Significance
Autophagy is an orderly degradation mechanism in eukaryotic cells. It is often dysregulated in a range of human diseases, including cancer. Some cancers rely on autophagy to survive and develop. In particular, tumor cells can use autophagy to cope with the cytotoxicity of certain anticancer drugs. Inhibition of autophagy is a promising strategy for tumor therapy. However, there are currently no effective autophagy inhibitors available for clinical use. We show that targeting the central regulatory transcription factor EB (TFEB) can effectively inhibit autophagy at the transcriptional level. And we found that the United States Food and Drug Administration (FDA)-approved drug Eltrombopag is actually a potent autophagy inhibitor targeting TFEB. This finding may help accelerate effective inhibition of autophagy in clinical practice.
Keywords: autophagy, transcription factor EB, high-throughput screen, Eltrombopag, cancer therapy
Abstract
Autophagy supports the fast growth of established tumors and promotes tumor resistance to multiple treatments. Inhibition of autophagy is a promising strategy for tumor therapy. However, effective autophagy inhibitors suitable for clinical use are currently lacking. There is a high demand for identifying novel autophagy drug targets and potent inhibitors with drug-like properties. The transcription factor EB (TFEB) is the central transcriptional regulator of autophagy, which promotes lysosomal biogenesis and functions and systematically up-regulates autophagy. Despite extensive evidence that TFEB is a promising target for autophagy inhibition, no small molecular TFEB inhibitors were reported. Here, we show that an United States Food and Drug Administration (FDA)-approved drug Eltrombopag (EO) binds to the basic helix-loop-helix-leucine zipper domain of TFEB, specifically the bottom surface of helix-loop-helix to clash with DNA recognition, and disrupts TFEB-DNA interaction in vitro and in cellular context. EO selectively inhibits TFEB’s transcriptional activity at the genomic scale according to RNA sequencing analyses, blocks autophagy in a dose-dependent manner, and increases the sensitivity of glioblastoma to temozolomide in vivo. Together, this work reveals that TFEB is targetable and presents the first direct TFEB inhibitor EO, a drug compound with great potential to benefit a wide range of cancer therapies by inhibiting autophagy.
Autophagy is an evolutionarily conserved catabolic process by which cellular materials are delivered to lysosomes for degradation to support nutrient recycling and metabolic adaption (1–4). Notably, up-regulated autophagy in both the tumor and host cells plays a critical role to support tumor growth by providing nutrients, maintaining metabolic homeostasis, increasing systemic arginine levels, decreasing p53 and surface major histocompatibility complex I, etc. (4–7). In addition, cytotoxic chemotherapy, targeted therapy, and radio therapy can activate cytoprotective autophagy and drive tumor resistance in multiple cancer types (7). Autophagy inhibition can limit the growth of tumors and improve the response to cancer therapeutics (4, 8–11). Therefore, targeting autophagy is an attractive therapeutic strategy for a wide range of cancers (2, 5–7, 12).
The antimalarials chloroquine (CQ) and hydroxychloroquine (HCQ) are the only two compounds used to inhibit autophagy in clinical trials, showing encouraging antitumor results in selected patients. However, the low efficiency and unclear mechanism of CQ and HCQ on autophagy hinder their further clinical application (4). The search for novel autophagy targets and small molecular modulators has been intensified in recent years (SI Appendix, Table S1). Novel autophagy inhibitors, like small molecular inhibitors of kinase vacuolar protein sorting-associated protein 34 (VPS34), Unc-51-like kinase 1 (ULK1), or cysteine protease autophagy-related gene 4B (ATG4B), were developed to suppress autophagy at the nucleation, elongation, fusion, or degradation phases with significantly increased efficiency (2). Because these targets also have multiple autophagy-independent functions (13), it was reasonably questioned whether it is possible to identify fully selective autophagy regulators (14). The safety of potential side effects is an important factor to be evaluated while developing autophagy inhibitors against these targets. In addition, it is of fundamental importance to identify novel therapeutic targets for autophagy and to discover corresponding drug-like compounds that effectively inhibit autophagy.
Transcription factor EB (TFEB) is a central transcriptional regulator of autophagy. A palindromic GTCACGTGAC motif, known as the Coordinated Lysosomal Expression and Regulation (CLEAR) element, is present in the promoter region of most known lysosomal genes (15). TFEB specifically binds the CLEAR element through the basic helix-loop-helix-leucine zipper (bHLH-LZ) domain and promotes lysosomal biogenesis and function (15, 16). Therefore, TFEB has a systemic role in up-regulation of autophagy (17). Consistent with the role of autophagy in supporting tumor growth and therapy resistance, TFEB is highly expressed, constitutively activated in multiple cancer types and has been indicated as the potential pharmacodynamic biomarkers to evaluate activation of autophagy in cancer patients (7). TFEB is a promising target for autophagy inhibition, but no small-molecule TFEB inhibitors have been reported.
Here, we developed a high-throughput assay and screened a library of drug molecules to identify TFEB inhibitors. Eltrombopag (EO), an United States Food and Drug Administration (FDA)-approved drug for thrombocytopenia treatment, was discovered to be a potent TFEB inhibitor. In this study, we show EO treatment inhibits the TFEB transcriptional activity at the genomic scale, blocks the starvation-induced autophagy response, and increases the sensitivity of glioblastoma to temozolomide (TMZ) in vivo. These results demonstrate a novel and potent autophagy inhibitor EO with pharmaceutical properties and highlight TFEB as a druggable target for autophagy inhibitor development.
Results
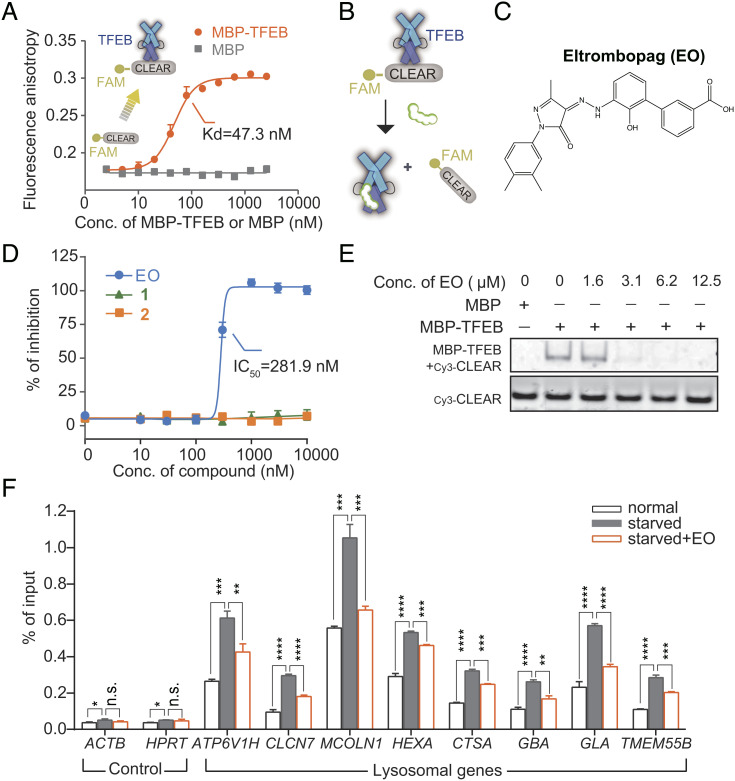
EO Inhibits TFEB-CLEAR DNA Interaction.
TFEB plays its role in autophagy regulation by recognizing CLEAR DNA. To screen for TFEB inhibitors, we designed a quantitative fluorescence anisotropy-based assay that monitoring the interaction between the bHLH-LZ domain of TFEB (TFEBbHLH-LZ) and the fluorophore FAM labeled CLEAR DNA (FAM-CLEAR DNA). The interaction of TFEBbHLH-LZ increased the fluorescence anisotropy of FAM-CLEAR DNA with a dissociation constant (Kd) of 47.3 nM (Fig. 1A). Unlabeled CLEAR DNA blocked the anisotropy signal with a medium inhibitory concentration (IC50) of 61.6 nM, while unlabeled mutant CLEAR DNAs had no effect (SI Appendix, Fig. S1A), indicating the validity of this assay. We screened a library of drug molecules that had been approved by the FDA or were undergoing clinical trials (Fig. 1B). The screening had a signal-to-background ratio of 25.41 and a Z’ factor of 0.83 (SI Appendix, Fig. S1 B and C).
Fig. 1.
EO binds to TFEB and inhibits TFEB’s recognition of CLEAR DNA in vitro and in cells. (A) Serial diluted (0 to 2,560 nM) MBP-TFEB or MBP control was titrated to 10 nM FAM labeled CLEAR DNA element (denoted as FAM-CLEAR), and the fluorescence anisotropy was measured. Binding with MBP-TFEB caused an increase in polarization of the fluorescently labeled DNA. The calculation formula of Kd is provided in the method section. Error bars represent the SDs of three repeats. (B) Design of the quantitative fluorescence anisotropy-based assay for agents that inhibit TFEB-CLEAR DNA interaction. (C) Chemical structure of EO. (D) Serial diluted (0 to 10 μM) EO or two substructures of EO (1,3-methyl-1-p-tolyl-5-pyrazolone; 2,3′-amino-2′-hydroxy-[1,1′-biphenyl]-3-carboxylic acid) was titrated to a solution containing 50 nM TFEB and 5 nM FAM-CLEAR DNA element, and the fluorescence anisotropy was measured. Error bars represent the SEMs of four repeats. (E) The disruption of the MBP-TFEB/Cy3-CLEAR complex by EO was examined by EMSA. (F) ChIP analysis of the effect of EO (10 µM) on the occupancy of TFEB on the promoters of lysosomal genes in HeLa cells. Error bars represent the SDs of three repeats. Student’s t test (unpaired); P* < 0.05, P** < 0.01, P*** < 0.001, P**** < 0.0001, and no significant (n.s.).
EO, an FDA-approved drug for thrombocytopenia treatment (18), was identified to inhibit the TFEB-CLEAR DNA interaction in the fluorescence anisotropy assay with an IC50 of 281.9 nM (Fig. 1 C and D). The two substructures bisected from EO (SI Appendix, Fig. S1D) were inactive with IC50 values exceeding 10 µM (Fig. 1D), indicating that the complete structure of EO is required for the activity. We further verified the activity of EO by electrophoretic mobility shift assays (EMSAs). The amount of TFEB-CLEAR DNA complexes decreased in a dose-dependent manner after the addition of EO (Fig. 1E and SI Appendix, Fig. S2), confirming that EO precluded TFEB from binding DNA. To assess if EO can disrupt the interaction between TFEB and CLEAR DNA in cells, we performed chromatin immunoprecipitation (ChIP) analysis. The addition of EO significantly disrupted the starvation-induced interaction between TFEB and the chromatin DNA (Fig. 1F). Together, these results indicate that EO inhibits the binding of TFEB to its target genes in both biochemical and cellular levels.
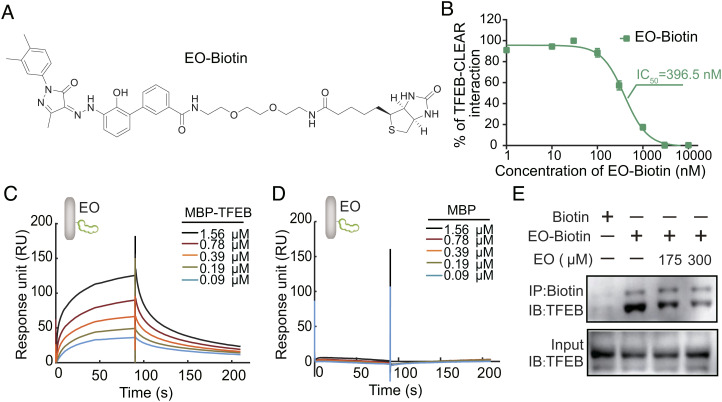
EO Directly Binds to TFEB Both In Vitro and in Cells.
To quantify the TFEB-EO interaction, we synthesized a biotin-labeled EO (EO-Biotin) which maintained the TFEB-CLEAR DNA disrupting ability (Fig. 2 A and B) and performed surface plasmon resonance (SPR) experiments with the EO immobilized on a chip through the Biotin tag and TFEB protein as the flow-through analyte. The results showed that maltose-binding protein (MBP)-TFEBbHLH-LZ interacted with EO with a Kd of 345.7 nM, while MBP control had no binding signal (Fig. 2 C and D). Using different truncations, we found that the interaction between EO and TFEB required the integrate HLH-LZ scaffold region of TFEB, but not the basic region (SI Appendix, Fig. S3).
Fig. 2.
Binding affinity between EO and TFEB was quantified by a biotinylated EO. (A) Chemical structure of biotinylated EO (EO-Biotin). (B) Serial diluted (0 to 10 μM) EO-Biotin was titrated to a solution containing 50 nM TFEB and 5 nM FAM-CLEAR DNA element. EO-Biotin inhibited the TFEB-CLEAR DNA interaction with an IC50 of 396.5 nM. Error bars represent the SEMs of four repeats. (C) EO-Biotin was immobilized on a SA chip. The affinity between the bHLH-LZ domain of TFEB and EO was measured by SPR experiment. MBP-TFEB bHLH-LZ interacts with EO with kon = 4.59E4 ± 5.60E2 (M s)−1, koff = 1.58E−2 ± 1.40E−4 s−1, and Kd = 345.7 nM. (D) The interaction between MBP control and EO cannot be detected in the SPR experiment. (E) Endogenous TFEB from the U87 cell lysate could be co-precipitated with EO-Biotin, while the competition of unlabeled EO reduced the co-precipitated TFEB. Abbreviations: IB, immunoblot; IP, immunoprecipitation.
To further validate the interaction between EO and endogenous TFEB, we performed pull-down assays with EO-Biotin in U87 cell lysate. The result showed that EO interacted with the endogenous TFEB but not with transcription factor binding to IGHM enhancer 3 (TFE3), both of which belong to the MI/TFE family (SI Appendix, Fig. S4). Unlabeled EO disrupted the interaction between TFEB and EO-Biotin in a dose-dependent manner, validating the specific engagement of EO with endogenous TFEB in the cellular environment (Fig. 2E). Together, these results confirm that small-molecule EO is capable of binding the HLH-LZ region of TFEB and preventing TFEB from binding to target DNAs.
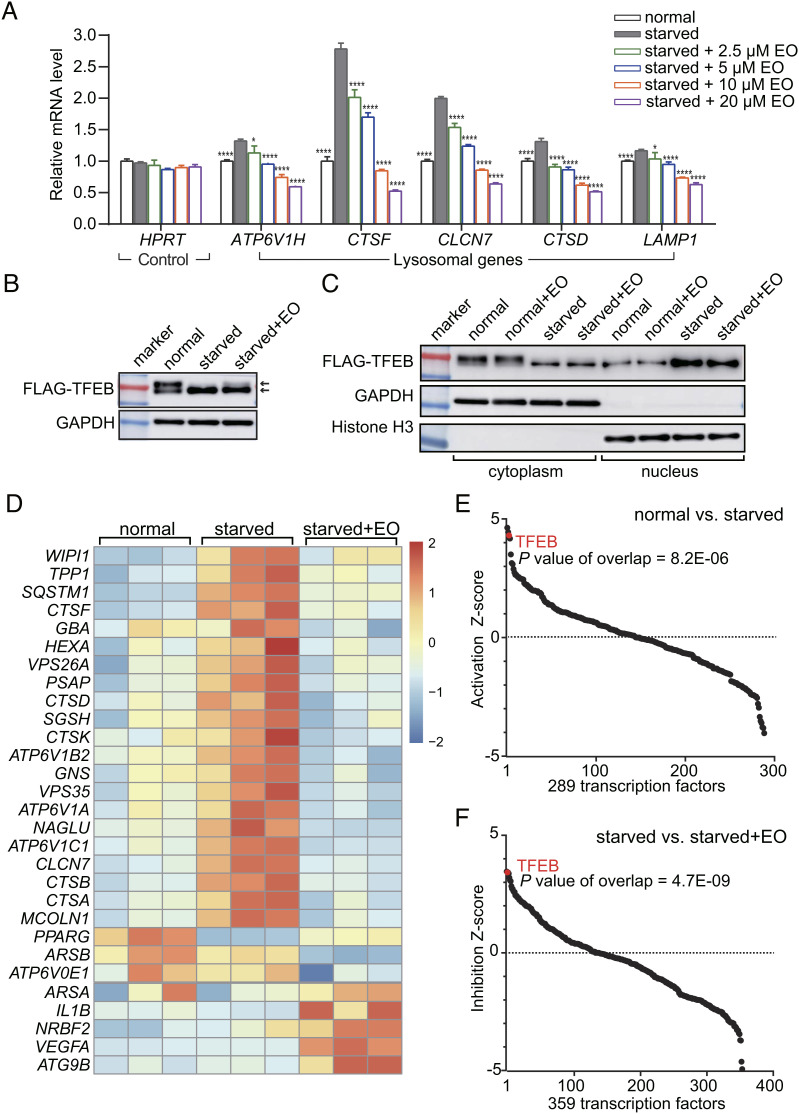
EO Selectively and Potently Inhibits TFEB Transcriptional Activity.
Next, we examined if EO could suppress autophagy-lysosomal genes expression. Using RT-PCR analysis, EO treatment dose-dependently reduced the mRNA levels of TFEB target lysosomal genes, which was activated by Earle’s Balanced Salt Solution (EBSS) starvation, but not the house keeping gene HPRT (Fig. 3A). Of note, the 2.5 to 10 µM EO treatment blocked lysosomal gene transcription to pre-starvation levels or even lower levels (Fig. 3A). The potency of EO to inhibit lysosomal gene expression was also observed in Hank’s Balanced Salt Solution-starved condition (SI Appendix, Fig. S5A). In addition, EO treatment also reduced the messenger RNA (mRNA) levels of TFEB downstream lysosomal genes under normal condition, implying that EO can affect the TFEB transcriptional activities under basal autophagy (SI Appendix, Fig. S5B). TFEB is reported to be regulated by multiple signaling pathways like the mammalian target of rapamycin (mTOR) and extracellular signal-regulated kinase (ERK) pathways. Pharmacological inhibition of mTOR by rapamycin promotes the TFEB transcriptional activity and further up-regulates autophagy (19). We also observed that EO treatment dose-dependently reduced the mRNA levels of TFEB target lysosomal genes under the condition of co-treatment with rapamycin (SI Appendix, Fig. S6). These results suggest that the TFEB inhibitor has systematic effects to inhibit autophagy under different signaling inputs.
Fig. 3.
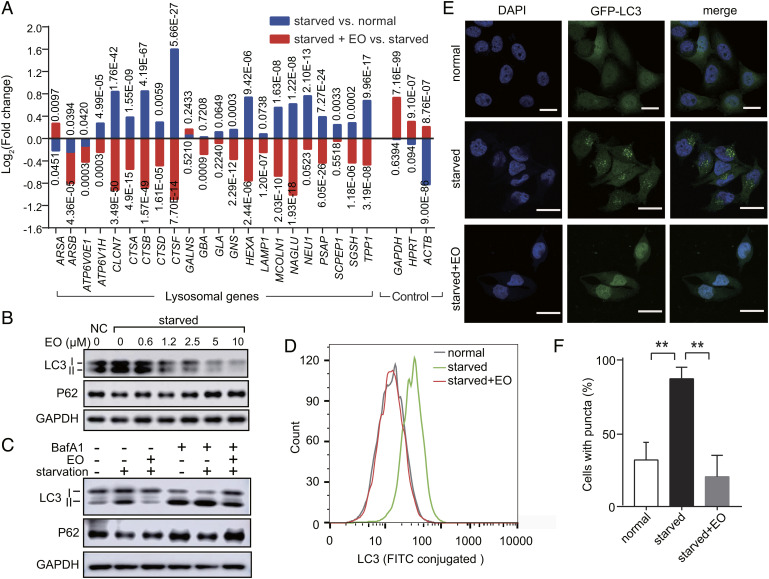
EO selectively inhibits starvation-induced TFEB transcriptional activity at a genome scale. (A) qPCR analysis of the mRNA levels of lysosomal genes from HeLa cells treated with normal medium (normal), nutrient-deprived EBSS medium with or without EO (concentration as indicated). The mRNA levels were normalized relative to GAPDH. HPRT is a representative housekeeping control gene. Error bars represent the SDs of four repeats. Significance relative to the starved group is calculated with Student’s t test (unpaired); P* < 0.05, P**** < 0.0001. (B) HeLa cells expressing 3 × FLAG-tagged TFEB were treated with normal medium (normal), nutrient-deprived EBSS medium (starved), or nutrient-deprived EBSS medium supplemented with 10 µM EO (starved + EO). The cell lysates were separated by electrophoresis and analyzed by western blot. (C) HeLa cells expressing 3 × FLAG-tagged TFEB were treated with or without 10 µM EO in normal medium or in nutrient-deprived EBSS medium. The cytosolic and nuclear fractions of these cells were isolated and analyzed by western blot. (D) RNA-Seq analysis of HeLa cells treated with normal medium, nutrient-deprived EBSS medium with or without EO (10 µM) (n = 3, N = 1). A heatmap for transcripts from 29 TFEB target genes according to IPA is shown. (E) Using IPA, TFEB was identified as the third significantly up-regulated transcription factor out of a total of 289 transcription factors after starving HeLa cells with EBSS (P = 8.2E−06). (F) Using IPA, TFEB was identified as the second significantly down-regulated transcription factor out of a total of 359 transcription factors after treating EBSS-starved HeLa cells with 10 µM EO (P = 4.7E−09).
Upon starvation, TFEB is dephosphorylated and translocates to nucleus for transcriptional activation (15, 16). It has been reported that changes in the phosphorylation state of TFEB are accompanied by shift of electrophoresis bands (16). And we found no significant shift of the electrophoresis band of TFEB in EO-treated cells, suggesting that EO treatment had no significant effect on the phosphorylation state of TFEB (Fig. 3B). And consistently, EO had no effect on the cytoplasmic and nuclear distribution of TFEB (Fig. 3C). Therefore, EO inhibits the expression of TFEB downstream genes primarily by preventing the binding of TFEB to DNA, rather than by altering its phosphorylation and subcellular localization. Because the phosphorylation and dephosphorylation of TFEB do not occur in the EO-binding region, the bHLH-LZ domain (20), EO may bind to both phosphorylated TFEB in the cytoplasm and dephosphorylated TFEB in the nucleus.
To assess the specificity of EO at the genome scale, we performed RNA sequencing (RNA-Seq) of the HeLa transcriptome. According to Ingenuity Pathway Analysis (IPA), 21 out of total 29 TFEB target genes were significantly up-regulated upon starvation (Fig. 3D). TFEB was the third significantly up-regulated transcription factor out of a total of 289 transcription factors in the starvation group compared to the normal group according to IPA analysis (P = 8.2E−06, Fig. 3E). Importantly, adding 10 µM EO into the starved condition, the increase of TFEB downstream genes was completely blocked (Fig. 3D). And TFEB was the second significantly down-regulated transcription factor out of a total of 359 transcription factors in the EO-treated group compared to the dimethylsulfoxide (DMSO)-treated group (P = 4.7E−09, Fig. 3F). Together, these results demonstrate that EO treatment systematically and selectively inhibits the TFEB transcriptional activity at the genomic scale.
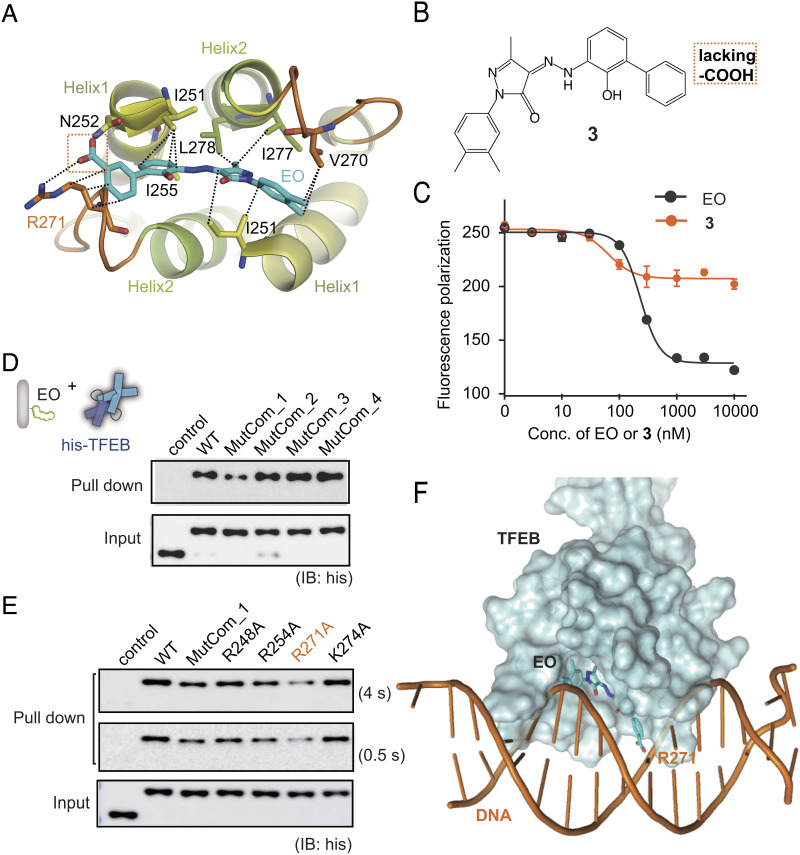
Mechanism of Action of EO on TFEB.
No crystal structure of TFEB has been previously reported. To explore the mode of action (MOA) of EO on TFEB, we solved the crystal structure of human HLH-LZ region of TFEB (residues 248 to 319, isoform 1, SI Appendix, Fig. S7A) to a resolution of 2.0 Å (SI Appendix, Table S2). The structure of TFEB contains two crystallographically independent molecules, chains A and B, to form a compact dimer. The first helix of TFEB started from R248 and ended at L261 (helix 1). Residues W272 to L319 formed an extended second helix consisting of helix 2 and the following leucine zipper (LZ) (SI Appendix, Fig. S7B). A hydrophobic four-helix bundle was constituted by residues on helix 1 and helix 2 from both chains (SI Appendix, Fig. S7C). The typical heptad repeats of leucine residues and other residues including I316, N309, H301, and E306 formed hydrophobic interactions or H-bonds in the LZ (SI Appendix, Fig. S7 D and E).
Then, we docked EO on the TFEB structure through the molecular dynamics simulations using Desmond. According to the docking model, EO binds to the bottom surface of the TFEB which contains a hydrophobic pocket formed by the helix 1-loop-helix 2 bundle (Fig. 4A). The I251, I255, V270, I277, and L278 formed hydrophobic interactions with EO. The carboxyl group of EO was recognized by N252 on helix 1 and R271 on the loop region through hydrogen bonds. In addition, the alkyl chain of R271 formed hydrophobic interactions with the benzene ring of EO (Fig. 4A). The model suggests that the carboxyl group of EO was crucial. To test this, we synthesized compound 3 which lacks carboxyl group compared with EO (Fig. 4B). Consistently, compound 3 significantly reduced the ability to disrupt TFEB-CLEAR interaction (Fig. 4C).
Fig. 4.
MOA of EO on TFEB. (A) EO is recognized of by TFEB through the HLH hydrophobic helix bundle according to molecular docking and dynamics simulation. (B) Chemical structure of compound 3 which lacks the carboxyl group compared with EO. (C) Compound 3 showed significantly lower inhibition activity against TFEB-DNA interaction than EO in the fluorescence anisotropy assay. Error bars represent the SEMs of four repeats. (D) Pull-down analysis of the interactions between EO and His-tagged TFEB WT, MutCom_1, MutCom_2, MutCom_3, and MutCom_4, using EO-Biotin immobilized on streptavidin beads. The precipitated TFEB proteins were analyzed by anti-His antibody. (E) Pull-down analysis of the interactions between EO and His-tagged TFEB WT, MutCom_1, R248A, R254A, R271A, and K274A. A representative result was shown with long exposure time (4 s) and short exposure time (0.5 s). (F) The CLEAR DNA is modeled onto the TFEB crystal structure using MITF-DNA complex (protein data bank code: 6KWI) as a reference. EO binding on the bottom surface of HLH region induces significant steric hindrance preventing further DNA binding.
We also performed orthogonal alanine scanning mutagenesis to explore the binding site of EO on TFEB. Since the carboxyl group of EO is required for TFEB interaction, we focused our mutagenesis studies on the basic arginines and lysines located on the outer surface of TFEB. Fifteen R/K residues were classified into four groups (MutCom_1 to 4) based on their distribution on the surface of TFEB (SI Appendix, Fig. S7 F–I). The interactions between EO and TFEB wild type (WT) were monitored using pull-down assays with EO immobilized on streptavidin beads through the biotin tag (SI Appendix, Fig. S8). The following pull-down assay between EO and TFEB mutants showed that only MutCom_1 lost the ability to bind EO, while mutation combos 2, 3, and 4 maintained the ability to bind EO as the TFEB WT (Fig. 4D). Next, we carried out point mutations within MutCom_1. R271A on the loop region is the most robust mutation to disrupt TFEB-EO interaction (Fig. 4E). Consistently, R271 is also the key residue to bind EO according to the docking model (Fig. 4A). Therefore, both molecular docking and alanine scanning mutagenesis converged to show that EO binds the bottom surface of HLH four-helix bundle, and the R271 on the loop is crucial for interaction. Superimposing the complex structure of DNA and microphthalmia associated transcription factor (MITF, which is the family member of TFEB) onto the TFEB, we observed that EO binding on the bottom surface of HLH region induces significant steric hindrance, preventing further DNA binding (Fig. 4F).
EO Blocks Starvation-Induced Autophagy in a Dose-Dependent Manner.
It’s known that knocking-down TFEB inhibited the starvation-induced autophagy response (15, 16). Consistently, EO treatment systematically reversed this starvation-induced transcriptional activation of lysosomal genes, indicating the ability of EO to inhibit autophagy (Fig. 5A). Light chain 3 (LC3)-II is the lipidated form of microtubule-associated protein 1A/1B LC3 and is accumulated during autophagy (16, 21). The adaptor protein sequestosome 1 (SQSTM1, also known as p62 protein) binds directly to LC3 and is also used as a marker to study autophagic flux, whose amount decreases with autophagy (22). EO dose-dependently induced p62 accumulation in starved HeLa cells upon EO treatment (Fig. 5B). Importantly, EO potently inhibited the formation of LC3-II in a dose-dependent manner with an IC50 less than 1.2 μM, and the formation of LC3 was completely blocked under the treatment of 10 μM EO (Fig. 5B). This efficiency is comparable to the VPS34 inhibitor—SAR405, a highly potent autophagy inhibitor being developed by Sanofi (23).
Fig. 5.
EO blocks the starvation-induced autophagy in a dose-dependent manner. (A) Expression analysis of lysosomal genes in HeLa cells treated with normal medium, nutrient-deprived EBSS medium with or without EO (n = 3, N = 1) according to RNA-Seq. Housekeeping genes GAPDHHPRT, and ACTB were used as controls. P values were marked accordingly. (B) Immunoblot analysis of LC3-I, LC3-II, and SQSTM1 (p62) in HeLa cells treated with normal medium control (NC), or nutrient-deprived EBSS medium with gradient EO (0 to 10 µM). (C) Immunoblot analysis of LC3-I, LC3-II and SQSTM1 (p62) in HeLa cells treated with normal medium, nutrient-deprived EBSS medium with or without EO (10 µM). BafA1 was used at a concentration of 200 nM. For B and C, GAPDH served as a loading control. One representative result of three independent experiments is shown. (D) Flow cytometric analysis of LC3 in HeLa cells treated with normal medium, nutrient-deprived EBSS medium with or without EO (10 µM). Using fluorescein isothiocyanate (FITC) conjugated LC3 antibody, the formation of lipidated LC3-II was quantified by flow cytometry. (E) Representative confocal images of HeLa cells stably expressing GFP-LC3-(ATG4 cleavage site)-RFP treated with normal medium, nutrient-deprived EBSS medium with or without 10 µM EO. Nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI). (Scale bars, 20 µm.) (F) Quantification of the percentage of LC3 puncta-positive cells in confocal images. Student’s t test (unpaired); P** < 0.01; error bars represent the SEMs of four repeats.
Bafilomycin A1 (BafA1) is a vacuolar-type ATPase (V-ATPase) inhibitor that prevents the re-acidification of synaptic vesicles, thereby blocking the fusion of autophagosomes and lysosomes. BafA1 treatment induced an accumulation of LC3-II in starved cells by disrupting autophagic flux (Fig. 5C). We found that EO treatment significantly inhibited the LC3-II formation in starved cells with or without BafA1, confirming that the attenuated LC3-II formation under EO treatment is caused by decreased autophagy, but not enhanced autophagic degradation (Fig. 5C). The ability of EO to inhibit the formation of LC3-II was further assessed through flow cytometry. Consistently, 10 µM treatment of EO reduced LC3-II in starved cells to the level of normal cells (Fig. 5D).
LC3-II exhibits puncta phenotype during autophagy, and the amount of LC3-II puncta reflects the number of autophagosomes (16, 21). To visualize the effect of EO to inhibit autophagy, we assessed the fluorescent signal generated by green fluorescent protein (GFP)-LC3 in HeLa cells (24). Upon starvation, GFP-LC3 formed puncta in cytoplasm indicating the formation of autophagosome (Fig. 5E). EO treatment blocked this starvation-induced LC3 puncta formation (Fig. 5 E and F) and induced a typical nucleus distribution of LC3 as observed in nutrient cells (Fig. 5E) (25, 26). These data confirm that EO is a potent autophagy inhibitor.
EO Increases the Sensitivity of Glioblastoma to TMZ Treatment.
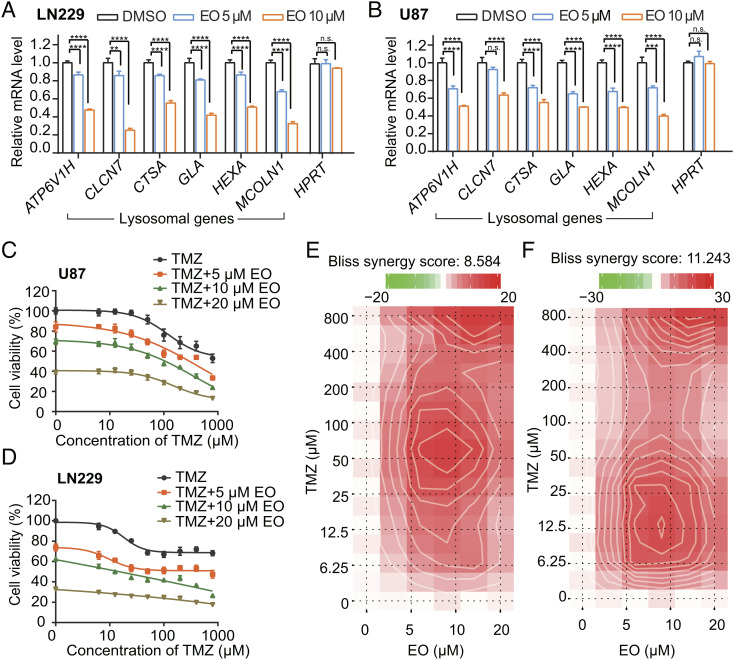
As a thrombopoietin receptor agonist to increase platelet number, EO has been employed to treat thrombocytopenia in patients with malignant hematologic disorders (27). With the potency to inhibit autophagy, EO may benefit a wide range of cancer treatments. We tested the potential usage of EO on glioblastoma, one of the most aggressive and intractable brain tumors (28). TMZ is the first-line chemotherapeutic drug to treat glioblastoma, while autophagy accounts for the high incidence of TMZ resistance and tumor recurrence (29). Suppressing autophagy to sensitize glioblastoma to TMZ treatment has been considered as a promising strategy (29). Here, we first assessed if EO is effective in LN229 and U87 glioblastoma cells. The result showed that treatment with EO decreased the mRNA levels of lysosomal genes in a dose-dependent manner, without affecting the house keeping gene HPRT (Fig. 6 A and B). The similar effect was also observed in U87 glioma cell line that overexpresses epidermal growth factor receptor variant III (U87-EGFRvIII), which confers radiation resistance (SI Appendix, Fig. S9A) (30). Consistently, the formations of LC3-II were attenuated by 5 to 20 μM EO treatment, indicating that EO decreased the autophagy levels in glioblastoma cells (SI Appendix, Fig. S9B). In the same concentration range, we observed that the sensitivity of glioblastoma cells U87 and LN229 to TMZ was significantly increased in the EO-treated cells (Fig. 6 C and D). Combining EO and TMZ showed a significant synergy with Bliss synergy scores of 8.58 and 11.24 in U87 and LN229 cells, respectively (Fig. 6 E and F). In contrast, CQ in the same concentration range had little effect on improving the sensitivity of glioblastoma cells to TMZ (SI Appendix, Fig. S10).
Fig. 6.
EO inhibits the autophagy level of glioblastoma cells and increases the sensitivity of glioblastoma cells to TMZ. (A and B) RT-PCR analysis of the mRNA levels of lysosomal genes normalized relative to GAPDH in glioblastoma cells LN229 (A) and U87 (B) treated with DMSO, 5 µM EO, and 10 µM EO, respectively. HPRT is a housekeeping control gene. Error bars represent the SDs of four repeats. Student’s t test (unpaired); P** < 0.01; P*** < 0.001; P**** < 0.0001, and no significant (n.s.). (C and D) Cytotoxicity of EO (0, 5, 10, and 20 µM) in combination with gradient TMZ (0 to 800 µM) on glioblastoma cells U87 (C) and LN229 (D) were assessed with the cell counting kit-8 assay. The cell viabilities were normalized relative to untreated samples. Error bars represent the SDs of five repeats. (E and F) Bliss model of EO (0, 5, 10, and 20 µM) and gradient TMZ (0 to 800 µM) synergy in U87 (E) and LN229 (F) cells. A positive score represents a synergistic effect.
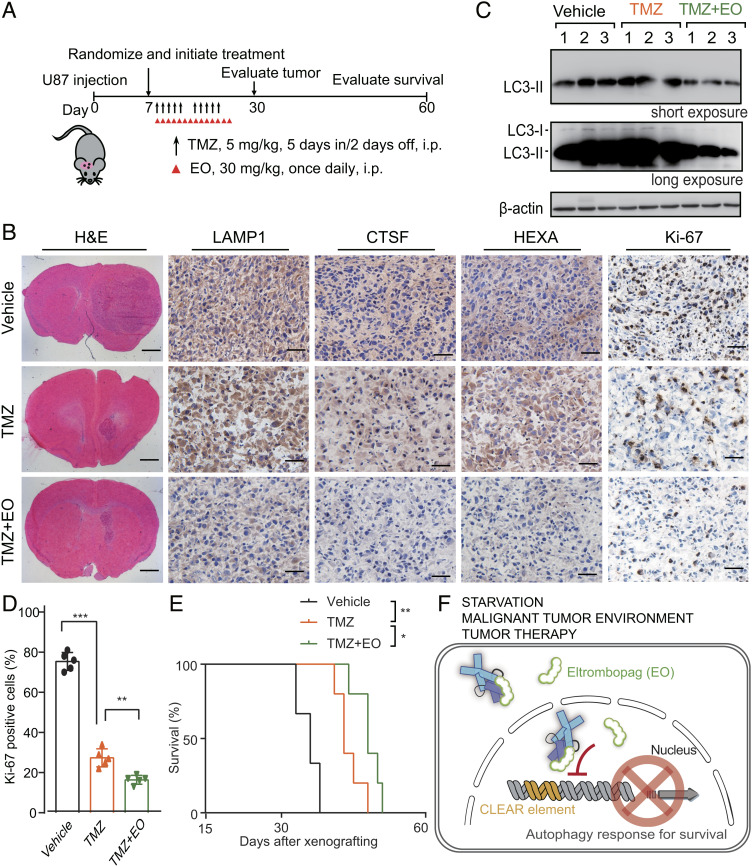
To evaluate the effect of EO in autophagy inhibition and tumor growth of glioblastoma in vivo, we transplanted U87 cells into the brains of athymic nude mice to establish an orthotopic glioblastoma xenograft model (Fig. 7A). EO combined with TMZ treatment significantly decreased the protein levels of the TFEB target genes LAMP1, CTSF, and HEXA, supporting that TFEB is directly engaged by EO in these xenografts (Fig. 7B). Of note, the LAMP1 is a classic lysosomal marker, and the decreasing of LAMP1 upon EO treatment indicates that EO impairs the lysosomal and autophagic pathways. The decreased LC3-II formation after EO treatment further confirmed that EO decreased the autophagy level in vivo (Fig. 7C). Consistent with the synergistic effects observed in cell experiments, Ki-67 staining showed that EO combined with TMZ treatment decreased tumor proliferation rate compared to TMZ alone (Fig. 7 B–D). Lastly, EO combined with TMZ significantly extended the survival time of mice bearing intracerebral glioblastoma (Fig. 7E). These results demonstrate the potency of EO to inhibit autophagy in vivo and the potential to benefit chemotherapy for glioblastoma treatment.
Fig. 7.
EO increases the sensitivity of glioblastoma to TMZ in a mouse model. (A) Treatment scheme for the evaluation of in vivo efficacy of EO in combination with TMZ in U87 tumor xenografts. (B) Representative images of H&E, LAMP1, CTSF, HEXA, and Ki-67 IHC of brain sections containing orthotopic tumors treated with TMZ, TMZ + EO, or vehicle after 30 d of injection. (Scale bars for H&E staining, 1 mm. Scale bars for LAMP1, CTSF, HEXA, and Ki-67 IHC, 50 μm.) (C) Quantification of Ki-67 positive cells in the brain sections. unpaired Student’s t test; P** < 0.01; P*** < 0.001; error bars represent the SDs of five repeats. (D) Immunoblot analysis of LC3-I, LC3-II in lysates of xenograft tumors resected from three of the brain sections in B under the microscope. The experiments were independently repeated three times with similar results. (E) The Kaplan–Meier survival curves of athymic nude mice bearing intracranial U87 gliomas treated with vehicle, TMZ, or TMZ + EO (vehicle, n = 6; TMZ, n = 5; TMZ + EO, n = 5). Statistical analysis was performed by log-rank test. P* < 0.05, P** < 0.01. (F) Schematic cartoon showing that the EO binds to TFEB to interfere CLEAR DNA recognition and inhibits autophagy response.
Discussion
In summary, the FDA-approved drug EO directly binds to the autophagy central transcriptional regulator TFEB, disrupts its interactions with CLEAR DNA without affecting its phosphorylation and nuclear localization, selectively inhibits TFEB’s transcriptional activity at a genomic scale, and eventually blocks autophagy (Fig. 7F).
EO inhibits the high autophagy level of glioblastoma in vitro and in vivo and increases the sensitivity of glioblastoma to TMZ. The cytotoxicity of EO alone against glioblastoma cells U87, LN229, and the non-cancerous normal human astrocytes (NHA) cell of the same origin is not significant, and the IC50s are around 20 to 30 μM (SI Appendix, Fig. S11). On the other hand, when the concentration was higher than 1.2 to 5 μM, EO showed significant inhibition of autophagy (Fig. 5B and SI Appendix, Fig. S9B). Therefore, the effect of EO is mainly manifested by inhibiting autophagy rather than selectively killing tumor cells. When it was used in combination with the chemotherapy drug TMZ, its inhibition on autophagy enhanced the therapeutic effect of TMZ (Fig. 7 B–E).
As an FDA-approved drug, EO has been used in patients with chronic immune thrombocytopenia for a long time. The safety of EO has been evaluated. Its side effects have been well-documented clinically, but no serious neurological side effects have been reported. This is an advantage of the clinical use of EO to inhibit autophagy. In the animal experiment, we also did not observe mental and behavioral abnormalities of mice in all groups except in the final course of diseases. The H&E staining on the other side of the brains from all groups showed normal neuronal appearances (SI Appendix, Fig. S12). There were no significant pathological changes such as cell body shrinkage, pyknotic nuclei, edema, or vacuolar alteration. These results suggest that EO is an effective and safe autophagy inhibitor and also highlights TFEB as a valuable target for autophagy inhibitor development.
Throughout the past decade, autophagy has attracted considerable attention for the development of new therapies (2, 31). Translational efforts to inhibit autophagy have predominantly focused on an antimalarial drug CQ or HCQ, which blocks lysosome acidification and autophagosome degradation (32). Extensive studies showed that autophagy inhibition with HCQ can augment the efficacy of DNA-damaging therapy, mTOR inhibition, histone deacetylases (HDAC) inhibition, and mitogen-activated protein kinase (MAPK) pathway inhibition; the combination therapies lead to enhancement of tumor shrinkage in patients with advanced solid tumors (8, 33–37). However, HCQ could only produce modest lysosomal inhibition observed in patients treated with the highest FDA-allowed dose (up to 1,200 mg/d) (38). Due to the limited efficiency and the related cytotoxicity, the identification of next-generation autophagy inhibitors is critical.
TFEB is the master regulator of lysosomal function and autophagy by orchestrating the expression of a broad range of genes involved in lysosome biogenesis and autophagic pathway, including autophagosome formation, autophagosome-lysosome fusion, and substrate degradation (15, 16, 39). TFEB inhibitor will have systematic and synergistic effects to inhibit autophagy.
Autophagy is central to the adaption to stresses induced by nutritional deprivation, infections, and metabolic, and physical and chemical challenges (1–3). Specifically, tumor cells are challenged by significantly elevated stresses from the deprivation of nutrient and oxygen and also the high metabolic demand of cell proliferation (32). As malignant tumors develop, autophagy is induced both in the tumor microenvironment and systemically in distant tissues to evade lethal metabolic stress and to maintain metabolic homeostasis (32, 40). Importantly, autophagy has also been considered as a key immune escape mechanism. Autophagy inhibition restricts cancer growth through its effects on the tumor microenvironment and on host immunity that myeloid cells found in the tumor microenvironment rely on autophagy to shut down the activity of anticancer T cells in the vicinity (41). Autophagy inhibition enhances programmed death-ligand 1 (PD-L1) expression in gastric cancer (42). Further, autophagy is up-regulated in response to chemotherapy and radiotherapy supporting the survival of the treatment resistant cancer cells (2). Using EO, a new potent and safety autophagy inhibitor, a goal of future research will be the development of combination strategy to benefit a wide variety of cancer treatments.
Materials and Methods
Protein Preparation.
The MBP tagged TFEB constructs were generated with the pHis-MBP-tobacco etch virus protease (TEV) vector. All the plasmids were induced to be expressed in the bacterial strain Rosetta and purified to homogeneity with a Ni-HiTrap affinity column and a size exclusion S200 column (GE Healthcare). Further details on protein preparation are provided in SI Appendix.
High-Throughput Screening.
A library of FDA-approved compounds was screened using the quantitative fluorescence anisotropy-based assay. The assay was performed using a 96-well plate. Each plate included negative control wells with signal that was generated by adding DMSO (1 %) to the system and positive control wells with signal that was generated by MBP. Compounds were tested in triplicate at a final concentration of 10 μM (1 % DMSO). A total of 54 plates were used for this assay.
Biochemical Experiments.
The measurements of the fluorescence anisotropy of FAM-CLEAR DNA were carried out using an EnVision® Multimode Plate Reader (PerkinElmer). EMSA was done by using Cy3 labeled CLEAR DNA. The DNA–protein complex and free DNA were separated by a 6% non-denaturing polyacrylamide gel. The gel was photographed with Amersham™ Imager 600 (GE Healthcare). To measure the affinity between EO and TFEB, a biotin conjugated EO was synthesized. The binding affinity between EO-Biotin and TFEB was measured using a Biocore T200 (GE Healthcare). EO analogs were synthesized to analyze the key pharmacophore. Complete protocols of chemical synthesis, fluorescence anisotropy assay, EMSA, and SPR assay are included in SI Appendix.
Cell Experiments.
HeLa, U87, and LN229 cell lines were purchased from the American Type Culture Collection. NHA cell line was kindly provided by Key Laboratory of Aging and Neurological Disorder Research of Zhejiang Province. HeLa cell line stably expressing GFP-LC3-(ATG4 cleavage site)-red fluorescent protein (RFP) was kindly provided by Lifeng Pan. U87 EGFRvIII cell line that overexpress exogenous EGFRvIII was established by transducing EGFRvIII into U87 cells and characterized by immunoblotting (43). These cells were cultured in Dulbecco’s Modified Eagle’s Medium (HyClone™), all supplemented with 10% of fetal bovine serum (Gibco) and 1% Penicillin-Streptomycin (BIOAGRIO). All cell culture was performed in a 37 °C and 5% CO2 incubator. Complete protocols of ChIP, pull-down, RT-PCR, RNA-Seq, western blotting, flow cytometry, confocal microscopy, cell viability, and drug synergy assays are included in SI Appendix.
Structure Determination and Analysis.
Crystallization of TFEB_HLH-LZ was performed by the sitting-drop vapor-diffusion method at 18 °C. X-ray diffraction data were collected on beamline 19U1 at the Shanghai Synchrotron Radiation Facility using a PILATUS detector (44). Further details on crystallization, data collection, structure refinement, docking calculations, and molecular dynamics are provided in SI Appendix.
Tumorigenicity Studies.
All xenograft studies were approved by the Shanghai Jiao Tong University Institutional Animal Care and Use Committee. Athymic nude mice aged 5 to 6 wk (SLAC) were randomly divided into 5 to 6 per group. In total, 5 × 105 cells of U87 were stereotactically implanted into the mouse brain as previously described (45). Treatment schemes were described in the corresponding figure legends. Mice were euthanized when neuropathological symptoms developed. Individuals who were blinded measured tumor volumes as (W2 × L)/2, W < L.
Immunohistochemistry (IHC).
Expression of Ki-67, LAMP1, CTSF, and HEXA on xenograft tumors was evaluated on 6-μm-thick optimum cutting temperature (O.C.T.) compound-embedded fresh-frozen tissue sections. Briefly, the tissue sections were separately stained with hematoxylin and eosin (H&E) or antibodies against Ki-67 (Invitrogen, MA5-14520), LAMP1 (Proteintech, 21997-1-AP), CTSF (Proteintech, 11055-1-AP), and HEXA (Proteintech, 11317-1-AP) as previously described (45). Images were captured using an Olympus BX53 microscope equipped with an Olympus DP73 digital camera. Ki-67 were quantified by calculating the percentage of Ki-67 positive cells. Two individuals blinded to the slides examined and scored the samples.
Statistical Analysis.
The statistical significance in the ChIP experiment (Fig. 1F), RT-PCR experiment (Figs. 3A and 6 A and B and SI Appendix, Figs. S5, S6, and S9A), quantification of the percentage of LC3 puncta-positive cells (Fig. 5F), and quantification of Ki-67 positive cells (Fig. 7D) was assessed by unpaired Student’s t test (two-tailed). The statistical significance of Kaplan–Meier survival curves in animal experiments (Fig. 7E) was assessed by log-rank test.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
The study is supported by the National Natural Science Foundation of China, Grants 21977108 and 22277134 (J.W.), 21977107 and 22277132 (P.F.), and 21621002 (B.Y.); the Shanghai Science and Technology Committee (20S11901100 to J.W. and 22ZR1475000 to P.F.); the Strategic Priority Research Program of the Chinese Academy of Sciences Grant No. XDB20000000; One-Hundred Talents Program of Chinese Academy of Sciences; Shanghai Pujiang Program; the State Key Laboratory of Bioorganic and Natural Products Chemistry (J.W.); a 1000-talent young investigator award (P.F.); and a Program of Shanghai Academic/Technology Research Leader (21XD1403100 to H.F.).
Author contributions
K.D., Z.W., H.F., B.Y., P.F., and J.W. designed research; Y.L., Q.S., G.Y., F.S., Y.Z., T.W., P.X., P.L., Z.L., H.S., and Z.Z. performed research; Y.L., Q.S., P.X., Z.W., H.F., B.Y., P.F., and J.W. analyzed data; and Y.L., P.F., and J.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Haizhong Feng, Email: fenghaizhong@sjtu.edu.cn.
Biao Yu, Email: byu@sioc.ac.cn.
Pengfei Fang, Email: fangpengfei@sioc.ac.cn.
Jing Wang, Email: Jwang@sioc.ac.cn.
Data, Materials, and Software Availability
The reported crystal structure has been deposited in the Protein Data Bank under the accession number 7Y62.
Supporting Information
References
- 1.Green D. R., Levine B., To be or not to be? How selective autophagy and cell death govern cell fate. Cell 157, 65–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galluzzi L., Bravo-San Pedro J. M., Levine B., Green D. R., Kroemer G., Pharmacological modulation of autophagy: Therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 16, 487–511 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinsztein D. C., Codogno P., Levine B., Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709–730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaravadi R. K., Kimmelman A. C., Debnath J., Targeting autophagy in cancer: Recent advances and future directions. Cancer Discov. 9, 1167–1181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N., Levine B., Autophagy in human diseases. N. Engl. J. Med. 383, 1564–1576 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Kimmelman A. C., White E., Autophagy and tumor metabolism. Cell Metab. 25, 1037–1043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy J. M. M., Towers C. G., Thorburn A., Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy J. M., et al. , Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 4, 773–780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strohecker A. M., et al. , Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 3, 1272–1285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang A., et al. , Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 8, 276–287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karsli-Uzunbas G., et al. , Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 4, 914–927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo J. Y., Xia B., White E., Autophagy-mediated tumor promotion. Cell 155, 1216–1219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galluzzi L., Green D. R., Autophagy-independent functions of the autophagy machinery. Cell 177, 1682–1699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitmarsh-Everiss T., Laraia L., Small molecule probes for targeting autophagy. Nat. Chem. Biol. 17, 653–664 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Sardiello M., et al. , A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Settembre C., et al. , TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuervo A. M., Cell biology. Autophagy’s top chef. Science 332, 1392–1393 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Zimmer J., Hentges F., Andres E., Eltrombopag in thrombocytopenia. N. Engl. J. Med. 358, 1072–1073 (2008). [PubMed] [Google Scholar]
- 19.Roczniak-Ferguson A., et al. , The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puertollano R., Ferguson S. M., Brugarolas J., Ballabio A., The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 37, e98804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabeya Y., et al. , LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjorkoy G., et al. , Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 452, 181–197 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Ronan B., et al. , A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10, 1013–1019 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Kaizuka T., et al. , An autophagic flux probe that releases an internal control. Mol. Cell 64, 835–849 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Huang R., Liu W., Identifying an essential role of nuclear LC3 for autophagy. Autophagy 11, 852–853 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang R., et al. , Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Pathak S., Roth M., Verma A., Steidl U., Eltrombopag for the treatment of thrombocytopenia in patients with malignant and non-malignant hematologic disorders. Expert Opin. Drug Metab. Toxicol. 9, 1667–1675 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Thomas A. A., Brennan C. W., DeAngelis L. M., Omuro A. M., Emerging therapies for glioblastoma. JAMA Neurol. 71, 1437–1444 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Yan Y., et al. , Targeting autophagy to sensitive glioma to temozolomide treatment. J. Exp. Clin. Cancer Res. 35, 23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee B., et al. , EGFRvIII and DNA double-strand break repair: A molecular mechanism for radioresistance in glioblastoma. Cancer Res. 69, 4252–4259 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolgin E., Anticancer autophagy inhibitors attract “resurgent” interest. Nat. Rev. Drug Discov. 18, 408–410 (2019). [DOI] [PubMed] [Google Scholar]
- 32.White E., DiPaola R. S., The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 15, 5308–5316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahalingam D., et al. , Combined autophagy and HDAC inhibition: A phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy 10, 1403–1414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rangwala R., et al. , Combined MTOR and autophagy inhibition: Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 10, 1391–1402 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barnard R. A., et al. , Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy 10, 1415–1425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rangwala R., et al. , Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 10, 1369–1379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collisson E. A., et al. , A central role for RAF–>MEK–>ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2, 685–693 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaud M., et al. , Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334, 1573–1577 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Napolitano G., Ballabio A., TFEB at a glance. J. Cell Sci. 129, 2475–2481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mancias J. D., Kimmelman A. C., Targeting autophagy addiction in cancer. Oncotarget 2, 1302–1306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunha L. D., et al. , LC3-associated phagocytosis in myeloid cells promotes tumor immune tolerance. Cell 175, 429–441.e416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., et al. , Autophagy inhibition enhances PD-L1 expression in gastric cancer. J. Exp. Clin. Cancer Res. 38, 140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lv D., et al. , TRIM24 is an oncogenic transcriptional co-activator of STAT3 in glioblastoma. Nat. Commun. 8, 1454 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W. Z., et al. , The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl. Sci. Tech. 30, 170 (2019). [Google Scholar]
- 45.Feng H., et al. , EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis. J. Clin. Invest. 124, 3741–3756 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The reported crystal structure has been deposited in the Protein Data Bank under the accession number 7Y62.