Abstract
In most low-G+C gram-positive bacteria, the phosphoryl carrier protein HPr of the phosphoenolpyruvate:sugar phosphotransferase system (PTS) becomes phosphorylated at Ser-46. This ATP-dependent reaction is catalyzed by the bifunctional HPr kinase/P-Ser-HPr phosphatase. We found that serine-phosphorylated HPr (P-Ser-HPr) of Lactococcus lactis participates not only in carbon catabolite repression of an operon encoding a β-glucoside-specific EII and a 6-P-β-glucosidase but also in inducer exclusion of the non-PTS carbohydrates maltose and ribose. In a wild-type strain, transport of these non-PTS carbohydrates is strongly inhibited by the presence of glucose, whereas in a ptsH1 mutant, in which Ser-46 of HPr is replaced with an alanine, glucose had lost its inhibitory effect. In vitro experiments carried out with L. lactis vesicles had suggested that P-Ser-HPr is also implicated in inducer expulsion of nonmetabolizable homologues of PTS sugars, such as methyl β-d-thiogalactoside (TMG) and 2-deoxy-d-glucose (2-DG). In vivo experiments with the ptsH1 mutant established that P-Ser-HPr is not necessary for inducer expulsion. Glucose-activated 2-DG expulsion occurred at similar rates in wild-type and ptsH1 mutant strains, whereas TMG expulsion was slowed in the ptsH1 mutant. It therefore seems that P-Ser-HPr is not essential for inducer expulsion but that in certain cases it can play an indirect role in this regulatory process.
HPr is one of the four proteins (or domains) forming the phosphorylation cascade of the phosphoenolpyruvate (PEP):sugar phosphotransferase system (PTS), which in gram-positive and gram-negative bacteria catalyzes the uptake and phosphorylation of numerous carbohydrates (for a review, see reference 28). During PTS-mediated carbohydrate uptake and phosphorylation, HPr becomes phosphorylated by PEP and enzyme I at the Nδ1 position of His-15. P-His-HPr transfers its phosphoryl group to one of several sugar-specific EIIAs usually present in bacterial cells. P∼EIIAs donate their phosphoryl group to the corresponding EIIB, from where the phosphoryl group is finally transferred to the carbohydrate bound to the membrane-integrated EIIC. After phosphorylation by P∼EIIB, the phosphorylated sugar is released into the cytoplasm. In enzyme I and EIIAs, the phosphoryl group is attached to the Nɛ2 position of a histidyl residue, whereas in EIIBs the phosphoryl group can be bound either to the Nδ1 position of a histidyl residue or to a cysteyl residue (28).
In gram-positive bacteria, HPr functions not only as a phosphoryl carrier within the PTS phosphorylation cascade but also as the central regulator of carbohydrate metabolism. For example, P∼His-HPr phosphorylates not only EIIAs but also histidyl residues in non-PTS proteins such as glycerol kinase (6), antiterminators, transcriptional activators (32), and non-PTS transporters (containing an EIIAGlc domain) (17). In most cases, P∼His-HPr-mediated phosphorylation of non-PTS proteins leads to a stimulation of their activity. In addition, HPr of gram-positive bacteria is also phosphorylated at the regulatory serine-46 (9, 11). This reaction requires ATP and is catalyzed by the metabolite-controlled bifunctional HPr kinase/P-Ser-HPr phosphatase (5, 12, 14, 19, 21, 29). The resulting P-Ser-HPr functions as a corepressor in carbon catabolite repression (CCR) or as a coactivator in carbon catabolite activation (CCA) by interacting with catabolite control protein A (CcpA) (8, 20), a member of the LacI/GalR repressor family (18). The P-Ser-HPr/CcpA complex binds to specific operator sites (13) called catabolite response elements (cre) (39). The cre's of catabolite-activated genes or operons are located in front of the promoter. By contrast, in the case of catabolite-repressed genes and operons, the cre's either overlap the promoter or are located downstream of it (for reviews see references 7 and 33).
Based on in vitro results, P-Ser-HPr has been suggested to participate also in inducer exclusion in Lactobacillus brevis (44, 47). This concept was further supported by in vivo experiments with a Streptococcus salivarius Ile47Thr ptsH mutant (16), which had lost the preferential uptake and metabolism of glucose over lactose. The participation of P-Ser-HPr in inducer exclusion has been established by in vivo experiments with Lactobacillus casei ptsH1 and hprK mutants, which are not able to form P-Ser-HPr. Maltose uptake, which was completely inhibited by glucose in a wild-type strain, was not affected by glucose in the ptsH1 (37) and hprK mutants (12).
In vitro results had suggested that P-Ser-HPr would also participate in inducer expulsion in Lactococcus lactis (42, 43). Addition of glucose to cells which had accumulated the nonmetabolizable methyl β-d-thiogalactoside (TMG) or 2-deoxy-d-glucose (2-DG) caused rapid expulsion of the nonmetabolizable sugar analogues from L. lactis wild-type cells (35) or vesicles (43). Both sugar analogues are taken up by the PTS and are therefore accumulated as 6-P derivatives. In the first step of inducer expulsion, an intracellular sugar-P phosphohydrolase dephosphorylates the accumulated P-sugars before the unphosphorylated sugars are expelled from the cells in the second step (30). In several gram-positive bacteria, including L. lactis, a sugar-P phosphohydrolase has been described which was activated by P-Ser-HPr (41, 45) and which was thought to catalyze the first step of inducer expulsion. In addition, electroporation of Bacillus subtilis Ser46Asp mutant HPr, which structurally resembles P-Ser-HPr (40), into L. lactis vesicles was reported to lead to stronger inducer expulsion than electroporation of Ser46Ala mutant HPr (42). It was therefore concluded that P-Ser-HPr would participate in inducer expulsion. However, recent in vivo experiments with L. casei ptsH1 and hprK mutants had established that in this organism inducer expulsion does not require P-Ser-HPr. Following the addition of glucose, ptsH1 and hprK mutants, which are not able to form P-Ser-HPr, expelled preaccumulated TMG at a rate similar to that observed with an L. casei wild-type strain (12, 37).
We here report the construction of an L. lactis ptsH1 mutant strain synthesizing Ser46Ala mutant HPr with the aim to test whether P-Ser-HPr-dependent inducer exclusion is a widespread phenomenon in gram-positive bacteria and is present also in L. lactis. In addition, we carried out in vivo expulsion experiments with TMG and 2-DG in order to test whether P-Ser-HPr participates in this regulatory process, as has been proposed based on the reported stimulation of a sugar-P phosphohydrolase by P-Ser-HPr (45) and on in vitro expulsion experiments carried out with L. lactis vesicles, into which purified B. subtilis wild-type and mutant HPrs had been electroporated (42, 43).
MATERIALS AND METHODS
Strains, culture conditions, and transformation procedures.
L. lactis MG5267, an MG1363 derivative (15) carrying the lactose operon integrated in the chromosome (36), was used in this study. MG5267 and its derivatives were grown at 30°C under static conditions in M17 medium supplemented with either 0.5 or 0.8% of the indicated carbohydrates. Escherichia coli NM522 (Stratagene) was used as a host for the cloning experiments. It was grown in Luria-Bertani medium at 37°C under agitation. The antibiotics chloramphenicol and erythromycin were used at a concentration of 5 μg/ml for L. lactis. For E. coli, chloramphenicol was used at 10 μg/ml and ampicillin was used at 100 μg/ml. Solid media were prepared by adding 1.5% agar to the liquid media. For α-complementation in E. coli, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside was used at a concentration of 20 μg/ml. For the electroporation experiments with L. lactis cells, strains were grown in M17 medium supplemented with 0.5% glucose, 0.5 M sucrose, and 1% glycine to an optical density at 600 nm (OD600) of 0.5. The cells were subsequently washed twice with cold 0.5 M sucrose containing 10% glycerol and finally resuspended in this solution (1% of initial volume). A 50-μl aliquot was electroporated at 2.5 kV, 200 Ω, and 25 μF in cuvettes with a 0.2-cm distance between the electrodes (Bio-Rad Gene-Pulser). Five milliliters of M17 medium containing 0.5% glucose, 0.5 M sucrose, 20 mM MgCl2, and 2 mM CaCl2 was rapidly added to the electroporated cells, which were subsequently incubated for 2 h at 30°C before aliquots were plated on selective media.
Construction of plasmids and strains.
Chromosomal DNA from L. lactis MG5267 was isolated as previously described (24) and was used to amplify by PCR a 3-kb DNA fragment containing the complete ptsHI operon and its upstream sequence. The PCR was carried out with Pfu DNA polymerase (Promega) and oligonucleotides PTS3 (5′-GACCTGCAGTACAAAGTTATC-3′) and PTS4 (5′-TAAGGATCCTATTATAGCTAAACAG-3′) as primers. The resulting 3-kb DNA fragment was digested with BamHI (restriction site indicated in italics in PTS4) and cloned into SmaI-BamHI-digested pNEB193 (New England Biolabs). In order to replace serine 46 in HPr with an alanine, the obtained plasmid pTSHI was used as a template in a PCR amplification together with the two divergent primers S46A (5′-CCTTAAAGCAATCATGGGTGT-3′) and S46A2 (5′-TTTACTGATTTACCTTTGTAT-3′). The altered codon 46 leading to the Ser46Ala replacement in ptsH (TCA to GCA) is underlined in the sequence of primer S46A. The resulting PCR product was phosphorylated with T4 polynucleotide kinase, ligated, and used to transform E. coli NM522. The presence of the ptsH1 mutation (Ser46Ala replacement) and the absence of other mutations were verified by sequencing the insert of plasmids isolated from several clones using a Perkin-Elmer Abiprism 373 automated sequencer. One plasmid, called pTS46A, carried the correct mutant ptsH1 allele and wild-type ptsI and was used for further experiments. Plasmid pTS46A was digested with KpnI and made blunt ended with Klenow DNA polymerase. After digestion with BamHI, the 3-kb fragment obtained was cloned into the thermosensitive pGhostCm vector (2) digested with EcoRV and BamHI, thus providing pGhostS46A.
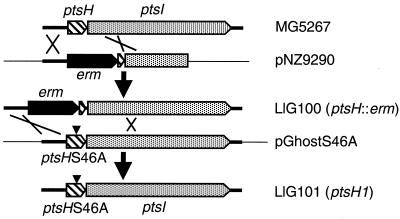
Strain LlG100 (ptsH::erm) was constructed by transforming L. lactis MG5267 with pNZ9290 (26) and selecting for erythromycin-resistant clones (Fig. 1). Strains carrying the plasmid inserted by a double crossover, which leads to the inactivation of ptsH, were identified by their inability to grow on PTS sugars, and in one such strain, LlG100, the insertion of the erythromycin resistance cassette into ptsH was confirmed by PCR amplification with appropriate primers.
FIG. 1.
Construction of an L. lactis strain expressing Ser46Ala mutant HPr. The wild-type strain MG5267 was transformed with plasmid pNZ9290 (26), which contains the 5′ part of the L. lactis ptsl gene and several hundred base pairs of the upstream region of the ptsHI operon. The ptsH gene located in front of ptsI is partly deleted and the deleted region is replaced with an erythromycin resistance cassette. After a double-crossover recombination, a ptsH::erm strain (LlG100) was obtained. This strain exhibited a pts-negative phenotype and was transformed with the thermosensitive plasmid pGhostS46A, which carries the ptsH1 allele (the position of the Ser46Ala mutation is indicated with a triangle). After two successive recombination events, a pts+ strain carrying the ptsH1 allele (LlG101) was obtained.
For the construction of the chromosomal ptsH1 mutant LlG101, LlG100 was transformed with pGhostS46A (Fig. 1). One chloramphenicol-resistant transformant was grown overnight at 38°C in the absence of antibiotics and subsequently plated on chloramphenicol-containing solid M17 medium and incubated at 38°C. Since pGhost is not replicated at 38°C, this procedure forced the integration of pGhostS46A into the chromosome of LlG100 by homologous recombination. One strain resistant to chloramphenicol and erythromycin during growth at 38°C was subsequently grown for several generations at 30°C in the absence of antibiotics and then plated on M17 medium, incubated at 30°C, and replica plated on M17 medium containing chloramphenicol and erythromycin. LlG101 was selected as a chloramphenicol- and erythromycin-sensitive strain in which the disrupted ptsH gene was replaced with the ptsH1 mutant allele by a second crossover event. The presence of this mutation in LlG101 was confirmed by sequencing appropriate PCR products.
Western blotting.
L. lactis strains were grown in 25 ml of glucose-containing M17 medium to an OD600 between 0.6 and 0.7 before the pH of the culture was rapidly lowered to 4.5 by adding concentrated HCl. After centrifugation at 4°C, the cell pellets were resuspended in 1 ml of 20 mM sodium acetate, pH 4.5, and cells were broken in a Fast-prep apparatus (Biospec) using 0.1-mm glass beads and three cycles of 30 s at maximum speed. The low pH and temperature were used to minimize changes in the HPr phosphorylation state potentially caused by enzyme I and HPr kinase/P-Ser-HPr phosphatase present in the extracts. The cell lysates were clarified by centrifugation, and proteins were separated on a 15% nondenaturing polyacrylamide gel. After electrophoretic transfer of the proteins onto nitrocellulose membranes, the blots were probed with rabbit polyclonal antibodies raised against B. subtilis HPr and developed by using anti-rabbit immunoglobulin G conjugated with alkaline phosphatase (Promega) as a second antibody.
β-Glucoside transport and 6-P-β-glucosidase assays.
Strains MG5267 and LlG101 were grown in 20 ml of M17 medium supplemented with either 0.8% salicin or 0.8% salicin plus 0.8% glucose to an OD600 between 0.4 and 0.5. Cells grown in medium containing only salicin exhibited an about 1.8-fold slower growth rate than cells grown on salicin plus glucose. The cells were washed twice with 50 mM Tris-HCl buffer, pH 7.4, and resuspended in 200 μl of the same buffer. 6-P-β-Glucosidase activities were determined in 50-μl assay mixtures containing 50 mM Tris-HCl, pH 7.4, 40 μl of the cell suspension, 5 mM p-nitrophenyl-β-d-glucopyranoside, and 5 mM MgCl2. The assay mixtures were incubated for 30 min at 30°C before the reaction was stopped by adding 800 μl of 10% sodium carbonate. After centrifugation, the OD405 was measured in the samples. Control experiments carried out with salicin-grown wild-type cells confirmed that there was a linear correlation between the measured OD405 and either the incubation time or the amount of cells used for the assay. Enzyme activities are expressed in nanomoles of p-nitrophenol formed per minute per milliliter of cell culture exhibiting an OD600 of 0.5. To determine whether glucose exerts an exclusion effect on p-nitrophenyl-β-d-glucopyranoside uptake, the above-described assay was carried out with salicin-grown wild-type and ptsH1 mutant cells in the presence of 10 mM glucose.
Sugar transport, inducer exclusion, and inducer expulsion.
Sugar transport studies and inducer exclusion experiments in the presence of 10 mM glucose were performed using the rapid-filtration method (37). Cells used for transport studies were grown in M17 medium containing different carbohydrates (at a concentration of 0.5%). Glucose-promoted expulsion experiments with cells which had accumulated the lactose analogue TMG or the glucose analogue 2-DG were carried out as previously described (12). 14C-radiolabeled sugars were purchased from Isotopchim (Ganagobie-Peyrus, France) and used at a final concentration of 1 mM (at a specific radioactivity of 0.5 mCi/mmol).
Thin-layer chromatography was used to separate phosphorylated and nonphosphorylated [14C]TMG. After cells had taken up [14C]TMG, they were washed twice with 1 ml of transport buffer before 10 mM glucose was added to one-half of the suspension (500 μl) and expulsion was allowed to proceed for 5 min. Subsequently, cells were kept for 10 min at 100°C and clarified by centrifugation, and 10-μl aliquots were separated by thin-layer chromatography on Silica Gel 60 plates (Merck) using a mixture of 1 M ammonium acetate, pH 5, 98% ethanol, and 0.1 M EDTA, pH 8 (70:29:1), as the solvent. The approximate amounts of TMG and TMG-6-P were determined by autoradiography (4 days of exposure with a Biomax MR film [Kodak]). The migration position of TMG was determined with untreated [14C]TMG.
RESULTS
Construction of an L. lactis ptsH1 mutant strain.
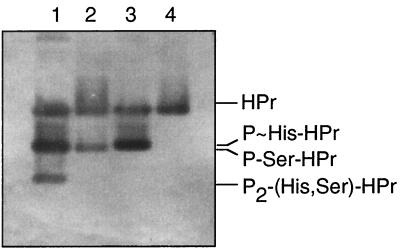
To study the role of P-Ser-HPr in the regulation of carbon metabolism in L. lactis, we constructed a ptsH1 mutant strain in which the phosphorylatable Ser-46 of HPr was replaced with an alanine. The L. lactis ptsHI operon encoding enzyme I and HPr of the PTS has recently been cloned and characterized (26). After insertion of an erythromycin resistance gene at the 3′ end of the ptsH gene of strain MG5267, the antibiotic resistance cassette and the wild-type ptsH were replaced with the Ser46Ala ptsH allele (ptsH1) present on the integrative plasmid pGhostS46A as described in Materials and Methods (Fig. 1). The expected absence of P-Ser-HPr in the resulting ptsH1 mutant strain LlG101 was confirmed by Western blotting. Polyacrylamide gel electrophoresis was performed under nondenaturing conditions with crude extracts prepared from the L. lactis wild-type and the ptsH1 mutant strains grown in glucose-containing M17 medium. This allowed us to separate HPr, P∼His-HPr/P-Ser-HPr, and doubly phosphorylated HPr and to get an estimate of their ratios in the cell. The approximate amounts of the different forms of HPr were detected with polyclonal antibodies directed against B. subtilis HPr. Heating an aliquot of the crude extract to 65°C allowed us to distinguish between P∼His-HPr and P-Ser-HPr, which migrate to nearly identical positions. P∼His-HPr is rapidly hydrolyzed at 65°C (38), whereas P-Ser-HPr is stable under these conditions. According to the results presented in lanes 1 and 2 of Fig. 2, glucose-grown L. lactis wild-type cells were estimated to contain a considerable amount of P-Ser-HPr and somewhat less HPr and P∼His-HPr, whereas only a small amount of doubly phosphorylated HPr was present. By contrast, glucose-grown ptsH1 mutant cells contained neither P-Ser-HPr nor doubly phosphorylated HPr and the major part of HPr was present as P∼His-HPr (Fig. 2, lanes 3 and 4).
FIG. 2.
Western blot with L. lactis crude extracts prepared from glucose-grown wild-type and ptsH1 mutant strains and separated on a nondenaturing polyacrylamide gel. The various forms of HPr were detected with antibodies raised against B. subtilis HPr. Crude extracts from L. lactis wild-type MG5267 (lanes 1 and 2) and L. lactis ptsH1 mutant LlG101 (lanes 3 and 4) are shown. Extracts separated in lanes 2 and 4 were heated for 10 min at 65°C before they were loaded onto the gel.
Glucose transport in L. lactis wild-type and ptsH1 mutant strains and glucose-mediated exclusion of PTS sugars.
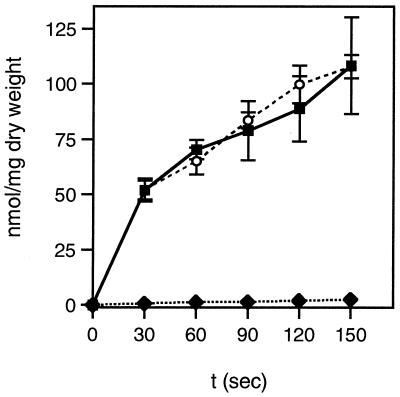
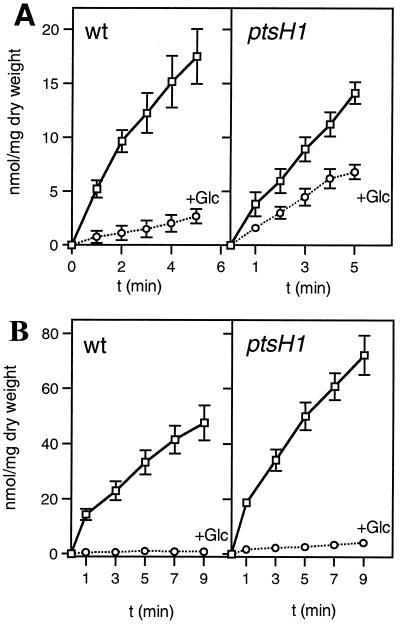
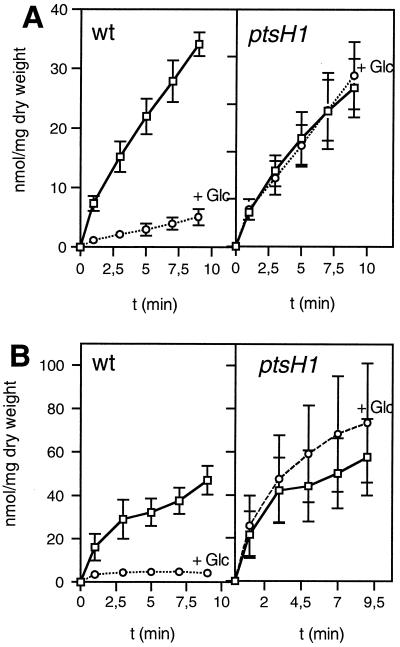
Since the main function of HPr is to act as phosphocarrier protein during PTS-catalyzed sugar transport and phosphorylation, we tested whether the ptsH1 mutation would influence PTS-catalyzed sugar uptake. Glucose transport activities were found to be very similar for the wild-type and the ptsH1 mutant strains (Fig. 3). By contrast, the ptsH disruption strain LlG100 had completely lost the capacity to transport glucose at the concentration used in the transport assay (1 mM). However, strain LlG100 was able to slowly grow in M17 medium containing 25 mM glucose, indicating the presence of a non-PTS transporter capable of transporting glucose with low affinity. Slow growth of an L. lactis ptsH strain on glucose has also been reported by Luesink et al. (26). In L. lactis MG5267, TMG is taken up by a lactose-specific chromosome-encoded PTS and accumulated as TMG-6-P (see Fig. 6). Compared to the wild-type strain, PTS-catalyzed TMG uptake by the ptsH1 mutant was slightly slower (Fig. 4A). In addition, the inhibition exerted by glucose on TMG uptake in the wild-type strain (sevenfold) was much weaker in the ptsH1 mutant (only about twofold).
FIG. 3.
Transport of [14C]glucose (1 mM) by the L. lactis wild-type strain MG5267 (filled squares), the ptsH::erm disruption strain LlG100 (filled rhombs), and the ptsH1 mutant LlG101 (open circles). Cells were grown in M17 medium containing 0.5% glucose.
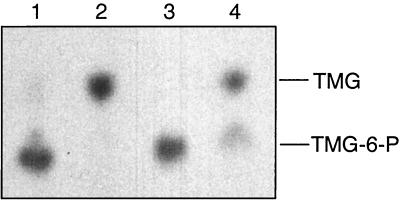
FIG. 6.
Autoradiogram showing the amounts of [14C]TMG and [14C]TMG-6-P present in L. lactis cells and in the medium before and after inducer expulsion. [14C]TMG and [14C]TMG-6-P were separated by thin-layer chromatography. Lanes 1 and 3, [14C]TMG-6-P accumulated in wild-type and ptsH1 mutant cells; lanes 2 and 4, [14C]TMG present in cells and in the medium after 5 min of expulsion. Expulsion experiments were carried out with the wild-type strain MG5267 (lanes 1 and 2) and the ptsH1 mutant LlG101 (lanes 3 and 4). The cells were grown in 0.5% lactose-containing M17 medium.
FIG. 4.
Transport of 14C-labeled TMG and mannitol and their exclusion by glucose in the L. lactis MG5267 (wild-type [wt]) and LlG101 (ptsH1 mutant) strains. (A) TMG transport with cells grown in M17 medium containing 0.5% lactose; (B) mannitol transport with cells grown in M17 medium containing 0.5% mannitol. Transport assays were carried out in the absence of glucose (squares) or with 10 mM glucose added 1 min prior to adding the radiolabeled sugar (circles). In panel B, the error bars for the experiments carried out in the presence of glucose were too small to be drawn by the program.
The inability of strain LlG100 to grow on mannitol suggested that this sugar might also be transported by the PTS. This assumption was supported by the finding that the genome of L. lactis IL1403 contains an operon (mtlARFD) encoding proteins with high sequence similarity to EIICBMtl, MtlR, EIIAMtl, and mannitol-1-P dehydrogenase from other organisms (3, 4) (see also http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/framik?db=Genome&gi=171). Compared to the wild-type strain, mannitol uptake via the PTS was almost twofold greater in the ptsH1 mutant. Interestingly, the presence of glucose completely inhibited mannitol uptake in the wild-type strain. Glucose inhibition of mannitol transport was only slightly relieved in the ptsH1 mutant strain (Fig. 4B).
CCR of aryl-β-d-glucoside metabolism.
L. lactis strain IL1403 was found to be capable of growing on aryl-β-d-glucosides such as esculin, salicin, and arbutin (1). Similarly, the wild-type strain MG5267 was able to grow on salicin, whereas the ptsH::erm strain LlG100 had lost this capacity, confirming that in L. Lactis aryl-β-d-glucosides are transported and phosphorylated by a PTS before they are split by a 6-P-β-d-glucosidase into glucose-6-P and the aglycon. The EII necessary for aryl-β-d-glucoside transport by MG5267 cells is probably encoded by the homologue of the ptbA gene located at kb 1482 of the L. lactis IL1403 chromosome (3, 4). This EIIBCA exhibits strong similarity to the EIIBCABgl (BglP) of B. subtilis (23). p-Nitrophenyl-β-d-glucopyranoside was found to be a substrate for the PtbA of L. lactis, since it was taken up by salicin-grown MG5267 cells and subsequently split into glucose-6-P and p-nitrophenol. These activities were repressed sixfold in cells grown in the presence of salicin and glucose (Table 1). The repressive effect of glucose had disappeared in the ptsH1 mutant strain. p-Nitrophenyl-β-d-glucopyranoside transport and hydrolysis experiments carried out in the presence of glucose with salicin-grown wild-type and ptsH1 mutant cells showed that glucose exerts no inducer exclusion effect on p-nitrophenyl-β-d-glucopyranoside uptake. The presence of glucose in the assay mixtures even stimulated p-nitrophenyl-β-d-glucopyranoside uptake and its subsequent hydrolysis about 1.5-fold in both wild-type and ptsH1 mutant strains (Table 1).
TABLE 1.
CCR of 6-P-β-glucosidase
| Strain | Mean ± SD of 6-P-β-glucosidase activitya for cells grown in M17 medium with:
|
||
|---|---|---|---|
| Salb | Sal + Glcb | Sal (Glc)c | |
| MG5267 WTd | 98 ± 7 | 16 ± 5 | 168 ± 12 |
| LIG101 ptsH1 | 104 ± 12 | 112 ± 9 | 177 ± 9 |
6-P-β-glucosidase activity is expressed in nanomoles of p-nitrophenol formed per minute and milliliter of cell culture with an OD600 of 0.5. Results from three experiments are presented.
Sal, salicin; Sal + Glc, salicin plus glucose.
6-P-β-glucosidase assays were carried out with salicin-grown cells with glucose (10 mM) present in the assay mixture.
WT, wild type.
The ptsH1 mutation prevents inducer exclusion of non-PTS sugars maltose and ribose.
In L. lactis, maltose and ribose are probably taken up by ATP-binding cassette (ABC) transport systems (MalE, MalF, and MalG; and RbsA, RbsC, and RbsD, respectively) (3, 4). For unknown reasons, the ptsH disruption strain LlG100 was unable to grow on ribose, although it grew normally on maltose. In the wild-type strain, the uptake of both carbohydrates was strongly inhibited when glucose was present during the transport reaction (Fig. 5A and B), suggesting that an inducer exclusion mechanism was operative. Interestingly, the inhibitory effect of glucose on the uptake of non-PTS carbohydrates ribose and maltose had disappeared in the ptsH1 mutant LlG101 (Fig. 5A and B), although it transported glucose at a rate identical to that observed with the wild-type strain (Fig. 3).
FIG. 5.
Transport of the 14C-labeled non-PTS sugars ribose (A) and maltose (B) and their exclusion by 10 mM glucose in L. lactis MG5267 (wild-type) and LlG101 (ptsH1 mutant) strains. Transport assays were carried out in the absence of glucose (squares) or with 10 mM glucose added 1 min prior to adding the radiolabeled sugar (circles). Cells were grown in M17 medium containing 0.5% ribose (A) or 0.5% maltose (B).
P-Ser-HPr is not essential for inducer expulsion in L. lactis.
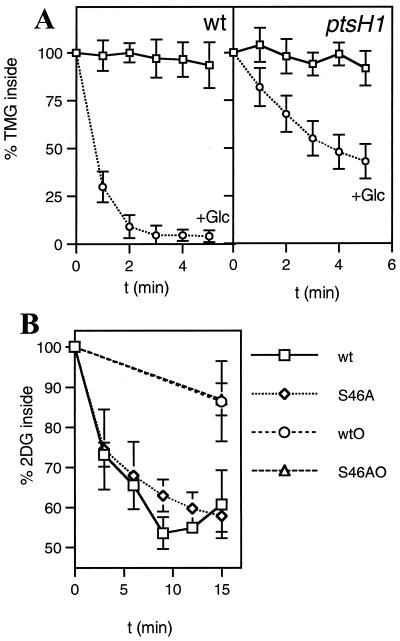
In vitro results obtained with L. lactis vesicles had suggested that P-Ser-HPr participates in inducer expulsion by stimulating the activity of a sugar-P phosphohydrolase catalyzing the first step of inducer expulsion (42, 43). To test whether P-Ser-HPr is indeed implicated in this regulatory process, we measured TMG expulsion in an L. lactis wild-type strain and a ptsH1 mutant strain. L. lactis strain MG5267 takes up [14C]TMG via the lactose-specific PTS and accumulates it as [14C]TMG-6-P, which cannot be further metabolized (Fig. 6, lane 1). When glucose was added to MG5267 cells preloaded with [14C]TMG-6-P, the nonmetabolizable sugar was rapidly expelled from the cells (Fig. 7A). In the first step, the presence of glucose has been shown to initiate the intracellular dephosphorylation of TMG-6-P (30) before TMG is expelled from the cells in the second step, probably via EIICBLac (31). In agreement with this model, only TMG and no TMG-6-P was found to be expelled from MG5267 cells preloaded with [14C]TMG-6-P (Fig. 6, lane 2). The ptsH1 mutant was also capable of accumulating [14C]TMG-6-P (Fig. 6, lane 3). However, expulsion of [14C]TMG occurred at a significantly slower rate and was not yet completed after 5 min (Fig. 7A). Similar to what was observed with the wild-type strain, [14C]TMG-6-P was dephosphorylated during the expulsion process (Fig. 6, lane 4). After 5 min of incubation in the presence of glucose, about two-thirds of the accumulated [14C]TMG-6-P was expelled from the ptsH1 mutant and dephosphorylated, whereas one-third remained in the cell as [14C]TMG-6-P (Fig. 7A).
FIG. 7.
Expulsion of accumulated [14C]TMG-6-P (A) and [14C]2-DG-6-P (B) in the L. lactis wild-type strain MG5267 and the ptsH1 mutant LlG101. Cells grown in the presence of 0.5% lactose or 0.5% glucose were preloaded with [14C]TMG or [14C]2-DG, respectively. The amount of labeled sugar remaining inside the cells during a 5-min (for TMG) or 15-min (for 2-DG) incubation period at 37°C in the presence or absence of glucose was determined by withdrawing aliquots at the indicated time intervals and analyzing them by the rapid-filtration method (37). Squares, no sugar added; circles, 10 mM glucose added at time zero. S46AO and wtO, no glucose added to cells preloaded with [14C]2-DG-6-P. For the latter samples, aliquots were withdrawn only at the beginning and at the end of the experiments. Leakage levels of [14C]2-DG-6-P from the cells were found to be nearly identical for the wild-type and the ptsH1 mutant strains, explaining why the lines for the two strains coincide.
Expulsion experiments were also carried out with cells which had taken up [14C]2-DG. Like TMG, [14C]2-DG is accumulated by L. lactis cells as the phospho derivative. After glucose was added, [14C]2-DG-6-P was first intracellularly dephosphorylated and subsequently expelled as unphosphorylated [14C]2-DG (34). Compared to TMG expulsion, glucose-activated expulsion of [14C]2-DG occurred at a slower rate (Fig. 7B). But almost no difference of [14C]2-DG expulsion could be observed between L. lactis wild-type and ptsH1 mutant strains.
DISCUSSION
HPr is the major regulator of carbon metabolism in gram-positive bacteria. The implication of P-Ser-HPr in CCR and CCA has been well established for B. subtilis (7), Staphylococcus xylosus (19), and L. casei (12, 37) and has also been suggested for L. lactis. Expression of the L. lactis las operon encoding several glycolytic enzymes was stimulated by the presence of glucose, whereas activation of las operon expression was absent in a ccpA mutant (27) as well as a ptsH disruption mutant transformed with a plasmid containing the ptsH1 allele (encoding Ser46Ala HPr) (26). The participation of P-Ser-HPr in catabolite regulation was confirmed by constructing a chromosomal ptsH1 mutant. Like B. subtilis, L. lactis possesses an aryl-β-glucoside-specific EII (PtbA) and a 6-P-β-glucosidase (BglH) (1, 4). In the wild-type strain, the synthesis of these enzymes was strongly repressed by glucose, but it was relieved from CCR in the ptsH1 mutant. In B. subtilis, the bgl operon is regulated by two CCR mechanisms (22): one involves P-Ser-HPr/CcpA and a cre present in the promoter region, and the other involves the antiterminator LicT, which is activated by P∼His-HPr-mediated phosphorylation (25). BglR, a homologue of LicT, is controlling β-glucoside metabolism in L. lactis (1), and a potential cre is preceding the ptbA gene encoding EIICBABgl (4), suggesting that CCR mechanisms similar to those described for the B. subtilis bgl operon might be operative for the ptbA-bglH operon in L. lactis.
We observed that in L. lactis glucose exerted a strong exclusion or expulsion effect on other PTS carbohydrates. The uptake of TMG and mannitol was almost completely inhibited when glucose was present. A much weaker inhibitory effect of glucose or fructose on mannitol uptake has been observed with B. subtilis (10, 46). In the latter organism, competition for the common phosphoryl donor P∼His-HPr seemed to be the reason for this inhibitory effect, since inhibition of mannitol transport was almost completely relieved in a B. subtilis ptsH1 mutant (10). Since Ser46A mutant HPr cannot be phosphorylated by HprK/P, a ptsH1 mutant contains more P∼His-HPr for the phosphoryl group transfer within the PTS phosphorylation cascade than does a wild-type strain (Fig. 2). TMG uptake in L. lactis seems to be regulated in a manner similar to that of mannitol uptake in B. subtilis, since the inhibitory effect of glucose on TMG uptake was much weaker in a ptsH1 mutant. The slowed TMG expulsion in the ptsH1 mutant (Fig. 7A) could also be responsible for the weaker inhibitory effect of glucose on TMG-6-P accumulation. Glucose-mediated inhibition of mannitol transport in L. lactis follows a different mechanism, since glucose exerted similarly strong inhibitory effects on mannitol transport in both wild-type strain MG5267 and ptsH1 mutant LlG101.
P-Ser-HPr has recently been shown to participate in inducer exclusion of non-PTS carbohydrates in gram-positive bacteria. The strong inhibitory effect of glucose on the uptake of the non-PTS sugar maltose observed with L. casei wild-type cells was absent in ptsH1 and hprK mutants (12, 37). In order to find out whether this recently established mechanism of inducer exclusion of non-PTS sugars, which so far has not been detected in B. subtilis, is widespread within gram-positive bacteria, we tested whether it was also operative in L. lactis. We found that in L. lactis the uptake of maltose and ribose was subject to inducer exclusion. The two corresponding transport activities were strongly inhibited by the presence of glucose. Similar to what is observed for L. casei, inducer exclusion in L. lactis is mediated via P-Ser-HPr, since the strong repressive effect of glucose had completely disappeared in the ptsH1 mutant. Interestingly, both non-PTS transport systems submitted to inducer exclusion in L. lactis are ABC transporters (3, 4). Since it is likely that in L. casei maltose is also taken up by an ABC transporter, there arises the question of whether P-Ser-HPr-mediated inducer exclusion in gram-positive bacteria affects only ABC transporters.
In previous reports, a participation of P-Ser-HPr in inducer expulsion by L. lactis has been suggested, whereas the results obtained during this study argue against an implication of P-Ser-HPr in this regulatory process. In the previous inducer expulsion studies, B. subtilis wild-type or Ser46Asp mutant HPr and glycolytic intermediates were electroporated into L. lactis vesicles, and the results suggested an involvement of P-Ser-HPr in the expulsion of both TMG and 2-DG (42, 43). P-Ser-HPr has been proposed to stimulate the first step of inducer expulsion, the dephosphorylation of accumulated nonmetabolizable PTS sugars, since it has been reported to activate in vitro a P-sugar phosphohydrolase (45). In contrast, in vivo experiments carried out with L. casei wild-type, ptsH1, and hprK mutant strains had allowed us to establish that in this organism P-Ser-HPr does not participate in inducer expulsion (12, 37). An almost identical expulsion of TMG was observed in the L. casei wild-type strain and the two mutant strains unable to form P-Ser-HPr. Constructing a ptsH1 mutant was expected to allow us to either confirm or refute the proposed participation of P-Ser-HPr in inducer expulsion of L. lactis. Since in the L. lactis ptsH1 mutant, 2-DG expulsion by glucose was not affected and TMG expulsion was still operative, although at a lower rate than in the wild-type strain, P-Ser-HPr is not necessary for inducer expulsion. A P-Ser-HPr-independent inducer expulsion mechanism must be operative in L. lactis and L. casei (12, 37) and probably in other gram-positive organisms. In the studies suggesting a role of P-Ser-HPr in inducer expulsion, the use of L. lactis vesicles, the electroporation of HPr and metabolites into these vesicles, and the use of a heterologous system (B. subtilis wild-type or Ser46Asp mutant HPr instead of L. lactis HPr or P-Ser-HPr) could have yielded misleading results. In comparison, the results obtained in this study seem to be more reliable, since the experiments were carried out in vivo with intact cells of an L. lactis wild-type strain and a ptsH1 mutant. The latter strain synthesized HPr, which was active in PTS transport but which, due to the replacement of Ser-46 with an alanine, could not be phosphorylated by HprK/P. Nevertheless, according to the Western blots (Fig. 2), the total amounts of the various forms of HPr present in the two strains were very similar.
What could have been the reason for the results of the vesicle studies suggesting an involvement of P-Ser-HPr in inducer expulsion? Ser46Asp mutant HPr might not exactly mimic P-Ser-HPr, and by using electroporation, the amount of HPr present in the vesicles was probably difficult to control. In addition, the reduced rate of TMG expulsion observed with the L. lactis ptsH1 mutant suggested an indirect role of HPr in this regulatory process. Evidence has previously been provided that expulsion of TMG in Streptococcus pyogenes is catalyzed by EIICBLac (31). In a wild-type strain growing in glucose-containing medium, only about 25% of the HPr is present as P∼His-HPr. Under these conditions, EIICBLac might be unphosphorylated or only slightly phosphorylated. By contrast, in a ptsH1 mutant growing in glucose-containing medium, the about threefold-greater amount of P∼His-HPr might allow a more efficient phosphorylation of EIICBLac. Phosphorylated EIICBLac probably does not expel TMG from the cells but rather transports and rephosphorylates it and might therefore be responsible for the slowed TMG expulsion observed with the ptsH1 mutant. Elevated phosphorylation of EIICBLac in vesicles, into which Ser46Ala mutant HPr had been electroporated, might also be the reason why these vesicles exhibited reduced inducer expulsion. By contrast, coelectroporation of glycolytic intermediates with either wild-type HPr, which under the experimental conditions employed was partly converted to P-Ser-HPr in the vesicles (42), or Ser46Asp mutant HPr, which like P-Ser-HPr is very slowly phosphorylated at His-15 by enzyme I and PEP, probably led to inefficient phosphorylation of EIICBLac and therefore caused no reduction or only a slight reduction of TMG expulsion.
ACKNOWLEDGMENTS
This research was supported by the CNRS, the INRA, the INA-PG, and the MENRT. V. Monedero was a recipient of an FPU fellowship from the Ministerio de Educación y Cultura of Spain.
We thank Manuel Zúñiga for providing us with a protocol for electroporation of L. lactis cells.
REFERENCES
- 1.Bardowski J, Ehrlich S D, Chopin A. BglR protein, which belongs to the BglG family of transcriptional antiterminators, is involved in β-glucoside utilization in Lactococcus lactis. J Bacteriol. 1994;176:5681–5685. doi: 10.1128/jb.176.18.5681-5685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas I, Gruss A, Ehrlich S D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. The complete genome sequence of the lactic acid bacterium Lactococcus lactis. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 5.Brochu D, Vadeboncoeur C. The HPr(Ser) kinase of Streptococcus salivarius: purification, properties, and cloning of the hprK gene. J Bacteriol. 1999;181:709–717. doi: 10.1128/jb.181.3.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charrier V, Buckley E, Parsonage D, Galinier A, Darbon E, Jaquinod M, Forest E, Deutscher J, Claiborne A. Cloning and sequencing of two enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate dependent, phosphotransferase system-catalyzed phosphorylation of a single histidyl residue. J Biol Chem. 1997;272:14166–14174. doi: 10.1074/jbc.272.22.14166. [DOI] [PubMed] [Google Scholar]
- 7.Deutscher, J., A. Galinier, and I. Martin-Verstraete. Carbohydrate transporters and regulation of carbohydrate uptake and metabolism. In A. L. Sonenschein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its relatives: from genes to cells, in press. American Society for Microbiology, Washington, D.C.
- 8.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 9.Deutscher J, Pevec B, Beyreuther K, Kiltz H-H, Hengstenberg W. Streptococcal phosphoenolpyruvate-sugar phosphotransferase system: amino acid sequence and site of ATP-dependent phosphorylation of HPr. Biochemistry. 1986;25:6543–6551. doi: 10.1021/bi00369a031. [DOI] [PubMed] [Google Scholar]
- 10.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutscher J, Saier M H., Jr ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci USA. 1983;80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dossonnet V, Monedero V, Zagorec M, Galinier A, Pérez-Martínez G, Deutscher J. Phosphorylation of HPr by the bifunctional HPr kinase/P-Ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion, but not inducer expulsion. J Bacteriol. 2000;182:2582–2590. doi: 10.1128/jb.182.9.2582-2590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 14.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M-C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;151:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauthier M, Brochu D, Eltis L D, Thomas S, Vadeboncoeur C. Replacement of isoleucine-47 by threonine in the HPr protein of Streptococcus salivarius abrogates the preferential metabolism of glucose and fructose over lactose and melibiose but does not prevent the phosphorylation of HPr on serine-46. Mol Microbiol. 1997;25:695–705. doi: 10.1046/j.1365-2958.1997.4981870.x. [DOI] [PubMed] [Google Scholar]
- 17.Gunnewijk M G W, Postma P W, Poolman B. Phosphorylation and functional properties of the IIA domain of the lactose transport protein of Streptococcus thermophilus. J Bacteriol. 1999;181:632–641. doi: 10.1128/jb.181.2.632-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 19.Huynh P L, Jankovic I, Schnell N F, Brückner R. Characterization of an HPr kinase mutant of Staphylococcus xylosus. J Bacteriol. 2000;182:1895–1902. doi: 10.1128/jb.182.7.1895-1902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones B E, Dossonnet V, Küster E, Hillen W, Deutscher J, Klevit R E. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1997;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 21.Kravanja M, Engelmann R, Dossonnet V, Blüggel M, Meyer H E, Frank R, Galinier A, Deutscher J, Schnell N, Hengstenberg W. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol Microbiol. 1999;31:59–66. doi: 10.1046/j.1365-2958.1999.01146.x. [DOI] [PubMed] [Google Scholar]
- 22.Krüger S, Gertz S, Hecker M. Transcriptional analysis of bglPH expression in Bacillus subtilis: evidence for two distinct pathways mediating carbon catabolite repression. J Bacteriol. 1996;178:2637–2644. doi: 10.1128/jb.178.9.2637-2644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Coq D, Lindner C, Krüger S, Steinmetz M, Stülke J. New β-glucoside (bgl) genes in Bacillus subtilis: the bglP gene product has both transport and regulatory functions similar to those of BglF, its Escherichia coli homolog. J Bacteriol. 1995;177:1527–1535. doi: 10.1128/jb.177.6.1527-1535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leenhouts K J, Kok J, Venema G. Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl Environ Microbiol. 1990;56:2726–2735. doi: 10.1128/aem.56.9.2726-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindner C, Galinier A, Hecker M, Deutscher J. Regulation of the activity of the Bacillus subtilis antiterminator LicT by multiple PEP-dependent, enzyme I- and HPr-catalysed phosphorylation. Mol Microbiol. 1999;31:995–1006. doi: 10.1046/j.1365-2958.1999.01262.x. [DOI] [PubMed] [Google Scholar]
- 26.Luesink E J, Beumer C M A, Kuipers O P, De Vos W M. Molecular characterization of the Lactococcus lactis ptsHI operon and analysis of the regulatory role of HPr. J Bacteriol. 1999;181:764–771. doi: 10.1128/jb.181.3.764-771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luesink E J, van Harpen R E M A, Grossiord B P, Kuipers O P, de Vos W M. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 28.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reizer J, Bachem S, Reizer A, Arnaud M, Saier M H, Jr, Stülke J. Novel phosphotransferase system genes revealed by genome analysis—the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology. 1999;145:3419–3429. doi: 10.1099/00221287-145-12-3419. [DOI] [PubMed] [Google Scholar]
- 30.Reizer J, Novotny M J, Panos C, Saier M H., Jr Mechanism of inducer expulsion in Streptococcus pyogenes: a two-step process activated by ATP. J Bacteriol. 1983;156:354–361. doi: 10.1128/jb.156.1.354-361.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reizer J, Saier M H., Jr Involvement of lactose Enzyme II of the phosphotransferase system in rapid expulsion of free galactosides from Streptococcus pyogenes. J Bacteriol. 1983;156:236–242. doi: 10.1128/jb.156.1.236-242.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 33.Stülke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annu Rev Microbiol. 2000;54:849–880. doi: 10.1146/annurev.micro.54.1.849. [DOI] [PubMed] [Google Scholar]
- 34.Thompson J, Chassy B M. Novel phosphoenolpyruvate-dependent futile cycle in Streptococcus lactis: 2-deoxyglucose uncouples energy production from growth. J Bacteriol. 1982;151:1454–1465. doi: 10.1128/jb.151.3.1454-1465.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J, Saier M H., Jr Regulation of methyl-β-d-thiogalactopyranoside-6-phosphate accumulation in Streptococcus lactis by exclusion and expulsion mechanisms. J Bacteriol. 1981;146:885–894. doi: 10.1128/jb.146.3.885-894.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rooijen R J, de Vos W M. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J Biol Chem. 1990;265:18499–18503. [PubMed] [Google Scholar]
- 37.Viana R, Monedero V, Dossonnet V, Vadeboncoeur C, Perez-Martinez G, Deutscher J. Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol Microbiol. 2000;36:570–584. doi: 10.1046/j.1365-2958.2000.01862.x. [DOI] [PubMed] [Google Scholar]
- 38.Waygood E B, Erickson E, El-Kabbani O A L, Delbaere L T J. Characterization of phosphorylated histidine-containing protein (HPr) of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Biochemistry. 1985;24:6938–6945. doi: 10.1021/bi00345a028. [DOI] [PubMed] [Google Scholar]
- 39.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wittekind M, Reizer J, Deutscher J, Saier M H, Klevit R E. Common structural changes accompany the functional inactivation of HPr by seryl phosphorylation or by serine to aspartate substitution. Biochemistry. 1989;28:9908–9912. doi: 10.1021/bi00452a005. [DOI] [PubMed] [Google Scholar]
- 41.Ye J-J, Minarcik J, Saier M H., Jr Inducer expulsion and the occurrence of an HPr(Ser-P)-activated sugar-phosphate phosphatase in Enterococcus faecalis and Streptococcus pyogenes. Microbiology. 1996;142:585–592. doi: 10.1099/13500872-142-3-585. [DOI] [PubMed] [Google Scholar]
- 42.Ye J-J, Reizer J, Cui X, Saier M H., Jr Inhibition of the phosphoenolpyruvate:lactose phosphotransferase system and activation of a cytoplasmic sugar-phosphate phosphatase in Lactococcus lactis by ATP-dependent metabolite-activated phosphorylation of serine 46 in the phosphocarrier protein HPr. J Biol Chem. 1994;269:11837–11844. [PubMed] [Google Scholar]
- 43.Ye J-J, Reizer J, Saier M H., Jr Regulation of 2-deoxyglucose phosphate accumulation in Lactococcus lactis vesicles by metabolite-activated, ATP-dependent phosphorylation of serine-46 in HPr of the phosphotransferase system. Microbiology. 1994;140:3421–3429. doi: 10.1099/13500872-140-12-3421. [DOI] [PubMed] [Google Scholar]
- 44.Ye J-J, Saier M H., Jr Allosteric regulation of the glucose:H+ symporter of Lactobacillus brevis: cooperative binding of glucose and HPr(ser-P) J Bacteriol. 1995;177:1900–1902. doi: 10.1128/jb.177.7.1900-1902.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J-J, Saier M H., Jr Purification and characterization of a small membrane-associated sugar phosphate phosphatase that is allosterically activated by HPr(Ser-P) of the phosphotransferase system in Lactococcus lactis. J Biol Chem. 1995;270:16740–16744. doi: 10.1074/jbc.270.28.16740. [DOI] [PubMed] [Google Scholar]
- 46.Ye J J, Saier M H., Jr Regulation of sugar uptake via the phosphoenolpyruvate-dependent phosphotransferase systems in Bacillus subtilis and Lactococcus lactis is mediated by ATP-dependent phosphorylation of seryl residue 46 in HPr. J Bacteriol. 1996;178:3557–3563. doi: 10.1128/jb.178.12.3557-3563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye J-J, Reizer J, Cui X, Saier M H., Jr ATP-dependent phosphorylation of serine-46 in the phosphocarrier protein HPr regulates lactose/H+ symport in Lactobacillus brevis. Proc Natl Acad Sci USA. 1994;91:3102–3106. doi: 10.1073/pnas.91.8.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]