction of Reassortment.
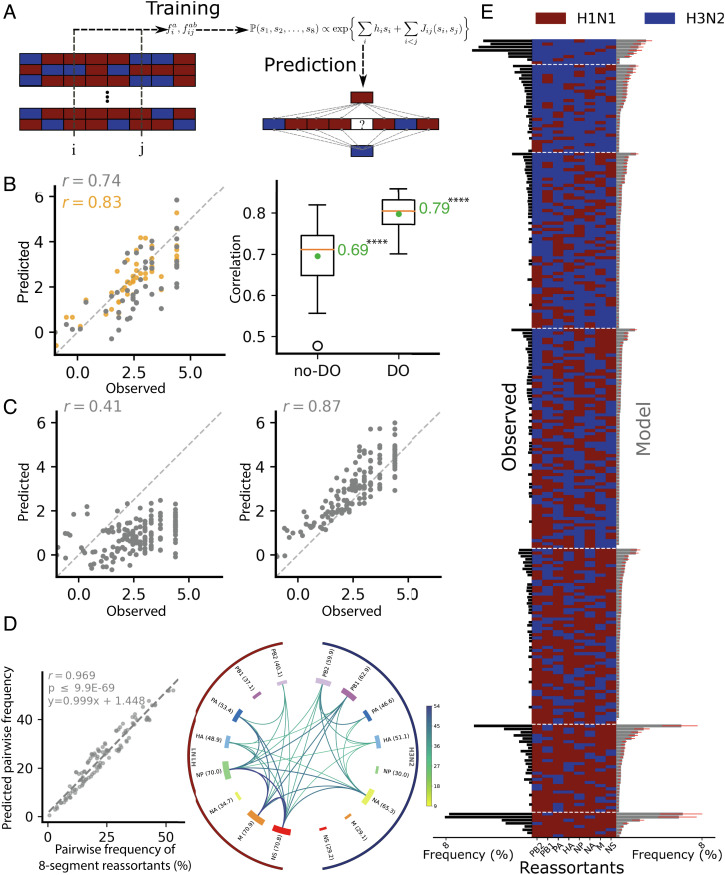
To better understand and predict reassortment statistics, we applied direct coupling analysis (DCA) to our datasets. DCA is a machine learning technique that trains a probabilistic model of high-order distributions from pairwise frequencies (58). It has been shown to predict sequence-to-activity relationships in enzymes (59) and is applicable to any relationships given measured pairwise frequencies. We applied a DCA model to learn parameters associated with single segments and segment pairs by maximizing the likelihood of the observed distribution of all segment combinations (Fig. 4A). This way, DCA can generate eight-segment combination frequencies learning from the observed occurrences of lower-order combinations down to pairs. As we show below, this allows us to exploit incomplete but numerous data from dropout measurements and increases the prediction quality for genotype frequencies, including those not present in the experimental data on which the model is trained.
Fig. 4.
Modeling and prediction of reassortment. (A) The probabilistic DCA model trained using the frequencies of H1N1p/H3N2 at single or pairs of segments. The probability of observing H1N1p, H3N2, or a dropout at segment i depends on what is observed at the other seven segments j. The interactions with the other segments are modeled by the Jij terms. (B) Model generalization using dropouts. Forty-eight random genotypes among the 159 of the eight-segment genotypes observed were reserved as the validation set. The Left panel shows the comparison of two models: a two-state model trained on the 70% remaining genotypes of the eight-segment dataset (111 genotypes, gray dots) and a three-state model trained on 70% of the 2- to 8-segment dataset (1,208 among the 1,726 2- to 8-segment genotypes remaining after the removal of the validation set, orange dots). The predicted (log) genotype frequencies are plotted against the measured (Observed) (log)frequencies of the remaining 30% reassortant genotypes [48] of the eight-segment dataset used as validation set. Pearson's correlation, r, is indicated. The box plots in the Right panel show the distribution of Pearson’s correlation for the model trained on the eight-segment dataset (no-DO) and trained on the 2- to 8-segment dataset (DO), each obtained for 30 randomly drawn training and validation sets. The median values are indicated as a line at the center of the boxes. All correlations obtained have P ≤ 10−3. The average gain (green dot) of predicted power resulting from using the 2- to 8-segment dataset is significant by t test (two-sample and one-tailed) on the two samples (****P < 10−4). (C) Prediction quality of the three-state pairwise model. For the model trained on the complete 2- to 8-segment (i.e., including dropout genomes) dataset (no training/validation split), the interaction terms Jij (Right) improved the Pearson’s correlation, r, for predicted (log)frequencies vs. observed frequencies of the validation set compared to the model without terms Jij (Left). (D) Prediction of pairwise segment frequencies by the three-state DCA model trained on the complete 2- to 8-segment dataset. Left: correlation between pairwise segment frequencies predicted by the DCA model (for eight-segment genomes) and observed frequencies in the eight-segment dataset (r = 0.969, P <10−5). Right: chord plot showing the pairwise segment frequencies predicted by the DCA model. (E) Histogram of observed genotype frequencies from the eight-segment dataset vs. predicted frequencies of reassortant genotypes (N = 254) in the merged 2- to 8-segment dataset. The genotypes are shown as colored lines (red: H1N1p-derived segment, blue: H3N2-derived segment) and are ordered according to the number of H1N1p-derived segments. Observed and predicted frequencies are represented with black bars on the left and gray bars on the right, respectively. Red lines indicate the 25 to 75% confidence intervals obtained from the bootstrap experiment.
To extract the statistical signal from dropouts, we considered a three-state DCA model (each segment can be H1N1p, H3N2, or missing) in which the probability to observe dropout segments is explicitly taken into account. Such a model has 240 free parameters, whereas a two-state model that only considers H1N1p and H3N2 segment probabilities has 90 free parameters. These additional parameters allow the dropout biases to be absorbed without requiring preliminary normalizations. We compared a two-state DCA model trained on the nonnormalized dataset comprising eight-segment reassortants (2,592 droplets) and a three-state DCA model trained on the nonnormalized dataset comprising 2- to 8-segment reassortants (9,438 droplets). We assessed the quality of the models by bootstrapping, where 30% of the genotypes among all observed eight-segment combinations were reserved for later validation (in practice, a random subset of 48 genotypes among the 159 observed ones). The two-state model was first trained on the frequencies measured for the 70% of genotypes distinct from the 48 genotypes of the validation set, while the three-state model was trained on 70% of the 2- to 8-segment dataset after the removal of the validation set.
Validation was then performed by computing the correlation between the genotype frequencies predicted by the model and those measured for the validation set (SI Appendix, Supplementary Methods). We found that the three-state DCA model trained on the dropout data is more predictive, as the correlation between the model and the validation set improved significantly, from Pearson’s correlation of 0.74 (P ≤ 10−3) without dropout genomes to 0.83 (P ≤ 10−3) including dropout genomes (Fig. 4 B, Left). This process was repeated on 30 random training and validation sets for each model, showing on average an increase from 0.69 to 0.79 of the correlation coefficient (Fig. 4 B, Right). Taking into account dropout genomes, genotypes were predicted with better confidence than without dropout genomes for frequencies lower than 2.7 × 10−3 (Student’s t test, P ≤ 10−3, SI Appendix, Fig. S16A). For both two-state and three-state models, the correlation converged when varying the size of the training dataset including dropout genomes, but the correlation was less noisy and higher with the three-state model (SI Appendix, Fig. S16 B and C). The model did not overfit as fitting on 58% of the dataset with 2- to 8-segments (4,532 reassortant genomes) was sufficient to predict the 36% eight-segment reassortants (921 reassortant genomes) left for validation with Pearson’s correlation of > 0.80 and P ≤ 10−5 (SI Appendix, Fig. S16C).
A three-state model trained on the complete 2- to 8-segment dataset that only considers independent single-segment frequencies poorly reproduces reassortant genome frequencies observed in the merged dataset containing 2,592 reassortant genomes with all eight-segments (Fig. 4 C, Left), consistent with the existence of physical interactions between them, as previously reported for certain segments (11, 13, 19, 37). In contrast, taking into account pairwise terms in the DCA model leads to a Pearson’s correlation of 0.87 with P ≤ 10−5 between predicted and measured reassortant genome frequencies, as good as the correlation between experimental replicates (ranging from correlation of 0.72 to 0.94 with P ≤ 10−5 (Fig. 4 C, Right and SI Appendix, Table S6).
The pairwise segment cosegregation frequencies generated by the DCA model trained on 9,438 reassortants with 2- to 8-segments closely resembled the data from eight-segment genomes (Fig. 4D, r = 0.969, P ≤ 10−68, compare to Fig. 3D), measured pairwise segment frequencies in the 2- to 8-segment reassortants (SI Appendix, Fig. S16D, r = 0.997 with P ≤ 10−288, and normalized data from reassortant genomes with ≥4 segments (SI Appendix, Fig. S16E, r = 0.972, P ≤ 10−70). Furthermore, it allowed the prediction of reassortant genotype frequencies, which match quite closely the frequency of experimentally observed eight-segment genotypes (Fig. 4E and SI Appendix, Fig. S17). However, the model also predicts the frequency of genotypes that were not observed in the eight-segment data (Fig. 4E). Indeed, all genotypes are predicted to occur with nonzero frequencies, the lowest being 2.95 × 10−5. The missing reassortant genotypes in the experimental data from the eight-segment genome appear to be an effect of the sample size. To test this, we subsampled 2,592 genomes from the 254 possible reassortant genotypes using the probabilities given by the DCA model: 95% of the reassortant genotypes not observed in the subsamples had frequencies of <0.07% in the model. Accordingly, 95% of the reassortant genotypes not observed in the eight-segment dataset had frequencies of <0.0038% in the model, which is below the threshold obtained by subsampling.
Discussion
We developed the µFlu system to analyze reassortment events occurring between different strains of IAV in a high-throughput manner (Fig. 1). We applied this system to sequence single cells infected with single progeny virions derived from coinfections of cultured cells with two human seasonal IAVs of H1N1p and H3N2 subtypes. Viral particles obtained from “producer” cells coinfected at high m.o.i. with the two IAV strains were used to infect “indicator” cells at low m.o.i. (so that cells are unlikely to be infected by more than one virus). The latter cells were then analyzed by adapting a droplet microfluidic system (46) to use custom cDNA primers specifically targeting vRNAs and to rapidly analyze larger numbers of cells. The beads carried 85 million different BCs (vs. 150,000 (46)) potentially permitting analysis of up to 1.7 million cells (vs. 3,000 (46)) with 99% unique labeling. Use of smaller hydrogel beads and droplets (120 pL vs. 1 to 5 nL) and a dual nozzle device for cell encapsulation at ~240 s−1 (vs. 1 to 10 cells s−1) allowed 0.5 × 105 to 1 × 105 low m.o.i. infected cells (vs. ~1,000 cells (46)) to be coencapsulated with beads in <10 min for each experiment. Finally, data analysis required the development of a robust, tailored bioinformatics pipeline. Single-cell RNA-seq is most often based on poly(A) capture of messenger RNAs (mRNAs). However, IAV vRNAs are not poly-adenylated and although the nature of vRNAs can be inferred from their corresponding mRNAs, traditional poly(A)-RNAseq could, in our case, be disadvantageous in several respects: i) Depending on the strain, viral mRNA reads within an infected cell represent 10 to 50% of the total reads and the concomitant cellular mRNA reads would result in a substantial loss of sequencing depth; ii) recently published data (60), based on strand-specific RT-qPCR amplification of viral mRNAs and vRNAs, reveal more heterogeneity among all eight mRNAs compared to the eight vRNAs (this observation holds true until 12 h post-infection, close to the time-point we chose for the analysis of the “indicator” cells (14 h), with mRNA levels generally lower than vRNA levels); and iii) incomplete viral gene expression observed during infection (61, 62) could be due to defects at the mRNA level. Of note, targeted scRNA-seq using custom-made and commercial droplet microfluidic systems is becoming more common, for example, for targeted antibody and T cell receptor sequencing ((63); 10X Genomics Single Cell Immune Profiling system (64)). Targeted scRNA-seq is also being achieved through transcript enrichment prior to poly(A) capture and sequencing (65–67). Our experimental design demonstrated high technical and biological reproducibility of the procedure (SI Appendix, Table S6) and permitted the analysis of 2,592 complete (with eight segments) reassortant genomes (Dataset S2), vastly exceeding the number of genotypes analyzed by standard methods (≤72) (35, 42, 43). With the microfluidic device being able to encapsulate 0.5 × 105 to 1 × 105 “indicator” cells in < 10 min, there is virtually no limit to the number of reassortant genomes that can be analyzed—it is simply a tradeoff with cost, which is largely driven by sequencing.
Focusing on single cells where all eight segments were identified, our data demonstrate that reassortment of segments between IAV strains is far from random (SI Appendix, Table S7). In this dataset, 159 of 254 possible reassortant genotypes were observed as a result of reassortment between the H1N1p and H3N2 strains (Dataset S1 and Fig. 3B). Their distribution approximates a negative exponential, with very few genotypes present at high frequencies (maximum 8.45%) and a large number at very low frequencies (minimum 0.039%) (Fig. 3B). Based on pairwise combination statistics, all 112 possible pairs of segments were detected, albeit at very different frequencies ranging from 0.5 to 41% (SI Appendix, Fig. S9). Based on control experiments in which reassortment is impossible (SI Appendix, Fig. S5B), the frequency of false reassortants is below 1% (0.93% for No-MixCtrl, 0.14% for Lib-MixCtrl, and 0.68% for Premix 1 + Premix2). The most frequent false reassortant genotype identified (an eight-segment genotype identified four times among the 9,546 eight-segment containing BCs of Premix2) represents 0.042% of total BCs (cells) identified (Dataset S2 and SI Appendix, Table S4), which constitutes a practical lower limit for the detection of rare genotypes. Hence, only the genotypes resulting from coinfection that were detected once or twice (41 genotypes, 0.020 and 0.040%, respectively, of all 4,952 eight-segment genotypes) fall below this limit. Inclusion of an additional 6,846 incomplete reassortant genomes (“dropout genomes”) that, due to imperfect vRNA capture, contained missing (and nonidentified) segments (“dropout segments”), increased the power to determine cosegregation frequencies (SI Appendix, Fig. S10 compared to SI Appendix, Fig. S13) but also introduced a minor bias in the data. Normalized frequencies of pairwise combinations of segments from the 9,324 reassortant genomes containing 4 to 8 segments (Fig. 3 G and H) closely matched those found in the 2,592 reassortant genomes of the eight-segment dataset (Fig. 3 D and E), as exemplified by the high Pearson correlation coefficient (r = 0.95728, P < 4.24 × 10−60 (Fig. 3I), justifying the use of dropout genomes. Of note, µFlu only detects viruses that are able to efficiently infect the “indicator” cells, efficiently replicate their vRNAs inside the newly infected cells, and efficiently express, at the cell-surface, the HA and M2 proteins used to sort the indicator cells by FACS. Such viruses must possess an active polymerase complex, i.e., PB2, PB1, PA, and NP proteins encoded by full-length PB2, PB1, PA, and NP segments and able to cooperate for transcription and replication. Among the 6,846 dropout genomes, 4,313 (63%) are missing at least one of the PB2, PB1, PA, or NP genes. We therefore assume that most dropout genomes arise from technical issues rather than from a large proportion of incomplete virions (61). This assumption is supported by scanning transmission electron microscopy tomography analysis of viral stocks, which revealed that at most 20% of virions among 50 virions of laboratory strains and 10% of virions of clinically isolated strains package less than eight viral RiboNucleoParticles (vRNPs), with no virions containing less than five vRNPs (65). Incomplete patterns of viral protein expression in infected cells, the observation of which led to the concept that a large proportion of virions might carry less than eight vRNA segments (61), may result from defects in, e.g., uncoating and nuclear transport of vRNPs, vRNA transcription, and/or viral protein synthesis, instead of the absence of one or several vRNAs in the viral particle. Finally, predictions from a DCA model, trained on data from reassortant genomes with ≥2 segments, showed that pairwise segment cosegregation frequencies could be reproduced, and full reassortant genotype frequencies could be inferred (Fig. 4). As, based on control experiments in which reassortment is impossible (SI Appendix, Fig. S5B), the frequency of false reassortants in the training set is below 1%, predicted frequencies are little influenced by false reassortants and it is expected that the DCA model should be able to reliably predict the frequency of even very rare reassortant genomes. Indeed, the DCA model suggested that analysis of ~1 × 105 genomes (SI Appendix, Supplementary Methods) would be needed to detect the rarest reassortant genotypes (P = 0.95).
Mechanistically, reassortment between genetically distant viruses is thought to be limited by incompatibilities between viral proteins and incompatibilities at the genomic level between packaging signals on vRNAs (38, 40). Packaging signals, close to the extremities of the vRNAs, have mainly been investigated for the lab-adapted A/WSN/33 and A/Puerto Rico/8/34 (PR8) strains (16, 27). In the case of the H1N1p strain, the importance of the 3′ and 5′ extremities of the HA and NA segments for packaging has been established (66). Concerning H3N2 strains, an in vitro-established vRNA–vRNA network, mostly involving terminal packaging signals and bringing together all eight vRNAs of the seasonal A/Moscow/10/99 (H3N2) strain, was identified (9). In addition, copackaging of the NA and PB1 segments of H3N2 strains (seasonal and A/Udorn/72) with six heterologous segments from the egg-adapted H1N1 virus PR8 was observed during vaccine seed production (36). Cosegregation between H3N2-NA and -PB1 in an engineered recombinant virus was found to be driven by an RNA–RNA interaction that was mapped at the nucleotide level on both segments (13). The latter study also confirmed the existence of numerous, potentially functionally redundant intersegmental vRNA–vRNA interactions which differ from one strain to another, making the prediction of RNA–RNA interactions that drive genome packaging and in turn genetic reassortment, a challenge. Our study reveals that 159 out of 254 of all possible genotypes were observed upon coinfection, and DCA modeling further predicts that all possible genotypes can occur, even if some will occur only at very low frequencies. The frequently observed incorporations of H3N2-NA, H3N2-PB1, or H3N2-(NA+PB1) into the H1N1p-background are consistent with the above-mentioned data (13, 36). Entry of H3N2-NA, H3N2-PB1, and H3N2-(NA+PB1) into an H1N1p-background was also observed after single- and multicycle replication on MDCK cells (42), but numbers were low and replicates only poorly correlated. Notably, most segments of the circulating seasonal H3N2 strains are inherited from an H1N1 (“PR8-like”) ancestor except for the NA segment that was acquired by reassortment in 1957 and the HA and PB1 segments that were acquired by reassortment in 1968. Thus, from an evolutionary point-of-view, the H3N2-HA, -NA and -PB1 segments already have a history of successful introduction into a “PR8-like” background. After several decades of circulation/evolution in the human population, they are still prone to introduction into a PR8 variant background (egg-adapted PR8 background) (36) but also into a quite genetically distinct H1N1p background (our experiments and ref. 42), possibly suggesting some intrinsic capacity to be incorporated in a heterologous background. Accordingly, the PB1 catalytic core subunit of the influenza polymerase sustains efficient activity in a heterologous background, as measured in a minireplicon assay (42).
In the human population, H3N2 viruses have cocirculated with H1N1 viruses from 1977 to 2009 and with H1N1p viruses since 2009. Although reassortment readily occurs between H3N2, H1N1, and H1N1p subtypes (67–69), only a few cases of H3N2-gene introduction, i.e., incorporation of the H3N2-NA segment into the H1N1p-background, have been reported (70–72), whilst reassortants harboring the H3N2-(PB1+NA) gene combination in an H1N1p background have not been detected in natural infections. In the H3N2-background, individual incorporations of H1N1p-segments, namely M, NS, NP, and PB1, are the most frequent in our experimental setting, in agreement with data from Phipps et al. (42). In nature, reassortant strains harboring the H1N1-HA in an H3N2 background emerged in 1989 in China (73) and worldwide in 2000 (74–76). After 2009, two cases of introduction of H1N1p-HA and H1N1p-(HA+NS) into the H3N2-bakground were reported, in India in 2009 (77) and in the Netherlands in 2018 (78), respectively. Coinfections in humans have been reported, with frequencies varying from 1 to 3% (79–85), with one exception of mixed infections reaching 15% (86). Coinfection frequencies are in the same range in the swine population (87), while frequencies of ~25% have been reported among poultry or wild bird populations (88, 89). While the minimum infectious dose of an H2N2 virus administered by aerosol to seronegative subjects was estimated to be around 3 TCID50 (90), the estimated range of in vivo burst ranges from 10 to 300 TCID50/producer cell, presumably resulting in a high amount of virus in the vicinity of neighboring cells and consequently, a high m.o.i. (91). Viral titers measured in nasopharyngeal washes at the peak of infection can reach >106 TCID50/mL (92). Other factors such as the thick layer of mucus in the respiratory tract that facilitates aggregation of viral particles and collective transfer might also promote high m.o.i. infection (93). Hence, experimental systems such as ours, with cultured cells coinfected at high m.o.i., are, at least in part, biologically relevant. Interestingly, even though IAVs do not necessarily rely on coinfection for productive viral replication, a recent study shows that infection at high m.o.i. is advantageous for an avian IAV to overcome the host barrier and efficiently replicate in mammalian hosts in the absence of genetic adaptation and leads to high levels of reassortment, which in turn can also facilitate adaptation to a new host (34). Limited or no detection in nature of reassortants that were detected in our experiments could be due to insufficient or biased sampling of natural reassortants or due to different selection pressures in MDCK cells vs. the human airway.
Cosegregation of vRNAs due to optimal compatibilities between encoded viral proteins is expected to be most prominent within the polymerase complex, encoded by PB1, PB2, and PA, together with the nucleoprotein (NP), which are collectively required for vRNA replication, and between the HA and NA proteins, responsible for a regulated balance between cellular attachment and release, respectively. In our eight-segment dataset, we find that 19.29% of all reassortant genomes (8.8% of all reassortant genotypes) carry the homologous H3N2-(PB1-PB2-PA-NP) gene combination, while 14.35% of all reassortants (7.5% of all reassortant genotypes) harbor the equivalent H1N1p combination, suggesting some compatibilities between nonhomologous polymerase subunits. Interestingly, 86% of all reassortant genomes (66.7% of all reassortant genotypes) carry a homologous (PB2-PA) combination, indicating strong coevolution between these two segments for optimal function. Overall, these results are consistent with previously published data regarding the effectiveness of polymerase activity of PB2-PA, PB2-NP, and NP-PA pairs in a minigenome assay (42). In a novel phylogenetic approach, assessing parallel evolution between vRNA segments of H3N2 and H1N1 (pre- and post-pandemic), that proposes to untangle coevolving viral protein–protein relationships from vRNA–vRNA relationships (94), the authors highlight an anticipated parallel evolution of the H3N2-PB1 and -PA segments. The H3N2-PB1 and -NA relationship that we and others (13, 36, 37) observed is also made apparent using this tool. Surprisingly, a strong relationship between PB2 and NA, which are not expected to cooperate in the viral replication cycle, was detected and supported by colocalization of the PB2 and NA vRNAs, inferring putative vRNA-vRNA interaction (94). Accordingly, we also observed significant cosegregation of the H3N2-PB2 and -NA segments in an otherwise H1N1p-background. The highest degree of parallel evolution at the protein level, coupled with the lowest parallel evolution at the vRNA level, was found between the H3N2-NA and -HA proteins (94). In our eight-segment dataset, the H3N2-(HA+NA) combination represents 71.9% of all reassortant genomes in an H3N2 background whilst the H1N1p-(HA+NA) represents 34.2% of the reassortant genomes in the H1N1p background. In the case of the H1N1p strain, the strongest RNA–RNA coevolutions (94) did not correspond to the most frequently cosegregated genes that we observed in a heterologous H3N2-background.
To conclude, we used state-of-the-art droplet microfluidics techniques to achieve high-throughput sequencing of individual reassortant influenza viral genomes at an unprecedented scale, enabling systematic and exhaustive monitoring of the reassortment potential of IAVs. Higher performance might be achieved in the future by reducing the frequency of droplets containing more than one infected cell (“doublets”), for example, by reducing formation of cell aggregates after FACS using “indicator” cells that are less prone to aggregation, by addition of nonionic surfactants that reduce cellular adherence, and/or reducing changes in cell density due to sedimentation during injection by better density matching between cells and medium. The µFlu system opens up a world of possibilities to explore questions related to the reassortment between IAV strains. Different combinations of strains can be examined, including those with pandemic potential to aid vaccine development. It will also allow scrutinization of reassortment between zoonotic animal viruses and human seasonal viruses, to identify those that represent a threat to human health providing an additional predictive parameter to complement existing Pandemic Influenza Risk Assessment Tools. In parallel, the choice of host cell (e.g., different species, different genetic backgrounds) could be modified to study how host background influences patterns of reassortment. Finally, additional passaging steps can be added to measure fitness levels of reassortant viral progeny: viruses that meet the sorting threshold during FACS in the µFlu procedure may vary in their capacity to replicate and produce infectious progeny virions. As a direct extension of this work and any future reassortment analysis, characterization and comparison of the replication capacity of frequently vs. rarely observed reassortants will be of interest.
To date, the database of genomic sequences compiles over 300,000 influenza virus strains. In recent years, efforts have been made to develop automated methods to identify reassortant strains (95–99), allowing the documentation of previously unrecognized reassortment events. Experimental data generated with the µFlu system combined with predictive methods such as DCA and with data mining using new algorithms (95–99) should enable a better understanding of the rules underlying IAV reassortment and prediction of the outcome of genetic reassortment. Finally, our work demonstrates that droplet microfluidics techniques originally developed for single-cell RNA-seq can be adapted to become a powerful tool to study viral evolution, with many applications beyond the field of influenza viruses
Methods
Cells and Viruses.
Madin-Darby Canine Kidney (MDCK) SIAT cells, kindly provided by M. Matrosovich (Philipps Universität, Marburg, Germany) (53), were propagated at 37 °C with 5% (v/v) CO2 in Modified Eagle’s medium (MEM, Gibco) supplemented with 5% (v/v) heat-inactivated fetal bovine serum and 1% penicillin-streptomycin. The human seasonal viruses H1N1 A/Bretagne/7608/2009 (H1N1p) and A/Centre/1003/2012(H3N2) were isolated by the Centre National de Référence at Institut Pasteur and kindly provided to us. Viruses were amplified on MDCK cells (m.o.i. 0.001 pfu/cell) and titrated on MDCK-SIAT cells as described in Matrosovich et al. (53).
Production of Reassortant Viruses.
Reassortant viral stocks were prepared by coinfecting “producer” MDCK-SIAT cells. The percentage of infected and coinfected cells were evaluated by flow cytometry. The supernatants were titrated by plaque assay on MDCK-SIAT cells. For details, see SI Appendix, Supplementary Methods.
Low m.o.i. Infection.
MDCK SIAT “indicator” cells were infected at a m.o.i. of 0.01 pfu/cell, as detailed in the SI Appendix, Supplementary Methods.
Cell Fixation, Antibody Staining, and Cell Sorting.
Cells were fixed, labeled with anti H1-HA and anti M2 antibodies and sorted using a FACSArial™ cell sorter (BD biosciences). For details, see SI Appendix, Supplementary Methods.
Microfluidic Device Fabrication and Microfluidics Operations.
Microfluidic devices (SI Appendix, Fig. S18) were microfabricated in poly(dimethyl siloxane) (PDMS) using soft lithography (100) as described (101). Fluids were driven using syringe pumps (Nemesys low pressure, CETONI). The continuous phase for all droplet microfluidics experiments was Novec HFE-7500 fluorinated oil (3M) containing 2% or 3% w/w 008-FluoroSurfactant (RAN Biotechnologies). For details, see SI Appendix, Supplementary Methods.
Preparation of Hydrogel Beads and Addition of Barcoded Primers to Hydrogel Beads by Split-and-Pool Ligation.
Polyacrylamide hydrogel beads were produced essentially as in ref. 54. For details, see SI Appendix, Supplementary Methods. Barcoded primers were constructed on the hydrogel beads by split-and-pool ligation essentially as in ref. 55. For details and quality control, see SI Appendix, Supplementary Methods.
Encapsulation.
Single cells were encapsulated with single hydrogel beads carrying barcoded primers and RT/cell lysis reagents essentially as in ref. 54. A single encapsulator device (SI Appendix, Fig. S18B) was used for the Premix control experiment and a double encapsulator device (SI Appendix, Fig. S18C) was used for all other experiments. The flow rate for beads, enzyme mix, cells, and oil were set as 3, 6, 6, and 35 µL/min, respectively, and resulted in an average, λ, of 0.1 cells per droplet. Droplets of ~120 pL volume were generated at a rate of 2,400 Hz. The use of this two-nozzle device allowed the encapsulation at a rate of ~ 4.3 × 105 to 8.6 × 105 cells/h, and ~ 50,000 to 100,000 cells were encapsulated in <10 min for each reassortment experiment.
RT and Library Preparation.
RT in droplets or in bulk, emulsion breaking, cDNA purification, second-strand synthesis, in vitro transcription, secondary RT reaction, and library preparation were performed essentially as described (54). For details, see SI Appendix, Supplementary Methods.
Processing of scRNA-seq Data.
Sequencing reads were mapped to the combined H1N1p-H3N2 reference genomes. For each cell-containing droplet, expression of each viral segment were determined using a statistical framework. Potential doublets (i.e., cells containing evidence for expression of one or more segments from both viral strains) and low-quality cells were excluded from downstream analyses. For details, see SI Appendix, Supplementary Methods. Scripts can be found at http://github.com/ONeillMB1/uFlu.
Dropout Normalization.
The data on pairwise segment cosegregation frequencies from single-cell genomes, termed "dropout genomes," that contained one or more missing (and nonidentified) segments, termed "dropout segments," were normalized using the pairwise frequencies of the eight-segment reassortant genomes of individual experimental replicates (R1.1, R1.2, R1.3, R2.1, R2.2, and R2.3) For details, see SI Appendix, Supplementary Methods.
Data Modeling.
DCA was used to model the probability of genotypes P(s1,s2,...,s8) where si is the segment identity in {H1N1, H3N2} at position i in {PB1...NA}. The probabilistic model is parameterized by two types of terms: the hi(si) that controls the propensity of segment type si at i and Jij(si, sj) that controls the propensity of the pair of segments. For details, see SI Appendix, Supplementary Methods. Scripts can be found at: http://github.com/ONeillMB1/uFlu.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (TXT)
Dataset S02 (XLSX)
Operation of the double encapsulator microfluidic chip.
Co-encapsulation of single cells, single hydrogel beads, lysis buffer and reverse transcription reagents in droplets.
Acknowledgments
We are grateful to Drs. R. Marquet and P. Dumas (Institut de Biologie Moléculaire et Cellulaire, Strasbourg), Dr. G. Simon (ANSES, Ploufragan, France) for insightful discussions. We thank the Genotyping and sequencing core facility at the Institut du Cerveau-ICM (Paris, France) and the Biomics Platform at the Institut Pasteur (Paris, France) for Next Generation Sequencing and S. Behillil and V. Enouf (National Reference Center for Respiratory Viruses, Institut Pasteur, Paris, France) for providing the virus samples used in this study. This work is supported by grants from the Agence Nationale de la Recherche (ANR 18 CE18 0026 01 FLU_REASSORT; ANR-10-LABX-62-IBEID; ANR-10-IDEX-0001-02 PSL; ANR-10-LABX-31 Institut Pierre-Gilles de Gennes). We also thank the Hospices de Nuits-Saint-Georges for their financial support.
Author contributions
N.N., A.G., and C.I. designed research; K.-Y.C., J.K., M.B.O., V.O., M.B., S.F., P.I., and C.I. performed research; L.Q.-M., T.O., and S.v.d.W. contributed new reagents/analytic tools; K.-Y.C., J.K., M.B.O., V.O., M.B., S.F., P.N., N.N., A.G., and C.I. analyzed data; L.Q.-M. and S.v.d.W. provided expertise and feedback and A.G. and C.I. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Nadia Naffakh, Email: Nadia.naffakh@pasteur.fr.
Andrew Griffiths, Email: Andrew.griffiths@espci.fr.
Catherine Isel, Email: Catherine.isel-griffiths@pasteur.fr.
Data, Materials, and Software Availability
Next generation sequencing data ara available at https://www.ncbi.nlm.nih.gov/bioproject/923733. All other study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Wright P. F., Neumann G., Kawaoka Y., “Orthomyxoviruses 6th Edition” in Fields Virology, Knipe D. M., et al., Eds. (Lippincott Williams & Wilkins, Philadelphia, PA, USA, ed. 6, 2013), vol. 1, pp. 1186–1243. [Google Scholar]
- 2.Ma W., Kahn R. E., Richt J. A., The pig as a mixing vessel for influenza viruses: Human and veterinary implications. J. Mol. Genet. Med. 3, 158–166 (2008). [PMC free article] [PubMed] [Google Scholar]
- 3.Kahn R. E., Ma W., Richt J. A., Swine and influenza: A challenge to one health research. Curr. Top. Microbiol. Immunol. 385, 205–218 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kilbourne E. D., Influenza pandemics of the 20th century. Emerg. Infect. Dis. 12, 9–14 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith G. J. D., et al. , Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122–1125 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Garten R. J., et al. , Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325, 197–201 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu A., et al. , Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe 14, 446–452 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Sun H., et al. , Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc. Natl. Acad. Sci. U.S.A. 117, 17204–17210 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fournier E., et al. , A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 40, 2197–2209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gavazzi C., et al. , An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: Comparison with a human H3N2 virus. Nucleic Acids Res. 41, 1241–1254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavazzi C., et al. , A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc. Natl. Acad. Sci. U.S.A. 110, 16604–16609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber M., Isel C., Moules V., Marquet R., Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol. 22, 446–455 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Dadonaite B., et al. , The structure of the influenza A virus genome. Nat. Microbiol. 4, 1781–1789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamoto S., Noda T., In vitro vRNA-vRNA interactions in the H1N1 influenza A virus genome. Microbiol. Immunol. 64, 202–209 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Le Sage V., et al. , Mapping of influenza virus RNA-RNA interactions reveals a flexible network. Cell Rep. 31, 107823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Gu M., Zheng Q., Gao R., Liu X., Packaging signal of influenza A virus. Virol. J. 18, 36 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seshimo E., Momose F., Morikawa Y., Identification of the 5’-terminal packaging signal of the H1N1 influenza a virus neuraminidase segment at single-nucleotide resolution. Front. Microbiol. 12, 709010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noda T., Selective genome packaging mechanisms of influenza A viruses. Cold Spring Harb. Perspect. Med. 11, a038497 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyamoto S., et al. , Contribution of RNA-RNA interactions mediated by the genome packaging signals for the selective genome packaging of influenza A virus. J. Virol. 96, e0164121 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noda T., et al. , Importance of the 1 + 7 configuration of ribonucleoprotein complexes for influenza A virus genome packaging. Nat. Commun. 9, 54 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatada E., Hasegawa M., Mukaigawa J., Shimizu K., Fukuda R., Control of influenza virus gene expression: Quantitative analysis of each viral RNA species in infected cells. J. Biochem. 105, 537–546 (1989). [DOI] [PubMed] [Google Scholar]
- 22.McGeoch D., Fellner P., Newton C., Influenza virus genome consists of eight distinct RNA species. Proc. Natl. Acad. Sci. U.S.A. 73, 3045–3049 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergmann M., Muster T., The relative amount of an influenza A virus segment present in the viral particle is not affected by a reduction in replication of that segment. J. General Virol. 76, 3211–3215 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Chou Y., et al. , One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc. Natl. Acad. Sci. U.S.A. 109, 9101–9106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inagaki A., Goto H., Kakugawa S., Ozawa M., Kawaoka Y., Competitive incorporation of homologous gene segments of influenza A virus into virions. J. Virol. 86, 10200–10202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giese S., Bolte H., Schwemmle M., The feat of packaging eight unique genome segments. Viruses 8, E165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakob C., Paul-Stansilaus R., Schwemmle M., Marquet R., Bolte H., The influenza A virus genome packaging network - complex, flexible and yet unsolved. Nucleic Acids Res. 50, 9023–9038 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noda T., et al. , Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat. Commun. 3, 639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreira É. A., et al. , A conserved influenza A virus nucleoprotein code controls specific viral genome packaging. Nat. Commun. 7, 12861 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall N., Priyamvada L., Ende Z., Steel J., Lowen A. C., Influenza virus reassortment occurs with high frequency in the absence of segment mismatch. PLoS Pathog. 9, e1003421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao H., Steel J., Lowen A. C., Intrahost dynamics of influenza virus reassortment. J. Virol. 88, 7485–7492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao H., Li L., White M. C., Steel J., Lowen A. C., Influenza A virus coinfection through transmission can support high levels of reassortment. J. Virol. 89, 8453–8461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richard M., Herfst S., Tao H., Jacobs N. T., Lowen A. C., Influenza A virus reassortment is limited by anatomical compartmentalization following coinfection via distinct routes. J. Virol. 92, e02063–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phipps K. L., et al. , Collective interactions augment influenza A virus replication in a host-dependent manner. Nat. Microbiol. 5, 1158–1169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganti K., et al. , Influenza A virus reassortment in mammals gives rise to genetically distinct within-host sub-populations. Nat. Commun. 13, 6846 (2022), 10.1101/2022.02.08.479600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cobbin J. C. A., et al. , Influenza Virus PB1 and Neuraminidase Gene Segments Can Cosegregate during Vaccine Reassortment Driven by Interactions in the PB1 Coding Region. J. Virol. 88, 8971–8980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbertson B., et al. , Influenza NA and PB1 Gene Segments Interact during the Formation of Viral Progeny: Localization of the Binding Region within the PB1 Gene. Viruses 8, E238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowen A. C., Constraints, drivers, and implications of influenza A virus reassortment. Annu. Rev. Virol. 4, 105–121 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Steel J., Lowen A. C., Influenza A virus reassortment. Curr. Top. Microbiol. Immunol. 385, 377–401 (2014). [DOI] [PubMed] [Google Scholar]
- 40.White M. C., Lowen A. C., Implications of segment mismatch for influenza A virus evolution. J. Gen. Virol. 99, 3–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isel C., Munier S., Naffakh N., Experimental approaches to study genome packaging of influenza A viruses. Viruses 8, E218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phipps K. L., et al. , Pandemic H1N1 influenza A viruses reassort efficiently but produce attenuated progeny. J. Virol. 91, e00830–17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trifkovic S., et al. , Gene segment interactions can drive the emergence of dominant yet suboptimal gene constellations during influenza virus reassortment. Front. Microbiol. 12, 683152 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding Y., Howes P. D., deMello A. J., Recent advances in droplet microfluidics. Anal. Chem. 92, 132–149 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Clausell-Tormos J., et al. , Droplet-based microfluidic platforms for the encapsulation and screening of Mammalian cells and multicellular organisms. Chem. Biol. 15, 427–437 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Klein A. M., et al. , Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macosko E. Z., et al. , Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell A. B., Trapnell C., Bloom J. D., Extreme heterogeneity of influenza virus infection in single cells. eLife 7, e32303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C., Cell-to-cell variation in defective virus expression and effects on host responses during influenza virus infection. mBio 11, e02880–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun J., et al. , Single cell heterogeneity in influenza A virus gene expression shapes the innate antiviral response to infection. PLoS Pathog. 16, e1008671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Neill M. B., et al. , Single-cell and bulk RNA-sequencing reveal differences in monocyte susceptibility to influenza a virus infection between Africans and Europeans. Front. Immunol. 12, 768189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loveday E. K., Sanchez H. S., Thomas M. M., Chang C. B., Single cell infection with influenza A virus using drop-based microfluidics. Microbiol. Spectr. 10, e0099322 (2021), 10.1128/spectrum.00993-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matrosovich M., Matrosovich T., Carr J., Roberts N. A., Klenk H.-D., Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J. Virol. 77, 8418–8425 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zilionis R., et al. , Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 12, 44–73 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Grosselin K., et al. , High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 51, 1060–1066 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Barnard G. A., A new test for 2 × 2 tables. Nature 156, 177–177 (1945). [Google Scholar]
- 57.Pearson K., Galton VII F., Note on regression and inheritance in the case of two parents. Proc. R. Soc. Lond. 58, 240–242 (1895). [Google Scholar]
- 58.Morcos F., et al. , Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U.S.A. 108, E1293–E1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russ W. P., et al. , An evolution-based model for designing chorismate mutase enzymes. Science 369, 440–445 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Phan T., et al. , Segment-specific kinetics of mRNA, cRNA and vRNA accumulation during influenza infection. J. Virol. 95, e02102–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brooke C. B., et al. , Most influenza a virions fail to express at least one essential viral protein. J. Virol. 87, 3155–3162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobs N. T., et al. , Incomplete influenza A virus genomes occur frequently but are readily complemented during localized viral spread. Nat. Commun. 10, 3526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gérard A., et al. , High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics. Nat. Biotechnol. 38, 715–721 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Goldstein L. D., et al. , Massively parallel single-cell B - cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun. Biol. 2, 304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakatsu S., et al. , Complete and incomplete genome packaging of influenza A and B viruses. mBio 7, e01248–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawahara M., Wada T., Momose F., Nobusawa E., Morikawa Y., Cell-based influenza A/H1N1pdm09 vaccine viruses containing chimeric hemagglutinin with improved membrane fusion ability. Vaccines 8, 458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson M. I., et al. , Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 4, e1000012 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nelson M. I., et al. , Molecular epidemiology of A/H3N2 and A/H1N1 influenza virus during a single epidemic season in the United States. PLoS Pathog. 4, e1000133 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westgeest K. B., et al. , Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011. J. Virol. 88, 2844–2857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kolosova N. P., et al. , Severe cases of seasonal influenza and detection of seasonal A(H1N2) in Russia in 2018–2019. Arch. Virol. 165, 2045–2051 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Wiman Å., et al. , Novel influenza A(H1N2) seasonal reassortant identified in a patient sample, Sweden, January 2019. Euro. Surveill. 24, 1900124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trebbien R., et al. , A case of reassortant seasonal influenza A(H1N2) virus, Denmark, April 2019. Euro Surveill. 24, 1900406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo Y. J., Xu X. Y., Cox N. J., Human influenza A (H1N2) viruses isolated from China. J. Gen. Virol. 73, 383–387 (1992). [DOI] [PubMed] [Google Scholar]
- 74.Gregory V., et al. , Emergence of influenza A H1N2 reassortant viruses in the human population during 2001. Virology 300, 1–7 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Chen M.-J., et al. , Genetic and phylogenetic analysis of multi-continent human influenza A(H1N2) reassortant viruses isolated in 2001 through 2003. Virus Res. 122, 200–205 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Komadina N., McVernon J., Hall R., Leder K., A historical perspective of influenza A(H1N2) virus. Emerg. Infect. Dis. 20, 6–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mukherjee T. R., Agrawal A. S., Chakrabarti S., Chawla-Sarkar M., Full genomic analysis of an influenza A (H1N2) virus identified during 2009 pandemic in Eastern India: Evidence of reassortment event between co-circulating A(H1N1)pdm09 and A/Brisbane/10/2007 - like H3N2 strains. Virol. J. 9, 233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meijer A., et al. , Case of seasonal reassortant A(H1N2) influenza virus infection, the Netherlands, March 2018. Euro. Surveil. L. 23, 18-00160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kendal A. P., et al. , Laboratory-based surveillance of influenza virus in the United States during the winter of 1977–1978. I. Periods of prevalence of H1N1 and H3N2 influenza A strains, their relative rates of isolation in different age groups, and detection of antigenic variants. Am. J. Epidemiol. 110, 449–461 (1979). [DOI] [PubMed] [Google Scholar]
- 80.Yamane N., Arikawa J., Odagiri T., Sukeno N., Ishida N., Isolation of three different influenza A viruses from an individual after probable double infection with H3N2 and H1N1 viruses. Jpn. J. Med. Sci. Biol. 31, 431–434 (1978). [DOI] [PubMed] [Google Scholar]
- 81.Falchi A., et al. , Dual infections by influenza A/H3N2 and B viruses and by influenza A/H3N2 and A/H1N1 viruses during winter 2007, Corsica Island, France. J. Clin. Virol. 41, 148–151 (2008). [DOI] [PubMed] [Google Scholar]
- 82.Lee N., Chan P. K. S., Lam W., Szeto C. C., Hui D. S. C., Co-infection with pandemic H1N1 and seasonal H3N2 influenza viruses. Ann. Intern. Med. 152, 618–619 (2010). [DOI] [PubMed] [Google Scholar]
- 83.Myers C. A., et al. , Dual infection of novel influenza viruses A/H1N1 and A/H3N2 in a cluster of Cambodian patients. Am. J. Trop. Med. Hyg. 85, 961–963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peacey M., et al. , Pandemic (H1N1) 2009 and seasonal influenza A (H1N1) co-infection, New Zealand, 2009. Emerg. Infect. Dis. 16, 1618–1620 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poon L. L. M., et al. , Quantifying influenza virus diversity and transmission in humans. Nat. Genet. 48, 195–200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu W., et al. , Mixed infections of pandemic H1N1 and seasonal H3N2 viruses in 1 outbreak. Clin. Infect. Dis. 50, 1359–1365 (2010). [DOI] [PubMed] [Google Scholar]
- 87.Hervé S., et al. , Virological and epidemiological patterns of swine influenza A virus infections in France: Cumulative data from the RESAVIP surveillance network, 2011–2018. Vet. Microbiol. 239, 108477 (2019). [DOI] [PubMed] [Google Scholar]
- 88.Peng Y., et al. , Epidemiological surveillance of low pathogenic avian influenza virus (LPAIV) from poultry in Guangxi Province, Southern China. PLoS One 8, e77132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dugan V. G., et al. , The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 4, e1000076 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alford R. H., Kasel J. A., Gerone P. J., Knight V., Human influenza resulting from aerosol inhalation. Exp. Biol. Med. 122, 800–804 (1966). [DOI] [PubMed] [Google Scholar]
- 91.Brooke C. B., Biological activities of “noninfectious” influenza A virus particles. Future Virol. 9, 41–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baccam P., Beauchemin C., Macken C. A., Hayden F. G., Perelson A. S., Kinetics of influenza A virus infection in humans. J. Virol. 80, 7590–7599 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sanjuán R., Collective properties of viral infectivity. Curr. Opin. Virol. 33, 1–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jones J. E., et al. , Parallel evolution between genomic segments of seasonal human influenza viruses reveals RNA-RNA relationships. eLife 10, e66525 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yurovsky A., Moret B. M. E., FluReF, an automated flu virus reassortment finder based on phylogenetic trees. BMC Genomics 12, S3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Silva U. C., Tanaka H., Nakamura S., Goto N., Yasunaga T., A comprehensive analysis of reassortment in influenza A virus. Biol. Open 1, 385–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ding X., Yuan X., Mao L., Wu A., Jiang T., FluReassort: A database for the study of genomic reassortments among influenza viruses. Brief Bioinform. 21, 2126–2132 (2020). [DOI] [PubMed] [Google Scholar]
- 98.Müller N. F., Stolz U., Dudas G., Stadler T., Vaughan T. G., Bayesian inference of reassortment networks reveals fitness benefits of reassortment in human influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 117, 17104–17111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gong X., et al. , Reassortment network of influenza A virus. Front. Microbiol. 12, 793500 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duffy D. C., McDonald J. C., Schueller O. J. A., Whitesides G. M., Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984 (1998). [DOI] [PubMed] [Google Scholar]
- 101.Mazutis L., et al. , Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 8, 870–891 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (TXT)
Dataset S02 (XLSX)
Operation of the double encapsulator microfluidic chip.
Co-encapsulation of single cells, single hydrogel beads, lysis buffer and reverse transcription reagents in droplets.
Data Availability Statement
Next generation sequencing data ara available at https://www.ncbi.nlm.nih.gov/bioproject/923733. All other study data are included in the article and/or SI Appendix.