Abstract
A gene encoding superoxide dismutase (SOD), sodM, from S. aureus was cloned and characterized. The deduced amino acid sequence specifies a 187-amino-acid protein with 75% identity to the S. aureus SodA protein. Amino acid sequence comparisons with known SODs and relative insensitivity to hydrogen peroxide and potassium cyanide indicate that SodM most likely uses manganese (Mn) as a cofactor. The sodM gene expressed from a plasmid rescued an Escherichia coli double mutant (sodA sodB) under conditions that are otherwise lethal. SOD activity gels of S. aureus RN6390 whole-cell lysates revealed three closely migrating bands of activity. The two upper bands were absent in a sodM mutant, while the two lower bands were absent in a sodA mutant. Thus, the middle band of activity most likely represents a SodM-SodA hybrid protein. All three bands of activity increased as highly aerated cultures entered the late exponential phase of growth, SodM more so than SodA. Viability of the sodA and sodM sodA mutants but not the sodM mutant was drastically reduced under oxidative stress conditions generated by methyl viologen (MV) added during the early exponential phase of growth. However, only the viability of the sodM sodA mutant was reduced when MV was added during the late exponential and stationary phases of growth. These data indicate that while SodA may be the major SOD activity in S. aureus throughout all stages of growth, SodM, under oxidative stress, becomes a major source of activity during the late exponential and stationary phases of growth such that viability and growth of an S. aureus sodA mutant are maintained.
Staphylococcus aureus is a gram-positive facultative anaerobe that typically resides on the skin and mucous membranes of approximately 30% of healthy individuals and up to 90% of health care workers (42). Therefore, it is not surprising that of the estimated 2 million hospitalizations each year that result in a nosocomial infection, S. aureus is one of the most common causative agents (5, 15). S. aureus has the capacity to produce more than 30 secreted proteins in the form of enzymes, immunotoxins, and cytotoxins and numerous cell surface-associated factors that promote adherence to various tissues and prevent attack by the host's defenses (17, 36). Consequently, S. aureus causes numerous different kinds of infections, ranging from skin abscesses to life-threatening endocarditis, meningitis, and pneumonia as well as toxemias such as scalded skin and toxic shock syndromes (42).
The skin and mucous membranes serve as the primary line of defense against infection by S. aureus (41). However, when this organism is introduced into the underlying tissues, the primary defense mechanism is the professional phagocyte (41). Polymorphonuclear leukocytes and macrophages use toxic reactive oxygen intermediates such as superoxide and hydrogen peroxide to aid in the killing of phagocytized bacteria (11, 22, 35). In addition, these same oxygen species are produced during aerobic respiration and have the potential to damage DNA, protein, and lipids (12, 18).
In order to detoxify these reactive oxygen intermediates, bacteria produce several classes of superoxide dismutases (SODs) and catalases that convert superoxide to hydrogen peroxide and hydrogen peroxide to water and oxygen (for reviews, see references 13 and 40). SODs are metalloenzymes classified by the type of metal cofactor utilized (13, 40). In bacteria, manganese (Mn) and iron (Fe) SODs are localized in the cytoplasm and are believed to be important in protecting nucleic acid, proteins, and lipids from the damaging effects of superoxide (13, 40). In gram-negative bacteria, copper-zinc SODs reside in the periplasm, where they are hypothesized to act upon exogenous superoxide (13, 40). Recently, a nickel-containing SOD was isolated and the gene subsequently cloned from Streptomyces coelicolor (20, 21).
The function of SOD in S. aureus has been presumed to be similar to that of other bacterial SODs. However, the only studies attempting to correlate a role for this enzyme in staphylococcal disease have generated conflicting information. Mandell (27) demonstrated that SOD activity in clinical isolates, whether high or low, did not correlate with lethality in a mouse model of infection. Furthermore, the differences in SOD activity did not impair the ability of polymorphonuclear leukocytes to kill intracellular staphylococci. In contrast, Kanafani and Martin (19) demonstrated that virulent strains of S. aureus from patients with confirmed staphylococcal disease exhibited significantly higher levels of SOD activity than nonvirulent isolates from patients who exhibited no staphylococcal disease. In addition, when these strains were compared in a neonatal mouse model, the mice inoculated with virulent strains demonstrated significantly lower weight gain than mice inoculated with nonvirulent strains (19).
Recently, Watson et al. (43) reported the isolation of a number of transposon mutants of S. aureus with an impaired ability to survive long-term starvation. One of these mutations was determined to be within a gene belonging to the Mn family of SODs, sodA (43). Upon further examination, it was determined that S. aureus produces three bands of SOD activity, as assessed by nondenaturing polyacrylamide gel electrophoresis (7). The two lowest-migrating bands of activity were absent in the sodA transposon mutant. Amino acid sequence analysis, along with a demonstrated dependence upon Mn for activity and relative resistance to hydrogen peroxide, led to the conclusion that the SodA of S. aureus is most likely a Mn-SOD (7). Previous to the Clements et al. (7) study, Poyart et al. (33) used degenerate primers designed from conserved regions of several gram-positive Mn-SOD genes to isolate a PCR product from S. aureus that appeared to represent ∼85% of a putative sod gene. The nucleic acid sequence of this gene, showed only 71% identity with the sodA gene isolated by Clements et al. (7), indicating that S. aureus most likely contains two SOD genes.
In this study, we report the cloning and characterization of a second gene for SOD activity in S. aureus. The gene has been designated sodM, due to its amino acid similarities to the Mn family of SODs and its insensitivity to hydrogen peroxide and potassium cyanide, a characteristic of Mn-SODs. Results from this study indicate that the two sod genes in S. aureus account for three distinct SOD activities; SodM, SodA, and a hybrid SodM-SodA form, which represents a SOD profile not previously recognized in gram-positive bacteria. Expression of sodM was greatest under high-aeration growth conditions during the late exponential and postexponential phases of growth, and SodM provided protection from oxidative stress for a sodA mutant strain of S. aureus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. Strains were routinely grown overnight (15 to 18 h) in tryptic soy broth (TSB; Difco Laboratories, Detroit, Mich.) at 37°C with rotary aeration (180 rpm) or on TSA plates (TSB containing 1.5% agar). To examine the effects of high and low aeration, S. aureus was grown essentially as described by Clements et al. (7). High aeration was achieved by using a flask-to-volume ratio of 25 with rotary aeration (225 rpm), while low aeration was achieved by using a flask-to-volume ratio of 2.5 with rotary aeration (125 rpm). Strains of Escherichia coli were routinely grown at 37°C with rotary aeration (225 rpm) in either Luria-Bertani (LB) broth or M63 minimal medium (30) with the appropriate antibiotic selection. Solid media consisted of either LB or M63 containing 1.5% agar. Antibiotic-resistant S. aureus strains were selected with and maintained on either erythromycin, tetracycline, or chloramphenicol (Sigma Chemical Co., St. Louis, Mo.) at 5 μg/ml, while antibiotic-resistant E. coli strains were grown in the presence of carbenicillin (Sigma) at 100 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype and/or relevant characteristic(s) | Reference and/or source |
|---|---|---|
| S. aureus | ||
| RN4220 | Nitrosoguanidine-induced restriction mutant | 32; J. J. Iandolo, University of Oklahoma Health Science Center |
| RN6390 | Prototypic strain | 32; M. S. Smeltzer, University of Arkansas for Medical Sciences |
| NTH205 | RN6390, sodM::erm | This study |
| NTH247 | RN6390, sodA::tet | This study |
| NTH248 | RN6390, sodM::erm sodA::tet | This study |
| E. coli | ||
| INVαF′ | F′ endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 φ80lacZΔM15(lacZYA-argF)U169 λ− | Invitrogen |
| HB101 | F−leuB6 supE44 hsdS20(rB− mB−) recA13 ara-14 proA2 galK2 lacY1 rpsL20 xyl-5 mtl-1 | Laboratory stock |
| QC779 | Δ(argF-lac)169 λ− φ(sodB-kan)1-Δ2IN(rrnD-rrnE)1 rpsL179(Str), sodA25::MudPR13 | 4; D. Touati, Institut Jacques Monod, Paris, France |
| MG1655 | Prototypic strain | Tony Romeo, University of North Texas Health Science Center |
| NTH212 | QC779, containing pBA23::sodC | This study; plasmid (26) obtained from R. M. Roop, Louisiana State University Health Science Center |
Cloning of sodM. (i) Amplification by PCR.
Chromosomal DNA was isolated from S. aureus using the method of Dyer and Iandolo (10). Oligonucleotide primers (5′-TTAATTCTCTTTAAAAGCGGGAAA-3′ and 5′-GGGACATTCATCAACTTTTATCAG-3′) were designed using sequences from the S. aureus DNA databases maintained by The Institute for Genomic Research (http://www.tigr.org) and the University of Oklahoma's Advanced Center for Genome Technology (http://www.genome.ou.edu/staph.html) and used to amplify a 787-bp contiguous region from S. aureus RN6390 by PCR. The PCR product was ligated into pCR2.1 (Invitrogen, Carlsbad, Calif.) and transformed into E. coli INVαF′, and transformants were selected as recommended by the manufacturer (Invitrogen). Plasmid DNA from antibiotic-resistant transformants was isolated using a plasmid miniprep kit (Bio-Rad Laboratories, Richmond Calif.) and digested with EcoRI to verify the presence of an approximately 800-bp insert. Plasmid DNA containing the desired fragment (pCR2.1sodM) was sequenced at the University of Arkansas for Medical Sciences DNA Sequencing Core Facility (Little Rock, Ark.) using a DNA sequencer (Perkin-Elmer Biosystems, Foster City, Calif.).
(ii) Complementation of an E. coli sodA sodB mutant.
The E. coli strain QC779 (sodA sodB) was transformed with the pCR2.1sodM construct by electroporation, as recommended by the manufacturer (Bio-Rad Laboratories), and selected on M63 agar plates containing antibiotic. Plates were incubated at 37°C for 2 to 3 days before colonies were visible. In addition, E. coli QC779 was transformed with pBA23sodC (pUC9 containing the sodC gene from Brucella abortus) and selected on LB agar plates containing antibiotic. Plasmid DNA from several antibiotic-resistant transformants was isolated to verify the presence of either pCR2.1sodM or pBA23sodC, and transformants containing the appropriate plasmid were procured for further study.
Construction of sod mutations in S. aureus.
The erythromycin resistance marker (erm) of plasmid pDG647 (14) (kindly provided by Ken Bayles, University of Idaho, Moscow, Idaho) was isolated as a 1.6-kbp BamHI fragment and gel purified. The 5′ overhangs of the fragment were filled in using the Klenow fragment of DNA polymerase (Promega Corp., Madison, Wis.) and subcloned into the single SnaBI site located approximately in the middle of the sodM gene in pCR2.1sodM. The resulting construct, pCR2.1sodM::erm, was verified by restriction analysis. The sodM::erm cassette, residing on the 2.4-kbp EcoRI fragment of pCR2.1sodM::erm, was gel purified and ligated into the temperature-sensitive shuttle vector, pCL10 (34) (kindly provided by Chia Lee, University of Kansas Medical Center, Kansas City, Kans.). The resulting construct, pCL10sodM::erm, was transformed into S. aureus RN4220 by electroporation as described by Kraemer and Iandolo (23), and erythromycin-resistant transformants were selected at 30°C. A single plasmid-containing colony was chosen and grown overnight at 30°C in TSB containing erythromycin. Portions of the overnight culture were diluted 1,000-fold and plated as 0.1-ml aliquots onto TSA plates containing erythromycin. Plates were incubated for 36 h at 43°C, which is nonpermissive for plasmid replication. Erythromycin-resistant colonies growing at 43°C were analyzed for loss of SOD activity as described in the next section. The sodM::erm mutation in S. aureus RN4220 was moved into S. aureus RN6390 by φ11-mediated transduction (29), and the erythromycin-resistant transductants were analyzed for loss of SOD activity. Southern analysis using the sodM PCR product as the probe was performed to verify the disruption of sodM. The sodA mutant was generated in a similar manner except that the tetracycline resistance marker from pDG1515 (14) (kindly provided by Ken Bayles, University of Idaho) was inserted as a blunt-ended, 2.1-kbp EcoRI fragment into the SnaBI site of sodA. The sodA::tet mutation generated in S. aureus RN4220 was transduced into S. aureus RN6390 and S. aureus RN6390 containing the sodM::erm mutation. Southern analysis and SOD activity gels were used to confirm the mutations.
The PCR product containing the sodM gene was also subcloned into the expression shuttle vector pCL15 (kindly provided by Chia Lee at the University of Kansas Medical Center), which contains an inducible lac promoter. The pCL15sodM construct was transformed into RN4220 and subsequently moved into the S. aureus RN6390 double (sodM sodA) mutant by transduction (29). A chloramphenicol-resistant transductant was inoculated into TSB containing chloramphenicol and grown under high-aeration growth conditions. Expression of sodM was induced with IPTG (isopropyl-β-d-thiogalactopyranoside, 0.5 mM; Fisher Scientific, Fairlawn, N.J.) 1 h postinoculation, and cells were harvested 5 h later. Whole-cell lysates were assessed for SOD activity as described below. In addition, viability of the pCL15sodM-containing transformants was determined in the presence of methyl viologen (MV; Sigma). Sampling and determination of viability were performed as described below.
Preparation of cell lysates and SOD activity assay.
Whole-cell lysates from strains of S. aureus and E. coli were prepared using the procedure of Blevins et al. (3). Briefly, cells from broth cultures of either S. aureus or E. coli were harvested by centrifugation (12,000 × g for 10 min at 4°C), washed with an equal volume of TEG buffer (25 mM Tris, 25 mM EGTA [pH 8.0]), and suspended in 0.4 ml of TEG buffer. The cell suspensions were pipetted into 2.0-ml Fast Prep Blue tubes (Bio 101, Vista, Calif.) containing acid-washed, RNase-free 0.1-mm silica beads. The tubes were placed into a high-speed reciprocator (Bio 101) and agitated at 6 m/s for 40 s. The tubes were cooled on ice for 15 min, and the lysates were cleared by centrifugation (16,170 × g, for 10 min at 4°C). The supernatant was recovered as 0.2-ml portions and stored at −20°C until needed. Total protein of whole-cell lysates was determined using the Bradford assay (Bio-Rad Laboratories).
Equal amounts of cell protein (5 μg) were loaded onto 15% (wt/vol) nondenaturing polyacrylamide gels and separated by electrophoresis in buffer lacking sodium dodecyl sulfate (25). SOD activity was determined using the nitroblue tetrazolium negative staining method of Beauchamp and Fridovich (2). To determine the sensitivity of SOD to either hydrogen peroxide (H2O2) or potassium cyanide (KCN), gels were exposed to 5 mM H2O2 (Sigma) for 30 min, washed twice in deionized, glass-distilled H2O, and stained for SOD activity as described by Clare et al. (6). Sensitivity to KCN (Sigma) was determined by exposing gels to 10 mM KCN for 15 min prior to negative staining for SOD activity (26).
SOD activity of resolved bands was quantified by densitometry using the AlphaImager 2000 (Alpha Innotech Corp., San Leandro, Calif.) imaging system. Final values were calculated from the linear region of a standard curve of activity as a function of protein.
RNA isolation and Northern analysis.
Total RNA was isolated from S. aureus as described by Hart et al. (17). RNA (A260/A280 = 1.9 to 2.0) was diluted in diethylpyrocarbonate (Sigma)-treated water to a final concentration of 1 μg/ml and verified by comparing the intensities of the rRNA bands. RNAs, serially diluted twofold, were denatured in the presence of glyoxal (Eastman Kodak Co., Rochester, N.Y.) and dimethyl sulfoxide (Fisher Scientific) at 50°C for 1 h, electrophoresed through a 1.4% GTG agarose gel (BioWhittaker Molecular Applications, Rockland, Maine), and transferred by passive diffusion onto neutral nylon (MagnaGraph; Micron Separations, Inc., Westborough, Mass.). Membranes were hybridized overnight (18 to 24 h) at 65°C with DNA probes specific for the cloned sodM gene and 16S rRNA (37). Probes were randomly labeled with digoxigenin-11-UTP (Roche Molecular Biochemicals, Indianapolis, Ind.) and the Klenow fragment of DNA polymerase (Promega). Hybridized probes were detected by autoradiography with alkaline phosphatase-conjugated anti-digoxygenin F(ab′)2 antibody fragments (Roche Molecular Biochemicals) and the chemiluminescent substrate CDP-Star (Roche Molecular Biochemicals).
MV treatment.
The S. aureus parent and the sod mutant strains were tested for susceptibility to the internal oxygen radical generator MV. Overnight (15 to 18 h) cultures were used to inoculate 500-ml flasks containing 20 ml of TSB to an initial optical density of approximately 0.05 at 550 nm. Cultures were incubated at 37°C with rotary aeration (225 rpm), and growth was monitored spectrophotometrically until cells reached early exponential phase (1.5 h, optical density at 550 nm of ca. 0.2). Freshly prepared MV was added to a final concentration of 10 mM at 1.5 h and at times in growth that corresponded to late exponential (6 h) and postexponential (12 h) phases of growth. Growth was assessed spectrophotometrically, and cell viability was determined by taking aliquots at various times points, diluting and plating on TSA. Plates were incubated overnight at 37°C before colonies were counted. Cell viability values are geometric means (×/÷ standard errors) of two to five independent determinations. Statistical significance was determined by analysis of variance followed by Fisher protected least significant difference multigroup comparison using StatView (SAS Institute Inc., Cary, N.C.) with a P value of ≤0.05.
Nucleotide sequence accession number.
The sodM nucleotide sequence has been assigned the accession number AF273269 by GenBank.
RESULTS AND DISCUSSION
Identification and isolation of the S. aureus sodM gene.
The incomplete sequence of the sod gene reported by Poyart et al. (33) was used to perform a BLAST search of the genomic DNA databases for S. aureus COL (The Institute for Genomic Research) and 8325 (University of Oklahoma Advanced Center for Genome Technology). The search revealed a match within both databases which included putative Shine-Dalgarno and promoter sequences and an open reading frame containing 187 codons, predicting a protein of 21.5 kDa (data not shown). Oligonucleotide primers were designed using sequence from the S. aureus COL database, and a region encompassing the open reading frame and its upstream region was amplified from S. aureus RN6390 by PCR. The product was cloned into pCR2.1 and verified by restriction analysis and nucleic acid sequencing. The relatedness of the deduced amino acid sequence of the gene ranged from 45% identity to the Fe-SOD of Legionella pneumophila to 64% identity to the Mn-SOD of Bacillus subtilis (Fig. 1), and 75% identity to the S. aureus SodA protein reported by Clements et al. (7). In addition, the amino acid alignment also identified key amino acids used to differentiate between Mn- and Fe-SODs (31). Nineteen of the 21 potential amino acid discriminators were consistent with Mn-SOD types (Fig. 1). The glycines at positions 76 and 77, the phenylalanine at position 84, and the glutamine and aspartate at positions 146 and 147, respectively, were particularly important in predicting a Mn-SOD due to their conservation among SODs that utilize Mn as a cofactor (Fig. 1).
FIG. 1.
Amino acid sequence alignments of bacterial SODs. The SodM (AF273269) and SodA (7) (AF121672) proteins of S. aureus and the SOD proteins from Bacillus subtilis (Bs-MnSOD, D86856), Listeria monocytogenes (Lm-MnSOD, M80526), E. coli (Ec-MnSOD, AE000465; Ec-FeSOD, AE000261), Pseudomonas putida (Pp-FeSOD, U64798), and Legionella pneumophila (Lp-FeSOD, D12922) are shown. Discriminating amino acids (31) used to differentiate between Mn- and Fe-SODs are in bold. The underlined region represents the partial SodM amino acid sequence identified by Poyart et al. (33).
S. aureus sodM rescues SOD deficiency in E. coli.
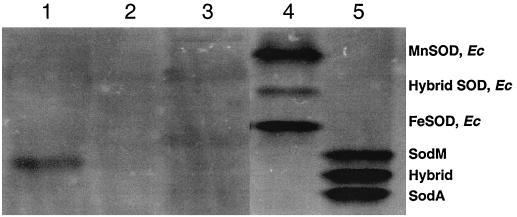
E. coli QC779 (sodA sodB) is unable to grow on minimal medium under aerobic conditions due in part to the superoxide-sensitive dehydratases needed for branched-chain amino acid synthesis (4, 24). The E. coli sodA sodB mutant was transformed by electroporation with either pCR2.1 or pCR2.1sodM and plated on minimal medium (M63 agar) containing carbenicillin. Plates were incubated at 37°C for 2 to 3 days before colonies transformed with pCR2.1sodM appeared. No transformants were recovered when the E. coli sodA sodB mutant was transformed with pCR2.1. SOD activity gels of whole-cell lysates from one of the transformants grown in LB broth revealed a single band of activity (Fig. 2, lane 1), which migrated between the upper and middle bands of activity from S. aureus RN6390 (Fig. 2, lane 5). No activity was observed with either the E. coli double sod mutant or the E. coli double sod mutant containing pCR2.1 under identical conditions (Fig. 2, lanes 2 and 3, respectively). At present, it is unknown why the recombinant SOD (Fig. 2, lane 1) does not migrate to the same location as the upper band of activity observed with S. aureus RN6390 (Fig. 2, lane 5).
FIG. 2.
Activity gel analysis of E. coli (Ec) and S. aureus SODs. Lane 1, E. coli (sodA sodB) containing pCR2.1sodM; lane 2, E. coli (sodA sodB); lane 3, E. coli (sodA sodB) containing pCR2.1; lane 4, E. coli MG1655; and lane 5, S. aureus RN6390. Stained gels were scanned using the AlphaImager 2000 (Alpha Innotech Corp.) imaging system, and the inverse image was generated using NIH Image software.
In wild-type E. coli, three bands of activity can be detected under aerobic conditions (9). The uppermost band of SOD activity represents the Mn-SOD, the lowermost band represents the Fe-SOD, and the middle band of activity represents a hybrid protein consisting of subunits of the Mn- and Fe-SOD (6, 9). The E. coli wild-type strain (MG1655) exhibited the expected three bands of activity (Fig. 2, lane 4).
SodM insensitivity to H2O2 and KCN.
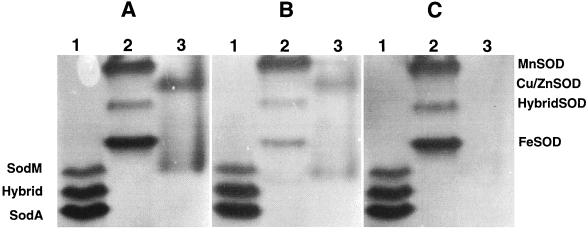
Inactivation by H2O2 and KCN has been used to predict the metal cofactor requirement of SODs (1, 8, 26). Given that the amino acid sequences of the staphylococcal sodM and sodA (7) genes indicate that these SODs are of the Mn variety, we examined the relative sensitivity of the staphylococcal SODs to H2O2 and KCN. As Fe-containing SODs are inactivated by H2O2 (1), polyacrylamide gels were treated with 5 mM H2O2 for 15 min prior to negative staining for SOD activity. All three SOD activities of S. aureus were relatively resistant to H2O2 (Fig. 3A and B, compare lanes 1) while the Fe-containing SOD activity of the wild-type E. coli strain was reduced considerably (Fig. 3A and B, compare lanes 2), suggesting that the three SOD activities of S. aureus do not utilize Fe as a cofactor. This is in contrast to the findings of Clements et al. (7), who reported that the uppermost band of activity is sensitive to H2O2. Currently, these differences are unexplained. Clements et al. (7) showed that metal depletion of cell lysates abolished all three bands of SOD activity and that only the lower two bands of activity were restored when Mn was added back to the cell lysate. Likewise, Fe did not restore the upper band of activity. These data suggest that the SodM protein may utilize some cofactor other than Mn (7).
FIG. 3.
SOD sensitivity to hydrogen peroxide and potassium cyanide. Whole-cell lysates prepared from S. aureus RN6390 (lane 1), E. coli MG1655 (lane 2), and E. coli (sodA sodB) containing plasmid pBA23sodC (lane 3) were resolved by nondenaturing polyacrylamide gel electrophoresis and treated with either hydrogen peroxide (B) or potassium cyanide (C) prior to negative staining for SOD activity. (A) Untreated control. The gel was analyzed and the image generated as described for Fig. 2.
Potassium cyanide inactivates SODs containing Cu-Zn metal cofactors (8, 26). Treatment of gels containing whole-cell lysates from S. aureus with 10 mM KCN resulted in no loss of activity from any of the three bands of SOD activity (Fig. 3A and C, compare lanes 1), while the B. abortus Cu-Zn-SOD, expressed by a recombinant plasmid in the E. coli double sod mutant, was inactivated (Fig. 3A and C, compare lanes 3).
These data taken collectively indicate that the SODs encoded by sodM and sodA are most likely of the Mn type. However, sensitivity to H2O2 and amino acid sequence similarity have been misleading in the characterization of certain SODs. For example, the SOD from the anaerobic archaebacterium Methanobacterium thermoautotrophicum is a Fe-containing enzyme, although its amino acid sequence and resistance to H2O2 suggest that it is of the Mn variety (38, 39). Conclusive evidence of the specific metal cofactor of the staphylococcal SODs will require protein purification and some means of ion detection.
SOD activity and sodM expression under low- and high-aeration conditions.
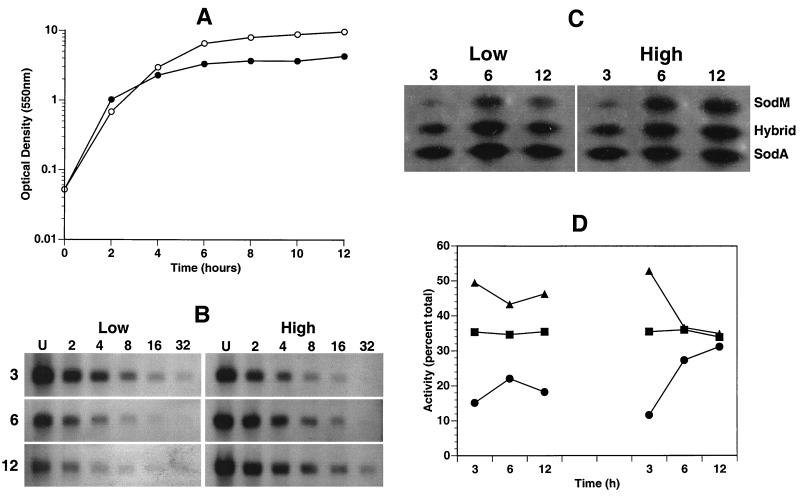
In order to determine when expression of sodM and the production of SOD occur in S. aureus, whole-cell lysates and total RNA were isolated from S. aureus RN6390 following 3, 6, and 12 h of growth under low- and high-aeration conditions (Fig. 4). Optical density readings indicate that while growth rate under low- and high-aeration conditions appears to be the same, growth under low-aeration conditions results in lower overall yield (Fig. 4A). Northern analysis using a DNA probe specific for sodM detected a single RNA species of 0.7 kb, approximately the size of the transcript predicted from the nucleic acid sequence of the sodM gene, indicating that sodM is transcribed monocistronically. Message levels for sodM under low-aeration growth conditions were most abundant at the 3-h time point, which corresponded to the transitional period prior to the postexponential phase of growth (Fig. 4B). After 6 and 12 h of growth, times that corresponded to the early and late stationary phases, message levels were reduced approximately two- and fourfold with respect to the 3-h time point (Fig. 4B). In contrast, under high-aeration growth conditions, message levels were observed to increase throughout this time course. Levels at 6 (late exponential phase) and 12 (stationary phase) h of growth were two- and fourfold elevated with respect to the levels at the 3-h time point (Fig. 4B).
FIG. 4.
Growth-phase-dependent SOD activity and sodM expression under conditions of low and high aeration. (A) Representative growth curve of S. aureus RN6390 grown under conditions of low (●) and high (○) aeration. Whole-cell lysates and total RNA were isolated under low- and high-aeration conditions at 3, 6, and 12 h of growth. (B) Northern analysis of total RNA hybridized with a sodM-specific probe. RNA concentrations were standardized according to A260 values and loaded as either undiluted (U) or twofold serially diluted (numerical values) samples. (C) Nondenaturing polyacrylamide gel of whole-cell lysates stained for SOD activity. The gel was analyzed and the image was generated as described for Fig. 2. (D) Activity of SodM (●), the hybrid (■), and SodA (▴) under low (left)- and high (right)-aeration conditions. Percentages were determined from values generated by quantitative densitometric analysis as described in Materials and Methods.
Whole-cell lysates from S. aureus RN6390 at 3, 6, and 12 h of low- and high-aeration growth were separated by electrophoresis on nondenaturing polyacrylamide gels and stained for SOD activity (Fig. 4C). Under low aeration, all three bands of activity were most abundant at 6 h of growth but decreased approximately 50% by 12 h (Fig. 4C). In contrast, under high-aeration conditions, all three bands of activity increased by 6 h of growth and remained high during the stationary phase of growth (12 h) (Fig. 4C).
The relative contribution of each SOD to the overall SOD activity was assessed by quantitatively comparing each band of activity by densitometry and determining the percentage of each with respect to total SOD activity (Fig. 4D). These data indicate that while SodA is the most abundant of the three SOD activities, the increase in total activity as cells entered the late exponential and postexponential phases of growth under high-aeration conditions appears to be due to an increase in SodM activity (Fig. 4C and D).
Data obtained from our study agree qualitatively with results obtained by Clements et al. (7). Using the pyrogallol spectrophotometric assay for total SOD activity (28), Clements et al. (7) determined that total SOD activity under low- and high-aeration conditions increased 10- and 18-fold, respectively, as cells entered the postexponential phase of growth. In addition, Clements et al. (7) also noted that total activity during the stationary phase of growth decreased under low-aeration conditions and remained the same under high-aeration conditions.
SOD activity in S. aureus sod mutants.
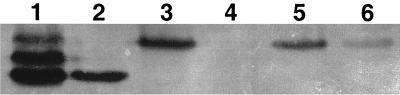
To verify the loss of SOD activity in each of the mutants, whole-cell lysates from S. aureus RN6390 and its isogenic sod mutant strains were electrophoresed on nondenaturing polyacrylamide gels and stained for SOD activity (Fig. 5). As expected, the parental strain RN6390 exhibited three closely migrating bands of activity (Fig. 5, lane 1) while the sodM mutant strain exhibited only a single band of activity that corresponded to the lowest-migrating band exhibited by the parent (Fig. 5, lane 2). As previously demonstrated by Clements et al. (7), who used a sodA transposon mutant, whole-cell lysates from our sodA::tet mutant contained only a single band of activity that corresponded to the upper most band exhibited by the parent strain (Fig. 5, lane 3). No bands of SOD activity were detected in cell lysates prepared from the sod double mutant (Fig. 5, lane 4). However, when the sodM gene was expressed from plasmid pCL15 in the sod double mutant, the uppermost band of activity was restored (Fig. 5, lanes 5 and 6). Maximal activity was observed when sodM was induced from the lac promoter with IPTG (Fig. 5, lane 5). It is not clear at present why the sodM gene is poorly expressed from its own putative promoter in a moderate-copy-number plasmid (Chia Lee, personal communication). Sequence analysis of the sodM gene revealed a putative promoter region and Northern analysis clearly demonstrates that the sodM message is approximately the size of the transcript predicted from this nucleic acid sequence. In addition, no other hybridizing bands were observed by Northern analysis (data not shown). Furthermore, sequence analysis a thousand base pairs upstream of the sodM open reading frame revealed no obvious sequences that might suggest a cis-acting element. Currently, experiments are under way to determine the precise transcriptional start site of sodM and whether expression involves additional cis-acting elements upstream of sodM.
FIG. 5.
SOD activities from S. aureus sod mutants. Lane 1, RN6390; lane 2, sodM mutant; lane 3, sodA mutant; lane 4, double (sodM sodA) mutant; lane 5, double mutant containing pCL15sodM induced with IPTG; lane 6, double mutant containing pCL15sodM uninduced.
These data demonstrate that the three bands of SOD activity observed for S. aureus RN6390 are encoded by two distinct genes, sodM and sodA. Because the middle band of activity observed for the parent strain is lost in either the sodM or sodA mutant, the middle band of activity is proposed to result from the formation of a hybrid protein composed of SodM and SodA. The SOD activity profile exhibited by S. aureus is similar to the pattern of SODs in E. coli where subunits of the Mn- and Fe-SOD form a hybrid band of activity that migrates between the Mn- and Fe-SOD (6, 9) (Fig. 2, lane 4). Even though Clare et al. (6) concluded that the formation of a hybrid SOD in E. coli is most likely due to subunit exchange between two enzymes with comparable catalytic activities and extensive amino acid sequence homology, a functional role for the hybrid SOD in E. coli has not been investigated. Likewise, whether the hybrid SOD band observed in S. aureus possesses a particular function in the physiology of the bacterium remains to be determined.
To the best of our knowledge, S. aureus represents the first gram-positive bacterium reported to contain two or more bands of SOD activity. The gram-positive bacteria studied thus far demonstrate a single band of SOD activity as determined by nondenaturing polyacrylamide electrophoresis and staining for SOD activity (40). In addition, several of these SODs are said to be cambialistic, capable of utilizing either Mn or Fe as the metal cofactor (40). Why S. aureus possesses two sod genes that account for three bands of activity is an intriguing question, particularly when one considers that whole-cell lysates from several coagulase-negative staphylococci, including Staphylococcus epidermidis, exhibit only one band of SOD activity, with an electrophoretic mobility identical to that of the S. aureus SodA (M. W. Valderas and M. E. Hart, unpublished data). Perhaps, these differences in SOD profiles between S. aureus and the coagulase-negative staphylococci represent an important divergence in the evolution of these species relevant to the environmental niches that these species primarily occupy.
Viability in the presence of MV.
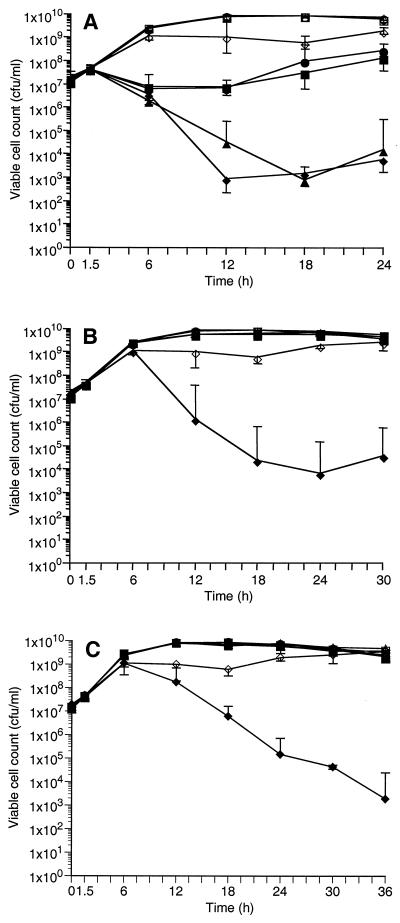
To assess the contribution of each of the SODs to resistance against an internal source of oxidative stress, MV was added to high-aeration broth cultures of S. aureus at 1.5, 6, and 12 h of growth, times that corresponded to early exponential, late exponential, and stationary phases of growth. Growth was monitored spectrophotometrically, and viability in the presence and absence of MV was determined at various times during growth (Fig. 6). Only the sod double mutant was affected when grown in the absence of MV, exhibiting a statistically significant reduction (P < 0.001) in the total number of cells compared to the parent strain and to the sodM and sodA single mutants (Fig. 6). When MV was added to early-exponential-phase growing cultures (1.5 h), cell viability of all cultures was reduced at 6 h to levels just below the cell concentration at zero hour (Fig. 6A). However, only the sodA mutant and the sod double mutant continued to lose viability over the next 6 to 12 h. An approximately 104-fold reduction in viability was observed for these mutants (Fig. 6A). While growth of the parent and sodM mutant was reduced approximately 10-fold by 6 h, no additional loss of viability was observed and cell number began to increase by 18 h (Fig. 6A). Interestingly, an increase in viability was also observed for the sodA mutant and the sod double mutant at the later time points. Whether these cells are the result of suppressor mutations is currently under investigation.
FIG. 6.
Viable cell count of S. aureus RN6390 (○, ●), the sodM mutant (□, ■), the sodA mutant (▵, ▴), and the double (sodM sodA) mutant (◊, ⧫) grown in the absence (open symbols) and presence (closed symbols) of MV. MV was added at 1.5 (A), 6 (B), and 12 (C) h of growth. Values are means ×/÷ standard errors.
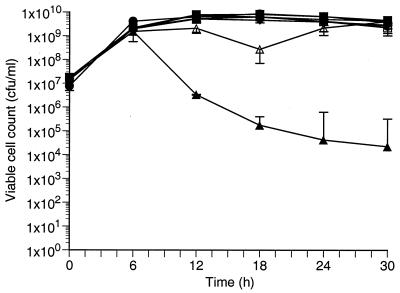
When MV was added to cells late in the exponential (6 h) or stationary (12 h) phases of growth, the viability of the parent and sodM mutant was unaffected (Fig. 6B and C). Surprisingly, the sodA mutant was unaffected (Fig. 6B and C). In fact, both the sodM and sodA mutants and the parent strain exhibited identical viability in the presence or absence of MV. In contrast, when MV was added to the sod double mutant at 6 or 12 h of growth, a significant reduction (105-fold) in viable cells was observed (Fig. 6B and C). These data indicate that SodA levels alone are sufficient during early growth to protect against oxidative stress (1.5 h) (Fig. 4C and D). In contrast, maximum expression of SodM is delayed until cells reach the late exponential to postexponential phases of growth (6 h) (Fig. 4C). Consequently, the sodA mutant is sensitive to oxidative stress caused by MV addition at 1.5 h of growth, and its viability is significantly reduced (Fig. 6A). However, MV is not inhibiting when added after SodM has been produced in the cell (6 or 12 h), and the viability of the sodA mutant is unaffected due to the compensatory levels of SodM (Fig. 6B and C). These results indicate that while SodA may be the major SOD activity in the exponential growth phase of S. aureus, SodM may play an important role in maintaining cell viability during the stationary phase of growth. To verify that SodM can function in this capacity, the sod double mutant containing pCL15sodM was grown in the presence of MV (Fig. 7). Cultures in the early exponential phase of growth (1 h) were induced to express sodM by the addition of IPTG. When cultures reached the late exponential phase of growth (6 h), MV was added. As previously demonstrated, MV added to either the parent or the sodA mutant strain at 6 h of growth had no effect on their viability (Fig. 7). However, when MV was added to the sod double mutant, a drastic reduction (104-fold) in viability was observed by 12 h. In contrast, the sod double mutant containing the pCL15sodM construct was unaffected by the addition of MV at 6 h. Growth of this strain was identical to that of the parent and sodA mutant strains (Fig. 7).
FIG. 7.
Viable cell count of S. aureus RN6390 (○, ●), the sodA mutant (□, ■), the double mutant (▵, ▴), and the double mutant containing pCL15sodM (◊, ⧫) grown in the absence (open symbols) and presence (closed symbols) of MV. Values are means ×/÷ standard errors.
In summary, results from this study clearly demonstrate that S. aureus possesses three bands of SOD activity accounted for by two genes, sodM and sodA. The third band of activity is most likely a hybrid SOD consisting of subunits of SodM and SodA. While this study has shown that SodM levels in the late exponential to postexponential phases of growth can protect an S. aureus sodA mutant from oxidative stress, it does not seem likely that S. aureus would employ a two-SOD system to protect cells in the event that one of the SODs became nonfunctional. The fact that viability of the sodM mutant is not adversely affected even in the presence of MV suggests an alternative role for this SOD. Perhaps the differences in SodM and SodA activities observed in this study using in vitro growth conditions suggest a more important role for one or both of these SODs in the host, particularly after phagocytosis by neutrophils. Studies addressing this question are currently in progress. In addition, studies that include the definitive determination of the metal cofactors used by both SODs and the environmental stimuli that control the expression of these genes should also provide helpful insight in determining the significance of each SOD in S. aureus.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI-36934 from the National Institute of Allergy and Infectious Diseases and a Faculty Research Grant from the University of North Texas Health Science Center.
We are indebted to Tony Romeo (UNTHSC) and Jerry Simecka (UNTHSC) for helpful discussions, critical reading of the manuscript, and continuous encouragement throughout this work. We are also indebted to Ken Bayles, John Iandolo, Chia Lee, Marty Roop, and Danièle Touati for strains, plasmids, and helpful discussions. A special thanks goes to Allen Gies of the University of Arkansas for Medical Sciences DNA Sequencing Core Facility for sequencing the sodM clones. Preliminary sequence data were obtained from The Institute for Genomic Research and the University of Oklahoma's Advanced Center for Genome Technology.
REFERENCES
- 1.Asada K, Yoshikawa K, Takahashi M-A, Maeda Y, Enmanji K. Superoxide dismutases from a blue-green alga, Plectonema boryanum. J Biol Chem. 1975;250:2801–2807. [PubMed] [Google Scholar]
- 2.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 3.Blevins J S, Gillaspy A F, Rechtin T M, Hurlburt B K, Smeltzer M S. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol Microbiol. 1999;33:317–326. doi: 10.1046/j.1365-2958.1999.01475.x. [DOI] [PubMed] [Google Scholar]
- 4.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. National nosocomial infection surveillance system report: data summary from October 1986–April 1996. U.S. Atlanta, Ga: Department of Health and Human Services; 1996. [Google Scholar]
- 6.Clare D A, Blum J, Fridovich I. A hybrid superoxide dismutase containing both functional iron and manganese. J Biol Chem. 1984;259:5932–5936. [PubMed] [Google Scholar]
- 7.Clements M O, Watson S P, Foster S J. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol. 1999;181:3898–3903. doi: 10.1128/jb.181.13.3898-3903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donahue J L, Okpodu C M, Cramer C L, Grabau E A, Alscher R G. Responses of antioxidants to paraquat in pea leaves. Plant Physiol. 1997;113:249–257. doi: 10.1104/pp.113.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dougherty H W, Sadowski S J, Baker E E. A new iron-containing superoxide dismutase from Escherichia coli. J Biol Chem. 1978;253:5220–5223. [PubMed] [Google Scholar]
- 10.Dyer D W, Iandolo J J. Rapid isolation of DNA from Staphylococcus aureus. Appl Environ Microbiol. 1983;46:283–285. doi: 10.1128/aem.46.1.283-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsbach P, Weiss J. Phagocytic cells: oxygen-independent antimicrobial systems. In: Gallin J I, Godstein I M, Snyderman R, editors. Inflammation: basic principles and clinical correlates. New York, N.Y: Raven Press; 1988. pp. 445–470. [Google Scholar]
- 12.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 14.Guérout-Fleury A-M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 15.Haley R W, Culver D H, White J W, Morgan W M, Emori T G. The nationwide nosocomial infection rate: a new need for vital statistics. Am J Epidemiol. 1985;121:159–167. doi: 10.1093/oxfordjournals.aje.a113988. [DOI] [PubMed] [Google Scholar]
- 16.Hart M E, Smeltzer M S, Iandolo J J. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J Bacteriol. 1993;175:7875–7879. doi: 10.1128/jb.175.24.7875-7879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iandolo J J. The genetics of staphylococcal toxins and virulence factors. In: Iglewski B H, Clark V L, editors. Molecular basis of bacterial pathogenesis. New York, N.Y: Academic Press, Inc.; 1990. pp. 399–426. [Google Scholar]
- 18.Imlay J A, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 19.Kanafani H, Martin S E. Catalase and superoxide dismutase activities in virulent and nonvirulent Staphylococcus aureus isolates. J Clin Microbiol. 1985;21:607–610. doi: 10.1128/jcm.21.4.607-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E-J, Chung H-J, Suh B, Hah Y C, Roe J-H. Transcriptional and post-transriptional regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Müller. Mol Microbiol. 1998;27:187–195. doi: 10.1046/j.1365-2958.1998.00674.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim E J, Kim H P, Hah Y C, Roe J-H. Differential expression of superoxide dismutases containing Ni and Fe/Zn in Streptomyces coelicolor. Eur J Biochem. 1996;241:178–185. doi: 10.1111/j.1432-1033.1996.0178t.x. [DOI] [PubMed] [Google Scholar]
- 22.Klebanoff S J. Myeloperoxidase: occurrence and biological function. In: Everse J, Everse K E, Grisham M B, editors. Peroxidases in chemistry and biology. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 1–35. [Google Scholar]
- 23.Kraemer G R, Iandolo J J. High-frequency transformation of Staphylococcus aureus by electroporation. Curr Microbiol. 1990;21:373–376. [Google Scholar]
- 24.Kuo C F, Mashino T, Fridovich I. α,β-Dihydroxyisovalerate dehydratase: a superoxide-sensitive enzyme. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- 25.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Latimer E, Simmers J, Sriranganathan N, Roop II R M, Schurig G G, Boyle S M. Brucella abortus deficient in copper/zinc superoxide dismutase is virulent in BALB/c mice. Microb Pathog. 1992;12:105–113. doi: 10.1016/0882-4010(92)90113-3. [DOI] [PubMed] [Google Scholar]
- 27.Mandell G L. Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. J Clin Invest. 1975;55:561–566. doi: 10.1172/JCI107963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marklund S, Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 29.McNamara P J, Iandolo J J. Genetic instability of the global regulator agr explains the phenotype of the xpr mutation in Staphylococcus aureus KSI9051. J Bacteriol. 1998;180:2609–2615. doi: 10.1128/jb.180.10.2609-2615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Parker M W, Blake C C F. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 1988;229:377–382. doi: 10.1016/0014-5793(88)81160-8. [DOI] [PubMed] [Google Scholar]
- 32.Peng H-L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequence of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poyart C, Berche P, Trieu-Cuot P. Characterization of superoxide dismutase genes from gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett. 1995;131:41–45. doi: 10.1016/0378-1097(95)00232-t. [DOI] [PubMed] [Google Scholar]
- 34.Sau S, Sun J, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal A W. The electron transport chain of the microbicidal oxidase of phagocytic cells and its involvement in the molecular pathology of chronic granulomatous disease. J Clin Invest. 1989;83:1785–1793. doi: 10.1172/JCI114083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smeltzer M S. Characterization of staphylococcal adhesins for adherence to host tissues. In: An Y H, Friedman R J, editors. Handbook of bacterial adhesion, principles, methods and applications. Totowa, N. J.: Humana Press; 2000. pp. 411–444. [Google Scholar]
- 37.Snodgrass J L, Mohamed N, Ross J M, Sau S, Lee C Y, Smeltzer M S. Functional analysis of the Staphylococcus aureus collagen adhesin B domain. Infect Immun. 1999;67:3952–3959. doi: 10.1128/iai.67.8.3952-3959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takao M, Oikawa A, Yasui A. Characterization of a superoxide dismutase gene from the archaebacterium Methanobacterium thermoautotrophicum Arch. Biochem Biophys. 1990;283:210–216. doi: 10.1016/0003-9861(90)90633-a. [DOI] [PubMed] [Google Scholar]
- 39.Takao M, Yasui A, Oikawa A. Unique characteristics of superoxide dismutase of a strictly anaerobic archaebacterium Methanobacterium thermoautotrophicum. J Biol Chem. 1991;266:14151–14154. [PubMed] [Google Scholar]
- 40.Touati D. Superoxide dismutases in bacteria and pathogen protists. In: Scadalios J G, editor. Oxidative stress and the molecular biology of antioxidant defenses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 447–493. [Google Scholar]
- 41.Verhoef J. Host defense against infection. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 213–232. [Google Scholar]
- 42.Waldvogel F A. Staphylococcus aureus (including toxic shock syndrome) In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone; 1995. pp. 1754–1777. [Google Scholar]
- 43.Watson S P, Antonio M, Foster S J. Isolation and characterization of Staphylococcus aureus starvation-induced, stationary-phase mutants defective in survival or recovery. Microbiology. 1998;144:3159–3169. doi: 10.1099/00221287-144-11-3159. [DOI] [PubMed] [Google Scholar]