Significance
HSPCs have the capacities of both self-renewal and multilineage differentiation and hold great importance for lifelong blood supply. The developmental signals ensure the production of HSPCs through endothelial-to-hematopoietic transition during vertebrate embryogenesis. However, how these signals are accurately regulated to execute cell fate transition remains elusive. In this study, we explore the role of Cpeb1b in HSPC production and uncover its function via cytoplasmic polyadenylation of shha mRNA in this process. We show that notochord-expressed Cpeb1b is required for HSPC development by regulating Hedgehog signaling. Cpeb1b interacts with shha mRNA in the liquid-like condensates and promotes its cytoplasmic polyadenylation. Cytoplasmic polyadenylation of shha mRNA enhances its own translation efficiency and therefore increases the Shha protein level, which facilitates Hedgehog signaling activation.
Keywords: hematopoietic stem and progenitor cell, cytoplasmic polyadenylation, Cpeb1b, Hedgehog signaling, translational control
Abstract
During vertebrate embryogenesis, hematopoietic stem and progenitor cell (HSPC) production through endothelial-to-hematopoietic transition requires suitable developmental signals, but how these signals are accurately regulated remains incompletely understood. Cytoplasmic polyadenylation, which is one of the posttranscriptional regulations, plays a crucial role in RNA metabolism. Here, we report that Cpeb1b-mediated cytoplasmic polyadenylation is important for HSPC specification by translational control of Hedgehog (Hh) signaling during zebrafish early development. Cpeb1b is highly expressed in notochord and its deficiency results in defective HSPC production. Mechanistically, Cpeb1b regulates hemogenic endothelium specification by the Hedgehog–Vegf–Notch axis. We demonstrate that the cytoplasmic polyadenylation element motif-dependent interaction between Cpeb1b and shha messenger RNA (mRNA) in the liquid-like condensates, which are induced by Pabpc1b phase separation, is required for cytoplasmic polyadenylation of shha mRNA. Intriguingly, the cytoplasmic polyadenylation regulates translation but not stability of shha mRNA, which further enhances the Shha protein level and Hh signal transduction. Taken together, our findings uncover the role of Cpeb1b-mediated cytoplasmic polyadenylation in HSPC development and provide insights into how posttranscriptional regulation can direct developmental signals with high fidelity to translate them into cell fate transition.
During vertebrate embryonic development, the hematopoiesis occurs through two waves: the primitive wave only generates erythrocytes and myelocytes, and the definitive wave can produce hematopoietic stem and progenitor cells (HSPCs), which have the capacity to give rise to all lineages of blood cells and maintain the entire blood system (1–3). During the definitive hematopoiesis period, a subset of endothelial cells (ECs) receive cell-intrinsic and cell-extrinsic signals and acquire hemogenic potential to become definitive hemogenic endothelium (HE) cells in the mammalian aorta-gonad-mesonephros region or the zebrafish ventral wall of the dorsal aorta (VDA) region. Then, these HE cells gradually change their shape and bud from the dorsal aorta (DA) to be converted into HSPCs through the endothelial-to-hematopoietic transition (4–7). However, how these ECs are precisely controlled to undergo EC-to-HSPC switch remains incompletely understood.
Several developmental signals have been reported to be critical for hematopoiesis (2, 3, 8). Among these signals, Hedgehog (Hh) signaling is shown to be involved in HSPC production in zebrafish and mammals (9–11). In the Hh signaling transduction process, the secreted signal moiety, which is generated by auto-cleavage of Hh precursor, binds to Patched receptor, leading to disinhibition and primary cilium tip location of Smoothened (Smo). Then, the activated Smo facilitates the dissociation of Gli2/3-Cos-SuFu complex, resulting in release of the full-length forms of Gli2/3, which function as activators for Hh downstream gene transcription. Hh downstream genes can indirectly induce Notch signaling through upregulation of Vegf signaling (1, 10, 11). Upon Notch activation, Notch intracellular domain (NICD) enters the nucleus to activate transcription of Notch target genes, which are required for HE specification (12–14). However, how these signals are accurately regulated during HSPC production should be further explored.
The posttranscriptional regulation, including alternative splicing, noncoding RNA-mediated regulation, RNA modification, poly(A) tail length, codon usage, and RNA structure, plays a crucial role in the regulation of gene expression and signaling specificity through controlling metabolism of RNA molecules (15–20). Recently, the role of posttranscriptional regulation in hematopoiesis continues to be reported. The posttranscriptional regulators, such as ADAR1 (21), miR-142a-3p (18), Rbm15 (22), Regnase-1 (23), and m6A modification (16), are involved in HSPC production or maintenance through different influences on RNA metabolism. Cytoplasmic polyadenylation as one of the posttranscriptional regulations is mainly regulated by the cytoplasmic polyadenylation element binding protein (CPEB) family proteins (24, 25). CPEB specifically binds to the U-rich cytoplasmic polyadenylation element (CPE) in 3′ untranslated region (UTR) of target mRNAs and nucleates a cytoplasmic complex containing the noncanonical poly(A) polymerase Tent2/Papd4/Gld2. Tent2/Papd4/Gld2 can add successive adenosine 5'-monophosphate monomers to the 3′-end of mRNAs for poly(A) tail formation, further facilitating mRNA circularization and translation initiation (20, 25–27). CPEB proteins are involved in regulation of diverse biological processes, including synaptic plasticity and long-term potentiation (28–31), germ cell differentiation (32), cellular senescence (33), muscle stem cell activation (34), and immune response (35). However, it remains unknown whether CPEB proteins play a role in regulating developmental signals that affect embryonic HSPC production in vertebrates.
Due to genome duplication in teleost fish, there are two CPEB1 orthologs in zebrafish, cpeb1a and cpeb1b (36). Here, we have demonstrated that Cpeb1b-mediated cytoplasmic polyadenylation is important for HSPC development in zebrafish. Cpeb1b deficiency leads to HE specification defect. Further mechanistic studies show that Cpeb1b interacts with shha mRNA in the liquid-like condensates, which are induced by Pabpc1b phase separation. This interaction is CPE motif-dependent and crucial for cytoplasmic polyadenylation of shha mRNA. Cpeb1b-regulated cytoplasmic polyadenylation of shha mRNA promotes its translation and further facilitates Hedgehog–Vegf–Notch signaling axis activation, which are required for HSPC production.
Results
Cpeb1b is Important for HSPC Production during Early Development.
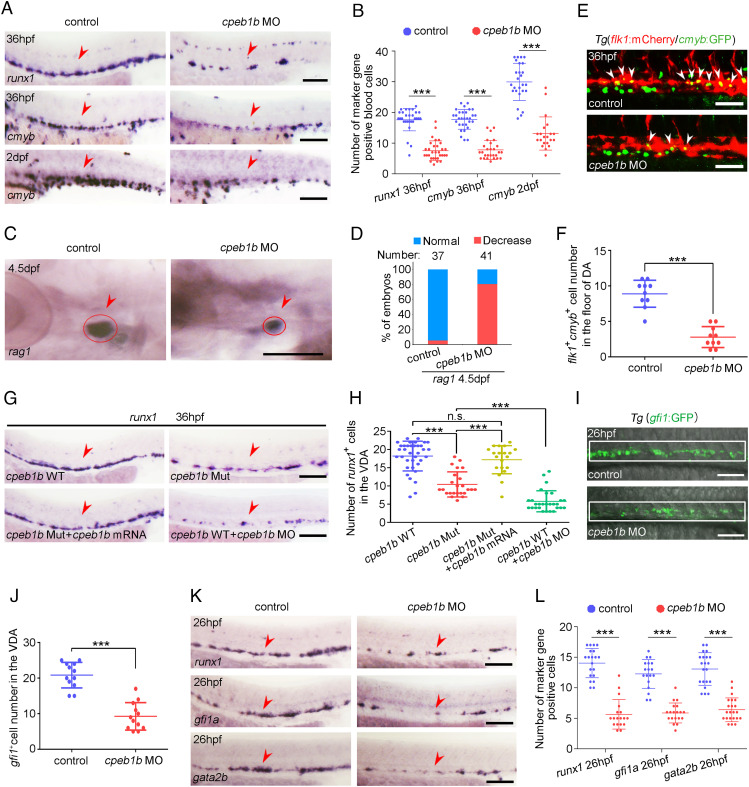
To examine the expression pattern of cpeb1b during zebrafish early development, we performed whole-mount in situ hybridization (WISH) and found that cpeb1b displayed maternal expression and was highly expressed in the notochord region from 14 to 24 h post fertilization (hpf) (SI Appendix, Fig. S1A). Moreover, double fluorescence in situ hybridization (FISH) was carried out with cpeb1b and a well-known notochord-specific marker gene shha. FISH result showed that the green cpeb1b signals were colocalized with the red shha signals in the notochord at 14 and 16 hpf (SI Appendix, Fig. S1B), confirming the notochord-enriched expression of cpeb1b. To determine whether Cpeb1b plays a potential role in zebrafish embryonic hematopoiesis, we utilized the knockdown approach by injection of cpeb1b anti-sense ATG morpholino (MO) (SI Appendix, Fig. S2A), whose effectiveness was verified by western blot (WB) (SI Appendix, Fig. S2 B and C). WISH revealed that the primitive hematopoiesis marked by gata1 (erythroid) and spi1 (myeloid) expression at 14 hpf was normal (SI Appendix, Fig. S2 D and E), and time-lapse imaging showed that the blood flow at 32 hpf was not altered in the cpeb1b morphants (MO-injected embryos) compared with control (Movie S1). However, cpeb1b morphants exhibited impaired HSPC production in definitive hematopoiesis marked by the expression of HSPC markers runx1 and cmyb in VDA at 36 hpf (Fig. 1 A and B). The number of cmyb-marked HSPCs in CHT at 2 days post fertilization (dpf) and HSPC-derived lymphocytes in thymus at 4.5 dpf was also evidently reduced in the cpeb1b morphants (Fig. 1 A–D). Notably, overexpression of cpeb1b mRNA without the cpeb1b MO binding site (to avoid inhibition by its antisense MO) (SI Appendix, Fig. S2 F and G) rescued this HSPC defect (SI Appendix, Fig. S2 H and I), indicating that the defective phenotype resulted from cpeb1b loss-of-function. To further validate the HSPC phenotype upon cpeb1b knockdown, we observed the living definitive hematopoietic precursors by using the Tg(flk1:mCherry/cmyb:GFP) double-transgenic line and found that the number of flk1 and cmyb double-positive precursors within the VDA region was markedly decreased in the cpeb1b morphants (Fig. 1 E and F). We performed further flow cytometry analysis and found similar results (SI Appendix, Fig. S2 J and K). In addition, we generated a cpeb1b-null frameshift mutant by CRISPR/Cas9 technology to confirm the cpeb1b loss-of-function phenotype. The zygotic mutant embryos were obtained by cross-mating heterozygous mutants (SI Appendix, Fig. S2L), however, we failed to obtain the maternal-zygotic mutant embryos due to the defective germ cell and ovary development in zygotic mutant adult zebrafish, which are similar to that in mouse (32). WISH showed that the HSPC production was impaired in cpeb1b zygotic mutant, and this phenotype was also rescued by cpeb1b mRNA overexpression (Fig. 1 G and H), supporting that the phenotype is not due to the off-target effect of CRISPR/Cas9. However, the HSPC phenotype in cpeb1b zygotic mutant was milder than that in the cpeb1b morphants (Fig. 1 G and H). Previous studies showed that the maternal effect in the zygotic mutant may alleviate the phenotype, and thus, suppressing this maternal effect by a low dose of MO injection leads to more severe phenotype (37, 38). Based on the finding of cpeb1b maternal expression, we wondered whether the maternal effect may lead to alleviative phenotype in cpeb1b zygotic mutant. To test this hypothesis, we injected low-dose (LD) cpeb1b MO into wild-type (WT) and cpeb1b zygotic mutants and observed severe HSPC defect, which is similar to that in the cpeb1b morphants, in the cpeb1b mutants injected with LD cpeb1b MO, but not in the wildtype (SI Appendix, Fig. S2 M and N), indicating that maternal effect indeed led to the alleviation of phenotype. Previous studies have demonstrated that HE specification is a prerequisite for HSPC generation during early development (2, 4, 8); therefore, we wondered whether HE specification was affected by Cpeb1b-deficiency. We observed the HE cells by using the Tg(gfi1:GFP) transgenic line and found that the number of gfi1 positive HE cells within the VDA region was decreased in the cpeb1b morphants (Fig. 1 I and J). The flow cytometry analysis showed similar results (SI Appendix, Fig. S2 O and P). Furthermore, WISH showed that there were fewer HE cells in the cpeb1b morphants compared to control (Fig. 1 K and L), indicating the importance of Cpeb1b in HE specification. Taken together, these results suggest that Cpeb1b is required for HE specification, and Cpeb1b-deficiency–induced HE defect eventually leads to HSPC number reduction.
Fig. 1.
Cpeb1b deficiency impairs HSPC production. (A) Examination of the expression of runx1 and cmyb in the control and cpeb1b morphants by WISH. The red arrowheads denote HSPCs. (Scale bar, 100 μm.) (B) Statistical analysis of the WISH. Error bar, mean ± SD, ***P < 0.001. (C) Examination of the rag1 expression in the control and cpeb1b morphants by WISH. (D) Quantification of the WISH. (E) Confocal imaging shows the flk1+cmyb+ definitive hematopoietic precursors in the VDA region of control and cpeb1b morphants at 36 hpf. White arrowheads denote flk1+cmyb+ precursors. (Scale bars, 100 μm.) (F) Statistical analysis of flk1+cmyb+ definitive hematopoietic precursors. Error bar, mean ± SD. The P value was calculated by Student’s t test, ***P < 0.001. (G) Examination of the runx1 expression in WT, cpeb1b mutant, cpeb1b mRNA-overexpressed cpeb1b mutant, and cpeb1b MO-injected WT embryos by WISH. In WISH experiments using cpeb1b mutant embryos, the embryos were obtained by cross-mating cpeb1b adult mutants and individually subjected to genotyping after WISH for identifying homozygous mutants. (Scale bar, 100 μm.) (H) Statistical analysis of the WISH. Error bar, mean ± SD, n.s.: no significance, ***P < 0.001. (I) Confocal imaging shows the gfi1+ HE cells in the VDA region of control and cpeb1b morphants at 26 hpf. (Scale bars, 100 μm.) (J) Statistical analysis of gfi1+ HE cells. Error bar, mean ± SD. The P value was calculated by Student’s t test, ***P < 0.001. (K) Examination of the HE marker runx1, gfi1a, and gata2b expression in control and cpeb1b morphants at 26 hpf by WISH. (Scale bars, 100 μm.) (L) Statistical analysis of the WISH. Error bar, mean ± SD, ***P < 0.001.
Cpeb1b Regulates HSPC Development through Hh Signaling.
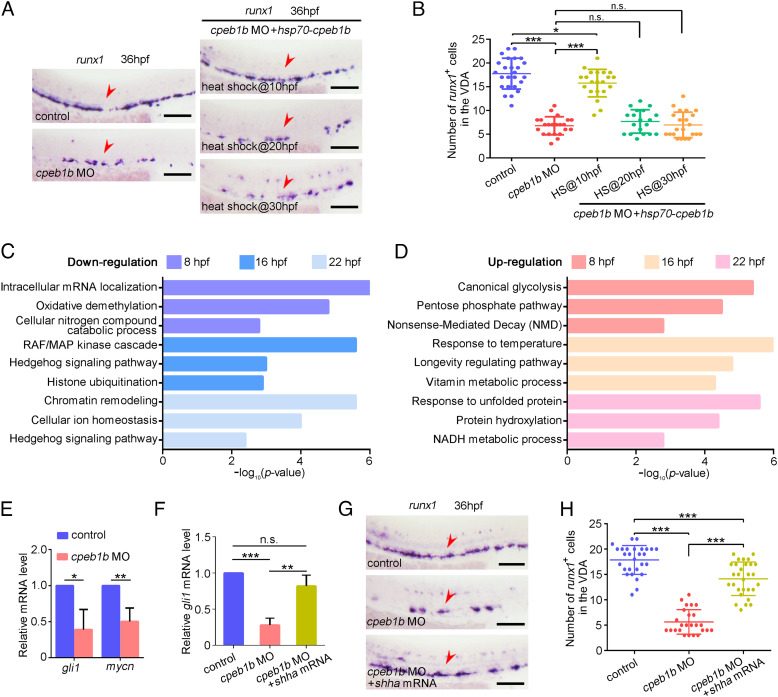
To further determine the timing of Cpeb1b function, we applied hsp70 promoter-mediated heat-shock (HS) inducible Cpeb1b overexpression system. Strong green fluorescence of green fluorescent protein (GFP)-Cpeb1b was detected from 2 h post heat shock at indicated time points to at least 20 h post heat shock (SI Appendix, Fig. S3A). WISH showed that heat shock induction of Cpeb1b at 10 hpf, but not at 20- or 30 hpf, rescued the HSPC defect in the cpeb1b morphants (Fig. 2 A and B), suggesting that Cpeb1b functions in HSPC production at the somitic period. To investigate the mechanism upon Cpeb1b-deficiency, we collected WT and cpeb1b mutants at different developmental stages (8 hpf, 16 hpf, and 22 hpf) for RNA-seq analysis. Gene ontology (GO) analysis revealed that the downregulated and upregulated genes in Cpeb1b-deficient embryos were enriched in various signaling pathways, such as canonical glycolysis, mitogen-activated protein kinase cascade, Hh signaling pathway, histone ubiquitination, cellular ion homeostasis, vitamin metabolic process, and others (Fig. 2 C and D and Dataset S1). Among these altered signaling pathways in GO analysis, the Hh signaling, which was downregulated in Cpeb1b-deficient embryos at 16 and 22 hpf, has been previously demonstrated to regulate HSPC generation in zebrafish and mouse (10, 11). Combined with the finding that the Hh ligand and cpeb1b were highly coexpressed in the notochord, we speculated that Hh signaling might be involved in Cpeb1b deficiency-induced HSPC defects. qRT-PCR results showed that the expression of Hh signaling downstream genes gli1 and mycn was reduced in the cpeb1b morphants (Fig. 2E). To determine whether Hh signaling induction can rescue the HSPC defect in cpeb1b-deficient embryos, we performed overexpression of shha, which is important for Hh signaling activation in zebrafish. The shha overexpression efficiency and its function on Hh signaling induction were confirmed (SI Appendix, Fig. S3 B–D). qRT-PCR showed that the reduced expression of Hh downstream gene gli1 in the cpeb1b morphants was rescued by shha overexpression (Fig. 2F), and WISH showed that the HSPC defect was also rescued (Fig. 2 G and H). Furthermore, we treated the cpeb1b morphants with the well-known Hh signaling agonist purmorphamine and found that purmorphamine treatment also provided a rescue effect (SI Appendix, Fig. S3 E and F). On the contrary, inhibition of the Hh signaling by its antagonist cyclopamine, which directly binds to and inhibits Hh signaling coreceptor and transducer Smo (39), impaired HSPC production (SI Appendix, Fig. S3 G and H), confirming a critical role of Hh signaling in HSPC development. In addition, we noticed that the Cpeb1b-deficiency–induced phenotype was weaker than that in embryos with cyclopamine treatment (SI Appendix, Fig. S3 G and H), implying that not all the Hh member-mediated signaling is inactivated in Cpeb1b-deficient embryos. Taken together, these results above indicate that Cpeb1b regulates HSPC development through Hh signaling.
Fig. 2.
Cpeb1b regulates HSPC development through Hh signaling. (A) Examination of the runx1 expression in the control, cpeb1b morphants, and Cpeb1b-overexpressed cpeb1b morphants by WISH. Cpeb1b overexpression was carried out by hsp70-GFP-cpeb1b HS at 10 hpf or 20 hpf or 30 hpf. The red arrowheads denote HSPCs. (Scale bar, 100 μm.) (B) Statistical analysis of the WISH. Error bar, mean ± SD, n.s.: no significance, *P < 0.05, ***P < 0.001. (C) The LD cpeb1b MO injected WT and cpeb1b mutant embryos at different developmental stages were collected for RNA-seq. The GO analysis showing the downregulated signaling pathways in cpeb1b mutant embryos. (D) GO analysis showing the upregulated signaling pathways in cpeb1b mutant embryos. (E) Relative mRNA level of Shh signaling downstream gene gli1 and mycn in the control and cpeb1b morphants at 16 hpf examined by qRT-PCR. Error bar, mean ± SD. The P value was calculated by Student’s t test, *P < 0.05, **P < 0.01. (F) Relative mRNA level of gli1 in the control, cpeb1b morphants, and shha mRNA-overexpressed cpeb1b morphants at 16 hpf examined by qRT-PCR. Error bar, mean ± SD. The P value was calculated by Student’s t test, n.s.: no significance, **P < 0.01, ***P < 0.001. (G) Examination of the runx1 expression in the control, cpeb1b morphants, and shha mRNA-overexpressed cpeb1b morphants at 36 hpf by WISH. (Scale bar, 100 μm.) (H) Statistical analysis of the WISH. Error bar, mean ± SD, ***P < 0.001.
Previous studies showed that Vegf signaling acts downstream of Hh signaling and upstream of the Notch signaling during early development, and this Hedgehog–Vegf–Notch signaling axis is crucial for HSPC generation (10, 40). Besides, Notch signaling is essential for HE specification, which is a prerequisite for HSPC generation (12, 13). To explore whether this signaling axis is involved in Cpeb1b-deficiency–induced HSPC defect, we performed the following assays. WISH showed that the expression of vegfa was decreased in the cpeb1b morphants compared with control (SI Appendix, Fig. S4 A and B), indicating that Cpeb1b-deficiency leads to downregulation of Vegf signaling. WB showed that the protein level of NICD was decreased in the cpeb1b morphants (SI Appendix, Fig. S4 C and D), and qRT-PCR showed that the expression of Notch signaling downstream genes hey1 and hey2 was also decreased in the cpeb1b morphants (SI Appendix, Fig. S4E). Next, we took advantage of the Notch reporter line Tg(fli1a:EGFP/tp1:mCherry) to examine Notch activity in ECs and found that the number of fli1a+tp1+ cells in the VDA region was reduced (SI Appendix, Fig. S4 F and G), demonstrating that Cpeb1b-deficiency resulted in attenuated Notch activity. Furthermore, WISH showed that the expression of Notch signaling-related artery genes efnb2a and dll4 was also decreased in the cpeb1b morphants at 26 hpf (SI Appendix, Fig. S4 H and I). By contrast, the expression of another arterial marker tbx20 was relatively normal in the cpeb1b morphants (SI Appendix, Fig. S4 H and I), suggesting that the dorsal aorta still developed in the Cpeb1b-deficient embryos, which is consistent with previous studies in that Notch signaling alteration does not necessarily result in loss of dorsal aorta (13, 41, 42). However, a complete inhibition of Hh signaling by cyclopamine led to nearly absent expression of tbx20 in the trunk region (SI Appendix, Fig. S4 J and K), implying that the phenotype severity might be dependent on the Hh signaling level. To investigate whether Notch signaling activation can rescue the HSPC phenotype, we utilized a system for HS–inducible overexpression of NICD, which is a dominant activator of Notch signaling. The function of NICD overexpression on Notch signaling activation was confirmed (SI Appendix, Fig. S4L). qRT-PCR showed that the reduced expression of Notch downstream gene hey2 in the cpeb1b morphants was rescued by NICD overexpression (SI Appendix, Fig. S4M), and WISH showed that the HSPC phenotype in the cpeb1b morphants was also rescued by NICD overexpression (SI Appendix, Fig. S4 N and O). Combined with the previous data, these results suggest that the Hedgehog–Vegf–Notch signaling axis is involved in Cpeb1b-mediated regulation of HSPC development.
Cpeb1b Interacts with shha mRNA in the Liquid-Like Condensates.
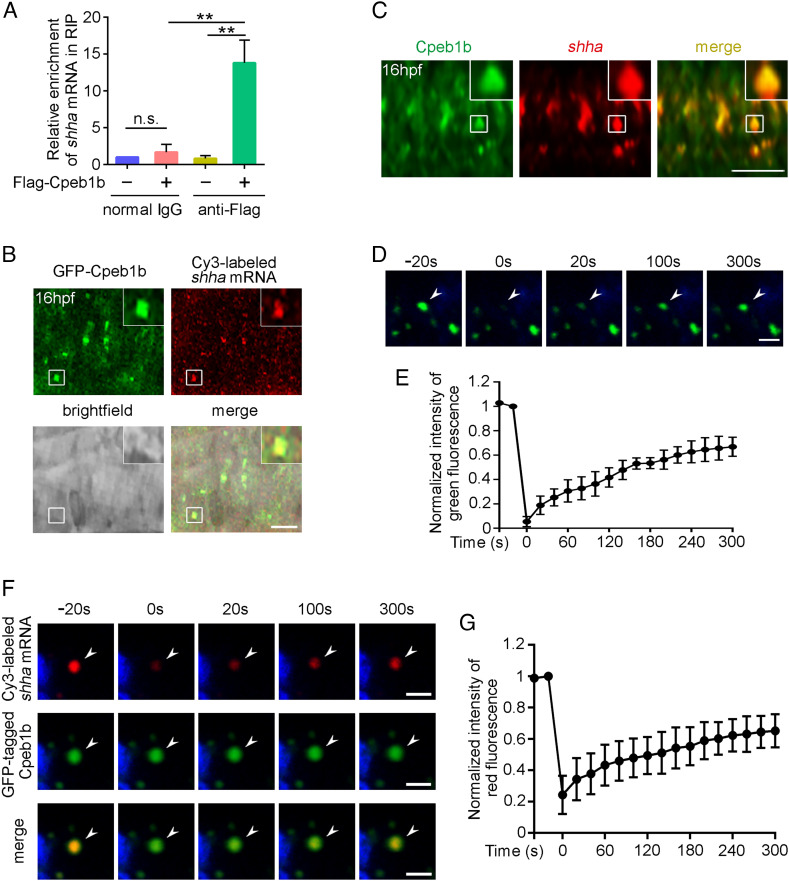
To further explore the molecular mechanism of Cpeb1b in HSPC development, we performed RNA immunoprecipitation (RIP)-seq by using Flag-Cpeb1b to identify target mRNAs of Cpeb1b in embryos. Intriguingly, the result showed that Cpeb1b coimmunoprecipitated with shha (Dataset S2), which is important for Hh signaling induction (10, 40). A previous study has demonstrated that the definitive hematopoiesis is defective in the zebrafish shha mutant, sonic-you (syu) (10). Consistently, we knocked down Shha by injection of shha MO and found that the runx1 expression was severely reduced in the shha morphants (SI Appendix, Fig. S5 A and B), which is similar to that in the shha mutant. Together with the observation that cpeb1b was colocalized with the shha in the notochord region of 16 hpf zebrafish embryo by FISH, we thus focused on shha mRNA for subsequent analysis. The interaction between Cpeb1b and shha mRNA in RIP-seq was further confirmed by RIP-qPCR assay (Fig. 3A). To observe the subcellular localization of Cpeb1b and shha mRNA, we cotransfected GFP-cpeb1b plasmid and Cy3-labeled shha mRNA into HEK293T cells. Confocal imaging showed that GFP-Cpeb1b colocalized with Cy3-labeled shha mRNA in a granular pattern in the cytoplasm of HEK293T cells (SI Appendix, Fig. S5C). To determine whether this also occurs in zebrafish embryo, we coexpressed GFP-Cpeb1b and Cy3-shha in embryos and found that GFP-Cpeb1b colocalized with Cy3-shha in a similar granular pattern in the notochord region of 16 hpf embryos (Fig. 3B). Furthermore, we detected the endogenous shha mRNA by FISH and utilized the immunofluorescence (IF) to detect the endogenous Cpeb1b. The confocal imaging showed similar result (Fig. 3C). Previous studies showed that condensation has different effects on movement and behavior of molecules (43–45), then we wondered whether the molecules in these Cpeb1b-positive condensates were trapped or dynamic. Fluorescence recovery after photobleaching (FRAP) assay was carried out in HEK293T cells by using a 488-nm laser, and the result showed recovery of GFP-Cpeb1b fluorescence within the bleached region (Fig. 3 D and E and Movie S2), suggesting that Cpeb1b molecules in these condensates could diffuse and exchange with surrounding dilute solution. Moreover, pairs of the GFP-Cpeb1b droplets could fuse and combine into one droplet (SI Appendix, Fig. S5D and Movie S3), further demonstrating liquid-like characteristics of these condensates. In addition, we performed FRAP on Cy3-shha by using a 561-nm laser and found recovery of Cy3-shha fluorescence within the bleached region (Fig. 3 F and G and Movie S4), suggesting that Cy3-shha in these condensates could also freely exchange with surrounding dilute solution. Taken together, these results above illustrate that Cpeb1b interacts with shha mRNA in the liquid-like condensates. Previous studies reported that some of the CPEB family members can undergo liquid–liquid phase separation by their prion-like domains (PLDs) (30, 46–48), then we wondered whether Cpeb1b also has this character. We analyzed the PLDs of zebrafish CPEB family members by using the prion-like amino acid composition (PLAAC) algorithm (49) and surprisingly found that there were no any PLDs within Cpeb1b, which was different from other CPEB family members (SI Appendix, Fig. S5E). Moreover, we performed in vitro cell-free phase separation assay by using purified Cpeb1b protein, which was labeled with Alexa488, and found that Cpeb1b did not form condensates (SI Appendix, Fig. S5F), indicating no phase separation ability of Cpeb1b itself. Considering that CPEB family proteins interact with poly(A)-binding protein (PABP) family proteins within cytoplasmic polyadenylation complex (24, 25), and many PABP family proteins have PLDs (50), we next asked whether formation of these condensates relies on the PABP family. Through searching the previous zebrafish gene expression pattern data, we found that pabpc1b was highly expressed in the notochord region during somitic period (51), which is consistent with cpeb1b expression. This finding was also confirmed by our WISH (SI Appendix, Fig. S6A). To reveal the subcellular localization of Pabpc1b, we cotransfected mCherry-pabpc1b and GFP-cpeb1b plasmids into HEK293T cells. Confocal imaging showed that mCherry-Pabpc1b colocalized with GFP-Cpeb1b in the cytoplasmic condensates (SI Appendix, Fig. S6B). Consistently, mCherry-Pabpc1b also colocalized with GFP-Cpeb1b in punctate pattern in the notochord region of 16 hpf embryos (SI Appendix, Fig. S6C). We analyzed the PLDs of Pabpc1b using the PLAAC algorithm and found that there indeed existed PLDs, which was similar to the ubiquitously expressed human homologous gene PABPC1 (SI Appendix, Fig. S6D). Then, we performed in vitro phase separation assay by using Alexa488-labeled Pabpc1b protein. The imaging showed that Pabpc1b formed spherical condensates in 150 mM but not 1 M KCl condition (SI Appendix, Fig. S6E), and FRAP showed recovery of Alexa488-Pabpc1b fluorescence within the bleached droplets (SI Appendix, Fig. S6 F and G and Movie S5). These results suggest that Pabpc1b was able to undergo phase separation, and the condensates formed by Pabpc1b were sensitive to high ionic strength, which can severely affect electrostatic interactions. To examine the intracellular Pabpc1b phase separation, we transfected mCherry-pabpc1b plasmid into HEK293T cells and found that Pabpc1b alone formed very few of condensates (SI Appendix, Fig. S6 H and I). By contrast, cotransfection of pabpc1b and cpeb1b led to much more condensates (SI Appendix, Fig. S6 H and I), implying that Cpeb1b might promote intracellular Pabpc1b phase separation. In addition, we knocked down Pabpc1b by pabpc1b MO injection and found that the runx1 expression at 36 hpf was reduced in the pabpc1b morphants compared with control (SI Appendix, Fig. S6 J and K), suggesting that Pabpc1b is also important for HSPC production.
Fig. 3.
Cpeb1b interacts with shha mRNA in the liquid-like condensates of cytoplasm. (A) Relative mRNA level of shha in normal IgG and anti-Flag groups examined by qRT-PCR. Error bar, mean ± SD. The P value was calculated by Student’s t test, n.s.: no significance, **P < 0.01. (B) Confocal imaging shows colocalization of GFP-Cpeb1b and Cy3-labeled shha mRNA in the notochord of 16 hpf embryos. (Scale bar, 10 μm.) (C) The endogenous shha mRNA was detected by FISH, and the endogenous Cpeb1b was detected by IF using an antibody that can specifically recognize the antigen (444 to 468 amino acids) in Cpeb1b. (Scale bar, 10 μm.) (D) The representative images of fluorescence recovery of GFP-Cpeb1b droplets. (Scale bar, 2 μm.) (E) Relative quantification of fluorescence recovery kinetics of GFP-Cpeb1b droplets. Three condensates were tested, and three condensates were recovered. The lapsed time was 320 s. The black curve shows the mean ± SD (n = 3). (F) The representative images of fluorescence recovery of Cy3-labeled shha mRNA. (Scale bar, 2 μm.) (G) Relative quantification of fluorescence recovery kinetics of Cy3-labeled shha mRNA. Three condensates were tested, and three condensates recovered. The lapsed time was 320 s. The black curve shows the mean ± SD (n = 3).
The CPE Motif in shha mRNA Is Crucial for Cpeb1b Binding.
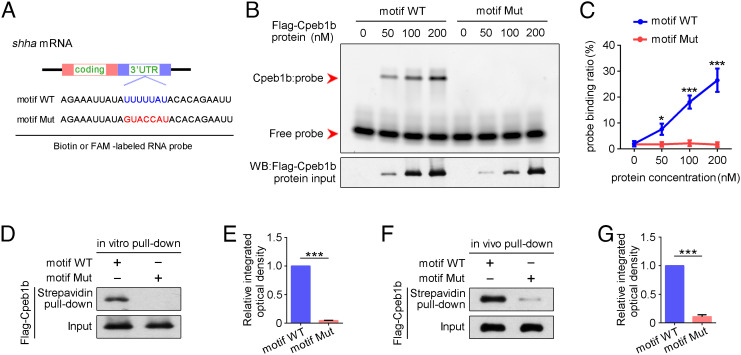
Previous studies demonstrated that the specificity of CPEB protein binding relies on the 3′ UTR-located U-rich CPE motif (consensus sequence UUUUUAU) (24, 25), which was also found in shha mRNA (Fig. 4A), then we wondered whether this CPE motif is required for Cpeb1b binding. To examine whether Cpeb1b recognizes the CPE motif in 3′ UTR of shha mRNA, we designed RNA probes with WT and mutant CPE motif (Fig. 4A) and performed electrophoretic mobility shift assay (EMSA). The purified Flag-Cpeb1b protein was mixed with an FAM-labeled RNA probe that contained WT or mutant CPE motif, and the protein–probe complexes were analyzed on a non–denaturing polyacrylamide gel. The EMSA showed retardation of the WT motif probe but not the mutant probe (Fig. 4 B and C), indicating that CPE motif of shha is important for Cpeb1b binding. We performed in vitro streptavidin-biotin pull-down assay by using purified Flag-Cpeb1b protein and biotin-labeled probes and found that Flag-Cpeb1b was only pulled down by probe containing WT CPE motif (Fig. 4 D and E). Moreover, the in vivo pull-down assay was carried out by using the extracts from Flag-cpeb1b mRNA-injected embryos, and the result was consistent with the in vitro pull-down assay (Fig. 4 F and G), further confirming specific interaction between Cpeb1b and the CPE motif of shha mRNA.
Fig. 4.
The CPEB motif in shha mRNA is important for Cpeb1b binding. (A) Diagram of biotin or FAM-labeled CPEB motif probe. The WT CPEB binding motif is in blue color, and the mutant CPEB binding motif is in red color. The sequence of motif WT and motif Mut probes is listed. (B) EMSA measuring the Cpeb1b binding with FAM-labeled motif WT and motif Mut probes. The protein concentration ranged from 0 nM to 200 nM. (C) Quantification of each band by gray analysis (Gel-Pro analyzer). The probe binding ratio at each group was determined by (RNA-protein)/[(free RNA) + (RNA-protein)]. GraphPad Prism was used for statistical analysis with Student’s t test. Statistic data were shown as mean ± SD. The P value was used for significance evaluation, *P < 0.05, ***P < 0.001. (D) WB showing the purified Flag-Cpeb1b protein pulled down with biotin-labeled CPEB motif probe. (E) Quantification of relative Flag-Cpeb1b protein level using gray analysis. Error bar, mean ± SD. The P value was calculated by Student’s t test, ***P < 0.001. (F) WB showing the Flag-Cpeb1b protein from Flag-cpeb1b mRNA-injected embryo extracts pulled down with biotin-labeled CPEB motif probe. (G) Quantification of relative Flag-Cpeb1b protein level using gray analysis. Error bar, mean ± SD. The P value was calculated by Student’s t test, ***P < 0.001.
Cpeb1b Regulates Cytoplasmic Polyadenylation of shha mRNA.
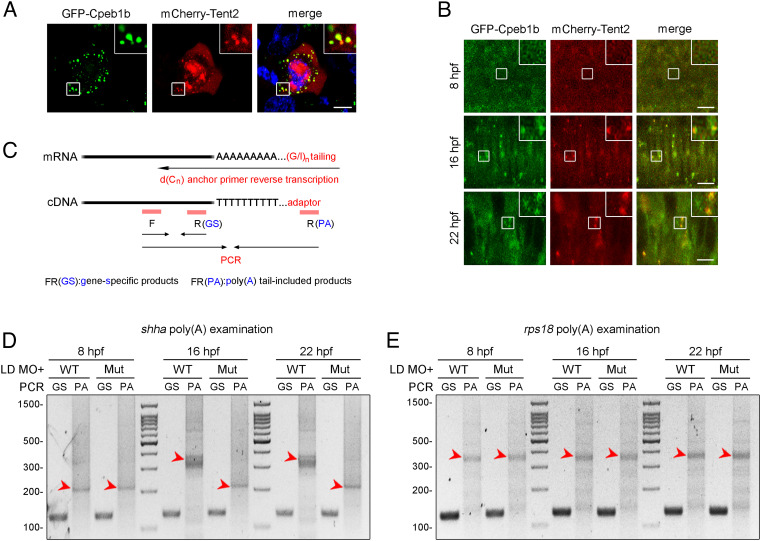
In Xenopus and mammals, CPEB-regulated cytoplasmic polyadenylation is reported to rely on the CPEB partner Tent2, which as a noncanonical poly(A) polymerase is response for cytoplasmic poly(A) lengthening (26, 27). Based on these previous reports, we asked whether Cpeb1b interacts with Tent2 to facilitate cytoplasmic polyadenylation. We cotransfected GFP-cpeb1b and mCherry-tent2 plasmids into HEK293T cells. Confocal imaging showed that GFP-Cpeb1b colocalized with mCherry-Tent2 in the cytoplasmic condensates (Fig. 5A), suggesting interaction between these two proteins. We further examined the localization of these proteins in embryos at different developmental stages and observed that GFP-Cpeb1b colocalized with mCherry-Tent2 in the condensates in the notochord of 16 and 22 hpf embryos, but not in the hypoblast of 8 hpf embryos (Fig. 5B). To further address whether Cpeb1b, Pabpc1b, Tent2, and shha mRNA form a complex together, we utilized a multicolor fluorescent labeling system. The confocal imaging showed the colocalization of GFP-Cpeb1b, mCherry-Pabpc1b, BFP-Tent2, and Cy5-shha mRNA in the cytoplasmic condensates of HEK293T cells (SI Appendix, Fig. S7A), suggesting that these molecules can form a complex together. Then, we examined the poly(A) length of shha mRNA in embryos at different developmental stages by PCR poly(A) test (PAT) assay (Fig. 5C) (52). Gel electrophoresis showed that the poly(A) length of shha at 8 hpf was shorter than that at 16 or 22 hpf, and the poly(A) length between 16 and 22 hpf was similar (SI Appendix, Fig. S7B), suggesting an important poly(A) length shift between 8 and 16 hpf. Next, we investigated the Cpeb1b-regulated cytoplasmic polyadenylation. The PAT assay revealed that the poly(A) tail length of shha mRNA in the cpeb1b mutants was decreased at 16 and 22 hpf, but not at 8 hpf, compared with that in the WT (Fig. 5D). By contrast, there was no difference in the poly(A) tail length of rps18 mRNA, which was not the Cpeb1b target transcript, between the WT and mutant embryos at different developmental stages (Fig. 5E). Taken together, these results indicate that Cpeb1b regulates cytoplasmic polyadenylation of shha mRNA.
Fig. 5.
Cpeb1b regulates cytoplasmic polyadenylation of shha mRNA. (A) Confocal imaging shows colocalization of GFP-Cpeb1b and mCherry-Tent2 in the cytoplasm of HEK293T cells. (Scale bar, 10 μm.) (B) Confocal imaging shows that GFP-Cpeb1b colocalizes with mCherry-Tent2 in the condensates in the notochord of 16 and 22 hpf embryos but not in the hypoblast of 8 hpf embryos. (Scale bar, 10 μm.) (C) Outline of the PAT assay. Cytoplasmic RNA extracted from the embryos was subjected to G/I tailing. The complementary DNA was synthesized by reverse transcription using the poly(G/I) tailed RNA as template. Then, the specific PCR products were synthesized by specific primers and detected by gel electrophoresis. GS: gene-specific products, PA: poly(A) tail-included products. (D) Examination of the poly(A) length of shha mRNA in the LD cpeb1b MO-injected WT and cpeb1b mutant embryos (elimination of maternal effects) at different developmental stages by PAT assay. The PCR products were detected by gel electrophoresis. The red arrowheads denote smear bands from poly(A) PCR products. (E) Examination of the poly(A) length of rps18 mRNA in the LD cpeb1b MO-injected WT and cpeb1b mutant embryos at different developmental stages. The red arrowheads denote smear bands from poly(A) PCR products.
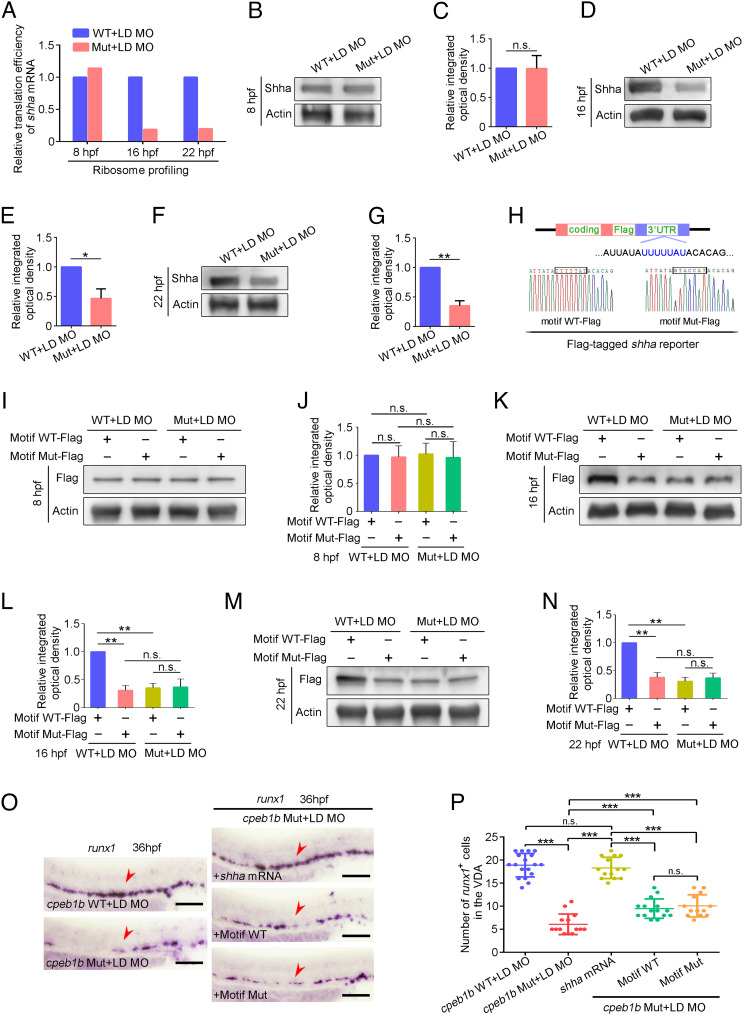
Cpeb1b Facilitates Shha Protein Expression through Translational Control.
Previous studies showed that polyadenylation selectively regulates mRNA stability or translation and further affects protein level in different biological processes (20), then we asked whether Cpeb1b-mediated cytoplasmic polyadenylation is involved in regulation of mRNA metabolism and protein level during HSPC development. We first examined the expression level of shha mRNA in the control and cpeb1b morphants at 16 hpf by WISH and surprisingly found no difference between the two groups (SI Appendix, Fig. S7 C and D). To examine the stability of shha mRNA, we treated the embryos with α-amanitin for inhibition of polymerase II-mediated nascent mRNA transcription and detected the relative level of shha mRNA in the control and cpeb1b morphants at the indicated time intervals by qRT-PCR. The 18s rRNA, which is transcribed by polymerase I and not sensitive to α-amanitin, was used as an internal control. The qRT-PCR result revealed that Cpeb1b deficiency did not alter the stability of shha mRNA (SI Appendix, Fig. S7E). Next, to explore whether the translational efficiency of shha mRNA was affected, we performed ribosome profiling assays and found that the global and shha mRNA translational efficiency in the cpeb1b mutants was decreased at 16 and 22 hpf, but not at 8 hpf (Fig. 6A, SI Appendix, Fig. S7 F–H, and Dataset S3). Consistently, WB analysis showed a decreased endogenous Shha protein level (Fig. 6 B–G). Furthermore, to determine whether the shha mRNA translation is CPE motif-dependent, we designed shha-Flag reporter mRNAs with WT and mutant CPE motif (Fig. 6H). After validating the reporter mRNA stability (SI Appendix, Fig. S7I), we showed that in WT embryos, the Shha-Flag expression was reduced in mutant CPE motif groups at 16 and 22 hpf, but not at 8 hpf, compared with that in the WT CPE motif groups (Fig. 6 I–N). However, in the cpeb1b mutant embryos, there was no difference between the two groups that contained different reporters (Fig. 6 I–N). In addition, we synthesized another Cpeb1b nontarget and polyadenylated shha mRNA as control. This mRNA, whose 3′UTR is from SV40 Poly(A) terminator, was in vitro polyadenylated. We respectively overexpressed these mRNAs in the cpeb1b mutant embryos and found that the polyadenylated shha mRNA almost fully rescued the HSPC defect (Fig. 6 O and P). However, the CPE motif WT and motif Mut mRNA had only a little but similar rescue effect (Fig. 6 O and P). Collectively, these results indicate that Cpeb1b regulates the Shha protein level though translational control of shha mRNA.
Fig. 6.
Cpeb1b regulates translation of shha mRNA. (A) Relative translation efficiency of shha mRNA in the LD cpeb1b MO injected WT and cpeb1b mutant embryos examined by ribosome profiling. (B) Protein level of endogenous Shha and Actin in the LD cpeb1b MO-injected WT and cpeb1b mutant embryos at 8 hpf examined by WB. (C) Quantification of relative Shha protein level using gray analysis. Two replicates, error bar, mean ± SD. The P value was calculated by Student’s t test, n.s.: no significance. (D) Protein level of endogenous Shha and Actin in the LD cpeb1b MO-injected WT and cpeb1b mutant embryos at 16 hpf examined by WB. (E) Quantification of relative Shha protein level using gray analysis. Two replicates, error bar, mean ± SD. The P value was calculated by Student’s t test, **P < 0.01. (F) Protein level of endogenous Shha and Actin in the LD cpeb1b MO-injected WT and cpeb1b mutant embryos at 22 hpf examined by WB. (G) Quantification of relative Shha protein level using gray analysis. Two replicates, error bar, mean ± SD. The P value was calculated by Student’s t test, *P < 0.05. (H) Diagram of Flag-tagged shha reporter. The CPEB binding motif is in blue color, and partial sequence of the motif WT and motif Mut reporters is listed. (I) Protein level of Shha-Flag and Actin in Motif WT-Flag or Motif Mut-Flag overexpressed WT and cpeb1b mutant embryos at 8 hpf examined by WB. Elimination of maternal effects was carried out by the LD cpeb1b MO injection. (J) Quantification of relative Shha-Flag protein level using gray analysis. Two replicates, error bar, mean ± SD. The P value was calculated by Student’s t test, n.s.: no significance. (K) Protein level of Shha-Flag and Actin in Motif WT-Flag or Motif Mut-Flag overexpressed WT and cpeb1b mutant embryos at 16 hpf examined by WB. (L) Quantification of relative Shha-Flag protein level using gray analysis. Two replicates, error bar, mean ± SD. The P value was calculated by Student’s t test, n.s.: no significance, **P < 0.01. (M) Protein level of Shha-Flag and Actin in Motif WT-Flag or Motif Mut-Flag overexpressed WT and cpeb1b mutant embryos at 22 hpf examined by WB. (N) Quantification of relative Shha-Flag protein level using gray analysis. Two replicates, error bar, mean ± SD. The P value was calculated by Student’s t test, n.s.: no significance, **P < 0.01. (O) Examination of the expression of runx1 by WISH. The red arrowheads denote HSPCs. LD: a low dose of cpeb1b MO injection. In HSPC phenotype rescue experiments using cpeb1b mutant embryos, these embryos were obtained by cross-mating cpeb1b adult mutants and individually subjected to genotyping after WISH experiment for identifying homozygous mutants. (Scale bar, 100 μm.) (P) Statistical analysis of the WISH. Error bar, mean ± SD, n.s.: no significance, ***P < 0.001.
Cpeb1b Deficiency Mildly Affects the Development of Nervous System and Somite, but Not Pectoral Fin.
In consideration of the role of Hh signaling is broad and not restricted to HSPC development (10, 53), we performed additional experiments to explore whether Cpeb1b-deficiency affects other organs and systems. WISH showed that the expression of muscle cell marker myod (54, 55) at 18 and 24 hpf was slightly reduced in the cpeb1b morphants compared with control (SI Appendix, Fig. S8 A and B), suggesting a mild somite developmental defect induced by Cpeb1b-deficiency. In addition, the fkd4, which is another downstream gene of Hh signaling (54, 56), expression in the hindbrain and ventral neural tube of cpeb1b morphants was also slightly reduced (SI Appendix, Fig. S8 C and D), suggesting a mild nervous system developmental defect. However, the pectoral fin development, which is also regulated by Hh signaling (54), was normal in the cpeb1b morphants (SI Appendix, Fig. S8 E and F).
Discussion
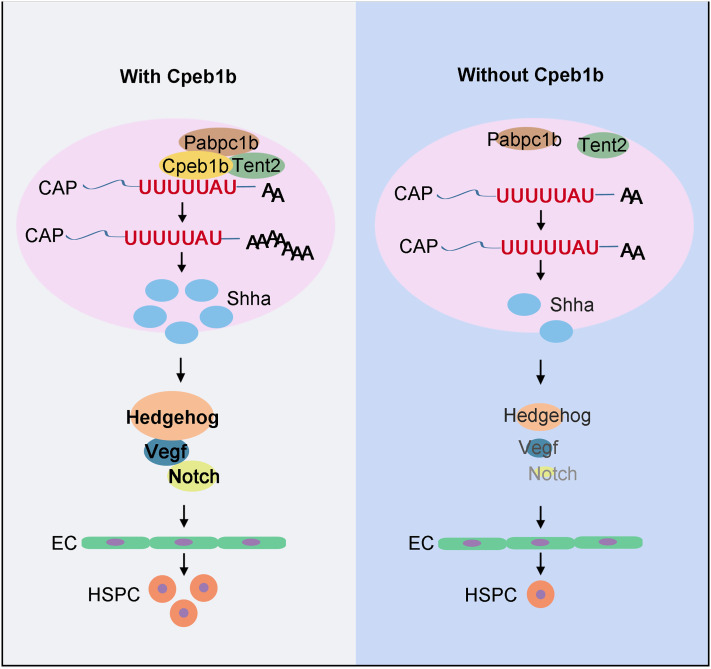
During HSPC development, how the signals are elaborately regulated to ensure the in vivo cell fate transition is a critical biological question. In this study, we have investigated the role of Cpeb1b in HSPC production and demonstrated the function of cytoplasmic polyadenylation in this developmental process in the zebrafish model. We find that the cytoplasmic polyadenylation regulator Cpeb1b is highly expressed in the notochord region during somitic period and is required for HSPC emergence. The Cpeb1b-deficiency leads to downregulation of the Hedgehog–Vegf–Notch signaling axis, further resulting in HE specification defect. Mechanistically, Cpeb1b interacts with shha mRNA through the CPE motif in the liquid-like condensates, and Cpeb1b-deficiency inhibits cytoplasmic polyadenylation of shha mRNA, further impairing its translation. Altogether, these findings suggest that Cpeb1b-mediated cytoplasmic polyadenylation plays an important role in HSPC development through translational control of Hh signaling (Fig. 7).
Fig. 7.
Schematic diagram showing that Cpeb1b modulates HSPCs development via cytoplasmic polyadenylation of shha mRNA. Cpeb1b-mediated cytoplasmic polyadenylation of shha mRNA is important for enhancing its translation efficiency and therefore increases the Shha protein level. An increase in the number of Shha protein is required for definitive hematopoiesis by regulating Hedgehog–Vegf–Notch signaling axis. In the absence of Cpeb1b, the Shha protein level is reduced, which results in downregulation of Hedgehog–Vegf–Notch signaling axis, further leading to definitive hematopoiesis defect.
In vertebrates, Hh signaling, as a crucial developmental signal, is regulated by various factors to improve the signal transduction accuracy. At the transcriptional level, HNF3β transcription factor binds to the specific HNF3β motif in shh promoter and activates shh transcription, and the misexpression of HNF3β results in shh ectopic expression (57–59). Moreover, the retinoic acid receptors and retinoid X receptors heterodimers bind to the RA response element in shh promoter and can respond to retinoic acid (RA) stimulation, further inducing RA-dependent shh transcription (59, 60). At the posttranslational level, the covalent modification by cholesterol at the N terminus of Hh protein is essential for restricting its spatial deployment, subsequently affecting cellular response (61). Besides, palmitoylation modification at the N terminus of Hh protein, which is promoted by the ski/HHAT acyl transferase, facilitates membrane association and secretion of Hh ligand (62). In addition to these transcriptional and posttranslational regulations of Hh signaling, we discover in this HSPC developmental study that Cpeb1b-mediated cytoplasmic polyadenylation may supply another layer of posttranscriptional regulation of Hh signaling. We show that 1) the expression of cpeb1b is enriched in notochord region during somitic period and colocalizes with shha, which contributes to spatial and temporal specificity at the tissue level; 2) the interaction between Cpeb1b and shha mRNA is dependent on the particular CPE motif and is important for regulating translation but not stability of shha mRNA, contributing to specificity at the molecular level. Although there are many other mRNAs binding to Cpeb1b in the RIP-seq data, based on a series of experiments on phenotype and signaling pathway analysis, we speculate that the shha should be the main and direct target responsible for the HSPC defect in the Cpeb1b-deficient embryos. Moreover, multiple signals besides Hh were altered in Cpeb1b-deficient embryos in our GO analysis. The signals, such as canonical glycolysis, histone ubiquitination, cellular ion homeostasis, response to unfolded protein, and vitamin metabolic process, are involved in physiological-biological activities of glucose homeostasis, histone modification, ion transport, unfolded protein response, and vitamin metabolism regulation. Whether these signals are indeed regulated by Cpeb1b as well as being involved in HSPC development awaits further investigation.
In addition, we show that Cpeb1b-deficiency mildly affects the development of nervous system and somite, which is known to be regulated by Hh signaling (54–56), suggesting that the role of Cpeb1b is not strictly restricted to HSPC development, and Cpeb1b-deficiency–induced HSPC defect is part of a more widespread developmental defects in different organs. Surprisingly, we find that the pectoral fin development, which is also reported to be regulated by Hh signaling (54), is not affected by Cpeb1b-deficiency. In consideration of the inactivation of Hh signaling in Cpeb1b-deficient embryos is incomplete and the development of different tissues depends on distinct levels of Hh signaling activity (53, 63), we speculate that zebrafish HSPC production may be more sensitive to Hh signaling alteration than pectoral fin development and then could be affected by Cpeb1b-deficiency more easily.
Biomolecular condensates as important cellular biochemistry organizers can not only influence fundamental properties of biomolecules but also provide membrane-less compartments for precise biochemical reaction, which may contribute to ensuring accuracy of signal transduction (43–45). Previous studies report that some proteins of CPEB family have the prion-like characters and can undergo aggregation, which is nonpathological but critical for their physiological functions. In Aplysia, the multimerization of ApCPEB, which is induced by its N-terminal PLD, is enhanced by serotonin stimulation and is essential for the maintenance of long-term facilitation by upregulation of local protein synthesis in neurons (46, 64). Similarly, the oligomerization of Drosophila CPEB Orb2 is critical for its function in translational control and persistence of long-term memory (30, 47). In addition, the PLD-mediated aggregation of mouse CPEB3 promotes the translation of AMPA receptors and facilitates persistence of memory (65). In this study, we show that Cpeb1b colocalizes with shha mRNA and poly(A) polymerase Tent2 in the cytoplasmic condensates under normal conditions. Based on the results of FRAP and granule fusion assays, we find that the molecules in these condensates are dynamic, suggesting liquid-like but not pathological or gel-like characteristics of these condensates. Intriguingly, Cpeb1b does not have PLDs and cannot undergo aggregation by itself, which is different from other CPEB members. Nevertheless, Cpeb1b colocalizes with Pabpc1b and efficiently promotes Pabpc1b’s phase separation ability, which further induces the condensate formation. Although this condensation phenomenon in Cpeb1b-mediated cytoplasmic polyadenylation is evident, the detailed molecular mechanism remains elusive. At the subcellular level, condensation changes the spatial distribution and local quantity of biomolecules (43–45, 66). We speculate that the condensation in cytoplasmic polyadenylation may increase the local concentration of specific enzymes and mRNAs, further improving the accuracy and efficiency of catalytic reaction. However, more studies are awaited to demonstrate this point.
Previous study showed that in Hodgkin’s lymphoma-derived cell line, the CPEB1-coordinated alternative polyadenylation and splicing with translational regulation are involved in regulation of genes important for hematologic malignancy progression (67). Moreover, the CPEB1 expression was reported to be significantly reduced in multiple myeloma cells (68), implying that CPEB1 may be related to myeloma progression. However, the role of CPEB1 in normal hematopoiesis in animal models has not been elucidated thus far. Our study shows that Cpeb1b-mediated cytoplasmic polyadenylation is important for zebrafish HSPC production through translational control during early embryogenesis. The appropriate cytoplasmic polyadenylation of shha mRNA facilitates its own translation in the notochord region and therefore activates the Hh signaling during the critical phase of HSPC development. These findings may provide some clues to answer how developmental signals are precisely controlled by posttranscriptional regulation and direct ECs to execute fate transition to HSPCs.
Materials and Methods
The information of ethics statement, zebrafish husbandry, MOs, vector construction, mRNA synthesis, microinjection, WISH, double FISH, generation of mutant by Crispr/Cas9, quantitative RT-PCR, WB, cell culture and transfection, protein purification, in vitro condensates formation assay, microscopy imaging, FRAP assay, EMSA, in vitro RNA pulldown assay, in vivo RNA pulldown assay, PAT assay, RNA-seq, GO analysis, RIP experiment, and ribosome profiling experiment are in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
The blood flow in the control and Cpeb1b-deficient embroys. Time-lapse imaging was performed to observe the blood flow in the control and Cpeb1b morphants. Scale bar, 100 μm.
Fluorescence signal recovery of GFP-Cpeb1b droplet. Time-lapse imaging was performed to observe the fluorescence signal recovery of GFP-Cpeb1b droplet in HEK293T cells. A bleaching 488 nm laser pulse was applied. In this representative movie, one condensate was tested, and one condensate recovered. The lapsed time was 420 s. Scale bar, 10 μm.
The fusion of GFP-Cpeb1b droplet. Time-lapse imaging was performed to observe the fusion of GFP-Cpeb1b droplet in HEK293T cells. In this representative movie, twenty condensates were showed, and four condensates fused. The lapsed time was 180 s. Scale bar, 2 μm.
Fluorescence signal recovery of Cy3-labeled shha mRNA. Time-lapse imaging was performed to observe the fluorescence signal recovery of Cy3-labeled shha mRNA in HEK293T cells. A bleaching 561 nm laser pulse was applied. In this representative movie, one condensate was tested, and one condensate recovered. The lapsed time was 420 s. Scale bar, 10 μm.
Fluorescence signal recovery of Alexa488-labeled Pabpc1b in vitro. Time-lapse imaging was performed to observe the in vitro fluorescence signal recovery of Alexa488-labeled Pabpc1b. A bleaching 488 nm laser pulse was applied. In this representative movie, one condensate was tested, and one condensate recovered. The lapsed time was 420 s. Scale bar, 2 μm.
Acknowledgments
We thank Lingfei Luo for providing us shha MO and Lu Wang, Suwei Gao, and Jun Xia for their assistance. This work was supported by grants from the National Natural Science Foundation of China (31830061, 32030032, 32100667, and 32121001), National Key Research and Development Program of China (2018YFA0800200, 2018YFA0801000, and 2019YFA0110900), the Strategic Priority Research Program of the Chinese Academy of Sciences, China (XDA16010207), and the China Postdoctoral Science Foundation (2021M703205).
Author contributions
J.H., Y.-G.Y., and F.L. designed research; J.H., B.S., J.-Y.Z., Y.Z., and D.M. performed research; J.H., B.S., and J.-Y.Z. analyzed data; and J.H., B.S., and F.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Jian Heng, Email: hengjian@ioz.ac.cn.
Yun-Gui Yang, Email: ygyang@big.ac.cn.
Feng Liu, Email: liuf@ioz.ac.cn.
Data, Materials, and Software Availability
The RNA-seq, RIP-seq, and ribosome profiling sequencing data supporting the conclusions of this article have been deposited in the Gene Expression Omnibus database (GEO: GSE207906).
Supporting Information
References
- 1.Clements W. K., Traver D., Signalling pathways that control vertebrate haematopoietic stem cell specification. Nat. Rev. Immunol. 13, 336–348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orkin S. H., Zon L. I., Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132, 631–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dzierzak E., Speck N. A., Of lineage and legacy: The development of mammalian hematopoietic stem cells. Nat. Immunol. 9, 129–136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boisset J. C., et al. , In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Bertrand J. Y., et al. , Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kissa K., Herbomel P., Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Chen M. J., Yokomizo T., Zeigler B. M., Dzierzak E., Speck N. A., Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887–891 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dzierzak E., Bigas A., Blood development: Hematopoietic stem cell dependence and independence. Cell Stem Cell 22, 639–651 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Peeters M., et al. , Ventral embryonic tissues and Hedgehog proteins induce early AGM hematopoietic stem cell development. Development 136, 2613–2621 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gering M., Patient R., Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev. Cell 8, 389–400 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Kim P. G., et al. , Signaling axis involving Hedgehog, Notch, and Scl promotes the embryonic endothelial-to-hematopoietic transition. Proc. Natl. Acad. Sci. U.S.A. 110, E141–150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigas A., Robert-Moreno A., Espinosa L., The Notch pathway in the developing hematopoietic system. Int. J. Dev. Biol. 54, 1175–1188 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Burns C. E., Traver D., Mayhall E., Shepard J. L., Zon L. I., Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19, 2331–2342 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanz D., Konantz M., Alghisi E., North T. E., Lengerke C., Endothelial-to-hematopoietic transition: Notch-ing vessels into blood. Ann. N. Y. Acad. Sci. 1370, 97–108 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Shi B., et al. , RNA structural dynamics regulate early embryogenesis through controlling transcriptome fate and function. Genome Biol. 21, 120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C., et al. , m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Komeno Y., et al. , SRSF2 is essential for hematopoiesis, and its myelodysplastic syndrome-related mutations dysregulate alternative Pre-mRNA splicing. Mol. Cell Biol. 35, 3071–3082 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X., et al. , miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res. 23, 1356–1368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishima Y., Tomari Y., Codon usage and 3’ UTR length determine maternal mRNA stability in Zebrafish. Mol. Cell 61, 874–885 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Passmore L. A., Coller J., Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell Biol. 23, 93–106 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartner J. C., Walkley C. R., Lu J., Orkin S. H., ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 10, 109–115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao N., et al. , Ott1 (Rbm15) regulates thrombopoietin response in hematopoietic stem cells through alternative splicing of c-Mpl. Blood 125, 941–948 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kidoya H., et al. , Regnase-1-mediated post-transcriptional regulation is essential for hematopoietic stem and progenitor cell homeostasis. Nat. Commun. 10, 1072 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter J. D., CPEB: A life in translation. Trends Biochem. Sci. 32, 279–285 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Mendez R., Richter J. D., Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2, 521–529 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Barnard D. C., Ryan K., Manley J. L., Richter J. D., Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119, 641–651 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Kwak J. E., Wang L., Ballantyne S., Kimble J., Wickens M., Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. U.S.A. 101, 4407–4412 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu L., et al. , CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron 21, 1129–1139 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Si K., et al. , A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell 115, 893–904 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Majumdar A., et al. , Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 148, 515–529 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Udagawa T., et al. , Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol. Cell 47, 253–266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tay J., Richter J. D., Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev. Cell 1, 201–213 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Groisman I., et al. , Control of cellular senescence by CPEB. Genes Dev. 20, 2701–2712 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng W., et al. , CPEB1 directs muscle stem cell activation by reprogramming the translational landscape. Nat. Commun. 13, 947 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivshina M., Alexandrov I. M., Vertii A., Doxsey S., Richter J. D., CPEB regulation of TAK1 synthesis mediates cytokine production and the inflammatory immune response. Mol. Cell Biol. 35, 610–618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postlethwait J. H., et al. , Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10, 1890–1902 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Wang L., et al. , Epigenetic regulation of left-right asymmetry by DNA methylation. EMBO J. 36, 2987–2997 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z., et al. , Primary cilia regulate hematopoietic stem and progenitor cell specification through Notch signaling in zebrafish. Nat. Commun. 10, 1839 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heretsch P., Tzagkaroulaki L., Giannis A., Cyclopamine and hedgehog signaling: Chemistry, biology, medical perspectives. Angew. Chem. Int. Ed Engl. 49, 3418–3427 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Lawson N. D., Vogel A. M., Weinstein B. M., Sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev. Cell 3, 127–136 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Konantz M., et al. , Evi1 regulates Notch activation to induce zebrafish hematopoietic stem cell emergence. EMBO J. 35, 2315–2331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heng J., et al. , Rab5c-mediated endocytic trafficking regulates hematopoietic stem and progenitor cell development via Notch and AKT signaling. PLoS Biol. 18, e3000696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Y., Protter D. S., Rosen M. K., Parker R., Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hyman A. A., Weber C. A., Julicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Si K., Choi Y. B., White-Grindley E., Majumdar A., Kandel E. R., Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 140, 421–435 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Khan M. R., et al. , Amyloidogenic oligomerization transforms Drosophila Orb2 from a translation repressor to an activator. Cell 163, 1468–1483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drisaldi B., et al. , SUMOylation is an inhibitory constraint that regulates the prion-like aggregation and activity of CPEB3. Cell Rep. 11, 1694–1702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lancaster A. K., Nutter-Upham A., Lindquist S., King O. D., PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riback J. A., et al. , Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thisse B., Thisse C., Fast release clones: A high throughput expression analysis. ZFIN Direct Data Submission (http://zfin.org) (2004). [Google Scholar]
- 52.Kusov Y. Y., Shatirishvili G., Dzagurov G., Gauss-Muller V., A new G-tailing method for the determination of the poly(A) tail length applied to hepatitis A virus RNA. Nucleic Acids Res. 29, E57–E67 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolff C., Roy S., Ingham P. W., Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 13, 1169–1181 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Schauerte H. E., et al. , Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development 125, 2983–2993 (1998). [DOI] [PubMed] [Google Scholar]
- 55.van Eeden F. J., et al. , Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development 123, 153–164 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Tyurina O. V., et al. , Zebrafish Gli3 functions as both an activator and a repressor in Hedgehog signaling. Dev. Biol. 277, 537–556 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Ruiz i Altaba A., Jessell T. M., Roelink H., Restrictions to floor plate induction by hedgehog and winged-helix genes in the neural tube of frog embryos. Mol. Cell Neurosci. 6, 106–121 (1995). [DOI] [PubMed] [Google Scholar]
- 58.Hynes M., Poulsen K., Tessier-Lavigne M., Rosenthal A., Control of neuronal diversity by the floor plate: Contact-mediated induction of midbrain dopaminergic neurons. Cell 80, 95–101 (1995). [DOI] [PubMed] [Google Scholar]
- 59.Chang B. E., Blader P., Fischer N., Ingham P. W., Strahle U., Axial (HNF3beta) and retinoic acid receptors are regulators of the zebrafish sonic hedgehog promoter. EMBO J. 16, 3955–3964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riddle R. D., Johnson R. L., Laufer E., Tabin C., Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401–1416 (1993). [DOI] [PubMed] [Google Scholar]
- 61.Beachy P. A., et al. , Multiple roles of cholesterol in hedgehog protein biogenesis and signaling. Cold Spring Harb. Symp. Quant. Biol. 62, 191–204 (1997). [PubMed] [Google Scholar]
- 62.Ingham P. W., Nakano Y., Seger C., Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Aanstad P., et al. , The extracellular domain of smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr. Biol. 19, 1034–1039 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miniaci M. C., et al. , Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron 59, 1024–1036 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fioriti L., et al. , The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron 86, 1433–1448 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Shi B., et al. , Phase separation of Ddx3xb helicase regulates maternal-to-zygotic transition in zebrafish. Cell Res. 32, 715–728 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bava F. A., et al. , CPEB1 coordinates alternative 3’-UTR formation with translational regulation. Nature 495, 121–125 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Heller G., et al. , Genome-wide transcriptional response to 5-aza-2’-deoxycytidine and trichostatin a in multiple myeloma cells. Cancer Res. 68, 44–54 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
The blood flow in the control and Cpeb1b-deficient embroys. Time-lapse imaging was performed to observe the blood flow in the control and Cpeb1b morphants. Scale bar, 100 μm.
Fluorescence signal recovery of GFP-Cpeb1b droplet. Time-lapse imaging was performed to observe the fluorescence signal recovery of GFP-Cpeb1b droplet in HEK293T cells. A bleaching 488 nm laser pulse was applied. In this representative movie, one condensate was tested, and one condensate recovered. The lapsed time was 420 s. Scale bar, 10 μm.
The fusion of GFP-Cpeb1b droplet. Time-lapse imaging was performed to observe the fusion of GFP-Cpeb1b droplet in HEK293T cells. In this representative movie, twenty condensates were showed, and four condensates fused. The lapsed time was 180 s. Scale bar, 2 μm.
Fluorescence signal recovery of Cy3-labeled shha mRNA. Time-lapse imaging was performed to observe the fluorescence signal recovery of Cy3-labeled shha mRNA in HEK293T cells. A bleaching 561 nm laser pulse was applied. In this representative movie, one condensate was tested, and one condensate recovered. The lapsed time was 420 s. Scale bar, 10 μm.
Fluorescence signal recovery of Alexa488-labeled Pabpc1b in vitro. Time-lapse imaging was performed to observe the in vitro fluorescence signal recovery of Alexa488-labeled Pabpc1b. A bleaching 488 nm laser pulse was applied. In this representative movie, one condensate was tested, and one condensate recovered. The lapsed time was 420 s. Scale bar, 2 μm.
Data Availability Statement
The RNA-seq, RIP-seq, and ribosome profiling sequencing data supporting the conclusions of this article have been deposited in the Gene Expression Omnibus database (GEO: GSE207906).