Abstract
Glucose-6-phosphate isomerase (phosphoglucose isomerase [PGI]) (EC 5.3.1.9) from the hyperthermophilic archaeon Pyrococcus furiosus was purified 500-fold to homogeneity. The enzyme had an apparent molecular mass of 43 kDa and was composed of a single type of subunit of 23 kDa indicating a homodimeric (α2) structure. Kinetic constants of the enzyme were determined at the optimal pH 7 and at 80°C. Rate dependence on both substrates followed Michaelis-Menten kinetics. The apparent Km values for glucose-6-phosphate and fructose-6-phosphate were 8.7 and 1.0 mM, respectively, and the corresponding apparent Vmax values were 800 and 130 U/mg. The enzyme had a temperature optimum of 96°C and showed a significant thermostability up to 100°C, which is in accordance with its physiological function under hyperthermophilic conditions. Based on the N-terminal amino acid sequence of the subunit, a single open reading frame (ORF; Pf_209264) was identified in the genome of P. furiosus. The ORF was characterized by functional overexpression in Escherichia coli as a gene, pgi, encoding glucose-6-phosphate isomerase. The recombinant PGI was purified and showed molecular and kinetic properties almost identical to those of the native PGI purified from P. furiosus. The deduced amino acid sequence of P. furiosus PGI did not reveal significant similarity to the conserved PGI superfamily of eubacteria and eucarya. This is the first description of an archaeal PGI, which represents a novel type of PGI.
Glucose-6-phosphate isomerase, or phosphoglucose isomerase (PGI) (EC 5.3.1.9), catalyzes the reversible isomerization of glucose-6-phosphate (G-6-P) to fructose-6-phosphate (F-6-P). PGI plays a central role in the sugar metabolism of members of the domains Bacteria and Eucarya, both in glycolysis via the Embden-Meyerhof (EM) pathway and in gluconeogenesis, where the enzyme operates in the reverse direction (29, 31).
In the Archaea domain, PGI was first demonstrated to be part of the gluconeogenetic pathways in various species of lithoautotrophic methanogens and in the sulfur-reducing lithoautotrophic Thermoproteus species (15, 35, 43, 51). In recent years, the pathways of sugar degradation have been studied in various hyperthermophilic archaea (44), such as the Euryarchaeota Pyrococcus furiosus and Thermococcus celer and the Crenarchaeota Desulfurococcus amylolyticus and Thermoproteus tenax. These hyperthermophiles have been found to degrade glucose, maltose, cellobiose, and starch via modified versions of the EM pathway. The modified pathways differ from the conventional EM pathway by the involvement of novel kinases, such as ADP-dependent hexokinase and ADP-dependent 6-phosphofructokinase (6-PFK), in P. furiosus and T. celer and of unusual enzymes of glyceraldehyde-3-phosphate oxidation, such as glyceraldehyde:ferredoxin oxidoreductase, in P. furiosus, T. celer, and D. amylolyticus and nonphosphorylating NAD+-reducing glyceraldehyde 3-phosphate dehydrogenase in T. tenax (5, 11, 17, 22, 38, 42). However, all these modified EM pathways involve the activity of a PGI catalyzing the isomerization of glucose-6-phosphate to fructose-6-phosphate.
PGIs have been purified and biochemically characterized for a variety of eucarya and bacteria. The genes encoding PGIs from various species have been cloned and sequenced, and crystal stuctures have been determined for the eukaryotic PGIs from pig, rabbit, and the bacterium Bacillus stearothermophilus (27, 29, 31, 41, 45). Multiple alignments of PGIs ranging from bacteria to mammals revealed two regions of conserved amino acids. These were assigned as signature patterns for the PGI superfamily (3).
To date, a PGI and its coding gene from the domain of archaea have not been characterized. During our studies of the sugar metabolism of the hyperthermophilic archaeon P. furiosus, high PGI activities have been detected both in maltose-grown cells and in pyruvate-grown cells, which perform gluconeogenesis (36, 37, 40).
Despite the fact that P. furiosus contained high activities of PGI, a gene showing significant similarity to the conserved PGI superfamily of eubacteria and eucarya could not be identified in the complete sequenced genomes of P. furiosus (website of Center of Marine Biotechnology UMBI, University of Maryland, for Blast archaeal genome sequences [http://combdna.umbi.umd.edu/bags.html#PfurInfo]), Pyrococcus horikoshii (21), and Pyrococcus abyssi (Pyrococcus abyssi home page at Genoscope [http://www.genoscope.cns.fr./Pab/]). This finding suggests that the PGI of P. furiosus might be significantly different from all known PGIs analyzed so far.
In this paper we report the purification and characterization of PGI from P. furiosus. The encoding gene, pgi, was identified and functionally expressed in Escherichia coli. The data indicate that this first characterized PGI from the domain of archaea represents a novel type of PGI, which is not related to the conserved PGI superfamily of bacteria and eucarya.
MATERIALS AND METHODS
Growth of the organism.
P. furiosus (DSM 3638) (13) was grown anaerobically at 90°C in a 100-1 Biostat fermentor on a complex medium containing starch as the carbon and energy source. The medium contained (per liter) 1 g of yeast extract, 5 g of peptone, 2 g of starch, 19.45 g of NaCl, 12.6 g of MgCl2 · 6 H2O, 0.16 g of NaHCO3, 3.24 g of Na2SO4, 2.38 g of CaCl2 · 2 H2O, 0.56 g of KCl, 0.1 g of sulfur, 0.5 g of cystein, 0.5 g of Na2S · 9 H2O, and 2 ml of trace element solution. The trace element solution contained (per liter) 40 g of KBr, 28.6 g of SrCl2 · 6 H2O, 11 g of H3BO3, 2 g of Na2SiO3 · 5 H2O, 1.2 g of NaF, 0.8 g of KNO3, and 5 g of Na2HPO4 · 2 H2O. Cells were grown and harvested (after 23 h) at the late exponential growth phase. About 120 g (wet weight) of cells were obtained from the 100-1 Biostat fermentor.
Preparation of cell extracts and purification of PGI.
Since the enzyme was not sensitive to oxygen, all steps of the purification procedure were carried out under oxic conditions at 4°C. Cell extracts were prepared from 20 g (wet weight) of frozen cells, which were suspended in 60 ml of 50 mM Tris-HCl, pH 7.5 (buffer A), containing 20 mM NaCl, 2 mM DTE, 2 mM EDTA, and the protease inhibitors (each 2 mg) aprotinin, leupeptin, and phenylmethylsulfonyl fluoride. Cells were disrupted by passing through a French pressure cell at 1.3 × 108 Pa. Cell debris and unbroken cells were removed by centrifugation for 60 min at 100,000 × g at 4°C.
The 100,000 × g supernatant was applied to a Q Sepharose Hiload column (10 by 2.6 cm) that had been equilibrated with buffer A. Protein was eluted at a flow rate of 2 ml/min with 250 ml of 20 mM piperazine (pH 5.8; 25°C; buffer B) as well as with two linear NaCl gradients in buffer B: 0 to 0.5 M NaCl (500 ml) and 0.5 to 1 M NaCl (70 ml). Fractions containing the highest PGI activity (42 ml, 0.1 to 0.2 M NaCl) were pooled and adjusted by the addition of both solid (NH4)2SO4 and solid Tris to a final 2 M (NH4)2SO4 and pH 7.5. The protein solution (42 ml) was applied to a phenyl-Sepharose Hiload column (10 by 2.6 cm) equilibrated with 20 mM Tris-HCl, pH 7.5 (buffer C), and was washed with 85 ml of buffer C. Protein was desorbed at a flow rate of 2 ml/min with two decreasing gradients, from 2 to 1.5 M and 1.5 to 0 M (NH4)2SO4, in buffer C (600 ml). The fractions containing the highest PGI activity [45 ml, 1 to 0.8 M (NH4)2SO4] were pooled and concentrated to a volume of 2 ml by ultrafiltration (exclusion size, 10 kDa). The concentrated protein solution was applied to a Superdex 200 gel filtration column (60 by 1.6 cm) equilibrated with 50 mM Tris-HCl, pH 7.5, containing 100 mM NaCl. Protein was eluted at a flow rate of 1 ml/min. The PGI-containing fractions were recovered between 77.5 and 85.5 ml and were pooled and applied to a Uno Q1 column (1 ml) equilibrated with 20 mM Tris-HCl, pH 9 (buffer D). Protein was eluted at a flow rate of 1 ml/min with a linear gradient of 0 to 0.5 M NaCl (15 ml). The fractions containing the highest PGI activity were eluted between 0.23 and 0.28 M NaCl. At this stage PGI was essentially pure. Purified PGI was stored in 1-ml fractions at −20°C. Under these conditions enzyme activity remained nearly constant for several months.
Analytical assays.
The purity of the preparations was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 14% gels followed by staining with Coomassie brilliant blue R 250 according to standard procedures (23). Protein concentrations were determined by the method of Bradford (4) with bovine serum albumin as the standard. Gel filtration chromatography was carried out at ambient temperature on a Superdex 200 column (50 mM Tris-HCl, 150 mM NaCl, pH 7.0; 1 ml/min).
Determination of N-terminal amino acid sequence.
The purified protein was analyzed on a 13% polyacrylamide gel in the presence of 6 M urea following the procedure of Schaegger and von Jagow (34). Blotting onto a polyvinylidene difluoride membrane and N-terminal microsequencing on a model 473A sequencer (Applied Biosystems) were carried out as previously described (26).
Enzyme assays and determination of kinetic parameters.
Since the enzyme activity was not sensitive to oxygen, all assays were performed under oxic conditions. The PGI activity (G-6-P ⇌ F-6-P) was determined in both directions, using either a discontinous assay at 50 to 96°C or a continuous assay at 50°C or below. With both assay systems similar results were obtained at 50°C.
(i) Discontinuous assays.
In all discontinuous assays initial velocities were investigated in parallel assays stopped at different time intervals. The velocity remained linear for a ΔE up to 1.0 to 1.5 (except assays above 50°C and those at low substrate concentrations). One unit of PGI activity is defined as either the conversion of 1 μmol of G-6-P to F-6-P (forward reaction) or the formation of 1 μmol of F-6-P from G-6-P (reverse reaction). The auxiliary enzymes in all assays were routinely tested to ensure that they were not rate limiting; the units given were according to the specifications of the manufacturer, except for ADP-PFK.
The formation of F-6-P was measured by coupling it to the oxidation of NADH via PFK, F-1,6-BP aldolase, triosephosphate isomerase (TIM), and glycerol-phosphate-dehydrogenase. The standard assay mixture (250 μl) contained 50 mM Tris-HCl (pH 7.0; 80°C), 20 mM G-6-P, 2.5 mM ADP, 5 mM MgCl2, and 5 U of ADP-PFK (80°C). After preincubation at 80°C, the reaction was started with an aliquot of PGI (0.18 μg of enzyme), which was incubated for 1 to 4 min and stopped by rapid addition of 750 μl of ice-cold stop solution (50 mM Tris-HCl [pH 7 at 25°C], 0.5 mM NADH, 0.9 U of F-1,6-BP aldolase, 5 U of TIM, 1.5 U of glycerol-phosphate-dehydrogenase), and the oxidation of NADH at 365 nm was measured exactly 5 min later. The formation of G-6-P was investigated at 80°C by coupling it to the reduction of NADP+ via glucose 6-phosphate dehydrogenase (GPDH). The standard mixture (250 μl) contained 50 mM Tris-HCl (pH 7.0, 80°C). After preincubation at 80°C, the reaction was started with an aliquot of PGI (0.18 to 0.5 μg of enzyme), was incubated for 0.25 to 8 min, and was stopped by rapid addition of 750 μl of ice-cold stop solution (50 mM Tris-HCl [pH 7 at 25°C], 0.5 mM NADP+, 0.2 U of GPDH) to a final volume of 1 ml. The reduction of NADP+ was measured at 365 nm exactly 5 min later.
(ii) Continuous assays.
The formation of F-6-P from G-6-P was determined by measuring NADH oxidation in an assay mixture containing 100 mM Tris-HCl (pH 7.0), 40 mM G-6-P, 3 mM ATP, 5 mM MgCl2, 0.5 mM NADH, 1 U of PFK, 1 U of FBP aldolase, 50 U of TIM, and 9 U of glycerol-3-phosphate dehydrogenase. The formation of G-6-P from F-6-P was determined by monitoring the reduction of NADP+ in an assay mixture containing 100 mM Tris-HCl (pH 7), 10 mM F-6-P, 0.5 mM NADP+, and 0.3 U of GPDH. This assay was used to determine PGI activity during the purification procedure.
pH dependence and substrate specificity.
The pH dependence of the enzyme was measured in the direction of both G-6-P formation from F-6-P (10 mM) or F-6-P formation from G-6-P (10 mM) between 5.4 and 9.3 at 50°C in the coupled assays described above using either morpholineethanesulfonic acid (pH 5.4 to 5.8), bis-Tris-propane (pH 6.2 to 7.0), Tris-HCl (pH 6.8 to 8.5), or glycine (pH 8.3 to 9.3) at a concentration of 100 mM each. For the test of substrate specificity for sugars, F-6-P and G-6-P were exchanged for fructose and glucose.
Temperature dependence and thermal stability.
The temperature dependence of the enzyme activity was measured between 20 and 100°C in 50 mM potassium phosphate, pH 7.0, at the respective temperature. The activity was measured in the direction of G-6-P formation by using 10 mM F-6-P and 0.35 μg of PGI. The thermostability of the purified enzyme (36 μg in 50 μl of 50 mM potassium phosphate, pH 7.0) was tested in sealed vials, which were incubated at temperatures between 70 and 100°C up to 180 min, as indicated. The vials were then cooled on ice for 3 min, and the remaining enzyme activity was tested at 50°C by using 10 mM F-6-P in the continuous enzyme assay and was compared to that of unheated controls.
Identification and cloning of ORF encoding PGI from P. furiosus.
Based on the N-terminal amino acid sequence, one open reading frame (ORF) was identified by a BLAST search (2) in the complete sequenced genome of P. furiosus (UMBI database [see above]) (26 out of 28 amino acids were identical). The ORF was characterized as the gene, pgi, encoding PGI by cloning and functional overexpression in E. coli as follows. The putative pgi gene was amplified from the genomic DNA of P. furiosus as a template by PCR by using Pwo polymerase with the primers 5′CTCGTGGTGCATATGTATAAGGAACTTTT3′ (forward) and 5′TAACATTGTCCAGTTAACTACTTT-TTCCACC3′ (reverse). For the addition of 5′T overhangs, the PCR product was incubated with Taq polymerase for 5 min at 72°C and then was cloned into pBAD via a linearized vector activated with topoisomerase I. The resulting vector pBAD-pgi contained an additional 18-amino-acid N-terminal leader sequence (MGSGSGNNNNKLALLVVH). The vector pBAD-gpi was transformed into E. coli BL21(DH10B) cells. The inserted gene sequence, as well as its orientation, was confirmed on each strand by the Sanger method (33).
Functional overexpression of the pgi gene in E. coli and purification of recombinant P. furiosus PGI.
Cells were grown in 400 ml of Luria-Bertani medium at 37°C to an optical density at 600 nm of 0.8, and PGI expression was initiated by induction of the araC promotor following the addition of 0.2% l-arabinose. After 4 h of further growth, the cells were harvested by centrifugation at 4°C and were washed in 50 mM Tris-HCl, pH 7.0, containing 50 mM NaCl. The pellet was frozen at −20°C. Cell extracts were prepared by French press treatment of cell suspensions in buffer E (50 mM NaCl, 50 mM Tris-HCl [pH 7.0, 80°C]). After centrifugation (100,000 × g for 60 min), the solution was heat precipitated at 80°C for 30 min and centrifuged again. Homogeneous enzyme preparation was achieved by chromatography on phenyl-Sepharose, a Superdex gel filtration column, and Uno Q1, as described below for the purification of native PGI from P. furiosus.
Sources of materials.
All commercially available chemicals used were of reagent grade and were obtained from Merck (Darmstadt, Germany), Fluka (Buchs, Switzerland), or Sigma (Deisenhofen, Germany). Yeast extract and peptone were from Difco (Stuttgart, Germany). Enzymes and coenzymes were from Roche Diagnostics (Mannheim, Germany), peQlab (Erlangen, Germany), and GIBCO BRL Life Technologies (Eggenstein, Germany). Gases were from Linde (Hamburg, Germany). P. furiosus (DSM 3638) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). All fast protein liquid chromatography material (Q Sepharose Hiload, phenyl-Sepharose Hiload, and Uno Q) and columns used were from Pharmacia (Freiburg, Germany), and Bio-Rad (Munich, Germany). Linearized pBAD vector activated with topoisomerase I as well as E. coli BL21(DH10B) were purchased from Invitrogen (Groningen, The Netherlands).
RESULTS
Purification of PGI.
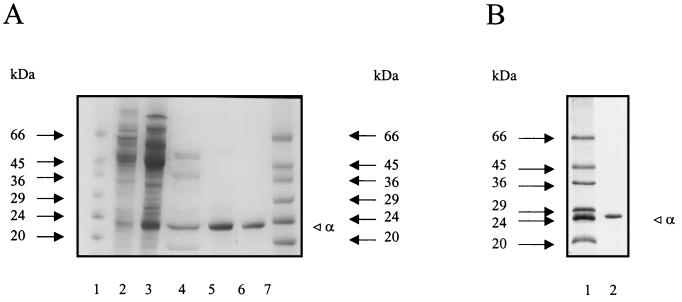
PGI was purified aerobically from cell extracts of P. furiosus by four purification steps involving anion exchange chromatography on Q Sepharose, hydrophobic interaction chromatography on phenyl-Sepharose, gel filtration on Superdex 200, and anion exchange chromatography on Uno Q1. By this procedure the enzyme was purified about 500-fold to a specific activity of 35 U/mg (at 50°C; fructose-6-phosphate as substrate) with a yield of 33% (Table 1). The purified protein was electrophoretically homogeneous as judged by denaturing SDS-PAGE (Fig. 1A). Thus, PGI represents about 0.2% of the cellular protein of P. furiosus.
TABLE 1.
Purification of PGI from P. furiosusa
| Purification step | Protein (mg) | Activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 1,118 | 78.3 | 0.07 | 100 | 1 |
| Q Sepharose | 176 | 65.3 | 0.37 | 83 | 5 |
| Phenyl-Sepharose | 9.9 | 38.6 | 3.9 | 49 | 56 |
| Superdex 200 | 1.8 | 31.2 | 17.7 | 34 | 253 |
| Uno Q1 | 0.7 | 25.8 | 34.8 | 33 | 497 |
Enzyme activity was measured in a continuous assay at 50°C in the direction of glucose-6-phosphate formation (see Materials and Methods).
FIG. 1.
Purification of PGI from P. furiosus (A) and of recombinant PGI from transformed E. coli (B) as analyzed by SDS-PAGE. Protein was denatured in SDS and separated in 14% (A) or 12% (B) slab gels (8 by 7 cm) (23), which were stained with Coomassie brilliant blue R 250. (A) Lanes 1 and 7, molecular mass standards (Sigma), in kilodaltons; lanes 2 to 6, analysis of PGI after various steps of the purification procedure (lane 2, 100,000 × g supernatant; lane 3, Q Sepharose; lane 4, phenyl-Sepharose; lane 5, Superdex 200; lane 6, Uno Q1). (B) Lane 1, molecular mass standards (Sigma); lane 2, purified recombinant PGI.
Molecular and catalytic properties.
The apparent molecular mass of native GPI was determined by gel filtration on Superdex 200 and was approximately 41 kDa. SDS-PAGE revealed only one subunit with an apparent molecular mass of 23 kDa (Fig. 1A), indicating a homodimeric (α2) structure of the native enzyme.
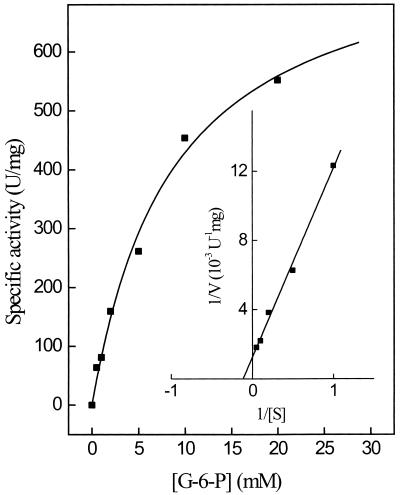
Kinetic constants of purified PGI were determined for both reaction directions (glucose-6-phosphate and fructose-6-phosphate). The rate dependence of the enzyme on G-6-P and on F-6-P followed Michaelis-Menten kinetics. At 80°C the apparent Km values for G-6-P and F-6-P were 8.7 mM (Fig. 2) and 1.0 mM; the corresponding apparent Vmax values were 800 and 130 U/mg, respectively. The pH optimum of PGI was at pH 7 for both reaction directions. About 50% of the activity was found at pH values 6 and 8. Purified PGI did not utilize glucose and fructose as substrates, indicating that the enzyme is specific for the phosphorylated hexoses.
FIG. 2.
Rate dependence of PGI purified from P. furiosus on the glucose-6-phosphate concentration at 80°C. The insert shows a double reciprocal plot of the rates versus the corresponding substrate concentrations. Enzyme activity was measured in the discontinuous assay system (see Materials and Methods). The assay mixture contained 0.18 μg of enzyme.
Temperature optimum and stability.
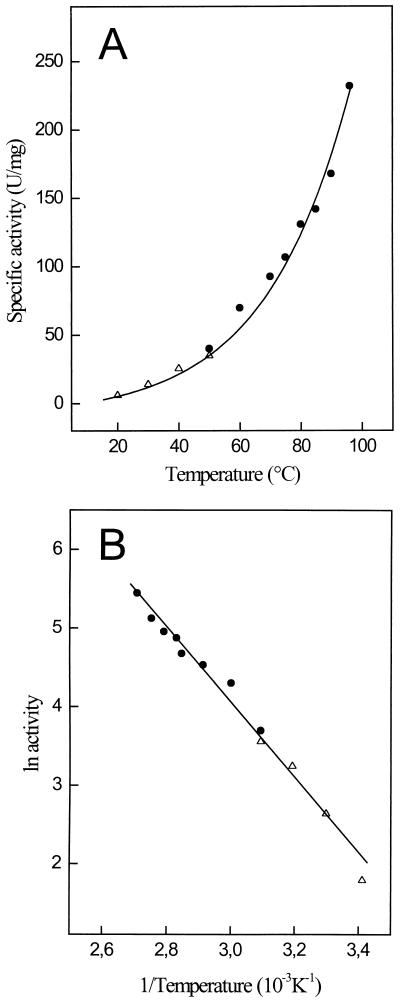
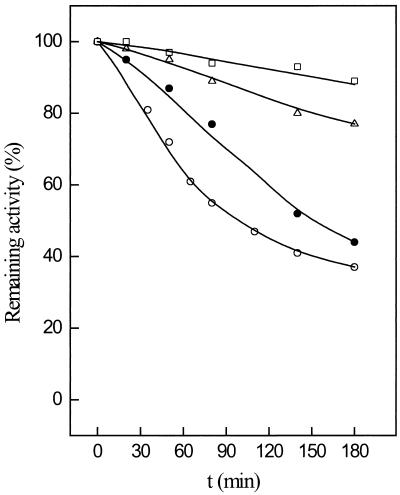
The temperature dependence of PGI is shown in Fig. 3. At 40°C the enzyme showed little activity, which, however, increased exponentially above 55°C, showing an optimum at 96°C (Fig. 3A). Enzyme activities at higher temperatures could not be measured accurately due to the decomposition of the substrate F-6-P. From the linear part of the Arrhenius plot between 20 and 96°C (Fig. 3B), an activation energy of 50 kJ/mol was calculated. The temperature stability of PGI was tested between 70 and 100°C in 50 mM potassium phosphate buffer (pH 7) by incubating the enzyme up to 180 min. The enzyme exhibited extremely high stability against thermal inactivation under these conditions. At 70 and 80°C PGI did not lose significant activity during incubation up to 180 min. Even at 100°C the enzyme had a half-life of about 90 min (Fig. 4).
FIG. 3.
Effect of temperature on the specific activity of the PGI purified from P. furiosus. (A) Temperature dependence of the specific activity. (B) Arrhenius plot of the same data. Enzyme activity was measured in the direction of glucose-6-phosphate formation using either the continuous assay for temperatures below 50°C (▵) or the discontinuous assay system for temperatures above 50°C (●) (see Materials and Methods). The assay mixture contained 0.35 μg of enzyme.
FIG. 4.
Thermostability of PGI purified from P. furiosus enzyme (1.8 μg) was incubated in 50 μl of 50 mM potassium phosphate buffer, pH 7.0, at 70°C (□), 80°C (▵), 90°C (●), and 100°C (○). At the times indicated, 15-μl aliquots were withdrawn and assayed for remaining activity at 55°C in the direction of glucose-6-phosphate formation. One hundred percent activity corresponded to the specific activity of PGI of 30 U/mg.
Identification and cloning of gene encoding PGI from P. furiosus and functional overexpression in E. coli.
Based on the N-terminal amino acid sequence determined for the 23-kDa subunit, MYKEPFGVKVDFETGIIEGAKKXVRRLP, a single ORF (Pf_209264) was identified in the genome of P. furiosus which exactly matches the 28 N-terminal amino acid residues with the exception of the two amino acids underlined. The ORF was characterized as the gene, pgi, encoding PGI from P. furiosus by its functional overexpression in E. coli (see below). The pgi gene (Pf_209264) contains 570 bp coding for a polypeptide of 189 amino acids with a calculated molecular mass of 23.3 kDa. The coding sequence starts with ATG and stops with TAG (Fig. 5). Immediately upstream of the initiation codon of the pgi gene, a putative ribosome binding site with the sequence TGGTGA was found. An archaeal promoter site, TATA boxes (18), could not be detected.
FIG. 5.
Multiple sequence alignment of deduced amino acid sequences of PGI from P. furiosus and of putative PGI from P. horikoshii (21) (PH1956) and P. abysii (Genoscope database; see above) (PAB1199). The sequence of PH1956 (192 amino acids) was truncated by 3 amino acids due to the identification of a putative ribosome binding site (TGGTGA) between bp −3 to −8 upstream of the start codon ATG.
The coding function of the ORF (Pf_209264) as pgi gene was proved by functional overexpression in E. coli. The ORF was amplified by PCR and cloned into vector pBAD-pgi and transformed into E. coli BL21(DH10B). After induction with 0.2% arabinose, a polypeptide of 25 kDa was overexpressed, showing thermoactive PGI activity. The recombinant PGI was purified from transformed E. coli about 50-fold by heat treatment and chromatography on phenyl-Sepharose, Superdex, and Uno Q1. As shown in Fig. 1B and Table 2, PGI purified from both P. furiosus and transformed E. coli showed almost identical molecular and kinetic properties. The apparent molecular mass of the subunit of recombinant PGI (25 kDa), as judged by SDS-PAGE (Fig. 1B), was 2 kDa larger than that of the native enzyme due to the presence of 18 additional N-terminal amino acids in the expression vector used (see Materials and Methods). Gel filtration of recombinant PGI revealed a single peak at about 45 kDa, indicating a dimeric structure. The temperature optimum (96°C) and thermostability as well as the kinetic constants (Km, Vmax) for G-6-P and F-6-P were virtually identical for both native and recombinant PGI.
TABLE 2.
Biochemical properties of glucose-6-phosphate isomerase from P. furiosus and E. coli (recombinant)a
| Enzyme isolated from: | Apparent molecular mass (kDa)
|
Oligomeric structure | Optimum
|
Arrhenius activation energy, 20–96°C (kJ·mol−1) | Apparent Km (mM)
|
Apparent Vmax (U/mg) for formation of:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Native enzyme | Subunit | Calculated | pH | Temp (°C) | G-6-P | F-6-P | G-6-P | F-6-P | |||
| P. furiosus | 41 | 23 | 23.257 | α2 | 7.0 | 96 | 50 | 8.7 | 1.0 | 130 | 800 |
| E. coli (recombinant) | 43 | 25 | 25.4 | α2 | 7 | 96 | 50 | 7.9 | 1.2 | 120 | 800 |
The molecular mass of native enzyme was determined by gel filtration of subunits by SDS-PAGE. The apparent Km values and Vmax values were determined at 80°C in a discontinuous assay in the direction of F-6-P formation and G-6-P formation.
DISCUSSION
In this report we describe the purification and characterization of the first archaeal PGI and its encoding gene from the hyperthermophile P. furiosus. The enzyme represents a novel type of PGI, probably forming a separate branch of PGI evolution.
The P. furiosus PGI has a native molecular mass of about 43 kDa and was composed of a single 23-kDa subunit, indicating a homodimeric structure. This oligomeric structure is a typical property of most characterized PGIs from eubacteria and eucarya. Exceptions are the PGIs from two thermophilic Bacillus species, B. stearothermophilus and Bacillus caldotenax, which have been shown to be homotetrameric enzymes (see Table 3).
TABLE 3.
Molecular properties of glucose-6-phosphate isomerases from bacteria, eucarya, and the archaeon P. furiosus
| Organism | Apparent molecular mass (kDa)
|
Reference(s) | |
|---|---|---|---|
| Native enzyme | Subunite | ||
| Archaea | |||
| P. furiosus | 41a (α2)c | 23.3* | This work |
| Bacteria | |||
| B. stearothermophilus (A, B) | 172b (α4) | 50.3*, 50.1* | 28, 47 |
| B. caldotenax | 202a (α4) | 50.6 | 46 |
| E. coli | 120a (α2) | 61.5* | 14, 39 |
| Lactobacillus casei | 67a (α2) | 34 | 30 |
| Eucarya | |||
| Saccharomyces cerevisiae | 119b (α2) | 61.2* | 25, 48 |
| Aspergillus niger | 118a (α2) | 60 | 32 |
| Pig | 132b (α2) | 63.0* | 1, 7, 9 |
| Mouse | 120a (α2) | 62.6* | 8, 16 |
| Rabbit | 125a (α2) | 64* | 24 |
| Human | NDd (α2) | 63.0* | 12, 49 |
Determined by gel filtration.
Determined by analytical ultracentrifugation.
The proposed subunit composition of the native enzyme is shown in parentheses.
ND, not determined.
Apparent molecular masses determined by SDS-PAGE. Asterisks (*) indicate molecular masses calculated from amino acid sequences.
However, the PGI from P. furiosus differs significantly from all known PGIs in various aspects. First, the subunit size of 23 kDa is significantly smaller than that of other PGIs, which are composed of subunits ranging from 34 to 64 kDa (Table 3). Second, PGI from P. furiosus showed a temperature optimum at 96°C, which is the highest value of all PGIs characterized so far. Furthermore, the enzyme exerts a high thermostability: it did not lose significant activity (<20%) upon incubation at 80°C for 120 min. In contrast, PGIs from the thermophilic Bacillus strains, B. stearothermophilus and B. caldotenax, had temperature optima of 70 and 77°C, respectively; both PGIs were inactivated by about 50% upon incubation at 65°C for 120 min (28, 46). The extremely high temperature optimum of activity and thermostability of P. furiosus PGI is in accordance with its function under the hyperthermophilic growth conditions of P. furiosus (13).
Third, the amino acid sequence of P. furiosus PGI did not show significant similarity to all known PGI sequences of eubacteria and eucarya. The amino acid sequence was deduced from the encoding pgi gene, which was identified via the following procedure. Based on the N-terminal amino acid sequence of the subunit of purified PGI, a single ORF was detected in the P. furiosus genome (UMBI database [see above]). The function of this ORF as a gene coding for PGI in P. furiosus was proved by its heterologous overexpression in E. coli. Biochemical properties of PGI isolated from P. furiosus and from transformed E. coli were almost identical (Table 2).
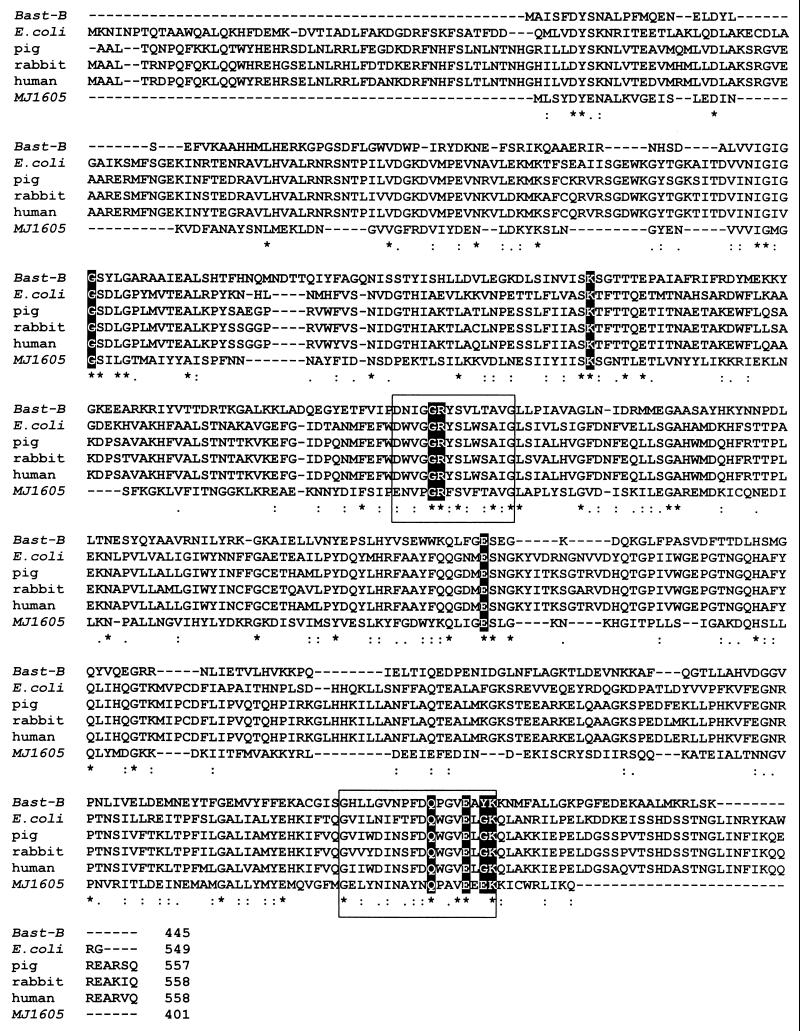
To date, a variety of PGI sequences are known from both bacteria and eucarya. Sequence comparison revealed that two consensus patterns of amino acids, [DENS]-X-[LIVM]-G-G-R-[FY]-S-[LIVMT]-X-[STA]-[PSAC]-[LIVMA]-G- and [GS]- X-[LIVM]-[LIVMFYW]-XXXX-[FY]-[DN]-Q-X-G-V-E-X- X-K, are almost completely conserved within all PGIs and have therefore been assigned as signature patterns of a PGI superfamily (3, 19). A multiple sequence alignment of selected PGIs from the bacteria B. stearothermophilus B and E. coli, as well as from the eucarya, pig, rabbit, and human, is given in Fig. 6. The two consensus patterns are highlighted by boxes. Those amino acid residues which represent putative substrate-binding sites, as concluded from the crystal structures of the B. stearothermophilus PGI (45), are shaded. This alignment includes the deduced amino acid sequence of a hypothetical PGI from the archaeon Methanococcus jannaschii (401 amino acids) deduced from the ORF (MJ1605) in the genome of M. jannaschii (6). This hypothetical PGI showed a similarity of 48% to the PGI of B. stearothermophilus and contained two amino acid sequences, which were almost identical to the signature patterns of the PGI superfamily. Therefore, this putative PGI might represent an archaeal member of the PGI superfamily. However, a final proof of this hypothetical PGI as an active enzyme in the sugar metabolism of M. jannaschii remains to be shown.
FIG. 6.
Multiple sequence alignment of the PGI superfamily of bacteria and eucarya. Deduced amino acid sequences of B. stearothermophilus B (47), E. coli (14), pig (7), rabbit (20), and human (12) are aligned. In addition, the amino acid sequence of the hypothetical PGI of the archaeon M. jannaschii (MJ1605) is included. The two PGI signature patterns [DENS]-X-[LIVM]-G-G-R-[FY]-S-[LIVMT]-X-[STA]-[PSAC]-[LIVMA]-G- and [GS]-X-[LIVM]-[LIVMFYW]-XXXX-[FY]-[DN]-Q-X-G-V-E-X-X-K are highlighted by boxes. Amino acids that constitute putative substrate-binding sites in accordance with the crystal structure of B. stearothermophilus PGI (45) are shaded.
The PGI sequence of P. furiosus (Fig. 5) did not show significant overall similarity (9 to 12%) or identity (3 to 6%) to the PGIs characterized so far. Furthermore, the P. furiosus PGI did not contain the two typical amino acid signature sequences of the PGI superfamily (3, 19), and putative substrate- binding sites (45) could not be identified (Fig. 6). Thus, the PGI of P. furiosus is significantly different from the enzymes of the PGI superfamily. In addition, BLAST search analysis did not reveal hypothetical proteins with similarity to the P. furiosus PGI in other archaea for which genome sequences are available to date. The P. furiosus PGI sequence does, however, show a high degree of identity (84 to 89%) with deduced hypothetical proteins identified by a BLAST search from the other Pyrococcus species (UMBI database [see above] and Genoscope database [see above]) P. horikoshii (PH1956; 89%) and P. abysii (PAB1199; 84%). This indicates the presence of homologous genes coding for PGI in these Pyrococcus strains. The unusual amino acid sequence of the Pyrococcus PGI explains why in a previous study (10) a PGI could not be annotated in the P. horikoshii genome by sequence comparison with known PGIs.
In summary, the unique amino acid sequence of P. furiosus PGI indicates that the enzyme constitutes a novel type of PGI possibly forming a separate branch of PGI evolution. Currently, studies of this novel isomerase are in progress, including crystallization of the enzyme and analysis of the structure-function relationship, to elucidate the reaction mechanism in comparison with the established PGIs for which high-resolution crystal stuctures are available, i.e., from B. stearothermophilus (45) and from rabbit (20).
Recent data indicate that the enzymes of the PGI superfamily exhibit a high degree of structural and functional relationship to several important proteins from eukaryotes, e.g., neuroleukins, certain cytokines, and maturation factors, which are involved in cell functions such as cell growth and differentiation (12, 50). Recently, functional homology of a prokaryotic PGI, from B. stearothermophilus, has been demonstrated by showing PGI-induced stimulation of the cell motility of cancer cells (45). Thus, the known PGIs constitute a multifunctional protein family exhibiting a broader spectrum of enzyme functions than their established catalytic activity in sugar metabolism. It will be interesting to test the effect of the novel type of PGI from P. furiosus, which is not related to the PGI superfamily, with respect to its effect on the above-mentioned eukaryotic cell functions.
ACKNOWLEDGMENTS
We thank R. Schmid (Mikrobiologie, Universität Osnabrück) for performing the N-terminal amino acid sequencing and H. Preidel for mass culturing P. furiosus. The expert technical assistance of K. Lutter-Mohr is also gratefully acknowledged.
This work was supported by grants of the Fonds der Chemischen Industrie.
REFERENCES
- 1.Achari A, Marshall S E, Muirhead H, Palmieri R H, Noltmann E A. Glucose-6-phosphate isomerase. Philos Trans R Soc Lond B Biol Sci. 1981;293:145–157. doi: 10.1098/rstb.1981.0068. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1995. Nucleic Acids Res. 1996;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Brunner N A, Brinkmann H, Siebers B, Hensel R. NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase from Thermoproteus tenax. The first identified archaeal member of the aldehyde dehydrogenase superfamily is a glycolytic enzyme with unusual regulatory properties. J Biol Chem. 1998;273:6149–6156. doi: 10.1074/jbc.273.11.6149. [DOI] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L X, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Chaput M, Claes V, Portetelle D, Cludts I, Cravador A, Burny A, Gras H, Tartar A. The neurotrophic factor neuroleukin is 90% homologous with phosphohexose isomerase. Nature. 1988;332:454–455. doi: 10.1038/332454a0. [DOI] [PubMed] [Google Scholar]
- 8.Charles D J, Lee C Y. Biochemical characterization of phosphoglucose isomerase and genetic variants from mouse and Drosophila melanogaster. Mol Cell Biochem. 1980;29:11–21. doi: 10.1007/BF00230952. [DOI] [PubMed] [Google Scholar]
- 9.Claes V, Taquet A N, Kettmann R, Burny A. Sequence analysis of the pig phosphoglucose isomerase gene promoter region. Biochim Biophys Acta. 1990;1087:339–340. doi: 10.1016/0167-4781(90)90009-q. [DOI] [PubMed] [Google Scholar]
- 10.Cordwell S J. Microbial genomes and “missing” enzymes: redefining biochemical pathways. Arch Microbiol. 1999;172:269–279. doi: 10.1007/s002030050780. [DOI] [PubMed] [Google Scholar]
- 11.de Vos W M, Kengen S W, Voorhorst W G, van der Oost J. Sugar utilization and its control in hyperthermophiles. Extremophiles. 1998;2:201–205. doi: 10.1007/s007920050061. [DOI] [PubMed] [Google Scholar]
- 12.Faik P, Walker J I, Redmill A A, Morgan M J. Mouse glucose-6-phosphate isomerase and neuroleukin have identical 3′ sequences. Nature. 1988;332:455–457. doi: 10.1038/332455a0. [DOI] [PubMed] [Google Scholar]
- 13.Fiala G, Stetter K O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 14.Froman B E, Tait R C, Gottlieb L D. Isolation and characterization of the phosphoglucose isomerase gene from Escherichia coli. Mol Gen Genet. 1989;217:126–131. doi: 10.1007/BF00330951. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs G, Stupperich E. Carbon assimilation pathways in archaebacteria. Syst Appl Microbiol. 1986;7:364–369. [Google Scholar]
- 16.Gurney M E, Heinrich S P, Lee M R, Yin H S. Molecular cloning and expression of neuroleukin, a neurotrophic factor for spinal and sensory neurons. Science. 1986;234:566–574. doi: 10.1126/science.3764429. [DOI] [PubMed] [Google Scholar]
- 17.Hansen T, Schonheit P. Purification and properties of the first-identified, archaeal, ATP-dependent 6-phosphofructokinase, an extremely thermophilic non-allosteric enzyme, from the hyperthermophile Desulfurococcus amylolyticus. Arch Microbiol. 2000;173:103–109. doi: 10.1007/s002039900114. [DOI] [PubMed] [Google Scholar]
- 18.Hethke C, Geerling A C, Hausner W, de Vos W M, Thomm M. A cell-free transcription system for the hyperthermophilic archaeon Pyrococcus furiosus. Nucleic Acids Res. 1996;24:2369–2376. doi: 10.1093/nar/24.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffery C J, Bahnson B J, Chien W, Ringe D, Petsko G A. Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator. Biochemistry. 2000;39:955–964. doi: 10.1021/bi991604m. [DOI] [PubMed] [Google Scholar]
- 21.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 22.Kengen S W, Stams A J, de Vos W M. Sugar metabolism of hyperthermophiles. FEMS Microbiol Rev. 1996;18:119–137. [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Chirgwin J M. Rabbit phosphoglucose isomerase/neuroleukin/autocrine motility factor: cloning via interspecies identity. Biochim Biophys Acta. 2000;1476:363–367. doi: 10.1016/s0167-4838(99)00258-7. [DOI] [PubMed] [Google Scholar]
- 25.Lowe S L, Reithel F J. The subunit structure of phosphoglucose isomerase from bakers' yeast. J Biol Chem. 1975;250:94–99. [PubMed] [Google Scholar]
- 26.Meyer C, Schmid R, Scriba P C, Wehling M. Purification and partial sequencing of high-affinity progesterone-binding site(s) from porcine liver membranes. Eur J Biochem. 1996;239:726–731. doi: 10.1111/j.1432-1033.1996.0726u.x. [DOI] [PubMed] [Google Scholar]
- 27.Muirhead H, Shaw P J. Three-dimensional structure of pig muscle phosphoglucose isomerase at 6 Å resolution. J Mol Biol. 1974;89:195–203. doi: 10.1016/0022-2836(74)90170-3. [DOI] [PubMed] [Google Scholar]
- 28.Muramatsu N, Noso Y. Purification and characterization of glucose-6-phosphate isomerase from Bacillus stearothermophilus. Arch Biochem Biophys. 1971;144:245–252. doi: 10.1016/0003-9861(71)90475-9. [DOI] [PubMed] [Google Scholar]
- 29.Noltmann E A. Aldose-ketose isomerases. In: Boyer P D, editor. The enzymes. New York, N.Y: Academic Press; 1972. pp. 271–354. [Google Scholar]
- 30.Pradhan P G, Nadkarni G B. Functional multiplicity of phosphoglucose isomerase from Lactobacillus casei. Biochim Biophys Acta. 1980;615:465–473. doi: 10.1016/0005-2744(80)90512-4. [DOI] [PubMed] [Google Scholar]
- 31.Rose I A. Mechanism of the aldose-ketose isomerase reactions. Adv Enzymol Relat Areas Mol Biol. 1975;43:491–517. doi: 10.1002/9780470122884.ch6. [DOI] [PubMed] [Google Scholar]
- 32.Ruijter G J, Visser J. Characterization of Aspergillus niger phosphoglucose isomerase. Use for quantitative determination of erythrose 4-phosphate. Biochimie. 1999;81:267–272. doi: 10.1016/s0300-9084(99)80061-3. [DOI] [PubMed] [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaegger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separations of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 35.Schäfer S, Barkowski C, Fuchs G. Carbon assimilation by the autotrophic thermophilic archaebacterium Thermoproteus neutrophilus. Arch Microbiol. 1986;146:301–308. [Google Scholar]
- 36.Schäfer T, Schönheit P. Maltose fermentation to acetate, CO2 and H2 in the anaerobic hyperthermophilic archaeon Pyrococcus furiosus: evidence for the operation of a novel sugar fermentation pathway. Arch Microbiol. 1992;158:188–202. [Google Scholar]
- 37.Schäfer T, Schönheit P. Gluconeogenesis from pyruvate in the hyperthermophilic archaeon Pyrococcus furiosus: involvement of reactions of the Embden-Meyerhof pathway. Arch Microbiol. 1993;159:354–363. [Google Scholar]
- 38.Schönheit P, Schäfer T. Metabolism of hyperthermophiles. World J Microbiol Biotechnol. 1995;11:26–57. doi: 10.1007/BF00339135. [DOI] [PubMed] [Google Scholar]
- 39.Schreyer R, Bock A. Phosphoglucose isomerase from Escherichia coli K 10: purification, properties and formation under aerobic and anaerobic condition. Arch Microbiol. 1980;127:289–298. doi: 10.1007/BF00427206. [DOI] [PubMed] [Google Scholar]
- 40.Selig M, Xavier K B, Santos H, Schönheit P. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch Microbiol. 1997;167:217–232. doi: 10.1007/BF03356097. [DOI] [PubMed] [Google Scholar]
- 41.Shaw P J, Muirhead H. Crystallographic structure analysis of glucose 6-phosphate isomerase at 3–5 Å resolution. J Mol Biol. 1977;109:475–485. doi: 10.1016/s0022-2836(77)80025-9. [DOI] [PubMed] [Google Scholar]
- 42.Siebers B, Klenk H P, Hensel R. PPi-dependent phosphofructokinase from Thermoproteus tenax, an archaeal descendant of an ancient line in phosphofructokinase evolution. J Bacteriol. 1998;180:2137–2143. doi: 10.1128/jb.180.8.2137-2143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprott D G, Ekiel I, Patel G B. Metabolic pathways in Methanococcus jannaschii and other methanogenic bacteria. Appl Environ Microbiol. 1993;59:1092–1098. doi: 10.1128/aem.59.4.1092-1098.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stetter K O. Extremophiles and their adaptation to hot environments. FEBS Lett. 1999;452:22–25. doi: 10.1016/s0014-5793(99)00663-8. [DOI] [PubMed] [Google Scholar]
- 45.Sun Y J, Chou C C, Chen W S, Wu R T, Meng M, Hsiao C D. The crystal structure of a multifunctional protein: phosphoglucose isomerase/autocrine motility factor/neuroleukin. Proc Natl Acad Sci USA. 1999;96:5412–5417. doi: 10.1073/pnas.96.10.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takama M, Nosoh Y. Purification and some properties of 6-phosphoglucose isomerase from Bacillus caldotenax. J Biochem (Tokyo) 1980;87:1821–1827. doi: 10.1093/oxfordjournals.jbchem.a132927. [DOI] [PubMed] [Google Scholar]
- 47.Tao W, Wang L, Shen R, Sheng R. Complete nucleotide sequences of two phosphoglucoisomerase isoenzymes from Bacillus stearothermophilus. Nucleic Acids Res. 1989;17:10107–10108. doi: 10.1093/nar/17.23.10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tekamp-Olson P, Najarian R, Burke R L. The isolation, characterization and nucleotide sequence of the phosphoglucoisomerase gene of Saccharomyces cerevisiae. Gene. 1988;73:153–161. doi: 10.1016/0378-1119(88)90321-6. [DOI] [PubMed] [Google Scholar]
- 49.Walker J I, Faik P, Morgan M J. Characterization of the 5′ end of the gene for human glucose phosphate isomerase (GPI) Genomics. 1990;7:638–643. doi: 10.1016/0888-7543(90)90212-d. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe H, Takehana K, Date M, Shinozaki T, Raz A. Tumor cell autocrine motility factor is the neuroleukin/phosphohexose isomerase polypeptide. Cancer Res. 1996;56:2960–2963. [PubMed] [Google Scholar]
- 51.Yu J P, Ladapo J, Whitman W B. Pathway of glycogen metabolism in Methanococcus maripaludis. J Bacteriol. 1994;176:325–332. doi: 10.1128/jb.176.2.325-332.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]