Abstract
Muscular dystrophy due to dystrophin deficiency in humans is phenotypically divided into a severe Duchenne and milder Becker type. Dystrophin deficiency has also been described in a few animal species, and few DMD gene variants have been identified in animals. Here, we characterize the clinical, histopathological, and molecular genetic aspects of a family of Maine Coon crossbred cats with clinically mild and slowly progressive muscular dystrophy. Two young adult male littermate cats exhibited abnormal gait and muscular hypertrophy with macroglossia. Serum creatine kinase activities were highly increased. Histopathologically, dystrophic skeletal muscle exhibited marked structural changes including atrophic, hypertrophic, and necrotic muscle fibers. Immunohistochemistry showed irregularly reduced expression of dystrophin but the staining of other muscle proteins such as β- and γ-sarcoglycans as well as desmin was also diminished. Whole genome sequencing of one affected cat and genotyping of the littermate found both to be hemizygous mutant at a single DMD missense variant (c.4186C>T). No other protein-changing variants in candidate genes for muscular dystrophy were detected. In addition, one clinically healthy male littermate was hemizygous wildtype, while the queen and one female littermate were clinically healthy, but heterozygous. The predicted amino acid exchange (p.His1396Tyr) resides in a conserved central rod spectrin domain of dystrophin. Various protein modeling programs did not predict major disruption of the dystrophin protein by this substitution, but the altered charge of the region may still affect protein function. This study represents the first genotype-to-phenotype correlation of Becker-type dystrophin deficiency in companion animals.
Keywords: Felis catus, feline, myopathy, creatine kinase, Duchenne, mutation, hereditary disease, X-linked, precision medicine
1. Introduction
Dystrophin deficiency was discovered in 1986 as the major cause of X-linked recessive muscular dystrophy in humans and thereafter also described in several domestic animal species [1,2,3,4]. Dystrophin-deficient dogs exhibit very similar clinical features, and thus became the favorite animal model to investigate this disorder and assess novel therapies at the preclinical stage (Online Mendelian Inheritance in Animals [OMIA] #001081-9615) [5,6,7]. In contrast to dystrophin-deficient humans and dogs, cats with X-linked muscular dystrophy exhibit predominantly muscular hypertrophy rather than dystrophy (OMIA #001081-9685) [8,9,10,11,12,13,14,15]. More recently, several juvenile pigs with dystrophin deficiency or insufficiency have been described (OMIA #001081-9823 [Duchenne]; #001888-9823 [Becker]) and few became useful disease models [16,17,18,19,20,21,22,23]. However, naturally occurring and genetically engineered models in mice [24,25,26,27], rats [28], rabbits [29], and pigs [16,17,20,22] did not or only selectively show the classic muscular dystrophy and atrophy as observed in human and canine patients.
Based on disease onset, progression, and clinical phenotype, dystrophin deficiency in humans is differentiated between a severe Duchenne- and milder Becker-type muscular dystrophy [30,31]. Moreover, the discovery of likely pathogenic DMD variants revealed that the clinical features of X-linked muscular dystrophy correlate well with the DMD genotype in humans. Human patients with Duchenne muscular dystrophy have typically nonsense or frameshift DMD variants with complete loss of dystrophin protein function. In contrast, those with Becker type have missense and other milder DMD variants allowing for varied dystrophin expression and function [32,33,34,35].
Domestic animals with dystrophin deficiency mostly exhibit a severe phenotype with the juvenile onset of, and the rapidly progressive myopathy which corresponds to, the Duchenne-type muscular dystrophy [3,4,17,25]. Only a few clinicopathological reports in domestic animals suggest the presence of a milder disease course in dogs and pigs and thus potentially reflect Becker-type muscular dystrophy [18,23,36,37,38]. We report here on adult male domestic cats with a clinically mild-to-moderate and slowly progressive dystrophic myopathy course. Histopathology, histochemistry, and immunohistochemistry revealed moderate-to-severe structural changes and irregular expression of dystrophin and other proteins, and a missense DMD variant likely causing X-linked recessive muscular dystrophy, which mimics Becker-type muscular dystrophy in human patients.
2. Results
2.1. Clinical Manifestations
A 2½-year-old castrated male domestic cat (index case, cat #1) and its male littermate (cat #2) were presented to the Tierärztliche Klinik für Kleintiere, Neu-Anspach, Germany, because of clumsy gait, difficulty jumping and grooming, and protrusion of the tongue tip (video in supplementary materials). These cats lived in- and outdoors in a suburban neighborhood without any major physical impediments according to their owners. No information of prior physical examinations, medical histories, and laboratory test results was available.
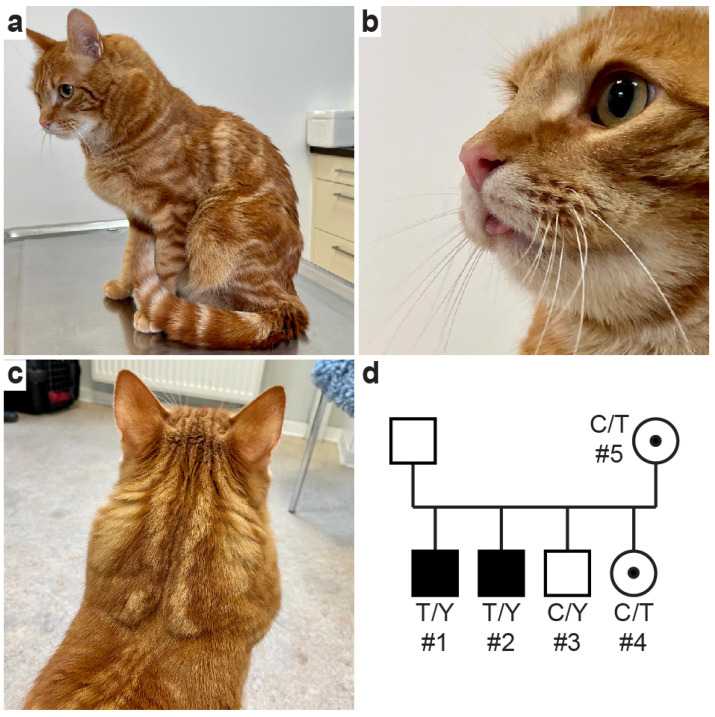
Physical examinations of the two affected male littermates revealed a normal body condition score but marked generalized muscular hypertrophy, particularly of the neck and upper limbs, as well as macroglossia and a more forceful breathing pattern (Figure 1). There was no muscle cramping and dimpling, making a congenital myopathy unlikely. Imaging of both the body cavities and heart of cat #1 and cat #2 did not reveal any abnormalities besides the systemic skeletal muscular hypertrophy. There were no cardiac murmurs auscultated, pulse rate and quality appeared normal, and echocardiogram parameters were in normal reference intervals (Figure S1), thus providing no clinical evidence of a cardiomyopathy. Furthermore, there was neither clinical nor radiographic evidence of megaesophagus in either of the affected cats.
Figure 1.
(a–c) Clinical images of affected cat #1, with muscle hypertrophy and protrusion of the tongue tip; (video in supplementary materials), (d) Pedigree of Maine Coon crossbred domestic cat family with muscular dystrophy. The affected males are depicted as black squares; clinically unaffected males are shown as empty symbols. The unaffected female littermate cat #4 and queen cat #5 were heterozygous for the mutant allele (C/T); symbols with black dots indicate carrier status for an X-linked trait. Genotypes for the DMD:c.4186C>T variant are indicated; the “Y” indicates the hemizygous genotype in males with either a mutant or wildtype X-chromosomal allele. The queen’s mother was a Maine Coon cat but not available for study. The litter’s tom was also not available for clinical examination and genotyping.
While the tom of this litter remains unknown, one male (cat #3) and one female littermate (cat #4) and the queen (cat #5) of the affected litter—all living in the same neighborhood—showed neither muscular hypertrophy nor other clinical abnormalities on physical examination. According to the owner, the queen was a Maine Coon x domestic shorthair crossbred cat, and the breed of the tom remained unknown. Compared to the healthy appearing littermates, the two affected males appeared to have longer hair coats, bushy tails, and ear tufts suggesting Maine Coon breed ancestry. The results of a commercial feline DNA panel of cat #1 revealed a 26% Maine Coon contribution, a blood type A by CMAH genotyping, and no known disease-causing variants tested for (Mars Veterinary, Portland, OR, USA) [39,40]. The conditions of both affected males only marginally progressed clinically, and the two healthy appearing littermates as well as the litter’s queen also remained unchanged over the eight-month observation period.
2.2. Blood Test Results
The serum creatine kinase activities of the two male affected cats were extremely high at initial examination as well as after eight months compared to the other male and female littermate, queen, and reference intervals (Table 1). In addition, the serum alanine (ALT) and aspartate aminotransferase (AST) activities were moderately increased in both affected males consistent with a dystrophic myopathy. All other routine blood parameters including complete blood cell count and serum chemistry results of both affected males were within reference intervals. Moreover, serum creatine kinase activities of the male and female littermates and queen of the litter were within the reference interval (Table 1).
Table 1.
Select serum enzyme activities and DMD missense genotyping results in a family of Maine Coon crossbred domestic cats with X-linked muscular dystrophy.
| Cat | Phenotype and Relationship | Age (Years) |
Serum Enzyme Activities (IU/L) | DMD:c.4186C>T Genotype | ||
|---|---|---|---|---|---|---|
| CK | AST | ALT | ||||
| #1 | Index Case, Affected Male | 2.5 | 15,597 | 364 | 364 | T/Y |
| 3.2 | 23,085 | 477 | 369 | |||
| #2 | Affected Male Littermate | 2.7 | 30,939 | 608 | 365 | T/Y |
| 3.2 | 38,814 | 732 | 472 | |||
| #3 | Unaffected Male Littermate | 2.5 | 136 | ND | ND | C/Y |
| #4 | Unaffected Female Littermate | 2.5 | 177 | ND | ND | C/T |
| #5 | Unaffected Queen | 3.5 | 235 | ND | ND | C/T |
| Reference Interval | Adults | 52–250 | 14–71 | 37–175 | C/Y or C/C | |
Serum enzyme activities: CK, creatine kinase; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ND, not determined; T/Y, hemizygous mutant; C/Y, hemizygous wildtype; C/T and C/C, heterozygous and homozygous wildtype females, respectively.
2.3. Histopathology and Immunohistochemistry
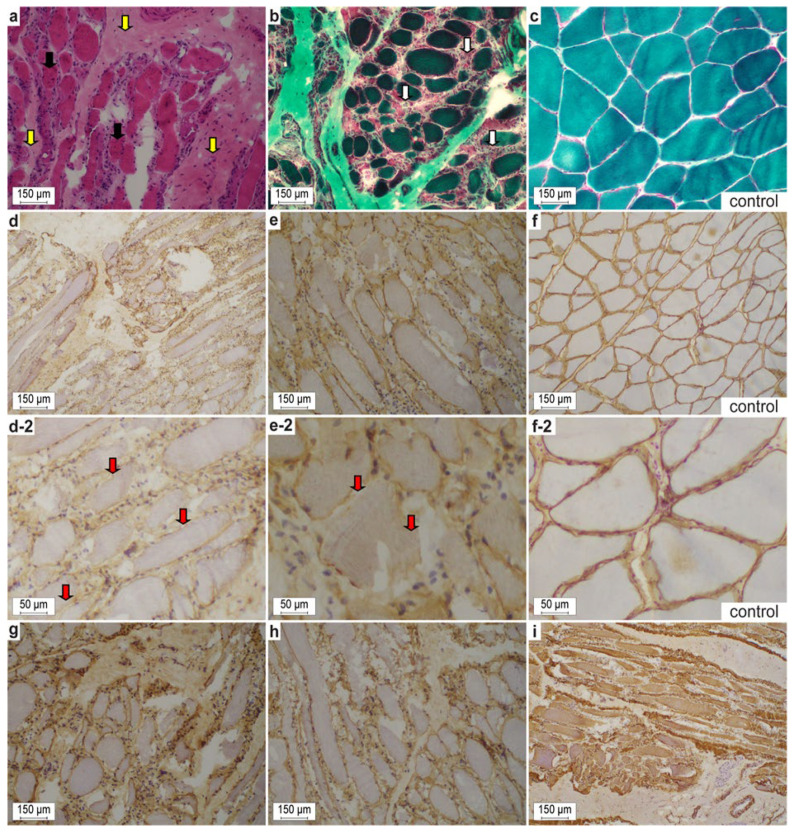
Transverse and longitudinal sections of gastrocnemius muscle obtained from cat #1 showed structural changes consistent with a dystrophic myopathy (Figure 2): Bimodal pathological fiber size variations, comprising multiple enlarged and atrophic myocytes, as well as muscle fiber necrosis, were present. Furthermore, chronic diffuse myofibrosis, nuclear internalization, and interstitial lymphocytic infiltration were seen. No ragged-red myofibers, cones, rods, cores, and targets were seen, but there were myocytes with increased fibrils and clumping of the myotubular apparatus. Very mild hypomyelination was noted in endomysial nerve fibers (Figure 2a,b). Enzyme histochemistry and myosin heavy chain immunohistochemistry revealed normal type 1 and 2 fiber distribution. Mild increases in subsarcolemmal and interfibrillar lipid droplets were found in a few muscle fibers. Positive acid phosphatase activity highlighted necrotizing myofibers. Together, these histopathological features were consistent with a dystrophic myopathy as previously described in domestic cats with X-linked muscular dystrophy [8,9,10,11,12,13,14,41]
Figure 2.
(a,b,d,e,g–i) Representative cryosections of the affected gastrocnemius muscle from the dystrophic Maine Coon crossbred domestic cat #1. (a,b) Noteworthy are the variability of myofiber sizes, degenerative (groups of necrotic fibers with myophagocytosis, white arrows) and regenerative muscle features (small fibers with prominent vesicular central nuclei; multiple nuclear internalization, black arrows), and endomysial fibrosis (yellow arrows) in affected cat #1. (d,e) On immunohistochemistry, membranous dystrophin often is discontinuous or even absent in between individual fibers (red arrows). Moreover, γ- and β-sarcoglycans (g,h) and desmin (i) were diminished and interrupted. (c,f) control sections from a European domestic shorthair cat. (a) H&E stain. (b,c) Gomori trichrome stain according to Engel. (d,f) Dystrophin 1 (1:100, DYS1). (e) Dystrophin 2 (1:100, DYS2) at regular and increased magnification. (g) γ-sarcoglycan (1:100, γ-SARC). (h) β-sarcoglycan (1:50, β-SARC). (i) desmin (1:50, Desmin Clone D33 M0760). (c,f) Control sections of a domestic short hair cat.
Immunohistochemistry of muscle from cat #1 revealed irregular and partially deficient expression of both DYS1 (rod domain) and DYS2 (carboxy terminus) (Figure 2d,e,d-2,e-2). Moreover, antibody reactivity of β- and γ-sarcoglycans (Figure 2g,h) as well as β-dystroglycan was diminished and uneven compared to the feline controls. Furthermore, the muscular intermediate filament desmin was severely interrupted (Figure 3). Despite the severe structural muscle fiber disturbances, the dystrophin remains identifiable in the dystrophin-associated muscle protein complex. The antibody reactions against DYS3 (amino terminus), ß-dystroglycan, δ-sarcoglycan, and also α2-laminin were weak or negative against the control as well as cat #1 muscle proteins, thus precluding any interpretation.
Figure 3.
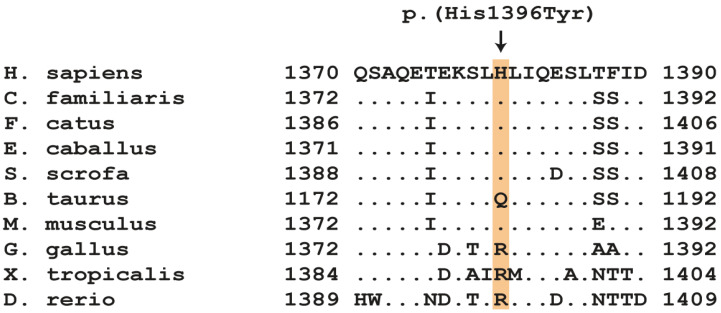
Amino acid alignment of a part of dystrophin’s spectrin 10 domain in several species surrounding the p.(His1396Tyr) variant. The DMD variant affects a histidine residue, which is mostly conserved among mammals. Accession numbers: human (H. sapiens) NP_003997.1; dog (C. familiaris) NP_001003343.1; cat (F. catus) XP_044906722.1; horse (E. caballus) XP_023489578.1; domestic pig (S. scrofa) XP_020935202.1; domestic cattle (B. taurus) XP_002700291.2.; mouse (M. musculus) NP_031894.1; chicken (G. gallus) NP_990630.1; western clawed frog (X. tropicalis) XP_031752861.1; zebrafish (D. rerio) NP_001313616.1.
2.4. Molecular Genetics
The whole genome of dystrophic cat #1 was sequenced and searched for private homozygous and hemizygous variants that were not present in the genome sequences of 74 control cats (Table 2 and Table S2).
Table 2.
Filtering for private protein-changing variants in dystrophic cat #1 against 74 control genomes. Only homozygous or hemizygous variants are reported (also Table S3).
| Filtering Steps for Dystrophic Cat | Variants in Dystrophic Cat #1 | |
|---|---|---|
| Autosomes | X-Chromosome | |
| All Variants | 4,339,302 | 180,936 |
| Protein-Changing Variants | 35,548 | 305 |
| Private Variants | 1881 | 2367 |
| Private Protein-Changing Variants | 8 | 10 |
| Private Variants in Muscle Genes | 0 | 1 |
Analysis of the detected variants revealed only 18 private protein-changing variants in 13 genes (Table S3). Seventeen did not affect known musculoskeletal genes and thus were unlikely responsible for the myopathy in the index case. One single nucleotide variant (SNV) was in the DMD gene representing a prime candidate gene for a hereditary dystrophic myopathy. This DMD missense variant in exon 30 constitutes a G>A exchange at position 27,988,938 on the short arm of the X-chromosome, XM_045050787.1:c.4186C>T. This variant is predicted to result in a histidine-to-tyrosine substitution in the DMD protein, XP_044906722.1:p.(His1396Tyr). The predicted amino acid substitution lies within the spectrin 10 domain of the protein, which is conserved among mammals (Figure 3). The His1396Tyr substitution was not predicted to be pathogenic by several in silico prediction tools (Table 3). A homologous amino acid exchange, NP_003997.2:His1380Tyr, was observed once in the DMD gene of a hemizygous adult male person according to the gnomAD database but without further clinicopathological information. The gnomAD ID for the variant is “X-32429964-G-A” [42].
Table 3.
Output of different in silico protein prediction tools for the feline DMD:p.(His1396Tyr) substitution.
| Prediction Tool | Variant Score/Accuracy | Score for Deleterious Prediction |
|---|---|---|
| PredictSNP [43] | 74% benign | n.a. * |
| PROVEAN [44] | −0.670 | <−2.5 ** |
| MutPred2 [45] | 0.101 | >0.8 *** |
* PredictSNP takes output from different prediction tools and calls for either a benign or a deleterious prediction with an associated accuracy of the prediction. ** Specificity 80% and sensitivity 79% at this cutoff. *** False positive rate of 5% at this cutoff.
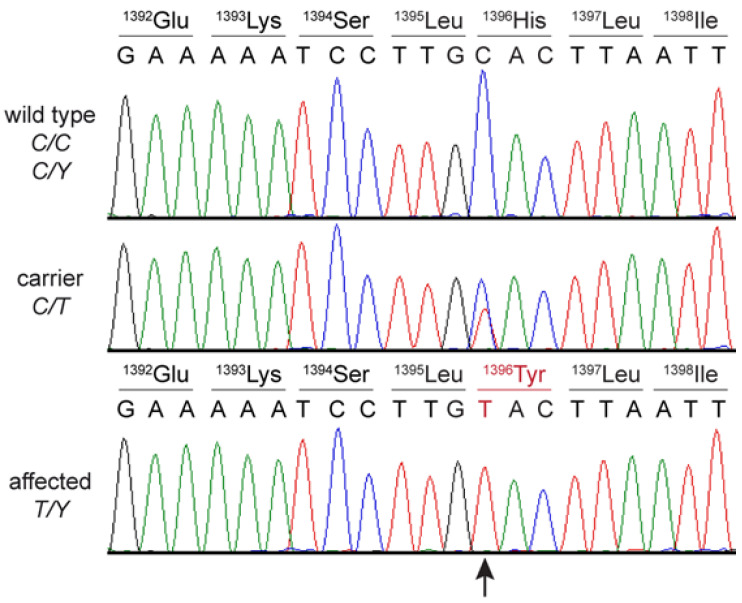
The presence of the DMD missense variant was confirmed by Sanger sequencing in both dystrophic male cats (Figure 4). The unaffected male littermate was hemizygous wildtype, while the female littermate and queen were heterozygous for the mutant allele (Figure 1d). The genotype–phenotype correlation was consistent for an X-linked recessive mode of inheritance. The variant was additionally genotyped in 277 control Main Coon cats and 320 additional control cats of diverse other breeds with similar sex distribution. None of these cats carried the mutant allele.
Figure 4.
Sanger sequencing electropherograms of a wildtype, female carrier cat #4, and dystrophic male cat #2 surrounding the DMD:c.4186C>T variant. Amino acid changes in carrier female and the dystrophic cat #2 are shown.
3. Discussion
Duchenne-type muscular dystrophy, characterized by an early severe disease course and complete dystrophin deficiency caused by DMD null variants, has been previously reported in different breeds of domestic dogs (OMIA #001081-9615) [5,6,46,47,48,49,50], domestic shorthair as well as Maine Coon cats (OMIA #001081-9685) [2,3,4,8,9,11,41,51], pigs [16,17,19,20], mice [25,26], rats [28], and rabbits [29]. In contrast, Becker-type muscular dystrophy has been rarely reported in domestic animals, e.g., in dogs [36,37,38] and pigs (OMIA #001888-9823) [18,23]. The here-reported two male Maine Coon crossbred cats with an adult onset and slowly progressive disease course, histopathologically a dystrophic myopathy with diminished but positive dystrophin staining and thus positive cross reacting material (CRM+), and a single DMD missense variant, support a diagnosis of Becker-type muscular dystrophy.
Based upon the whole genome sequencing of one of the two dystrophic cats, with a specific focus on musculoskeletal genes, the only private protein-changing variant in a known functional candidate gene was identified in the DMD gene. The single nucleotide exchange (c.4186C>T) is predicted to lead to an amino acid substitution (His1396Tyr), which resides in the center rod spectrin 10 domain—a conserved domain among all mammals except cattle (Figure 3). The amino acid substitution is predicted to remove a single positive charge, but protein structure and function may not be seriously affected based upon the various functional prediction programs applied. According to the gnomAD variant database [42], a missense variant leading to an exchange from histidine to tyrosine at the homologous position in the human DMD protein (NP_003997.2:His1380Tyr) has been once observed in an older man, but there are no descriptions whether or not this person had a dystrophic phenotype. Many DMD variants associated with Becker-type muscular dystrophy are missense variants and show only minor impacts with protein modeling on dystrophin protein structure and function [32,33,52]. No protein function studies were performed to prove causality of the missense variant of dystrophic cats described here in vitro or in vivo.
When genotyping the available family members for the DMD:c.4186C>T SNV, the other affected male was also hemizygous for the mutant allele, the queen and a female littermate were heterozygous and a male littermate was hemizygous wildtype at the SNV. The expected co-segregation of genotypes with the observed dystrophy was consistent with an X-linked recessive mode of inheritance. The tom of the litter and mother of the queen were not available for genotyping. The queen as well as affected males had features of the Maine Coon breed, and indeed cat #1 had 26% SNV alleles seen in Maine Coon cats [39]. The other littermates appeared more like regular domestic shorthair cats. Interestingly, a DMD nonsense variant causing Duchenne-type muscular dystrophy in a Maine Coon family was just reported [51].
The identification of a likely candidate causal variant would allow genetic testing to reliably identify additional carrier females. However, the mutant DMD allele was absent from 597 unrelated control cats mostly sampled in Switzerland, including 277 Maine Coon cats. It is unclear how widely the mutant DMD allele may have spread in the regional cat population in Germany, but it is not anticipated that this DMD missense variant is widespread.
Consistent with an amino acid substitution in dystrophin, which is not predicted to drastically change the protein structure and stability, immunohistochemistry for dystrophin protein was CRM+. All previously reported cats with dystrophin deficiency were CRM- including the recently reported two Maine Coon cats with Duchenne muscular dystrophy [51]. Accurate quantification would have been helpful but could not be performed because no samples were available. Dogs with muscular dystrophy show muscle dystrophy and atrophy with sometimes macroglossia [4], and various deleterious DMD variants have been described in several canine breeds (OMIA #001081-9615) [6,46,47,49]. While in most affected dogs, dystrophin is absent by immunohistology, few cases have been described having a partially truncated dystrophin and a slower disease progression [36,37,38]. For instance, there was one nine-year-old male Border terrier where the immunohistochemical staining of the amino-terminal dystrophin protein was present but the carboxy terminal was patchy and reduced, suggesting an 80 kDa truncated protein. However, no molecular genetic studies were pursued to define the DMD variant [38].
The staining for dystrophin and other musculoskeletal proteins was noticeably reduced in the affected cat studied here. None of the musculoskeletal proteins appeared completely deficient in the affected cat #1 of this report and no protein altering DNA variants were found in any of the corresponding genes except DMD. It is well recognized that dystrophin deficiency and dysfunction are associated with impaired stability of the entire dystrophin-associated protein complex, and thus reduced amounts of any of its muscle proteins may be expected [53]. Furthermore, previously studied dystrophin-deficient cats also exhibited reduced complex proteins [12]. All previously reported cats were classified as having Duchenne-type muscular dystrophy, including the recently described dystrophic Maine Coon cat [51]. Likewise, human patients with Duchenne-type as well as Becker-type muscular dystrophy have varying degrees of reduced muscle dystrophin-associated complex proteins [54,55].
Extremely high serum CK activities are a hallmark finding in all dystrophin-deficient humans and domestic animals. The dystrophic cats reported here had high serum CK activity, elevated 30–70× compared to the upper limit of normal reference intervals as previously described with other dystrophic cats [8,9,13,41] (except one reported cat with an unexplained normal serum CK activity [14]). In contrast, human patients and animals including cats with laminin and other dystrophin-associated complex protein deficiencies have only mildly to moderately increased serum CK activities [56,57,58]. The increase in serum CK activities does not appear to allow differentiation between Duchenne and Becker type in cats particularly as cats may massively increase their serum CK activity with anesthesia and stress.
A unique feature of all thus-far reported dystrophin-deficient cats, including the two males of this study, is that on clinical examinations they phenotypically exhibit predominantly muscle hypertrophy rather than muscle atrophy [3,8,9,13,14,15,41]. In that vein, the phenotype of a dystrophic cat may be confused with disorders such as myotonia congenita. However, those also exhibit muscle cramps and dimpling and have only mildly increased CK values [59]. The appendicular muscle hypertrophy and macroglossia were recognized at juvenile age in all previously described cats but were not noted until adulthood in the dystrophic cats reported here. The later onset is consistent with the adult onset of human patients with Becker-type muscular dystrophy [31]. Becker-type dystrophy has an approximate onset of 30 years in human patients and progressing slowly, while Duchenne type is typically evident in boys in early childhood and progressing rapidly [31]. Over the eight-month observation period, only mild clinical progression was noted in the two young adult dystrophic cats reported here.
There are various potential genetic and environmental modifiers affecting the clinical manifestations of dystrophin deficiency in humans and animals [60,61]. Interestingly, the cytidine monophospho-N-acetylneuraminic acid hydroxylase (CMAH) activity has been associated with protecting against severe clinical signs of dystrophin deficiency. To that end, DMD and CMAH double knockout mice exhibit muscular dystrophy, while those with only dystrophin deficiency remain clinically mostly unaffected [61]. In humans, the CMAH gene contains a large deletion and thus is a dysfunctional pseudogene, which may in part explain the more severe clinical manifestations in dystrophin deficient human patients [62]. In contrast, in domestic cats CMAH activity is determining the clinically important AB(C) blood group [40]. Most cats are type A and have a functional CMAH enzyme converting an N-acetyl into a N-glycolyl-neuraminic acid group. The two dystrophic cats have blood type A by phenotypic evaluation and did not harbor any of the four variants screened for in purebred cats to cause blood type B and AB(C) [40]. The blood types of the other described dystrophic cats in the literature were not reported but were also likely having type A blood as it is the most common blood type in domestic cats [40]. It remains unknown if the phenotype of the dystrophic cats could be more severe with non-functional CMAH and blood type B.
Dystrophin deficiency also affects cardiac muscles and leads to dystrophic cardiomyopathy in dystrophic human patients [63] and large animal models such as dogs [64], pigs [16], and cats [65]. No cardiac murmurs were noted in either of the dystrophic cats in this report and cardiac imaging studies did not indicate any cardiac abnormalities even at 3.5 years of age. However, these cats may still develop a cardiomyopathy as they age. Likewise, the two dystrophic cats in this report had no clinical and imaging evidence of megaesophagus reported in other dystrophic cats of the Duchenne type [3].
Among the dystrophic animal models, the dystrophic golden retrievers were the first model that phenotypically most closely mimics the Duchenne muscular dystrophy in human patients. This animal-disease model has been extensively used to study pathogenesis as well as to evaluate the safety and efficacy of novel therapies over the past three decades [5,66].
Pigs are the only other domestic species in which their naturally occurring muscular dystrophy has been well characterized [18]. Recently, a 6-month-old pig with macroglossia at a slaughterhouse was found to have severely reduced dystrophin staining and a pseudoexon insertion was reported. While those authors suggested this to be a Becker-type dystrophy, the early onset, near complete lack of dystrophin staining and DMD insertion predicting a CRM- pathology makes this more likely a Duchenne-type dystrophin deficiency [23]. Interestingly, a couple of decades ago, young piglets thought to have porcine stress syndrome at slaughterhouses had reduced dystrophin protein and DMD missense mutations, thereby resembling Becker-type dystrophin deficiency [67]. Finally, porcine models of Duchenne muscular dystrophy have been engineered by targeted deletion of DMD exon 52 in porcine kidney cells followed by somatic cell nuclear transfer to generate affected male DMDY/- piglets, in the first run, and breeding with newly generated female DMD+/- carriers on the second. DMDY/- piglets appear to be a close homologue for severe Duchenne muscular dystrophy in humans based upon clinical, biochemical, and pathological examination, including cardiomyopathy [16,17,19,20,22]. The majority of dystrophin-deficient piglets die within the first weeks of life, which might be extended with intense neonatal care to a few months of age. Since there is only one reproducible large animal model available yet for Becker-type dystrophy [18,19], clinical cases of subtotal dystrophin deficiency in companion animals may narrow the gap offering additional insights into the pathobiology of Becker-type dystrophin deficiency.
When applying the guidelines for the interpretation of DNA sequence variants in human genetics [68] to the herein investigated cats, four arguments lead to our classification of the DMD:c.4186C>T variant. One pathogenic strong (PS3), one pathogenic moderate to supporting (PM2), as well as two pathogenic supporting (PP1 and PP4) criteria taken together are enough to classify the sequence variant as “likely pathogenic” [68].
4. Materials and Methods
4.1. Animals and Samples
A family of random-bred domestic cats with partial Maine Coon ancestry, two of which were adult male cats with muscular hypertrophy, were clinicopathologically and genetically investigated. The clinical features, blood test results, and pathological examinations were performed as part of the routine diagnostic approach and for clinical management. The molecular genetic studies with DNA samples were part of an approved research study (the Cantonal Committee for Animal Experiments Bern; permit BE 71/19). Routine physical and cardiac examinations, hematology and serum chemistry tests, as well genetic testing for breed, hereditary disease, and other genetic traits (e.g, ABC blood typing) (Mars Veterinary, Portland, OR, USA) [39], were performed.
A biopsy from gastrocnemius muscle (~10 × 8 × 3 mm) was surgically obtained from one affected male cat (index case cat #1) at 2½ years of age and sent to the neuromuscular referral laboratory (Institute for Pathology, Klinikum Leverkusen, Germany) for diagnostic examination.
Ethylenediaminetetraacetate (EDTA)-anticoagulated blood samples for genetic analyses were obtained from index case cat #1 and were also available for family members (cat #2-5). In addition, archived genomic DNA samples from 597 domestic cats without evidence of muscular dystrophy of different breeds from the Vetsuisse Biobank were screened for the herein discovered DMD missense variant. Cheek swabs were used for commercial feline genetic marker screening (Mars Veterinary, Portland, OR, USA) [39].
4.2. Histological and Immunohistological Skeletal Muscle Investigations
Histopathological, histochemical, as well as immunohistochemical studies were performed as described [69,70,71]. Briefly, cryosections (10 μm) and paraffin-embedded sections (4–6 μm) of longitudinal and transverse skeletal muscle biopsies were stained with hematoxylin & eosin (H&E), Engel’s modified Gomori, Masson-Trichrome Elastica van Gieson (EvG), oil red O, periodic acid Schiff and/or Tibor Pap silver impregnation stains.
Antibodies for immunohistochemical stains of dystrophin 1 (1:100, DYS1), dystrophin 2 (1:100, DYS2), β-sarcoglycan (1:50, β-SARC), and γ-sarcoglycan (1:100, γ-SARC) from Novocastra Laboratory, Newcastle upon Tyne, UK, and desmin (clone D33M0760 from Agilent Dako, Santa Clara, CA, USA) were applied. All tissue sections were examined by a veterinary pathologist (T.B.) with brightfield microscopy.
4.3. Molecular Genetic Studies
Genomic DNA was isolated with standard protocols and the genome of cat #1 was sequenced at approximately 28.2× coverage with Illumina 2 × 150 bp paired end reads (Nova Seq 6000, Illumina Inc., San Diego, CA, USA). The data were analyzed as described previously [72] and submitted to the European Nucleotide Archive under sample accession no. SAMEA14502951.
Variant calling was performed using GATK Haplotype Caller [73] in gVCF mode [72]. Functional effects of called variants were predicted with the SnpEff software [74], together with the F. catus_Fca126_mat1.0 reference genome assembly and annotation release 105. For variant filtering, genomes from 74 control cats were included in the analysis (Table S2). PredictSNP [43], PROVEAN [44], and MutPred2 [45] were used to predict biological consequences of the discovered protein variant. Numbering in the feline DMD gene used here corresponds to the NCBI RefSeq accession numbers XM_045050787.1 (mRNA) and XP_044906722.1 (protein).
Available family members of the affected cat #1 and 597 other cats from the Vetsuisse Biobank with similar even sex distribution and without evidence of myopathy were genotyped, including 277 Maine Coon cats. Genomic DNA was amplified using AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific, Reinach, Switzerland). Primers 5′-TCC ATA CAA TGA GCC GCA TA-3′ (Primer F) and 5′-ACC GCG AGT AAG CAT TTC AC-3′ (Primer R) were used for generation of an amplicon containing the DMD:c.4186C>T variant. Direct Sanger sequencing of the PCR amplicons on an ABI 3730 DNA Analyzer (Thermo Fisher Scientific) was performed after treatment with exonuclease I and alkaline phosphatase (BioConcept Ltd., Allschwil, Switzerland). The Sanger sequences were analyzed using the Sequencher 5.1 software (Gene Codes, Ann Arbor, MI, USA).
5. Conclusions
In conclusion, based upon the likely X-linked recessive inheritance, the later and milder clinical muscle manifestations, the persistently high serum CK activities, the histopathological muscular dystrophy, and a single DMD missense variant, we believe this family represents the first clinical-to-molecular genetic characterization of a Becker type and thus cross-reactive material positive (CRM+) dystrophin deficiency in cats.
Acknowledgments
We thank the Next Generation Sequencing Platform of the University of Bern for performing the high-throughput experiments and the Interfaculty Bioinformatics Unit of the University of Bern for providing high-performance computing infrastructure. We would also like to thank all cat owners and veterinarians of dystrophic cats and family members as well as of control cats for sending blood samples for storage and research at the Vetsuisse Biobank (University of Bern, Bern, Switzerland). Finally, we acknowledge Kaspar Matiasek, Section of Clinical and Comparative Neuropathology, Institute of Veterinary Pathology, Centre for Clinical Veterinary Medicine, LMU Munich, Germany for reading and commenting on the manuscript.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24043192/s1.
Author Contributions
Conceptualization, S.H., T.L. and U.G.; investigations, S.H., M.C., T.B. and U.G.; resources, S.H.; data curation, S.H., M.C. and V.J.; writing—original draft preparation, S.H., M.C. and U.G.; writing—review and editing, S.H., M.C., V.J., T.B., T.L. and U.G.; visualization, S.H., M.C., T.B. and U.G.; supervision, T.L. and U.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Written informed consent was obtained from the owners, and the study was approved by the Cantonal Committee for Animal Experiments approved the animal experiments in this study (Canton of Bern; permit BE 71/19).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The accessions for the sequence data reported in this study are listed in Table S2.
Conflicts of Interest
The authors declare no conflict of interest regarding the research, authorship, and publication of this article.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. Complete cloning of the duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Gaschen F., Jaggy A., Jones B. Congenital diseases of feline muscle and neuromuscular junction. J. Feline Med. Surg. 2004;6:355–366. doi: 10.1016/j.jfms.2004.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shelton G.D., Engvall E. Muscular dystrophies and other inherited myopathies. Vet. Clin. N. Am. Small Anim. Pract. 2002;32:103–124. doi: 10.1016/S0195-5616(03)00081-0. [DOI] [PubMed] [Google Scholar]
- 4.Shelton G.D., Engvall E. Canine and feline models of human inherited muscle diseases. Neuromuscul. Disord. 2005;15:127–138. doi: 10.1016/j.nmd.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Kornegay J.N. The golden retriever model of Duchenne muscular dystrophy. Skelet. Muscle. 2017;7:9. doi: 10.1186/s13395-017-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinkmeyer-Langford C., Kornegay J. Comparative Genomics of X-linked Muscular Dystrophies: The Golden Retriever Model. Curr. Genom. 2013;14:330–342. doi: 10.2174/13892029113149990004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson K., Faelan C., Patterson-Kane J.C., Rudmann D.G., Moore S.A., Frank D., Charleston J., Tinsley J., Young G.D., Milici A.J. Duchenne and Becker Muscular Dystrophies: A Review of Animal Models, Clinical End Points, and Biomarker Quantification. Toxicol. Pathol. 2017;45:961–976. doi: 10.1177/0192623317734823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter J.L., Hoffman E.P., Romanul F.C., Kunkel L.M., Rosales R.K., Ma N.S., Dasbach J.J., Rae J.F., Moore F.M., McAfee M.B. Feline muscular dystrophy with dystrophin deficiency. Am. J. Pathol. 1989;135:909–919. [PMC free article] [PubMed] [Google Scholar]
- 9.Gaschen F.P., Hoffman E.P., Gorospe J.R.M., Uhl E.W., Senior D.F., Cardinet G.H., Pearce L.K. Dystrophin deficiency causes lethal muscle hypertrophy in cats. J. Neurol. Sci. 1992;110:149–159. doi: 10.1016/0022-510X(92)90022-D. [DOI] [PubMed] [Google Scholar]
- 10.Blunden A.S., Gower S. Hypertrophic feline muscular dystrophy: Diagnostic overview and a novel immunohistochemical diagnostic method using formalin-fixed tissue. Vet. Rec. 2011;168:510-510. doi: 10.1136/vr.d1119. [DOI] [PubMed] [Google Scholar]
- 11.Gambino A.N., Mouser P.J., Shelton G.D., Winand N.J. Emergent Presentation of a Cat with Dystrophin-Deficient Muscular Dystrophy. J. Am. Anim. Hosp. Assoc. 2014;50:130–135. doi: 10.5326/JAAHA-MS-5973. [DOI] [PubMed] [Google Scholar]
- 12.Remmers G., Hayden D.W., Jaeger M.A., Ervasti J.M., Valberg S.J. Postanesthetic Death in a Cat With Myopathy. Vet. Pathol. 2015;52:186–188. doi: 10.1177/0300985814524797. [DOI] [PubMed] [Google Scholar]
- 13.Kohn B., Guscetti F., Waxenberger M., Augsburger H. Muskeldystrophie bei einer Katze. Tierärztliche Prax. 1993;21:451–457. [PubMed] [Google Scholar]
- 14.Van Soens I., Mols N., Van Meervenne S., Bilzer T., Waelbers T., Binst D., Tshamala M., Verhaeghe A., Saunders J., Van Ham L. A case of hypertrophic feline muscular dystrophy in a Belgian domestic shorthair cat. Vlaams Diergeneeskd. Tijdschr. 2009;78:111–115. [Google Scholar]
- 15.Gaschen F., Burgunder J.-M. Changes of skeletal muscle in young dystrophin-deficient cats: A morphological and morphometric study. Acta Neuropathol. 2001;101:591–600. doi: 10.1007/s004010000299. [DOI] [PubMed] [Google Scholar]
- 16.Stirm M., Fonteyne L.M., Shashikadze B., Lindner M., Chirivi M., Lange A., Kaufhold C., Mayer C., Medugorac I., Kessler B., et al. A scalable, clinically severe pig model for Duchenne muscular dystrophy. Dis. Model. Mech. 2021;14:dmm049285. doi: 10.1242/dmm.049285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stirm M., Fonteyne L.M., Shashikadze B., Stöckl J.B., Kurome M., Keßler B., Zakhartchenko V., Kemter E., Blum H., Arnold G.J., et al. Pig models for Duchenne muscular dystrophy—From disease mechanisms to validation of new diagnostic and therapeutic concepts. Neuromuscul. Disord. 2022;32:543–556. doi: 10.1016/j.nmd.2022.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Hollinger K., Yang C.X., Montz R.E., Nonneman D., Ross J.W., Selsby J.T. Dystrophin insufficiency causes selective muscle histopathology and loss of dystrophin-glycoprotein complex assembly in pig skeletal muscle. FASEB J. 2014;28:1600–1609. doi: 10.1096/fj.13-241141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selsby J.T., Ross J.W., Nonneman D., Hollinger K. Porcine Models of Muscular Dystrophy. ILAR J. 2015;56:116–126. doi: 10.1093/ilar/ilv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klymiuk N., Blutke A., Graf A., Krause S., Burkhardt K., Wuensch A., Krebs S., Kessler B., Zakhartchenko V., Kurome M., et al. Dystrophin-deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle. Hum. Mol. Genet. 2013;22:4368–4382. doi: 10.1093/hmg/ddt287. [DOI] [PubMed] [Google Scholar]
- 21.Horiuchi N., Aihara N., Mizutani H., Kousaka S., Nagafuchi T., Ochiai M., Ochiai K., Kobayashi Y., Furuoka H., Asai T., et al. Becker Muscular Dystrophy-Like Myopathy Regarded as So-Called “Fatty Muscular Dystrophy” in a Pig: A Case Report and Its Diagnostic Method. J. Vet. Med. Sci. 2014;76:243–248. doi: 10.1292/jvms.13-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Echigoya Y., Trieu N., Duddy W., Moulton H.M., Yin H., Partridge T.A., Hoffman E.P., Kornegay J.N., Rohret F.A., Rogers C.S., et al. A Dystrophin Exon-52 Deleted Miniature Pig Model of Duchenne Muscular Dystrophy and Evaluation of Exon Skipping. Int. J. Mol. Sci. 2021;22:13065. doi: 10.3390/ijms222313065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aihara N., Kuroki S., Inamuro R., Kamiya Y., Shiga T., Kikuchihara Y., Ohmori E., Noguchi M., Kamiie J. Macroglossia in a pig diagnosed as Becker muscular dystrophy due to dystrophin pseudoexon insertion derived from intron 26. Vet. Pathol. 2022;59:455–458. doi: 10.1177/03009858221079669. [DOI] [PubMed] [Google Scholar]
- 24.Bulfield G., Siller W.G., Wight P.A., Moore K.J. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc. Natl. Acad. Sci. USA. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura A., Takeda S. Mammalian Models of Duchenne Muscular Dystrophy: Pathological Characteristics and Therapeutic Applications. J. Biomed. Biotechnol. 2011;2011:184393. doi: 10.1155/2011/184393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willmann R., Possekel S., Dubach-Powell J., Meier T., Ruegg M.A. Mammalian animal models for Duchenne muscular dystrophy. Neuromuscul. Disord. 2009;19:241–249. doi: 10.1016/j.nmd.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Araki E., Nakamura K., Nakao K., Kameya S., Kobayashi O., Nonaka I., Kobayashi T., Katsuki M. Targeted Disruption of Exon 52 in the Mouse Dystrophin Gene Induced Muscle Degeneration Similar to That Observed in Duchenne Muscular Dystrophy. Biochem. Biophys. Res. Commun. 1997;238:492–497. doi: 10.1006/bbrc.1997.7328. [DOI] [PubMed] [Google Scholar]
- 28.Larcher T., Lafoux A., Tesson L., Remy S., Thepenier V., François V., Le Guiner C., Goubin H., Dutilleul M., Guigand L., et al. Characterization of Dystrophin Deficient Rats: A New Model for Duchenne Muscular Dystrophy. PLoS ONE. 2014;9:e110371. doi: 10.1371/journal.pone.0110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sui T., Lau Y.S., Liu D., Liu T., Xu L., Gao Y., Lai L., Li Z., Han R. A novel rabbit model of Duchenne muscular dystrophy generated by CRISPR/Cas9. Dis. Model. Mech. 2018;11:dmm032201. doi: 10.1242/dmm.032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman E.P., Kunkel L.M. Dystrophin abnormalities in Duchenne/Becker muscular dystrophy. Neuron. 1989;2:1019–1029. doi: 10.1016/0896-6273(89)90226-2. [DOI] [PubMed] [Google Scholar]
- 31.Darras B.T., Urion D.K., Ghosh P.S. Rosenberg’s Molecular and Genetic Basis of Neurological and Psychiatric Disease. Volume 35 Academic Press; New York, NY, USA: 1993. Dystrophinopathies. [Google Scholar]
- 32.Aartsma-Rus A., Van Deutekom J.C.T., Fokkema I.F., Van Ommen G.-J.B., Den Dunnen J.T. Entries in the Leiden Duchenne muscular dystrophy mutation database: An overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 33.Muntoni F., Torelli S., Ferlini A. Dystrophin and mutations: One gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/S1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 34.Tuffery-Giraud S., Béroud C., Leturcq F., Yaou R.B., Hamroun D., Michel-Calemard L., Moizard M.-P., Bernard R., Cossée M., Boisseau P., et al. Genotype-phenotype analysis in 2405 patients with a dystrophinopathy using the UMD-DMD database: A model of nationwide knowledgebase. Hum. Mutat. 2009;30:934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 35.Ferlini A., Neri M., Gualandi F. The medical genetics of dystrophinopathies: Molecular genetic diagnosis and its impact on clinical practice. Neuromuscul. Disord. 2013;23:4–14. doi: 10.1016/j.nmd.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Cooper B.J., Winand N.J., Stedman H., Valentine B.A., Hoffman E.P., Kunkel L.M., Scott M.-O., Fischbeck K.H., Kornegay J.N., Avery R.J., et al. The homologue of the Duchenne locus is defective in X-linked muscular dystrophy of dogs. Nature. 1988;334:154–156. doi: 10.1038/334154a0. [DOI] [PubMed] [Google Scholar]
- 37.Jones B.R., Brennan S., Mooney C.T., Callanan J.J., McAllister H., Guo L.T., Martin P.T., Engvall E., Shelton G.D. Muscular dystrophy with truncated dystrophin in a family of Japanese Spitz dogs. J. Neurol. Sci. 2004;217:143–149. doi: 10.1016/j.jns.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Jeandel A., Garosi L.S., Davies L., Guo L.T., Salgüero R., Shelton G.D. Late-onset Becker-type muscular dystrophy in a Border terrier dog. J. Small Anim. Pract. 2019;60:514–517. doi: 10.1111/jsap.12824. [DOI] [PubMed] [Google Scholar]
- 39.Anderson H., Davison S., Lytle K.M., Honkanen L., Freyer J., Mathlin J., Kyöstilä K., Inman L., Louviere A., Chodroff Foran R., et al. Genetic epidemiology of blood type, disease and trait variants, and genome-wide genetic diversity in over 11,000 domestic cats. PLoS Genet. 2022;18:e1009804. doi: 10.1371/journal.pgen.1009804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kehl A., Mueller E., Giger U. CMAH genotyping survey for blood types A, B and C (AB) in purpose-bred cats. Anim. Genet. 2019;50:303–306. doi: 10.1111/age.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winand N.J., Edwards M., Pradhan D., Berian C.A., Cooper B.J. Deletion of the dystrophin muscle promoter in feline muscular dystrophy. Neuromuscul. Disord. 1994;4:433–445. doi: 10.1016/0960-8966(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 42.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bendl J., Stourac J., Salanda O., Pavelka A., Wieben E.D., Zendulka J., Brezovsky J., Damborsky J. PredictSNP: Robust and Accurate Consensus Classifier for Prediction of Disease-Related Mutations. PLoS Comput. Biol. 2014;10:e1003440. doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi Y., Chan A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pejaver V., Urresti J., Lugo-Martinez J., Pagel K.A., Lin G.N., Nam H.-J., Mort M., Cooper D.N., Sebat J., Iakoucheva L.M., et al. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 2020;11:5918. doi: 10.1038/s41467-020-19669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mata López S., Hammond J.J., Rigsby M.B., Balog-Alvarez C.J., Kornegay J.N., Nghiem P.P. A novel canine model for Duchenne muscular dystrophy (DMD): Single nucleotide deletion in DMD gene exon 20. Skelet. Muscle. 2018;8:16. doi: 10.1186/s13395-018-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nghiem P.P., Bello L., Balog-Alvarez C., López S.M., Bettis A., Barnett H., Hernandez B., Schatzberg S.J., Piercy R.J., Kornegay J.N. Whole genome sequencing reveals a 7 base-pair deletion in DMD exon 42 in a dog with muscular dystrophy. Mamm. Genome. 2017;28:106–113. doi: 10.1007/s00335-016-9675-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shrader S.M., Jung S., Denney T.S., Smith B.F. Characterization of Australian Labradoodle dystrophinopathy. Neuromuscul. Disord. 2018;28:927–937. doi: 10.1016/j.nmd.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Brunetti B., Muscatello L.V., Letko A., Papa V., Cenacchi G., Grillini M., Murgiano L., Jagannathan V., Drögemüller C. X-Linked Duchenne-Type Muscular Dystrophy in Jack Russell Terrier Associated with a Partial Deletion of the Canine DMD Gene. Genes. 2020;11:1175. doi: 10.3390/genes11101175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shelton G.D., Minor K.M., Vieira N.M., Kunkel L.M., Friedenberg S.G., Cullen J.N., Guo L.T., Zatz M., Mickelson J.R. Tandem duplication within the DMD gene in Labrador retrievers with a mild clinical phenotype. Neuromuscul. Disord. 2022;32:836–841. doi: 10.1016/j.nmd.2022.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beckers E., Cornelis I., Bhatti S.F.M., Smets P., Shelton G.D., Guo L.T., Peelman L., Broeckx B.J.G. A Nonsense Variant in the DMD Gene Causes X-Linked Muscular Dystrophy in the Maine Coon Cat. Animals. 2022;12:2928. doi: 10.3390/ani12212928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monaco A.P., Bertelson C.J., Liechti-Gallati S., Moser H., Kunkel L.M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 53.Ervasti J., Campbell K. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deburgrave N., Daoud F., Llense S., Barbot J.C., Récan D., Peccate C., Burghes A.H.M., Béroud C., Garcia L., Kaplan J.-C., et al. Protein- and mRNA-based phenotype-genotype correlations in DMD/BMD with point mutations and molecular basis for BMD with nonsense and frameshift mutations in the DMD gene. Hum. Mutat. 2007;28:183–195. doi: 10.1002/humu.20422. [DOI] [PubMed] [Google Scholar]
- 55.Ervasti J.M. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim. Biophys. Acta–Mol. Basis Dis. 2007;1772:108–117. doi: 10.1016/j.bbadis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 56.Poncelet L., Résibois A., Engvall E., Shelton G.D. Laminin α2 deficiency-associated muscular dystrophy in a Maine coon cat. J. Small Anim. Pract. 2003;44:550–552. doi: 10.1111/j.1748-5827.2003.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 57.Awamura Y., Uchida K., Arikawa-Hirasawa E. Long-term follow-up of laminin α2 (merosin)-deficient muscular dystrophy in a cat. J. Feline Med. Surg. 2008;10:274–279. doi: 10.1016/j.jfms.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salvadori C., Vattemi G., Lombardo R., Marini M., Cantile C., Shelton G.D. Muscular Dystrophy with Reduced β-Sarcoglycan in a Cat. J. Comp. Pathol. 2009;140:278–282. doi: 10.1016/j.jcpa.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Hickford F.H., Jones B.R., Gething M.A., Pack R., Alley M.R. Congenital myotonia in related kittens. J. Small Anim. Pract. 1998;39:281–285. doi: 10.1111/j.1748-5827.1998.tb03651.x. [DOI] [PubMed] [Google Scholar]
- 60.Brinkmeyer-Langford C., Balog-Alvarez C., Cai J.J., Davis B.W., Kornegay J.N. Genome-wide association study to identify potential genetic modifiers in a canine model for Duchenne muscular dystrophy. BMC Genom. 2016;17:665. doi: 10.1186/s12864-016-2948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allen D.G., Whitehead N.P., Froehner S.C. Absence of Dystrophin Disrupts Skeletal Muscle Signaling: Roles of Ca 2+, Reactive Oxygen Species, and Nitric Oxide in the Development of Muscular Dystrophy. Physiol. Rev. 2016;96:253–305. doi: 10.1152/physrev.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Irie A., Koyamat S., Kozutsumi Y., Kawasaki T., Suzuki A. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J. Biol. Chem. 1998;273:15866–15871. doi: 10.1074/jbc.273.25.15866. [DOI] [PubMed] [Google Scholar]
- 63.Ferlini A., Sewry C., Melis M.A., Mateddu A., Muntoni F. X-linked dilated cardiomyopathy and the dystrophin gene. Neuromuscul. Disord. 1999;9:339–346. doi: 10.1016/S0960-8966(99)00015-2. [DOI] [PubMed] [Google Scholar]
- 64.Guo L.-J., Soslow J.H., Bettis A.K., Nghiem P.P., Cummings K.J., Lenox M.W., Miller M.W., Kornegay J.N., Spurney C.F. Natural History of Cardiomyopathy in Adult Dogs With Golden Retriever Muscular Dystrophy. J. Am. Heart Assoc. 2019;8:e012443. doi: 10.1161/JAHA.119.012443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chetboul V., Blot S., Sampedrano C.C., Thibaud J.-L., Granger N., Tissier R., Bruneval P., Gaschen F., Gouni V., Nicolle A.P., et al. Tissue Doppler imaging for detection of radial and longitudinal myocardial dysfunction in a family of cats affected by dystrophin-deficient hypertrophic muscular dystrophy. J. Vet. Intern. Med. 2006;20:640–647. doi: 10.1111/j.1939-1676.2006.tb02909.x. [DOI] [PubMed] [Google Scholar]
- 66.Amoasii L., Hildyard J.C.W., Li H., Sanchez-Ortiz E., Mireault A., Caballero D., Harron R., Stathopoulou T.-R., Massey C., Shelton J.M., et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nonneman D.J., Brown-Brandl T., Jones S.A., Wiedmann R.T., Rohrer G.A. A defect in dystrophin causes a novel porcine stress syndrome. BMC Genom. 2012;13:233. doi: 10.1186/1471-2164-13-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Volk H.A., Shihab N., Matiasek K. Neuromuscular Disorders in the Cat: Clinical Approach to Weakness. J. Feline Med. Surg. 2011;13:837–849. doi: 10.1016/j.jfms.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brands J., Steffen F., Spennes J., Leeb T., Bilzer T. COL6A1 related muscular dystrophy in Landseer dogs: A canine model for Ullrich congenital muscular dystrophy. Muscle Nerve. 2021;63:608–616. doi: 10.1002/mus.27162. [DOI] [PubMed] [Google Scholar]
- 71.Salvadori C., Vattemi G., Guglielmi V., Marini M., Tomelleri G., Cantile C. Protein Expression of Canine and Feline Muscular Dystrophies. Top. Companion Anim. Med. 2021;42:100500. doi: 10.1016/j.tcam.2020.100500. [DOI] [PubMed] [Google Scholar]
- 72.Jagannathan V., Drögemüller C., Leeb T., Aguirre G., André C., Bannasch D., Becker D., Davis B., Ekenstedt K., Faller K., et al. A comprehensive biomedical variant catalogue based on whole genome sequences of 582 dogs and eight wolves. Anim. Genet. 2019;50:695–704. doi: 10.1111/age.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accessions for the sequence data reported in this study are listed in Table S2.