Abstract
A new open reading frame, encoding a putative integrase-like protein, was detected downstream from the six genes of the vanD glycopeptide resistance cluster in Enterococcus faecium BM4339 (B. Casadewall and P. Courvalin, J. Bacteriol. 181:3644–3648, 1999). In this cluster, genes coding for the VanRD-VanSD two-component regulatory system were cotranscribed from the PRD promoter, whereas transcription of the vanYD, vanHD, vanD, vanXD, and intD genes was initiated from the PYD promoter located between vanSD and vanYD (the D subscript indicates that the gene is part of the vanD operon). The VanRD-VanSD regulatory system is likely to activate transcription of the resistance genes from the promoter PYD. Glycopeptide-susceptible derivatives of BM4339 were obtained by trans complementation of the frameshift mutation in the ddl gene, restoring functional d-alanine:d-alanine ligase activity in this strain. The glycopeptide-susceptible transformant BM4409, producing only d-alanyl-d-alanine-terminating peptidoglycan precursors, did not express the resistance genes encoding the VanYD d,d-carboxypeptidase, the VanHD dehydrogenase, the VanD ligase, the VanXD d,d-dipeptidase, and also the IntD integrase, although the regulatory region of the vanD cluster was still transcribed. In BM4409, the absence of VanRD-VanSD, apparently dependent, transcription from promoter PYD correlated with the lack of d-alanyl-d-lactate-terminating precursors. The vanXD gene was transcribed in BM4339, but detectable amounts of VanXD d,d-dipeptidase were not synthesized. However, the gene directed synthesis of an active enzyme when cloned on a multicopy plasmid in Escherichia coli, suggesting that the enzyme was unstable in BM4339 or that it had very low activity that was detectable only under conditions of high gene dosage. This activity is not required for glycopeptide resistance in BM4339, since this strain cannot synthesize d-alanyl-d-alanine.
Glycopeptides inhibit the late stages of peptidoglycan assembly (27, 34). Since these large, hydrophilic molecules cannot penetrate the cytoplasmic membrane of the cell, their antibacterial action results from forming complexes with the d-alanyl-d-alanine (d-Ala-d-Ala) C termini of peptidoglycan precursors on the external side of the membrane (9). The formation of these complexes prevents the cross-linking reactions catalyzed by transglycosylases, d,d-transpeptidases, and d,d-carboxypeptidases and prevents cell wall assembly.
Resistance to glycopeptides in enterococci is mediated by the synthesis of modified peptidoglycan precursors (9). Two types of precursors have been identified: (i) those ending in the depsipeptide d-alanyl-d-lactate (d-Ala-d-Lac), which exhibit a 1,000-fold-lower binding affinity for vancomycin, and (ii) precursors terminating with d-alanyl-d-serine, whose reduced affinity (7-fold) results from steric hindrance. Resistance to glycopeptides by production of d-Ala-d-Lac-terminating precursors can be categorized into three types: VanA, VanB, and VanD (9, 31). Although all three types involve genes encoding related enzymatic functions, they are distinguishable by the location of those genes, either on plasmids or on the chromosome or both, and by the different mechanisms of gene expression and regulation (9). Enterococci belonging to the VanA type are inducibly resistant to high levels of both vancomycin and teicoplanin, whereas VanB-type enterococci show inducible resistance to various levels of vancomycin only. The VanD type is unique and is characterized by constitutively (30, 31) or inducibly (29) expressed resistance to moderate levels of vancomycin (MICs, 16 to 256 mg/liter) and teicoplanin (MICs, 2 to 64 mg/liter). These three types of glycopeptide resistance are mediated by the gene clusters vanA, vanB, and vanD. Each cluster consists of five essential genes and one or two additional genes encoding functions that are not required to achieve resistance (9). In susceptible strains, the chromosomally encoded d-Ala:d-Ala ligase (Ddl) synthesizes the dipeptide incorporated into peptidoglycan precursors that are the target of glycopeptides. In resistant enterococci, synthesis of peptidoglycan precursors utilizes an alternative pathway that avoids these d-Ala-d-Ala precursors and relies instead on d-Ala:d-Lac ligases (VanA, VanB, and VanD). These enzymes are capable of synthesizing modified precursors that compete with the normal ones for their incorporation into the cell wall but escape glycopeptide binding. The dehydrogenases VanH, VanHB, and VanHD convert pyruvate into d-Lac, which is in turn used by the d-Ala:d-Lac ligases (6, 12). In the VanA and VanB types, two d,d-peptidases (VanX and VanY, VanXB and VanYB) increase resistance by sequentially eliminating the normal peptidoglycan precursors (1). First, the VanX and VanXB d,d-dipeptidases hydrolyze the dipeptide d-Ala-d-Ala (16, 35). This action is supplemented by the hydrolysis of the residual pentapeptide precursors by the VanY and VanYB d,d-carboxypeptidases that are not essential but increase the levels of resistance to glycopeptides (5, 16). Genes encoding a d,d-dipeptidase and a d,d-carboxypeptidase have also been identified in the vanD cluster, but their roles in resistance are unclear (14). All three clusters include genes for a two-component regulatory system, vanR-vanS, vanRB-vanSB, and vanRD-vanSD (7, 14, 16). The mechanisms by which these systems regulate expression of resistance genes have been elucidated for both the VanA and VanB types. The VanS and VanSB histidine protein kinases are sensor proteins that, in the presence of both vancomycin and teicoplanin or only in the presence of vancomycin, control the phosphorylation level of their cognate VanR and VanRB response regulators, which in turn trigger transcription of the resistance genes from the promoters PR and PH (7, 20) and PRB and PYB (16, 38). Additional genes have been identified in both the vanA and vanB clusters. Expression of vanZ is responsible for low-level teicoplanin resistance in VanA-type enterococci (3), whereas the function of the vanW gene product in VanB-type strains is still unknown (16).
Enterococcus faecium BM4339 was the first clinical isolate shown to harbor the vanD gene cluster (14, 31). The gene organization (14) and chromosomal location of the cluster (30) in this strain have been elucidated, and expression of resistance has been partially studied (31). Unlike VanA- and VanB-mediated resistance, VanD-type resistance is expressed constitutively, proposed to be the result of nonstringent control of the phosphorylation level of VanRD by the putative phosphatase activity of VanSD (14, 31). In addition, determination of the sequence of the chromosomal ddl gene in BM4339 revealed a frameshift mutation, likely to generate an inactive product that would be responsible for the lack of peptidoglycan precursors ending in d-Ala-d-Ala (14, 31). Constitutive expression of resistance accounts for the lack of dependence on glycopeptides for growth of BM4339. The presence of an apparently intact vanXD gene has been reported (14). However, d,d-dipeptidase activity was not detected in cytoplasmic or membrane extracts from BM4339 (31). Although such an activity is not essential in a genetic background lacking the ability to synthesize d-Ala-d-Ala, these results prompted us to probe the mechanisms responsible for expression of glycopeptide resistance in BM4339.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are described in Table 1. Unless specified, Escherichia coli TB1 (Focus, Life Technologies Inc., Gaithersburg, Md.) and E. coli DH5α (43) were used as host strains in cloning experiments. Bacteria were cultured in brain heart infusion broth or agar (Difco Laboratories, Detroit, Mich.) at 37°C. The method of Steers et al. (40) was used to determine the MICs of antibiotics with 105 CFU per spot on Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France) after 24 h of incubation.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TB1 | JM83 hsdR(rK− mK+) | Life Technologies |
| DH5α | recA1 gyrA (Nalr) Δ(lacIZYA-argF) [φ80dlacΔ(lacZ)M15] | 43 |
| E. faecalis JH2-2 | Fusr Rifr | 21 |
| E. faecium | ||
| BM4339 | Vmr Ter | 31 |
| BM4409 | BM4339/pAT662 | 14 |
| BM4458 | BM4339::pAT665 | This work |
| BM4459 | BM4339::pAT665 | This work |
| Plasmids | ||
| pUC18 | Apr, lacZα vector | 42 |
| pBGS18+ | Kmr, lacZα vector | 39 |
| pAT78 | Spr, oriR from pAMβ1, oriR from pUC, oriT from RK2, lacZα cat | 7 |
| pAT79 | Spr, oriR from pAMβ1, oriR from pUC, oriT from RK2, lacZα P2cat | 7 |
| pAT113/Sp | Spr Mob+ (IncP), oriR from pACYC184, att Tn1545 lacZα | 15 |
| pAT145 | Kmr Mob+, oriR from pAMβ1, oriR from pBR322, oriT from RK2, int-Tn1545 | 41 |
| pAT632 | Cmr derivative of pAT145 ΔoriT RK2 | This work |
| pAT654 | 5.3-kb Sau3AI fragment (vanSD′vanYDHDDXD) of BM4339 cloned into pUC18 | 14 |
| pAT655 | 2.85-kb XmaI-HindIII fragment (vanXD′intD) of BM4339 cloned into pUC18 | This work |
| pAT659 | 0.7-kb PCR fragment (vanXD) of pAT654 cloned into pUC18 | This work |
| pAT660 | 1.15-kb PCR fragment (vanYD) of pAT654 cloned into pBGS18+ | This work |
| pAT662 | 1.2-kb SacI-XbaI fragment (ddl) of BM4147 cloned into pAT79 | 14 |
| pAT665 | 1.45-kb EcoRI fragment (P2ddl) of pAT662 cloned into pAT113/Sp | This work |
| pAT666 | 160-bp PCR fragment (PYD) of pAT654 cloned into pAT78 | This work |
Recombinant DNA techniques.
Plasmid DNA isolation, cleavage of DNA with restriction endonucleases (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England, and Gibco BRL-Life Technologies Inc.), purification of restriction fragments from agarose gel, dephosphorylation of vector DNA with calf intestinal phosphatase (Amersham Pharmacia Biotech), and ligation with T4 DNA ligase (Amersham Pharmacia Biotech) were performed by standard methods (36).
Plasmid construction.
The plasmids were constructed as follows (Fig. 1B).
FIG. 1.
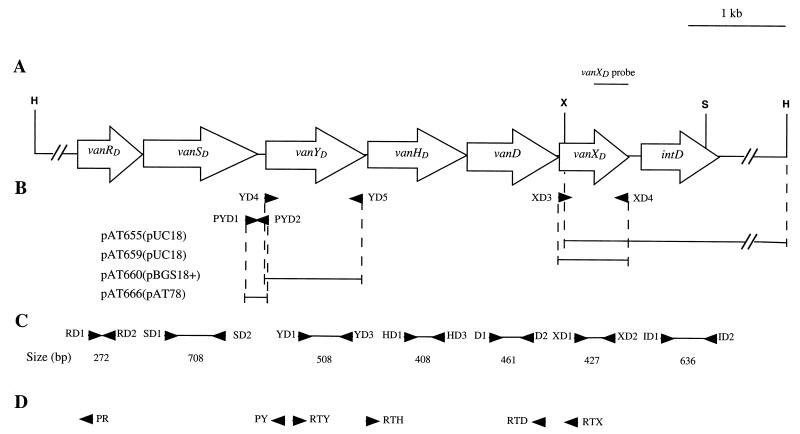
Schematic representation of the vanD gene cluster in BM4339. (A) Map of the 8.7-kb region containing the vanRD, vanSD, vanYD, vanHD, vanD, vanXD, and intD ORFs comprising the vanD gene cluster. Open arrows indicate sense of transcription. The PCR fragment internal to vanXD used as a probe in hybridization experiments is indicated. Abbreviations: H, HindIII; S, Sau3AI; X, XmaI. (B) Inserts in recombinant plasmids. The inserts are represented by solid lines, and the vectors are indicated in parentheses. (C) Probes used in Northern hybridization and in RT-PCR. (D) Oligodeoxynucleotides used in RT-PCR and in primer extension. Arrowheads indicate positions and orientations of primers.
(i) Plasmid pAT655.
To complete the sequence downstream from the vanXD gene, E. faecium BM4339 total DNA was digested with XmaI and HindIII and the size of the fragment hybridizing with a 304-bp probe corresponding to the 3′ end of the vanXD gene (Fig. 1A) was estimated (36). To identify recombinant plasmids, clones were screened by colony hybridization (36) with the same probe. Plasmid pAT655 contained a 2.85-kb XmaI-HindIII insert.
(ii) Plasmids pAT659 and pAT660.
The vanXD and vanYD genes were amplified using primer pairs XD3-XD4 and YD4-YD5, respectively, and plasmid pAT654 DNA (Table 1) as a template. Oligodeoxynucleotide XD3 (5′cgacgcgaattCGGTTTTACGCTTTCTGA) contained an EcoRI site (underlined) and 18 bases complementary to the sequence upstream from vanXD(in uppercase letters). Oligodeoxynucleotide XD4 (5′ agcggaagcTTCATTTCTTCAGGCTC) harbored a HindIII site (underlined) and 17 bases complementary to the sequence downstream from vanXD (in uppercase letters). Primer YD4 (5′ gagtgcgaattcttgATATCAGGAGGGGCGAT) contained an EcoRI site (underlined), a translation stop codon (italicized), and 17 bases complementary to the sequence upstream from vanYD (in uppercase letters). Primer YD5 (5′ gcagcgaagcttCTCCATTCATTTCCTCCTT) included a HindIII site (underlined) and 19 bases complementary to the sequence downstream from vanYD (in uppercase letters). The vanXD and vanYD PCR products were digested with EcoRI and HindIII and cloned into pUC18 and pBGS18+, respectively. Sequencing of the inserts and of the flanking regions confirmed that the two genes were under the control of the lac promoter of pUC18 and pBGS18+.
(iii) Plasmid pAT666.
The PYD promoter region of the vanD gene cluster was amplified by PCR using primers PYD1 and PYD2 and plasmid pAT654 DNA as a template. Oligodeoxynucleotide PYD1 (5′agagtcgaattcTTGAGGTTACATTGCCCG) harbored an EcoRI site (underlined) and 18 bases complementary to the 3′ end of vanSD (in uppercase letters). Oligodeoxynucleotide PYD2 (5′ gcactcgagctcAAAAAAATCGCCCCTCCT) contained a SacI site (underlined) and 18 bases complementary to the sequence just upstream from the 5′ extremity of vanYD (in uppercase letters). After digestion of the 160-bp PCR product by EcoRI and SacI, the fragment was cloned into similarly digested pAT78 DNA. The recombinant plasmid pAT666 was introduced into Enterococcus faecalis JH2-2 and E. faecium BM4339 by electrotransformation. Transformants selected with spectinomycin (60 μg/ml) and chloramphenicol (10 μg/ml), respectively, were screened for the presence of pAT666 DNA.
(iv) Plasmid pAT632.
The cat gene, encoding chloramphenicol acetyltransferase (CAT), along with the enterococcal constitutive P2 promoter, was amplified by PCR using primers 79-1 and 79-2 and plasmid pAT79 (P2cat) DNA as a template. Oligodeoxynucleotide 79-1 (5′ cacggtatgcatGTAAAACGACGGCCAGT) contained an NsiI site (underlined) and 17 bases corresponding to the universal primer −20 (in uppercase letters) (Amersham Pharmacia Biotech). Oligodeoxynucleotide 79-2 (5′ggagcgatgcatCAGGAAACAGCTATGAC) included an NsiI site (underlined) and 17 bases corresponding to the universal primer Reverse (in uppercase letters) (Amersham Pharmacia Biotech). The 1-kb P2cat PCR product was digested by NsiI and cloned into the 10-kb PstI fragment of pAT145, replacing the portion of pAT145 containing oriT from RK2. Plasmid pAT632 (Cmr Int-Tn) was introduced into E. faecium BM4339 by electrotransformation, and transformants were selected on chloramphenicol (10 μg/ml). The presence of pAT632 was confirmed by plasmid DNA extraction (36).
(v) Plasmid pAT665.
During the cloning steps for the construction of pAT662, an additional EcoRI site was incorporated between the 3′ end of the ddl gene and the XbaI site used to generate the plasmid (14). Plasmid pAT665 was constructed by inserting the P2ddl fragment, obtained by digestion of pAT662 with EcoRI, in pAT113/Sp integrative vector DNA cleaved with EcoRI.
Strain construction.
The integrative plasmid pAT665 (P2ddl) was introduced into E. faecium BM4339/pAT632 by electrotransformation, and transformants resulting from integration of pAT665 by illegitimate recombination mediated by the integrase of Tn1545 were selected with spectinomycin (120 μg/ml). The spontaneous loss of pAT632 (Cmr Int-Tn) was obtained by subculturing transformants for ca. 30 generations in chloramphenicol-free medium. Total DNA from 12 clones was digested with HindIII, KpnI, NdeI, NsiI, and SspI and analyzed by Southern hybridization (36) using pAT113/Sp- and pAT665-labeled DNA and the P2ddl fragment purified from pAT665 as probes. The data obtained (not shown) indicated the presence of a single chromosomal copy of pAT665 (P2ddl) in at least two clones, BM4458 and BM4459. These transformants were shown to harbor a copy of pAT665 integrated in different loci in the chromosome and were selected for further studies.
Nucleotide sequencing.
DNA sequencing was performed by the dideoxynucleotide chain termination method (37) with α-35S-dATP (Amersham Pharmacia Biotech) and the T7 Sequenase Version 2.0 DNA sequencing kit (Amersham Pharmacia Biotech). The plasmid DNA used as a template was extracted with the commercial Wizard Plus Minipreps DNA Purification System (Promega, Madison, Wis.).
RNA techniques. (i) Extraction of total RNA.
Enterococcal strains were grown to an optical density at 600 nm of 0.7. Suspensions were disrupted with a Mickle disintegrator using 3.5-g glass beads (106 μm) (Sigma Chemical Co., St. Louis, Mo.) in the presence of 0.25 ml of 10% sodium dodecyl sulfate, 1 ml of 2% Macaloïd (National Lead Co., New York, N.Y.), and 3 ml of phenol (19). The mixtures were shaken three times for 1 min at 4°C and centrifuged for 15 min at 8,500 × g. Supernatants were extracted with phenol and chloroform. Total RNA was precipitated by addition of 0.1 volume of 3 M sodium acetate, pH 5.2, and 3 volumes of ice-cold 100% ethanol. RNA pellets were resuspended in sterile water.
(ii) Northern analysis.
Equal amounts of total RNA (20 μg each) were separated under denaturing conditions in a 1.2% agarose-formaldehyde-morpholinepropanesulfonic acid gel, stained with ethidium bromide, and blotted onto Hybond N+ membranes (Amersham Pharmacia Biotech) (11, 36). Filters were prehybridized and hybridized as described previously (11). PCR products were amplified using total DNA from E. faecium BM4339 as a template and primers RD1, SD1 and SD2, YD1, HD1, and XD2 (30), primers D1 and D2 (31), whose positions from the first base of the vanRD gene are 4401 to 4421 and 4862 to 4842, respectively, and primers listed in Table 2, as indicated in Fig. 1C. The PCR fragments were labeled with [α-32P]dCTP (3,000 Ci/mmol; Amersham Pharmacia Biotech) by using the Megaprime DNA Labelling System (Amersham Pharmacia Biotech) and used as probes. Washes were performed as described previously (23).
TABLE 2.
Oligodeoxynucleotides used in RNA experimentsa
| Primer | Nucleotide sequence | Positionb |
|---|---|---|
| RD2 | 5′-AACCGTTCACCCCATAC | 575–558 |
| YD3 | 5′-TTTCTCCGTCCATCACC | 2940–2923 |
| HD3 | 5′-AACGAAACATAGTCTGC | 3611–3594 |
| XD1 | 5′-AGAAGATGGCGGAGAAGCTA | 5203–5223 |
| ID1 | 5′-GTAAAGGCCCAGACAGT | 5971–5988 |
| ID2 | 5′-ATTCAAGATCCGCTCGTG | 6607–6589 |
| PR | 5′-CACTTTAGTGCATCCTTGCCATTA | 122–98 |
| PY | 5′-GCAAACAAAGTGTGACACATGCAGCCCTG | 2064–2035 |
| RTD | 5′-AGCGGCTGTAGGAAGTAA | 4958–4940 |
| RTH | 5′-TAATGGATGCCGGTGTG | 3182–3199 |
| RTX | 5′-CCGTCCCATAAGAGCAAACCA | 5249–5228 |
| RTY | 5′-AATCTTTGGAGTTGTGC | 2077–2094 |
Oligodeoxynucleotides were synthesized in the Unité de Chimie Organique, Institut Pasteur, Paris, France.
Nucleotide numbering begins at the first base of the vanRD gene.
(iii) RT-PCR experiments.
Total RNA samples were digested with DNase I (5 U of RNA/μg) (Amersham Pharmacia Biotech) in a final volume of 1 ml for 10 min at 37°C. Samples were treated with proteinase K (0.2 mg/ml) (Boehringer, Mannheim, Germany), extracted with phenol-chloroform, and precipitated with ethanol. Reverse transcription was carried out with 2 μg of purified RNA in a 20-μl final volume containing 1× enzyme buffer (Promega), 50 mM magnesium chloride, 0.1 mg of bovine serum albumin (New England Biolabs Inc., Beverly, Ma.)/ml, 1 mM concentrations each of four deoxyribonucleoside triphosphates (Amersham Pharmacia Biotech), 50 pmol of the primer RTX (Fig. 1D; Table 2), 20 U of RNase inhibitor (RNAguard, Amersham Pharmacia Biotech), and 200 U of Moloney murine leukemia virus reverse transcriptase RNase H− (Promega). Samples were incubated for 30 min at 37°C, and Moloney murine leukemia virus reverse transcriptase was inactivated by incubation for 5 min at 95°C. The DNA products were amplified by PCR in an 80-μl reaction volume containing the previous 20-μl samples, 50 pmol each of the RTD and RTH or RTY primers (Fig. 1D; Table 2), 1× enzyme buffer (Amersham Pharmacia Biotech), and 2 U of Taq DNA polymerase (Amersham Pharmacia Biotech). PCR (30 cycles) was performed in a GeneAmp PCR system 2400 (Perkin-Elmer Cetus, Norwalk, Conn.). For Southern hybridization, PCR products were transferred from agarose gel to a Hybond N+ membrane (Amersham Pharmacia Biotech) (36). Hybridizations were performed using D1-D2 (31), HD1 (30)-HD3 (Table 2), and YD1 (30)-YD3 (Table 2) probes (Fig. 1C).
(iv) Primer extension analysis.
The synthetic oligodeoxynucleotides PR and PY (Fig. 1D; Table 2) were 5′-end labeled with [γ-32P]ATP (4,500 Ci/mmol; Amersham Pharmacia Biotech) and T4 polynucleotide kinase (Amersham Pharmacia Biotech). After phenol-chloroform extraction, labeled primers were precipitated with ethanol and redissolved in sterile water to a final concentration of 1 pmol/μl. Labeled primers (1 pmol) were annealed to 50 μg of total RNA at 65°C for 3 min, and extension was performed in a 20-μl final volume with 40 U of avian myeloblastosis virus reverse transcriptase (Boehringer) for 30 min at 42°C. After addition of 5 μl of stop solution (Amersham Pharmacia Biotech) and heat denaturation, 5-μl samples were immediately loaded onto 6% polyacrylamide–urea sequencing gels for electrophoresis. Sequencing reactions using the same primers and appropriate plasmid DNA templates were run in parallel to allow determination of the endpoints of extension products.
Analysis of peptidoglycan precursors.
Extraction and analysis of peptidoglycan precursors were carried out as described previously (26, 35). Enterococci were grown in the presence of appropriate antibiotics (10 μg of chloramphenicol/ml or 100 μg of spectinomycin/ml). Results were expressed as the percentages of total late peptidoglycan precursors represented by UDP-MurNAc-tripeptide, UDP-MurNAc-tetrapeptide, UDP-MurNAc-pentapeptide, and UDP-MurNAc-pentadepsipeptide that were determined from the integrated peak areas.
Enzyme assays.
CAT, VanX, and VanY activities in bacterial fractions were assayed as described previously (4, 7).
For determination of CAT production, enterococcal strains JH2-2, JH2-2/pAT666, BM4339, and BM4339/pAT666 were grown in the presence of spectinomycin (60 μg/ml) or chloramphenicol (10 μg/ml), with or without vancomycin (1 or 8 μg/ml). CAT activity in S100 extracts was assayed at 37°C as described previously (7).
To determine d,d-dipeptidase and d,d-carboxypeptidase activities for E. coli, strains were grown in brain heart infusion broth containing ampicillin (100 μg/ml) or kanamycin (50 μg/ml). The supernatant (S100) and resuspended pellet (C100) were collected and assayed for d,d-peptidase activities by measuring the d-Ala released from substrate hydrolysis (d-Ala-d-Ala or l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala) through coupled indicator reactions using d-amino acid oxidase and horseradish peroxidase (4). Specific activity was defined as the number of nanomoles of product formed at 37°C per minute per milligram of protein contained in the extracts.
Nucleotide sequence accession number.
The 1,400-bp fragment containing the intD gene was submitted to GenBank and assigned accession no. AF288684.
RESULTS AND DISCUSSION
Identification of the intD gene.
To clone the region flanking the 3′ end of the vanXD gene, total DNA from E. faecium BM4339 was digested with XmaI and HindIII and cloned into E. coli, and transformants were screened by hybridization with a probe corresponding to the 3′ portion of vanXD (Fig. 1A). The recombinant plasmid pAT655 (vanXD′intD) carried a 2.85-kb insert (Fig. 1B), with approximately half of the insert overlapping that in pAT654 (vanSD′vanYDHDDXD) (Table 1 and Fig. 1A) (14). Analysis of the sequence revealed an open reading frame (ORF) with the same orientation as that of the genes in the vanD cluster (Fig. 1A). The deduced sequence displayed 28% identity with the Tn4430 transposon-encoded TnpI integrase from Bacillus thuringiensis (24), 27% identity with the XerD (or XprD) recombinase from E. coli (22), and 23% identity with the XerC integrase of Haemophilus influenzae (17). The 278-amino-acid putative product of this new ORF was named IntD. IntD contained the conserved tetrad R-H-R-Y (Arg137, His224, Arg227, Tyr259), which is a hallmark for the integrase family of site-specific recombinases (Fig. 2) (28). This motif was located in the C-terminal domain of the protein (Fig. 2). The intD gene was not associated with another ORF in the 1.3-kb region downstream from intD in the pAT655 (vanXD′intD) insert. Northern hybridization indicated that intD was part of the vanD gene cluster in BM4339 (see below).
FIG. 2.
Partial alignment of the deduced sequences of IntD from E. faecium BM4339, TnpI from B. thuringiensis H1.1 (accession no. P10020) (24), XerC from H. influenzae RD/KW20 (accession no. P44818) (17), and XerD from E. coli K-12 (accession no. P21891) (22). Tetrads of residues conserved in all the members of the Int family of site-specific recombinases (28) are indicated in boldface. The numbers of residues separating the different segments containing the conserved amino acids are indicated for each protein.
Transposition is known to play an important role in the dissemination of acquired glycopeptide resistance in enteroccocci. The vanA gene cluster is included in Tn1546, a Tn3-related transposon (8), and the vanB operon is part of transposons Tn1547 (32) and Tn1549-Tn5382 (13, 18). Whether the IntD integrase is involved in the mobility of the vanD resistance gene cluster remains unknown.
Transcriptional analysis of the vanD gene cluster.
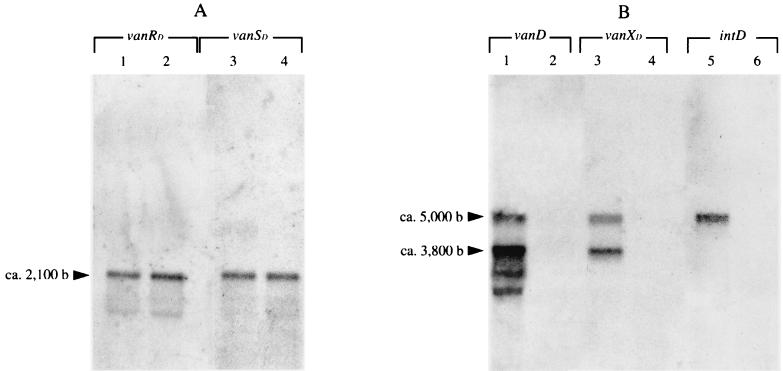
Studies of expression of the vanA and vanB clusters have shown that vanH, vanA, and vanX and at least vanYB, vanW, and vanHB are cotranscribed from their respective promoters, PH and PYB, located between the vanS and vanH genes and between the vanSB and vanYB genes, respectively (7, 16). The promoters PR and PRB, controlling transcription of the genes for the two-component regulatory system, have been identified by characterizing the activity of VanRS (2, 20) and of VanRBSB (38) on expression of the resistance genes, respectively. The similarity between the vanA, vanB, and vanD clusters, particularly in gene organization (14), suggests that the transcriptional start points in the vanD cluster would be located in the same regions as those in the vanA and vanB operons. Northern hybridization and reverse transcription-PCR (RT-PCR) were carried out to test this hypothesis, and primer extension was done to locate the vanD promoters precisely. Glycopeptide-susceptible E. faecalis JH2-2 was used as a negative control in Northern hybridization. Total RNA was extracted from exponentially growing JH2-2 and BM4339 and analyzed by Northern hybridization with probes internal to the seven genes in the vanD cluster, including intD (Fig. 1C). Hybridization was performed in duplicate in two independent experiments. As expected, RNA from JH2-2 failed to hybridize with the probes (data not shown). Three major transcripts were detected in BM4339. A transcript with the expected size of ca. 2,100 nucleotides hybridized with the vanRD and the vanSD probes (Fig. 3A, lanes 1 and 3), indicating that the genes for the two-component system are cotranscribed from a start point upstream from vanRD. Two transcripts were detected with the vanYD, vanHD, vanD, and vanXD probes, one of ca. 3,800 nucleotides and the other of ca. 5,000 nucleotides (Fig. 3B, lanes 1 and 3, and data not shown). The presence of degradation products did not prevent observation of these two transcripts with the probes internal to vanYD, vanHD, and vanD (Fig. 3B, lane 1, and data not shown). The hybridizing degradation products increased going from vanD to vanYD (data not shown), suggesting that they result from a contaminating 3′ to 5′ exonuclease (33). The ca. 5,000-bp product was also observed using the intD probe (Fig. 3B, lane 5), and the size of this transcript was in agreement with that predicted for a single mRNA, including vanYD , vanHD, vanD, vanXD, and intD. The size of the ca. 3,800-bp transcript correlated with a mRNA that would include the vanYD, vanHD, vanD, and vanXD genes. No inverted repeats, likely to form a hairpin structure for termination of transcription, were identified between the vanXD and intD genes.
FIG. 3.
Analysis of the vanD gene cluster transcription by Northern hybridization. Total RNA from BM4339 (lanes 1, 3, and 5) and BM4409 (lanes 2, 4, and 6) was hybridized with the vanRD (lanes 1 and 2) and the vanSD (lanes 3 and 4) probes (A) and the vanD (lanes 1 and 2), the vanXD (lanes 3 and 4), and the intD (lanes 5 and 6) probes (B) (see Fig. 1C). The size of the transcripts was determined according to RNA molecular weight marker I (Boehringer) (not shown).
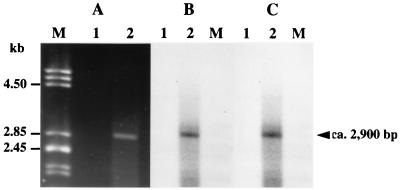
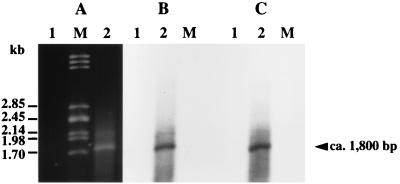
To confirm that the vanYD, vanHD, vanD, and vanXD genes were contranscribed, reverse transcription with purified total RNA from BM4339 and primer RTX internal to vanXD was performed (Fig. 1D). The complementary DNA was amplified by PCR using primer pairs internal to vanD, vanYD, and vanHD (Fig. 1D). Amplification resulted in the expected 2.9- and 1.8-kb products that cohybridized with probes specific for vanD and vanYD (Fig. 1C and 4) and for vanD and vanHD (Fig. 1C and 5), respectively. These results, along with those obtained by Northern hybridization, indicate that the vanYD, vanHD, vanD, and vanXD genes are cotranscribed and that the approximately 3,800-nucleotide resulting messenger starts upstream from vanYD.
FIG. 4.
Analysis of the transcription of the vanYD and vanD genes. Electrophoresis of the products obtained by RT-PCR using the primers RTY and RTD (see Fig. 1D and Table 2) (A) and corresponding Southern hybridization (B and C). Incubations were carried out in the absence (lanes 1) or presence (lanes 2) of reverse transcriptase. Lanes M contained DNA from bacteriophage lambda digested with PstI as a marker. (B) Hybridization with a vanD probe (see Fig. 1C). (C) Hybridization with a vanYD probe (see Fig. 1C).
FIG. 5.
Analysis of the transcription of the vanHD and vanD genes. Electrophoresis of the products obtained by RT-PCR using the primers RTH and RTD (see Fig. 1D and Table 2) (A) and corresponding Southern hybridization (B and C). Incubations were carried out in the absence (lanes 1) or presence (lanes 2) of reverse transcriptase. Lanes M contained DNA from bacteriophage lambda digested with PstI as a marker. (B) Hybridization with a vanD probe (see Fig. 1C). (C) Hybridization with a vanHD probe (see Fig. 1C).
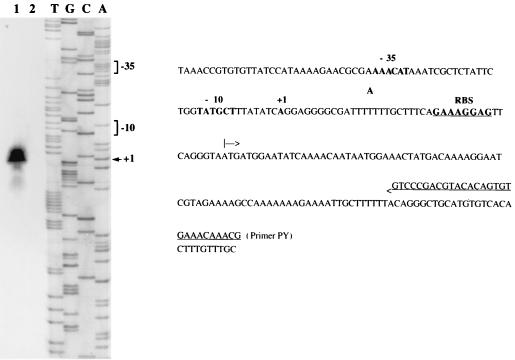
Primer extension was used to explore the region upstream from vanRD and the vanSD-vanYD intergenic region for transcriptional start sites using the oligodeoxynucleotides PR and PY (Fig. 1D) complementary to the 5′ end of vanRD and vanYD respectively, as primers. This allowed the positioning of transcriptional start sites PRD (Fig. 6) and PYD (Fig. 7). These results were confirmed by using two other oligodeoxynucleotides complementary to sequences close to PR and PY but located closer to the mapped start sites (data not shown). Promoters PRD and PYD contained the −35 and −10 regions corresponding to the ς70 recognition sequences which were separated by 17 bp.
FIG. 6.
Identification of the transcriptional start site for the vanRD and vanSD genes in BM4339 by primer extension analysis. Left panel, lane 1, primer elongation product obtained with oligodeoxynucleotide PR and 50 μg of total RNA from BM4339 (arrowhead); lane 2, control without RNA; lanes T, G, C, and A, results of sequencing reactions performed with the same primer. Right panel, sequence from nucleotide position −128 to +122 (numbering from the A of the ATG start codon of vanRD, negative in the 3′ to 5′ direction and positive in the 5′ to 3′ direction). The +1 transcriptional start site for the vanRD and vanSD mRNA in BM4339 and the −35 and −10 promoter sequences located upstream are in boldface. The ATG start codon of vanRD is indicated by an arrow, and the RBS is in boldface and underlined.
FIG. 7.
Identification of the transcriptional start site for the vanYD, vanHD, vanD, vanXD, and intD genes in BM4339 by primer extension analysis. Left panel, lane 1, primer elongation product obtained with oligodeoxynucleotide PY and 50 μg of total RNA from BM4339 (arrowhead); lane 2, control without RNA; lanes T, G, C, and A, results of sequencing reactions performed with the same primer. Right panel, sequence from nucleotide position −110 to +109 (numbering from the A of the ATG start codon of vanYD, negative in the 3′-to-5′ direction and positive in the 5′-to-3′ direction). The +1 transcriptional start site for the vanYD, vanHD, vanD, vanXD, and intD mRNA in BM4339 and the −35 and −10 promoter sequences located upstream are in boldface. The ATG start codon of vanYD is indicated by an arrow, and the RBS is in boldface and underlined.
Regulation of the PYD promoter by VanRD and VanSD.
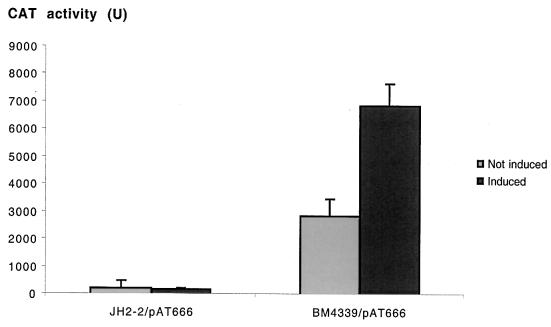
To investigate the role of the VanRD-VanSD two-component regulatory system in the control of transcription of the vanYD, vanHD, vanD, and vanXD genes, a DNA fragment containing the PYD promoter was cloned upstream from the cat reporter gene of the shuttle promoter probing vector pAT78(cat) to generate plasmid pAT666 (PYD cat) (Fig. 1B). E. faecalis JH2-2 was used as a control for expression of the cat gene under the control of the PYD promoter in the absence of vanRD-vanSD. No significant CAT activity was detected in E. faecalis JH2-2/pAT666(PYD cat) (Fig. 8); the cat gene carried by pAT666 (PYD cat) was transcribed in this strain at a low level similar to that detected with the promoterless pAT78 (cat) (data not shown). This basal level of transcription was not modified by the presence of vancomycin at 1 μg/ml (Fig. 8). In contrast, when pAT666 (PYD cat) was introduced into E. faecium BM4339, significant production of CAT was detected (Fig. 8), at a ca. 2,800-fold-higher level than that displayed by BM4339 or BM4339/pAT78 (cat) (data not shown).
FIG. 8.
CAT activity in cytoplasmic extracts from E. faecalis JH2-2 and from E. faecium BM4339 harboring plasmid pAT666 (PYD cat). Controls were performed without addition of vancomycin to the culture medium (“not induced” bars), and induction was achieved by adding 1 μg of vancomycin/ml to cultures of JH2-2/pAT666 and 8 μg of vancomycin/ml to cultures of BM4339/pAT666 (“induced” bars). Enzymatic activity was expressed as nanomoles of product formed per minute per milligram of protein in S100 extracts. Results are means ± standard deviations obtained from three independent extracts.
These observations indicate that the PYD promoter did not direct constitutive transcription but required a signal to promote gene expression. Promoter PYD was not 1066016.dhp active in E. faecalis JH2-2, owing to the absence of an inducing signal, which could be transmitted by the VanRD and VanSD regulatory partners or by some other regulatory factors specific for E. faecium strains.
According to the homology between the vanA, vanB, and vanD gene clusters, the chromosomally encoded VanRD-VanSD system could be responsible for trans activation of transcription from PYD in BM4339/pAT666 (PYD cat). If so, constitutive expression of the vanD gene cluster in BM4339 suggests that the VanRD protein is present in its phosphorylated form, thus activating transcription from the PYD promoter. This could be due to alteration in signal recognition or of the phosphatase activity of the cognate VanSD sensor, or to phosphorylation of VanRD by a nonpartner protein kinase (14).
The presence of vancomycin at a concentration of 8 μg/ml doubled transcription from PYD in BM4339 (Fig. 8). However, the construction used may not reflect the in vivo conditions, since the high-copy-number plasmid pAT666 (PYD cat) allows for high-level expression from the PYD promoter. Nevertheless, an inducing effect of vancomycin on the level of transcription of the chromosomal resistance genes cannot be excluded, since this would not necessarily make an impact on the level of translation or of protein activity in BM4339.
VanXD and VanYD d,d-peptidase activity in E. coli.
Strain BM4339 does not produce d-Ala-d-Ala-containing peptidoglycan precursors because of a frameshift mutation in the chromosomal ddl gene (14). Thus, no d,d-dipeptidase activity is required for glycopeptide resistance in this genetic background. In fact, BM4339 does not produce active VanXd (31), although vanXD is transcribed (Fig. 3B, lane 3) and the deduced sequence of VanXD does not contain mutations in the conserved residues known to be involved in zinc binding and catalysis (25). To test if vanXD and vanYD encode functional enzymes, the genes and their ribosome binding sites (RBS) were cloned into E. coli under the control of the Plac promoter of pUC18 and pBGS18+, respectively. Hydrolysis of d-Ala-d-Ala was detected in cytoplasmic extracts from E. coli harboring pAT659 (vanXD) but not pUC18 (Table 3), indicating that vanXD encodes a functional enzyme. It is possible that VanXD is translated in BM4339 but is unstable and is degraded. Alternatively, its activity may be too low to be measured in BM4339, in which there is a single copy of the vanXD gene, but is detectable when it is cloned in pUC18 in E. coli, which can harbor up to 700 copies of the plasmid.
TABLE 3.
d,d-dipeptidase and d,d-carboxypeptidase activities in E. coli TB1 harboring the indicated plasmids
| Plasmid harbored | Activity (nmol min−1 mg−1)a
|
||
|---|---|---|---|
| d,d-dipeptidaseb |
d,d-carboxypeptidasec
|
||
| Without penicillin G | With penicillin G (10 mM) | ||
| pUC18 | NDd | ND | NTe |
| pAT659 (pUC18ΩvanXD) | 810 ± 65 | ND | NT |
| pBGS18+ | NT | ND | ND |
| pAT660 (pBGS18+ΩvanYD) | NT | 3.39 ± 0.11 | ND |
Enzymatic activity was assayed on protein extracts from E. coli harboring the vectors pUC18 and pBGS18+ or plasmids pAT659 and pAT660 which carry the vanXD and the vanYD genes, respectively.
Activity in cytoplasmic extracts.
Activity in membrane extracts.
ND, not detectable.
NT, not tested.
d,d-carboxypeptidase activity was detected in membrane preparations from E. coli harboring pAT660 (vanYD), which was completely inhibited by the presence of 10 mM penicillin G (Table 3), as shown for BM4339 (31). The sequence of VanYD is homologous to those of penicillin-binding proteins (14), and the expression of the BM4339 vanYD gene in E. coli generates an enzymatically active d,d-carboxypeptidase. The mechanism by which this enzyme affects resistance remains to be elucidated, since, although VanYD presumably binds penicillins on the external surface of the cytoplasmic membrane, the tetrapeptide product formed by the d,d-carboxypeptidase is located in the cytoplasm. The location and properties of the VanYD protein are currently under investigation (P. E. Reynolds, B. Casadewall, and P. Courvalin, unpublished data).
Characterization of glycopeptide-susceptible derivatives of E. faecium BM4339.
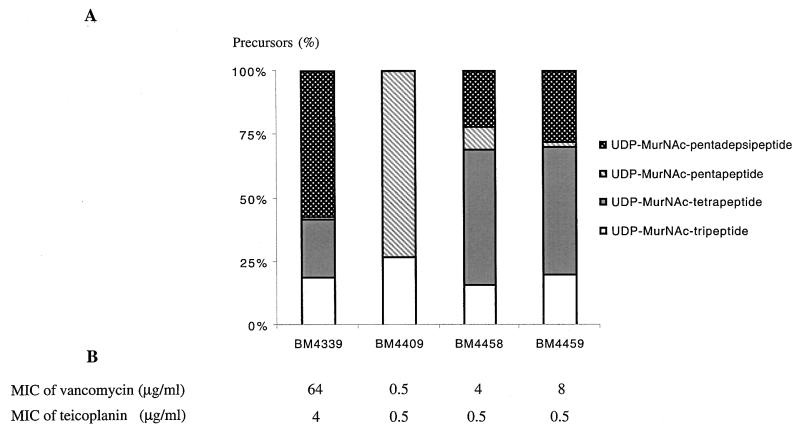
Complementation of the frameshift mutation in the BM4339 chromosomal ddl gene (14) was studied in two systems. Transformation of BM4339 with the high-copy-number plasmid pAT662 (P2ddl) restored glycopeptide susceptibility in E. faecium BM4409 by trans complementation (Fig. 9B). The effect of a single copy of the heterologous ddl gene in the chromosome of BM4339 was investigated, since cis complementation was anticipated to be more stable and more analogous to the natural situation. Transformants were obtained following integration of the suicide plasmid pAT665 (P2ddl) into the BM4339 chromosome. Glycopeptide-susceptible BM4458 and BM4459 [BM4339::pAT665 (P2ddl)] differed in their sites of insertion (data not shown), and the MICs of vancomycin for these strains were significantly, but unequally, decreased (Fig. 9B). The MICs of teicoplanin were also altered (Fig. 9B).
FIG. 9.
Analysis of derivatives of E. faecium BM4339 harboring a wild-type ddl gene on a high-copy-number plasmid (BM4409) or the same gene integrated as a single copy in the chromosome (BM4458 and BM4459). (A) Proportions of late soluble cytoplasmic peptidoglycan precursors accumulated in the presence of ramoplanin. (B) Levels of resistance to vancomycin and teicoplanin.
The amounts of late peptidoglycan precursors were analyzed in the BM4339 derivatives BM4409 [BM4339/pAT662 (P2ddl)], BM4458, and BM4459. Transformants BM4458 and BM4459 [BM4339::pAT665 (P2ddl)] contained different amounts of pentapeptides, consistent with the vancomycin MICs (Fig. 9A and B). This observation indicates that the chromosomal sequences flanking the heterologous ddl gene are likely to affect its expression. Although a larger proportion of pentapeptide was present in the peptidoglycan precursor pool of BM4458 and BM4459 in comparison with that in BM4339, tetrapeptide was the main component, being present at similar and high levels in both strains (Fig. 9A). The ratio of pentadepsipeptide to tetrapeptide in the two transformants was inverted in comparison with that in BM4339 (Fig. 9A). In BM4339, tetrapeptide originated mainly from pentadepsipeptide, with possibly a small amount generated from pentapeptide synthesized by the Ddl activity of VanD. However, the Ddl in BM4458 and BM4459, encoded by the single chromosomal copy of ddl, synthesized a greater amount of pentapeptide, and tetrapeptide results from hydrolysis of both pentapeptide and pentadepsipeptide. As already shown for VanY (1), further investigations have demonstrated that the d,d-carboxypeptidase of BM4339 has greater activity against pentapeptide than pentadepsipeptide (P. E. Reynolds, B. Casadewall, and P. Courvalin, unpublished data).
In addition to tripeptide precursors, BM4409 [BM4339/pAT662 (P2ddl)] contains only pentapeptide (Fig. 9A). No pentadepsipeptide was detected, as if the vanD gene cluster was no longer expressed (Fig. 9A). This result is in agreement with the high-level expression of a functional Ddl and with the low MICs of glycopeptides for BM4409 (Fig. 9B). To investigate the effect of a wild-type Ddl on the expression of the vanD gene cluster, Northern hybridization was performed with BM4409 and BM4339 total RNA, the latter being a control for transcription of the vanD cluster. Surprisingly, the vanRD-vanSD regulatory region was transcribed in BM4409 (Fig. 3A), whereas the vanYD, vanHD, vanD, vanXD, and intD genes were not (Fig. 3B and data not shown). Cell wall biosynthesis in BM4409 has apparently been switched from the production of d-Ala-d-Lac-ending precursors, which occurs constitutively in BM4339, to that of d-Ala-d-Ala-containing precursors. The lack of transcription of the genes controlled by PYD indicated an absence of activation of the promoter. This is consistent with the existence of a signal-transducing pathway in E. faecium BM4339, probably involving the VanRD-VanSD two-component system. One explanation for the silencing of transcription of vanYD, vanHD, vanD, vanXD, and intD could be that the high levels of d-Ala-d-Ala, synthesized by the heterologous Ddl in BM4409, disrupt transduction of the signal by preventing VanRD-phosphate from accumulating and activating transcription at the PYD promoter. To test this possibility, the effect of d-Ala-d-Ala in the culture medium on the level of vancomycin resistance in BM4339 was determined (6). If high levels of intracellular d-Ala-d-Ala prevent expression of the vanD resistance genes from PYD, BM4339 would be expected to become susceptible to vancomycin. The level of vancomycin resistance of BM4339 was unaffected by d-Ala-d-Ala added at final concentrations ranging from 0 to 40 mM (data not shown). Since the uptake of dipeptides is mediated by peptide permeases with broad specificity (10), the lack of an effect of d-Ala-d-Ala is unlikely to result from inefficient transport. Alternatively, the absence of transcription of the resistance genes in BM4409 may result from the lack of a signal by VanRD-VanSD, thus preventing transcription from the PYD promoter.
In conclusion, the vanD glycopeptide resistance gene cluster from E. faecium BM4339 comprises seven genes which are transcribed from two promoters, PRD for the vanRD and vanSD regulatory genes and PYD for the vanYD, vanHD, vanD, van XD, and intD genes. Expression of the latter five genes is likely to result from activation of transcription from PYD by the VanRD response regulator. The signal responsible for constitutive expression of the resistance genes remains to be established.
ACKNOWLEDGMENTS
We thank T. Msadek for help with RNA preparation and helpful discussions and J. Blanchard for critical reading of the manuscript. B.C. is grateful to F. Depardieu for construction of pAT632 and to S. Goussard and B. Périchon for constant technical advice.
This work was supported in part by a Bristol-Myers Squibb unrestricted biomedical research grant in infectious diseases and by the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires from the Ministère de l'Education Nationale, de la Recherche et de la Technologie. B.C. was the recipient of a grant from the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Arthur M, Depardieu F, Cabanié L, Reynolds P, Courvalin P. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol Microbiol. 1998;31:819–830. doi: 10.1046/j.1365-2958.1998.01114.x. [DOI] [PubMed] [Google Scholar]
- 2.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol. 1997;179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene. 1995;154:87–92. doi: 10.1016/0378-1119(94)00851-i. [DOI] [PubMed] [Google Scholar]
- 4.Arthur M, Depardieu F, Reynolds P, Courvalin P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol. 1996;21:33–44. doi: 10.1046/j.1365-2958.1996.00617.x. [DOI] [PubMed] [Google Scholar]
- 5.Arthur M, Depardieu F, Snaith H A, Reynolds P E, Courvalin P. Contribution of VanY d,d-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994;38:1899–1903. doi: 10.1128/aac.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur M, Molinas C, Bugg T D H, Wright G D, Walsh C T, Courvalin P. Evidence for in vivo incorporation of d-lactate into peptidoglycan precursors of vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1992;36:867–869. doi: 10.1128/aac.36.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 10.Atherton F R, Hall M J, Hassall C H, Lambert R W, Lloyd W J, Ringrose P S. Phosphonopeptides as antibacterial agents: mechanism of action of alaphosphin. Antimicrob Agents Chemother. 1979;15:696–705. doi: 10.1128/aac.15.5.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 12.Bugg T D H, Wright G D, Dutka-Malen S, Arthur M, Courvalin P, Walsh C T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- 13.Carias L L, Rudin S D, Donskey C J, Rice L B. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J Bacteriol. 1998;180:4426–4434. doi: 10.1128/jb.180.17.4426-4434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casadewall B, Courvalin P. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J Bacteriol. 1999;181:3644–3648. doi: 10.1128/jb.181.12.3644-3648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Celli J, Trieu-Cuot P. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol Microbiol. 1998;28:103–117. doi: 10.1046/j.1365-2958.1998.00778.x. [DOI] [PubMed] [Google Scholar]
- 16.Evers S, Courvalin P. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. 1996;178:1302–1309. doi: 10.1128/jb.178.5.1302-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, Fitzhugh W, Fields C A, Gocayne J D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 18.Garnier F, Taourit S, Glaser P, Courvalin P, Galimand M. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology. 2000;146:1481–1489. doi: 10.1099/00221287-146-6-1481. [DOI] [PubMed] [Google Scholar]
- 19.Glatron M F, Rapoport G. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie. 1972;54:1291–1301. doi: 10.1016/s0300-9084(72)80070-1. [DOI] [PubMed] [Google Scholar]
- 20.Holman T R, Wu Z, Wanner B L, Walsh C T. Identification of the DNA-binding site for the phosphorylated vanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry. 1994;33:4625–4631. doi: 10.1021/bi00181a024. [DOI] [PubMed] [Google Scholar]
- 21.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovett S T, Kolodner R D. Nucleotide sequence of the Escherichia coli recJ chromosomal region and construction of RecJ-overexpression plasmids. J Bacteriol. 1991;173:353–364. doi: 10.1128/jb.173.1.353-364.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnet S, Courvalin P, Lambert T. Activation of the cryptic aac(6′)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J Bacteriol. 1999;181:6650–6655. doi: 10.1128/jb.181.21.6650-6655.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahillon J, Lereclus D. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 1988;7:1515–1526. doi: 10.1002/j.1460-2075.1988.tb02971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCafferty D G, Lessard I A, Walsh C T. Mutational analysis of potential zinc-binding residues in the active site of the enterococcal d-Ala-d-Ala dipeptidase VanX. Biochemistry. 1997;36:10498–10505. doi: 10.1021/bi970543u. [DOI] [PubMed] [Google Scholar]
- 26.Messer J, Reynolds P E. Modified peptidoglycan precursors produced by glycopeptide-resistant enterococci. FEMS Microbiol Lett. 1992;94:195–200. doi: 10.1016/0378-1097(92)90608-q. [DOI] [PubMed] [Google Scholar]
- 27.Nagarajan R. Antibacterial activities and modes of action of vancomycin and related glycopeptides. Antimicrob Agents Chemother. 1991;35:605–609. doi: 10.1128/aac.35.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunes-Düby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostrowsky B E, Clark N C, Thauvin-Eliopoulos C, Venkataraman L, Samore M H, Tenover F C, Eliopoulos G M, Moellering R C, Jr, Gold H S. A cluster of VanD vancomycin-resistant Enterococcus faecium: molecular characterization and clinical epidemiology. J Infect Dis. 1999;180:1177–1185. doi: 10.1086/315030. [DOI] [PubMed] [Google Scholar]
- 30.Périchon B, Casadewall B, Reynolds P, Courvalin P. Glycopeptide-resistant Enterococcus faecium BM4416 is a VanD-type strain with an impaired d-alanine:d-alanine ligase. Antimicrob Agents Chemother. 2000;44:1346–1348. doi: 10.1128/aac.44.5.1346-1348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Périchon B, Reynolds P, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997;41:2016–2018. doi: 10.1128/aac.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quintiliani R, Jr, Courvalin P. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene. 1996;172:1–8. doi: 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 33.Rauhut R, Klug G. mRNA degradation in bacteria. FEMS Microbiol Rev. 1999;23:353–370. doi: 10.1111/j.1574-6976.1999.tb00404.x. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds P E. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1989;8:943–950. doi: 10.1007/BF01967563. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds P E, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol Microbiol. 1994;13:1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva J C, Haldimann A, Prahalad M K, Walsh C T, Wanner B L. In vivo characterization of the type A and B vancomycin-resistant enterococci (VRE) VanRS two-component systems in Escherichia coli: a nonpathogenic model for studying the VRE signal transduction pathways. Proc Natl Acad Sci USA. 1998;95:11951–11956. doi: 10.1073/pnas.95.20.11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spratt B G, Hedge P J, te Heesen S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 40.Steers E, Foltz E L, Graves B S, Rindel J. An inocula replicating apparatus for routine testing of bacterial susceptibility to antibiotics. Antibiot Chemother (Basel) 1959;9:307–311. [PubMed] [Google Scholar]
- 41.Storrs M J, Poyart-Salmeron C, Trieu-Cuot P, Courvalin P. Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon-encoded integrase. J Bacteriol. 1991;173:4347–4352. doi: 10.1128/jb.173.14.4347-4352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vieira J, Messing J. The pUC plasmids and M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 43.Woodcock D M, Crowther P J, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith S S, Michael M Z, Graham M W. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]