Significance
Gastrinreleasing peptide receptor (GRPR) is an attractive target for the cancer and itch therapy. We present an inactive-state crystal structure of GRPR complexed with non-peptide antagonist PD176252 and two active-state cryo-EM structures of GRPR complexed with natural peptide agonist GRP and synthetic BBN analog [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14). Our work shows the interactions critical for different ligands binding receptor activation, and Gq heterotrimers signaling. These structures provide a framework for rational design of drugs targeting GRPR for the treatments of cancer and pruritus.
Keywords: GRPR, cryo-EM, molecular mechanism
Abstract
Gastrin releasing peptide receptor (GRPR), a member of the bombesin (BBN) G protein-coupled receptors, is aberrantly overexpressed in several malignant tumors, including those of the breast, prostate, pancreas, lung, and central nervous system. Additionally, it also mediates non-histaminergic itch and pathological itch conditions in mice. Thus, GRPR could be an attractive target for cancer and itch therapy. Here, we report the inactive state crystal structure of human GRPR in complex with the non-peptide antagonist PD176252, as well as two active state cryo-electron microscopy (cryo-EM) structures of GRPR bound to the endogenous peptide agonist gastrin-releasing peptide and the synthetic BBN analog [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14), in complex with Gq heterotrimers. These structures revealed the molecular mechanisms for the ligand binding, receptor activation, and Gq proteins signaling of GRPR, which are expected to accelerate the structure-based design of GRPR antagonists and agonists for the treatments of cancer and pruritus.
In mammals, the bombesin (BBN) receptor family belongs to the peptide G protein-coupled receptors (GPCRs) and consists of three subtypes, the neuromedin B receptor (NMBR/BB1), the gastrin-releasing peptide receptor (GRPR/BB2), and an orphan BBN receptor subtype 3 (BRS-3/BB3) (1–3). These receptors are widely distributed in the central nerve system (CNS) and peripheral tissues with extensive effects on the physiological and pathological processes (4). Mammalian BBN receptors, especially GRPR, are of particular interest in oncology as their high expression in several human cancers, such as the prostate (5, 6), breast (7), and small-cell lung cancers (8), as well as in the CNS/neural tumors including gliomas, neuroblastomas, and medulloblastomas (9). BBN is a 14-residue peptide (Pyr-QRLGNQWAVGHLM-NH2) first isolated from the skin of the European fire-bellied toad Bombina bombina and its mammalian counterpart gastrin-releasing peptide (GRP), a 27-residue peptide, shares the same C terminus of Trp-Ala-Val-Gly-His-Leu-Met-NH2 with BBN (10). Many peptides derived from the C-terminal fragment of GRP/BBN with high affinity for the BBN receptor family, have been employed as motifs for tumor therapy and diagnosis (11, 12). The synthetic BBN analog, [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) has been shown to bind to all three human BBN receptors with high affinity, and thus could be especially useful for this approach (13). On the one hand, such peptides could be exploited to decrease the cancer proliferation for therapy, through coupled with anticancer agents including paclitaxel, doxorubicin, marine toxins, magainin II, and so on (14). On the other hand, extensive efforts have been devoted to develop the radiolabeled BBN compounds as GRPR imaging probes for the cancer diagnosis. However, these peptides exhibit diverse conformations and the short half-life due to their fast metabolic degradation with approximately 600 different proteases in the human body (15). PD176252 is the first non-peptide antagonist of GRPR with high affinity of Ki= 1.0 nM (16). This antagonist could inhibit the growth and proliferation of lung cancer cells in a dose-dependent manner with low toxicity (17). However, PD176252 and its derivatives exhibit poor solubility and selectivity because of its excessive conformational flexibility (18, 19). Thus, designing chemically stable GRP/BBN analogs with increased selectivity and affinity for GRPR is of paramount importance (20). It has been very challenging to design the GRP/BBN analogs due to lacking of the three-dimensional structure of GRPR.
Chronic itch is associated with the atopic dermatitis, allergic contact dermatitis, psoriasis, systemic diseases, neurological diseases, and drug side effect, and caused intense suffering in everyday life of human beings (21). The clinical studies indicated that most chronic pruritus was intractable to the antihistamine drugs and lack of safe and effective therapeutic medication (22, 23). GRPR has been regarded as an extremely versatile receptor in the spinal cord that was activated by GRP in dorsal root ganglion neurons to principally mediate the non-histaminergic itch (24–29). Ablation of GRPR neurons completely abolishes all types of itch transmission including mechanical itch without impacting pain transmission (27, 30). GRPR is crucial for mediating opioid-induced itch, a clinically significant side effect of opioid analgesia (31), via cross-talk with μ-opioid receptor (MOR) isoform 1D in mice and MOR1Y in humans (32, 33). In addition, the deletion of GRPR in the suprachiasmatic nucleus of mice abolishes contagious itch transmission (34, 35). GRPR has been shown to be upregulated in the spinal cord of various rodent and monkey models of chronic itch and a blockade of GRPR dramatically attenuates chronic itch (36–43). κ-opioid receptor (KOR) agonists display potent anti-nonhistaminergic pruritus effects and have been used in clinical trials to attenuate chronic itch (44). KOR agonists exert their anti-pruritus centrally by blocking GRPR function (45, 46). Importantly, GRP/GRPR is highly conserved across animal species (36, 47, 48), indicating that it is the principal receptor for nonhistaminergic itch in the spinal cord and trigeminal sensory neurons. Therefore, the structure-based design of GRPR non-peptide antagonists is promising to usher in a new avenue of potential strategies for treating itch.
To illustrate the molecular mechanism of the ligand binding and receptor activation, we determined two active-state cryo-electron microscopy (cryo-EM) structures of GRPR in complex with the peptide agonists, the natural ligand GRP and synthetic BBN analog [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14). Moreover, we determined the inactive-state crystal structure of GRPR in complex with the non-peptide antagonist PD176252. Taken together, these structures throw a light on the molecular mechanisms for binding the peptide agonists and non-peptide antagonist of GRPR. And these findings are expected to accelerate the structure-guided efforts for developing more selective and stable GRPR agonists and antagonists with potential therapeutic effects for cancer and non-histaminergic itch.
Results
Crystal Structure Determination of GRPR in Complex with Non-Peptide Antagonist.
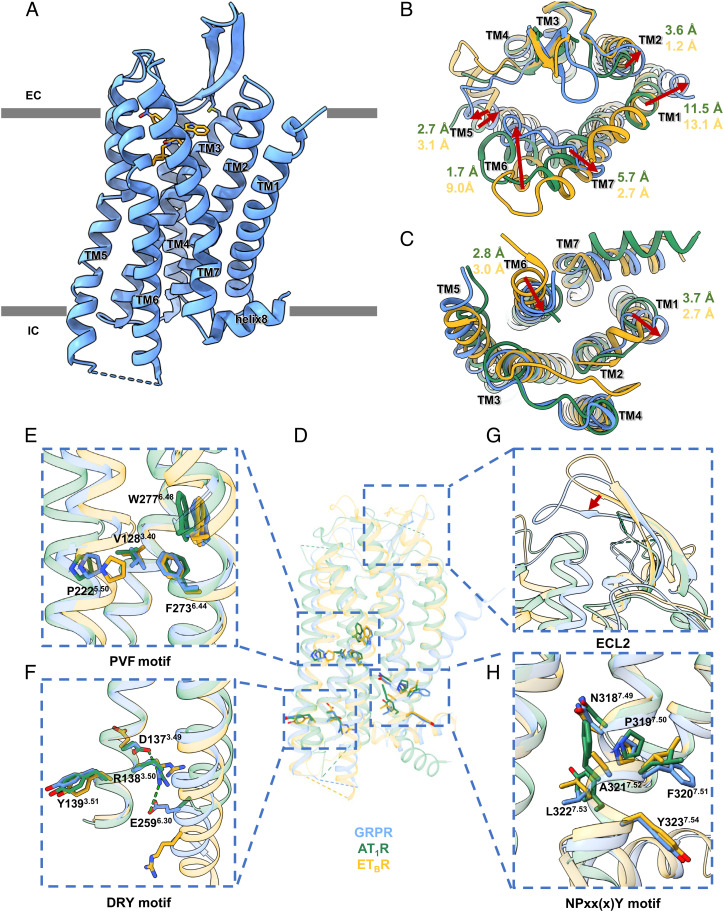
The crystal structure of human GRPR-PD176252 was solved at 2.95 Å resolution, with N-terminal 23-residue and C-terminal 29-residue truncations, the Pyrococcus abysii glycogen synthase (PGS) (49) insertion between A2415.69 and K2546.25 at the intracellular loop 3 (ICL3), and mutations of S1273.39K, I1574.42A, and R2596.30E [superscripts indicate Ballesteros-Weinstein numbers (50)], to improve the stability and facilitate the crystallization (SI Appendix, Fig. S1A and Table S1). The determined GRPR structure exhibited a canonical seven-transmembrane helical bundle (TMs1-7) architecture of GPCRs (Fig. 1A). The extracellular loop 2 (ECL2) of GRPR formed a β-hairpin with extended anti-parallel β-strands, which was observed in many peptide GPCRs (51–56) (Fig. 1A). This β-hairpin structure was further stabilized by the conserved disulfide bond between C1133.25 and C196ECL2 connecting the TM3 and ECL2 (Fig. 1A).
Fig. 1.
(A) Overall structure of GRPR-PD176252, with GRPR in blue cartoons and PD176252 in orange sticks. (B–H) Structural comparisons among GRPR (in blue), AT1R (in green), and ETBR (in goldenrod) from the extracellular view (B) and the intracellular view (C), with enlarged views of the conserved P5.50V(I)3.40F6.44 motif (E), D3.49R3.50Y3.51 motif (F), ECL2 (G), and N7.49P7.50xx(x)Y7.54 motif (H).
Among the class A peptide GPCRs, GRPR showed high sequence identity of 24% and 33% with the angiotensin receptor AT1R (57) and the endothelin receptor ETBR (58), respectively. Compared with the inactive structures of AT1R and ETBR, GRPR exhibited the overall Cα RMSD of 1.08 Å and 1.10 Å, respectively (Fig. 1D). From the extracellular view, compared to AT1R, TM5, and TM6 of GRPR shifted inward by 2.7 Å and 1.7 Å (Cα of P2075.35 and H2906.61), and TM2 and TM7 of GRPR moved outward by 3.6 Å and 5.7 Å (Cα of L1022.66 and M2987.29) (Fig. 1B). Compared with ETBR, the extracellular parts of TM6 of GRPR was observed a dramatic inward movement by 9.0 Å (Cα of H2906.61) and TMs2/5/7 shifted outward by 1.2 Å, 3.1 Å, and 2.7 Å (Cα of L1022.66, P2075.35, and M2987.29) (Fig. 1B). Surprisingly, the extracellular tilt of TM1 in GRPR exhibited a significant outward shift by 11.5 Å and 13.1 Å (Cα of I391.31) compared to AT1R and ETBR (Fig. 1B). Considering that TM1 was not involved in the ligand-binding, its unusual conformation might result from the crystal packing by interacting with TM1 and TM5 of GRPR in the asymmetry unit (Fig. 1B and SI Appendix, Fig. S1C). The β-hairpin of ECL2 formed a lid conformation and moved closer to the ligand-binding pocket to stabilize the non-peptide antagonist, compared to AT1R and ETBR (Fig. 1G).
From the intracellular side, TM1 and TM5 of GRPR shifted outward by 3.7 Å and 2.5 Å (Cα of T671.59 and N2365.64) and TM6 exhibited an inward movement by 2.8 Å (Cα of K2606.31), compared to AT1R. With a comparison of ETBR, TM6 of GRPR moved inward by 3.0 Å (Cα of K2606.31) and TM1 shifted outward by 2.7 Å (Cα of T671.59) (Fig. 1C). Additionally, a short α helix in ICL2 was observed in GRPR, which was not seen in AT1R or ETBR (Fig. 1C). The helices 8 of GRPR and ETBR extended in parallel to the membrane, which was different from that of AT1R (Fig. 1D).
The toggle switch W2776.48 and P5.50-I(V)3.40-F6.44 motif have been characterized as the conserved microswitches in class A GPCRs (Fig. 1E). Moreover, R2596.30E could stabilize GRPR by an additional polar interaction with R1383.50 of the D3.49R3.50Y3.51 motif, besides the ionic lock between D1373.49 and R1383.50, which induced an inward rearrangement of TM6 by 2.8 Å and 3.0 Å compared to the inactive AT1R and ETBR, respectively (Fig. 1F). Additionally, unlike the classic N7.49P7.50xxY7.53 motif conserved in AT1R, GRPR showed the N7.49P7.50xx(x)Y7.54 motif (NPFALY7.54) similar to ETBR (NPIALY7.54), with F3207.51 of GRPR forming an additional π–π interaction with Y3237.54 (Fig. 1H). Thus, the conformations of all microswitches indicated that the determined GRPR crystal structure was in an inactive state.
Non-Peptide Antagonist Binding to GRPR.
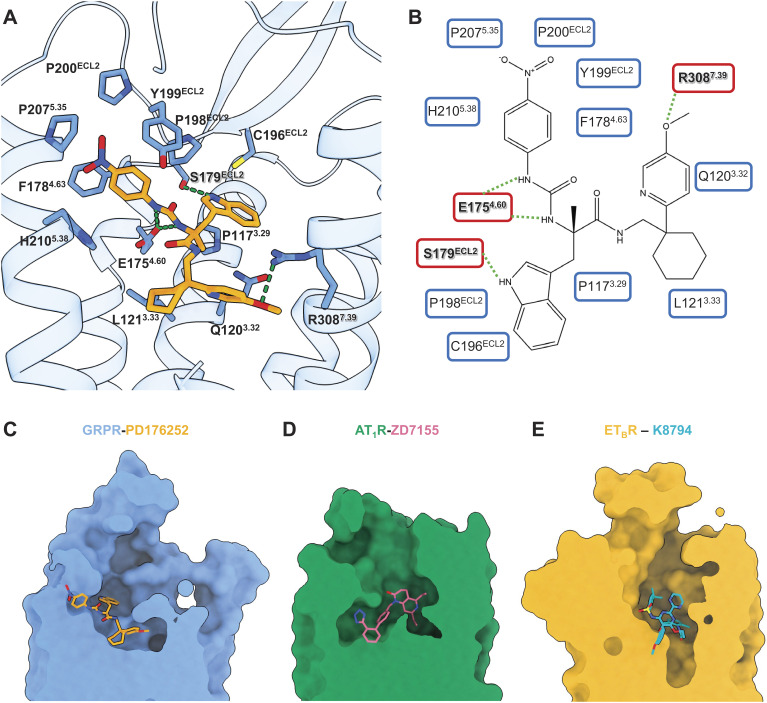
The GRPR-PD176252 complex structure showed unambiguous electron density for the non-peptide antagonist PD176252 with extensive interactions in the ligand-binding pocket (Fig. 2A and SI Appendix, Fig. S1E). PD176252 is comprised of four moieties of a para-nitrophenyl, a cyclohexyl, a para-methoxy-2-pyridine, and an indole (Fig. 2B). PD176252 occupied the typical ligand-binding pocket of GRPR, comparable to AT1R and ETBR (Fig. 2 C–E). Specially, the para-nitrophenyl of PD176252 extended to the extracellular region and was responsible for a series of hydrophobic interactions with ECL2 and the top end of TM5, which showed an obviously different binding orientation compared to the non-peptide antagonists ZD7155 of AT1R and K-8794 of ETBR (Fig. 2 C–E).
Fig. 2.
Non-peptide antagonist binding to GRPR. (A) Detailed interactions of PD176252 with GRPR. The receptor backbone was shown in blue ribbons and the GRPR residues involved in interactions were displayed as blue sticks. PD176252 was displayed in orange sticks, with oxygen atoms in red and nitrogen atoms in blue. The hydrogen bonds were indicated as green dash lines. (B) Alternative 2D diagrams showing direct interactions between GRPR and PD176252. The GRPR residues responsible for important interactions are shown in red background. The hydrogen bonds were indicated as green dash lines. (C–E) Comparisons of the ligand-binding pockets in surface among GRPR (in blue, PD17625 in orange) (C), AT1R (in green, ZD7155 in pink, PDB ID: 4YAY) (D), and ETBR (in goldenrod, K-8794 in cyan, PDB ID: 5X93) (E).
The 1-nitrogen atom of the para-nitrophenyl and the amide nitrogen atom of PD176252 next to the para-nitrophenyl moiety formed two hydrogen bonds with E1754.60 (Fig. 2 A and B). Indeed, E1754.60A could significantly decrease the GRP-induced calcium signaling by more than 10-fold (SI Appendix, Fig. S6 and Table S2). Remarkably, there was a hydrogen bond linking the oxygen atom of the para-methoxy-2-pyridine of PD176252 and R3087.39 of GRPR (Fig. 2 A and B). This arginine of TM7 mutated to alanine could dramatically reduce the GRP activation by 66-fold, indicating the importance of this hydrogen bond between PD176252 and GRPR (SI Appendix, Fig. S6 and Table S2). It was also supported by previous structure-activity relationship (SAR) studies that substitution of the methoxy group of the para-methoxy-2-pyridine with a hydrogen atom, as another non-peptide antagonist PD168368, caused a 17-fold reduction for GRPR binding (16). In addition to the interactions with TMs of GRPR, PD176252 used its indole ring to form a hydrogen bond with S179ECL2, pulling the ECL2 inward and stabilizing the ligand-binding (Fig. 2 A and B). Besides, the para-nitrophenyl and the indole moieties at the extracellular side formed extensive hydrophobic interactions with P1173.29, C196ECL2, P198ECL2, Y199ECL2, P200ECL2, P2075.35, and H2105.38 of GRPR (Fig. 2 A and B). Both the para-methoxy-2-pyridine and cyclohexyl groups bound deeply into the bottom of the pocket by hydrophobic contacts with Q1203.32, L1213.33, F1784.63, N2806.51, H2816.52, and Y2846.55 (Fig. 2 A and B).
Cryo-EM Structure Determination of GRPR-Gq Signaling Complex.
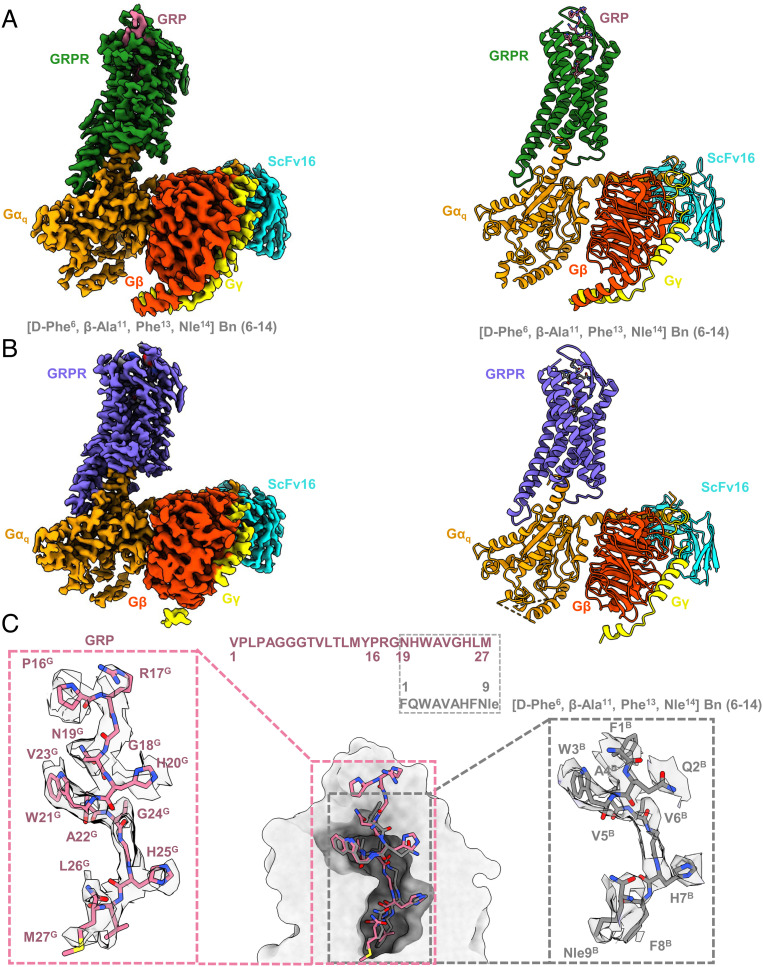
Human GRPR, Gαq, Gβ1, and Gγ2 were used to assemble the complexes with two peptide agonists. To improve the stability and homogeneity, the NanoBiT tethering strategy was applied in which the C terminus of GRPR was fused to the large part of NanoBiT (LgBiT), and the C terminus of Gβ1 was fused to the renovated 13-amino acid peptide of NanoBiT (HiBiT) (SI Appendix, Fig. S2A) (59). In addition, the maltose-binding protein (MBP) tag was fused at the N terminus of GRPR with the mutation I1574.42A to increase the protein expression and thermostability (SI Appendix, Fig. S2A) (55). The GRPR construct used for cryo-EM structure determination showed similar signaling property to the wild-type (WT) GRPR measured in the GRP-induced calcium mobilization assays (SI Appendix, Table S3). The chimera Gαq was generated by replacing the N-terminal 35-residue with the N-terminal 29-residue of Gi to facilitate the binding of a single-chain variable fragment scFv16, and introducing two dominant-negative mutations R183Q and Q209L, thereby further stabilizing the GRPR-Gαq complex for structural studies (60, 61). Compared to the WT-Gq, the R183Q mutation attenuated the ability of the engineered Gq to activate the downstream calcium signaling by disrupting the hydrogen bonds between R183 and GDP molecule (SI Appendix, Table S3) (61). The structures of GRPR-Gq heterotrimers complexes with GRP and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) were determined using the single-particle cryo-EM both at a global nominal resolution of 3.0 Å. The cryo-EM maps allowed modeling the majority of the receptor residues, ligands, and G-proteins (Fig. 3 and SI Appendix, Fig. S3 and Table S5). The overall structures of GRP and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14)-bound GRPR were identical with Cα RMSD values of 0.04 Å for the whole complex, 0.15 Å for GRPR, 0.03 Å for Gαq, 0.04 Å for Gβ1, and 0.14 Å for Gγ2 (Fig. 3 A and B). [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) shared a conserved C-terminal segment largely overlapped with the C-terminal nine residues of GRP, both of which were well resolved in the current structures (Fig. 3 and SI Appendix, Fig. S4). However, the N-terminal 16-residue from M1G to Y15G of GRP could not be clearly modeled due to the poor density (Fig. 3A), which was consistent with the previous results that the N terminus of GRP was not essential for its high affinity binding to GRPR (62–64). As GRP and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) resembled in their chemical scaffolds, they induced similar conformational changes of GRPR (Fig. 3). Thus, the structural analysis was mainly focused on the GRP-bound GRPR-Gq complex structure, unless otherwise specified (Fig. 3C).
Fig. 3.
Cryo-EM structure determination of GRPR-Gq signaling complex. (A and B) Orthogonal views of the maps (Left) and models (Right) for the GRP (pale violet red) or [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) (dark grey) bound GRPR-Gq complexes, with GRPR was colored in forest green and medium slate blue, respectively. Gαq, Gβ, Gγ, and ScFv16 were colored in orange, orange red, yellow, and cyan, respectively. (C) Binding poses and sequence alignment of GRP and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14). The density maps of ligands were highlighted.
GRPR shares the sequence identity of 24% and 27% to the neurokinin-1 receptor (NK1R) and cholecystokinin A receptor (CCK1R), respectively, whose structures were also solved in complex with Gq proteins, all belonging to the neuropeptide GPCRs. Compared to the Gq-coupled NK1R and CCK1R, GRPR adopted a similar overall conformation, with Cα RMSD of 1.08 Å and 1.16 Å, respectively (SI Appendix, Fig. S5). However, an extra kink was found in the last cytoplasmic α-helix of TM6 in the Gq-coupled GRPR (SI Appendix, Fig. S5). At the intracellular side, TM6 of GRPR displayed 2.6 Å outward movement in comparison with NK1R and CCK1R (measured at the Cα of the residue at position 6.25) (SI Appendix, Fig. S5). At the extracellular side, ECL2 of GRPR showed 6.3 Å closer to the ligand-binding pocket compared to CCK1R (measured at the Cα of the residues T189ECL2 in GRPR and N188ECL2 in the CCK1R) (SI Appendix, Fig. S5). Besides, there was a 7.8 Å inward shift of TM7 in comparison with NK1R and CCK1R (measured at the Cα of the residue at position 7.29) (SI Appendix, Fig. S5).
Peptide Agonists Binding to GRPR.
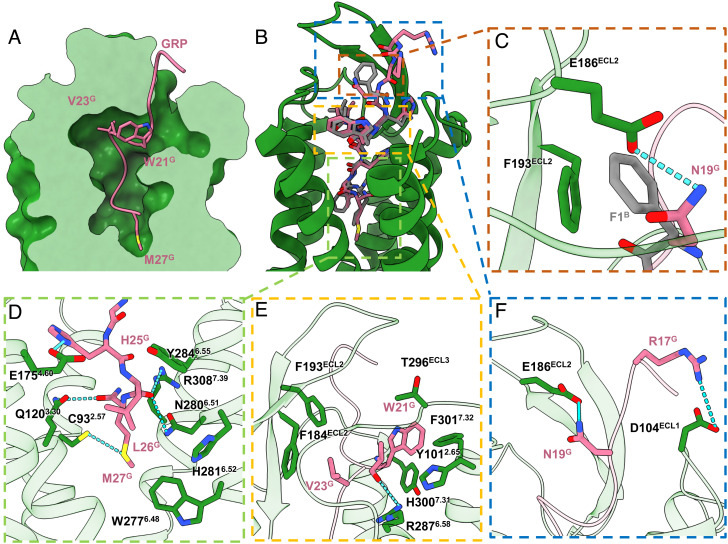
GRP and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) occupied the same orthosteric ligand-binding pocket of GRPR, with their C-termini inserting deeply into the helical bundle involving all ECLs and TMs except TM1 and TM5, and the N-termini pointing outward the ligand binding cavity (Fig. 4 A and B). The GRP could be divided into three regions binding to the bottom pocket, the hydrophobic cavities, and extending to the extracellular side (Fig. 4).
Fig. 4.
Peptide agonists binding to GRPR. (A) Cross-section of the peptide-binding pocket of GRPR was shown as forest green surface, with GRP as pink ribbons and side chains of W21G, V23G, and M27G displayed as pink sticks. (B and C) Structural alignment of GRP (green) and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) (gray), with enlarged views of detailed interactions at N19G and F1B.(D) The TM bottom pocket. (E) The hydrophobic cavities around the helical fragment of GRP. (F) The extracellular loops.
At the bottom pocket, the C terminus of GRP, H25G, L26G, and M27G penetrated into the bottom of pocket, stabilized by extensive hydrogen bonds and hydrophobic interactions (Fig. 4D). The M27G maintained the bottom location of GRP through hydrogen bonds with C932.57 and Q1203.30 which was supported by the calcium mobilization assays that alanine mutations in the residues C932.57 and Q1203.30 decreased the GRP-induced GRPR activation with the EC50 values of 12-fold and 25-fold higher than the WT-GRPR (SI Appendix, Fig. S6 and Table S2). Moreover, the main chain carbon moiety of L26G of GRP was hydrogen-bonded with the side chains of both N2806.50 and R3087.39 (Fig. 4D). Mutating R3087.39 to alanine produced significant decrease by 66-fold in GRPR activity, while mutating N2806.50 diminished the activities by only sixfold (SI Appendix, Fig. S6 and Table S2). The TM bottom binding moiety of GRP was further stabilized by a hydrogen bond between H25G and E1754.60(Fig. 4D). Mutation of E1754.60 resulted in the Gq signaling decreasing by 14-fold (SI Appendix, Fig. S6 and Table S2). Additionally, L26G formed multiple hydrophobic interactions with H2816.51 and Y2846.55, while M27G hydrophobically contacted with W2776.48 (Fig. 4D). Substitutions of H2816.51, Y2846.55, and W2776.48 with alanine displayed diminished receptor activation compared with WT-GRPR (SI Appendix, Fig. S6 and Table S2).
In the hydrophobic cavities, the helical fragment from W21G to V23G of GRP formed extensive hydrophobic interactions of V23G with F184ECL2 and F193ECL2, and W21G with Y1012.65, H3007.31, F3017.32, and T296ECL3 (Fig. 4E). Indeed, mutations on these residues resulted in decrease in Gq signaling (SI Appendix, Fig. S6 and Table S2). In addition to the hydrophobic interactions, the backbone carbonyl group of W21G formed a hydrogen bond with the side chain of R2876.58 to further stabilize the helical conformation (Fig. 4E). R2876.58A mutation significantly diminished the activities by 51-fold (SI Appendix, Fig. S6 and Table S2).
At the extracellular side, two hydrogen bonds existed between N19G and E186ECL2, as well as R17G and D104ECL1(Fig. 4C). Different from GRP, the synthetic peptide agonist [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) formed a π-π interaction with F193ECL2 through the N-terminal F1B (Fig. 4C). Mutating E186ECL2 decreased the GRPR activation by 10-fold on the GRP induced calcium mobilization (SI Appendix, Fig. S6 and Table S2). On the contrary, mutating E186ECL2 increased the GRPR activation by twofold on the [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) induced calcium mobilization (SI Appendix, Table S4). Taken together, the ECL2 might play a critical role for different peptide agonists recognition.
Gq Binding to GRPR.
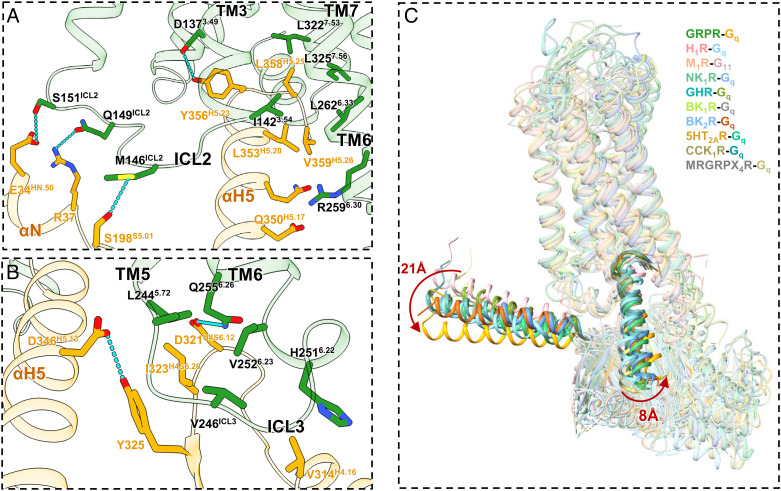
The Gq-coupling modes were almost identical between the two peptide agonists (Fig. 3 A and B). The engagements of Gq to GRPR were mainly mediated by the αH5 helix through key interactions with TMs3/6/7. The C terminus of αH5 was stabilized by the hydrogen bond between Y356H5.23 and D1373.49 (Fig. 5A). Additionally, the side chains of D346H5.13 and Q350H5.17 engaged in polar interactions with the main chain carbonyl of R2596.30 (Fig. 5A). Furthermore, the residues I1423.54, L2626.33, L3227.53, and L3257.56 formed hydrophobic contacts with L353H5.20, L358H5.25, and V359H5.26 (Fig. 5A). Besides, the ICL2/αN interaction was also observed in the binding interface (Fig. 5A). The side chains of M146ICL2, Q149ICL2, and S151ICL2 were hydrogen-bonded with the side chains of S198S5.01, E34HN.50, and R37 of αN-β1 junction, respectively (Fig. 5A).
Fig. 5.
Gq binding to GRPR. (A) Detailed key interactions between GRPR (green) and Gq (orange), with the hydrogen bonds shown as blue dashed lines. (B) An extra patch of interactions in the TM5-ICL3-TM6 region. (C) Structural comparisons on the G proteins shifts among GRPR-Gq (forest green/orange), H1R-Gq (medium purple/light coral, PDB ID: 7DFL), M1R-G11 (light salmon/light pink, PDB ID: 6OJI), NK1R-Gq (medium aquamarine/cornflower blue, PDB ID: 7RMG), GHR-Gq (medium turquoise/olive drab, PDB ID: 7F9Y), BK1R-Gq (yellow green/dark grey, PDB ID: 7EIB), BK2R-Gq (royal blue/chocolate, PDB ID: 7F6H), 5HT2AR-Gq (goldenrod/medium sea green, PDB ID: 6WHA), CCK1R-Gq (dark khaki/cadet blue, PDB ID: 7EZM), and MRGRPX4R-Gq complex structures (dark gray/dark khaki, PDB ID: 7S8P).
The overall structures of the GRPR-Gq complexes displayed the prototypical GPCR-G protein interface (Fig. 5) (60, 65–73). However, an extra patch of interactions with Gq was found in the TM5-ICL3-TM6 region, where the hydrophobic residue I323H4S6.20 intruded into a hydrophobic pocket formed by L2445.72, V246ICL3, and H2516.22 of GRPR (Fig. 5B). Besides, H2516.22 was in close contacts with the hydrophobic residues V314H4.16 and L310H4.12. Specially, unlike the only hydrophobic patch found in other GRPR-Gq/11 complexes (71), the side chain of Q2556.26 of GRPR formed a unique hydrogen bond with the side chain of D321H4S6.12 of Gq (Fig. 5B). Thus, these interactions might induce the large displacement of αH5 through Y325 and F339 H5.06 of Gq in the GRPR-Gq complex, eventually leading to the dramatic shift of the entire Gq heterotrimers by 21 Å of αN and 8 Å of αH5 in GRPR-Gq compared to MRGRPX4R-Gq (Fig. 5 B and C). In addition, these contacts also contributed to the conformational changes of the cytoplasmic segment of TM6, followed by the activation of GRPR (Fig. 5 and SI Appendix, Fig. S5). Therefore, our findings indicated that the TM5-ICL3-TM6 junction of GRPR also contributed to the Gq protein coupling.
Activation Mechanisms of GRPR.
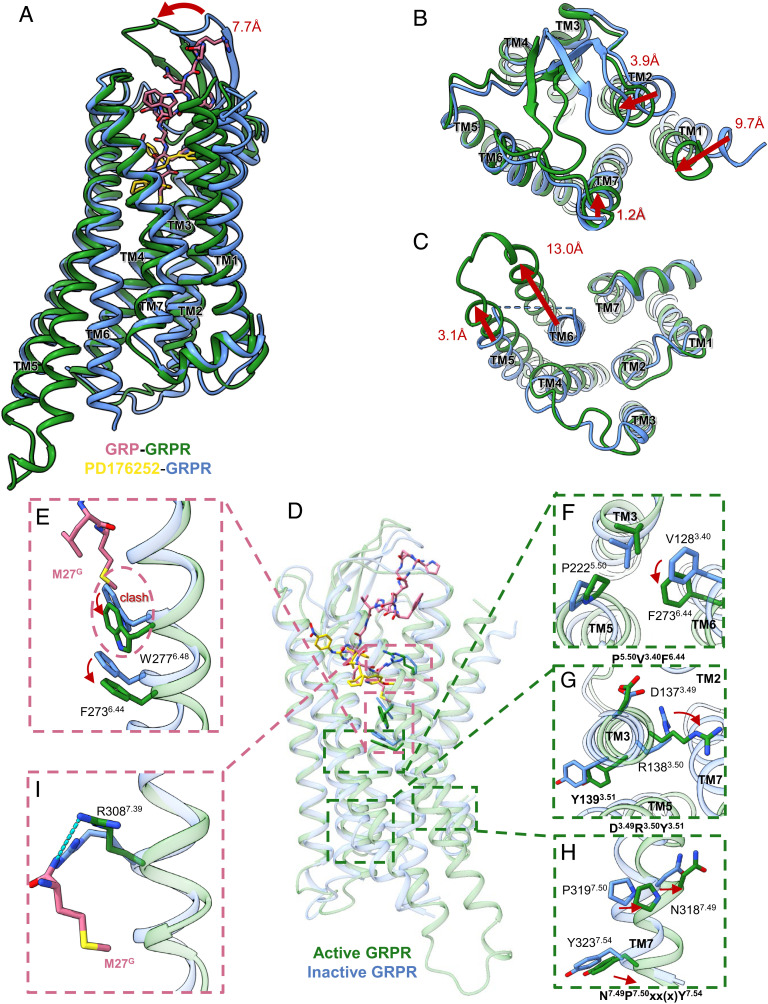
Structural superpositions between our inactive and active states GRPR structures showed remarkable conformational rearrangements in both TMs and ECL2 (Fig. 6 A–C). The peptide agonists occupied the orthosteric ligand-binding pocket with their N-termini interacting with TM2, TM7, and ECLs (Fig. 6A). The residues C932.57 and Y1012.65 in TM2, and H3007.31, F3017.32, and R3087.39 in TM7 were all involved in the peptide agonist-binding but not the non-peptide antagonist-binding (Fig. 4). From the extracellular view, the ECL2 β-hairpin with a lid-like architecture moved toward the orthosteric pocket by 7.7 Å upon activation (Fig. 6A). Besides, the TM1, TM2, and TM7 of the active GRPR moved inward by 9.7, 3.9, and 1.2 Å, respectively, compared to the inactive structure (Fig. 6B).
Fig. 6.
Activation mechanisms of GRPR. (A–C) Structural superposition of the active (green) and inactive (blue) GRPR structures from side view (A), extracellular view (B), and intracellular view (C), with red arrows indicating the TM shifts upon receptor activation. (D) Conformational differences between the active (green) and inactive (blue) GRPR structures at the ligand-binding sites, with enlarged views of F2736.44 and W2776.48 (E), P5.50V3.40F6.44 motif (F), D3.49R3.50Y3.51 motif (G), N7.49P7.50xx(x)Y7.54 motif (H), and R3087.39 (I).
Different from the antagonist, the C-terminal M27G of agonists bound deeply into the pocket surrounded by TM3 and TM6, forming hydrophobic contacts with the indole ring of W2776.48 (Fig. 6 D and E). In class A GPCRs, the conserved W6.48 was suggested to be a rotamer toggle switch to control the GPCRs transitioning between active and inactive states (74). Agonist-binding induced a large displacement of the side chain of W2776.48, acting as a pivot for the outward movement and rotation of TM6, which initiated the cascade of the conformational changes for receptor activation (Fig. 6). Additionally, the displacement of W2776.48 also led to the swing of F2736.44 of the conserved P5.50V3.40F6.44 motif for GRPR activation, followed by the D3.49R3.50Y3.51 motif undergoing active-like conformational changes (Fig. 6 F and G). Besides, the C-terminal M27G formed a hydrogen bond with R3087.39, which further triggered the active state of the conserved N7.49P7.50xx(x)Y7.54 motif for GRPR activation (Fig. 6 H and I). Thus, the P5.50V3.40F6.44 and N7.49P7.50xx(x)Y7.54 motifs connected the ligand-binding pocket and G protein-coupling interface through the translocation of F6.44 in the P5.50V3.40F6.44 motif and Y7.54 in N7.49P7.50xx(x)Y7.54 motif. Due to the rearrangements of these microswitches, TM5 and TM6 in the active-state GRPR showed significant outward movements of 3.1 Å and 13.0 Å for the C-terminal helix of Gαq to engage the receptor core from the intracellular side (Fig. 6C), which was conserved for GPCR activation (75). Therefore, these findings suggested that the GRP adopts the “message-address” frame like other neuropeptides (76). The extracellular GRP interactions with the ECLs acted as the “address” part for the peptide recognition, while the C terminus of GRP interacting with the bottom pocket played the essential roles as the “message” part for GRPR signaling by activating the conserved motifs.
Discussion
GRPR is one of the most commonly overexpressed receptors in prostate (5, 6), breast (7) and lung cancer (8). And GRPR mediates the transmission of non-histaminergic itch in the dorsal horn of the spinal cord (25). Agonists and antagonists with high affinity and selectivity with GRPR have great potentials for treatments of GRPR-related cancers and itch. In the current research, we determined the inactive state crystal structure of human GRPR in complex with the first non-peptide antagonist PD176252, together with two active state cryo-EM structures of GRPR-Gq complexes bound to the endogenous peptide agonist GRP and a synthetic BBN analogue [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14). These structures revealed the ligand-binding pocket, where the non-peptide antagonist occupied the bottom of pocket, and the para-nitrophenyl of PD176252 extended to the extracellular region of GRPR. GRP and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) occupied the same orthosteric binding pocket, with the N terminus pointing outward and the C terminus binding deeply into the helical bundle core. Especially, the ECL2 of GRPR played critical roles as the lid-like architecture for different peptide agonist recognition. In combination with the site-directed mutagenesis, the key residues for different ligand binding were determined. Consistent with previous SAR studies (16, 77), the methoxyl of the para-methoxy-2-pyridine of PD176252 formed a hydrogen bond with R3087.39 of GRPR and showed prominent effects on the high affinity and selectivity of non-peptide antagonist.
Structural comparisons also revealed the molecular mechanisms for GRPR activation. The peptide agonist binding induced the outward movements of the side-chain of W2776.48 and consequently initiated the outward movement and rotation of TM6. Meanwhile, the displacement of W2776.48 led to the swing of F2736.44 in the conserved P5.50V(I)3.40F6.44 motif, followed by the rearrangements of the D3.49R3.50Y3.51 and N7.49P7.50xx(x)Y7.54 motifs, and then opened the intracellular cleft for the insertion of αH5 of Gq. Therefore, the GRPR activation was initiated by the extracellular interactions of GRP and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14), as the “address” part, for the peptide agonist recognition, while the C-terminal peptides binding the bottom of pocket, as the “message” part, induced the activation of conserved motifs and TMs for GRPR signaling to Gq proteins.
Furthermore, the GRPR-Gq displayed a special GPCR-G proteins interface, where Gq formed substantial interactions with TM3/6/7 and ICLs2/3 of GRPR. Specially, the TM5-ICL3-TM6 junction was found to interact with αH5 and the Ras domain of the Gq, eventually leading to the remarkable shift of the entire Gq heterotrimers.
Therefore, these novel findings from our determined GRPR structures with different ligands and Gq heterotrimers revealed comprehensive molecular details in the ligand-binding pocket, activation motifs, and Gq coupling interface, which are expected to help better understand the GRPR structure–function relationships and guide the structure-based design of novel GRPR agonists and antagonists with better affinity and selectivity for the treatments of GRPR-involved cancers and pruritus.
Materials and Methods
Generation of GRPR Constructs for Crystallography and Cryo-EM.
A DNA fragment encoding the human gastrin-releasing peptide receptor amino acid sequence (Uniprot ID: P30550) was codon-optimized for the insect cell expression (Synbio Tech) and cloned into a modified pFastBac1 baculovirus expression vector. For crystallization construct, it contained a hemagglutinin signal sequence, a Flag-tag and 10 His-tag followed by a TEV protease cleavage site at the N terminus. To assist crystal formation in lipidic cubic phase (LCP), the N terminus residues M1-S23 and the C terminus residues T356-V384 were truncated. And the 196-amino acids from residues G218 to S413 of PGS (PDB:2BFW) (49) were inserted into the ICL3 between A241 and K254 of GRPR by overlap PCR. S1273.39K and I1574.42A could lead to higher stability and yields for many GPCRs (55, 78). According to GPCRdb (79), R2596.30E could increase the melting temperature (Tm) of the GRPR bound to PD176252.
For the cryo-EM study, the WT human GRPR truncated with N-terminal residues 1 to 23 and C-terminal residues 342 to 384 to increase the expression levels, and fused with an N-terminal MBP and a C-terminal LgBiT, were cloned into pFastBac1. Before MBP, there were haemagglutinin(HA) and FLAG tags followed by a 10 His-tag, as well as a tobacco etch virus (TEV) cleavage site. A 15-residue linker of GSSGGGGSGGGGSSG was inserted between the GRPR and LgBiT. One mutation I1574.42A was introduced to increase the thermostability and homogeneity of the GRPR (55). The engineered human Gαq was generated by replacing the N-terminal 35 residues with the N-terminal 29 amino acids of Gαi and introduced two mutations R183Q and Q209L to stabilize the GRPR-Gq complex (59, 61). The human Gβ1 with a N-terminal 10 His-tag was followed by the C-terminal HiBiT connected with a 15-residue linker (GSSGGGGSGGGGSSG). The Gβ1γ2 subunits and Gαq-Ric8A (resistance to inhibitors of cholinesterase 8A) were integrated into the pFastBac-Dual vector (Invitrogen), respectively. The scFv16 gene was cloned into a modified expression vector pFastBac1 with a N-terminal GP67 signaling peptide and a C-terminal 10 His-tag.
Expression for GRPR-PD176252 and GRPR-Gq Complexes.
The high-titer recombinant baculovirus (>109 viral particle per mL) was generated by the Bac-to-Bac Baculovirus Expression System (Invitrogen). After the recombinant bacmid transfected for 96 h at 27 °C, P0 viral stock was harvested by centrifugation and used the supernatant to produce high-titer P1 baculovirus stock by infection of 40 mL of 2.1 106 Spodoptera frugiperda (Sf9) cells/mL and incubation for 48 h. GRPR was expressed in 1 L volume of biomass for crystallization according to infect Sf9 cells at a density of 2 to 3 cells/mL with P1 baculovirus stock at a multiplicity of infection of 5.48 h after infection at 27 °C, cells were collected by centrifugation and stored at −80 °C until further use. For the GRPR-Gq complexes, the GRPR, GαqRic-8A, Gβ1γ2, and scFv16 were co-expressed in Sf9 cells at density of 2.5 106 cells/mL in the ratio of 4:4:4:1. After infected by 48 h at 27 °C, the cells were harvested by centrifugation and stored at −80 °C until further use.
Purification of GRPR-PD176252 and GRPR-Gq Complexes.
For the GRPR-PD176252, insect cells were thawed on ice and washed firstly in hypotonic buffer containing 10 mM HEPES, pH7.5, 10 mM MgCl2, 20 mM KCl, and 1 homemade protease inhibitors cocktail. Soluble and membrane proteins were isolated by repeated dounce homogenization and ultracentrifugation at 4 °C and 58,000 g for 30 min in hypotonic buffer (twice) and hypertonic buffer (three times) containing 10 mM HEPES, pH7.5, 10 mM MgCl2, 20 mM KCl, 1,000 mM NaCl, and 1 homemade protease inhibitors cocktail. The washed membranes were resuspended in the presence of 2 mg/mL iodoacetamide (Sigma) and 1 cocktail at 4 °C for 30 min. Subsequently, GRPR was extracted from the membrane slowly and entirely for 3 h at 4 °C in solubilization buffer consisting of 50 mM HEPES, pH7.5, 500 mM NaCl, 10% (v/v) glycerol, 0.5% (w/v) n-dodecyl-β-Dmaltoside (DDM, Antrance), 0.1% (w/v) cholesterol hemisuccinate (CHS, Sigma), and 10 mM imidazole. The solubilized proteins were separated by high-speed centrifugation in 58,000 g for 1 h at 4 °C and incubated with the TALON IMAC resin (Takara) at 4 °C overnight. The receptor-bound resin was washed on a gravity column (Bio-Rad) with 10 column volumes (CV) of wash buffer I (50 mM HEPES, pH7.5, 500 mM NaCl, 5% (v/v) glycerol, 0.1/0.02% (w/v) DDM/CHS, 20 mM imidazole, 10 mM MgCl2, and 8 mM ATP (Sigma) followed the detergent exchanged to Lauryl-maltose neopentyl glycol (LMNG, Antrance) by 10 CV of wash buffer II (50 mM HEPES, pH7.5, 500 mM NaCl, 5% (v/v) glycerol, 0.1/0.01% (w/v) LMNG/CHS, 20 mM imidazole, 10 mM MgCl2, and 8 mM ATP). Wash buffer III (20 mM HEPES, pH7.5, 500 mM NaCl, 5% (v/v) glycerol, 0.05/0.005% (w/v) LMNG/CHS, and 25 mM imidazole) was used to further improve the purity. The protein was eluted with 5 CV of elution buffer (10 mM HEPES, pH7.5, 100 mM NaCl, 5% (v/v) glycerol, 0.02/0.002% (w/v) LMNG/CHS, and 300 mM imidazole). Then the sample was concentrated using a 100 kDa molecular weight cutoff (MWCO) concentrator (Sartorius) and applied to a PD MiniTrap G-25 desalting column (GE Healthcare) equilibrated with buffer containing 10 mM HEPES, pH7.5, 100 mM NaCl, 5% (v/v) glycerol, and 0.002/0.0002% (w/v) LMNG/CHS to remove imidazole, MgCl2, and ATP, followed by incubating with 200 μM PD176252 for 1 h at 4 °C. The purified GRPR-PD176252 proteins were concentrated to ~30 mg/mL with a 100 kDa MWCO concentrator for the crystallization. Protein purity and monodispersity were assessed by SDS-PAGE and analytical size-exclusion chromatography via a Sepax Nanofilm SEC-300 column (SI Appendix, Fig. S1B).
For the GRPR-Gq complex, cells were lysed by dounce homogenization in 20 mM HEPES, pH7.5, 50 mM NaCl, 10 mM MgCl2. The lysate was incubated at room temperature for 3 h supplemented with 5 mM β-ME (Sigma-Aldrich), 100 mU/mL apyrase, and 1 μM ligands (GRP or [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14), GenScript). The supernatant was then centrifuged at 30,000 g for 30 min to collect the membranes. The washed membranes were resuspended in 20 mM HEPES, pH7.5, 50 mM NaCl, 10 mM MgCl2, 1 μM ligands [GRP or [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14)], 100 mU/mL apyrase (Sigma-Aldrich), 5 mM β-ME, and protease inhibitors, and incubated at 4 ˚C for 1 h. After incubation, 0.5% (w/v) LMNG with 0.1% CHS were used for solubilization at 4 ˚C for 3.5 h. Insoluble materials were removed by centrifugation at 58,000 g for 1 h and the solubilized complex was incubated at 4 ˚C overnight with TALON IMAC resin (Takara) containing 20 mM imidazole. The resin was collected by centrifugation at 800 g for 5 min, loaded onto a gravity flow column, and washed with 20 CV of wash buffer I containing 20 mM HEPES, pH7.5, 100 mM NaCl, 10 mM MgCl2, 10 mM imidazole, 1 μM ligands [GRP or (D-Phe6, β-Ala11, Phe13, Nle14) Bn (6–14)], 0.1% (w/v) LMNG, 0.02% (w/v) CHS, and 20 CV of wash buffer II containing 20 mM HEPES, pH7.5, 100 mM NaCl, 10 mM MgCl2, 5 mM CaCl2, 30 mM imidazole, 1 μM ligands [GRP or (D-Phe6, β-Ala11, Phe13, Nle14) Bn (6–14)], 0.01% LMNG (w/v) ,0.01% (w/v) glyco-diosgenin (GDN, Anatrace) and 0.02% (w/v) CHS. The protein was then eluted with 5 CV of elution buffer containing 20 mM HEPES, pH7.5, 100 mM NaCl, 300 mM imidazole, 2 mM MgCl2, 0.01% LMNG, 0.01% GDN, 0.02% CHS, and 10 μM ligands [GRP or (D-Phe6, β-Ala11, Phe13, Nle14) Bn (6–14)]. The protein was incubated at 4 °C for 2 h supplemented with 5 mM β-ME, 100 mU/mL apyrase, and then incubated with amylose resin for 4 h at 4 °C. The amylose column was washed with a buffer of 20 mM HEPES, pH 7.5, 100 mM NaCl, 10 μM ligands [GRP or (D-Phe6, β-Ala11, Phe13, Nle14) Bn (6–14)], and 0.01% GDN, and then eluted with the same buffer with additional 10 mM maltose. The elution was concentrated and loaded onto a Superdex 200 10/300 GL column (GE Healthcare) with running buffer containing 20 mM HEPES, pH7.5, 50 mM NaCl, 2 mM MgCl2, 0.3 μM ligands [GRP or (D-Phe6, β-Ala11, Phe13, Nle14) Bn (6–14)], and 0.006% (w/v) GDN. The fractions of monomeric protein complexes were collected and concentrated to 5 mg/mL for the cryo-EM studies.
Crystallization in LCP for GRPR-PD176252.
For the crystallization, the GRPR-PD176252 was reconstituted into the LCP by mixing 40% protein with 60% molten lipid containing 90% monoolein and 10% (v/v) cholesterol using a mechanical syringe mixer (80). Crystallization trials were carried out on 96-well glass sandwich plates using an automated crystallization robot (Gryphon) by overlaying 40 nL of mesophase with 800 nL precipitant solution following by sealed with cover glasses as soon as possible. Crystals of GRPR-PD176252 appeared in 48 h and grew to a maximum size of 40 μm within 1 to 2 wk at 20 °C in a series of conditions (SI Appendix, Fig. S1D). The diffraction quality crystals were obtained in 100 mM MES, pH6.8, 250 to 350 mM ammonium phosphate monobasic, 25 to 30% (v/v) polyethylene glycol 400 (PEG400), and 3 to 7% Polypropylene glycol P 400. Crystals were harvested from LCP using 50 μm micromounts (MiTeGen) and flash-frozen in liquid nitrogen.
Data Collection and Crystal Structure Determination of GRPR-PD176252.
X-ray diffraction data of GRPR-PD176252 crystals were collected at the beam line BL18U1 of the Shanghai Synchrotron Radiation Facility (SSRF) at a wavelength of 1.0332 Å using Pilatus3 6M detector. Crystals were exposed with a 20 μm 20 μm beam for 1 s to an unattenuated beam and 1° oscillation per frame, and 60 frames per crystal. XDS (81, 82) was applied to indexing, integrating, scaling, and merging data from four crystals of GRPR-PD176252. Initial phase information was acquired using molecular replacement with Phaser (83, 84) using a model of ETB receptor (PDB ID: 5X93) (85) and PGS (PDB ID: 2BWF) (86) as search models. All refinement was carried out using ccp4 suite (87) followed by manual examination and regulation of the refined structures in COOT (88) with both 2|Fo|-|Fc| and |Fo|-|Fc| maps. The final model of the structure contained 288 residues of GRPR (residue G38-A241 and K254-L337) and 196 residues of PGS (residues G1218-S1413). The statistics of data collection and structure refinement were shown in SI Appendix, Table S1. Coordinates and structure factors have been submitted in the PDB under accession code 7W41.
Grid Preparation and Cryo-EM Data Collection for GRPR-Gq Complexes.
For the cryo-EM grids preparation, 3 μL of purified GRP-bound GRPR complex at 4 mg/mL and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14)-bound GRPR complex at 6 mg/mL were loaded individually onto a glow-discharged holey carbon grid (Quantifoil, Cu300 R1.2/1.3) and vitrified using a Vitrobot Mark IV (FEI) at 4 °C and 100% humidity with blot time of 10 s and blot force of 19 s followed by being flash-frozen in liquid ethane. Cryo-EM imaging was performed on a Titan Krios at 300 kV accelerating voltage in the Center of cryo-EM (CCEM), Zhejiang University. Micrographs were recorded using a Gatan K2 Summit direct electron detector in counting mode with a nominal magnification of 29,000. The resulting image stacks have a pixel size of 1.014 Å in counting mode for GRPR-GRP complex and 0.507 Å in super resolution mode for GRPR- [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) complex. Movies were obtained using serial-EM at a dose rate of about 64 electrons per Å2 per second with a defocus ranging from −1.10 to −1.30 μm for GRPR-GRP complex and −1.10 to −1.25 μm for GRPR- [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) complex. The total exposure time was 8 s and intermediate frames were recorded in 0.2 s intervals, resulting in an accumulated dose of 62 electrons per Å2 and a total of 40 frames per micrograph. A total of 2,784 and 1,912 movies were collected for the GRP-bound and [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14)-bound GRPR complexes, respectively.
Cryo-EM Data Processing and Cryo-EM Structure Determination of GRPR-Gq Complexes.
Dose-fractionated image stacks for GRPR-GRP and GRPR-[D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) were subjected to the beam-induced motion correction using Motion-Cor2.1 (89). Contrast transfer function (CTF) parameters were estimated by Ctffind4 (90). Data processing was performed using RELION-3.0 (91). For particle selection, 2D and 3D classifications were performed on a binned dataset with a pixel size of 3.042 Å. For the GRP-bound GRPR-Gq complex, about 1,859,252 particles were auto-picked and subjected to 50 rounds of reference-free 2D classifications, producing 1,035,568 particle projections for further processing. For the [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14)-bound GRPR-Gq complex, particle selection yielded 1,380,329 particle projections that were subjected to 50 rounds of reference-free 2D classifications, producing 1,119,475 particle projections for further processing. Particles selected from 2D classification were subjected to 142 rounds of 3D classifications, resulting in two well-defined subsets with 577,397 particles for GRPR-GRP complex and 384,207 particles for GRPR- [D-Phe6, β-Ala11, Phe13, Nle14] Bn (6–14) complex. Further 3D refinement, CTF refinement and Bayesian polishing processing generated density maps with an indicated global resolution of 3.0 Å at a Fourier shell correlation of 0.143.
Model Building and Refinement.
The GRPR structure prediction with Alpha-Fold2 (92) was used as an initial model for model rebuilding and refinement against the electron microscopy maps of GRPR-Gq complexes. The coordinate of BK2-Gq (PDB ID: 7F6G) was used to generate the initial models for Gαq. Models were docked into the EM density map using UCSF Chimera X (93), followed by iterative manual adjustment and rebuilding in COOT (94). The generated final model was refined in Phenix (95). The statistics for data collection and refinement were provided in SI Appendix, Table S5. Structural figures were prepared in UCSF Chimera X (93).
Calcium Mobilization Assay.
The WT GRPR gene was subcloned into the pcDNA3.1 vector with an N-terminal HA and FLAG tag. Mutations were introduced by QuickChange PCR. All of the constructs were verified by DNA sequencing. Chinese hamster ovary(CHO) cells transfected with WT or mutant GRPRs were seeded into 96-well black plates at a density of 30,000 cells per well and incubated for 24 h. On next day, cells were loaded with reagents from Calcium-5 Assay Kit (Molecular Devices) for 45 min at 37 °C in 5% CO2 according to the manufacturer’s protocol. Cells were treated with varied concentrations of ligands and detected with Flexstation three Multi-Mode Microplate Reader (Molecular Devices) with excitation at 485 nm and emission at 525 nm. Data were presented as mean SEM of three independent experiments. EC50 and Emax values for each curve were calculated by Prism 5.0 software.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
H.Z. is supported by National Key R&D Program of China (2018YFA0508100), National Natural Science Foundation of China (81722044, 91753115, 21778049, 81861148018), and National Science and Technology Major Project of China (2018ZX09711002). We thank S.C. and X.Z. in the CCEM, Zhejiang University, for their technical assistance on Cryo-EM data collection. We thank W.Q. and Q.X. from BL18U1 beamline of National Facility for Protein Science in Shanghai at Shanghai Synchrotron Radiation Facility, for their technical assistance on crystallographic data collection. We thank W.L. and M.L. in the Shanghai Yuyao Biotech Ltd. for the assistance on the calcium mobilization assays.
Author contributions
H.Z. designed research; S.P., Y.Z., D.Z., L.R., and A.C. performed research; S.P., Y.Z., Z.-F.C., and H.Z. analyzed data; and S.P., Y.Z., Z.-F.C., and H.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. R.R. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes: EMD-32297 (GRPR-Gq-GRP) and EMD-32298 [GRPR-Gq-(D-Phe6, β-Ala11, Phe13, Nle14) Bn (6–14)] (96–97). The atomic coordinates have been deposited in the Protein Data Bank under accession codes: 7W3Z (GRPR-Gq-GRP), 7W40 [GRPR-Gq-(D-Phe6, β-Ala11, Phe13, Nle14) Bn (6–14)], and 7W41 (GRPR-PD176252) (98–100).
Supporting Information
References
- 1.Battey J., Wada E., Two distinct receptor subtypes for mammalian bombesin-like peptides. Trends Neurosci. 14, 524–528 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Gorbulev V., Akhundova A., Buchner H., Fahrenholz F., Molecular cloning of a new bombesin receptor subtype expressed in uterus during pregnancy. Eur. J. Biochem. 208, 405–410 (1992). [DOI] [PubMed] [Google Scholar]
- 3.Ryan R. R., et al. , Ability of various bombesin receptor agonists and antagonists to alter intracellular signaling of the human orphan receptor BRS-3. J. Biol. Chem. 273, 13613–13624 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Ramos-Alvarez I., et al. , Insights into bombesin receptors and ligands: Highlighting recent advances. Peptides 72, 128–144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baratto L., Jadvar H., Iagaru A., Prostate cancer theranostics targeting gastrin-releasing peptide receptors. Mol. Imaging Biol. 20, 501–509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Case T. C., et al. , Blocking GRP/GRP-R signaling decreases expression of androgen receptor splice variants and inhibits tumor growth in castration-resistant prostate cancer. Transl. Oncol. 14, 101213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michalski K., et al. , Gastrin-releasing peptide receptor antagonist [(68)Ga]RM2 PET/CT for staging of pre-treated, Metastasized Breast Cancer. Cancers (Basel) 13, 6106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akbar M. J., Lukasewicz Ferreira P. C., Giorgetti M., Stokes L., Morris C. J., Bombesin receptor-targeted liposomes for enhanced delivery to lung cancer cells. Beilstein J. Nanotechnol. 10, 2553–2562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moody T. W., et al. , Bombesin receptor family activation and CNS/neural tumors: Review of evidence supporting possible role for novel targeted therapy. Front. Endocrinol. (Lausanne) 12, 728088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen R. T., Battey J. F., Spindel E. R., Benya R. V., International union of pharmacology. LXVIII. Mammalian bombesin receptors: Nomenclature, distribution, pharmacology, signaling, and functions in normal and disease states. Pharmacol. Rev. 60, 1–42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H., et al. , Bombesin analogue-mediated delivery preferentially enhances the cytotoxicity of a mitochondria-disrupting peptide in tumor cells. PLoS One 8, e57358 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno P., Ramos-Alvarez I., Moody T. W., Jensen R. T., Bombesin related peptides/receptors and their promising therapeutic roles in cancer imaging, targeting and treatment. Expert Opin. Ther. Targets 20, 1055–1073 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uehara H., et al. , The molecular basis for high affinity of a universal ligand for human bombesin receptor (BnR) family members. Biochem. Pharmacol. 84, 936–948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C., et al. , Breast cancer targeted chemotherapy based on doxorubicin-loaded bombesin peptide modified nanocarriers. Drug Deliv. 23, 2697–2702 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Tornesello A. L., Tornesello M. L., Buonaguro F. M., An overview of bioactive peptides for in vivo imaging and therapy in human diseases. Mini Rev. Med. Chem. 17, 758–770 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Ashwood V., et al. , PD 176252–the first high affinity non-peptide gastrin-releasing peptide (BB2) receptor antagonist. Bioorg. Med. Chem. Lett. 8, 2589–2594 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Moody T. W., Leyton J., Garcia-Marin L., Jensen R. T., Nonpeptide gastrin releasing peptide receptor antagonists inhibit the proliferation of lung cancer cells. Eur. J. Pharmacol. 474, 21–29 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Lacivita E., et al. , Structure-activity relationship study towards non-peptidic positron emission tomography (PET) radiotracer for gastrin releasing peptide receptors: Development of [(18)F] (S)-3-(1H-indol-3-yl)-N-[1-[5-(2-fluoroethoxy)pyridin-2-yl]cyclohexylmethyl]-2-me thyl-2-[3-(4-nitrophenyl)ureido]propionamide. Bioorg. Med. Chem. 25, 277–292 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Ryan R. R., et al. , Comparative pharmacology of the nonpeptide neuromedin B receptor antagonist PD 168368. J. Pharmacol. Exp. Ther. 290, 1202–1211 (1999). [PubMed] [Google Scholar]
- 20.Hoppenz P., et al. , A selective carborane-functionalized gastrin-releasing peptide receptor agonist as boron delivery agent for boron neutron capture therapy. J. Org. Chem. 85, 1446–1457 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Ikoma A., Steinhoff M., Stander S., Yosipovitch G., Schmelz M., The neurobiology of itch. Nat. Rev. Neurosci. 7, 535–547 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Kini S. P., et al. , The impact of pruritus on quality of life: The skin equivalent of pain. Arch. Dermatol. 147, 1153–1156 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Wang F., Kim B. S., Itch: A paradigm of neuroimmune crosstalk. Immunity 52, 753–766 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Barry D. M., et al. , Exploration of sensory and spinal neurons expressing gastrin-releasing peptide in itch and pain related behaviors. Nat. Commun. 11, 1397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y. G., Chen Z. F., A gastrin-releasing peptide receptor mediates the itch sensation in the spinal cord. Nature 448, 700–703 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Chen Z. F., A neuropeptide code for itch. Nat. Rev. Neurosci. 22, 758–776 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y. G., et al. , Cellular basis of itch sensation. Science 325, 1531–1534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiguchi N., et al. , GRP receptor and AMPA receptor cooperatively regulate itch-responsive neurons in the spinal dorsal horn. Neuropharmacology 170, 108025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan L., et al. , Distinct roles of NMB and GRP in itch transmission. Sci. Rep. 7, 15466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S., et al. , A spinal neural circuitry for converting touch to itch sensation. Nat. Commun. 11, 5074 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szarvas S., Harmon D., Murphy D., Neuraxial opioid-induced pruritus: A review. J. Clin. Anesth. 15, 234–239 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Liu X. Y., Ginosar Y., Yazdi J., Hincker A., Chen Z. F., Cross-talk between human spinal cord mu-opioid receptor 1Y isoform and gastrin-releasing peptide receptor mediates opioid-induced scratching behavior. Anesthesiology 131, 381–391 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X. Y., et al. , Unidirectional cross-activation of GRPR by MOR1D uncouples itch and analgesia induced by opioids. Cell 147, 447–458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Y. Q., Barry D. M., Hao Y., Liu X. T., Chen Z. F., Molecular and neural basis of contagious itch behavior in mice. Science 355, 1072–1076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao F., et al. , A non-canonical retina-ipRGCs-SCN-PVT visual pathway for mediating contagious itch behavior. Cell Rep. 41, 111444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nattkemper L. A., et al. , Overexpression of the gastrin-releasing peptide in cutaneous nerve fibers and its receptor in the spinal cord in primates with chronic itch. J. Invest. Dermatol. 133, 2489–2492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X. T., et al. , Spinal GRPR and NPRA contribute to chronic itch in a murine model of allergic contact dermatitis. J. Invest. Dermatol. 140, 1856–1866 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Kiguchi N., Saika F., Fukazawa Y., Matsuzaki S., Kishioka S., Critical role of GRP receptor-expressing neurons in the spinal transmission of imiquimod-induced psoriatic itch. Neuropsychopharmacol. Rep. 40, 287–290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Z. Q., et al. , Chronic itch development in sensory neurons requires BRAF signaling pathways. J. Clin. Invest. 123, 4769–4780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Z. Q., et al. , Descending control of itch transmission by the serotonergic system via 5-HT1A-facilitated GRP-GRPR signaling. Neuron 84, 821–834 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koga K., et al. , Sensitization of spinal itch transmission neurons in a mouse model of chronic itch requires an astrocytic factor. J. Allergy Clin. Immunol. 145, 183–191.e10 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Shiratori-Hayashi M., et al. , STAT3-dependent reactive astrogliosis in the spinal dorsal horn underlies chronic itch. Nat. Med. 21, 927–931 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Lagerstrom M. C., et al. , VGLUT2-dependent sensory neurons in the TRPV1 population regulate pain and itch. Neuron 68, 529–542 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inan S., Cowan A., Antipruritic effects of kappa opioid receptor agonists: Evidence from rodents to humans. Handb. Exp. Pharmacol. 271, 275–292 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Munanairi A., et al. , Non-canonical opioid signaling inhibits itch transmission in the spinal cord of mice. Cell Rep. 23, 866–877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H. P., et al. , 100 Hz electroacupuncture alleviated chronic itch and GRPR expression through activation of kappa opioid receptors in spinal dorsal horn. Front. Neurosci. 15, 625471 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takanami K., et al. , Comparative anatomy of gastrin-releasing peptide pathways in the trigeminal sensory system of mouse and the asian house musk shrew suncus murinus. Acta Histochem. Cytochem. 49, 181–190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takanami K., et al. , Characterization of the expression of gastrin-releasing peptide and its receptor in the trigeminal and spinal somatosensory systems of Japanese macaque monkeys: Insight into humans. J. Comp. Neurol. 530, 2804–2819 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin J., Mobarec J. C., Kolb P., Rosenbaum D. M., Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature 519, 247–250 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Ballesteros J. A., Weinstein H., [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995). [Google Scholar]
- 51.White J. F., et al. , Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkatakrishnan A. J., et al. , Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Shihoya W., et al. , Activation mechanism of endothelin ETB receptor by endothelin-1. Nature 537, 363–368 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Chen S. H., et al. , Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography. Nat. Commun. 10, 638 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waltenspuhl Y., Schoppe J., Ehrenmann J., Kummer L., Pluckthun A., Crystal structure of the human oxytocin receptor. Sci. Adv. 6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X. F., et al. , Structures of the human cholecystokinin receptors bound to agonists and antagonists. Nat. Chem. Biol. 17, 1230–1237 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Zhang H., et al. , Structure of the angiotensin receptor revealed by serial femtosecond crystallography. Cell 161, 833–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shihoya W., et al. , X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog. Nat. Struct. Mol. Biol. 24, 758–764 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Duan J., et al. , Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat. Commun. 11, 4121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maeda S., Qu Q. H., Robertson M. J., Skiniotis G., Kobilka B. K., Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martins L., et al. , Computational analysis for GNAQ mutations: New insights on the molecular etiology of Sturge-Weber syndrome. J. Mol. Graph. Model. 76, 429–440 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Uehara H., et al. , The molecular basis for high affinity of a universal ligand for human bombesin receptor (BnR) family members. Biochem. Pharmacol. 84, 936–948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sancho V., et al. , Pharmacology of putative selective hBRS-3 receptor agonists for human bombesin receptors (BnR): Affinities, potencies and selectivity in multiple native and BnR transfected cells. Peptides 31, 1569–1578 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tokita K., et al. , Molecular basis for selectivity of high affinity peptide antagonists for the gastrin-releasing peptide receptor. J. Biol. Chem. 276, 36652–36663 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Kim K., et al. , Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell 182, 1574–1588.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maeda S., Qu Q., Robertson M. J., Skiniotis G., Kobilka B. K., Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao C., et al. , Structure, function and pharmacology of human itch GPCRs. Nature 600, 170–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harris J. A., et al. , Selective G protein signaling driven by substance P-neurokinin receptor dynamics. Nat. Chem. Biol. 18, 109–115 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., et al. , Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor. Nat. Commun. 12, 5064 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia R., et al. , Cryo-EM structure of the human histamine H1 receptor/Gq complex. Nat. Commun. 12, 2086 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin Y. L., et al. , Molecular basis for kinin selectivity and activation of the human bradykinin receptors. Nat. Struct. Mol. Biol. 28, 755–761 (2021). [DOI] [PubMed] [Google Scholar]
- 72.Liu Q., et al. , Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor. Nat. Chem. Biol. 17, 1238–1244 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen J., et al. , Cryo-EM structures of human bradykinin receptor-Gq proteins complexes. Nat. Commun. 13, 714 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W., Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 30, 249–259 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Schoneberg T., Schultz G., Gudermann T., Structural basis of G protein-coupled receptor function. Mol. Cell Endocrinol. 151, 181–193 (1999). [DOI] [PubMed] [Google Scholar]
- 76.Schwyzer R., ACTH: A short introductory review. Ann. N. Y. Acad. Sci. 297, 3–26 (1977). [DOI] [PubMed] [Google Scholar]
- 77.Lacivita E., et al. , Structure-activity relationship study towards non-peptidic positron emission tomography (PET) radiotracer for gastrin releasing peptide receptors: Development of [(18)F] (S)-3-(1H-indol-3-yl)-N-[1-[5-(2-fluoroethoxy)pyridin-2-yl]cyclohexylmethyl]-2-methyl-2-[3-(4-nitrophenyl)ureido]propionamide. Bioorg. Med. Chem. 25, 277–292 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Yasuda S., et al. , Hot-spot residues to be mutated common in G protein-coupled receptors of class A: Identification of thermostabilizing mutations followed by determination of three-dimensional structures for two example receptors. J. Phys. Chem. B. 121, 6341–6350 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Munk C., et al. , An online resource for GPCR structure determination and analysis. Nat. Methods 16, 151–162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caffrey M., Cherezov V., Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McCoy A. J., Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shihoya W., et al. , X-ray structures of endothelin ET(B) receptor bound to clinical antagonist bosentan and its analog. Nat. Struct. Mol. Biol. 24, 758–764 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Horcajada C., Guinovart J. J., Fita I., Ferrer J. C., Crystal structure of an archaeal glycogen synthase: Insights into oligomerization and substrate binding of eukaryotic glycogen synthases. J. Biol. Chem. 281, 2923–2931 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 95.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhan Y., Peng S., Zhang H., Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Gastrin Releasing Peptide and Gq heterotrimers. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-32297. Deposited 26 Nov 2022.
- 97.Zhan Y., Peng S., Zhang H., Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Bombesin (6-14) [D-Phe6, beta-Ala11, Phe13, Nle14] and Gq heterotrimers. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-32298. Deposited 26 Nov 2022.
- 98.Zhan Y., Peng S., Zhang H., Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Gastrin Releasing Peptide and Gq heterotrimers. Protein Data Bank. https://www.rcsb.org/structure/7W3Z. Deposited 26 Nov 2022.
- 99.Zhan Y., Peng S., Zhang H., Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Bombesin (6-14) [D-Phe6, beta-Ala11, Phe13, Nle14] and Gq heterotrimers. Protein Data Bank. https://www.rcsb.org/structure/7W40. Deposited 26 Nov 2022.
- 100.Peng S., Zhan Y., Zhang H., Crystal Structure of Human Gastrin Releasing Peptide Receptor in complex with the antagonist PD176252. Protein Data Bank. https://www.rcsb.org/structure/7W41. Deposited 26 Nov 2022
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes: EMD-32297 (GRPR-Gq-GRP) and EMD-32298 [GRPR-Gq-(D-Phe6, β-Ala11, Phe13, Nle14) Bn (6–14)] (96–97). The atomic coordinates have been deposited in the Protein Data Bank under accession codes: 7W3Z (GRPR-Gq-GRP), 7W40 [GRPR-Gq-(D-Phe6, β-Ala11, Phe13, Nle14) Bn (6–14)], and 7W41 (GRPR-PD176252) (98–100).