Abstract
(1) Background: The HIV-1 latent reservoir harboring replication-competent proviruses is the major barrier in the quest for an HIV-1 infection cure. HIV-1 infection at all stages of disease progression is associated with immune activation and dysfunctional production of proinflammatory soluble factors (cytokines and chemokines), and it is expected that during HIV-1 infection, different immune components and immune cells, in turn, participate in immune responses, subsequently activating downstream biological pathways. However, the functional interaction between HIV-1 integration and the activation of host biological pathways is presently not fully understood. (2) Methods: In this work, I used genes targeted by proviruses from published datasets to seek enriched immunologic signatures and host biological pathways alongside HIV-1 infections based on MSigDb and KEGG over-representation analysis. (3) Results: I observed that different combinations of immunologic signatures of immune cell types and proinflammatory soluble factors appeared alongside HIV-1 infections associated with antiretroviral therapy. Moreover, enriched KEGG pathways were often related to “cancer specific types”, “immune system”, “infectious disease viral”, and “signal transduction”. (4) Conclusions: The observations in this work suggest that the gene sets harboring provirus integration sites may define specific immune cells and proinflammatory soluble factors during HIV-1 infections associated with antiretroviral therapy.
Keywords: human immunodeficiency virus type 1 (HIV-1), HIV-1 integration sites, Molecular Signatures Database (MSigBb), virus–host interaction, infection and immunity
1. Introduction
HIV-1 infection has a dynamic disease progression associated with a combination of immune cells, immune components, and various cellular restriction factors aimed at battling infections. It is known that HIV-1 infection induces a plethora of proinflammatory soluble factors that recruit and activate innate immune-related cells to the site of HIV-1 infection to restrict the replication and spread of viruses. Even though these defense mechanisms begin at the early stage of HIV-1 infections, HIV-1 still manages to complete its life cycle and establish latent reservoirs.
Several host genes, so-called recurrent integration genes [1,2,3], have been demonstrated to be frequently targeted by HIV-1 in infected individuals. However, the mechanism that leads HIV-1 integration sites to these recurrent integration genes and the biological meaning of this phenomenon remains unclear. Nevertheless, a remarkable study attempted to answer this question by showing the interaction between the selection of HIV-1 integration sites and functional pathways assigned by different biological processes in the host cells [4]. Their findings implied that the possible mechanism leading to HIV-1 integration in hot spot genes can happen at the level of the functional genome property formed by a group of genes rather than a gene per se.
In the continuation of this concept, in this work, I sought any functional linkage between provirus-targeted genes and host immunity alongside HIV-1 infections. I used the host genes targeted by proviruses [5,6] as an input gene list. This list of genes included integration sites retrieved from elite controllers [6] and HIV-1-infected individuals subjected to antiretroviral therapy (ART) [5]. Of note, the integration sites published by Einkauf et al. (2022) [5] were longitudinally collected from HIV-1-infected individuals, thereby allowing us to follow how the HIV-1 reservoir evolves during the period of ART. Based on the over-representation analysis, I observed that different immunologic signatures were enriched alongside HIV-1 infections associated with ART. In contrast, few immunologic signatures were enriched in elite controllers. In addition, enriched gene sets were mainly involved in biological pathways related to “cancer specific types”, “immune system”, “infectious disease viral”, and “signal transduction”. Finally, I found that enriched immunologic signatures were contributed by provirus-targeted genes that are associated with gene products of HIV-1 or affect HIV-1 replication and infectivity. Altogether, these findings appeared to resemble the concept of network theory [7,8,9] that provirus-targeted genes engaged in similar biological functions may form a community (refer to a “signature” in this study) to satisfy the need for host immunity during different periods of HIV-1 infections associated with ART. Based on this logic, I proposed a hypothesis that there could be a dynamic interplay between HIV-1 integration sites and the host functional genome property: HIV-1 integration frequency might be used as a surrogate for gene sets, which may define specific immune cell types and proinflammatory soluble factors during HIV-1 infection.
2. Materials and Methods
2.1. Acquisition and Procession of Public Datasets
HIV-1 integration sites were collected from Jiang et al. (2020) [6] for elite controllers and Einkauf et al. (2022) [5] for pretreatment HIV-1-infected individuals (viremic) and patients subjected to a short (1 year) and a long period (7.5–12 years) of ART. A total of 232 integration sites from 64 elite controllers applied by Jiang et al. (2020) [6] were analyzed in this study. Among them, 103 sites were inserted by intact proviruses. Integration sites retrieved from elite controllers were overlaid on Human_genome_GENCODE_v32.bed released by the GENCODE project [10] to determine if the insertion of each HIV-1 integration is genic or non-genic by executing the command intersect with default options provided in bedtools [11]. Since multiple proviruses were observed to integrate into the same genomic position, in the end, 104 provirus-targeted genes were released; among them, 22 genes were targeted by intact proviruses. A total of 1270 provirus-targeted genes from 6 patients (P1: 468 integration sites; P2: 149 integration sites; P3: 128 integration sites; P4: 94 integration sites; P5: 305 integration sites; P6: 125 integration sites) used in Einkauf et al. (2022) [5] were selected for this study. In total, 317 integration sites that fall into the category of pretreatment HIV-1-infected individuals (viremic) yielded 121 provirus-targeted genes, 396 integration sites that fall into the category of patients subjected to a short period (1 year) of ART yielded 149 provirus-targeted genes, and 557 integration sites that fall into the category of patients subjected to a long period (7.5–12 years) of ART yielded 176 provirus-targeted genes. Among them, 26 intact provirus-targeted genes were found in pretreatment HIV-1-infected individuals, 6 intact provirus-targeted genes were found in patients subjected to a short period of ART, and 14 intact provirus-targeted genes were found in patients subjected to a long period of ART. Lists of the input genes used for the analysis are shown in Table 1; the genes targeted by intact proviruses are summarized in Table S26.
Table 1.
Input gene list.
| Pretreatment (ut) | Short Period of ART (st) | Long Period of ART (lt) | Elite Controllers |
|---|---|---|---|
| ACOX1 | AAK1 | ABCC1 | ABCA11P |
| ACSF3 | AC004791.2 | ACTR3 | AGPAT3 |
| ADARB1 | ACAP2 | ADGRE5 | ANKRD11 |
| ADTRP | ADD3 | AHR | APBA2 |
| AIM1 | AJ003147.9 | ANKRD11 | ARHGAP17 |
| ANKLE2 | ANKRD11 | APC | ARHGAP31 |
| ANKRD31 | ARHGAP24 | ARHGEF12 | ARHGEF18 |
| ARHGAP35 | ARIH2 | ARHGEF3 | ASXL1 |
| ARID4B | ASAP1 | ARID1A | ATR |
| ASNA1 | ASH1L | ARNT | BACH2 |
| BRD1 | ATXN2L | ASH1L | CEACAM21 |
| BUB1B | BOLA2 | ASPSCR1 | COX10-AS1 |
| C10orf76 | BRD4 | ATN1 | CREBBP |
| CCDC26 | C16orf52 | AUH | DCLRE1C |
| CCDC57 | C17orf62 | B3GALT4 | DECR1 |
| CD58 | C4B | BACH2 | DHTKD1 |
| CEP85 | C6orf89 | BCL7C | DIP2A |
| CHAF1A | CHD6 | BRD4 | DIP2A-IT1 |
| CLIP1 | CLN5 | C16orf72 | DLGAP1 |
| CNN2 | CLUAP1 | C4orf22 | ELMO1 |
| CNOT1 | CNOT6L | CAMK2D | ENSG00000231731 |
| CNOT4 | CNTRL | CCDC30 | ENSG00000260989 |
| CRTC3 | COL5A1 | CCDC57 | ENSG00000265987 |
| CSNK1G1 | COPB1 | CD84 | ENSG00000267179 |
| CSNK1G2 | CPNE1 | CENPC | ENSG00000267552 |
| CTCF | CREBBP | CHMP4B | ENSG00000281016 |
| CTSA | CTSS | CLIP4 | ENSG00000283515 |
| CYTH1 | CYLD | CNKSR2 | ENSG00000285816 |
| DAB1 | DARS-AS1 | COTL1 | ENSG00000288715 |
| DAZAP1 | DGKA | CRKL | ENTR1 |
| DNM2 | DIAPH1 | CUL5 | ERC1 |
| EHMT1 | DNAJC18 | CYTH1 | EVL |
| ETFA | DPF2 | DAP3 | FCHSD2 |
| FANCA | DYNC1H1 | DAPK2 | GRB2 |
| FBXL12 | EHD4 | DAZAP1 | HLA-DQA1 |
| FGD3 | EP400 | DCLRE1C | HSF5 |
| FOXK2 | ERCC6L2 | DCUN1D1 | IFT140 |
| FRMD4B | FAM102A | DLG1 | IWS1 |
| FUS | FAM102B | DNAH1 | JMJD1C |
| GBF1 | FAM134B | DNAJC18 | KANSL2 |
| GRSF1 | FANCA | DNMT1 | KDM2A |
| HSPBAP1 | FBXL20 | DPP8 | KIAA0319L |
| IARS | FGD3 | E2F3 | KMT2C |
| IFNAR1 | FOXK2 | EED | LHPP |
| IGSF6 | FUT8 | EIF4G3 | LINC00298 |
| IL12RB1 | GIMAP8 | EPB41 | LMTK2 |
| IP6K1 | GPR89A | ERCC6L2 | MGMT |
| KIAA2026 | GRB2 | ERMP1 | MYDGF |
| KIF21A | GTF2E2 | ESCO1 | MYO1F |
| KMT2A | HIST1H2AC | EVI5 | NELL2 |
| LINC | HMGCS1 | EXOC7 | NF1 |
| LINC00499 | HN1L | FAM13A | NPHP3 |
| LINC00536 | HNRNPC | FAM214A | NPHP3-ACAD11 |
| MACF1 | HTT | FNBP1 | NUMA1 |
| MALAT1 | IKZF3 | FRY | P4HB |
| METTL15 | IRF2 | GATAD2A | PACS1 |
| METTL8 | IRGQ | GGA3 | PARP4 |
| MFSD12 | ITPKB | GON4L | PCNT |
| MROH1 | ITPR3 | GRAP2 | PIK3IP1 |
| MTG1 | KIAA0753 | HBS1L | POLA2 |
| NAA38 | KXD1 | HECTD4 | PPM1G |
| NCOA7 | LCOR | HFE | PPP4R3A |
| NCOR1 | LINC00869 | HIVEP3 | PRDM2 |
| NLRC5 | MACF1 | IL27RA | RAB14 |
| NPLOC4 | MALT1 | IL2RB | RABEP1 |
| NT5C2 | MAN1A2 | IQGAP1 | RAD54L2 |
| NUDT3 | MAPK14 | IQGAP2 | RAI1 |
| NUP98 | MARS | ITGA6 | RAP1GDS1 |
| PARPBP | MAST2 | ITGAL | RNF157 |
| PBX3 | MCF2L2 | JAK1 | RPL15P4 |
| PDK3 | MDS2 | KANSL3 | RPTOR |
| PDXDC2P | MED15 | KDM2A | SMG7 |
| PGM2L1 | METTL15 | KDM3A | SPEN |
| PPFIA1 | MKL1 | KIAA0430 | SPPL3 |
| PPP2CA | MORC3 | KIAA0753 | STK38 |
| PRDM2 | MRPS6 | KIF13B | STX16 |
| PRKAG1 | MYH9 | LDLRAD4 | STX16-NPEPL1 |
| PRRC1 | NACC2 | LEF1 | TBCD |
| PSMB2 | NCAPD2 | LPP | TMEM245 |
| PTPN1 | NCBP3 | LSM14A | TNRC6A |
| RAPGEF2 | NDC80 | LUZP1 | TPTE2P6 |
| RBX1 | NDUFA6 | MALAT1 | TRAPPC14 |
| RN7SKP127 | NDUFAF2 | MAP3K3 | TRIM37 |
| RNF214 | NMD3 | MCOLN2 | TRIT1 |
| RNF216 | NPEPPS | MEGF8 | UBE2O |
| RNF34 | NR2C2 | MICB | UBE3C |
| RPTOR | OS9 | MIR3681HG | UBR2 |
| RRN3 | PACS1 | MROH1 | UTP20 |
| RUNX2 | PIBF1 | MYCBP2 | ZMYM4 |
| SARS | PIK3R1 | NDUFA6 | ZNF160 |
| SCMH1 | PLEC | NFKBIL1 | ZNF180 |
| SECISBP2L | PPP3CC | NSD1 | ZNF225 |
| SEPT9 | PPP6R3 | NUDT3 | ZNF274 |
| SETD3 | PRKCA | NUDT4 | ZNF350 |
| SMARCC1 | PRKCQ | NVL | ZNF350-AS1 |
| SPG11 | PRR19 | OGDH | ZNF407 |
| STAU1 | PSME4 | PATL1 | ZNF430 |
| TAF1 | PSMF1 | PBRM1 | ZNF544 |
| TBC1D4 | PTPN4 | PCBP2 | ZNF607 |
| TGFB1 | QRICH1 | PHACTR4 | ZNF700 |
| THRAP3 | RAB11FIP3 | PHC1 | ZNF720 |
| TMEM87A | RAC2 | PHF12 | ZNF721 |
| UBAP1 | RAD51 | PIP4K2A | ZSCAN16 |
| UBE2D2 | RALGDS | PITPNC1 | ZSCAN16-AS1 |
| UBE2G1 | RAPGEF2 | PLA2G16 | |
| UMAD1 | RB1 | PPP3CC | |
| UNC13B | RBL1 | PPP6R1 | |
| USP3 | RCOR1 | PRR19 | |
| VAV1 | REV1 | PUM1 | |
| VDR | RNF157 | PXK | |
| VPS13B | RPRD2 | RAD18 | |
| WARS2 | RPTOR | RANBP9 | |
| ZAN | SACM1L | RAPGEF2 | |
| ZBED5 | SAE1 | RAPGEF6 | |
| ZNF226 | SCYL1 | RBM15 | |
| ZNF557 | SGMS1 | RFFL | |
| ZNF701 | SKAP1 | RIMKLB | |
| ZNF737 | SMCHD1 | RPS19 | |
| ZNF850 | SMG6 | RPS6KA3 | |
| ZNF91 | SNX25 | RPTOR | |
| ZSCAN22 | SPECC1L | RSRC2 | |
| STK11 | SAE1 | ||
| TARBP1 | SCNN1A | ||
| TATDN2 | SERPINH1 | ||
| TBK1 | SETX | ||
| TEX2 | SF3B2 | ||
| TMEM65 | SFMBT1 | ||
| TNRC6C | SHCBP1 | ||
| TOM1L2 | SLC23A2 | ||
| TPM3 | SLC25A40 | ||
| TRAF3 | SLC30A7 | ||
| TRPC4AP | SLC35A3 | ||
| TUBA8 | SLC44A2 | ||
| TYW1 | SLC9A6 | ||
| UBOX5 | SLX4IP | ||
| USP25 | SNTB2 | ||
| USP49 | SPEN | ||
| USP6NL | ST13 | ||
| USP9Y | ST7 | ||
| VCL | STAT5B | ||
| VRK1 | SUMO2 | ||
| WASH1 | TAB1 | ||
| WBP1L | TAF2 | ||
| WHSC1L1 | TANGO6 | ||
| WNK1 | TAP2 | ||
| ZBED4 | TBC1D2B | ||
| ZC3H18 | TCF3 | ||
| ZNF564 | TMBIM6 | ||
| ZNF609 | TNFSF12 | ||
| TNRC6B | |||
| TNRC6C | |||
| TOM1L2 | |||
| TP53TG5 | |||
| TRAT1 | |||
| TSEN54 | |||
| TSG101 | |||
| UBA6 | |||
| UBE2Q2 | |||
| UBTD2 | |||
| UPF1 | |||
| USP34 | |||
| VARS | |||
| VAV1 | |||
| VPS13D | |||
| VPS8 | |||
| WASF2 | |||
| XPO6 | |||
| Y_RNA | |||
| YWHAE | |||
| ZCCHC11 | |||
| ZCCHC7 | |||
| ZKSCAN8 | |||
| ZNF140 | |||
| ZNF451 | |||
| ZNF529 | |||
| ZNRF1 |
2.2. Data and Bioinformatic Analyses
2.2.1. MsigDb Over-Representation Analysis
Enriched immunologic signatures were computed using the R package clusterProfiler (Version 4.4.1) [12,13] with the function enricher and default options. Over-representation analysis [14] was performed using C7 immunologic signature gene sets curated in the Molecular Signatures Database (MsigDb) [15,16,17] as a reference. Only the enriched immunologic signatures with p-values (adjusted using the Benjamini–Hochberg method) of less than 0.05 were selected. Readouts of over-representation analyses generated in this study were all curated in Supplementary Materials. Rich factors [13] were further calculated to represent the enrichment score for every enriched immunologic signature (Figure 1c) by dividing GeneRatio by BgRatio with the command lines described below.
>MsigDb_output_file$GeneRatio <-as.numeric(gsub(“(\\d+)/(\\d+)”, “\\1”, MsigDb_output_file$GeneRatio, perl = T))/as.numeric(gsub(“(\\d+)/(\\d+)”, “\\2”, MsigDb_output_file$GeneRatio, perl = T))
# Convert GeneRatio to numerical variables.
>MsigDb_output_file$BgRatio <-as.numeric(gsub(“(\\d+)/(\\d+)”, “\\1”, MsigDb_output_file$BgRatio, perl = T))/as.numeric(gsub(“(\\d+)/(\\d+)”, “\\2”, MsigDb_output_file$BgRatio, perl = T))
# Convert BgRatio to numerical variables.
>MsigDb_output_file <-MsigDb_output_file %>% dplyr::mutate(rich_factor = GeneRatio/BgRatio)
# Calculate rich factors.
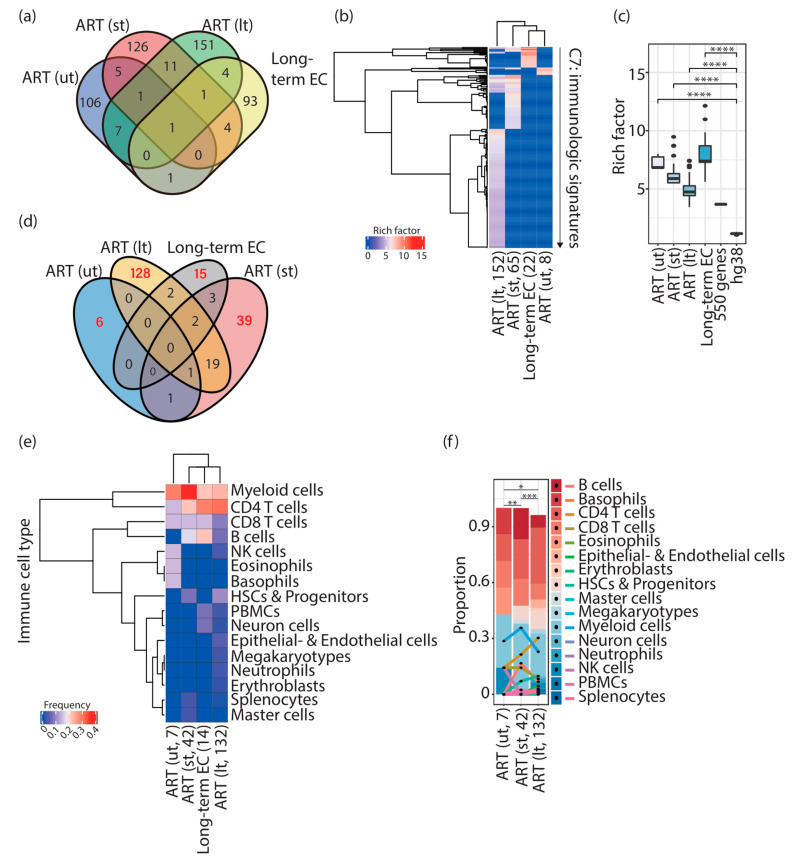
Figure 1.
Distinct immunologic signatures were enriched in HIV-1-infected individuals and elite controllers. (a) Venn diagram representing the overlap of input genes in HIV-1-infected individuals and elite controllers. Genes shared among different groups of input genes are listed in Table S27. (b) Cluster heatmap representing immunologic signatures enriched in HIV-1-infected individuals and elite controllers. Parentheses placed under the column indicate the status of ART (“lt” refers to HIV-1-infected individuals subjected to a long period of ART; “st” refers to HIV-1-infected individuals subjected to a short period of ART; “ut” refers to pretreatment HIV-1-infected individuals) followed by the quantity of enriched immunologic signatures. The color scale represents the magnitude of the enrichment represented by rich factors. (c) Box plots representing the enrichment of immunologic signatures represented by rich factors. Commands executed to calculate rich factors are described in the Materials and Methods. Statistical significance was calculated using the Wilcoxon test in R with default options. (d) Venn diagram representing the overlap of enriched immunologic signatures in HIV-1-infected individuals and elite controllers. Red numbers indicate the quantity of unique enriched immunologic signatures. (e) Cluster heatmap representing the frequency of immune cell types present in unique enriched immunologic signatures in HIV-1-infected individuals and elite controllers. Parentheses placed under the column indicate the number of immune cell types counted in the enriched immunologic signatures. (f) Stacked bar chart representing the proportion of the different compositions of immune cell types in pretreatment HIV-1-infected individuals and patients receiving ART. Parentheses placed under the column indicate the number of immune cell types counted in the enriched immunologic signatures. Statistical significance was calculated using Pearson’s chi-square test in R with default options, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001. ART; antiretroviral therapy. EC; elite controllers. HSCs; hematopoietic stem cells. NK cells; natural killer cells. PBMCs; peripheral blood mononuclear cells.
Notably, after performing the MsigDB over-representation analysis, terms of immune cell types and proinflammatory soluble factors shown in the standard names of the enriched immunologic signatures were collected for further statistical calculation. Based on the Blood Cell Lineage chart published in the NIH website’s SEER Training Modules (https://training.seer.cancer.gov/leukemia/anatomy/lineage.html (accessed on 10 September 2022)), immune cell types were classified into 16 groups, namely, (1) B cells, including plasma cells, (2) basophils, (3) CD4 T cells, (4) CD8 T cells, (5) eosinophils, (6) epithelial and endothelial cells, (7) erythroblasts, (8) hematopoietic stem cells (HSCs) and progenitors, (9) master cells, (10) megakaryocytes, (11) myeloid cells, (12) neuron cells, including microglia, (13) neutrophils, (14) Natural Killer (NK) cells, (15) peripheral blood mononuclear cells (PBMCs), and (16) splenocytes. Group (8) hematopoietic stem cells (HSCs) and progenitors included stem cells and thymocytes; group (11) myeloid cells included bone-marrow-derived dendritic cells, bone-marrow-derived macrophages, dendritic cells, monocyte-derived macrophages, and macrophages. The percentages or abundances of signatures of immune cell types and proinflammatory soluble factors from the total number of enriched immunologic signatures were represented as cluster heatmaps generated using the R package ComplexHeatmap [18] with default options. All the plots presented in this manuscript were generated in R with default options.
To obtain immunologic signatures enriched by randomly selected genes and the whole genome, protein-coding genes in humans were retrieved from Human_genome_GENCODE_v32.bed released by the GENCODE project [10], and the R command sample_n() (with replacement) was executed to randomly select 150, 250, 350, 450, 550, 650, and 1000 human genes, or all protein-coding genes were used to perform the MSigDb over-representation analysis described above.
2.2.2. KEGG Pathway Over-Representation Analysis
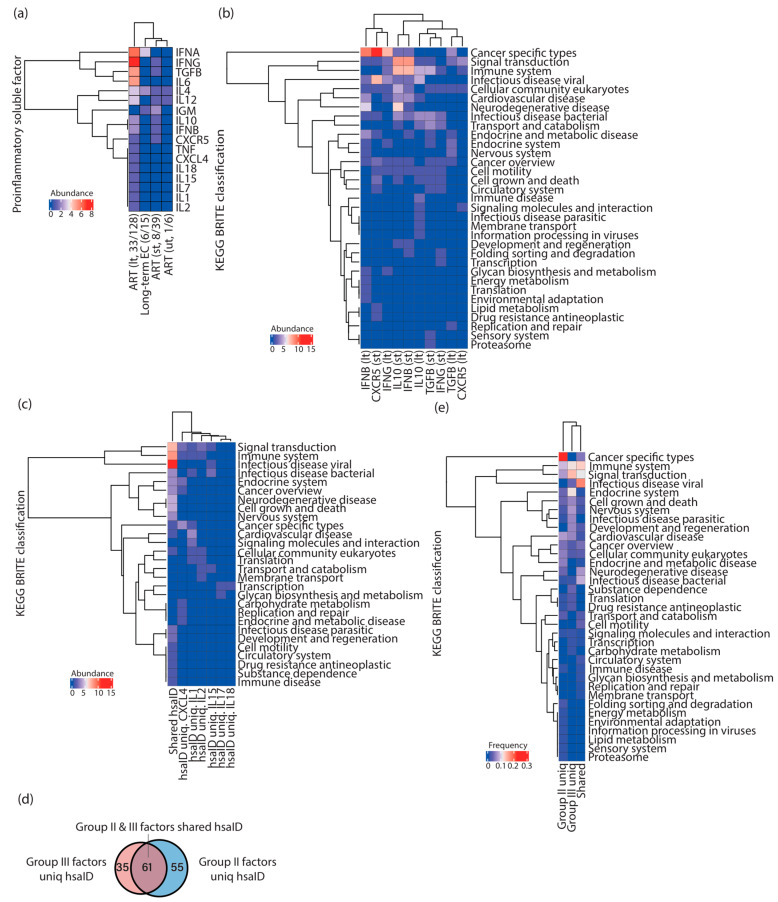
Enriched KEGG pathways were computed by conducting KEGG pathway over-representation analysis using the R package clusterProfiler [12] with the p-value cutoff equal to 0.5. Based on KEGG BRITE hierarchies documented at https://www.genome.jp/kegg/brite.html, accessed on 2 October 2022, every enriched pathway was assigned to its corresponding KEGG BRITE classification to generate the plots in Figure 2b–d. Of note, to create Figure 2c, enriched KEGG pathways that were found in all group III factors (so-called “Shared hsaID” in the figure panel) were first separated from those that were only present in individual group III factors, and then the cluster heatmap was created.
Figure 2.
Different compositions of proinflammatory soluble factor signatures were present alongside HIV-1 infections associated with ART. (a) Cluster heatmap representing the abundance of proinflammatory soluble factors present in unique immunologic signatures enriched in HIV-1-infected individuals and elite controllers. Parentheses placed under the column indicate the number of proinflammatory soluble factors from the total signatures. (b) Cluster heatmap representing the abundance of each KEGG BRITE classification harboring enriched pathways related to group II factors in HIV-1-infected individuals after a short (st) and a long (lt) period of ART. (c) Cluster heatmap representing the abundance of each KEGG BRITE classification harboring enriched pathways related to group III factors in HIV-1-infected individuals after a long period of ART. Enriched pathways that were found in all group III factors (so-called “Shared hsaID”) were separated from other enriched pathways unique to individual group III factors to create the cluster heatmap. (d) Venn diagram showing the overlap of enriched KEGG pathways represented by hsaID between group II and group III factors. (e) Cluster heatmap representing the frequency of KEGG BRITE classifications harboring enriched pathways between group II and group III factors. ART; antiretroviral therapy. EC; elite controllers. “hsaID”; Homo sapiens (human) ID.
2.2.3. Determination of Provirus-Targeted Genes Interacting with HIV-1
The HIV-1 Human Interaction Database (https://www.ncbi.nlm.nih.gov/genome/viruses/retroviruses/hiv-1/interactions/, accessed on 30 October 2022) was used to verify whether provirus-targeted genes interact with HIV-1 or not. This database documents all known interactions of HIV-1 and human genes that have been reported to affect HIV-1 replication and infectivity as well as interactions between HIV-1 gene products and host cell proteins, or proteins from disease organisms associated with HIV-1/AIDS [19,20,21].
2.2.4. Statistics
All statistical tests were performed in R with default options. Details are provided where appropriate in the main text.
3. Results
3.1. Different Immunologic Signatures Were Enriched Alongside HIV-1 Infections Associated with ART
In total, 121, 149, and 176 genes from pretreatment HIV-1-infected individuals (ut), HIV-1-infected individuals subjected to a short period of ART (st), and those subjected to a long period of ART (lt) [5], respectively, and 104 genes from elite controllers [6] (Table 1) were employed to perform over-representation analysis. A few of the input genes were shared among ut, st, lt, and elite controllers (Figure 1a and Table S27).
In total, 8 (Table S1), 65 (Table S2), 152 (Table S3), and 22 (Table S4) immunologic signatures were enriched in pretreatment HIV-1-infected individuals, HIV-1-infected individuals subjected to a short period of ART, those subjected to a long period of ART, and elite controllers, respectively (Figure 1b). It is essential to stress that due to a lack of expression data corresponding to integration sites used in this work, I therefore calculated rich factors [13] (see Materials and Methods), rather than net enrichment scores, to represent the enrichment intensity. I used 150, 250, 350, 450, 550, 650, and 1000 randomly selected genes (data not shown) and the whole genome as controls in the over-representation analysis to validate the significance of the score of the rich factor (Figure 1c). None of the immunologic signatures was enriched using 150, 250, 350, 450, 650, and 1000 genes (data not shown), and one immunologic signature was enriched using 550 randomly selected genes (rich factor: 3.681). Although 4872 immunologic signatures were enriched using the whole genome (rich factor: median, 1.162; mean, 1.154), rich factors measured in HIV-1-infected individuals (both pretreatment and receiving ART) and in elite controllers showed significance compared to controls (Figure 1c, controls: 550 genes and hg38 labeled in the figure panel), indicating that the immunologic signatures detected in this study were significantly enriched.
3.2. Different Combinations of Immune Cell Signatures Were Observed Alongside HIV-1 Infections Associated with ART
In line with the finding that provirus-targeted genes enriched specific immunologic signatures alongside HIV-1 infections associated with ART, I continuously investigated whether these enriched signatures could assign specific immune cell types and proinflammatory soluble factors that are stated in the hypothesis of this work. I retrieved immunologic signatures uniquely enriched in pretreatment HIV-1-infected individuals (ut, 6 signatures, 75%), HIV-1-infected individuals subjected to a short period of ART (st, 39 signatures, 60%), HIV-1-infected individuals subjected to a long period of ART (lt, 128 signatures, 84%), and elite controllers (15 signatures, 68%) (Figure 1d). From them, I counted the appearance of names of immune cell types in the standard names of the enriched immunologic signatures one by one (Figure 1e,f). I further classified immune cell types into 16 groups based on the Blood Cell Lineage chart published in the NIH website’s SEER Training Modules (subtypes of cells in each cell category are described in the Materials and Methods). It is important to note that ART interrupts HIV-1/AIDS progression; the composition of immune cell types observed here was in physiological HIV-1 infection conditions associated with ART. First, an increased variety of immune cell types in HIV-1-infected individuals subjected to a long period of ART (lt, found 132 terms of immune cells) were found (Figure 1e,f); among them, CD4 T cells (found 40 times, 30.3%) showed the highest frequency, followed by myeloid cells (found 35 times, 26.5%) (Figure 1e,f). This composition of immune cell signatures showed a difference between pretreatment HIV-1-infected individuals (ut, found 7 times) and patients subjected to a short period of ART (st, found 42 times) (Figure 1e,f, Pearson’s chi-square test). The composition of the immune cell signatures in elite controllers (found 14 times) was unique in pretreatment HIV-1-infected individuals and ART-treated patients (Figure 1e). Of note, the proportion of the group “B cells” was highly enriched in elite controllers compared to ART-treated patients (found 3 times, 21.4%) (Figure 1e); whereas, in ART-treated patients, it showed a decrease in HIV-1-infected individuals subjected to a short (found 7 times, 16.7%) and a long period of ART (found 9 times, 6.8%) (Figure 1e,f). The group “B cells” was not found in unique enriched immunologic signatures in pretreatment HIV-1-infected individuals (Figure 1e,f). CD4 T cells, the main HIV-1 reservoir cell type, showed an increase in their proportion alongside HIV-1 infections associated with ART (ut, found 1 time, 14.3%; st, found 9 times, 21.4%; lt, found 40 times, 30.3%). A regression of the presence of CD8 T cells (ut, found 1 time, 14.3%; st, found 6 times, 14.3%; lt, found 11 times, 8.3%) was, however, observed after a long period of ART (Figure 1e,f). The highest peak of the proportion of the myeloid cell signature was found in HIV-1-infected individuals subjected to a short period of ART (found 15 times, 35.7%). Additionally, the proportion of myeloid cells between pretreatment HIV-1-infected individuals (found 2 times, 28.6%) and patients subjected to a long period of ART (found 35 times, 26.5%) was comparable (Figure 1f). Intriguingly, the presence of the NK cell signature was observed in pretreatment HIV-1-infected individuals (found 1 term, 14.3%) and HIV-1-infected individuals subjected to a long period of ART (found 4 times, 3%) (Figure 1f). Collectively, these findings suggest that different combinations of immune cells with an increasing variety were present alongside HIV-1 infections with ART.
3.3. Different Combinations of Proinflammatory Soluble Factor Signatures Were Observed Alongside HIV-1 Infections Associated with ART
In the continuation of the previous logic, I also counted the appearances of the names of proinflammatory soluble factors, including cytokines and chemokines, in each unique enriched immunologic signature (Figure 2a), and found that 1, 8, 33, and 6 unique immunologic signatures contained the names of proinflammatory soluble factors in pretreatment HIV-1-infected individuals, HIV-1-infected individuals subjected to a short period of ART, those subjected to a long period of ART, and elite controllers, respectively (Figure 2a). I further grouped these proinflammatory soluble factors based on their appearance in HIV-1-infected individuals: group I factors, including IL4 and IL12, were present in pretreatment HIV-1-infected individuals and throughout the whole infection process (Figure 2a); group II factors, including CXCR5, IFNG, IFNB, IL10, and TGFB, were present in HIV-1-infected individuals subjected to ART irrespective of the period (Figure 2a); and group III factors, including CXCL4, IFNA, IL1, IL2, IL6, IL7, IL15, IL18, and TNF, appeared only after a long period of ART (Figure 2a). To further investigate each group of proinflammatory soluble factors, I sought KEGG pathways enriched by the genes retrieved from unique enriched immunologic signatures with corresponding proinflammatory soluble factors (Figure 2b,c). No clear separation of enriched KEGG pathways related to group II factors between HIV-1-infected individuals subjected to a short and a long period of ART was observed (Figure 2b), implying that the enrichment of these pathways was irrelevant to the period of therapy. Nevertheless, enriched pathways in “cancer specific types” were often associated with IFNB (lt), CXCR5 (st), and IFNG (lt); enriched pathways in “signal transduction” and “immune system” were more related to IL10 (st) and IFNB (st); enriched pathways in “infectious disease viral” were more related to CXCR5 (st) (Figure 2b). As for group III factors, pathways in “signal transduction”, “immune system”, and “infectious disease viral” were prevalent for every proinflammatory soluble factor in this group (Figure 2c, the first column from the left). Pathways in “membrane transport” (hsaID uniq. IL12), “glycan biosynthesis and metabolism” (hsaID uniq. IL17), “signaling molecules and interaction” (hsaID uniq. IL1), “carbohydrate metabolism” (hsaID uniq. CXCL4), “replication and repair” (hsaID uniq. CXCL4), and “endocrine and metabolic disease” (hsaID uniq. CXCL4) became specific to the indicated proinflammatory soluble factors (Figure 2c). As I compared the enriched KEGG pathways between group II and group III factors, more than half (61 pathways) of the pathways were common (Figure 2d). Among them, “infectious disease viral”, “immune system”, and “signal transduction” were the top three KEGG BRITE classifications shared in both groups of factors (Figure 2e). Enriched pathways in the classification “cancer specific types” were only found in group II factors (Figure 2e). Collectively, these findings suggest that different combinations of proinflammatory soluble factors participated in immunity alongside HIV-1 infections associated with ART.
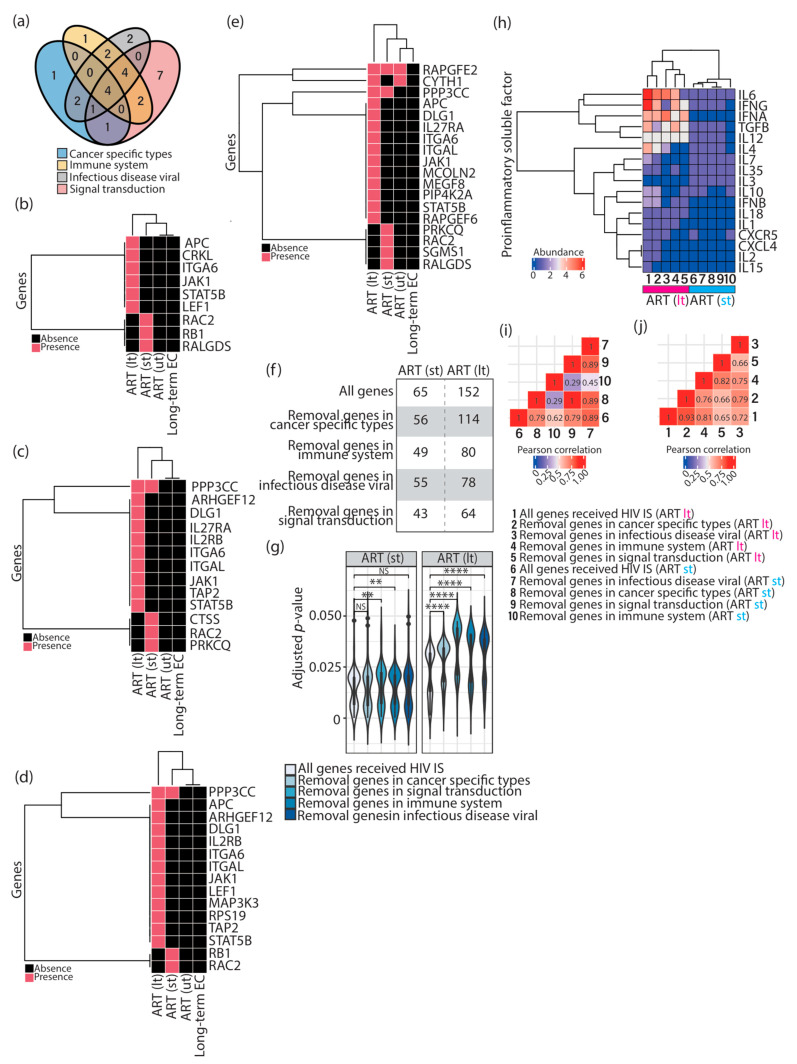
To verify whether enriched pathways involved in “cancer specific types”, “immune system”, “signal transduction”, and “infectious disease viral” were truly required during HIV-1 infections, I withdrew all genes present in these four KEGG BRITE classifications from the input gene list (Table 1) and repeated the over-representation analysis performed in Figure 1b. First, I observed that the majority of genes present in “cancer specific types”, “immune system”, and “infectious disease viral” are common, whereas the genes present in “signal transduction” appeared to be distinct from the other three classifications (Figure 3a). Intriguingly, genes present in these four KEGG BRITE classifications were restricted to HIV-1-infected individuals receiving ART and specific to different periods of treatment (Figure 3b–e), except the genes rapgfe2 and ppp3cc. The gene rapgfe2 was observed throughout the whole period of HIV-1 infections (Figure 3e), and ppp3cc appeared after patients received ART (Figure 3c–e). The gene cyth1 was found in pretreatment HIV-1-infected individuals and patients subjected to a long period of ART; whether the absence of cyth1 in HIV-1-infected individuals subjected to a short period of ART (Figure 3e) is due to technical reasons or is biologically relevant will require further investigation. Furthermore, a decreased number of enriched immunologic signatures with minor significance were obtained as genes in these four KEGG BRITE classifications, and were withdrawn (Figure 3f–g) (Tables S5–S12). Additionally, in HIV-1-infected individuals subjected to a long period of ART, several proinflammatory soluble factor signatures (IL4, IL6, IL7, IL15, IFNG) were infrequently present (Figure 3h), especially when the genes involved in “signal transduction” were taken away (Figure 3h, column 5). Consistently, the same pattern was present in the correlation plot as well (Figure 3j). However, signatures related to IL3, IL12, and IL18 were not affected (Figure 3h). Other signatures appeared to be either random (IFNA) or slightly affected (CXCL4, CXCR5, IFNB, IL1, IL2, IL10, IL35, TGFB) when genes in different KEGG BRITE classifications were withdrawn (Figure 3h, columns 1–5). The impact of the genes retrieved from HIV-1-infected individuals subjected to a short period of ART was minor (Figure 3h), thereby causing a clear separation of two main clusters between short and long periods of ART (Figure 3h). Nevertheless, a decreased correlation was still observed in HIV-1-infected individuals subjected to a short period of ART when genes in “immune system” were taken away (Figure 3i).
Figure 3.
Impact of enriched KEGG pathways involved in “cancer specific types”, “immune system”, “signal transduction”, and “infectious disease viral” in HIV-1-infected individuals subjected to ART. (a) Venn diagram representing the overlap of the genes present in enriched KEGG pathways involved in the four KEGG BRITE classifications. (b–e) Cluster heatmap representing the presence of the genes in enriched KEGG pathways related to “cancer specific types” (b), “immune system” (c), “infectious disease viral” (d), and “signal transduction” (e) in HIV-1-infected individuals before ART (ut) and after a short (st) or a long period (lt) of ART, and in elite controllers. (f) Numbers of immunologic signatures enriched by either a complete list of input genes (all genes) or a gene list with the removal of the genes in the four KEGG BRITE classifications in HIV-1-infected individuals after a short (st) or a long period (lt) of ART. (g) Violin plots representing the distribution of adjusted p-values corresponding to immunologic signatures enriched by either a complete list of the genes (all genes) or a gene list with the removal of the genes in the four KEGG BRITE classifications. Statistical significance was calculated using the Wilcoxon test in R with default options. (h) Cluster heatmap representing the abundance of proinflammatory soluble factor signatures enriched by either a complete list of the genes (all genes, column 1 and column 6) or a gene list with the removal of the genes in the four KEGG BRITE classifications in HIV-1-infected individuals after a short (st, columns 7–10) or a long period (lt, columns 2–5) of ART. (i,j) Correlation plots representing the Pearson correlation calculated based on the abundance of proinflammatory soluble factors in HIV-1-infected individuals after a short (i) or a long period (j) of ART. NS > 0.05, ** p ≤ 0.01, **** p ≤ 0.0001. ART; antiretroviral therapy. EC; elite controllers. HIV IS; HIV integration sites.
3.4. The Majority of Enriched Immunologic Signatures Resulted from Gene Sets That Interact with HIV-1
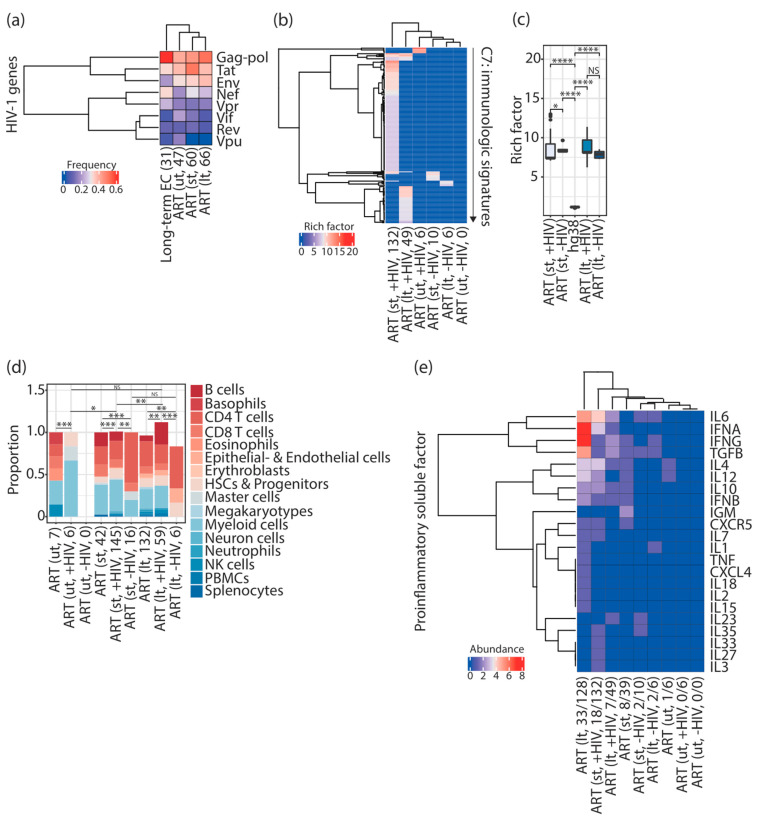
As I hypothesized that HIV-1 integration frequency might be used as a surrogate for gene sets to define specific immune cell types and proinflammatory soluble factors required during HIV-1 infections, I therefore tested whether any connection is present between HIV-1 integration and the targeted genes. If this is the case, I expect that the required immunologic signatures are enriched by the gene sets that either show protein interactions with gene products of HIV-1 or affect HIV-1 replication and infectivity; whereas, other gene sets that do not interact with HIV-1 are not responsible for the enrichment of any immunologic signature during HIV-1 infections. To verify this hypothesis, I separated input genes reported to interact with HIV-1 from others that do not interact with HIV-1 (see Materials and Methods), and observed that 38.8% (47 genes out of 121 genes), 40.5% (60 genes out of 148 genes), 37.5% (66 genes out of 176 genes), and 29.8% (31 genes out of 104 genes) of the genes have been reported to interact with HIV-1 in pretreatment HIV-1-infected individuals, HIV-1-infected individuals subjected to a short period of ART, those subjected to a long period of ART, and elite controllers, respectively. The majority of the genes showed interactions with HIV-1 gag-pol genes, followed by the tat gene, the env gene, and the nef gene; few HIV-1-targeted genes were found to interact with the HIV-1 vpr, vif, rev, and vpu genes (Figure 4a).
Figure 4.
HIV-1-targeted gene sets that interact with HIV-1 contributed to enriching immunologic signatures alongside HIV-1 infections associated with ART. (a) Cluster heatmap representing the frequency of provirus-targeted genes interacting with HIV-1. Parentheses placed under the column indicate the total number of provirus-targeted genes reported to interact with HIV-1. (b) Cluster heatmap representing enriched immunologic signatures in HIV-1-infected individuals and elite controllers. Inside the parentheses, +HIV and −HIV indicate provirus-targeted genes reported to interact with HIV-1 or not, respectively. The number indicates the number of enriched immunologic signatures. The color scale represents the intensity of the enrichment represented by rich factors. (c) Box plots representing the enrichment of immunologic signatures represented by rich factors. Commands executed to calculate rich factors are described in the Materials and Methods. Statistical significance was calculated using the Wilcoxon test in R with default options. (d) Stacked bar chart representing the proportion of immune cell types present in enriched immunologic signatures in HIV-1-infected individuals. Statistical significance was calculated using Pearson’s chi-square test in R with default options. Inside the parentheses, +HIV and −HIV indicate provirus-targeted genes reported to interact with HIV-1 or not, respectively. The number indicates the number of counted immune cell types in enriched immunologic signatures. (e) Cluster heatmap representing the abundance of proinflammatory soluble factors present in enriched immunologic signatures in HIV-1-infected individuals. Inside the parentheses, +HIV and −HIV indicate provirus-targeted genes reported to interact with HIV-1 or not, respectively. The number indicates the number of proinflammatory soluble factors from the total number of enriched immunologic signatures. NS > 0.05, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001. “ut”; pretreatment. “st”; short period of ART. “lt”; long period of ART. ART; antiretroviral therapy. EC; elite controllers. HSCs; hematopoietic stem cells. NK cells; natural killer cells. PBMCs; peripheral blood mononuclear cells.
I further performed over-representation analysis on both subsets of input genes (interact with HIV-1 or not) and found that the enrichment of the immunologic signatures was mainly contributed by the genes interacting with HIV-1 (Figure 4b) (Tables S14 and S16). Only a tiny proportion of immunologic signatures were enriched by the genes that do not interact with HIV-1 (Tables S15 and S17) (Figure 4b). Notably, genes interacting with HIV-1 in pretreatment HIV-1-infected individuals were only responsible for a small proportion of the enriched immunologic signatures (Figure 4b) (Table S13). None of the immunologic signatures can be enriched using the genes that do not interact with HIV-1 in pretreatment HIV-1-infected individuals (Figure 4b). Intriguingly, rich factors showed an increase in HIV-1-infected individuals subjected to ART, regardless of whether provirus-targeted genes were reported to interact with HIV-1 or not (Figure 4c). Furthermore, a variety of immune cell signatures were only observed using genes interacting with HIV-1 in patients receiving a short (145 terms) and a long period (59 terms) of ART (Figure 4d), resembling similar circumstances using a complete list of input genes. In contrast, a poor combination of immune cell types was found in enriched immunologic signatures using genes that do not interact with HIV-1 (Figure 4d). Consistently, the majority of the proinflammatory soluble factor signatures were present in enriched immunologic signatures using a complete list of input genes and the genes interacting with HIV-1 in infected individuals subjected to ART (Figure 4e). Either a tiny proportion of proinflammatory soluble factor signatures or no proinflammatory soluble factor signatures were observed using the genes that do not interact with HIV-1 and using the genes retrieved from pretreatment HIV-1-infected individuals (Figure 4e). Altogether, these findings indicate that enriched immunologic signatures in HIV-1-infected individuals subjected to ART mainly resulted from gene sets interacting with HIV-1.
4. Discussion
In this work, I focused on the possible interplay between host genes targeted by proviruses and the host functional genome property. Based on the over-representation analysis of the genes targeted by proviruses, I first observed that different unique immunologic signatures were enriched alongside HIV-1 infections associated with ART (Figure 1b). Furthermore, an increase in the variation in immune cell types was present in HIV-1-infected individuals subjected to a long period of ART (Figure 1e,f), reflecting an environment with complicated immune responses. Given that different kinds of innate immune-related cells are recruited to the site of HIV-1 infection over time, unveiling the alterations of immune cells alongside HIV-1 infections can thus be essential to understand the evolution of HIV-1 reservoirs. As shown in this work, different compositions of immune cell types were present in unique enriched immunologic signatures in pretreatment HIV-1-infected individuals, ART-treated HIV-1-infected patients, and elite controllers (Figure 1e,f). This observation can also be linked to different compositions of proinflammatory soluble factors alongside HIV-1 infections associated with ART, as observed in this work (Figure 2a).
This study demonstrated that different combinations of proinflammatory soluble factor signatures were present at different stages of ART. I observed that CXCR5, IFNB, IFNG, IL10, and TGFB were found in enriched immunologic signatures in HIV-1-infected individuals subjected to a short and a long period of ART (Figure 2a). CXCL4, IFNA, IL1, IL2, IL6, IL7, IL15, IL18, and TNF were observed only in enriched immunologic signatures in HIV-1-infected individuals subjected to a long period of ART (Figure 2a). Few proinflammatory soluble factors were observed in pretreatment HIV-1-infected individuals and elite controllers (Figure 2a). Indeed, it has been known for a long time that HIV-1 infections result in dysregulation of the cytokine profile [22]. Further investigations will be required to understand the immune strategy using different compositions of proinflammatory soluble factors to interfere with the evolution of an HIV-1 latent reservoir. KEGG pathways enriched by gene sets related to group II and III factors mainly fell into the KEGG BRITE classifications “cancer-specific types”, “infectious disease viral”, “immune system”, and “signal transduction” (Figure 2b–e). It is essential to note that gene sets from these four KEGG BRITE classifications were restricted to HIV-1-infected individuals subjected to a short and a long period of ART, and the majority of them were specific to different periods of treatment, indicating that the selection of proviral integration can be disease progression-specific (Figure 3b–e). According to the observation in this work, this selection is irrelevant to elite control (Figure 3b–e). Finally, I showed that the enriched immunologic signatures were mainly contributed by genes interacting with HIV-1 (Figure 4b), even though the genes reported to interact with HIV-1 did not represent the majority of the genes in the input list. Here, it is intriguing to point out that these founder genes analyzed in this work not only showed interaction with HIV-1 but also were invaded by the genome of the virus in physiological HIV-1 infection conditions. This observation may indicate that the affinity of the provirus-targeted genes towards gene products of HIV-1 can serve as a potential indicator to predict the required immunologic signatures during HIV-1 infections associated with ART. The following are open questions: What makes the genes reported to interact with HIV-1 different from others? What is the role of the provirus-targeted genes that do not interact with HIV-1?
Although it is presently workable to sequence the nearly full-length HIV-1 genome, the yield of retrieved sequences is limited to the range of 50 to 100 sequences in cells purified from a patient [23,24], thereby rendering accurate and quantitative investigation difficult. A better strategy to sequence the full-length HIV-1 genome of proviruses and elevate the yield of output sequences from clinical samples will be an absolute requisite because intact proviruses are responsible for viral rebounds after treatment cessation [24]. In total, 26, 6, 14, and 22 input genes were targeted by intact proviruses in pretreatment HIV-1-infected individuals, patients subjected to a short period of ART, those subjected to a long period of ART, and elite controllers, respectively (Table S26). Intriguingly, around one third to half of the intact provirus-targeted genes were associated with the signatures of immune cell types (10 genes in pretreatment HIV-1-infected individuals, 3 genes in patients subjected to a short period of ART, 7 genes in patients subjected to a long period of ART, and 11 genes in elite controllers) (Tables S18–S21). Conversely, intact provirus-targeted genes were not frequently associated with the signatures of proinflammatory soluble factors (2 genes in pretreatment HIV-1-infected individuals, 1 gene in patients subjected to a short period of ART, 4 genes in patients subjected to a long period of ART, and 4 genes in elite controllers) (Tables S22–S25). A more recent study published by Sun et al. (2023) [25] continued to strengthen this “immune selection hypothesis” proposed by Einkauf and colleagues (the term “immune selection hypothesis” was mentioned in Peer Review File available at https://static-content.springer.com/esm/art%3A10.1038%2Fs41586-022-05538-8/MediaObjects/41586_2022_5538_MOESM2_ESM.pdf (accessed on 25 January 2023)). They observed that HIV-1 reservoir cells harboring intact proviruses frequently express ensemble phenotypic signatures of surface markers (including the checkpoint marker PD-1) that confer resistance to immune-mediated killing by CD8+ T cells and NK cells [25]. In this study, two unique immunologic signatures, GSE26495_NAIVE_VS_PD1LOW_CD8_TCELL_DN [26] and GSE24026_PD1_LIGATION_VS_CTRL_IN_ACT_TCELL_LINE_DN [27], enriched in HIV-1-infected individuals subjected to a long period of ART were observed to be associated with PD-1 (Table S3).
In this work, I investigated whether the functional genome property also contributes to this immune selection alongside HIV-1 infections associated with ART and tested if genes targeted by proviruses can form different communities that are beneficial to the immunological process by recruiting immune cells and activating signaling pathways in different pathogenic stages of HIV-1 infections associated with ART. It is important to note that although the enriched immunologic signature gene sets represented less than 10% of the input genes (Tables S1–S4), the rich factors showed significant enrichment (Figure 1c and Figure 4c), making the results from the over-representation analysis robust. A previous study also demonstrated that over-representation of pathways is not correlated with the number of genes in the pathways [28]. Several important questions still need to be addressed regarding (1) whether host genes associated with innate immunity are more frequently targeted by proviruses, and (2) whether HIV-1 integration in genes relevant to innate immunity is due to a transcriptionally active status, thereby being favorable for HIV-1 integration. Among the input genes analyzed in this study, very few genes overlapped between ART-treated patients and elite controllers, and between pretreatment HIV-1-infected individuals and patients subjected to ART (Figure 1a and Table S27). The gene rptor is the only gene present in the four lists of input genes (Table S27). The gene product of rptor forms a stoichiometric complex with the mammalian target of rapamycin (mTOR) kinase. mTOR signaling largely regulates cell metabolism in activated T cells and is known to be required for HIV-1 replication and latency [29,30], highlighting the importance of immunometabolism involved in HIV-1 pathogenesis [31]. The gene rapgef2 was found throughout the infection course of HIV-1 in patients (Figure 3e and Table S27). The gene product of rapgef2 is responsible for RAS activation [32]. The Ras protein serves as a central intermediate in many signaling pathways, including NF-κB activation [33]. NF-κB is one of the critical transcription factors required for the initiation of HIV-1 transcription [34,35] and a therapeutic target activated by antiretroviral drugs [36]. Five common genes were found in pretreatment HIV-1-infected individuals and patients subjected to a short period of ART; however, little is known about their roles in HIV-1 pathogenesis (Table S27). Of note, the function of the gene product of fanca is involved in DNA repair. Recently, Kabi and Filion proposed their initial hypothesis that DNA repair may help flag integrated viruses, including HIV-1, allowing the host immunity to sense viral DNA [37]. If this hypothesis is tested to be true, it would open a new avenue of research on integrated viral immune sensing, where chromatin would play a role. Seven genes are identical between pretreatment HIV-1-infected individuals and patients subjected to a long period of ART (Table S27). The gene malat1, which produces a long non-coding RNA, has been shown to promote HIV-1 transcription and infection [38]. The gene product of vav1 and its mediated signaling pathway has been shown to interact with the HIV-1 Nef [39,40,41]. A total of 11 genes were shared between patients subjected to a short and a long period of ART (Table S27). Among them, the gene brd4 was observed. BRD4 is one of the essential epigenetic readers that regulate HIV-1 transcription [42,43,44] and has become a popular target in the design of antiretroviral drugs [45]. In one research article, the influence of ndufa6 gene expression on HIV-1-induced T-cell apoptosis was found [46]. Here, I have summarized input genes that are associated with signatures of either immune cell types or proinflammatory soluble factors in Table 2.
Table 2.
The number of provirus-targeted genes (from Table 1) associated with signatures of immune cell types and proinflammatory soluble factors.
| Patient Sample | Number of Provirus-Targeted Genes (Number of Enriched Immunologic Signatures) |
|
|---|---|---|
| Immune Cell Types | Proinflammatory Soluble Factors | |
| Pretreatment (ut) | 35 (6) | 7 (1) |
| Short period of ART (st) | 101 (37) | 37 (7) |
| Long period of ART (lt) | 164 (121) | 109 (33) |
| Elite controllers | 48 (14) | 25 (6) |
Intriguingly, more than 90% of the input genes were associated with immune cell type signatures in HIV-1-infected individuals subjected to a long period of ART, followed by HIV-1-infected individuals subjected to a short period of ART (67.7%), elite controllers (46.1%), and pretreatment HIV-1-infected individuals (28.9%) (Table 2). Consistently, 61.9% of the input genes were associated with the signatures of proinflammatory soluble factors in HIV-1-infected individuals subjected to a long period of ART, followed by HIV-1-infected individuals subjected to a short period of ART (24.8%), elite controllers (24%), and pretreatment HIV-1-infected individuals (5.7%) (Table 2). While overlapping with human cytokine genes (in total 1793 unique genes) available at ImmPort (https://www.immport.org (accessed on 27 January 2023)) [47,48,49], none of the provirus-targeted genes associated with proinflammatory soluble factors in pretreatment HIV-1-infected individuals belonged to cytokine genes. Five (ctss, nr2c2, pik3r1, prkcq, rac2), ten (bach2, eed, grap2, il27ra, il2rb, itga, jak1, micb, ppp3cc, tap2) and one (grb2) provirus-targeted genes associated with proinflammatory soluble factors belong to cytokine genes in st, lt, and elite controllers, respectively. The gene pik3r1 has been proposed to be one of the essential host factors targeted by HIV-1 viral proteins using genome-wide RNAi screening [50]. Several studies have shown that protein kinase C theta, the gene product of prkcq, is involved in controlling HIV-1 replication [51,52] and is associated with HIV-1 Nef which leads to activation of ERK1/2 [53,54]. The gene product of rac2 (Rac) and its relevant signaling pathway has been reported to be activated by HIV-1 Nef-associated protein complex [55] and involved in HIV-1 Nef-induced blockade of superoxide release in human macrophages [56]. The gene bach2 is presently one of the well-known cancer-related genes that are favorable for HIV-1 integration identified in HIV-1-infected individuals [1]. The gene eed encodes a member of the polycomb complex 2 that interacts with the HIV-1 matrix protein. Inhibition of EED allows HIV-1 latency reversal by decreasing the level of H3K27 tri-methylation [57]. The gene il17ra encodes interleukin 27 receptor subunit alpha. Recently, IL-27 signaling (IL-27 and its receptor) has been proposed to be involved in the pathogenesis of HIV-1 infection and immune reconstitution in infected individuals subjected to ART [58,59]. The gene il2rb encodes interleukin 2 receptor subunit beta. Expression of IL2RB required for the production of granulysin used by lymphocytes to kill tumors and microbial cells has been shown to be dysfunctional in HIV-1-infected individuals [60]. The gene jak1 is a key component of the interleukin-6/JAK1/STAT3 immune and inflammation response. JAK1/2 inhibitors, such as ruxolitinib and baricitinib, have been approved by U.S. Food and Drug Administration (FDA) as a potential therapeutic strategy to target the HIV-1 latent reservoir [61]. The gene micb encodes a heavily glycosylated protein which is a ligand for the natural killer group 2 member type II receptor; this receptor is responsible for host immunosurveillance and NK cell-mediated cytotoxicity during HIV-1 infections [62]. The gene tap2 is one of the MHC-II-encoded genes necessary for the generation of a cellular immune response. One study has shown that the heterozygous A/G and homozygous G/G polymorphisms on TAP2, rather than TAP1, were positively associated with an increased risk of HIV-1/AIDS infection [63]. Rom and colleagues (2011) have shown the interaction between the cellular factor Grb2 (the gene product of grb2) and the HIV-1 Tat protein subsequently impairs the activation of the Raf/MAPK pathway [64]. Although the discussion between ART-treated HIV-1-infected individuals and elite controllers is modest in the current study, several findings (Figure 1b, Figure 2a and Figure 3b–e) suggest the different propensities of enriched immunologic signatures between these two groups of people living with HIV-1. Given that the specific phenotype of elite control is most likely beneficial to host factors [65], further investigation of the functional interplay between elite controllers and the genome property could be expected to shed some light on the molecular basis of elite control.
At present, transcriptional datasets that correspond to the input gene lists used in this study are not yet available. It is presently unclear whether the transcriptional status of a gene may also be involved in the interaction between HIV-1 integration sites and the host functional genome property. Further orthogonal approaches and additional transcriptomic datasets on corresponding patient cells will be requisites to clarify whether integration site frequency is also dependent on gene expression levels. Another critical issue is that it is not clear whether integration sites of proviruses retrieved by different methods (FLIP-seq [66] and MIP-seq [67] used by Jiang et al. (2020) [6]; PRIP-seq used by Einkauf et al. (2022) [5]) may cause any bias while comparing results between HIV-1-infected individuals and elite controllers. In the future, experiments to retrieve (intact) provirus integration sites from both types of patients in parallel will be required to compare their differences systematically. Nevertheless, the observations in this study allow for proposing a new avenue of research on HIV-1 integration and the host functional genome property alongside HIV-1 infections associated with ART.
5. Conclusions
The experimental findings from Einkauf et al. (2022) [5] and Jiang et al. (2020) [6] strongly suggest the presence of immune selection pressure during ART in HIV-1-infected individuals and a long period of elite control, respectively. In this study, I performed the over-representation analysis on provirus-targeted genes from these two studies and demonstrated that unique immunologic signatures were enriched in pretreatment HIV-1-infected individuals, HIV-1-infected individuals subjected to a short period of ART, those subjected to a long period of ART, and elite controllers (Figure 1b). Furthermore, I observed that unique immunologic signatures associated with different combinations of immune cell types (Figure 1e,f) and proinflammatory soluble factors (Figure 2a) would be required to fulfill the host immune response alongside HIV-1 infections associated with ART and a consecutive period of elite control. KEGG pathways enriched by the provirus-targeted genes associated with the signature of proinflammatory soluble factors were frequently related to “cancer specific types”, “immune system”, “infectious disease viral”, and “signal transduction” (Figure 3). Most importantly, this study has shown that immunologic signatures mainly resulted from provirus-targeted genes that interact with HIV-1 (Figure 4). All these findings led us to hypothesize that host genes harboring HIV-1 integration may form different genomic communities, which may define specific immune cell types and proinflammatory soluble factors to support host immunity alongside HIV-1 infections associated with ART.
Acknowledgments
I would like to thank Ariberto Fassati, Chien-Kuo Lee, and Zeger Debyser (alphabetic order) for their critical reading of and constructive feedback on the manuscript.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/vaccines11020402/s1, Table S1. Immunologic signatures enriched in pretreatment HIV-1-infected individuals. A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from pretreatment HIV-1-infected individuals; Table S2. Immunologic signatures enriched in HIV-1-infected individuals subjected to a short period of ART. A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a short period of ART; Table S3. Immunologic signatures enriched in HIV-1-infected individuals subjected to a long period of ART. A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a long period of ART; Table S4. Immunologic signatures enriched in elite controllers. A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from elite controllers; Table S5. Immunologic signatures enriched in HIV-1-infected individuals subjected to a short period of ART (removal of the genes in the KEGG BRITE classification “cancer specific types”). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a short period of ART. The provirus-targeted genes present in the KEGG BRITE classification “cancer specific types” were withdrawn in this analysis; Table S6. Immunologic signatures enriched in HIV-1-infected individuals subjected to a short period of ART (removal of the genes in the KEGG BRITE classification “immune system”). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a short period of ART. The provirus-targeted genes present in the KEGG BRITE classification “immune system” were withdrawn in this analysis; Table S7. Immunologic signatures enriched in HIV-1-infected individuals subjected to a short period of ART (removal of the genes in the KEGG BRITE classification “infectious disease viral”). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a short period of ART. The provirus-targeted genes present in the KEGG BRITE classification “infectious disease viral” were withdrawn in this analysis; Table S8. Immunologic signatures enriched in HIV-1-infected individuals subjected to a short period of ART (removal of the genes in the KEGG BRITE classification “signal transduction”). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a short period of ART. The provirus-targeted genes present in the KEGG BRITE classification “signal transduction” were withdrawn in this analysis; Table S9. Immunologic signatures enriched in HIV-1-infected individuals subjected to a long period of ART (removal of the genes in the KEGG BRITE classification “cancer specific types”). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a long period of ART. The provirus-targeted genes present in the KEGG BRITE classification “cancer specific types” were withdrawn in this analysis; Table S10. Immunologic signatures enriched in HIV-1-infected individuals subjected to a long period of ART (removal of the genes in the KEGG BRITE classification “immune system”). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a long period of ART. The provirus-targeted genes present in the KEGG BRITE classification “immune system” were withdrawn in this analysis; Table S11. Immunologic signatures enriched in HIV-1-infected individuals subjected to a long period of ART (removal of the genes in the KEGG BRITE classification “infectious disease viral”). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a long period of ART. The provirus-targeted genes present in the KEGG BRITE classification “infectious disease viral” were withdrawn in this analysis; Table S12. Immunologic signatures enriched in HIV-1-infected individuals subjected to a long period of ART (removal of the genes in the KEGG BRITE classification “signal transduction”). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes retrieved from HIV-1-infected individuals subjected to a long period of ART. The provirus-targeted genes present in the KEGG BRITE classification “signal transduction” were withdrawn in this analysis; Table S13. Immunologic signatures enriched in pretreatment HIV-1-infected individuals (input genes reported to interact with HIV-1). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes interacting with HIV-1; Table S14. Immunologic signatures enriched in HIV-1-infected individuals subjected to a short period of ART (input genes reported to interact with HIV-1). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes interacting with HIV-1; Table S15. Immunologic signatures enriched in HIV-1-infected individuals subjected to a short period of ART (input genes that do not interact with HIV-1). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes that were not reported to interact with HIV-1; Table S16. Immunologic signatures enriched in HIV-1-infected individuals subjected to a long period of ART (input genes reported to interact with HIV-1). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes interacting with HIV-1; Table S17. Immunologic signatures enriched in HIV-1-infected individuals subjected to a long period of ART (input genes that do not interact with HIV-1). A table list, including ID and Description, GeneRatio, BgRatio, p-value, adjusted p-value, q-value, and gene ID of immunologic signatures enriched using over-representation analysis with provirus-targeted genes that were not reported to interact with HIV-1. Table S18. Provirus-targeted genes associated with immune cell type signatures in pretreatment HIV-1-infected individuals. A table list, including the gene symbol, gene name, and entrez gene id of input genes associated with immune cell type signatures in pretreatment HIV-1-infected individuals. Table S19. Provirus-targeted genes associated with immune cell type signatures in HIV-1-infected individuals subjected to a short period of ART. A table list, including the gene symbol, gene name, and entrez gene id of input genes associated with immune cell type signatures in HIV-1-infected individuals subjected to a short period of ART. Table S20. Provirus-targeted genes associated with immune cell type signatures in HIV-1-infected individuals subjected to a long period of ART. A table list, including the gene symbol, gene name, and entrez gene id of input genes associated with immune cell type signatures in HIV-1-infected individuals subjected to a long period of ART. Table S21. Provirus-targeted genes associated with immune cell type signatures in elite controllers. A table list, including the gene symbol, gene name, and entrez gene id of input genes associated with immune cell type signatures in elite controllers. Table S22. Provirus-targeted genes associated with proinflammatory soluble factor signatures in pretreatment HIV-1-infected individuals. A table list, including the gene symbol, gene name, and entrez gene id of input genes associated with proinflammatory soluble factor signatures in pretreatment HIV-1-infected individuals. Table S23. Provirus-targeted genes associated with proinflammatory soluble factor signatures in HIV-1-infected individuals subjected to a short period of ART. A table list, including the gene symbol, gene name, and entrez gene id of input genes associated with proinflammatory soluble factor signatures in HIV-1-infected individuals subjected to a short period of ART. Table S24. Provirus-targeted genes associated with proinflammatory soluble factor signatures in HIV-1-infected individuals subjected to a long period of ART. A table list, including the gene symbol, gene name, and entrez gene id of input genes associated with proinflammatory soluble factor signatures in HIV-1-infected individuals subjected to a long period of ART. Table S25. Provirus-targeted genes associated with proinflammatory soluble factor signatures in elite controllers. A table list, including the gene symbol, gene name, and entrez gene id of input genes associated with proinflammatory soluble factor signatures in elite controllers. Table S26. Input genes targeted by intact proviruses. A table indicates input genes harboring intact proviruses. Table S27. Overlap of provirus-targeted genes among different groups of patient samples. A table indicates the gene symbol and the number of genes shared among different groups of input gene lists.
Author Contributions
Conceptualization, H.-C.C.; methodology, H.-C.C.; validation, H.-C.C.; formal analysis, H.-C.C.; investigation, H.-C.C.; resources, H.-C.C.; data curation, H.-C.C.; writing—original draft preparation, H.-C.C.; writing—review and editing, H.-C.C.; visualization, H.-C.C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study.
Conflicts of Interest
The author declares no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ikeda T., Shibata J., Yoshimura K., Koito A., Matsushita S. Recurrent HIV-1 Integration at theBACH2Locus in Resting CD4 T Cell Populations during Effective Highly Active Antiretroviral Therapy. J. Infect. Dis. 2007;195:716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- 2.Maldarelli F., Wu X., Su L., Simonetti F.R., Shao W., Hill S., Spindler J., Ferris A.L., Mellors J.W., Kearney M.F., et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner T.A., McLaughlin S., Garg K., Cheung C.Y.K., Larsen B.B., Styrchak S., Huang H.C., Edlefsen P.T., Mullins J.I., Frenkel L.M. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhyvoloup A., Melamed A., Anderson I., Planas D., Lee C.-H., Kriston-Vizi J., Ketteler R., Merritt A., Routy J.-P., Ancuta P., et al. Digoxin reveals a functional connection between HIV-1 integration preference and T-cell activation. PLoS Pathog. 2017;13:e1006460. doi: 10.1371/journal.ppat.1006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Einkauf K.B., Osborn M.R., Gao C., Sun W., Sun X., Lian X., Parsons E.M., Gladkov G.T., Seiger K.W., Blackmer J.E., et al. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell. 2022;185:266–282.e15. doi: 10.1016/j.cell.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang C., Lian X., Gao C., Sun X., Einkauf K.B., Chevalier J.M., Chen S.M.Y., Hua S., Rhee B., Chang K., et al. Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature. 2020;585:261–267. doi: 10.1038/s41586-020-2651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pastor-Satorras R., Vázquez A., Vespignani A. Dynamical and correlation properties of the internet. Phys. Rev. Lett. 2001;87:258701. doi: 10.1103/PhysRevLett.87.258701. [DOI] [PubMed] [Google Scholar]
- 8.Girvan M., Newman M.E.J. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause J., Croft D.P., James R. Social network theory in the behavioural sciences: Potential applications. Behav. Ecol. Sociobiol. 2007;62:15–27. doi: 10.1007/s00265-007-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frankish A., Diekhans M., Ferreira A.-M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinlan A.R., Hall I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 19.Ptak R.G., Fu W., Sanders-Beer B.E., Dickerson J.E., Pinney J.W., Robertson D.L., Rozanov M.N., Katz K.S., Maglott D.R., Pruitt K.D., et al. Short Communication: Cataloguing the HIV Type 1 Human Protein Interaction Network. AIDS Res. Hum. Retrovir. 2008;24:1497–1502. doi: 10.1089/aid.2008.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu W., Sanders-Beer B.E., Katz K.S., Maglott D.R., Pruitt K.D., Ptak R.G. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 2009;37:D417–D422. doi: 10.1093/nar/gkn708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinney J.W., Dickerson J.E., Fu W., Sanders-Beer B.E., Ptak R.G., Robertson D.L. HIV-host interactions: A map of viral perturbation of the host system. AIDS. 2009;23:549–554. doi: 10.1097/QAD.0b013e328325a495. [DOI] [PubMed] [Google Scholar]
- 22.Clerici M., Shearer G.M. A TH1→TH2 switch is a critical step in the etiology of HIV infection. Immunol. Today. 1993;14:107–111. doi: 10.1016/0167-5699(93)90208-3. [DOI] [PubMed] [Google Scholar]
- 23.Hiener B., Horsburgh B.A., Eden J.-S., Barton K., Schlub T.E., Lee E., Stockenstrom S., Odevall L., Milush J.M., Liegler T., et al. Identification of Genetically Intact HIV-1 Proviruses in Specific CD4 T Cells from Effectively Treated Participants. Cell Rep. 2017;21:813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho Y.-C., Shan L., Hosmane N.N., Wang J., Laskey S.B., Rosenbloom D.I.S., Lai J., Blankson J.N., Siliciano J.D., Siliciano R.F. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun W., Gao C., Hartana C.A., Osborn M.R., Einkauf K.B., Lian X., Bone B., Bonheur N., Chun T.-W., Rosenberg E.S., et al. Phenotypic signatures of immune selection in HIV-1 reservoir cells. Nature. 2023 doi: 10.1038/s41586-022-05538-8. online ahead of print . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duraiswamy J., Ibegbu C.C., Masopust D., Miller J.D., Araki K., Doho G.H., Tata P., Gupta S., Zilliox M.J., Nakaya H.I., et al. Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults. J. Immunol. 2011;186:4200–4212. doi: 10.4049/jimmunol.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quigley M., Pereyra F., Nilsson B., Porichis F., Fonseca C., Eichbaum Q., Julg B., Jesneck J.L., Brosnahan K., Imam S., et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 2010;16:1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen K.-C., Wang T.-Y., Chan C.-H. Associations between HIV and human pathways revealed by protein-protein interactions and correlated gene expression profiles. PLoS ONE. 2012;7:e34240. doi: 10.1371/journal.pone.0034240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besnard E., Hakre S., Kampmann M., Lim H.W., Hosmane N.N., Martin A., Bassik M.C., Verschueren E., Battivelli E., Chan J., et al. The mTOR Complex Controls HIV Latency. Cell Host Microbe. 2016;20:785–797. doi: 10.1016/j.chom.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crater J.M., Nixon D.F., O’Brien R.L.F. HIV-1 replication and latency are balanced by mTOR-driven cell metabolism. Front. Cell Infect. Microbiol. 2022;12:1068436. doi: 10.3389/fcimb.2022.1068436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valle-Casuso J.C., Angin M., Volant S., Passaes C., Monceaux V., Mikhailova A., Bourdic K., Avettand-Fenoel V., Boufassa F., Sitbon M., et al. Cellular Metabolism Is a Major Determinant of HIV-1 Reservoir Seeding in CD4 T Cells and Offers an Opportunity to Tackle Infection. Cell Metab. 2019;29:611–626.e5. doi: 10.1016/j.cmet.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Rebhun J.F., Castro A.F., Quilliam L.A. Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction. J. Biol. Chem. 2000;275:34901–34908. doi: 10.1074/jbc.M005327200. [DOI] [PubMed] [Google Scholar]
- 33.Folgueira L., Algeciras A., MacMorran W.S., Bren G.D., Paya C.V. The Ras-Raf pathway is activated in human immunodeficiency virus-infected monocytes and particpates in the activation of NF-kappa B. J. Virol. 1996;70:2332–2338. doi: 10.1128/JVI.70.4.2332-2338.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerritsen M.E., Williams A.J., Neish A.S., Moore S., Shi Y., Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang B., Yang X.-D., Zhou M.-M., Ozato K., Chen L.-F. Brd4 Coactivates Transcriptional Activation of NF-κB via Specific Binding to Acetylated RelA. Mol. Cell. Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brogdon J., Ziani W., Wang X., Veazey R.S., Xu H. In Vitro effects of the small-molecule protein kinase C agonists on HIV latency reactivation. Sci. Rep. 2016;6:39032. doi: 10.1038/srep39032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabi M., Filion G.J. Chromatin and viral integration in immunity: The challenge of silencing non-self genes. Trends Immunol. 2022;43:449–458. doi: 10.1016/j.it.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Qu D., Sun W.-W., Li L., Ma L., Sun L., Jin X., Li T., Hou W., Wang J.-H. Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter. Nucleic Acids Res. 2019;47:3013–3027. doi: 10.1093/nar/gkz117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quaranta M.G., Mattioli B., Spadaro F., Straface E., Giordani L., Ramoni C., Malorni W., Viora M. HIV-1 Nef triggers Vav-mediated signaling pathway leading to functional and morphological differentiation of dendritic cells. FASEB J. 2003;17:2025–2036. doi: 10.1096/fj.03-0272com. [DOI] [PubMed] [Google Scholar]
- 40.Rauch S., Pulkkinen K., Saksela K., Fackler O.T. Human immunodeficiency virus type 1 Nef recruits the guanine exchange factor Vav1 via an unexpected interface into plasma membrane microdomains for association with p21-activated kinase 2 activity. J. Virol. 2008;82:2918–2929. doi: 10.1128/JVI.02185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarmady M., Dampier W., Tozeren A. Sequence- and interactome-based prediction of viral protein hotspots targeting host proteins: A case study for HIV Nef. PLoS ONE. 2011;6:e20735. doi: 10.1371/journal.pone.0020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jang M.K., Mochizuki K., Zhou M., Jeong H.-S., Brady J.N., Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z., Yik J.H.N., Chen R., He N., Jang M.K., Ozato K., Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 44.Bisgrove D.A., Mahmoudi T., Henklein P., Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. USA. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z., Guo J., Wu Y., Zhou Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013;41:277–287. doi: 10.1093/nar/gks976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ladha J.S., Tripathy M.K., Mitra D. Mitochondrial complex I activity is impaired during HIV-1-induced T-cell apoptosis. Cell Death Differ. 2005;12:1417–1428. doi: 10.1038/sj.cdd.4401668. [DOI] [PubMed] [Google Scholar]
- 47.Chaussabel D., Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat. Rev. Immunol. 2014;14:271–280. doi: 10.1038/nri3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S., Rouphael N., Duraisingham S., Romero-Steiner S., Presnell S., Davis C., Schmidt D.S., Johnson S.E., Milton A., Rajam G., et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhattacharya S., Dunn P., Thomas C.G., Smith B., Schaefer H., Chen J., Hu Z., Zalocusky K.A., Shankar R.D., Shen-Orr S.S., et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data. 2018;5:180015. doi: 10.1038/sdata.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie D., Han L., Luo Y., Liu Y., He S., Bai H., Wang S., Bo X. Exploring the associations of host genes for viral infection revealed by genome-wide RNAi and virus-host protein interactions. Mol. Biosyst. 2015;11:2511–2519. doi: 10.1039/C5MB00309A. [DOI] [PubMed] [Google Scholar]
- 51.Bermejo M., López-Huertas M.R., Hedgpeth J., Mateos E., Rodríguez-Mora S., Maleno M.J., Plana M., Swindle J., Alcamí J., Coiras M. Analysis of protein kinase C theta inhibitors for the control of HIV-1 replication in human CD4+ T cells reveals an effect on retrotranscription in addition to viral transcription. Biochem. Pharmacol. 2015;94:241–256. doi: 10.1016/j.bcp.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 52.López-Huertas M.R., Li J., Zafar A., Rodríguez-Mora S., García-Domínguez C., Mateos E., Alcamí J., Rao S., Coiras M. PKCθ and HIV-1 Transcriptional Regulator Tat Co-exist at the LTR Promoter in CD4(+) T Cells. Front. Immunol. 2016;7:69. doi: 10.3389/fimmu.2016.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith B.L., Krushelnycky B.W., Mochly-Rosen D., Berg P. The HIV Nef Protein Associates with Protein Kinase C Theta. J. Biol. Chem. 1996;271:16753–16757. doi: 10.1074/jbc.271.28.16753. [DOI] [PubMed] [Google Scholar]
- 54.Witte V., Laffert B., Gintschel P., Krautkrämer E., Blume K., Fackler O.T., Baur A.S. Induction of HIV transcription by Nef involves Lck activation and protein kinase Cθ raft recruitment leading to activation of ERK1/2 but not NFκB. J. Immunol. 2009;181:3327. doi: 10.4049/jimmunol.0990009. [DOI] [PubMed] [Google Scholar]
- 55.Janardhan A., Swigut T., Hill B., Myers M.P., Skowronski J. HIV-1 Nef binds the DOCK2-ELMO1 complex to activate rac and inhibit lymphocyte chemotaxis. PLoS Biol. 2004;2:E6. doi: 10.1371/journal.pbio.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olivetta E., Pietraforte D., Schiavoni I., Minetti M., Federico M., Sanchez M. HIV-1 Nef regulates the release of superoxide anions from human macrophages. Biochem. J. 2005;390:591–602. doi: 10.1042/BJ20042139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turner A.-M.W., Dronamraju R., Potjewyd F., James K.S., Winecoff D.K., Kirchherr J.L., Archin N.M., Browne E.P., Strahl B.D., Margolis D.M., et al. Evaluation of EED Inhibitors as a Class of PRC2-Targeted Small Molecules for HIV Latency Reversal. ACS Infect. Dis. 2020;6:1719–1733. doi: 10.1021/acsinfecdis.9b00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruiz-Riol M., Berdnik D., Llano A., Mothe B., Gálvez C., Pérez-Álvarez S., Oriol-Tordera B., Olvera A., Silva-Arrieta S., Meulbroek M., et al. Identification of Interleukin-27 (IL-27)/IL-27 Receptor Subunit Alpha as a Critical Immune Axis for HIV Control. J. Virol. 2017;91:e00441-17. doi: 10.1128/JVI.00441-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng Y.-H., Xiao S.-L., He B., He Y., Zhou H.-Y., Chen Z., Zheng L.-W., He M., Wang H.-Y., Lin Y.-H., et al. The Role of IL-27 and its Receptor in the Pathogenesis of HIV/AIDS and Anti-viral Immune Response. Curr. HIV Res. 2017;15:279–284. doi: 10.2174/1570162X15666170517130339. [DOI] [PubMed] [Google Scholar]
- 60.Zheng C.F., Jones G.J., Shi M., Wiseman J.C.D., Marr K.J., Berenger B.M., Huston S.M., John Gill M., Krensky A.M., Kubes P., et al. Late expression of granulysin by microbicidal CD4+ T cells requires PI3K- and STAT5-dependent expression of IL-2Rbeta that is defective in HIV-infected patients. J. Immunol. 2008;180:7221–7229. doi: 10.4049/jimmunol.180.11.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Armas L.R., Gavegnano C., Pallikkuth S., Rinaldi S., Pan L., Battivelli E., Verdin E., Younis R.T., Pahwa R., Williams S.L., et al. The Effect of JAK1/2 Inhibitors on HIV Reservoir Using Primary Lymphoid Cell Model of HIV Latency. Front. Immunol. 2021;12:720697. doi: 10.3389/fimmu.2021.720697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matusali G., Tchidjou H.K., Pontrelli G., Bernardi S., D’Ettorre G., Vullo V., Buonomini A.R., Andreoni M., Santoni A., Cerboni C., et al. Soluble ligands for the NKG2D receptor are released during HIV-1 infection and impair NKG2D expression and cytotoxicity of NK cells. FASEB J. 2013;27:2440–2450. doi: 10.1096/fj.12-223057. [DOI] [PubMed] [Google Scholar]
- 63.Abitew A.M., Sobti R.C., Sharma V.L., Wanchu A. Analysis of transporter associated with antigen presentation (TAP) genes polymorphisms with HIV-1 infection. Mol. Cell Biochem. 2020;464:65–71. doi: 10.1007/s11010-019-03649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rom S., Pacifici M., Passiatore G., Aprea S., Waligorska A., Del Valle L., Peruzzi F. HIV-1 Tat binds to SH3 domains: Cellular and viral outcome of Tat/Grb2 interaction. Biochim. Biophys. Acta. 2011;1813:1836–1844. doi: 10.1016/j.bbamcr.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fellay J., Shianna K.V., Ge D., Colombo S., Ledergerber B., Weale M., Zhang K., Gumbs C., Castagna A., Cossarizza A., et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee G.Q., Orlova-Fink N., Einkauf K., Chowdhury F.Z., Sun X., Harrington S., Kuo H.-H., Hua S., Chen H.-R., Ouyang Z., et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J. Clin. Invest. 2017;127:2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Einkauf K.B., Lee G.Q., Gao C., Sharaf R., Sun X., Hua S., Chen S.M.Y., Jiang C., Lian X., Chowdhury F.Z., et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Invest. 2019;129:988–998. doi: 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study.