Abstract
Arylamine N-acetyltransferase activity has been described in various bacterial species. Bacterial N-acetyltransferases, including those from bacteria of the gut flora, may be involved in the metabolism of xenobiotics, thereby exerting physiopathological effects. We characterized these enzymes further by steady-state kinetics, time-dependent inhibition, and DNA hybridization in 40 species, mostly from the human intestinal microflora. We report for the first time N-acetyltransferase activity in 11 species of Proteobacteriaceae from seven genera: Citrobacter amalonaticus, Citrobacter farmeri, Citrobacter freundii, Klebsiella ozaenae, Klebsiella oxytoca, Klebsiella rhinoscleromatis, Morganella morganii, Serratia marcescens, Shigella flexneri, Plesiomonas shigelloides, and Vibrio cholerae. We estimated apparent kinetic parameters and found that 5-aminosalicylic acid, a compound efficient in the treatment of inflammatory bowel diseases, was acetylated with a catalytic efficiency 27 to 645 times higher than that for its isomer, 4-aminosalicylic acid. In contrast, para-aminobenzoic acid, a folate precursor in bacteria, was poorly acetylated. Of the wild-type strains studied, Pseudomonas aeruginosa was the best acetylator in terms of both substrate spectrum and catalytic efficiency. DNA hybridization with a Salmonella enterica serovar Typhimurium-derived probe suggested the presence of this enzyme in eight proteobacterial and four gram-positive species. Molecular aspects together with the kinetic data suggest distinct functional features for this class of microbial enzymes.
The acetyl coenzyme A (AcCoA):arylamine N-acetyltransferases (NAT; EC 2.3.1.5.) catalyze the transfer of an acetyl group from AcCoA to the nitrogen or oxygen atom of primary arylamines, hydrazines, and their N-hydroxylated metabolites. They therefore play important roles in both the detoxification and metabolic activation of numerous xenobiotics. In the Salmonella enterica serovar Typhimurium mutation assays developed by Ames and coworkers (2), the carcinogenic aromatic amines are N-hydroxylated and subsequently O-acetylated before they can exert genotoxic properties. The O-acetylation step is mediated by the NAT of S. enterica serovar Typhimurium, which therefore governs the activity of many promutagens through N-hydroxyarylamine O-acetyltransferase activity (26).
NAT isoforms (28) have been detected in several vertebrate species, including human (6), rabbit (53), rat (21), mouse (37), hamster (23), and chicken (47). Partial data are also available for amphibian (30), helminth (12, 16), and insect (66) species. In prokaryotes, NATs were first described in S. enterica serovar Typhimurium (65) and then in several species of facultative and obligate anaerobes of the dog and human intestinal microflora. NAT activity has been reported in Escherichia coli, Bacteroides vulgatus, Clostridium sporogenes, Lactobacillus bifidus, Proteus vulgaris, Pseudomonas fluorescens, Enterococcus faecalis (48), Staphylococcus aureus (9), Helicobacter pylori (18), Klebsiella pneumoniae (32), Aeromonas hydrophila (15), Enterobacter aerogenes (61), Pseudomonas aeruginosa (33), Lactobacillus acidophilus (11), Citrobacter koseri (39), and Shigella sonnei (62). However, NAT genes have been cloned and their expression products characterized only for S. enterica serovar Typhimurium (57, 65), Mycobacterium smegmatis, and Mycobacterium tuberculosis (49).
The aminosalicylate isomers 5-aminosalicylic acid (5-ASA) and 4-aminosalicylic acid (4-ASA), both arylamine acceptor substrates of human NATs, are used to treat inflammatory bowel diseases such as ulcerative colitis and Crohn's disease (55, 59). 5-ASA is considered one of the most efficient therapies for inducing and maintaining remission in such diseases (50, 67). However, its use is associated with adverse side effects such as allergic rash, pancreatitis, nephropathy, and hepatitis (41). N-Acetylation activity for 5-ASA has been reported in both aerobic and anaerobic intestinal microflora in human (63). NATs from the bacteria of the intestinal microflora may therefore affect both the efficacy and side effects of this drug.
The catalytic residues of NATs from eukaryotic and prokaryotic species may be identical. Thus, the Cys69 residue in the NAT sequence of S. enterica serovar Typhimurium has been shown to be crucial for enzyme activity (65), and the homologous Cys68 in humans probably plays a similar role (22). The recent structure determination of S. enterica serovar Typhimurium NAT has revealed that the activation of the active-site cysteine requires the presence of a Cys69-His107-Asp122 catalytic triad (58). In addition, it has been suggested that basic residues highly conserved in all NAT sequences, at positions corresponding to Arg9 and Arg64 in the human sequences, contribute to conformational stability and are involved in the enzyme-substrate interactions of the human isoenzymes, NAT1 and NAT2 (20). It has also been suggested that amino acids 125 and 127 are important determinants of NAT1-type and NAT2-type acceptor substrate selectivity (24). Extending the study of conserved residues to NATs from various bacterial species could therefore provide new insight into the functional specificities of this class of microbial enzymes.
As NATs in the bacteria of the gut microflora may be involved in the activation of carcinogens and the metabolism of drugs, we used functional and molecular approaches in an attempt to further identify and characterize this class of enzymes. We experimentally studied 40 different bacterial species, most of which were indigenous to the human gut. These species belonged to 30 genera from four different taxonomic groups: gram-positive bacteria with high and low G+C contents, Bacteroides, and Proteobacteria. The Proteobacteria studied included the Enterobacteriaceae, Vibrionaceae, and Pasteurellaceae families. We characterized bacterial NATs biochemically and compared them by determining their reaction spectra with arylamine substrates known to be specific for the human NAT1 or NAT2 isoform. We studied these enzymes kinetically using aminosalicylate substrates. We also found some features that were clearly common and specific to bacterial NATs, including a higher level of activity for 5-ASA than for its isomer, 4-ASA.
MATERIALS AND METHODS
Chemicals.
5-ASA was purchased from Acros Organics. 4-ASA, p-aminobenzoic acid (PABA), 2-aminofluorene (2-AF), sulfamethazine (SMZ), procainamide (PA), iodoacetamide, AcCoA, and the AcCoA-regenerating system d,l-acetylcarnitine and carnitine acetyltransferase were all obtained from Sigma Chemical Co. All other chemicals were of the highest purity commercially available.
Bacterial strains and growth conditions.
Two types of strains were studied. First, six control strains were tested, including the recombinant Escherichia coli strains DMG100 and DMG200 (22), which overproduce the human NAT1 and NAT2 isoforms, respectively, and four S. enterica serovar Typhimurium tester strains originally constructed for genotoxicity quantitation assays. Of these strains, YG1024 (64) overexpresses multiple copies of the NAT gene cloned from the TA1538 strain (40), whereas the TA98 1,8/DNP6 strain (43) produces no functionally active NAT. The TA98 strain (40) differs from TA1538 only in its greater sensitivity to mutagenesis. Second, the species listed in Table 1 were also studied. H. pylori was cultured for 3 days on selective or sheep blood agar plates (BioMérieux) in microaerobiosis conditions using an Oxoid Unipath jar with a Campypack Plus microaerophilic generator (Becton Dickinson). All other strains were cultured at 37°C until the exponential growth phase, using a two-step procedure: 100 ml of medium was inoculated using 10 ml of an 18-h culture and incubated for 3 h with shaking (obligate and facultative aerobes) or for 72 h without shaking (obligate anaerobes). Bacteria were grown in Trypticase-yeast-glucose-cysteine or Rosenow broth (Bacteroides sp.), from Sanofi Diagnostics Pasteur or in Luria-Bertani medium from Difco Laboratories (all other strains). The identity of the wild-type strains in each frozen glycerol stab was routinely checked using API20E and/or ID32GN (for Enterobacteriaceae, Aeromonas hydrophila, Plesiomonas shigelloides, Vibrio cholerae, and P. aeruginosa), API20NE (for Alcaligenes xylosoxidans), or Rapid ID32A (for Bacteroides sp.) identification kits, with the ATB automated expression system (BioMérieux). The purity of working cultures obtained from these stabs was checked in each experiment by examining the morphology of colonies from solid-medium cultures.
TABLE 1.
Bacterial species studied
| Species | Strain no. | Sourcea |
|---|---|---|
| Proteobacteria | ||
| Enterobacteriaceae | ||
| Citrobacter amalonaticus | 82.89 | CIP |
| Citrobacter farmeri | 104553 | CIP |
| Citrobacter freundii | 57.32 | CIP |
| Citrobacter koseri | 82.94 | CIP |
| Escherichia coli | 54.8 | CIP |
| Escherichia coli | K-12 MG1655 | IJM |
| Klebsiella oxytoca | 103434 | CIP |
| Klebsiella ozaenae | 52.211 | CIP |
| Klebsiella pneumoniae | 82.91 | CIP |
| Klebsiella rhinoscleromatis | 52.210 | CIP |
| Morganella morganii | A.231 | CIP |
| Proteus vulgaris | 58.60 | CIP |
| Providentia alcalifaciens | 82.90 | CIP |
| Salmonella enterica serovar Typhimurium | 60.62 | CIP |
| Serratia marcescens | 103235 | CIP |
| Shigella flexneri | 82.48 | CIP |
| Vibrionaceae | ||
| Aeromonas hydrophila | 76.14 | CIP |
| Helicobacter pylori | 43579 | ATCC |
| Plesiomonas shigelloides | 63.5 | CIP |
| Vibrio cholerae | 54.315 | CIP |
| Pasteurellaceae | ||
| Haemophilus influenzae | 102514 | CIP |
| Pasteurella multocida | 103286 | CIP |
| Others | ||
| Acinetobacter baumanii | 70.34 | CIP |
| Alcaligenes xylosoxidans | 71.32 | CIP |
| Bordetella bronchiseptica | 55.110 | CIP |
| Neisseria lactamica | 72.17 | CIP |
| Pseudomonas aeruginosa | 100720 | CIP |
| Bacteroides sp. | 100 | CHU |
| High-G+C gram-positive | ||
| Corynebacterium urealyticum | 111 | CHU |
| Corynebacterium xerosis | 112 | CHU |
| Low-G+C gram-positive | ||
| Bacillus sp. | 110 | CHU |
| Clostridium difficile | 101 | CHU |
| Enterococcus faecalis | 102 | CHU |
| Lactobacillus sp. | 113 | CHU |
| Listeria monocytogenes | 114 | CHU |
| Micrococcus sp. | 115 | CHU |
| Staphylococcus aureus | 116 | CHU |
| Staphylococcus epidermidis | 9901 | CHU |
| Streptococcus A | 117 | CHU |
| Streptococcus pneumoniae | 118 | CHU |
| Streptococcus mitis | 9902 | CHU |
CIP, Collection de l'Institut Pasteur, Paris, France; IJM, Institut Jacques Monod, Université Paris 7, Paris, France; ATCC, American Type Culture Collection; CHU, Centre Hospitalier Universitaire de Brest, Brest, France.
Enzyme assays.
Bacterial cells were harvested by centrifugation (2,500 × g, 20 min, 4°C) and resuspended in 2 ml of Tris (pH 7.4)-EDTA-dithiothreitol-KCl buffer (22). The cells were lysed by sonication on ice twice for 30 s each, with a 30-s interval. The insoluble fraction was removed by centrifugation (20,000 × g, 5 min, 4°C), and the resulting supernatant was used for NAT activity assays within 20 min of lysis. The soluble fraction of bacterial lysates contained 2 to 8 g of total proteins per liter, as assessed by a dye-binding assay (7). Lysates from DMG100 and DMG200 were prepared as previously described (22). NAT activity was also determined in human feces collected from five unrelated healthy Caucasian donors. Informed consent was obtained from all participants in accordance with local ethical committee guidelines. Samples (1 to 2 g) of feces that had been stored at −80°C were suspended in 10 ml of Tris (pH 8.0)-EDTA-dithiothreitol-KCl buffer (22) supplemented with lysozyme (0.5 g/liter) and treated by sonication on ice six times for 30 s each with 30-s intervals between pulses, to facilitate lysis of gram-positive cells. The insoluble fraction was sedimented by two centrifugation steps (2,500 × g, 5 min, 4°C, followed by 20,000 × g, 5 min, 4°C), and the final supernatant was used in an assay of NAT activity.
Unless otherwise stated, N-acetylation reactions were performed at 37°C in the presence of 100 μM AcCoA and an AcCoA-regenerating system (22). The NAT activity of bacterial lysates was tested in two or more 30-min reactions performed in duplicate with 200 μM (for bacterial NATs) or lower half-saturating concentrations (for recombinant human NATs) of 2-AF, 5-ASA, 4-ASA, PABA, SMZ, or PA as the arylamine acceptor substrate. Arylamines and their acetylated products were quantified by reverse-phase high-pressure liquid chromatography as previously described (20, 25). 5-ASA and N-acetyl-5-ASA were eluted in a 7% (wt/wt) acetonitrile–20 mM sodium perchlorate (pH 2.5) mobile phase, with UV detection at 264 nm, resulting in retention times of 1.5 and 3.8 min, respectively. The apparent Km and Vmax (± standard error) values for 2-AF, 5-ASA, 4-ASA, PABA, and SMZ were determined using a nonlinear fit analysis. Steady-state kinetic parameters were determined after checking that rates of product formation were constant throughout the reaction. Kinetic studies were performed using arylamine substrate concentrations in the 0.1 to 10 Km range unless substrate solubility was limitated, as was the case for 2-AF. Maximum conversion rates of 10 to 20% of the initial substrate amount were obtained, if necessary by diluting the cell lysate in bovine serum albumin-containing buffer, as reported (20).
Inhibition studies.
To achieve irreversible inhibition by iodoacetamide, undiluted cell extract was first incubated for 2 to 20 min at 25°C with various inhibitor concentrations. Aliquots were then withdrawn, and residual NAT activity was measured in 5-min assays performed at 25°C in the presence of 200 μM 2-AF without the AcCoA-regenerating system. At the times indicated, the percent residual activity was calculated by comparison with a control assay with the same concentration of iodoacetamide but without prior incubation with inhibitor (set at 100%). For each inhibitor concentration, the logarithm of the percent residual activity was plotted against the length of time for which the cell extract was incubated with inhibitor.
Hybridization analysis.
Southern blotting was carried out on genomic DNA from 22 of the wild-type Proteobacteria species studied. Genomic DNA was digested with EcoRI, size fractionated by electrophoresis in a 1% agarose gel, and transferred to a nylon membrane by capillary blotting. Genomic DNA from Proteobacteria and gram-positive strains was amplified by PCR using the BACT5 and BACT3 degenerate primers in the following thermocycling procedure: 5 min of denaturation at 94°C, 2 min of hybridization at 46 or 50°C, and then 40 cycles including 1 min of denaturation at 94°C, 30 s of hybridization at 46 or 50°C, and 2 min of extension at 72°C.BACT5: 5′-CATGCTTACTTCACGAGA-3′ T T C CGTBACT3: 5′-CGCGGCCAGCTCCGCCTC-3′ A TGT
The MgCl2 concentration of the PCR mixture was 4 mM for hybridization at 46°C and 3 mM for hybridization at 50°C. To test for the presence of NAT-hybridizing sequences in genomic and PCR-amplified bacterial DNA, a 184-bp probe including the region encoding the active site of the S. enterica serovar Typhimurium NAT (positions 124 to 307 from the initiation codon, GenBank accession number D90301) was generated by PCR. Primers SALM5 (5′-GATGTGCTACTGCCTCGTGAA-3′) and SALM3 (5′-GTAAACTGGCGGGATGAGACA-3′) were used in the presence of 1.5 mM MgCl2 with the thermocycling procedure described above except that hybridization was performed at 60°C. The amplified DNA was labeled with [α-32P]dCTP using a random priming kit (Roche Diagnostics) and used in standard conditions to probe genomic or PCR-amplified DNA.
RESULTS
Comparison of substrate selectivity of bacterial NATs.
2-AF, which is efficiently acetylated by both human NAT1 and NAT2, was used for comparative screening of NAT activity levels in gram-negative species found in the human gut microflora, including 23 Proteobacteria and one Bacteroides species. The TA98, TA1538, and YG1024 (NAT-overproducing) S. enterica serovar Typhimurium strains were used as positive controls. Enzyme activity was also tested using the structurally related human NAT1-selective PABA, 4-ASA, and 5-ASA (Fig. 1) and the human NAT2-selective SMZ and PA arylamine substrates (Table 2). The E. coli control strains DMG100 and DMG200, which overproduce the recombinant human NAT1 and NAT2 isoenzymes, respectively, displayed the expected level of activity with the substrates tested.
FIG. 1.
Comparison of chemical structures of PABA, 4-ASA, and 5-ASA. 4-ASA is also called p-aminosalicylic acid in the literature.
TABLE 2.
N-Acetylation velocities of bacterial NATs for six arylamine substratesa
| Strainb | NAT activity (nmol min−1 [mg of protein]−1)
|
|||||
|---|---|---|---|---|---|---|
| 2-AF | 5-ASA | 4-ASA | PABA | SMZ | PA | |
| Control strains | ||||||
| DMG100 (human NAT1) | 520 | 1,170 | 1,090 | 580 | 8.30 | 5.68 |
| DMG200 (human NAT2) | 0.99 | 1.89 | 0.075 | 0.002 | 2.41 | 0.06 |
| S. enterica serovar Typhimurium YG1024 | 27.5 | 20.8 | <0.001 | 0.014 | <0.001 | <0.001 |
| S. enterica serovar Typhimurium TA98 1,8/DNP6 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| S. enterica serovar Typhimurium TA1538 | 0.04 | ND | <0.001 | <0.001 | <0.001 | <0.001 |
| S. enterica serovar Typhimurium TA98 | 0.06 | ND | <0.001 | <0.002 | <0.001 | <0.001 |
| Study strains | ||||||
| S. enterica serovar Typhimurium | 0.06 | 0.27 | <0.001 | 0.005 | <0.001 | |
| C. amalonaticus | 0.09 | 0.10 | <0.001 | 0.005 | <0.001 | 0.004 |
| C. farmeri | 0.10 | 0.33 | 0.006 | <0.001 | 0.002 | <0.001 |
| C. freundii | 0.14 | 0.16 | 0.005 | <0.001 | <0.001 | <0.002 |
| C. koseri | 0.32 | 0.59 | 0.008 | <0.001 | 0.003 | <0.001 |
| E. coli 54.8 | 0.05 | 0.09 | 0.004 | <0.001 | <0.003 | <0.001 |
| E. coli K-12 | 0.02 | <0.001 | 0.01 | <0.001 | 0.001 | <0.001 |
| K. pneumoniae | <0.001 | <0.001 | <0.001 | <0.001 | 0.006 | <0.001 |
| K. ozaenae | 0.007 | ND | ND | ND | ND | ND |
| K. oxytoca | 0.16 | 0.61 | <0.001 | <0.001 | 0.001 | <0.001 |
| K. rhinoscleromatis | 0.15 | ND | <0.002 | <0.002 | 0.006 | <0.002 |
| M. morganii | 0.002 | <0.001 | 0.02 | <0.001 | <0.001 | <0.001 |
| P. vulgaris | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| P. alcalifaciens | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| S. marcescens | 0.004 | 0.006 | <0.001 | <0.001 | 0.002 | <0.001 |
| S. flexneri | <0.001 | <0.001 | <0.001 | <0.001 | 0.003 | <0.006 |
| A. hydrophila | <0.001 | <0.001 | <0.001 | 0.002 | <0.001 | <0.001 |
| H. pylori | 0.01 | <0.002 | <0.001 | <0.001 | <0.001 | <0.001 |
| P. shigelloides | <0.001 | <0.002 | <0.001 | <0.001 | <0.001 | 0.006 |
| V. cholerae | 0.001 | <0.001 | 0.005 | <0.001 | <0.001 | <0.001 |
| P. multocida | <0.001 | ND | ND | ND | ND | ND |
| A. xylosoxidans | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| P. aeruginosa | 1.06 | 2.20 | 0.27 | 0.14 | <0.001 | <0.001 |
| Bacteroides sp. | <0.001 | 0.02 | <0.001 | 0.05 | <0.001 | <0.001 |
N-Acetylation activities were measured as described in Materials and Methods in the presence of 100 μM AcCoA and, unless otherwise stated, 200 μM arylamine substrate. Mean values from two or more experiments performed in duplicate are presented. ND, not determined. N-Acetylation activity was measured in the presence of a 20 μM (human NAT1 with 2-AF), 2 μM (human NAT2 with 2-AF), or 10 μM (human NAT1 with 5-ASA, 4-ASA, and PABA) concentration of the arylamine substrate.
Significant NAT activity was identified for the first time in 11 Proteobacteria species from seven different genera: C. amalonaticus, C. farmeri, C. freundii, K. ozaenae, K. oxytoca, K. rhinoscleromatis, M. morganii, S. marcescens, S. flexneri, P. shigelloides, and V. cholerae. Fifteen Proteobacteria species had detectable NAT activity with 2-AF. With the exception of M. morganii, H. pylori, and E. coli K-12, the species or genera that acetylated 2-AF also acetylated 5-ASA, whereas the Bacteroides sp. acetylated 5-ASA but not 2-AF. Of the arylamines tested, 5-ASA and, to a lesser extent, 2-AF were the most highly acetylated by bacteria in our experimental conditions, with velocities of 0.006 (S. marcescens) to 2.20 (P. aeruginosa) and of 0.002 (M. morganii) to 1.06 (P. aeruginosa) nmol min−1 (mg of protein)−1, respectively. P. aeruginosa, E. coli 54.8, C. freundii, C. koseri, and C. farmeri, which acetylated both aminosalicylate isomers studied, had 8, 22, 50, 65, and 65 times higher levels of activity, respectively, with 5-ASA than with 4-ASA.
Only 4 of the 22 species tested for N-acetylation of PABA exhibited significant activity, from 0.002 (A. hydrophila) to 0.14 (P. aeruginosa) nmol min−1 (mg of protein)−1. P. aeruginosa, C. amalonaticus, and S. enterica serovar Typhimurium YG1024 acetylated 5-ASA 11, 21, and 1,480 times more efficiently than PABA, respectively, whereas N-acetylation velocities were of the same order of magnitude for both compounds in the Bacteroides sp. With the exception of P. aeruginosa, which showed similar levels of activity with PABA and 4-ASA, the PABA-acetylating species did not acetylate 4-ASA.
The bacterial species studied displayed a low level of N-acetylation activity with SMZ and PA in our experimental conditions (from 0.001 to 0.006 nmol min−1 [mg of protein]−1). PA was acetylated only by C. amalonaticus, and P. shigelloides, P. vulgaris, P. alcalifaciens, and A. xylosoxidans had no detectable NAT activity with the arylamine substrates studied here, nor did the NAT-defective S. enterica serovar Typhimurium TA98/1,8 DNP6 strain used as a negative control. Overall, P. aeruginosa appeared from our comparative screening to be the most potent N-acetylator among the species studied, as it acetylated 2-AF and the three human NAT1-specific substrates with the highest velocity values.
NAT activity in human colonic content.
We estimated the overall N-acetylation activity of the colonic microflora by performing enzyme assays with lysates of human feces and 5-ASA or 2-AF as the acceptor substrate. Samples collected from the various donors displayed NAT activity with 5-ASA (0.062 ± 0.011, 0.063 ± 0.014, 0.079 ± 0.007, 0.113 ± 0.021, and 0.639 ± 0.049 nmol min−1 per g of initial sample). Conversely, the N-acetylation velocities by fecal lysates of 2-AF were lower than 0.015 nmol min−1 per g in our experimental conditions. Given the estimated bacterial content in feces (4) and in our bacterial cultures (optical density at 600 nm), it can be roughly estimated that the feces lysates were made from 10 to 100 times less bacteria than the pure-culture lysates.
Comparative kinetics of bacterial NATs.
The strains found to have the highest rates of enzyme activity in the reaction spectrum study were studied in more detail to characterize further and compare their N-acetylation activities. We determined apparent kinetic parameters for these strains (Tables 3 to 5). The five arylamine acceptor substrates studied can be assigned to three overlapping classes on the basis of their apparent Km: (i) 5-ASA (26 to 350 μM), 2-AF (44 to 405 μM), and SMZ (59 to 550 μM); (ii) 4-ASA (260 to 2,200 μM); and (iii) PABA (870 to 51,600 μM). Bacterial NATs had similar apparent affinities for 5-ASA, 2-AF, and SMZ, as shown by the weak differences between the corresponding Km values for a given strain. Conversely, bacterial N-acetylation of the aminosalicylate isomers studied displayed a higher affinity for 5-ASA. The Km of 4-ASA was 4 times higher than that of 5-ASA in C. koseri, 10 times higher in P. aeruginosa, 11 times higher in E. coli 54.8, 14 times higher in C. freundii, and 29 times higher in C. farmeri. Similarly, the Km of human NAT2 for 4-ASA was 41 times higher than that for 5-ASA, whereas human NAT1 acetylated both isomers with similar Michaelis constants. The Km values of bacterial NATs and human NAT2 were of the same order of magnitude for 5-ASA. Finally, of the arylamines studied, PABA was the substrate for which bacterial NATs had the lowest apparent affinity, as indicated in P. aeruginosa by Km values 5, 27, and 47 times higher than those for 4-ASA, 2-AF, and 5-ASA, respectively.
TABLE 3.
Kms of bacterial and human NATsa
| Strain | Mean Km (μM) ± SD
|
||||
|---|---|---|---|---|---|
| 2-AF | 5-ASA | 4-ASA | PABA | SMZ | |
| Control strains | |||||
| DMG100 (human NAT1) | 28 ± 2.8b | 6.7 ± 1.4 | 9.4 ± 1.2b | 13b | 5,600 ± 1,000b |
| DMG200 (human NAT2) | 2.25 ± 0.12b | 184 ± 32 | 7,590 ± 1,350b | —b | 110 ± 6.0b |
| S. enterica serovar Typhimurium YG1024 | 365 ± 46 | ND | — | 51,600 ± 5,275c | — |
| S. enterica serovar Typhimurium TA98 | 397 ± 69 | ND | — | — | — |
| S. enterica serovar Typhimurium TA1538 | 404 ± 56 | ND | — | — | — |
| Study strains | |||||
| S. enterica serovar Typhimurium 60.62 | 311 ± 39c | 208 ± 32c | — | — | 283 ± 26c |
| E. coli 54.8 | 127 ± 13∗∗∗∗ | 117 ± 17∗∗ | 1,240 ± 173c | — | 59 ± 8.1∗∗∗∗ |
| E. coli K-12 | 205 ± 27∗ | — | 1,160 ± 150 | — | 121 ± 3.3∗∗∗∗ |
| P. aeruginosa | 44 ± 5.1∗∗∗∗ | 26 ± 5.0∗∗∗∗ | 258 ± 63∗∗∗∗ | 1,200 ± 205∗∗∗∗ | — |
| C. koseri | 96 ± 7.8∗∗∗∗ | 210 ± 20 | 860 ± 83∗ | — | 115 ± 16∗∗∗∗ |
| C. farmeri | 64 ± 3.8∗∗∗∗ | 61 ± 12∗∗∗∗ | 1,790 ± 259 | — | 70 ± 10∗∗∗∗ |
| C. freundii | 165 ± 25∗∗∗ | 158 ± 22 | 2,200 ± 365∗∗ | — | — |
| C. amalonaticus | 82 ± 11∗∗∗∗ | 112 ± 7.2∗∗∗ | — | ND | — |
| K. rhinoscleromatis | 141 ± 41∗∗∗ | ND | — | — | 146 ± 21∗∗∗∗ |
| K. oxytoca | 158 ± 27∗∗∗ | 347 ± 30∗∗∗ | — | — | — |
| M. morganii | 117 ± 18∗∗∗∗ | — | ND | — | — |
| H. pylori | 106 ± 11∗∗∗∗ | — | — | — | — |
| S. marcescens | 79 ± 12∗∗∗∗ | ND | — | — | 550 ± 58∗∗∗∗ |
| Bacteroides sp. | — | ND | — | 870 ± 150∗∗∗∗ | — |
Km and Vmax were estimated for five arylamine substrates in two or more separate experiments. ND, not determined; —, no detectable activity.
Values determined in previous studies (18, 20). The values given for recombinant human NATs were not included in the statistical comparison.
Results obtained from S. enterica serovar Typhimurium or, by default (for the substrate 4-ASA), from E. coli and used as a reference for the statistical comparison with values obtained for the other bacterial NAT activities and for the same arylamine substrate, using Student's t test. ∗∗∗∗, P < 0.001; ∗∗∗, P < 0.01; ∗∗, P < 0.02; ∗, P < 0.05).
TABLE 5.
Vmax/Km results for bacterial NATsa
| Strain |
Vmax/Km (% of control value)
|
||||
|---|---|---|---|---|---|
| 2-AF | 5-ASA | 4-ASA | PABA | SMZ | |
| Control strains | |||||
| DMG100 (human NAT1) | 49b | 257 | 193b | 465b | 2.2 × 10−2b |
| DMG200 (human NAT2) | 1.04b | 1.6 × 10−2 | 5.1 × 10−4b | —b | 3.5 × 10−2b |
| S. enterica serovar Typhimurium YG1024 | 1.6 × 10−1 (39,480) | ND | — | 1.1 × 10−4 (100) | — |
| S. enterica serovar Typhimurium TA98 | 4.7 × 10−4 (112) | ND | — | — | — |
| S. enterica serovar Typhimurium TA1538 | 2.9 × 10−4 (69) | ND | — | — | — |
| Study strains | |||||
| S. enterica serovar Typhimurium 60.62 | 4.1 × 10−4 (100) | 3.8 × 10−3 (100) | — | — | 3.2 × 10−5 (100) |
| E. coli 54.8 | 4.8 × 10−4 (115) | 7.8 × 10−4 (20) | 2.9 × 10−5 (100) | — | 5.0 × 10−5 (155) |
| E. coli K-12 | 1.8 × 10−4 (42) | — | 4.2 × 10−5 (140) | — | 1.1 × 10−5 (33) |
| P. aeruginosa | 2.6 × 10−2 (6,375) | 9.5 × 10−2 (2,470) | 1.2 × 10−3 (4,300) | 9.1 × 10−4 (845) | — |
| C. koseri | 5.0 × 10−3 (1,200) | 6.4 × 10−3 (170) | 3.2 × 10−5 (110) | — | 7.2 × 10−6 (22) |
| C. farmeri | 2.0 × 10−3 (480) | 6.9 × 10−3 (180) | 1.1 × 10−5 (37) | — | 2.6 × 10−5 (81) |
| C. freundii | 1.4 × 10−3 (330) | 1.7 × 10−3 (44) | 3.9 × 10−5 (130) | — | — |
| C. amalonaticus | 1.7 × 10−3 (415) | 1.4 × 10−3 (37) | — | ND | — |
| K. rhinoscleromatis | 1.4 × 10−3 (335) | ND | — | — | 5.8 × 10−5 (180) |
| K. oxytoca | 1.5 × 10−3 (350) | 5.0 × 10−3 (130) | — | — | — |
| M. morganii | 2.5 × 10−5 (6) | — | ND | — | — |
| H. pylori | 2.4 × 10−4 (59) | — | — | — | — |
| S. marcescens | 1.3 × 10−4 (31) | ND | — | — | 1.3 × 10−6 (4) |
| Bacteroides sp. | — | ND | — | 3.1 × 10−4 (290) | — |
See Table 3, footnotes a, b, and c.
In any given strain, the apparent Vmax for 5-ASA acetylation by the bacterial NAT was higher than that for any other substrate (Table 4). Similarly, the highest catalytic efficiencies, Vmax/Km, were also measured for 5-ASA in bacteria (Table 5). In particular, Vmax/Km values were 27 (for E. coli 54.8) to 645 (for C. farmeri) times higher for 5-ASA than for 4-ASA, consistent with the Km data. Human recombinant NAT2 also had a Vmax/Km for 5-ASA that was 32 times higher than that for 4-ASA, whereas NAT1 had similar catalytic efficiencies with both isomers. Human recombinant NAT1 acetylated PABA with the highest Vmax/Km (22) (Table 5), whereas in P. aeruginosa, the catalytic efficiency of PABA acetylation was 1/100 that for 5-ASA and 1/30 that for 2-AF. In S. enterica serovar Typhimurium YG1024, the catalytic efficiency was 1,500 times higher for 2-AF than for PABA. Conversely, P. aeruginosa displayed similar Vmax/Km values for PABA and 4-ASA. Finally, the catalytic efficiencies for the acetylation of SMZ by bacterial NATs were of the same order of magnitude or lower than those for 4-ASA in a given species.
TABLE 4.
Vmaxs of bacterial and human NATSa
| Strain | Mean Vmax (nmol min−1 [mg of protein]−1) ± SD
|
||||
|---|---|---|---|---|---|
| 2-AF | 5-ASA | 4-ASA | PABA | SMZ | |
| Control strains | |||||
| DMG100 (human NAT1) | 870 ± 39b | 1,715 ± 57 | 1,810 ± 75b | 6,050b | 123 ± 19b |
| DMG200 (human NAT2) | 2.34 ± 0.04b | 2.98 ± 0.18 | 3.90 ± 0.14b | —b | 3.80 ± 0.07b |
| S. enterica serovar Typhimurium YG1024 | 60 ± 4.8∗∗∗∗ | ND | <0.001 | 5.58 ± 0.54c | <0.001 |
| S. enterica serovar Typhimurium TA98 | 0.18 ± 0.02∗ | ND | <0.001 | <0.002 | <0.001 |
| S. enterica serovar Typhimurium TA1538 | 0.12 ± 0.01 | ND | <0.001 | <0.001 | <0.001 |
| Study strains | |||||
| S. enterica serovar Typhimurium 60.62 | 0.13 ± 0.01c | 0.80 ± 0.08c | <0.001 | <0.001 | 91 (±5.4) × 10−4 |
| E. coli 54.8 | 0.06 ± 0.004∗∗∗∗ | 0.09 ± 0.005∗∗∗∗ | 0.04 ± 0.002c | <0.001 | 30 (±0.8) × 10−4∗∗∗∗ |
| E. coli K-12 | 0.04 ± 0.003∗∗∗∗ | <0.001 | 0.05 ± 0.004∗∗∗ | <0.001 | 13 (±0.1) × 10−4∗∗∗∗ |
| P. aeruginosa | 1.17 ± 0.06∗∗∗∗ | 2.44 ± 0.13∗∗∗∗ | 0.32 ± 0.02∗∗∗∗ | 1.09 ± 0.06∗∗∗∗ | <0.001 |
| C. koseri | 0.48 ± 0.02∗∗∗∗ | 1.35 ± 0.06∗∗∗∗ | 0.03 ± 0.001∗∗∗∗ | <0.001 | 8.3 (±0.3) × 10−4∗∗∗∗ |
| C. farmeri | 0.13 ± 0.003 | 0.42 ± 0.03∗∗∗∗ | 0.02 ± 0.001∗∗∗∗ | <0.001 | 18 (±0.5) × 10−4∗∗∗∗ |
| C. freundii | 0.23 ± 0.02∗∗∗∗ | 0.27 ± 0.02∗∗∗∗ | 0.08 ± 0.01∗∗∗∗ | <0.001 | <0.001 |
| C. amalonaticus | 0.14 ± 0.01 | 0.16 ± 0.004∗∗∗∗ | <0.001 | ND | <0.001 |
| K. rhinoscleromatis | 0.20 ± 0.03∗ | ND | <0.002 | <0.002 | 85 (±3.6) × 10−4 |
| K. oxytoca | 0.23 ± 0.03∗∗∗∗ | 1.73 ± 0.08∗∗∗∗ | <0.001 | <0.001 | ND |
| M. morganii | 0.003 ± 0.0002∗∗∗∗ | <0.001 | ND | <0.001 | <0.001 |
| H. pylori | 0.03 ± 0.0009∗∗∗∗ | <0.002 | <0.001 | <0.001 | <0.001 |
| S. marcescens | 0.01 ± 0.001∗∗∗∗ | ND | <0.001 | <0.001 | 7.3 (±0.4) × 10−4∗∗∗∗ |
| Bacteroides sp. | <0.001 | ND | <0.001 | 0.27 ± 0.02∗∗∗∗ | <0.001 |
See Table 3, footnotes a, b, and c.
Consistent with the results obtained for the reaction spectra of bacterial NATs, with the exception of the recombinant NAT-overproducing strains, P. aeruginosa was the most efficient acetylator among the species studied for each arylamine substrate tested. It exhibited Vmax/Km values 64, 25, and 8 times higher than those of S. enterica serovar Typhimurium for 2-AF, 5-ASA, and PABA, respectively. Its Vmax/Km for 4-ASA was also 43 times higher than that of E. coli 54.8. In contrast S. marcescens, H. pylori, and M. morganii displayed the lowest catalytic efficiencies for 2-AF in our experimental conditions.
Inhibition of bacterial NAT activity.
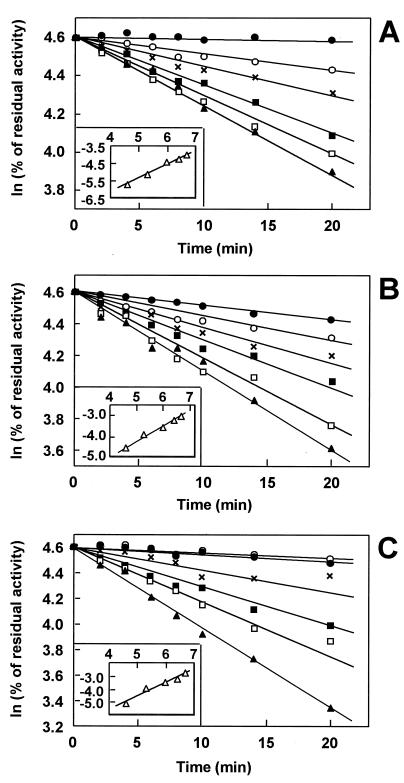
To obtain further insight into the nature of the putative active amino acid residue of bacterial NATs, we performed irreversible inhibition experiments with iodoacetamide. This compound irreversibly reacts with the thiol group of accessible Cys side chains in proteins to give a stable alkylated product (19). Bacterial lysates from S. enterica serovar Typhimurium YG1024, C. koseri, and P. aeruginosa were incubated with or without 100 to 800 mM iodoacetamide, corresponding to a 200 to 1,600-fold molar excess of the modifying agent over the estimated Cys residue content of bacterial lysates (45), and the residual NAT activity was measured in the presence of 2-AF. The inactivation of NAT depended on time and inhibitor concentration (Fig. 2). Apparent pseudo-first-order inactivation constants (Kobs) were obtained for each iodoacetamide concentration by linear regression of plots of the logarithm of the percentage of control activity versus time (38). Secondary plots of ln (Kobs) versus ln (iodoacetamide) were linear and had slopes of 0.81 for S. enterica serovar Typhimurium YG1024, 0.72 for C. koseri, and 1.07 for P. aeruginosa. Thus, the order of the inhibition reaction with iodoacetamide was probably similar for these three bacterial NAT activities. In each case, we estimated that a single functional Cys reacted with the modifying agent.
FIG. 2.
Time- and concentration-dependent inactivation of bacterial NATs with iodoacetamide. Cell extracts from S. enterica serovar Typhimurium YG1024 (A), C. koseri (B), and P. aeruginosa (C) were incubated without (●) or with iodoacetamide at a final concentration of 100 (○), 200 (×), 400 (■), 600 (□), or 800 (▴) mM, and residual N-acetylation activity with 2-AF was measured. Results are the means of two experiments performed in duplicate. Insets show the double-logarithmic plots of the apparent first-order inactivation rate (kobs) versus molar inhibitor concentration. A value of 0.81 (r = 0.99), 0.72 (r = 0.99) and 1.07 (r = 0.98) for the reaction order (n) with respect to iodoacetamide was calculated from the slopes for S. enterica serovar Typhimurium YG1024, C. koseri, and P. aeruginosa, respectively. The initial reaction rates without inhibitor were 11.23 ± 0.63, 0.42 ± 0.02, and 2.93 ± 0.03 nmol min−1 (mg of protein)−1 for S. enterica serovar Typhimurium YG1024, C. koseri, and P. aeruginosa, respectively.
Molecular hybridization of DNA from several bacterial species with an S. enterica serovar Typhimurium NAT probe.
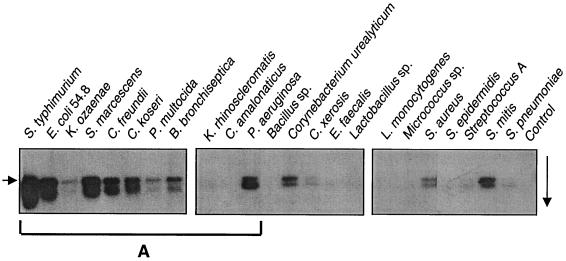
Having detected NAT activity in a number of bacterial species, we attempted to detect new NAT-encoding regions. Genomic DNA from various bacterial species was amplified by PCR at two different levels of stringency, using degenerate primers derived from the sequence of the S. enterica serovar Typhimurium NAT gene. The degenerate sense (BACT5) and antisense (BACT3) primers used hybridized to the coding sequence of the S. enterica serovar Typhimurium NAT gene at positions 16 to 33 and 797 to 804, respectively, resulting in a PCR product of 788 bp for S. enterica serovar Typhimurium. We checked the specificity of the amplified DNAs by Southern blotting of the PCR products, using a radiolabeled 184-bp DNA probe amplified by PCR from the putative active-site region of the S. enterica serovar Typhimurium NAT gene (i.e., spanning the Cys69 residue) (Fig. 3). All positive strains gave the expected band of approximately 790 bp. An unidentified 500-bp band was also detected. In addition to S. enterica serovar Typhimurium, 8 of the 22 proteobacteria (5 members of the Enterobacteriaceae [C. koseri, C. freundii, E. coli 54.8, K. ozaenae, and S. marcescens] and 3 species from other families [B. bronchiseptica, P. multocida, and P. aeruginosa]) as well as 4 of the 12 gram-positive species studied (2 with a high G+C content [Corynebacterium urealyticum and C. xerosis] and 2 with a low G+C content [S. aureus and S. mitis]) produced a signal. A faint signal was obtained for Streptococcus A and S. pneumoniae. Genomic DNA blots (EcoRI digests) from all Proteobacteria strains studied except E. coli K-12 and H. pylori (Table 1) were also incubated with the probe (data not shown). The presence or absence of hybridization signals was recorded with PCR-amplified as well as genomic DNA blots (Fig. 3). Of all the species studied only by genomic DNA hybridization, only K. pneumoniae and S. flexneri provided a faint hybridization signal.
FIG. 3.
Hybridization of PCR-amplified bacterial DNA with the S. enterica serovar Typhimurium NAT probe PCR products amplified from bacterial genomic DNA were blotted as described in the text. A negative control consisting of an 835-bp amplification product of the human NAT2 coding region was blotted in the control lane. The hybridization temperature and MgCl2 content used for PCR were 50°C and 3 mM (left blot) or 46°C and 4 mM (central and right blots). Similar hybridization results were obtained for both sets of PCR conditions for the S. enterica serovar Typhimurium DNA blot. In addition to the expected 790-bp band (left arrow), an unidentified 500-bp band was also detected. A similar doublet was found for S. enterica serovar Typhimurium at shorter exposure time. Some of the strains (group A) also gave a positive signal by hybridization of EcoRI-digested genomic DNA. The right arrow indicates the direction of electrophoretic migration.
DISCUSSION
In this study, we used molecular and functional approaches to identify and characterize bacterial NATs. Thirteen NAT sequences from nine eubacterial genera were retrieved from databases, using the S. enterica serovar Typhimurium NAT sequence as a query, and were multialigned (data not shown). These sequences showed a highly conserved Cys that aligned with Cys69 in S. enterica serovar Typhimurium (65), which has been suggested to be the active-site residue in four bacterial NATs (49). We inhibited the NAT activity from P. aeruginosa, C. koseri, and S. enterica serovar Typhimurium YG1024 by a large excess of the Cys-modifying agent iodoacetamide. We found that this inhibition followed a 1:1 stoichiometry, suggesting that one essential Cys residue is accessible to the inhibitor in these three bacterial enzymes (Fig. 2). The essential nature of Cys69 in S. enterica serovar Typhimurium has already been demonstrated by Watanabe and colleagues (65) from site-directed mutagenesis experiments. Thus, like vertebrate NATs (20), eubacterial isoforms seem to have a single Cys as the active-site residue. Our sequence alignment (data not shown) identified two strictly conserved residues aligning with His107 and Asp122 of the S. enterica serovar Typhimurium NAT. These residues have recently been described as forming part of its catalytic triad (58). We also identified two highly conserved basic residues aligning with Arg9 and Arg64 of human NATs. It has been suggested that the Arg9 and Arg64 residues are essential for enzyme-substrate interactions and conformational stabilization in vertebrate NATs (20). The conservation of these residues (Arg11 and Arg65 of S. enterica serovar Typhimurium NAT) suggests that similar mechanisms may operate for all NATs. The structural study of S. enterica serovar Typhimurium NAT conducted by Sinclair et al. (58) supports that hypothesis. In particular, it was found that Arg65 stabilizes the conformation of the Cys69 residue through a salt bridge interaction with the highly conserved Glu39.
We identified NAT activity for the first time in 11 proteobacterial species belonging to the Citrobacter, Klebsiella, Morganella, Serratia, Shigella, Plesiomonas, and Vibrio genera (Table 2). These enzyme activities were detected and some of them were kinetically characterized using one or several of six arylamine acceptor substrates chosen as NAT1-type (PABA, 4-ASA, and 5-ASA), NAT2-type (SMZ and PA), or mixed (2-AF) with regard to substrate selectivity of human NATs. The values obtained for the S. enterica serovar Typhimurium control strains showed that these activities were specific (Tables 3 to 5). It has to be noted that our Km values for 2-AF in P. aeruginosa, E. coli, and H. pylori lysates are 3 to 35 times lower than those reported by Chung et al. (8, 10, 33). These discrepancies may be related to differences in experimental conditions, such as AcCoA concentration and waiting time between bacterial sonication and NAT assay. We observed that above a 20-min interval between cell lysis and enzymatic assay, the apparent Km values could markedly increase, with loss in reproducibility. This might contribute to the variability in kinetic parameter values reported in distinct studies by Chung et al. (10, 13, 14, 17).
Bacterial NAT activities appear to have features similar to those of the human NAT1 and NAT2 isoenzymes. They are most similar to NAT1 in terms of reaction spectrum, as shown by their low N-acetylation activity with the NAT2-selective substrates SMZ and PA (Table 2) and by their high catalytic efficiency with 5-ASA and 2-AF compared to that with SMZ (Tables 3 to 5). This may be related, in 11 of the 13 bacterial sequences retrieved, to the presence of a conserved Phe residue aligning with Phe125 in the human NAT1 sequence (data not shown). This residue may be associated with the low NAT1-type substrate selectivity for SMZ (24). In contrast, most of the bacterial NATs studied had poor or undetectable activity with PABA, which makes them similar to human NAT2 (Table 2). The ranges of apparent Km values for SMZ, 5-ASA, and 4-ASA also place bacterial NATs closer to human NAT2 than to NAT1. Consistently, the NAT from M. smegmatis exhibits a low apparent Km (25 μM) for the NAT2 substrate isoniazid (49). We found that bacterial NATs share with human NAT2 a higher catalytic efficiency for 5-ASA than for its isomer, 4-ASA (Tables 3 to 5). The bacterial sequences studied have no conserved residue aligning with Arg127 in the human NAT1 sequence (data not shown). This residue may be associated with the high selectivity of human NAT1 for 4-ASA (24). Finally, bacterial NATs appear to have functional specificities that distinguish them from their eukaryotic counterparts, making them useful as a new model for study of the structure-function relationships of arylamine N-acetyltransferases.
Enterobacteriaceae species (Table 1) widely acetylated one or more of the tested arylamines. Most of them cross-reacted with the previously described rabbit antiserum raised against purified recombinant S. enterica serovar Typhimurium NAT (49) and gave an immunoblot signal comigrating with the S. enterica serovar Typhimurium NAT band (data not shown), consistent with other data (M. Payton, A. Mushtaq, J. Sinclair, J. Sandy, T.-W. Yu, M. Noble, and E. Sim, submitted for publication). However, unlike a previous study (48), we did not detect NAT activity in P. vulgaris, possibly due to differences in the strains used. The E. coli 54.8 and K-12 strains consistently differed in NAT activity in this study. This may be due to the high polymorphism within the E. coli species reflected by differences in genome length (5) or in metabolic phenotypes (36) between strains. P. alcalifaciens and A. xylosoxidans also had no detectable activity for the six arylamine substrates tested in our study. The low nutritive content of our bacterial growth medium is unlikely to play a significant role in this lack of enzyme activity because these species are members of the normal intestine microflora and the contents of the colonic lumen are considered nutritively poor. Although these three inactive species gave no signal in genomic DNA hybridization experiments, they gave a clear NAT-specific immunoreactivity signal (data not shown). This suggests that these organisms may contain NATs with a different and unknown substrate selectivity. The Pasteurellaceae species P. multocida did not acetylate 2-AF in our test conditions, but the positive results of DNA hybridization experiments (Fig. 3) suggest that this species contains a genomic sequence with some similarities to the S. enterica serovar Typhimurium NAT gene.
The four Vibrionaceae species studied were only weakly immunoreactive with the S. enterica serovar Typhimurium-specific antiserum (data not shown) and had marginal NAT activities. H. pylori acetylated 2-AF much less efficiently than in previous studies (10, 18) (Table 2). We were also unable to retrieve a putative NAT sequence from the H. pylori genome database even though two unrelated strains have been now completely sequenced (1, 60). Only a partial putative sequence was obtained from the V. cholerae genome (data not shown). Consistently, no genomic DNA hybridization signal was obtained for V. cholerae, A. hydrophila, and P. shigelloides (data not shown). This suggests that the NATs of the Vibrionaceae differ greatly in structure and reaction spectrum from those found in other proteobacteria. In contrast, within obligate anaerobes, we found significant NAT activity for PABA and 5-ASA in the Bacteroides sp. (Table 2) and a small activity with 2-AF and 5-ASA in C. difficile (data not shown), consistent with previous data for these genera (48). However, the absence of a detectable immunoblot signal for the Bacteroides sp. (data not shown) may be related to the absence of an epitope in common with S. enterica serovar Typhimurium NAT.
Among the Proteobacteria and Bacteroides species tested here, P. aeruginosa was the most effective N-acetylator in terms of both reaction spectrum and catalytic efficiency, since it exhibited the highest activity for 2-AF, 5-ASA, 4-ASA, and PABA (Tables 2, 3, 4, and 5). P. aeruginosa gave NAT-specific DNA hybridization (Fig. 3) and cross-immunoreactivity (data not shown) signals. A complete putative NAT sequence was also obtained from this species (data not shown). Moreover, the results of our DNA hybridization experiments (Fig. 3) and the identification of several putative NAT sequences (data not shown) suggest that NATs are widely present in gram-positive species with low and high G+C contents.
Microbial interactions and drug administration are determinants for the composition of the intestinal microflora. Therefore, the identification of bacterial NATs may serve as a means of understanding the enzyme activity in different clinical situations. The aminosalicylate 5-ASA is one of the most effective treatments for inflammatory bowel diseases in humans (50). It is mainly metabolized into N-acetyl-5-ASA, which is thought to be therapeutically inactive (35). We detected N-acetylation activity with 5-ASA in human feces, with a 1- to 10-fold interindividual variability in enzyme velocities (see Results). This activity mostly reflects that of obligate anaerobes, including gram-positives, as they constitute the great majority of the colonic microflora (4, 56). These results are consistent with the ability of the Bacteroides sp. (Table 2) and of the entire obligate anaerobic colonic microflora in culture (63) to N-acetylate 5-ASA. The absence of a detectable NAT activity for 2-AF in feces samples is also consistent with the lack of activity found with the Bacteroides sp. (Table 2). The endogenous NAT1 activity of intestinal mucosal cells (29) is unlikely to contribute to the observed N-acetylation of 5-ASA in feces, given that (i) maximal N-acetyl-5-ASA synthesis was reported for high dilutions of fecal material in humans (63) and (ii) the endogenous pancreatic enzymes present in the intestinal content are greatly inactivated by the microflora in rats (51). We detected NAT activity with 5-ASA in species of Proteobacteria and Bacteroides from the intestinal microflora, including members known to be overrepresented in inflammatory bowel diseases. These microbes include the aerobes E. coli, P. aeruginosa, and Klebsiella (31) and the obligate anaerobes Bacteroides (3, 46) and C. difficile (52). Of the various arylamines tested here, 5-ASA was the most efficiently acetylated by bacterial NATs (Tables 2, 3, 4, and 5). Bacterial N-acetylation may therefore be responsible, in addition to the intestinal metabolism (68), for 5-ASA inactivation. However, N-acetyl-5-ASA has itself been found to inhibit the growth of obligate anaerobes such as C. difficile (52). We also found that NATs from P. aeruginosa, E. coli, and various Citrobacter species had stronger activity for 5-ASA than for its isomer, 4-ASA (Tables 3, 4, and 5). Similar therapeutic benefits have been reported for both isomers in specific cases of ulcerative colitis, and patient tolerance of 4-ASA is higher due to the adverse effects of 5-ASA (42). The use of 4-ASA in inflammatory bowel disease patients presenting intolerance to 5-ASA may therefore be recommended, given its low level of inactivation by intestinal microflora. N-Acetyl-5-ASA may be unstable in the intestine and/or hydrolyzed by nonspecific amidases to regenerate 5-ASA. However, administered acetyl-5-ASA was reported not to be deacetylated, and acetyl-5-ASA is considered more stable than 5-ASA (35). Thus, we suggest that the bacterial acetylation of 5-ASA could modulate the concentration equilibrium between the two forms, promoting the acetylated form in the gut. Other works have highlighted the importance of the metabolism of intestinal bacteria in drug side effects (44). Consistently, NAT activity in Mycobacterium species was suggested to modulate the therapeutic response to isoniazid treatment of tuberculosis. It should be important to investigate such prokaryotic NATs for polymorphism and potential implications for drug resistance (54).
In contrast to previous reports (8, 18), only a small proportion of the bacterial species studied here acetylated PABA (Table 2). In addition, it was previously observed that bacterial NATs have lower activities with PABA than with 2-AF (33, 48), although with similar apparent Km (8, 10, 18). In our study, reactions with PABA had much lower catalytic efficiencies than those with 2-AF or with the structurally related 5-ASA, mainly due to the higher apparent Km values measured for PABA (Table 3). The results of reversible inhibition experiments suggested that PABA acts as a competitive inhibitor for the N-acetylation of 2-AF in S. enterica serovar Typhimurium 60.62 (data not shown). These data are consistent with the essential role of PABA as a precursor for folate synthesis in bacteria and suggest a double level of preservation of the PABA pool in the bacterial cell with regard to N-acetylation (34, 49).
Bacterial NATs have been shown to have the potential to generate strongly alkylating cationic compounds involved in carcinogenesis, through N- and/or O-acetylation of arylamine or hydroxyarylamine metabolites (27). Microflora and endogenous NATs may thus affect susceptibility to human bowel cancers, in addition to other bacterial carcinogen-producing pathways such as nitroreduction and azoreduction (56). The data presented here may ultimately make it possible to design new tester strains with higher efficiencies in mutagenicity tests. To this end, we suggest that the P. aeruginosa NAT should be further investigated at the molecular level. Indeed, this study shows that this enzyme is potentially useful in terms of both the substrate diversity and the activity levels required in such tests. However, further kinetic studies on purified enzyme are necessary, since differences in enzyme activities can be due to differences in catalytic properties but also to differences in expression levels.
ACKNOWLEDGMENTS
This work was supported by grants from the Ligue Nationale contre le Cancer (France).
We thank Philippe Quillardet (Laboratoire de Programmation Moléculaire et Toxicologie Génétique, Institut Pasteur, Paris, France) for kindly providing the S. enterica serovar Typhimurium Ames tester strains. We are grateful to Edith Sim (Department of Pharmacology, Oxford University, Oxford, England) for the kind gift of rabbit antiserum raised against purified recombinant S. enterica serovar Typhimurium NAT and for sharing preliminary data. We also thank Jacques Elion for constant encouragement during this work and Rajagopal Krishnamoorthy, Philippe Marteau, Mark Payton, and Sylvie Rabot for helpful discussion. We also thank Catherine Arnaudeau for technical assistance.
REFERENCES
- 1.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. . (Erratum, 397:719.) [DOI] [PubMed] [Google Scholar]
- 2.Ames B, Gold L S. Environmental pollution, pesticides, and the prevention of cancer: misconceptions. FASEB J. 1997;11:1041–1052. doi: 10.1096/fasebj.11.13.9367339. [DOI] [PubMed] [Google Scholar]
- 3.Bamba T, Matsuda H, Endo M, Fujiyama Y. The pathogenic role of Bacteroides vulgatus in patients with ulcerative colitis. J Gastroenterol. 1995;30:45–47. [PubMed] [Google Scholar]
- 4.Berg R D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 5.Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 6.Blum M, Grant D M, McBride O M, Heim M, Meyer U A. Human arylamine N-acetyltransferase genes: isolation, chromosomal localization and functional expression DNA. Cell Biol. 1990;9:193–203. doi: 10.1089/dna.1990.9.193. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Chang F C, Chung J G. Evidence for arylamine N-acetyltransferase activity in the Escherichia coli. Curr Microbiol. 1998;36:125–130. doi: 10.1007/pl00006755. [DOI] [PubMed] [Google Scholar]
- 9.Chang F C, Chung J G, Chang W C, Wu L T, Chen G W, Chang S H. Arylamine N-acetyltransferase activity in Staphylococcus aureus. J Microbiol Immunol Infect. 1997;30:170–181. [PubMed] [Google Scholar]
- 10.Chang S H, Chung J G, Huang L J, Chen S C, Kuo S C. Ibuprofen affects arylamine N-acetyltransferase activity in Helicobacter pylori from peptic ulcer patients. J Appl Toxicol. 1998;18:179–185. doi: 10.1002/(sici)1099-1263(199805/06)18:3<179::aid-jat494>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 11.Chen G W, Hung C F, Chang S H, Lin J G, Chung J G. Purification and characterization of an arylamine N-acetyltransferase from Lactobacillus acidophilus. Microbios. 1999;98:159–174. [PubMed] [Google Scholar]
- 12.Chung J G, Kuo H, Lin T, Ho C, Lee J, Lai J, Levy G, Weber W. Evidence for arylamine N-acetyltransferase in the nematode Anisakis simplex. Cancer Lett. 1996;106:1–8. doi: 10.1016/0304-3835(96)04288-7. [DOI] [PubMed] [Google Scholar]
- 13.Chung J G. The effects of vitamin E on arylamine N-acetyltransferase activity in strains of Helicobacter pylori from peptic ulcer patients. Food Chem Toxicol. 1999;37:655–661. doi: 10.1016/s0278-6915(99)00047-2. [DOI] [PubMed] [Google Scholar]
- 14.Chung J G. Inhibitory actions of ellagic acid on growth and arylamine N-acetyltransferase activity in strains of Helicobacter pylori from peptic ulcer patients. Microbios. 1998;93:115–127. [PubMed] [Google Scholar]
- 15.Chung J G. Purification and characterization of an arylamine N-acetyltransferase from the bacteria Aeromonas hydrophilia. Curr Microbiol. 1998;37:70–73. doi: 10.1007/s002849900341. [DOI] [PubMed] [Google Scholar]
- 16.Chung J G, Kuo H M, Wu L T, Lai J M, Lee J H, Hung C F. Evidence for arylamine N-acetyltransferase in Hymenolepis nana. J Microbiol Immunol Infect. 1997;30:1–17. [PubMed] [Google Scholar]
- 17.Chung J G, Tsou M F, Wang H H, Lo H H, Hsieh S E, Yen Y S, Wu L T, Chang S H, Ho C C, Hung C F. Rhein affects arylamine N-acetyltransferase activity in Helicobacter pylori from peptic ulcer patients. J Appl Toxicol. 1998;18:117–123. doi: 10.1002/(sici)1099-1263(199803/04)18:2<117::aid-jat486>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 18.Chung J G, Wang H H, Tsou M F, Hsieh S E, Lo H H, Yen Y S, Chang S S, Wu L T, Lee J H, Hung C F. Evidence for arylamine N-acetyltransferase activity in the bacterium Helicobacter pylori. Toxicol Lett. 1997;91:63–71. doi: 10.1016/s0378-4274(97)03870-8. [DOI] [PubMed] [Google Scholar]
- 19.Creighton T E. Proteins: structures and molecular properties. 2nd ed. New York, N.Y: W. H. Freeman and Co.; 1993. Amino acid residues; pp. 6–20. [Google Scholar]
- 20.Deloménie C, Goodfellow G H, Krishnamoorthy R, Grant D M, Dupret J-M. Study of the role of the highly conserved residues Arg9 and Arg64 in the catalytic function of human N-acetyltransferases NAT1 and NAT2 by site-directed mutagenesis. Biochem J. 1997;323:207–215. doi: 10.1042/bj3230207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doll M A, Hein D W. Cloning, sequencing and expression of NAT1 and NAT2 encoding genes from rapid and slow acetylator inbred rats. Pharmacogenetics. 1995;5:247–251. doi: 10.1097/00008571-199508000-00009. [DOI] [PubMed] [Google Scholar]
- 22.Dupret J-M, Grant D M. Site-directed mutagenesis of recombinant human arylamine N-acetyltransferase expressed in Escherichia coli. J Biol Chem. 1992;267:7381–7385. [PubMed] [Google Scholar]
- 23.Ferguson R, Doll M, Rustan T, Hein D. Cloning, expression and functional characterization of rapid and slow acetylator polymorphic N-acetyltransferase encoding genes of the syrian hamster. Pharmacogenetics. 1996;6:55–66. doi: 10.1097/00008571-199602000-00004. [DOI] [PubMed] [Google Scholar]
- 24.Goodfellow G H, Dupret J-M, Grant D M. Identification of amino acids imparting acceptor substrate selectivity in human arylamine acetyltransferases NAT1 and NAT2. Biochem J. 2000;348:159–166. [PMC free article] [PubMed] [Google Scholar]
- 25.Grant D M, Blum M, Beer M, Meyer U A. Monomorphic and polymorphic human arylamine N-acetyltransferases: a comparison of liver isozymes and expressed products of two cloned genes. Mol Pharmacol. 1991;39:184–191. [PubMed] [Google Scholar]
- 26.Grant D M, Josephy P D, Lord H L, Morrison L D. Salmonella typhimurium strains expressing human arylamine N-acetyltransferases: metabolism and mutagenic activation of aromatic amines. Cancer Res. 1992;52:3961–3964. [PubMed] [Google Scholar]
- 27.Hein D W, Doll M A, Rustan T D, Gray K, Feng Y, Ferguson R J, Grant D M. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis. 1993;14:1633–1638. doi: 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- 28.Hein D W, Grant D M, Sim E. Update on consensus arylamine N-acetyltransferase gene nomenclature. Pharmacogenetics. 2000;10:291–292. doi: 10.1097/00008571-200006000-00002. [DOI] [PubMed] [Google Scholar]
- 29.Hickman D, Pope J, Patil S D, Fakis G, Smelt V, Stanley L A, Payton M, Unadkat J D, Sim E. Expression of arylamine N-acetyltransferase in human intestine. Gut. 1998;42:402–409. doi: 10.1136/gut.42.3.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho C, Lin T, Lai Y, Chung J, Levy G, Weber W. Kinetics of acetyl coenzyme A-arylamine N-acetyltransferase from rapid and slow acetylator frog tissues. Drug Metab Dispos. 1996;24:137–141. [PubMed] [Google Scholar]
- 31.Horing E, Gopfert D, Schroter G, von Gaisberg U. Frequency and spectrum of microorganisms isolated from biopsy specimens in chronic colitis. Endoscopy. 1991;23:325–327. doi: 10.1055/s-2007-1010707. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh S E, Lo H H, Chen Y S, Chung J G. The effect of vitamin C on N-acetyltransferase activity in Klebsiella pneumoniae. Food Chem Toxicol. 1997;35:1151–1157. doi: 10.1016/s0278-6915(97)85467-1. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh S E, Lo H H, Chung J G. The characteristics of arylamine N-acetyltransferase in Pseudomonas aeruginosa. Curr Microbiol. 1998;36:353–360. doi: 10.1007/s002849900322. [DOI] [PubMed] [Google Scholar]
- 34.Ilett K F, Kadlubar F F, Minchin R F. 1998 international meeting on the arylamine N-acetyltransferases: synopsis of the workshop on nomenclature, biochemistry, molecular biology, interspecies comparisons, and role in human disease risk, Drug Metab. Dispos. 1999;27:957–959. [PubMed] [Google Scholar]
- 35.Ireland A, Priddle J D, Jewell D P. Comparison of 5-aminosalicylic acid and N-acetylaminosalicylic acid uptake by the isolated human colonic epithelial cell. Gut. 1990;33:1343–1347. doi: 10.1136/gut.33.10.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito H, Kido N, Arakawa Y, Ohta M, Sugiyama T, Kato N. Possible mechanisms underlying the slow lactose fermentation phenotype in Shigella spp. Appl Environ Microbiol. 1991;57:2912–2917. doi: 10.1128/aem.57.10.2912-2917.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly S L, Sim E. Arylamine N-acetyltransferase in BALB/c mice: identification of a novel mouse isoenzyme by cloning and expression in vitro. Biochem J. 1994;302:347–353. doi: 10.1042/bj3020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitz R, Wilson I B. Esters of methanesulfonic acid as irreversible inhibitors of acetylcholinesterase. J Biol Chem. 1962;237:3245–3249. [PubMed] [Google Scholar]
- 39.Lo H H, Chung J G. The effects of plant phenolics, caffeic acid, chlorogenic acid and ferulic acid on arylamine N-acetyltransferase activities in human gastrointestinal microflora. Anticancer Res. 1999;19:133–139. [PubMed] [Google Scholar]
- 40.Maron D M, Ames B N. Revised methods for the Salmonella mutagenicity test. Mutat Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 41.Marteau P, Cellier C. Effets indésirables de l'acide 5-aminosalicylique. Gastroenterol Clin Biol. 1997;21:377–386. [PubMed] [Google Scholar]
- 42.Marteau P, Halphen M. Comparative randomized open study of the efficacy and tolerance of enemas with 2 gr of 4-aminosalicylic acid (4-ASA) and 1 gr of 5-aminosalicylic acid (5-ASA) in distal forms of hemorrhagic rectocolitis. Gastroenterol Clin Biol. 1995;19:31–35. [PubMed] [Google Scholar]
- 43.McCoy E C, Rosenkranz H S, Mermelstein R. Evidence for the existence of a family of bacterial nitroreductases capable of activating nitrated polycyclics to mutagens. Environ Mutagen. 1981;3:421–427. doi: 10.1002/em.2860030403. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama H, Kinouchi T, Kataoka K, Akimoto S, Matsuda Y, Ohnishi Y. Intestinal anaerobic bacteria hydrolyse sorivudine, producing the high blood concentration of 5-(E)-(2-bromovinyl)uracil that increases the level and toxicity of 5-fluorouracil. Pharmacogenetics. 1997;7:35–43. doi: 10.1097/00008571-199702000-00005. [DOI] [PubMed] [Google Scholar]
- 45.Neidhardt F C. Chemical composition of Escherichia coli. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 3–6. [Google Scholar]
- 46.Neut C, Colombel J F, Guillemot F, Cortot A, Gower P, Quandalle P, Ribet M, Romond C, Paris J C. Impaired bacterial flora in human excluded colon. Gut. 1989;30:1094–1098. doi: 10.1136/gut.30.8.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohsako S, Ohtomi M, Sakamoto Y, Uyemura K, Deguchi T. Arylamine N-acetyltransferase from chicken liver: cloning of cDNA and expression in Chinese hamster ovary cells. J Biol Chem. 1988;263:7534–7538. [PubMed] [Google Scholar]
- 48.Okumura F, Ueda O, Kitamura S, Tatsumi K. N-Acetylation and N-formylation of carcinogenic arylamines and related compounds in dogs. Carcinogenesis. 1995;16:71–76. doi: 10.1093/carcin/16.1.71. [DOI] [PubMed] [Google Scholar]
- 49.Payton M, Auty R, Delgoda R, Everett M, Sim E. Cloning and characterization of arylamine N-acetyltransferase genes from Mycobacterium smegmatis and Mycobacterium tuberculosis: increased expression results in isoniazid resistance. J Bacteriol. 1999;181:1343–1347. doi: 10.1128/jb.181.4.1343-1347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prantera C, Scribano M L. Current treatment for prevention of relapse and recurrence in Crohn's disease. Ital J Gastroenterol Hepatol. 1999;31:515–518. [PubMed] [Google Scholar]
- 51.Reddy B S, Pleasants J R, Wostmann B S. Pancreatic enzymes in germfree and conventional rats fed chemically defined, water-soluble diet free from natural substrates. J Nutr. 1969;97:327–334. doi: 10.1093/jn/97.3.327. [DOI] [PubMed] [Google Scholar]
- 52.Sandberg-Gertzen H, Kjellander J, Sundberg-Gilla B, Jarnerot G. In vitro effects of sulphasalazine, azodisal sodium, and their metabolites on Clostridium difficile and some other faecal bacteria. Scand J Gastroenterol. 1985;20:607–612. doi: 10.3109/00365528509089704. [DOI] [PubMed] [Google Scholar]
- 53.Sasaki Y, Ohsako S, Deguchi T. Molecular and genetic analyses of arylamine N-acetyltransferase polymorphism of rabbit liver. J Biol Chem. 1991;266:13243–13250. [PubMed] [Google Scholar]
- 54.Sim E, Payton M, Noble M, Minchin R. An update on genetic structural and functional studies of arylamine N-acetyltransferases in eucaryotes and procaryotes. Hum Mol Genet. 2000;9:2435–2441. doi: 10.1093/hmg/9.16.2435. [DOI] [PubMed] [Google Scholar]
- 55.Simmonds N J, Millar A D, Blake D R, Rampton D S. Antioxidant effects of aminosalicylates and potential new drugs for inflammatory bowel disease: assessment in cell-free systems and inflamed human colorectal biopsies. Aliment Pharmacol Ther. 1999;13:363–372. doi: 10.1046/j.1365-2036.1999.00484.x. [DOI] [PubMed] [Google Scholar]
- 56.Simons G L, Gorbach S L. Intestinal flora in health and disease. In: Johnson L R, editor. Physiology of the gastrointestinal tract. New York, N.Y: Raven Press; 1981. pp. 1361–1380. [Google Scholar]
- 57.Sinclair J C, Delgoda R, Noble M E M, Jarmia S, Goh N K, Sim E. Purification, characterization, and crystallization of an N-hydroxyarylamine O-acetyltransferase from Salmonella typhimurium. Protein Expr Purif. 1998;12:371–380. doi: 10.1006/prep.1997.0856. [DOI] [PubMed] [Google Scholar]
- 58.Sinclair J C, Sandy J, Delgoda R, Sim E, Noble M E M. Structure of arylamine N-acetyltransferase reveals a catalytic triad. Nat Struct Biol. 2000;7:560–564. doi: 10.1038/76783. [DOI] [PubMed] [Google Scholar]
- 59.Thomson A B R, Williams C N. Trends in inflammatory bowel disease therapy. Can J Gastroenterol. 1999;13:775–776. [PubMed] [Google Scholar]
- 60.Tomb J F, White O, Kerlavage A R, Clayten R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. . (Erratum, 389:412.) [DOI] [PubMed] [Google Scholar]
- 61.Tsou M F, Chung J G, Wu L T, Cheng K S, Hung C F. Characterization of arylamine N-acetyltransferase in Enterobacter aerogenes. Microbios. 1998;94:133–143. [PubMed] [Google Scholar]
- 62.Tsou M F, Hung C F, Lu H F, Wu L T, Chang S H, Chang H L, Chen G W, Chung J G. Effects of caffeic acid, chlorogenic acid and ferulic acid on growth and arylamine N-acetyltransferase activity in Shigella sonnei (group D) Microbios. 2000;101:37–46. [PubMed] [Google Scholar]
- 63.van Hogezand R A, Kennis H M, van Schaik A, Koopman J P, van Hees P A, Tongeren J H. Bacterial acetylation of 5-aminosalicylic acid in faecal suspensions cultured under aerobic and anaerobic conditions. Eur J Clin Pharmacol. 1992;43:189–192. doi: 10.1007/BF01740669. [DOI] [PubMed] [Google Scholar]
- 64.Watanabe M, Ishidate M J, Nohmi T. Sensitive method for the detection of mutagenic nitroarenes and aromatic amines: new derivatives of Salmonella typhimurium tester strains possessing elevated O-acetyltransferase levels. Mutat Res. 1990;234:337–348. doi: 10.1016/0165-1161(90)90044-o. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe M, Sofuni T, Nohmi T. Involvement of Cys69 residue in the catalytic mechanism of N-hydroxyarylamine O-acetyltransferase of Salmonella typhimurium: sequence similarity at the amino acid level suggests a common catalytic mechanism of acetyltransferase for S. typhimurium and higher organisms. J Biol Chem. 1992;267:8429–8436. [PubMed] [Google Scholar]
- 66.Whitaker D P, Goosey M W. Purification and properties of the enzyme arylamine N-acetyltransferase from the housefly Musca domestica. Biochem J. 1993;295:149–154. doi: 10.1042/bj2950149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeitz M. Consequences of galenic considerations and clinical results for therapy of ulcerative colitis. Med Klin. 1999;94(Suppl. 1):41–43. doi: 10.1007/BF03042033. . (In German.) [DOI] [PubMed] [Google Scholar]
- 68.Zhou S Y, Fleisher D, Pao L H, Li C, Winward B, Zimmermann E M. Intestinal metabolism and transport of 5-aminosalicylate. Drug Metab Dispos. 1999;27:479–485. [PubMed] [Google Scholar]