Abstract
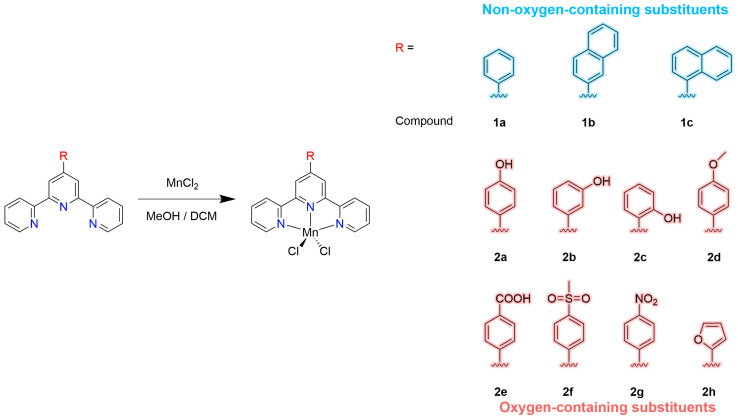
Eleven manganese 4′-substituted-2,2′:6′,2″-terpyridine complexes (1a–1c and 2a–2h) with three non-oxygen-containing substituents (L1a–L1c: phenyl, naphthalen-2-yl and naphthalen-1-yl, L1a–L1c) and eight oxygen-containing substituents (L2a–L2h: 4-hydroxyl-phenyl, 3-hydroxyl-phenyl, 2-hydroxyl-phenyl, 4-methoxyl-phenyl, 4-carboxyl-phenyl, 4-(methylsulfonyl)phenyl, 4-nitrophenyl and furan-2-yl) were prepared and characterized by IR, elemental analysis or single crystal X-ray diffraction. In vitro data demonstrate that all of these show higher antiproliferative activities than cisplatin against five human carcinoma cell lines: A549, Bel-7402, Eca-109, HeLa and MCF-7. Compound 2d presents the strongest antiproliferative effect against A549 and HeLa cells, with IC50 values being 0.281 μM and 0.356 μM, respectively. The lowest IC50 values against Bel-7402 (0.523 μM) Eca-109 (0.514 μM) and MCF-7 (0.356 μM) were obtained for compounds 2h, 2g and 2c, respectively. Compound 2g with a nitro group showed the best results on the whole, with relevantly low IC50 values against all the tested tumor cells. The DNA interactions with these compounds were studied by circular dichroism spectroscopic and molecular modeling methods. Spectrophotometric results revealed that the compounds have strong affinities in binding with DNA as intercalators, and the binding induces DNA conformational transition. Molecular docking studies indicate that the binding is contributed by the π–π stacking and hydrogen bonds. The anticancer activities of the compounds are correlated with their DNA binding ability, and the modification of oxygen-containing substituents significantly enhanced the anticancer activity, which could provide a new rationale for the future design of terpyridine-based metal complexes with antitumor potential.
Keywords: terpyridine, manganese complex, anticancer activity, DNA interaction, molecular docking
1. Introduction
Cancer is considered the most deadly disease impacting the different countries of the world [1,2,3]. According to WHO reports, as of 2018, cancer was the leading cause of death, with a global estimate of around 9.6 million, and one out of every six cases of death is due to cancer worldwide [4]. Metal-based drugs have been known and widely used as early as in ancient medicine. In modern times, a new era in oncology has been reopened using platinum anticancer agents [5]. Platinum drugs attracted attention in cancer treatment once the antineoplastic activity of cisplatin was discovered in the 1960s [6,7,8,9]. Despite the broad anticancer potential of platinum drugs, their clinical use has regularly been constrained by renal toxicity, low solubility and high cross-resistance [10]. To overcome these adverse reactions, the design and development of new drugs that have high efficacy, good bioavailability and weak cross-resistance are hotspots in the research of anti-tumor metal complexes [11,12,13].
As very popular ligands in coordination chemistry, terpyridine and its derivatives are a kind of multidentate N-donor ligands with a strong coordination ability to transition metals, and the planar aromatic ring structure is conducive to the interaction with DNA [14,15,16,17,18,19,20,21,22,23,24,25]. The metal complexes of terpyridine and its derivatives have rich photophysical and electrical properties. Therefore, they have important research and application prospects in the fields of luminescent materials, photodynamic therapy and chemical sensors [26,27,28,29,30,31,32]. After the success of cisplatin and carboplatin in cancer chemotherapy, other transition metal ions have been of great interest, since they are involved in many biological processes [33,34,35,36,37,38,39,40,41,42,43,44,45]. Among them, manganese is used to synthesize new complexes that exhibit excellent antibacterial and antitumor activity [46,47,48].
Due to their multi-faceted biological activity, a clear-cut target is not always easy to identify for metal or metalloid drugs [49]. Clinically approved platinum drugs are widely accepted to target DNA, and their activities are known to be influenced by DNA damage response pathways or respective repair mechanisms [50,51]. Therefore, DNA is considered the primary target for most anticancer and antiviral therapeutics [52,53,54,55,56]. DNA Topoisomerases (Topo) are ubiquitous nuclear enzymes involved in regulating the topological state of DNA, and in eukaryotic organisms, Topo can be classified into two structurally and functionally different main classes: Topo I and Topo II. Both these enzymes are the cellular target of clinically important anticancer and antibacterial drugs, and their inhibition has been considered an effective strategy for the design of many anticancer agents. At the same time, Topo I or II inhibitors show considerable wide spectrum antitumor activities, an important feature to be included in many chemotherapeutic protocols [57,58,59,60,61,62,63].
Although many terpyridine complexes have shown an interesting antiproliferative activity against tumor cells [21,22,23,24], the factors that govern the activity and the biological target are unknown. In this work, a series of manganese dichloride-substituted terpyridine complexes were synthesized by the reaction of manganese dichloride with the corresponding 4′-substituted-2,2′:6′,2″-terpyridine-bearing phenyl (L1a), naphthalen-2-yl (L1b), naphthalen-1-yl (L1c), 4-hydroxyl-phenyl (L2a), 3-hydroxyl-phenyl (L2b), 2-hydroxyl-phenyl (L2c), 4-methoxyl-phenyl (L2d), 4-carboxyl-phenyl (L2e), 4-(methylsulfonyl)phenyl (L2f), 4-nitrophenyl (L2g) or furan-2-yl (L2h), which were characterized by IR, elemental analysis and X-ray single-crystal diffraction. Their antiproliferative potentials against five tumor cell lines were studied. The ability of the complexes to bind to DNA was investigated by spectrophotometric studies and molecular modeling methods.
2. Results and Discussion
2.1. Synthesis and Characterization
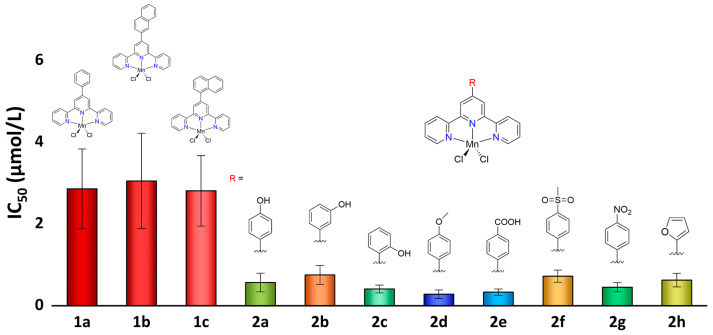
The complexes 4′-phenyl-terpyridine (L1a), 4′-(naphthalen-2-yl)-2,2′:6′,2″-terpyridine (L1b), 4′-(naphthalen-1-yl)-2,2′:6′,2″-terpyridine (L1c), 4′-(4-hydroxyl-phenyl)-2,2′:6′,2″-terpyridine (L2a), 4′-(3-hydroxyl-phenyl)-2,2′:6′,2″-terpyridine (L2b), 4′-(2-hydroxyl-phenyl)-2,2′:6′,2″-terpyridine (L2c), 4′-(4-methoxyl-phenyl)-2,2′:6′,2″-terpyridine (L2d), 4′-(4-carboxyl-phenyl)-2,2′:6′,2″-terpyridine (L2e), 4′-(4-(methylsulfonyl)phenyl)-2,2′:6′,2″-terpyridine (L2f), 4′-(4-nitrophenyl)-2,2′:6′,2″-terpyridine (L2g) and 4′-(furan-2-yl)-2,2′:6′,2″-terpyridine (L2h) were obtained using the general established protocol, and the syntheses of the ligands L1a and L2a–L2f were reported earlier [38,41]. The manganese compounds 1a–1c and 2a–2h (Figure 1) were synthesized by the reaction of L1a–L1c and L2a–L2h with MnCl2·4H2O in MeOH/DCM solution, giving yields in the range of 46–73%.
Figure 1.
Synthesis of compounds 1a–1c and 2a–2h.
The structures of the target compounds were confirmed by IR, elemental analysis and single-crystal X-ray crystallography. Their IR spectra (Figures S1–S11) display the expected bands [43,64,65]. Namely, multiple bands in the range of 1650–1401 cm−1 and 1263–1011 cm−1 were observed for the C=C stretch and for in-plane C–H bending vibrations, as well as bands in the range of 911–673 cm−1 for out-of-plane C–H bending vibrations. The hydroxyl group of 2a–2c gave a broad band in the range of 3300–3600 cm−1 for the O–H stretch, and the methoxyl group of 2d showed a band at 1240 cm−1 for the C–O–C stretch. The carboxyl group of 2e displayed a broad band in the range of 3400–3600 cm−1 with the O–H stretch and a band at 1726 cm−1 with the C=O stretch. The sulfonyl group of 2f gave strong bands at 1308 and 1297 cm−1 for SO2 stretching, a band at 793 cm−1 for the S–O stretch and two bands at 532 and 548 cm−1 for SO2 bending. The nitro group of 2g exhibited strong bands at 1519 and 1343 cm−1 for NO2 stretching, a band at 858 cm−1 for the CN stretch and a band at 754 cm−1 for CNO bending. The furyl group of 2h displayed bands at 790 and 782 cm−1 for C–H bending vibrations.
2.2. Single Crystal X-ray Crystallography
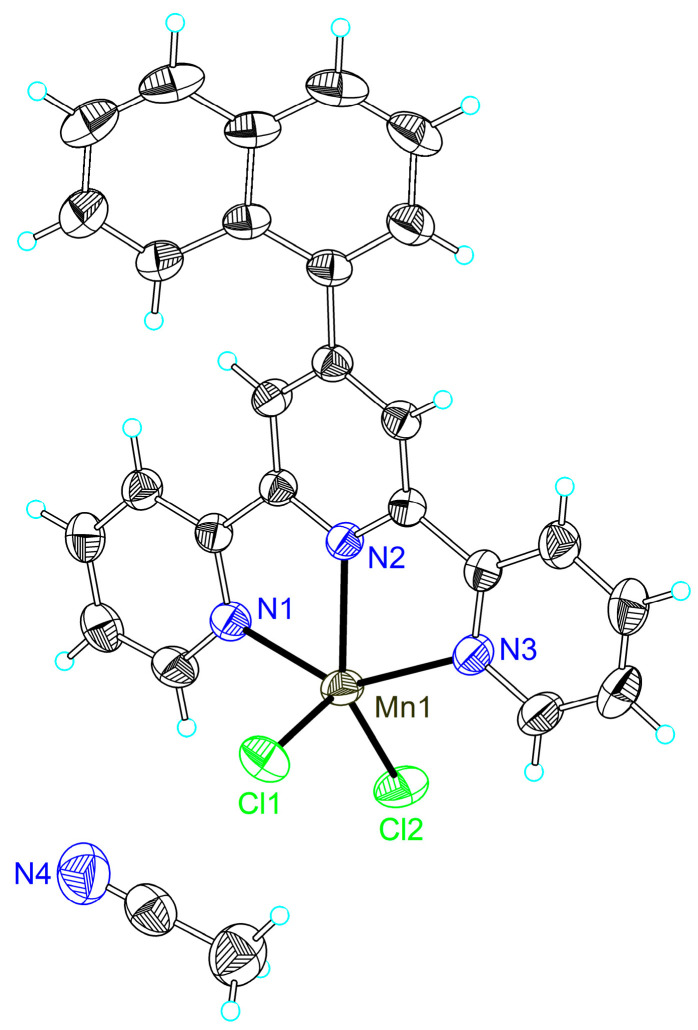
The single-crystal X-ray crystallography of compounds 1a, 1c, 2a–2c, 2f and 2g confirmed their formulation as [MnCl2Ln] (n = 1a, 1c, 2a–2c, 2f and 2g), and the manganese cation presented the common square pyramidal geometry. The crystal structures of compounds 1a and 2c have been reported [64,66]. Thermal ellipsoid plots and packing diagrams of 1a, 1c, 2a–2c, 2f and 2g are shown in Figure 2 and Figures S12–S24.
Figure 2.
Thermal ellipsoid plot, drawn at the 50% probability level, of [MnCl2L3]·CH3CN (3·CH3CN) with atomic numbering scheme. Selected bond lengths (Å) and angles (°): Mn(1)-N(2) 2.2028(13), Mn(1)-N(1) 2.2541(15), Mn(1)-N(3) 2.2595(15), Mn(1)-Cl(1) 2.3603(6), Mn(1)-Cl(2) 2.3558(5), N(2)-Mn(1)-N(1) 71.51(5), N(2)-Mn(1)-N(3) 71.71(5), N(1)-Mn(1)-N(3) 141.23(5), N(2)-Mn(1)-Cl(1) 108.08(4), N(1)-Mn(1)-Cl(1) 103.45(4), N(3)-Mn(1)-Cl(1) 99.15(4), N(2)-Mn(1)-Cl(2) 137.27(4), N(1)-Mn(1)-Cl(2) 100.29(4), N(3)-Mn(1)-Cl(2) 98.33(4), Cl(1)-Mn(1)-Cl(2) 114.57(2).
Compound 1c is a mononuclear neutral manganese complex that crystallized in the centric symmetric space group P-1, as shown in Figure 1. Each mononuclear ion is coordinated by the three N atoms of the 4′-(naphthalen-1-yl)-2,2′:6′,2″-terpyridine (L1c) ligand and two chlorine atoms as two auxiliary ligands, therefore forming an irregular square-based pyramid with a N3Cl2 coordination environment (τ = 0.07) [67]. Three hydrogen bonds exist in the structure, involving the chloride atom Cl(1), the hydrogens at the carbon atoms of the terpyridine ligand (C(12) and C(25)) and the chloride atom Cl(2) and the hydrogens at the carbon atoms of the terpyridine ligand (C(27)) with a range of distances in 2.73–2.80 Å. The structure of 1c shows two kinds of π-ring (Y-H...Cg) interactions, including one between one hydrogen at C27 and the ring formed by C(20)–C(21)–C(22)–C(23)–C(24)–C(25) of the naphthalen-1-yl, and the other one involving another hydrogen atom at C27 and the ring formed by C(16)–C(17)–C(18)–C(19)–C(20)–C(21) of the naphthalen-1-yl with atom-centroid distances (X..Cg) of 3.733(3) and 3.679(3) Å. Due to the specific arrangements of the ligands in 1c, the compound molecules present two kinds of intermolecular π–π interactions, including one between the two pyridyl rings of the ligand and the other one between the ring formed by Mn(1)–N(1)–C(5)–C(6)–N(2) and one terminal pyridyl ring with centroid distances of 3.641 and 3.687 Å.
Similar to compound 1c, complexes 1a, 2a–2c, 2f and 2g are also mononuclear species crystallized in the space groups P21/n for 1a, P21/n for 2a, P21/n for 2b, P-1 for 2c, P21/n for 2f and C2/c for 2g. The asymmetric units of the compounds present half of the molecule in 2g and one molecule in the other compounds, because of the symmetry of the molecules. The atoms Mn1, N2, C9, C10, C13 and N3 are located on the twofold axis in 2g. The coordination environment around the manganese ion in these compounds displays an irregular square-based pyramid (τ = 0.23 for 1a, 0.24 for 2a, 0.26 for 2b, 0.13 for 2c, 0.14 for 2f and 0.08 for 2g) [67], which results from the penta-coordination of manganese by three nitrogen atoms from the substituted terpyridine ligands and two chloride atoms as the auxiliary ligands in a monodentate mode. The average contacts between the central metal ion and the chloride atoms (Mn–Cl) is 2.360 Å in 1a, being longer than those in 1c (2.358 Å), 2c (2.352 Å), 2f (2.341 Å) and 2g (2.326 Å), but shorter than those in 2a (2.363 Å) and 2b (2.365 Å). Such bond lengths are not only affected by the electrophilicity of the substituents, but also by other factors, including hydrogen bonds or even spatial environments determined by the positions of the substituent groups at the phenyl rings.
Hydrogen bonds are observed in compounds 1a, 2a–2c, 2f and 2g, and the bond lengths are 2.81, 2.73, 2.80 and 2.78 Å for 1a; 2.33 Å for 2a; 2.31, 2.81 and 2.82 Å for 2b; 2.02, 1.83 and 1.82 Å for 2c; 2.57, 2.47, 2.45 and 2.68 Å for 2f; and 2.82, 2.59 and 2.60 Å for 2g. Several π-ring interactions exist in the structure of compound 9, but there was no such interaction in 2a–2c and 2g. In 2f, there are two kinds of π-ring (Y-X…Cg) interactions, one between an oxygen at S(1) of the methylsulfonyl and the rings formed by Mn(1)–N(1)–C(5)–C(6)–N(2) with atom-centroid distances (X…Cg) of 3.399 Å, and another between a hydrogen at C(22) of the methylsulfonyl and the terminal pyridyl ring N(1)–C(1)–C(2)–C(3)–C(4)–C(5) with atom-centroid distances (X…Cg) of 2.990 Å. In these structures, the different packing patterns lead to the different π–π stackings: two in 1a, 2a and 2b (in the range of 3.538–3.646, 3.572–3.736 and 3.561–3.718 Å, respectively), three in 2c (3.684–3.773 Å) and one for 2f and 2g (with a distance of 3.610 Å and 3.668 Å, respectively), involving the ring formed by Mn(1)–N(1)–C(5)–C(6)–N(2) and a terminal pyridyl ring or a terminal and a middle pyridyl ring.
2.3. Antiproliferative Properties against Tumor Cells
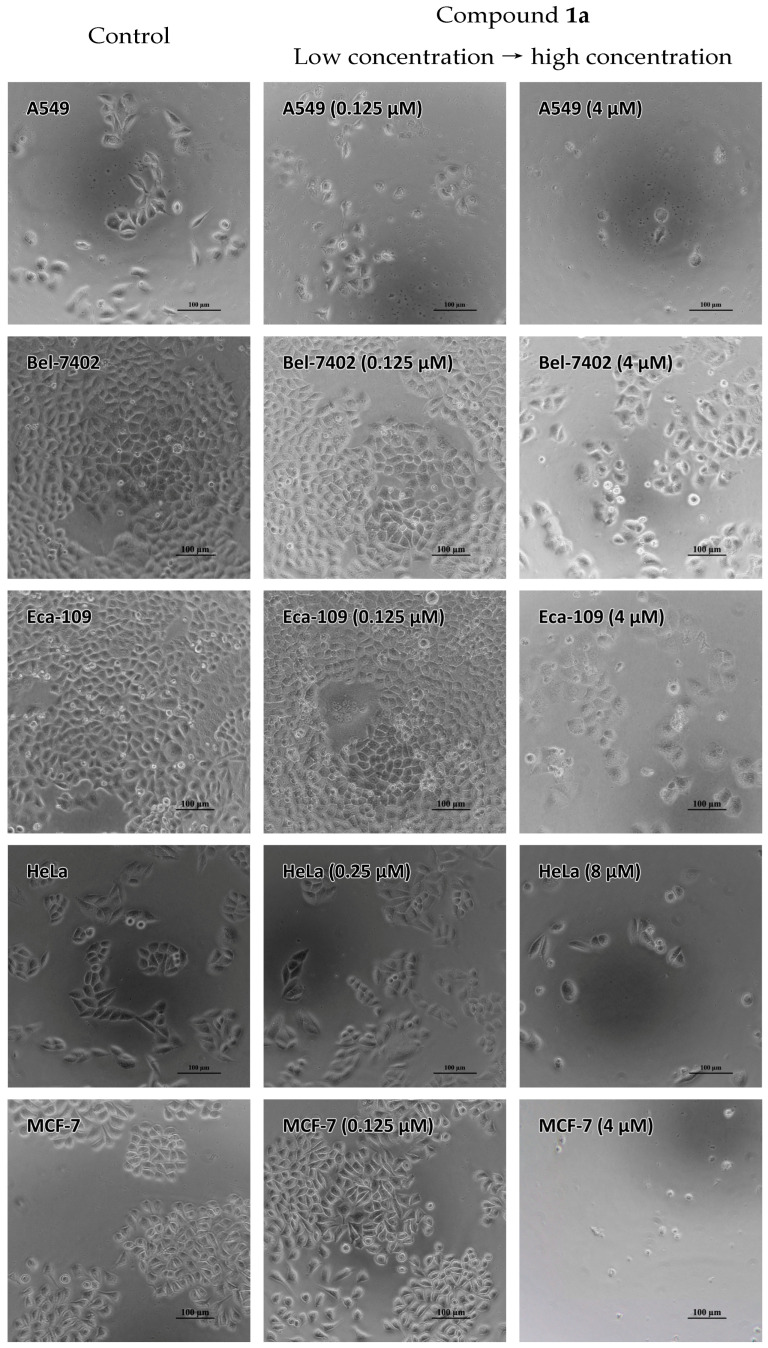
Five human carcinoma cell lines, including A549, Bel-7402, Eca-109, HeLa and MCF-7, were treated with various concentrations of compounds 1a–1c and 2a–2h (0.125–4 µM for A-549, Bel-7402, Eca-109 and MCF-7 cells or 0.25–8µM for HeLa cell line) to evaluate the in vitro antiproliferative activities of the eleven compounds. Figure 3 shows the live-cell images treated with different concentrations of compound 1a, and the microscopic photographs of the five cells treated with various concentrations of compounds 1b, 1c and 2a–2h were collected in the supporting information as Figures S25–S34. With the increasing concentrations of the compounds, the number of cancer cells observed by microscope decreased significantly. Notably, different kinds of cells respond differently to these compounds. For A549 cells, slight swelling of the cell body to nuclei shrinkage and blurring of cell boundaries were observed as the concentrations of the compounds increased, which showed the death of the cells. Regarding the Bel-7402 cell line, the obvious swelling of the cell body was observed at low concentrations of the compounds. As the concentrations of the compounds increased, a further increase in the volume of the single cell, followed by shrinkage of the nuclei and the fragmentation of the cells can be observed, suggesting the necrosis of Bel-7402 cells. Swelling of the cell body at low concentrations of the tested compounds was also observed for Eca-109 cells, with cell contraction, blurring of cell boundaries and fragmentating of the cell body being observed at high concentrations of the compounds. At low concentrations, there was no significant change in the cell morphology of HeLa cells. However, as the concentration of the compounds increased, some cells changed from a fusiform to a spherical shape with shrinkage of the nucleus. Significant swelling of the cell body was also observed in MCF-7 cells, but the higher concentration of the compounds led to cell contraction, the blurring of cell boundaries and nuclei shrinkage.
Figure 3.
Microscopic photographs of the A-549, Bel-7402, Eca-109, HeLa and MCF-7 cancer cells treated with increasing concentrations of compound 1a.
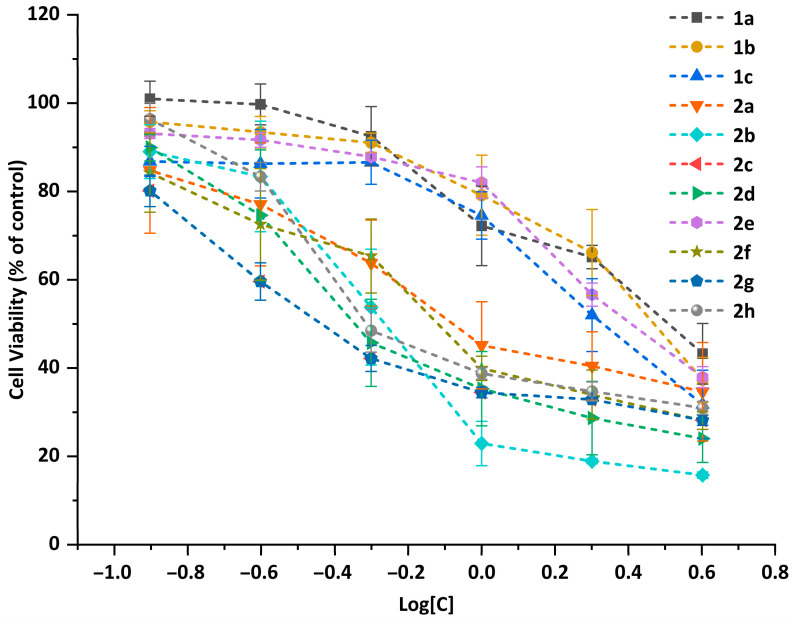
Cell viability of the five cell lines was determined by the CCK-8 assay. The plots of the cell viability vs. the concentration of compounds 1a–1c and 2a–2h against the Eca-109 cell line (Figure 4) showed that all the compounds exhibited a strong antiproliferative effect against the selected cell lines. The viability of Eca-109 cells decreases with the increase in the compound concentrations, exhibiting a dose-dependent manner. For the A549, Bel-7402, HeLa and MCF-7 cell lines, similar trends were observed and are shown in the curves of the antiproliferative activities (Figures S35–S38).
Figure 4.
Plots of cell viability vs. the concentration of compounds 1a–1c and 2a–2h in different concentrations against Eca-109 cell line.
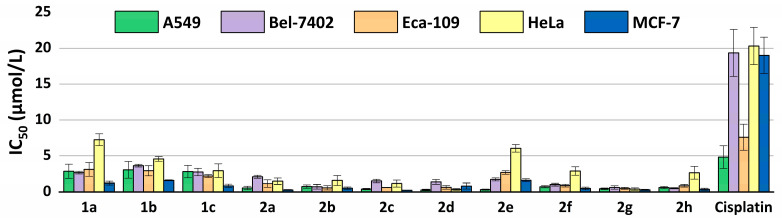
The half-maximal inhibitory concentrations (IC50) of the eleven compounds against the carcinoma cells were calculated and are listed in Table 1. The IC50 values of compounds 1a–1c and 2a–2h against the A549, Bel-7402, Eca-109, HeLa and MCF-7 cell lines were much lower than the widely used anti-tumor drug cisplatin. In the results, compound 2d presented the lowest IC50 value against both A549 and HeLa cells; the values were 0.281 μM and 0.356 μM, respectively. Compound 2h showed the best results (0.523 μM) on Bel-7402 cells, 2g gave the lowest IC50 value (0.514 μM) against Eca-109 cells and 2c had the lowest value (0.249 μM) on MCF-7 cells.
Table 1.
IC50 values (μM) from the dose-response assay of compounds 1a–1c and 2a–2h and the reference compound cisplatin in the A549, Bel-7402, Eca-109, HeLa and MCF-7 cell lines, after an incubation time of 48 h.
| Compounds | A549 | Bel-7402 | Eca-109 | HeLa | MCF-7 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (μmol/L) | 95% Confidence Intervals (μmol/L) | IC50 (μmol/L) | 95% Confidence Intervals (μmol/L) | IC50 (μmol/L) | 95% Confidence Intervals (μmol/L) | IC50 (μmol/L) | 95% Confidence Intervals (μmol/L) | IC50 (μmol/L) | 95% Confidence Intervals (μmol/L) | |
| 1a | 2.858 | 2.142–3.813 | 2.667 | 0.455–0.718 | 3.115 | 2.575–3.769 | 7.249 | 6.118–8.589 | 1.229 | 0.980–1.540 |
| 1b | 3.052 | 1.628–5.724 | 3.631 | 0.434–0.650 | 2.927 | 2.222–3.857 | 4.566 | 3.857–5.406 | 1.594 | 1.244–2.042 |
| 1c | 2.811 | 2.034–3.883 | 2.751 | 0.308–0.450 | 2.196 | 1.818–2.652 | 2.935 | 1.765–4.883 | 0.850 | 0.671–1.077 |
| 2a | 0.567 | 0.398–0.808 | 2.088 | 1.329–1.590 | 1.153 | 0.714–1.863 | 1.486 | 1.086–2.031 | 0.284 | 0.227–0.355 |
| 2b | 0.754 | 0.519–1.096 | 0.730 | 0.616–0.796 | 0.579 | 0.486–0.688 | 1.583 | 1.085–2.310 | 0.525 | 0.399–0.691 |
| 2c | 0.409 | 0.242–0.692 | 1.510 | 1.043–1.227 | 0.614 | 0.488–0.774 | 1.160 | 0.969–1.389 | 0.249 | 0.207–0.301 |
| 2d | 0.281 | 0.217–0.364 | 1.382 | 0.304–0.471 | 0.645 | 0.502–0.829 | 0.356 | 0.280–0.452 | 0.819 | 0.583–1.151 |
| 2e | 0.334 | 0.287–0.390 | 1.720 | 0.656–0.876 | 2.690 | 2.452–2.950 | 6.047 | 5.304–6.894 | 1.610 | 1.240–2.090 |
| 2f | 0.723 | 0.538–0.970 | 1.011 | 0.441–0.605 | 0.886 | 0.717–1.094 | 2.889 | 1.815–4.600 | 0.501 | 0.360–0.698 |
| 2g | 0.453 | 0.313–0.655 | 0.626 | 0.616–0.796 | 0.514 | 0.414–0.638 | 0.412 | 0.266–0.639 | 0.304 | 0.249–0.372 |
| 2h | 0.626 | 0.455–0.862 | 0.523 | 1.043–1.227 | 0.868 | 0.677–1.111 | 2.648 | 1.683–4.167 | 0.379 | 0.317–0.454 |
| Cisplatin | 4.832 | 3.501–6.669 | 19.34 | 16.27–22.98 | 7.594 | 5.879–9.809 | 20.30 | 17.87–23.06 | 19.00 | 16.19–22.30 |
A comparison of the antiproliferative activity of the compounds against different cell lines is shown in Figure 5. Among the five cell lines, A549, Bel-7402, MCF-7 and Eca-109 were more susceptible, and HeLa was more tolerant for the tested compounds. Among the eleven compounds, 2g with the nitro group performed the best antiproliferative activities against the five cell lines on the whole, usually showing much lower IC50 values in the tested cells than the others. Since the compounds differ from each other only in the 4-substituents at the terpyridyl group, the relationship between the anticancer activity of all the compounds and their structures was further analyzed.
Figure 5.
IC50 values (μM) from the dose-response assay of compounds 1a–1c and 2a–2h and the reference compound cisplatin in the A549, Bel-7402, Eca-109, HeLa and MCF-7 cell lines, after an incubation time of 48 h. The results shown are means ± SD of quadruplicate experiments.
As shown in Figure 6, when A549 cancer cells were used, the anticancer activity of the 2-naphthyl and 1-naphthyl-substituted terpyridine compounds was close to that of 4′-phenyl-terpyridine. When the substituent at terpyridine was an oxygen-containing group, more than four times higher anticancer activities were obtained in comparison with those of 4′-phenyl-terpyridine compounds. The electronegativity and stereochemical effects of the oxygen-containing substituents are critical for the anticancer activity of the complexes; they showed an increase in bioactivity with the increase in the electronegativity of the substituent and a decrease in bioactivity with the increase in steric hindrance, resulting in anticancer activity as 4-OMe-Ph– > 4-COOH-Ph– > 2-OH-Ph– > 4-NO2-Ph– > 4-OH-Ph > 4-(furan-2-yl)– > 4-Ms-Ph– > 3-OH-Ph. For the four other cell lines, Bel-7402, Eca-109, HeLa and MCF-7, the compounds with oxygen-containing substituents showed significantly better anticancer activity.
Figure 6.
IC50 values (μM) of compounds 1a–1c and 2a–2h against the A549 cell line and the structures of the compounds.
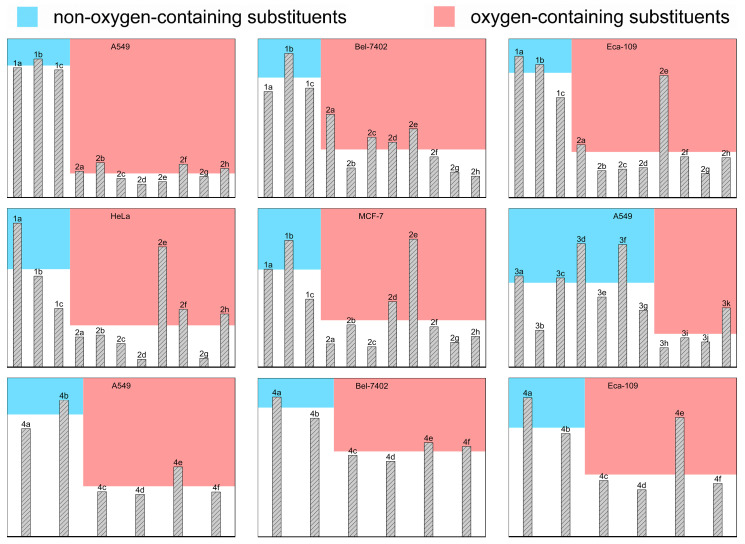
In order to clarify the effect of oxygen-containing substituent modification on the anticancer activity of terpyridine complexes, we summarized the anticancer activities of a total of 28 substituted terpyridine complexes against different cancer cells in Figure 7 from this work and previously reported works [44,65]. When the average activity of the non-oxygen-containing substituent-modified terpyridine complexes is compared with that of the terpyridine complexes modified with oxygen-containing substituents, we can clearly find that oxygen-containing substituents brought a significant increase in anticancer activity. Although the electronegativity, steric hindrance and ability to form hydrogen bonds of substituents can also affect the order of anticancer activity of compounds, modification with oxygen-containing substituents is still an effective strategy to enhance the anticancer activity of terpyridine complexes.
Figure 7.
The in vitro anticancer activity of substituted 2,2′:6′,2″-terpyridine(tpy) complexes 1a–1c, 2a–2h, 3a–3k [65] and 4a–4f [44] against different cancer cell lines, including A549, Bel-7402, Eca-109, HeLa and MCF-7. The heights of the blue and red blocks on the background represent the average performance of the anticancer activity of the non-oxygen-containing substituents and oxygen-containing substituent-modified complexes, respectively. Here, 3a = [Cu(tpy)Cl2], 3b = [Cu(4′-(4′-cyano-phenyl)-tpy)Cl2], 3c = [Cu(4′-(4′-iodo-phenyl)-tpy)Cl2], 3d = [Cu(4′-(4′-bromo-phenyl)-tpy)Cl2], 3e = [Cu(4′-(4′-chloro-phenyl)-tpy)Cl2], 3f = [Cu(4′-(4′-fluoro-phenyl)-tpy)Cl2], 3g = [Cu(4′-(p-hydroxyl-phenyl)-tpy)Cl2], 3h = [Cu(4′-(m-hydroxyl-phenyl)-tpy)Cl2], 3i = [Cu(4′-(o-hydroxyl-phenyl)-tpy)Cl2], 3j = [Cu(4′-(methoxyl-phenyl)-tpy)Cl2], 3k = [Cu(4′-(4′-methylsulfonyl-phenyl)-tpy)Cl2], 4a = [Zn(4′-(4′-methyl-phenyl)-tpy)Br2], 4b = [Zn(4′-(4′-methyl-phenyl)-tpy)I2], 4c = [Zn(4′-(4′-methylsulfonyl-phenyl)-tpy)Br2], 4d = [Zn(4′-(4′-methylsulfonyl-phenyl)-tpy)I2], 4e = [Zn(4′-(4′-methoxyl-phenyl)-tpy)Br2], 4f = [Zn(4′-(4′-methoxyl-phenyl)-tpy)I2].
2.4. Circular Dichroism Spectroscopic Studies
CD spectroscopy is one of the most sensitive techniques for monitoring structural changes in DNA in solution [68,69,70]. The reliance on CD spectroscopy to study DNA conformations has stemmed from the sensitivity and ease of CD measurements, the nondestructive nature of such measurements, the fact that conformations can be studied in solution and the requirement for relatively small amounts of material [71].
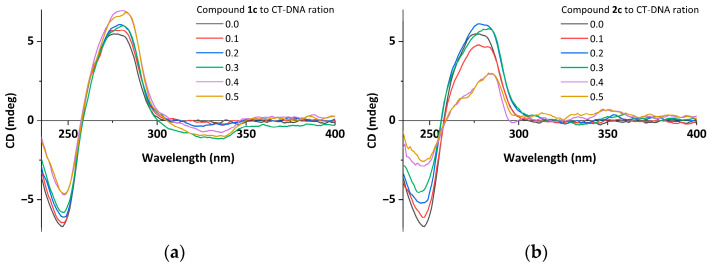
The CD spectra of CT-DNA with increased concentrations of compounds 1a–1c and 2a–2h were measured and are shown in Figure 8 and Figures S39–S47. For reference, when no compound was added, the CD spectrum of CT-DNA showed two peaks at 277 nm and 246 nm, respectively. The two bands are the known features of a right-handed B-form DNA. Specifically, the positive band at 277 nm is due to base stacking, and the negative band at 246 nm is from the right-handed helicity. When the different concentrations of the compounds were added and incubated with CT-DNA, obvious changes in both positive and negative bands were observed. All the CD spectral bands of the DNA with the different concentrations of the compounds are tabulated in Table S1.
Figure 8.
(a) Circular dichroism spectra of CT-DNA (6.0 × 10−4 mol/L) in the presence or absence of compound 1a in Tris-HCl buffer (pH 7.2) at 20 °C. (b) Circular dichroism spectra of CT-DNA (6.0 × 10−4 mol/L) in the presence or absence of compound 2c in Tris-HCl buffer (pH 7.2), at 20 °C.
As the concentrations of the compounds increase to 300 µmol/L, the intensities of the positive bands decrease by 12–62% compared to CT-DNA, with slight shifts in the λmax, and the intensities of the negative bands decrease by 5–46%. Notably, when compound 2h at a concentration of 60 µmol/L interacts with CT-DNA, the band attributed to base stacking (at 277 nm) shows a 10% increase in positive ellipticity with no considerable shift in its position. For compound 1c, the positive band shows a 26% increase as the compound concentration increases, which may be due to the participation and facilitation of the compounds with high planarity in the π–π stacking of the base pairs of DNA.
The changes in the ellipticity and wavelength of the CD signals around 277 nm are important for elucidating their ability to intercalate between DNA base pairs [72,73]. By comparing the hypochromism caused by compounds 1a–1c and 2a–2h at the same ratio, compound 2c exhibited the strongest intercalation ability. This indicates that the different substituents at the terpyridine significantly affect the interaction between the compound and DNA, and the o-hydroxyl group seems to promote the intercalation.
2.5. Molecular Docking Studies
Molecular docking techniques have shown great promise as a new tool in the discovery of novel small-molecule drugs for predicting the plausible interactions between the drug and nucleic acid in a non-covalent fashion. Most anti-tumor drugs have functions by incorporating into the base pairs of the DNA of tumor cells to interrupt their replication and transcription [74,75]. The syntheses of important proteins is terminated or disrupted, resulting in the termination of cell division and growth, cell swelling, cell necrosis, etc., which was observed in this study. In order to explore the mechanism of their anticancer activity, the DNA interaction with the compounds was studied using the molecular modeling method.
2.5.1. Molecular Docking with DNA
In this study, the AutoDock program was used to examine the compound–DNA interactions by investigating the potential binding modes and calculating the binding energies. At first, rigid molecular docking studies of compounds 1a–1c and 2a–2h with the DNA duplex of sequence d(CGCGAATTCGCG)2 dodecamer (PDB ID: 1BNA) were performed in order to predict the binding site along with the preferred orientation of the ligand. Detailed simulations of the compound–DNA interactions were carried out and are presented in Figure 9 and Figures S48–S57. The free energies of the binding were calculated and are shown in Table 2.
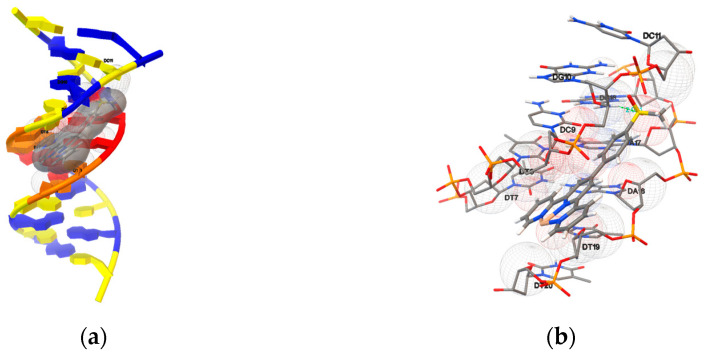
Figure 9.
(a) The most favorable orientation and (b) enlarged view of compound 2f bound with the minor groove of the B-DNA (PDB ID: 1BNA). The formed hydrogen bond and distance have been marked.
Table 2.
The calculated free energy of binding of compounds 1a–1c and 2a–2h with B-DNA (1BNA), oligonucleotide (4JD8M) and DNA-Topo I complex (1SC7).
| Compounds | Affinity (kcal/mol) | ||
|---|---|---|---|
| B-DNA (1BNA) |
Oligonucleotide (4JD8) |
DNA-Topo I Complex (1SC7) |
|
| 1a | −9.11 | −9.32 | −9.97 |
| 1b | −10.34 | −9.61 | −10.23 |
| 1c | −10.27 | −9.93 | −10.89 |
| 2a | −9.07 | −9.28 | −9.58 |
| 2b | −9.21 | −8.78 | −10.26 |
| 2c | −9.29 | −8.97 | −10.39 |
| 2d | −9.16 | −8.41 | −9.97 |
| 2e | −8.55 | −8.07 | −9.75 |
| 2f | −10.86 | −10.23 | −12.34 |
| 2g | −9.38 | −9.20 | −10.53 |
| 2h | −8.67 | −8.11 | −9.06 |
It was observed that the ligand fits into the minor groove perfectly, involving outside edge interactions without disrupting the double-helical structure of the DNA. The bindings were stabilized by van der Waals interaction and hydrophobic contacts with DNA functional groups, which define the stability of groove, and the binding energies were between –8.55 and –10.86 kcal mol−1. Hydrogen bonds were found between the compounds and the DNA, and the detailed bond distances and bond energies are listed in Table 3. Compounds 2a–2c, 2f and 2g formed hydrogen bonds with B-DNA benefitting from the presence of the substituent groups, whereas the other compounds showed no hydrogen bond formation in their most favorable orientations.
Table 3.
Hydrogen bond interactions for compounds 2a–2c, 2f and 2g.
| Compounds | Receptor | Bonds Formed | Bond Distance (Å) | Bond Energy (kcal/mol) |
|---|---|---|---|---|
| 2a | B-DNA (1BNA) | O–H…O (DA18) | 2.105 | −4.619 |
| Oligonucleotide (4JD8) | O–H…O (DT2) | 1.993 | −0.824 | |
| DNA-Topo I complex (1SC7) | O–H…O (ASN722) | 2.133 | −3.223 | |
| 2b | B-DNA (1BNA) | O–H…O (DT19) | 2.175 | −4.074 |
| Oligonucleotide (4JD8) | O–H…O (DT2) | 2.073 | −4.052 | |
| DNA-Topo I complex (1SC7) | O–H…O (ASN722) | 2.081 | −0.401 | |
| DNA-Topo I complex (1SC7) | O…H–N (ASN722) | 1.956 | −3.016 | |
| 2c | B-DNA (1BNA) | O–H…O (DT7) | 1.804 | −1.485 |
| Oligonucleotide (4JD8) | O–H…O (DG3) | 2.231 | −5.235 | |
| DNA-Topo I complex (1SC7) | O–H…O (TGP11) | 2.134 | −6.073 | |
| 2f | B-DNA (1BNA) | O…H–N(DG16) | 2.061 | −5.108 |
| Oligonucleotide (4JD8) | O…H–N(DA5) | 2.130 | −4.107 | |
| DNA-Topo I complex (1SC7) | O…H–N (MET428) | 1.990 | −4.015 | |
| DNA-Topo I complex (1SC7) | O…H–N (ALA351) | 2.204 | −1.600 | |
| 2g | B-DNA (1BNA) | O…H–N (DG14) | 2.079 | −1.999 |
| Oligonucleotide (4JD8) | O…H–N(DA5) | 2.201 | −2.900 | |
| DNA-Topo I complex (1SC7) | O…H–N (LYS425) | 1.877 | −4.568 |
Furthermore, compounds 1a–1c and 2a–2h were docked onto an oligonucleotide (ds(ATGCAT)2, PDB ID: 4JD8) to explore the potential binding mode and energy. The binding energies of the eleven compounds were calculated and are listed in Table 2. They were in the range between −8.07 and −10.23 kcal mol−1, and the detailed docking poses are presented in Figure 10 and Figures S58–S67. The results indicate that the docked compounds intercalate into the base pairs of the DNA, involving π–π stacking, van der Waals interaction and hydrophobic and hydrogen bonding. Compounds 2a–2c, 2f and 2g formed hydrogen bonds with the oligonucleotide by the hydroxyl group. No hydrogen bond was detected in the most favorable poses of 1a–1c, 2d, 2e and 2h.
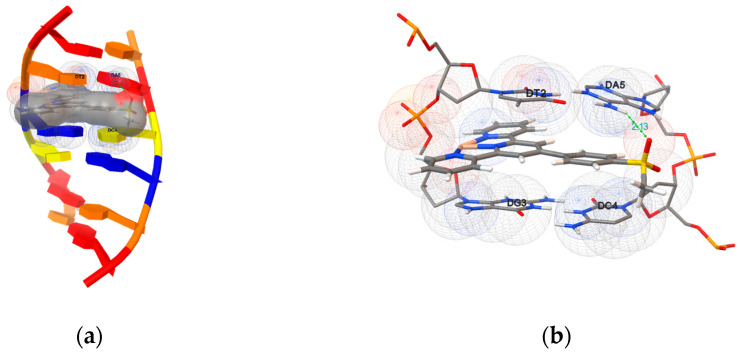
Figure 10.
(a) The most favorable orientation and (b) enlarged view of compound 2f intercalating with the DNA (4JD8). The formed hydrogen bond and distance have been marked.
2.5.2. Molecular Docking with Topoisomerase I
To elucidate the interaction and locate the exact binding site between the compounds 1a–1c and 2a–2h and Topo-I, molecular docking studies were performed using the Human-DNA Topo-I complex (PDB ID: 1SC7). As seen in Figure 11 and Figures S68–S77, the terpyridyl group was intercalated into the base pairs of the DNA, and the substituents at the terpyridyl group formed hydrogen bonds with the residues of Topo-I. An in silico molecular docking experiment revealed that the docked compounds fit into the Topo I-DNA complex perfectly and resulted in the binding energy between –9.06 and −12.34 kcal mol−1. No hydrogen bond was detected in the most favorable pose of compounds 1a–1c, 2d, 2e and 2h. Meanwhile, compounds 2a–2c, 2f and 2g showed a binding energy value from –9.58 to −12.34 kcal mol−1 with one or two hydrogen bonds. For compound 2a, the O–H formed a hydrogen bond with an oxygen of the ASN722, and the bond length was 2.133 Å. The hydroxyl of compound 2b formed two hydrogen bonds with the ASN722, and the O–H of 6 formed a hydrogen bond with an oxygen of the phosphate. For compound 2f, one N–H at the MET428 and one N–H at the ALA351 interacted with the two oxygen atoms at the methylsulfonyl group to form two hydrogen bonds with bond lengths of 1.990 and 2.204 Å, respectively. For 2g, one N–H at the LYS425 interacted with the oxygen at the nitro group and formed a hydrogen bond with a bond length of 1.877 Å. The details of the hydrogen bonds have been summarized in Table 3. The results proved that the substituents at the terpyridyl group contribute to forming a stable complex in the DNA–Topo I active site through π–π stacking with the purine ring of DNA, van der Waals, hydrophobic bonding and hydrogen bonding with the residues of Topo-I. It can be inferred from the present docking studies that some subtle change in the structure of the drug molecule alters the ligand-binding domain in the drug target. These phenomena are very interesting, as well as desirable for drug designing, because repeated application of the same drug/compound leads to the development of resistance to the action of the drug, due to unavoidable conformational modifications in the drug target.
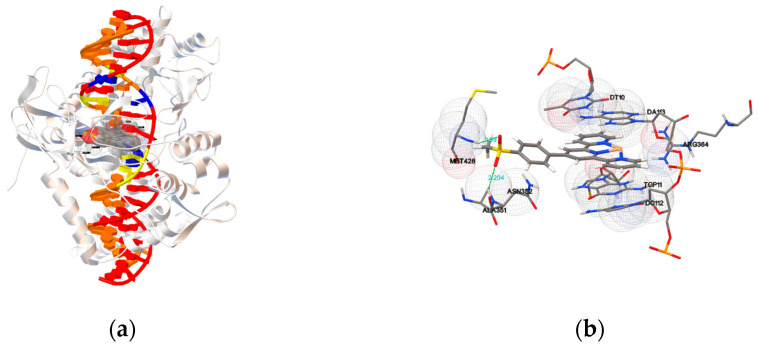
Figure 11.
(a) Molecular docking models and (b) enlarged view of 2f in the active site of DNA–Topo I complex (PDB ID: 1SC7). The formed hydrogen bond and distance have been marked.
3. Methods and Materials
3.1. Chemicals and Reagents
All common reagents employed in this work were of analytical grade.
3.2. Instruments and Apparatus
The infrared spectra were obtained with a Thermo Scientific Nicolet iS10 spectrophotometer, and elemental analyses (C, H, N) were performed on an Elementar vario EL cube.
3.3. Synthesis of the Compounds
The methanol solution of MnCl2 was added to dichloromethane solutions of the corresponding ligands, and the mixture was stirred for 24 h. The separation was performed by filtering the compound powders from the mother solution and drying them in a desiccator. Recrystallization from a methanol/acetonitrile solution upon slow evaporation led to the formation of crystals that were suitable for X-ray analysis of complexes 1a, 1c, 2a–2c, 2f and 2g.
[MnCl2L1a] (1a). Orange crystals. Yield: 0.30 g, 69%. Anal. calcd for C21H15Cl2N3Mn: C, 57.96; H, 3.47; N, 9.66%. Found: C 56.55, H 3.44, N 9.47%. IR (KBr disc, cm−1, s = strong, m = medium, w = weak): 3052 (m, νC–H), 2988 (m, νC–H), 2900 (w, νC–H), 1600 (s, νC=C), 1549 (m, νC=C), 1475 (s, νC=C), 1434 (m, νC=C), 1401 (m, νC=C), 1303 (m), 1232 (m, βC–H), 1163 (m, βC–H), 1077 (m, βC–H), 1066 (m, βC–H), 1014 (m, βC–H), 898 (m, γC–H), 886 (m, γC–H), 832 (m, γC–H), 791 (m, γC–H), 773 (m, γC–H), 746 (s, γC–H), 731 (m, γC–H), 695 (m, γC–H), 657 (m), 638 (m), 619 (m), 581 (m) and 560 (m).
[MnCl2L1b] (1b). Yield: 0.27 g, 56%. Anal. calcd for C25H17Cl2N3Mn·0.5H2O: C, 60.75; H, 3.67; N, 8.50%. Found: C 60.71, H 3.45, N 8.42%. IR (KBr disc, cm−1): 3057 (m, νC–H), 1608 (s, νC=C), 1570 (m, νC=C), 1545 (m, νC=C), 1475 (s, νC=C), 1440 (m, νC=C), 1417 (m, νC=C), 1250 (m, βC–H), 1160 (m, βC–H), 1131 (m, βC–H), 1067 (m, βC–H), 1012 (s, βC–H), 896 (m, γC–H), 869 (m, γC–H), 824 (m, γC–H), 794 (s, γC–H), 785 (m, γC–H), 769 (m, γC–H), 741 (s, γC–H), 728 (m, γC–H), 686 (m, γC–H), 657 (m), 638 (m), 604 (m), 566 (m) and 556 (m).
[MnCl2L1c] (1c). Yellow crystals. Yield: 0.28 g, 58%. Anal. calcd for C25H17Cl2N3Mn: C, 61.88; H, 3.53; N, 8.66%. Found: C 61.64, H 3.49, N 8.53%. IR (KBr disc, cm−1): 3099 (m, νC–H), 3065 (m, νC–H), 3022 (m, νC–H), 1606 (m, νC=C), 1595 (m, νC=C), 1567 (m, νC=C), 1541 (m, νC=C), 1468 (m, νC=C), 1439 (m, νC=C), 1413 (m, νC=C), 1293 (m), 1244 (m, βC–H), 1011 (m, βC–H), 911 (m, γC–H), 886 (m, γC–H), 868 (m, γC–H), 802 (s, γC–H), 791 (s, γC–H), 781 (s, γC–H), 741 (m, γC–H), 734 (m, γC–H), 689 (w, γC–H), 664 (m), 651 (m), 639 (m), 632 (m) and 589 (m).
[MnCl2L2a] (2a). Orange crystals. Yield: 0.30 g, 66%. Anal. calcd for C21H15Cl2N3Mn·H2O·CH2Cl2: C, 47.68; H, 3.46; N, 7.58%. Found: C 47.91, H 3.16, N 7.28%. IR (KBr disc, cm−1): 3419 (m, νO–H), 3273 (m, νC–H), 1598 (s, νC=C), 1586 (s, νC=C), 1573 (m, νC=C), 1547 (m, νC=C), 1522 (m, νC=C), 1475 (s, νC=C), 1442 (s, νC=C), 1410 (m, νC=C), 1362 (m, δO–H), 1279 (m), 1245 (s, νC=C), 1218 (m, βC–H), 1182 (m, νC–OH), 1120 (m, βC–H), 1068 (m, βC–H), 1012 (m, βC–H), 850 (m, γC–H), 824 (m, γC–H), 790 (m, γC–H), 748 (m, γC–H), 730 (m, γC–H), 658 (m), 638 (m), 623 (m), 573 (m).
[MnCl2L2b] (2b). Brown crystals. Yield: 0.28 g, 62%. Anal. calcd for C21H15Cl2N3OMn·2H2O·CH2Cl2: C, 46.18; H, 3.70; N, 7.34%. Found: C 46.03, H 3.63, N 7.05%. IR (KBr disc, cm−1): 3419 (s, νO–H), 3285 (m, νC–H), 2935 (m, νC–H), 2841 (m, νC–H), 1650 (m, νC=C), 1599 (s, νC=C), 1571 (m, νC=C), 1545 (m, νC=C), 1520 (m, νC=C), 1475 (s, νC=C), 1416 (s, νC=C), 1364 (m, δO–H), 1283 (m), 1243 (s, νC=C), 1187 (m, νC–OH), 1164 (m, βC–H), 1103 (m, βC–H), 1070 (m, βC–H), 1012 (s, βC–H), 892 (m, γC–H), 839 (m, γC–H), 791 (s, γC–H), 781 (s, γC–H), 729 (m, γC–H), 685 (m, γC–H), 658 (m), 638 (m) and 576 (m).
[MnCl2L2c] (2c). Orange crystals. Yield: 0.27 g, 60%. Anal. calcd for C21H15Cl2N3OMn·H2O·CH2Cl2: C, 47.68; H, 3.46; N, 7.58%. Found: C 47.41, H 3.16, N 7.76%. IR (KBr disc, cm−1): 3478 (s, νO–H), 2960 (m, νC–H), 2926 (m, νC–H), 2852 (m, νC–H), 1608 (s, νC=C), 1598 (s, νC=C), 1570 (m, νC=C), 1546 (m, νC=C), 1475 (m, νC=C), 1456 (m, νC=C), 1411 (s, νC=C), 1307 (m), 1263 (m, νC=C), 1244 (m, νC=C), 1163 (m, βC–H), 1097 (m, νC–OH), 1013 (s, βC–H), 889 (s, γC–H), 859 (m, γC–H), 789 (s, γC–H), 764 (s, γC–H), 750 (m, γC–H), 731 (m, γC–H), 657 (m), 637 (m), 621 (m) and 547 (m).
[MnCl2L2d] (2d). Yield: 0.34 g, 73%. Anal. calcd for C22H17Cl2N3OMn·H2O: C, 54.68; H, 3.96; N, 8.70%. Found: C 54.28, H 3.65, N 8.56%. IR (KBr disc, cm−1): 3058 (m, νC–H), 2937 (m, νC–H), 2840 (m, νC–H), 1598 (s, νC=C), 1545 (m, νC=C), 1520 (m, νC=C), 1475 (s, νC=C), 1433 (m, νC=C), 1406 (m, νC=C), 1363(m, δC–H), 1309 (m), 1285 (m), 1240 (s, νC–O–C), 1186 (m, βC–H), 1240 (m, βC–H), 1240 (m, βC–H), 1186 (m, βC–H), 1069 (m, βC–H), 1013 (m, βC–H), 891 (m, γC–H), 839 (m, γC–H), 791 (s, γC–H), 748 (m, γC–H), 729 (m, γC–H), 658 (m), 639 (m) and 576 (m).
[MnCl2L2e] (2e). Yield: 0.23 g, 48%. Anal. calcd for C22H15Cl2N3O2Mn·H2O·CH2Cl2: C, 47.45; H, 3.29; N, 7.22%. Found: C 47.39, H 3.25, N 7.55%. IR (KBr disc, cm−1): 3445 (s, νOH), 2987 (m, νC–H), 2970 (m, νC–H), 2901 (m, νC–H), 1726 (m, νC=O), 1609 (m, νC=C), 1598 (s, νC=C), 1571 (m, νC=C), 1544 (m, νC=C), 1476 (s, νC=C), 1429 (s, νC=C), 1397 (m, δO–H), 1251 (m, νC–O), 1187 (m, βC–H), 1121 (m, βC–H), 1105 (m, βC–H), 1068 (s, βC–H), 1013 (s, βC–H), 893 (m, γC–H), 857 (m, γC–H), 795 (s, γC–H), 769 (s, γC–H), 729 (m, γC–H), 694 (m, γC–H), 659 (m) and 638 (m).
[MnCl2L2f] (2f). Orange crystals. Yield: 0.28 g, 55%. Anal. calcd for C22H17Cl2N3O2SMn·H2O·1.5CH2Cl2: C, 42.85; H, 3.37; N, 6.38%. Found: C 42.56, H 3.07, N 6.33%. IR (KBr, cm−1): 3058 (w, νC–H), 2936 (w, νC–H), 2908(w, νC–H), 2841(w, νC–H), 1598 (s, νC=C), 1520 (m, νC=C), 1475 (s, νC=C), 1433 (m, νC=C), 1407 (m, νC=C), 1308 (s, νSO2), 1297 (s, νSO2), 1241 (m, βC–H), 1187 (m, βC–H), 1147 (s, νSO2), 1092 (m, βC–H), 1069 (m, βC–H), 1012 (m, βC–H), 965 (m, γC–H), 890 (m, γC–H), 835 (m, γC–H), 801 (s, γC–H), 793 (s, νS–O), 768 (m, γC–H), 729 (m, γC–H), 673 (m, γC–H), 657 (m), 638 (m), 577 (s, δSO2) and 549 (s, δSO2).
[MnCl2L2g] (2g). Orange crystals. Yield: 0.22 g, 46%. Anal. calcd for C21H14Cl2N4O2Mn·H2O: C, 50.63; H, 3.24; N, 11.25%. Found: C 50.90, H 3.09, N 11.19%. IR (KBr disc, cm−1): 3059 (m, νC–H), 2937 (w, νC–H), 2840 (m, νC–H), 1597 (s, νC=C), 1571 (m, νC=C), 1546 (m, νC=C), 1519 (s, νNO2), 1475 (s, νC=C), 1431 (m, νC=C), 1343 (s, νNO2), 1241 (m, βC–H), 1187 (m, βC–H), 1163 (m, βC–H), 1069 (m, βC–H), 1013 (s, βC–H), 892 (m, γC–H), 858 (m, γCN), 819 (m, γC–H), 791 (s, γC–H), 754 (m, γCNO), 730 (m, γC–H), 658 (m), 638 (m) and 576 (m).
[MnCl2L2h] (2h). Yield: 0.22 g, 52%. Anal. calcd for C19H13Cl2N3OMn·1.5H2O: C, 50.47; H, 3.57; N, 9.29%. Found: C 50.36, H 3.46, N 9.16%. IR (KBr disc, cm−1): 3094 (m, νC–H), 3060 (m, νC–H), 3032 (m, νC–H), 1615 (s, νC=C), 1600 (s, νC=C), 1544 (m, νC=C), 1589 (s, νC=C), 1459 (m, νC=C), 1427 (s, νC=C), 1383 (m), 1254 (m, βC–H), 1231 (m, βC–H), 1160 (m, βC–H), 1072 (m, βC–H), 1030 (m, βC–H), 1012 (s, βC–H), 1003 (s, βC–H), 925 (m), 883 (m, γC–H), 839 (m, γC–H), 790 (s, δC–H), 782 (s, δC–H), 743 (s, γC–H), 727 (s, γC–H), 682 (s, γC–H), 654 (m), 638 (m) and 584 (m).
3.4. Crystallography
Compound 1c was initially crystallized from acetonitrile, and the crystal structure was found to contain solvent molecules. For cell viability and DNA interaction studies, 1c was re-synthesized from MeOH/DCM to prevent the incorporation of solvent molecules into the structure of the complex. Similarly, the crystal structure of compound 2c was found to contain water molecules, and then the compound 2c was re-synthesized from MeOH/DCM solution. Single crystals of complexes 1a, 2a–2c and 2f were mounted on glass fibers, and 1c and 2g were mounted in loops and fixed with Fomblin® Y LVAC 25/6 oil. Intensity data were collected using a Bruker D8 Quest (for 1c), Bruker AXS KAPPA APEX II (for 2g) and Agilent SuperNova (for the other complexes) diffractometers with graphite monochromated Mo-Kα (λ = 0.71073 Å) radiation. The data were collected using phi and omega scans in a conventional (for 2g) and shutterless (for the other complexes) mode using 0.5° per frame (for the other complexes), and the full sphere of data were obtained. Cell parameters were retrieved and refined by Bruker SAINT (for 1c and 2g) and Agilent CrysAlisPro (for the other complexes) software on all of the observed reflections [76]. Absorption corrections were applied using SADABS [77] (for 1c and 2g) and ABSPACK [77] (for the other complexes). The structures were solved by direct methods using the SHELX-2018/3 [78] (for 1c and 2g) and SHELX–97 [79] (for the other complexes) packages and refined with SHELXL–97 [79]. The thermal ellipsoid plots were drawn by Diamond 3.2 [80]. The hydrogen atoms were inserted in calculated positions theoretically. Least square refinements with anisotropic thermal motion parameters for all of the non-hydrogen atoms and isotropic for the remaining atoms were employed. CCDC 2062271–2062277 for compounds 1a, 1c, 2a–2c, 2f and 2g contain the supplementary crystallographic data of this paper. These data can be obtained free of charge from the Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif, accessed on 1 December 2022. Crystal data and details of data collections are reported in Table 4.
Table 4.
Hydrogen bond interactions for compounds 1a, 1c, 2a–2c, 2f and 2g.
| Compound | 1a | 1c | 2a | 2b | 2c | 2f | 2g |
|---|---|---|---|---|---|---|---|
| Empirical formula | C21H15Cl2MnN3 | C27H20Cl2MnN4 | C21H15Cl2MnN3O | C21H15Cl2MnN3O | C21H19Cl2MnN3O | C22H17Cl2MnN3O2S | C21H14N4O2Cl2Mn |
| Formula weight | 435.20 | 526.31 | 451.2 | 451.2 | 455.23 | 513.29 | 480.2 |
| Temperature | 123(2) K | 293(2) K | 293(2) K | 293(2) K | 293(2) K | 293(2) K | 150(2) K |
| Crystal system | Monoclinic | Triclinic | Monoclinic | Monoclinic | Triclinic | Monoclinic | Monoclinic |
| Space group | P21/n | P-1 | P21/n | P21/n | P-1 | P21/n | C2/c |
| a (Å) | 12.1515(5) | 8.6474(6) | 12.0365(11) | 12.2520(15) | 8.9693(18) | 10.2754(15) | 11.1314(16) |
| b (Å) | 9.7548(4) | 11.0270(7) | 9.8576(10) | 9.7098(10) | 10.584(2) | 18.416(2) | 22.013(3) |
| c (Å) | 16.6109(6) | 13.3718(10) | 17.1166(17) | 17.083(2) | 12.780(3) | 12.7472(17) | 8.6607(11) |
| α (°) | 90.00 | 85.629(3) | 90 | 90 | 103.63(3) | 90 | 90 |
| β (°) | 106.581(2) | 74.006(3) | 107.487(11) | 107.349(14) | 108.46(3) | 110.752(16) | 110.471(8) |
| γ (°) | 90.00 | 87.326(3) | 90 | 90 | 96.63(3) | 90 | 90 |
| Volume (Å3) | 1887.10(13) | 1221.70(15) | 1937.1(3) | 1939.8(4) | 1094.4(4) | 2255.7(5) | 1988.1(5) |
| Z | 4 | 2 | 4 | 4 | 2 | 4 | 4 |
| Calculated density (Mg/m3) |
1.532 | 1.431 | 1.547 | 1.545 | 1.382 | 1.511 | 1.604 |
| Absorption coefficient (mm−1) | 0.993 | 0.782 | 0.974 | 0.973 | 0.863 | 0.939 | 0.96 |
| F(000) | 884 | 538 | 916 | 916 | 466 | 1044 | 972 |
| Crystal size (mm−1) | 0.41 × 0.33 × 0.29 | 0.40 × 0.36 × 0.36 | 0.48 × 0.33 × 0.31 | 0.48 × 0.46 × 0.26 | 0.41 × 0.23 × 0.16 | 0.45 × 0.40 × 0.38 | 0.26 × 0.22 × 0.15 |
| θmax, θmin (°) | 30.69, 1.85 | 26.45, 2.36 | 29.40, 3.24 | 29.47, 3.21 | 29.27, 2.84 | 29.54, 2.80 | 30.07, 2.75 |
| Index range h | –17 → 17 | –10 → 10 | –16 → 14 | –15 → 14 | –8 → 12 | –13 → 13 | –11 → 15 |
| K | –13 → 13 | –13 → 13 | –13 → 12 | –13 → 10 | –14 → 13 | –25 → 17 | –27 → 30 |
| L | –23 → 23 | –16 → 16 | –23 → 22 | –16 → 23 | –17 → 16 | –16 → 17 | –11 → 12 |
| Reflections collected /unique |
36,786/5834 [R(int) = 0.0378] |
41,099/5022 [R(int) = 0.0555] |
10,821/4560 [R(int) = 0.0214] |
11,912/4640 [R(int) = 0.0228] |
9493/5049 [R(int) = 0.0164] |
12,816/5327 [R(int) = 0.0248] |
10,177/2910 [R(int) = 0.0558] |
| Data/restraints/parameters | 5834/0/244 | 5022/0/376 | 4560/0/254 | 4640/0/253 | 5049/0/287 | 5327/0/280 | 2910/0/177 |
| GOF on F2 | 1.034 | 1.003 | 1.031 | 1.002 | 1.034 | 1.030 | 1.031 |
| Final R indices [I>2σ(I)] |
R1 = 0.0248 wR2 = 0.0683 |
R1 = 0.0304 wR2 = 0.0804 |
R1 = 0.0314 wR2 = 0.0777 |
R1 = 0.0320 wR2 = 0.0801 |
R1 = 0.0368 wR2 = 0.0949 |
R1 = 0.0364 wR2 = 0.0957 |
R1 = 0.0492 wR2 = 0.1123 |
| R indices (all data) |
R1 = 0.0283 wR2 = 0.0704 |
R1 = 0.0424 wR2 = 0.0899 |
R1 = 0.0423 wR2 = 0.0866 |
R1 = 0.0457 wR2 = 0.0910 |
R1 = 0.0476 wR2 = 0.1046 |
R1 = 0.0540 wR2 = 0.1078 |
R1 = 0.0862 wR2 = 0.1296 |
| Largest diff. peak and hole (e Å–3) |
0.41 and −0.25 | 0.19 and −0.32 | 0.26 and −0.27 | 0.20 and −0.29 | 0.38 and −0.41 | 0.79 and −0.34 | 0.93 and −0.64 |
| CCDC number | 2062271 | 2062273 | 2062274 | 2062275 | 2062276 | 2062277 | 2062272 |
3.5. Antiproliferative Activity against Tumor Cells
Five different cell lines, human lung carcinoma cell line (A549), human hepatocellular carcinoma cell line (Bel-7402), human esophageal squamous carcinoma cell line (Eca-109), human cervix carcinoma cell line (HeLa) and human breast cancer cell line (MCF-7), purchased from the American Type Culture Collection (ATCC), were used to evaluate the antiproliferative activity of the synthesized compounds. All the cells were cultured with a completed Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 U/mL streptomycin in a humidified atmosphere at 37 °C with 5% CO2. The cells were seeded in 96-well plates with 3000 cells per well. After 12 h, series concentrations of compounds 1a–1c and 2a–2h were added into the predefined wells and then incubated for 48 h. Cell morphology was observed and imaged with an inverted microscope (Nikon eclipses TS100) equipped with a Nikon digital camera (DXM 1200F). The cell viability was measured using CCK-8 assay (cell counting kit-8, Beyotime Biotechnology, China) following the manufacturer’s instructions. The cell viability (% of control) was expressed as the percentage of (ODtest − ODblank)/(ODcontrol − ODblank). GraphPad Prism V5.0 for Windows (Graphpad Software, San Diego, CA, USA) was used to calculate the 50% inhibitive concentration (IC50) of the tested compounds against tumor cells.
3.6. Circular Dichroism (CD) Spectropolarimetry
Circular dichroism spectra (differential absorption of left and right circularly polarized light) of CT-DNA in the presence or absence of compounds 1a–1c and 2a–2h at different concentrations were recorded using a Chirascan spectropolarimeter (Applied Photophysics, UK). The CD measurements were carried out using 2 mm Suprasil quartz cells from Hellma Analytics and maintained at a temperature of 20 °C using a TC125 temperature controller from Quantum Northwestern running on the Chirascan spectrophotometer. The spectra were recorded between 230 and 400 nm, with a bandwidth of 1 nm and time per point of 1 s. The spectra of 5 mM Tris-HCl and 50 mM NaCl buffer (pH 7.2) were used as the baselines, and they were automatically subtracted from the CD spectra of the samples.
3.7. Molecular Docking
The coordination sphere of the manganese compounds was generated from their X-ray crystal structures as CIF files. Subsequently, the CIF file was converted to the PDB format using Mercury software (http://www.ccdc.cam.ac.uk/, accessed on 1 December 2022). The X-ray crystallographic structure of B-DNA dodecamer d(CGCGAATTCGCG)2 (PDB ID: 1BNA) and human DNA–topoisomerase I complex (PDB ID: 1SC7) were retrieved and modified from the protein Data Bank (https://www.rcsb.org/, accessed on 1 December 2022). Topo I is bound to the oligonucleotide sequence 5′-AAAAAGACTTsX-GAAAATTTTT-3′, where ‘s’ is 5′-bridging phosphorothiolate of the cleaved strand, and ‘X’ represents any of the four bases A, G, C or T. The SH of G11 on the scissile strand was changed to OH, and the phosphoester bond of G12 in 1SC7 was rebuilt [81]. A molecular docking study was performed with AutoDock Tools (ADT) version 1.5.6 and AutoDock version 4.2.6 programs while using the implemented empirical free energy function and the Lamarckian Genetic Algorithm. The structures of the receptors were kept rigid during the docking, whereas the metal compound was allowed to have rotatable bonds. Prior to performing docking, all of the water molecules were charged, and polar hydrogen atoms were added. The size of the grid was set to 80 × 80 × 120 or 80 × 80 × 80 with a spacing of 0.375 Å. All of the other parameters were kept as the defaults. Among them, the conformation having the lowest energy was selected to depict the mode of interaction between the compounds and DNA. The results were visualized using the PyMol Molecular Viewer package [82,83].
4. Conclusions
Eight oxygen-containing substituent-modified terpyridine manganese complexes and three non-oxygen-containing substituent-modified terpyridine manganese complexes were synthesized, and their structures were characterized by IR, elemental analysis and single-crystal X-ray diffraction. The in vitro cell viability studies illustrated the anticancer potential of the compounds, with the lowest IC50 values of 0.281 μM (2d) against A549, 0.523 μM (2h) against Bel-7402, 0.514 μM (2g) against Eca-109, 0.356 μM (2d) against HeLa and 0.249 μM (2c) against MCF-7. They were more active than cisplatin against the five tested cell lines and, compound 2g with a nitro group had the best antiproliferative activities against five cell lines on the whole. When comparing the complexes with different types of substituents, we found that complexes modified with oxygen-containing substituents showed lower IC50 values against all five cell lines. Then, the circular dichroism spectroscopy studies revealed a strong affinity between the compounds and the DNA in an intercalative mode. The CD spectra showed that the secondary structure of the DNA was changed by the addition of the compounds, the different substituents at the terpyridine significantly affect the interaction between the compound and the DNA, and oxygen-containing substituents seem to promote the intercalation. The molecular docking studies confirmed the interaction between the compounds and DNA or DNA–topoisomerase I complex, and the models showed that the interactions were stabilized by π–π stacking and hydrogen bonding. The results of these experiments indicated that both the steric and electrostatic effects of the compounds had strong influences on their interactions with DNA. It appears that less steric hindrance and higher electronegativity can enhance the DNA-binding affinity of the compounds. The correlation between anticancer activity and DNA-binding affinity suggests that DNA binding is a plausible mechanism. Notably, in this study and some published anticancer activity data of complexes with similar structures, the anticancer activity of the non-oxygen-containing substituent-modified terpyridine complexes was significantly lower than that of the terpyridine complexes modified with oxygen-containing substituents. Combined with the results of circular dichroism and molecular docking studies, we believe that the introduction of oxygen-containing substituents enhances the binding affinity to DNA through hydrogen bonding and π–π stacking, promotes intercalation and thus enhances their anticancer activity. Accordingly, the current results propose an effective strategy to enhance the anticancer activity of terpyridine complexes by introducing oxygen-containing substituents, which form a promising base for future in vitro and in vivo investigations of anticancer manganese metallodrugs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24043903/s1.
Author Contributions
Conceptualization, J.L.; Funding acquisition, H.C., L.P. and A.J.L.P.; Investigation, J.L., M.C., J.J. and J.H.; Methodology, J.L.; Project administration, Z.M.; Resources, H.C., L.P. and A.J.L.P.; Supervision, H.C., L.P., D.S.N., A.J.L.P. and Z.M.; Visualization, J.L.; Writing—original draft, J.L.; Writing—review and editing, H.C., L.P. and Z.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Crystallographic data for the structures reported in this manuscript have been deposited with the Cambridge Crystallographic Data Centre under the CCDC numbers: 2062271 (Compound 1a), 2062273 (Compound 1c), 2062274 (Compound 2a), 2062275 (Compound 2b), 2062276 (Compound 2c), 2062277 (Compound 2f) and 2062272 (Compound 2g). Copies of these data can be obtained free of charge from https://www.ccdc.cam.ac.uk/structures/, accessed on 1 December 2022.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the National Natural Science Foundation of China (grant no. 31860245, 21261002, 31660251 and 31960203), the Foundation for Science and Technology (FCT), Portugal (projects UIDB/00100/2020, UIDP/00100/2020 and LA/P/0056/2020 of Centro de Química Estrutural and contract IST-ID/086/2018), central government direct special funds for local science and technology development projects (ZY1949015) and the Guangxi University Overseas Training Program (2017-Jiahe Li). This publication has been supported by the RUDN University Strategic Academic Leadership Program (recipient A.J.L.P, preparation).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ferlay J., Ervik M., Lam F., Colombet M., Mery L., Piñeros M., Znaor A., Soerjomataram I., Bray F. Global Cancer Observatory: Cancer Today. International Agency for Research on Cancer; Lyon, France: 2018. [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D., Piñeros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 4.Rajput A., Osmani R.A.M., Singh E., Banerjee R. Biosensor Based Advanced Cancer Diagnostics. Elsevier; Amsterdam, The Netherlands: 2022. Cancer: A sui generis threat and its global impact; pp. 1–25. [Google Scholar]
- 5.Musiol R., Malecki P., Pacholczyk M., Mularski J. Terpyridines as promising antitumor agents: An overview of their discovery and development. Expert Opin. Drug Discov. 2022;17:259–271. doi: 10.1080/17460441.2022.2017877. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg B., Van Camp L., Krigas T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature. 1965;205:698–699. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- 7.Lebwohl D., Canetta R. Clinical development of platinum complexes in cancer therapy: An historical perspective and an update. Eur. J. Cancer. 1998;34:1522–1534. doi: 10.1016/S0959-8049(98)00224-X. [DOI] [PubMed] [Google Scholar]
- 8.Muller C.M., Babak M.V., Kubanik M., Hanif M., Jamieson S.M., Hartinger C.G., Wright L.J. Pt (II) pyridinium amidate (PYA) complexes: Preparation and in vitro anticancer activity studies. Inorg. Chim. Acta. 2016;450:124–130. doi: 10.1016/j.ica.2016.05.025. [DOI] [Google Scholar]
- 9.Musumeci D., Platella C., Riccardi C., Merlino A., Marzo T., Massai L., Messori L., Montesarchio D. A first-in-class and a fished out anticancer platinum compound: Cis-[PtCl 2 (NH 3) 2] and cis-[PtI 2 (NH 3) 2] compared for their reactivity towards DNA model systems. Dalton Trans. 2016;45:8587–8600. doi: 10.1039/c6dt00294c. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., Ye Z.-w., Tew K.D., Townsend D.M. Cisplatin chemotherapy and renal function. Adv. Cancer Res. 2021;152:305–327. doi: 10.1016/bs.acr.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medrano M.Á., Álvarez-Valdés A., Perles J., Lloret-Fillol J., Muñoz-Galván S., Carnero A., Navarro-Ranninger C., Quiroga A.G. Oxidation of anticancer Pt (II) complexes with monodentate phosphane ligands: Towards stable but active Pt (IV) prodrugs. Chem. Commun. 2013;49:4806–4808. doi: 10.1039/c3cc38416k. [DOI] [PubMed] [Google Scholar]
- 12.Infante I., Azpiroz J.M., Blanco N.G., Ruggiero E., Ugalde J.M., Mareque-Rivas J.C., Salassa L. Quantum dot photoactivation of Pt (IV) anticancer agents: Evidence of an electron transfer mechanism driven by electronic coupling. J. Phys. Chem. C. 2014;118:8712–8721. doi: 10.1021/jp501447q. [DOI] [Google Scholar]
- 13.Liu W., Jiang J., Xu Y., Hou S., Sun L., Ye Q., Lou L. Design, synthesis and anticancer activity of diam (m) ine platinum (II) complexes bearing a small-molecular cell apoptosis inducer dichloroacetate. J. Inorg. Biochem. 2015;146:14–18. doi: 10.1016/j.jinorgbio.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Ahmadi R., Urig S., Hartmann M., Helmke B.M., Koncarevic S., Allenberger B., Kienhoefer C., Neher M., Steiner H.-H., Unterberg A.J.F.R.B. Medicine, Antiglioma activity of 2, 2′:6′, 2″-terpyridineplatinum (II) complexes in a rat model—Effects on cellular redox metabolism. Free Radic. Biol. Med. 2006;40:763–778. doi: 10.1016/j.freeradbiomed.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 15.Darabi F., Hadadzadeh H., Simpson J., Shahpiri A. A water-soluble Pd (II) complex with a terpyridine ligand: Experimental and molecular modeling studies of the interaction with DNA and BSA; and in vitro cytotoxicity investigations against five human cancer cell lines. New J. Chem. 2016;40:9081–9097. doi: 10.1039/C6NJ01880G. [DOI] [Google Scholar]
- 16.Mahendiran D., Kumar R.S., Viswanathan V., Velmurugan D., Rahiman A.K. Targeting of DNA molecules, BSA/c-Met tyrosine kinase receptors and anti-proliferative activity of bis (terpyridine) copper (II) complexes. Dalton Trans. 2016;45:7794–7814. doi: 10.1039/C5DT03831F. [DOI] [PubMed] [Google Scholar]
- 17.Lazić D., Arsenijević A., Puchta R., Bugarčić Ž.D., Rilak A. DNA binding properties, histidine interaction and cytotoxicity studies of water soluble ruthenium (II) terpyridine complexes. Dalton Trans. 2016;45:4633–4646. doi: 10.1039/C5DT04132E. [DOI] [PubMed] [Google Scholar]
- 18.Milutinović M.M., Rilak A., Bratsos I., Klisurić O., Vraneš M., Gligorijević N., Radulović S., Bugarčić Ž.D. New 4′-(4-chlorophenyl)-2,2′:6′,2″-terpyridine ruthenium (II) complexes: Synthesis, characterization, interaction with DNA/BSA and cytotoxicity studies. J. Inorg. Biochem. 2017;169:1–12. doi: 10.1016/j.jinorgbio.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Yu C., Chan K.H.-Y., Wong K.M.-C., Yam V.W.-W. Nucleic acid-induced self-assembly of a platinum (II) terpyridyl complex: Detection of G-quadruplex formation and nuclease activity. Chem. Commun. 2009:3756–3758. doi: 10.1039/b903080h. [DOI] [PubMed] [Google Scholar]
- 20.Ma Z., Lu W., Liang B., Pombeiro A.J. Synthesis, characterization, photoluminescent and thermal properties of zinc (II) 4′-phenyl-terpyridine compounds. New J. Chem. 2013;37:1529–1537. doi: 10.1039/c3nj41176a. [DOI] [Google Scholar]
- 21.Becker K., Herold-Mende C., Park J.J., Lowe G., Schirmer R.H. Human thioredoxin reductase is efficiently inhibited by (2,2′:6′,2″-terpyridine) platinum (II) complexes. Possible implications for a novel antitumor strategy. J. Med. Chem. 2001;44:2784–2792. doi: 10.1021/jm001014i. [DOI] [PubMed] [Google Scholar]
- 22.Lowe G., Droz A.S., Vilaivan T., Weaver G.W., Park J.J., Pratt J.M., Tweedale L., Kelland L.R. Cytotoxicity of 2,2′:6′,2″-Terpyridineplatinum (II) Complexes against Human Ovarian Carcinoma. J. Med. Chem. 1999;42:3167–3174. doi: 10.1021/jm991053y. [DOI] [PubMed] [Google Scholar]
- 23.Lowe G., Droz A.S., Vilaivan T., Weaver G.W., Tweedale L., Pratt J.M., Rock P., Yardley V., Croft S.L. Cytotoxicity of (2,2′:6′,2″-Terpyridine) platinum (II) Complexes to Leishmania d onovani, Trypanosoma c ruzi, and Trypanosoma b rucei. J. Med. Chem. 1999;42:999–1006. doi: 10.1021/jm981074c. [DOI] [PubMed] [Google Scholar]
- 24.Bonse S., Richards J.M., Ross S.A., Lowe G., Krauth-Siegel R.L. (2,2′:6′,2″-Terpyridine) platinum (II) Complexes Are Irreversible Inhibitors of Trypanosoma c ruzi Trypanothione Reductase But Not of Human Glutathione Reductase. J. Med. Chem. 2000;43:4812–4821. doi: 10.1021/jm000219o. [DOI] [PubMed] [Google Scholar]
- 25.McFadyen W.D., Wakelin L.P., Roos I.A., Leopold V.A. Activity of platinum (II) intercalating agents against murine leukemia L1210. J. Med. Chem. 1985;28:1113–1116. doi: 10.1021/jm00146a026. [DOI] [PubMed] [Google Scholar]
- 26.Chakrabarti S., Bhattacharyya D., Dasgupta D. Structural basis of DNA recognition by anticancer antibiotics, chromomycin A3, and mithramycin: Roles of minor groove width and ligand flexibility. Biopolym. Orig. Res. Biomol. 2000;56:85–95. doi: 10.1002/1097-0282(2000)56:2<85::AID-BIP1054>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Bugreev D., Buneva V., Sinitsyna O., Nevinsky G. The Mechanism of the Supercoiled DNA Recognition by the Eukaryotic Type I Topoisomerases: I. The Enzyme Interaction with Nonspecific Oligonucleotides. Russ. J. Bioorg. Chem. 2003;29:143–153. doi: 10.1023/A:1023208431839. [DOI] [PubMed] [Google Scholar]
- 28.Jacquemard U., Routier S., Dias N., Lansiaux A., Goossens J.-F., Bailly C., Mérour J.-Y. Synthesis of 2, 5-and 3, 5-diphenylpyridine derivatives for DNA recognition and cytotoxicity. Eur. J. Med. Chem. 2005;40:1087–1095. doi: 10.1016/j.ejmech.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Zimbron J.M., Sardo A., Heinisch T., Wohlschlager T., Gradinaru J., Massa C., Schirmer T., Creus M., Ward T.R. Chemo-Genetic Optimization of DNA Recognition by Metallodrugs using a Presenter-Protein Strategy. Chem.—Eur. J. 2010;16:12883–12889. doi: 10.1002/chem.201001573. [DOI] [PubMed] [Google Scholar]
- 30.Lin C., Mathad R.I., Zhang Z., Sidell N., Yang D. Solution structure of a 2: 1 complex of anticancer drug XR5944 with TFF1 estrogen response element: Insights into DNA recognition by a bis-intercalator. Nucleic Acids Res. 2014;42:6012–6024. doi: 10.1093/nar/gku219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong K.M.-C., Yam V.W.-W. Luminescence platinum (II) terpyridyl complexes—From fundamental studies to sensory functions. Coord. Chem. Rev. 2007;251:2477–2488. doi: 10.1016/j.ccr.2007.02.003. [DOI] [Google Scholar]
- 32.Wong K.M.-C., Yam V.W.-W. Self-assembly of luminescent alkynylplatinum (II) terpyridyl complexes: Modulation of photophysical properties through aggregation behavior. Acc. Chem. Res. 2011;44:424–434. doi: 10.1021/ar100130j. [DOI] [PubMed] [Google Scholar]
- 33.Liang X., Jiang J., Xue X., Huang L., Ding X., Nong D., Chen H., Pan L., Ma Z. Synthesis, characterization, photoluminescence, anti-tumor activity, DFT calculations and molecular docking with proteins of zinc (II) halogen substituted terpyridine compounds. Dalton Trans. 2019;48:10488–10504. doi: 10.1039/C8DT04924F. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y., Zhou P., Liang B., Huang L., Zhou Y., Ma Z. Effects of counterions of colorful sandwich-type zinc (II) 4′-phenyl-terpyridine compounds on photoluminescent and thermal properties. J. Mol. Struct. 2017;1146:504–511. doi: 10.1016/j.molstruc.2017.05.129. [DOI] [Google Scholar]
- 35.Xue X., Wang Q., Mai F., Liang X., Huang Y., Li J., Zhou Y., Yang D., Ma Z. Study on the Photoluminescent and Thermal Properties of Zinc Complexes with a N6O4 Macrocyclic Ligand. Molecules. 2018;23:1735. doi: 10.3390/molecules23071735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H., Liu C.-S., Bu X.-H., Yang M. Synthesis, crystal structure, cytotoxic activity and DNA-binding properties of the copper (II) and zinc (II) complexes with 1-[3-(2-pyridyl) pyrazol-1-ylmethyl] naphthalene. J. Inorg. Biochem. 2005;99:1119–1125. doi: 10.1016/j.jinorgbio.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Sheng X., Guo X., Lu X.-M., Lu G.-Y., Shao Y., Liu F., Xu Q. DNA binding, cleavage, and cytotoxic activity of the preorganized dinuclear zinc (II) complex of triazacyclononane derivatives. Bioconjugate Chem. 2008;19:490–498. doi: 10.1021/bc700322w. [DOI] [PubMed] [Google Scholar]
- 38.Ma Z., Cao Y., Li Q., da Silva M.F.C.G., da Silva J.J.F., Pombeiro A.J. Synthesis, characterization, solid-state photo-luminescence and anti-tumor activity of zinc (II) 4′-phenyl-terpyridine compounds. J. Inorg. Biochem. 2010;104:704–711. doi: 10.1016/j.jinorgbio.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Zhou P., Huang L., Zhang Y., Xue X., Zhou Y., Ma Z. Synthesis, characterization and photoluminescence of substituted terpyridine compounds and their molecular docking studies with bovine hemoglobin. J. Photochem. Photobiol. A Chem. 2018;358:17–25. doi: 10.1016/j.jphotochem.2018.02.028. [DOI] [Google Scholar]
- 40.Huang L., Liu R., Li J., Liang X., Lan Q., Shi X., Pan L., Chen H., Ma Z. Synthesis, characterization, anti-tumor activity, photo-luminescence and BHb/HHb/Hsp90 molecular docking of zinc (II) hydroxyl-terpyridine complexes. J. Inorg. Biochem. 2019;201:110790. doi: 10.1016/j.jinorgbio.2019.110790. [DOI] [PubMed] [Google Scholar]
- 41.Li J., Liu R., Jiang J., Liang X., Huang L., Huang G., Chen H., Pan L., Ma Z. Zinc (II) Terpyridine Complexes: Substituent Effect on Photoluminescence, Antiproliferative Activity, and DNA Interaction. Molecules. 2019;24:4519. doi: 10.3390/molecules24244519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang J., Li J., Liu C., Liu R., Liang X., Zhou Y., Pan L., Chen H., Ma Z. Study on the substitution effects of zinc benzoate terpyridine complexes on photoluminescence, antiproliferative potential and DNA binding properties. JBIC J. Biol. Inorg. Chem. 2020;25:311–324. doi: 10.1007/s00775-020-01763-6. [DOI] [PubMed] [Google Scholar]
- 43.Li J., Liu R., Jiang J., Liang X., Huang G., Yang D., Chen H., Pan L., Ma Z. Synthesis, characterization, photoluminescence, antiproliferative activity, and DNA interaction of cadmium (II) substituted 4′-phenyl-terpyridine compounds. J. Inorg. Biochem. 2020;210:111165. doi: 10.1016/j.jinorgbio.2020.111165. [DOI] [PubMed] [Google Scholar]
- 44.Liu R., Yan H., Jiang J., Li J., Liang X., Yang D., Pan L., Xie T., Ma Z. Synthesis, Characterization, Photoluminescence, Molecular Docking and Bioactivity of Zinc (II) Compounds Based on Different Substituents. Molecules. 2020;25:3459. doi: 10.3390/molecules25153459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Z., Xing Y., Yang M., Hu M., Liu B., da Silva M.F.C.G., Pombeiro A.J. The double-helicate terpyridine silver (I) compound [Ag2L2](SO3CF3) 2 (L= 4′-phenyl-terpyridine) as a building block for di-and mononuclear complexes. Inorg. Chim. Acta. 2009;362:2921–2926. doi: 10.1016/j.ica.2009.01.018. [DOI] [Google Scholar]
- 46.Li Z., Yan H., Liu K., Huang X., Niu M. Syntheses, structures, DNA/BSA binding and cytotoxic activity studies of chiral alcohol-amine Schiff base manganese (II/III) complexes. J. Mol. Struct. 2019;1195:470–478. doi: 10.1016/j.molstruc.2019.05.110. [DOI] [Google Scholar]
- 47.Reddy A.S., Mao J., Krishna L.S., Badavath V.N., Maji S. Synthesis, spectral investigation, molecular docking and biological evaluation of Cu (II), Ni (II) and Mn (II) complexes of (E)-2-((2-butyl-4-chloro-1H-imidazol-5-yl) methylene)-N-methylhydrazinecarbothioamide (C10H16N5ClS) and its DFT studies. J. Mol. Struct. 2019;1196:338–347. doi: 10.1016/j.molstruc.2019.06.085. [DOI] [Google Scholar]
- 48.Khalaf-Alla P.A. Antioxidant, Antimicrobial and Antitumor Studies of Transition Metal Complexes Derived from N-(2-Aminoethyl)-1, 3-Propanediamine with DFT Calculations and Molecular Docking Investigation. Appl. Organomet. Chem. 2020;34:e5628. doi: 10.1002/aoc.5628. [DOI] [Google Scholar]
- 49.Calladine C.R., Drew H. Understanding DNA: The Molecule and How It Works. Academic Press; Cambridge, MA, USA: 1997. [Google Scholar]
- 50.Valente A., Podolski-Renić A., Poetsch I., Filipović N., López Ó., Turel I., Heffeter P. Metal-and metalloid-based compounds to target and reverse cancer multidrug resistance. Drug Resist. Updates. 2021;58:100778. doi: 10.1016/j.drup.2021.100778. [DOI] [PubMed] [Google Scholar]
- 51.Makovec T. Cisplatin and beyond: Molecular mechanisms of action and drug resistance development in cancer chemotherapy. Radiol. Oncol. 2019;53:148–158. doi: 10.2478/raon-2019-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung Y., Lippard S.J. Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 53.Wang L.-D., Zheng K., Li Y.-T., Wu Z.-Y., Yan C.-W. Synthesis and crystal structure of a new copper (II) complex with N, N′-(4, 4′-bithiazole-2, 2′-diyl) diacetimidamide as ligand: Molecular docking, DNA-binding and cytotoxicity activity studies. J. Mol. Struct. 2013;1037:15–22. doi: 10.1016/j.molstruc.2012.11.035. [DOI] [Google Scholar]
- 54.Maity B., Roy M., Banik B., Majumdar R., Dighe R.R., Chakravarty A.R. Ferrocene-promoted photoactivated DNA cleavage and anticancer activity of terpyridyl copper (II) phenanthroline complexes. Organometallics. 2010;29:3632–3641. doi: 10.1021/om100524x. [DOI] [Google Scholar]
- 55.García-Giménez J.L., Hernández-Gil J., Martínez-Ruíz A., Castiñeiras A., Liu-González M., Pallardó F.V., Borrás J., Piña G.A. DNA binding, nuclease activity, DNA photocleavage and cytotoxic properties of Cu (II) complexes of N-substituted sulfonamides. J. Inorg. Biochem. 2013;121:167–178. doi: 10.1016/j.jinorgbio.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Li G.-Y., Du K.-J., Wang J.-Q., Liang J.-W., Kou J.-F., Hou X.-J., Ji L.-N., Chao H. Synthesis, crystal structure, DNA interaction and anticancer activity of tridentate copper (II) complexes. J. Inorg. Biochem. 2013;119:43–53. doi: 10.1016/j.jinorgbio.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 57.Lee S.K., Tan K.W., Ng S.W. Topoisomerase I inhibition and DNA cleavage by zinc, copper, and nickel derivatives of 2-[2-bromoethyliminomethyl]-4-[ethoxymethyl] phenol complexes exhibiting anti-proliferation and anti-metastasis activity. J. Inorg. Biochem. 2016;159:14–21. doi: 10.1016/j.jinorgbio.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Neves A.P., Pereira M.X., Peterson E.J., Kipping R., Vargas M.D., Silva-Jr F.P., Carneiro J.W.M., Farrell N.P. Exploring the DNA binding/cleavage, cellular accumulation and topoisomerase inhibition of 2-hydroxy-3-(aminomethyl)-1, 4-naphthoquinone Mannich bases and their platinum (II) complexes. J. Inorg. Biochem. 2013;119:54–64. doi: 10.1016/j.jinorgbio.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 59.Lo Y.-C., Ko T.-P., Su W.-C., Su T.-L., Wang A.H.-J. Terpyridine–platinum (II) complexes are effective inhibitors of mammalian topoisomerases and human thioredoxin reductase 1. J. Inorg. Biochem. 2009;103:1082–1092. doi: 10.1016/j.jinorgbio.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 60.Kwon H.-B., Park C., Jeon K.-H., Lee E., Park S.-E., Jun K.-Y., Kadayat T.M., Thapa P., Karki R., Na Y. A series of novel terpyridine-skeleton molecule derivants inhibit tumor growth and metastasis by targeting topoisomerases. J. Med. Chem. 2015;58:1100–1122. doi: 10.1021/jm501023q. [DOI] [PubMed] [Google Scholar]
- 61.Ma Z., Zhang B., Da Silva M.F.C.G., Silva J., Mendo A.S., Baptista P.V., Fernandes A.R., Pombeiro A.J. Synthesis, characterization, thermal properties and antiproliferative potential of copper (II) 4′-phenyl-terpyridine compounds. Dalton Trans. 2016;45:5339–5355. doi: 10.1039/C5DT02744F. [DOI] [PubMed] [Google Scholar]
- 62.Son J.-K., Zhao L.-X., Basnet A., Thapa P., Karki R., Na Y., Jahng Y., Jeong T.C., Jeong B.-S., Lee C.-S. Synthesis of 2, 6-diaryl-substituted pyridines and their antitumor activities. Eur. J. Med. Chem. 2008;43:675–682. doi: 10.1016/j.ejmech.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 63.Sun Y., Li J., Zhao H., Tan L. Ruthenium (II) polypyridyl complexes with 1, 8-naphthalimide group as DNA binder, photonuclease, and dual inhibitors of topoisomerases I and IIα. J. Inorg. Biochem. 2016;163:88–94. doi: 10.1016/j.jinorgbio.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 64.Long X.-J., Dai J.-W., Wu J.-Z. Synthesis and supramolecular networks of mono-and dinuclear manganese chloride complexes with 2-(2, 2′: 6′, 2 ”-terpyridin-4′-yl) phenol. J. Coord. Chem. 2012;65:316–324. doi: 10.1080/00958972.2011.652623. [DOI] [Google Scholar]
- 65.Li J., Yan H., Wang Z., Liu R., Luo B., Yang D., Chen H., Pan L., Ma Z. Copper chloride complexes with substituted 4′-phenyl-terpyridine ligands: Synthesis, characterization, antiproliferative activities and DNA interactions. Dalton Trans. 2021;50:8243–8257. doi: 10.1039/D0DT03989F. [DOI] [PubMed] [Google Scholar]
- 66.Chen H., Tagore R., Das S., Incarvito C., Faller J., Crabtree R.H., Brudvig G.W. General synthesis of di-μ-oxo dimanganese complexes as functional models for the oxygen evolving complex of photosystem II. Inorg. Chem. 2005;44:7661–7670. doi: 10.1021/ic0509940. [DOI] [PubMed] [Google Scholar]
- 67.Addison A.W., Rao T.N., Reedijk J., van Rijn J., Verschoor G.C. Synthesis, structure, and spectroscopic properties of copper (II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua [1, 7-bis (N-methylbenzimidazol-2′-yl)-2, 6-dithiaheptane] copper (II) perchlorate. J. Chem. Soc. Dalton Trans. 1984:1349–1356. doi: 10.1039/DT9840001349. [DOI] [Google Scholar]
- 68.Berova N., Nakanishi K., Woody R.W. Circular Dichroism: Principles and Applications. John Wiley & Sons; Hoboken, NJ, USA: 2000. [Google Scholar]
- 69.Nordén B., Kurucsev T. Analysing DNA complexes by circular and linear dichroism. J. Mol. Recognit. 1994;7:141–155. doi: 10.1002/jmr.300070211. [DOI] [PubMed] [Google Scholar]
- 70.Garbett N.C., Ragazzon P.A., Chaires J.B. Circular dichroism to determine binding mode and affinity of ligand–DNA interactions. Nat. Protoc. 2007;2:3166–3172. doi: 10.1038/nprot.2007.475. [DOI] [PubMed] [Google Scholar]
- 71.Gray D.M., Ratliff R.L., Vaughan M.R. [19] Circular dichroism spectroscopy of DNA. Methods Enzymol. 1992;211:389–406. doi: 10.1016/0076-6879(92)11021-a. [DOI] [PubMed] [Google Scholar]
- 72.Kashanian S., Askari S., Ahmadi F., Omidfar K., Ghobadi S., Tarighat F.A. In vitro study of DNA interaction with clodinafop-propargyl herbicide. DNA Cell Biol. 2008;27:581–586. doi: 10.1089/dna.2008.0740. [DOI] [PubMed] [Google Scholar]
- 73.Dehkordi M.N., Bordbar A.-K., Lincoln P., Mirkhani V. Spectroscopic study on the interaction of ct-DNA with manganese Salen complex containing triphenyl phosphonium groups. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012;90:50–54. doi: 10.1016/j.saa.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 74.Rohs R., Bloch I., Sklenar H., Shakked Z. Molecular flexibility in ab initio drug docking to DNA: Binding-site and binding-mode transitions in all-atom Monte Carlo simulations. Nucleic Acids Res. 2005;33:7048–7057. doi: 10.1093/nar/gki1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tabassum S., Zaki M., Afzal M., Arjmand F. Synthesis and characterization of Cu (II)-based anticancer chemotherapeutic agent targeting topoisomerase Iα: In vitro DNA binding, pBR322 cleavage, molecular docking studies and cytotoxicity against human cancer cell lines. Eur. J. Med. Chem. 2014;74:509–523. doi: 10.1016/j.ejmech.2013.12.046. [DOI] [PubMed] [Google Scholar]
- 76.Agilent C.P. CrysAlis PRO. Agilent Technologies; Yarnton, England: 2012. [Google Scholar]
- 77.Sheldrick G.M. SADABS Program for Empirical Absorption Correction for Area Detector Data. University of Göttingen; Göttingen, Germany: [Google Scholar]
- 78.Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sheldrick G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 80.Putz H., Brandenburg K. Diamond-Crystal and Molecular Structure Visualization. Crystal Impact-GbR, Kreuzherrenstr; Bonn, Germany: 2018. [Google Scholar]
- 81.Xiao X., Antony S., Pommier Y., Cushman M. On the Binding of Indeno [1, 2-c] isoquinolines in the DNA− Topoisomerase I Cleavage Complex. J. Med. Chem. 2005;48:3231–3238. doi: 10.1021/jm050017y. [DOI] [PubMed] [Google Scholar]
- 82.DeLano W. PyMOL Molecular Viewer. DeLano Scientific LLC; Palo Alto, CA, USA: 2008. [Google Scholar]
- 83.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.—Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallographic data for the structures reported in this manuscript have been deposited with the Cambridge Crystallographic Data Centre under the CCDC numbers: 2062271 (Compound 1a), 2062273 (Compound 1c), 2062274 (Compound 2a), 2062275 (Compound 2b), 2062276 (Compound 2c), 2062277 (Compound 2f) and 2062272 (Compound 2g). Copies of these data can be obtained free of charge from https://www.ccdc.cam.ac.uk/structures/, accessed on 1 December 2022.