Abstract
Two mutant strains of Lactococcus lactis in which the promoter of the las operon, harboring pfk, pyk, and ldh, were replaced by synthetic promoters were constructed. These las mutants had an approximately twofold decrease in the activity of phosphofructokinase, whereas the activities of pyruvate kinase and lactate dehydrogenase remained closer to the wild-type level. In defined medium supplemented with glucose, the growth rate of the mutants was reduced to 57 to 70% of wild-type levels and the glycolytic flux was reduced to 62 to 76% of wild-type levels. In complex medium growth was even further reduced. Surprisingly, the mutants still showed homolactic fermentation, which indicated that the limitation was different from standard glucose-limited conditions. One explanation could be that the reduced activity of phosphofructokinase resulted in the accumulation of sugar-phosphates. Indeed, when one of the mutants was starved for glucose in glucose-limited chemostat, the growth rate could gradually be increased to 195% of the growth rate observed in glucose-saturated batch culture, suggesting that phosphofructokinase does affect the concentration of upstream metabolites. The pools of glucose-6-phosphate and fructose-6-phosphate were subsequently found to be increased two- to fourfold in the las mutants, which indicates that phosphofructokinase exerts strong control over the concentration of these metabolites.
Lactococcus lactis plays an important role in dairy fermentations, mainly in the production of cheeses. During such fermentation processes, lactose is present at very high concentrations (50 g/liter) and is converted through glycolysis to primarily form lactic acid as well as minor amounts of other compounds (homolactic fermentation). The resulting low pH contributes to the texture and flavor of cheeses and inhibits the growth of other bacterial species. During homolactic fermentation more than 90% of the lactose consumed is recovered as by-products (42), which shows that the glycolytic pathway functions almost exclusively as a catabolic pathway to supply ATP to the cells. When sugar becomes limiting for growth, or in the presence of a less readily metabolized carbon source, the pattern of product formation switches from homolactic to mixed-acid fermentation, i.e., to the formation of formate, ethanol, and acetate with smaller amounts of lactate (43).
The mechanisms responsible for regulation of glycolytic flux and the shift between different fermentation modes in L. lactis have been studied intensively. The concentrations of intermediary metabolites and cofactors are affected by the external sugar concentration (8, 10, 37, 46). In the presence of excess sugar, the concentrations of fructose-1,6-bisphosphate, the triose-phosphates, and pyruvate and the NADH/NAD+ ratio are high, whereas the concentrations of phosphoenolpyruvate and inorganic phosphate are relatively low. The glycolytic flux was proposed to be regulated through the level of fructose-1,6-bisphosphate (43), which is known to activate both pyruvate kinase and lactate dehydrogenase. In contrast, when sugar is limiting, the level of fructose-1,6-bisphosphate decreases and the level of inorganic phosphate increases (8, 37, 46), which results in decreased pyruvate kinase activity and increased phosphoenolpyruvate concentration (28, 45, 49). Finally, phosphoenolpyruvate inhibits the activity of phosphofructokinase (7), which provides a mechanism for regulating the glycolytic flux.
Work has been performed to determine the factors that control the flux through glycolysis by applying metabolic control analysis (13, 22). Poolman et al. (35) showed that the activity of glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate mutase decreased 5- to 15-fold during lactose starvation. Based on inhibitor titration results, they suggested that glyceraldehyde-3-phosphate dehydrogenase had a large amount of control over the glycolytic flux during nongrowing conditions. However, it is still unclear whether the enzyme is therefore probably less important when the flux through glycolysis is high.
In L. lactis the genes pfk, pyk, and ldh, encoding the glycolytic enzymes phosphofructokinase, pyruvate kinase, and lactate dehydrogenase, respectively, are clustered in the las operon (25). This unique organization could in principle allow for coordinated regulation of the expression of these genes, and it was suggested that expression of the las operon might be involved in regulating energy production and lactate flux in L. lactis (5). Indeed, a fivefold up-regulation of expression of the las operon was recently demonstrated at the onset of the stationary growth phase (1). In another study it was suggested that the las operon might be activated by the CcpA protein (26). The physiological role of regulating expression of the las operon is not clear, and it is unknown whether a coordinated expression of glycolytic genes might be important.
In this study we constructed mutants of L. lactis in which expression of the first gene of the las operon, pfk, was altered. We show that a 50% reduction in the expression of pfk dramatically affects the physiology of L. lactis in an unexpected manner, an effect which appears to be caused by the accumulation of upstream metabolites.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| L. lactis | ||
| MG1363 | L. lactis subsp. cremoris is a prophage-cured and plasmid-free derivative of NCDO712; and regarded as a wild-type strain in this study | 11 |
| HWA217 | MG1363 las::CP25, obtained by double homologous recombination with pHWA182 | This study |
| HWA232 | MG1363 las::CP29, obtained by double homologous recombination with pHWA183 | This study |
| HWA254 | MG1363 transformed with pMU2916, Ermr | This study |
| HWA246 | HWA217 transformed with pMU2916, Ermr | This study |
| HWA258 | HWA232 transformed with pMU2916, Ermr | This study |
| E. coli | ||
| ABLE K | Cloning host which lowers the plasmid copy number; E. coli C lac(LacZω−) [KanrmcrA mcrCB mcrF mrr hsdR(rK− mK−)] [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pMU2916 | pMG36e harboring pfk on a 1.4-kb EcoRI fragment (−311→+1696) encoding pfk of the las operon in L. lactis subsp. lactis LM0230, accession no. L07920, Ermr | 25 |
| pRC1 | E. coli subcloning vector, Ermr | 23 |
| pUC18 | E. coli cloning vector, pBR322 ori, MCS in lacZ′, Ampr | New England Biolabs |
| pCI372 | Shuttle vector between E. coli and L. lactis, MCS from pUC except for SphI and HindIII, Ermr | 12 |
| pBluescript II SK | E. coli cloning vector, pBR322 ori, MCS in lacZ′, Ampr | Stratagene |
| pUC7erm | pUC7 derivative, Ampr, Ermr | Received from W. de Vos |
| pCP25 | pAK80 derivative carrying constitutive promoter CP25-lacLM, Ermr | 18 |
| pCP29 | pAK80 derivative carrying constitutive promoter CP29-lacLM, Ermr | 18 |
| pHWA58 | pRCI EcoRI::pMU2916 EcoRI, 1.4 kb (−311→+1696), Ermr | This study |
| pHWA71 | pUC18 SmaI::pHWA58 SspI-EcoRI (+26→+1696), Ampr | This study |
| pMW40 | pHWA71 ClaI-BamHI::internal pfk fragment 1.15 kb (+456→+1696) of MG1363, Ampr | Received from M. Willemoës |
| pHWA134 | Upstream region of the las operon (nagA gene) cloned in pCI372; pCI372 smaI::fragment 1.071 kb (−1087 → −17) of MG1363 obtained by inverse PCR, BsaAI, Camr | This study |
| pHWA149 | Upstream region cloned in pBluescript II SK BamHI-KpnI::pHWA134 BamHI-KpnI, 1.076 kb, Ampr | This study |
| pHWA151 | Truncated region of pfk cloned in the vector carrying the upstream region; pHWA149 SacI::pMW40 SacI, 870 bp, Ampr | This study |
| pHWA157 | Cloning of CP25 in the vector with upstream region and pfk′; pHWA151 BamHI-XbaI::CP25 promoter amplified with pAK80 and pAK80/erm from pCP25 BamHI-XbaI, 185 bp, Ampr | This study |
| pHWA158 | Cloning of CP29 in the vector with upstream region and pfk′; pHWA151 BamHI-XbaI::CP29 promoter amplified with pAK80 and pAK80/erm from pCP29 BamHI-XbaI, 171 bp, Ampr | This study |
| pHWA182 | erm gene cloned in the vector in which CP25 proceeds pfk′; pHWA157 NaeI::pUC7erm PvuII, 1.422 kb, Ampr, Ermr | This study |
| pHWA183 | erm gene cloned in the vector in which CP29 proceeds pfk′; pHWA158 NaeI::pUC7erm PvuII, 1.422 kb, Ampr, Ermr | This study |
The feature of plasmids are indicated first by the vector ligated (::) to the insert. The restriction endonuclease used for digestion is shown. PCR fragments were amplified by Pfu polymerase. Kilobases and base pairs denote the sizes of inserts. The coordinates in brackets are relative to the transcription start site of the las operon. Abbreviations: Ampr, ampicillin resistance; Camr, chloramphenicol resistance; Ermr, erythromycin-resistance.
Growth media and growth conditions.
Escherichia coli strains were grown aerobically at 37°C in Luria-Bertani broth (36). L. lactis strains were cultivated as batch cultures (flasks) without aeration in M17 broth (40) or defined SA medium (17). The cultures were supplemented with 0.2 or 1% glucose, 1% fructose, 2% mannose, or 2% maltose and incubated at 30°C. A slow stir with magnets was used in order to keep the culture homogenous. Growth experiments were also performed in the Biostat Q fermentor system (B. Braun Biotech International, Melsungen, Germany) as either batch or glucose-limited chemostat cultures. L. lactis strains were routinely cultivated in batch fermentation in 1-liter vessels with a working volume of 600 ml of SA medium supplemented with 0.25% glucose. The temperature was set to 30°C, and the cultures were stirred at 200 rpm. The pH was maintained at 6.8 by the addition of 2 M NaOH. The medium in the fermentor was inoculated with an exponentially growing preculture to an initial optical density at 600 nm (OD600) ranging from 0.02 to 0.04. Sampling for measurements of biomass, fermentation products, and enzyme activities were performed by pipetting. The cell density was correlated to the cell mass of L. lactis to be 0.36 g (dry weight)/liter of SA medium at an OD600 of 1 (32).
Chemostat cultures.
The medium used for the chemostat cultures was M17 with a glucose concentration of 0.25%.
Nongrowing cultures.
Precultures were grown as batch cultures in 100 ml of SA medium supplemented with 0.2% glucose. In the exponential growth phase at an OD600 of 0.5, the cultures were cooled on ice and harvested by centrifugation. The cultures were resuspended in buffer (SA medium without amino acids and vitamins) supplemented with 0.2% glucose to a final OD600 of 0.4.
Excision of integrated plasmids.
Counterselection was used to isolate las mutants in which the erythromycin resistance plasmid (pHWA182 or pHWA183) was crossed out of the chromosome as follows. A culture was inoculated to an OD600 of 0.03 in M17 medium supplemented with 1% glucose without antibiotic selection for the plasmid. At an OD600 of 0.150, erythromycin (2 μg/ml) was added, and at an OD600 of 0.300, ampicillin (100 μg/ml) was added to the culture. The culture was incubated 15 h before the cells were removed, washed, and diluted on M17 plates. The colonies were tested by replica plating with or without erythromycin. Clones which had lost erythromycin resistance were verified by Southern blot analysis (39) and DNA sequencing of the promoter area upstream of the pfk gene.
Antibiotics.
Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml (selection of pBluescript II SK derivatives, pUC7erm, and pMW40 in E. coli); erythromycin, 2 μg/ml (selection for maintenance of pMU2916 and for the recombination event of pHWA182 and pHWA183 in L. lactis) and 200 μg/ml (selection of pAK80 derivatives and pMU2916 in E. coli); chloramphenicol, 50 μg/ml (selection of pCI372 derivatives); and tetracycline, 10 μg/ml (selection of E. coli strain ABLE K).
DNA techniques, DNA isolation, and DNA sequence analysis.
All manipulations with recombinant DNA were carried out according to standard procedures as described in reference 36 unless otherwise stated. Chromosomal DNA from L. lactis was purified as previously described (21) with the following modifications. A final concentration of 15.5 mg of lysozyme/ml for 30 min was used for lysis, and the sample was incubated with sodium dodecyl sulfate for 10 min at 37°C followed by 10 min at 65°C. Plasmid DNA from E. coli for analytic purposes was isolated by using an alkaline lysis method and for preparative purposes by using a plasmid kit (Qiagen, Hilden, Germany). Restriction enzymes, T4 DNA ligase, and DNA polymerase Klenow fragment were purchased from Pharmacia Biotech and used as specified by the manufacturer. The restriction enzymes BsaAI and NaeI were purchased from New England Biolabs. Linearized cloning vectors were treated with shrimp alkaline phosphatase (Amersham Life Science) to avoid religation of the vector. PCR products for cloning purposes were amplified using Pfu DNA polymerase (Stratagene), and the verification of plasmid or chromosomal constructs by PCR was performed with Taq DNA polymerase (Pharmacia Biotech).
The sequence of the pfk gene in MG1363 and the two las mutants was verified by sequencing PCR products obtained from chromosomal DNA. These sequences and the sequence of the region upstream of the las operon were obtained with the 373A DNA sequencer (Applied Biosystems) using Dye Terminator labeling (Perkin-Elmer).
Rapid isolation of chromosomal DNA from L. lactis.
A rapid method for isolating high quality chromosomal DNA was necessary in the screening of the las mutants. The procedure is a modification of the method described by Jensen and Michelsen (19). A sample of 0.9 ml of culture was added to a 2-ml Eppendorf tube containing 0.9 ml of phenol (preheated to 80°C) and 0.6 g of fine glass beads (Sigma) and the mixture was vortexed for 10 s. The sample was placed at room temperature for 30 min. Cell debris was removed by centrifugation, and the water phase was extracted with 0.8 ml of chloroform. Subsequently, the DNA was precipitated by isopropanol precipitation (36).
Transformation.
L. lactis was transformed by electroporation as previously described (15) and plated on SR plates (31). E. coli was made competent using CaCl2 (36) or made electrocompetent with 10% (vol/vol) glycerol.
Plasmid constructions.
The plasmids constructed in this research are listed in Table 1. The Plas promoter region and the complete coding region of pfk were cloned from pMU2916 into pRC1 by digestion with EcoRI, resulting in pHWA58. This plasmid was digested with SspI and EcoRI (blunted), and the coding region of pfk was transferred to pUC18 digested with SmaI, resulting in pHWA71. Due to a mutation in the 3′ end of pfk in pMU2916, this DNA region in pHWA71 was replaced by a nonmutated DNA region from MG1363, resulting in pMW40 (kindly provided by M. Willemoës). The region upstream of the las operon in MG1363 was obtained by inverse PCR (36). The PCR product was digested with BsaAI giving a 1.071-kb fragment, which was cloned in pCI372 digested with SmaI, resulting in pHWA134. Subsequently, the upstream fragment was transferred from pHWA134 to pBluescript II SK by digestion with BamHI and KpnI, resulting in pHWA149. The DNA region upstream of pfk and a truncated pfk gene (870 bp) were combined by ligating pMW40 and pHWA149 digested with SacI, resulting in pHWA151. In this parental plasmid the synthetic promoters were inserted. Plasmids pCP25 and pCP29 were used as templates to amplify promoter-containing fragments with the primers pAK8/erm (5′−TTTCAACTGCCTGGCAC-3′) and pAK80 (5′-TCCTTTCAAAGTTACCC-3′). The products were digested with BamHI and XbaI, yielding fragments of 185 and 171 bp. These were inserted into pHWA151 digested with the same restriction enzymes, resulting in pHWA157 and pHWA158. Finally, the erythromycin resistance gene located on a PvuII fragment (from pUC7erm) was inserted in the NaeI site of pHWA157 and pHWA158, resulting in pHWA182 and pHWA183, respectively.
Quantification of glucose and fermentation products by HPLC.
Samples of 2 ml were withdrawn from the cultures at different time intervals, filtered through a 0.22-μm filter, and stored at −20°C until quantification of glucose, pyruvate, lactate, formate, acetoin, acetate, and ethanol. The high-pressure liquid chromatography (HPLC) equipment used was from Shimadzu Corporation, Kyoto, Japan, and the program Class VP 5.0 controlled the system. Products were measured as reported in reference 16 with the following modifications. Separation was performed with a Supelcogel C-610H column, 30 cm by 7.8 mm (Supelco, Inc., Bellafonte, Pa.). Supelcoguard C-610H, 5 cm by 4.5 mm, was used as a guard column. The columns were thermostated at 30°C. The mobile phase consisted of 5 mM H2SO4, and the flow rate was 0.5 ml/min. All products were detected on the refractive index detector, RID-10A. Pyruvate was detected on the diode array detector at 208 nm. SA medium contains a large amount of acetate, which was subtracted in the calculation of the amount of acetate produced. Detection of glucose by the refractive index detector is disturbed by the presence of pyruvate, which has the same retention time as glucose. This was corrected by measuring the amount of pyruvate in the sample at 208 nm. The catabolic carbon balance was calculated as the recovery of substrates converted into products in terms of molar concentrations (C-moles).
Enzymatic determination of sugar-phosphates.
Extracts were prepared from batch cultures at an OD600 of 0.3 to 0.9, using quenching in hot phenol; see “Rapid isolation of chromosomal DNA from L. lactis,” above. After chloroform extraction, the concentration of sugar-phosphates was measured by recording the increase in NADH fluorescense (48) with the following modification. The buffer contained 50 mM triethanolamine, pH 7.5, instead of imidazolehydrochloride. Glucose-6-phosphate dehydrogenase from yeast was obtained from Boehringer Mannheim GmbH, Mannheim, Germany, and phosphoglucose-isomerase was obtained from Roche Diagnostics GmbH, Mannheim, Germany. Intracellular concentrations of sugar-phosphates (44) were calculated by assuming that 1 g (dry weight) corresponds to 1.67 ml of intracellular volume and an OD600 of 1 corresponds to 0.36 g (dry weight) per liter (32).
Measurement of phosphofructokinase, pyruvate kinase, and lactate dehydrogenase activities.
Enzyme activities were measured in permeable cells using a method analogous to the standard procedure used to determine β-galactosidase activity (29) with the following modifications. Cells from a 5-ml culture at an OD600 of 0.5 were harvested and washed twice in 0.2% KCl. The cells were resuspended in 0.5 ml of extract buffer (45 mM Tris–15 mM tricarballylate buffer, pH 7.5, containing 20% glycerol, 4.5 mM MgCl2, and 1 mM dithiothreitol [10]). The cells were made permeable by adding 5 μl of 0.1% sodium dodecyl sulfate and 12.5 μl of chloroform and vortexing the mixture for 10 s. The cell extract was diluted in extract buffer and used for assaying phosphofructokinase, pyruvate kinase, and lactate dehydrogenase. The activity was determined from the rate of NADH oxidation at A340 at 28°C using a Specord M500 spectrophotometer (Zeiss, Zena, Germany). Phosphofructokinase was assayed according to reference 7 with the following modifications. The final concentrations of the components of the reaction mixture were 87 mM Tris-HCl (pH 8.0), 1 mM ATP, 1 mM fructose-6-phosphate, 10 mM MgCl2, 10 mM NH4Cl, and 0.2 mM NADH; a mixture of 0.3 U of triosephosphase isomerase, 1 U of glycerol-3-phosphate dehydrogenase, 0.3 U of aldolase, and 1 mM ATP was used to initiate the reaction. The pyruvate kinase activity was determined as described in reference 4. The final concentrations of the components of the reaction mixture were 40 mM triethanolamine–64 mM KCl buffer (pH 7.5), 10 mM MgCl2, 1 mM fructose-1,6-bisphosphate, 1 mM GDP, 0.2 mM NADH, and 6.3 U of lactate dehydrogenase; 1 mM phosphoenolpyruvate was used to initiate the reaction. Lactate dehydrogenase was measured by using a modified procedure described in reference 3. The final concentrations of the components of the reaction mixture were 50 mM triethanolamine (pH 7.5), 1 mM fructose-1,6-bisphosphate, and 0.2 mM NADH; 10 mM sodium pyruvate was used to initiate the reaction.
The cell mass was estimated as the OD600 value at the time of sampling according to Miller (29). The specific enzyme activity is given in micromoles per minute per cell density.
Nucleotide sequence accession number.
The sequence of the region upstream of the las operon and the 3′ end of the nagA gene has been deposited in the NCBI data bank with accession no. AY007718.
RESULTS
Construction of strains with altered expression of las genes.
To alter the expression of the las operon, the native promoter Plas was replaced by synthetic promoters by double homologous recombination (Fig. 1). The region upstream of the las operon (Fig. 1A) from L. lactis subsp. cremoris MG1363 (11) was obtained by inverse PCR and shown to contain a gene with 34% homology to the nagA gene in E. coli (33) encoding N-acetylglucosamine-6-phosphate deacetylase. The nagA gene is also found upstream of the las operon in the closely related L. lactis strain IL1403 (2). To construct plasmids for gene replacement, a fragment encoding the N-terminal part of phosphofructokinase without the Plas promoter was obtained from pMW40 and cloned together with the upstream region in an E. coli vector, resulting in pHWA151. Subsequently, the synthetic promoters CP25 and CP29 were inserted in addition to an erythromycin resistance gene, erm, which resulted in pHWA182 and pHWA183. A schematic presentation of pHWA182 is shown in Fig. 1B.
FIG. 1.
Strategy used to alter expression of las genes. (A) Schematic presentation of the chromosomal las operon in MG1363 and of the DNA fragments which have been cloned in pHWA182. The bent arrow indicates the native promoter Plas of the operon. The heavy black line indicates the DNA fragment upstream of the operon. The truncated pfk region is indicated by the light gray line. (B) The synthetic promoter CP25 was flanked by the upstream region of the las operon and the truncated region of pfk on the E. coli vector pHWA182. (C) After selection for erythromycin resistance and subsequent counterselection with ampicillin, the native promoter Plas was replaced by CP25 by double homologous recombination between pHWA182 and MG1363, resulting in HWA217.
pHWA182 and pHWA183 were integrated on the chromosome of MG1363 by transformation with selection for erythromycin resistance. After a second recombination event, the las mutants HWA217 and HWA232 were isolated and the synthetic promoters CP25 and CP29, respectively, transcribed the las operon. The ratio of strains in which the plasmid excised the wild type versus the las mutant was 6:1. The structure of the las locus in HWA217 is shown in Fig. 1C. Interestingly, HWA217 and HWA232 formed very small colonies on M17 plates supplemented with glucose.
las mutants have uncoordinated expression of pfk, pyk, and ldh relative to the wild type.
To determine the expression levels of the las genes in the las mutants, the phosphofructokinase, pyruvate kinase, and lactate dehydrogenase activities were measured (Table 2). HWA217 and HWA232 had 39 and 60% phosphofructokinase activity compared to MG1363, respectively. The activity of pyruvate kinase was slightly increased in HWA217 (122%) and close to the wild-type level in HWA232 (97%). The lactate dehydrogenase activity of HWA217 was 140%, and HWA232 had an activity near the wild-type level (104%). As seen in Table 2, the differences of activities of the las enzymes compared to those of MG1363 were more pronounced in strain HWA217. In strain HWA232, phosphofructokinase was the most strongly affected enzyme activity, whereas the activities of pyruvate kinase and lactate dehydrogenase were closer to their respective wild-type values.
TABLE 2.
Specific activities of phosphofructokinase, pyruvate kinase, and lactate dehydrogenase in las mutants
| Strain | Genotypea | Sp act of las enzymeb
|
|||||
|---|---|---|---|---|---|---|---|
| Phosphofructokinase
|
Pyruvate kinase
|
Lactate dehydrogenase
|
|||||
| U/OD600 | % of wt | U/OD600 | % of wt | U/OD600 | % of wt | ||
| MG1363 | wt | 5.56 | 100 (6) | 13.52 | 100 (1) | 23.30 | 100 (7) |
| HWA217 | wt las::CP25 | 2.18 | 39 (2) | 16.46 | 122 (21) | 32.58 | 140 (1) |
| HWA232 | wt las::CP29 | 3.34 | 60 (4) | 13.12 | 97 (6) | 24.14 | 104 (18) |
All mutant strains are MG1363 derivatives. The synthetic promoters transcribing the las genes are shown. wt, wild type.
Specific activities (U/OD600) are micromoles per minute per cell density. Values are averages of at least three independent measurements and within the experiments, the activities from each strain were determined in triplicate. The specific activities of the las enzymes of the constructed strains are shown as percentages of that of the wild-type strain MG1363, and standard deviations are shown in parentheses.
It was surprising that the expression of the three las genes in the las mutants was not changed to the same extent. This indicated that the coordination of expression of the las genes had been disrupted as a result of substituting the wild-type promoter for the synthetic promoters. One explanation could be that mutations had accidentally been introduced in the pfk gene during the cloning procedure. To rule out this possibility, the pfk gene was sequenced in HWA217, HWA232, and MG1363, and the pfk sequences in the three strains turned out to be identical.
Growth rate was strongly affected by altered expression of las genes.
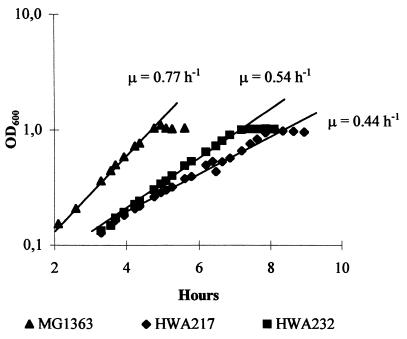
The growth of the two las mutants was observed in batch fermentation with glucose as a carbon source. A typical growth curve for cultures of MG1363, HWA217, and HWA232 in Fig. 2 shows that the las mutants had a lower growth rate than the wild-type strain MG1363. Strain HWA217 had a specific growth rate of 0.44 h−1, compared to 0.77 h−1 for MG1363. HWA232 grew slightly faster, with a specific growth rate of 0.54 h−1. Thus, the growth rates of HWA217 and HWA232 were reduced to 57 and 70% of the wild-type value. The yields of biomass per mole of glucose for all strains were approximately the same (Table 3).
FIG. 2.
Growth curves of las mutants HWA217 and HWA232 and wild-type control strain MG1363. The cultures were grown as batch fermentation cultures in SA medium supplemented with 0.25% glucose, and pH was kept constant at 6.8 by the addition of 2 M NaOH.
TABLE 3.
Glycolytic flux and flux to end products of las mutantsa
| Strain | Glycolytic flux (mmol · g [dry wt]−1 · h−1) | Biomass yield (g [dry wt]/mol glucose) | Flux to end products (mmol · g [dry wt]−1 · h−1)
|
||
|---|---|---|---|---|---|
| Lactate | Formate | Acetate | |||
| MG1363 | 23.5 (100) | 32.7 | 36.6 (100) | 1.8 (100) | n.d.b |
| HWA217 | 14.6 (62) | 30.2 | 24.9 (68) | 2.2 (122) | 1.0 |
| HWA232 | 17.5 (76) | 30.2 | 29.5 (81) | 2.0 (111) | 0.6 |
Values in parentheses are percentages of the values for MG1363.
n.d., not detectable.
Fermentation was mainly homolactic.
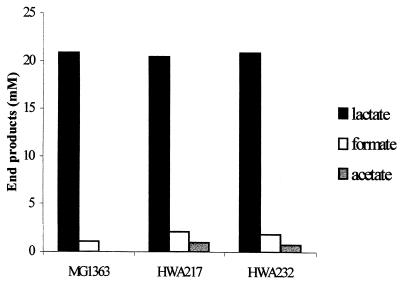
The fact that growth rate of the las mutants was decreased while the biomass yield on glucose remained unchanged indicated that altered expression of the las genes decreased the glycolytic flux. To investigate the pattern of fermentation in these strains, the end product formation of the las mutants grown in batch cultures of SA medium supplemented with 0.25% glucose was determined (Fig. 3). The main product produced by all strains was lactate (85%). HWA232 and HWA217 produced slightly larger amounts of formate than did the wild-type strain and small amounts of acetate. Trace amounts of acetate are normally produced by the wild-type strain, but this was not detected in this experiment due to the high background level of acetate in the SA medium. Other end products such as acetoin and ethanol were not detected. The same pattern of product formation was seen with 1% glucose (data not shown).
FIG. 3.
End-product formation of las mutants. The products produced by the las mutants and MG1363 during batch fermentation were analyzed by HPLC. The strains displayed a homolactic fermentation pattern. Samples were withdrawn from cultures in the stationary growth phase with an OD600 of approximately 1.1, immediately after the glucose had been exhausted. The C-mole recovery ranged between 80 and 83%.
Metabolic fluxes in the las mutants.
Samples for analysis by HPLC were withdrawn from a batch fermentation at different time points during growth (Fig. 2). The glycolytic flux was measured as the steady-state consumption rate of glucose. The lactate, formate, and acetate fluxes were measured as the steady-state production rate of the respective products (Table 3). The glycolytic flux was reduced to 62 and 76% of the wild-type level, and the lactate flux was reduced to 68 and 81% in HWA217 and HWA232, respectively. The formate flux was increased to 122% in HWA217 and to 111% in HWA232, with respect to MG1363, but still the flux through the pyruvate formate lyase branch accounted for less than 10% of the total flux to by-products.
The homolactic fermentation of the las mutants was unexpected because the strains had a lower glycolytic flux and therefore a lower rate of ATP synthesis. Typically, strains that are energy limited show a mixed-acid fermentation pattern (8, 43). Thus, the homolactic fermentation pattern indicated that the las mutants did not experience a typical energy limitation.
Growth characteristics on various sugars.
To investigate the reason for the altered physiology of the las mutants, the strains were grown as batch cultures (flasks) with different sugars (Table 4). As expected, the growth rate of the las mutants was reduced in SA medium supplemented with glucose. A similar phenomenon was seen with mannose, which utilizes the same uptake system as glucose, namely, PTSMan (47).
TABLE 4.
Growth rates of las mutants on various sugars
| Medium | Sugarb | Specific growth rate (h−1)a
|
||
|---|---|---|---|---|
| MG1363 | HWA217 | HWA232 | ||
| SA | Glucose | 0.74 (5) | 0.46 (2) | 0.56 (2) |
| Mannose | 0.73 (4) | 0.46 (8) | 0.55 (17) | |
| Fructose | 0.70 | 0.73 | 0.72 | |
| Maltose | 0.39 | 0.36 | 0.40 | |
| M17 | Glucose | 1.14 (1) | 0.57 (6) | — |
The specific growth rates (μ) are averages of at least two independent measurements, and standard deviations are shown in parentheses. —, not determined.
Experiments where fructose and maltose were used as substrates were performed one time. The cultures were grown as batch cultures (flasks).
Fructose circumvents the phosphofructokinase step since the transport of fructose is mediated via PTSFru (24, 27), resulting in the formation of intracellular fructose-1-phosphate. Fructose-1-phosphokinase subsequently converts this to fructose-1,6-bisphosphate, which enters the glycolytic pathway. When fructose was used as substrate, the growth rates of HWA217 and HWA232 were identical to that of MG1363. This result indicates that the reduced expression of pfk caused the lower growth rates of the las mutants HWA217 and HWA232 in medium with abundant glucose.
Growth on maltose is considered to be energy limited because the sugar is fed into glycolysis at a lower rate, resulting in mixed-acid fermentation (37). Maltose enters MG1363 through PTSMal as maltose-6-phosphate. The enzyme maltase hydrolyzes the phosphorylated form to glucose and glucose-6-phosphate, which enters the glycolytic pathway through the activity of phosphofructokinase. With maltose, both las mutants and MG1363 had a low growth rate, around 0.40 h−1. Apparently, under conditions where the glycolytic flux is much lower than on glucose, the growth of the mutants is not inhibited.
Although glycolysis is considered to be a catabolic pathway, a small flux of molecules is routed into anabolic pathways (30). In SA medium the strains have to synthesize, e.g., nucleotides (17). A limitation in reactions in the upper part of glycolysis might have caused a decrease in the concentration of downstream glycolytic intermediates required for anabolic reactions. To test this assumption, the strains were grown in complex M17 medium supplemented with glucose. This complex medium contains most building blocks not found in defined SA medium. However, in complex medium HWA217 showed an even more reduced growth rate (0.57 h−1) compared to MG1363, which had a growth rate of 1.14 h−1. This result suggests that the reduced growth rate of the las mutants is not due to a lack of building blocks.
Complementing the reduced activity of phosphofructokinase.
Out of the three enzymes encoded by the las operon, the activity of phosphofructokinase differed most in the las mutants and we therefore attempted to restore the phosphofructokinase activity by complementation. Strains MG1363, HWA217, and HWA232 were transformed with pMU2916, which carries the pfk gene transcribed from the native Plas promoter, resulting in strains HWA254, HWA256, and HWA258, respectively. These strains were grown as batch cultures in SA medium supplemented with 1% glucose, and the activities of phosphofructokinase and the growth rates were determined (Table 5). The pfk gene was overexpressed six- to ninefold in HWA254, HWA256, and HWA258, compared to the wild-type MG1363, and the strains had similar growth rates, close to 0.60 h−1, which was somewhat lower than the usual growth rate observed for MG1363 without any plasmid (Table 5). These experiments were performed in the presence of erythromycin in order to select for the plasmid pMU2916. The reduced growth rate of the transformed strains could be due either to high expression of the pfk gene or to the presence of erythromycin. Indeed, when grown without selection for erythromycin resistance, the wild-type strain carrying pMU2916 (HWA254) had a growth rate of 0.71 h−1 and the las mutants HWA256 and HWA258 had growth rates of 0.70 h−1 and 0.71 h−1, respectively. Thus, there is a significant increase in the growth rates of the mutants when the pfk gene is expressed in trans compared to growth rates of 0.46 and 0.56 h−1 without the plasmid. These observations, together with the results concerning growth on various sugars, indicate that the low activity of phosphofructokinase was the primary cause of slower growth and glycolytic flux in HWA217 and HWA232.
TABLE 5.
Complementation of reduced phosphofructokinase activity in las mutants by expression of pfk from a plasmid
| Strain | Genotypea | Growth rate (h−1)b
|
Sp act of phosphofructokinasec
|
||
|---|---|---|---|---|---|
| −Erm | +Erm | U/OD600 | % of wt | ||
| MG1363 | wt | 0.74 | 5.56 | 100 | |
| HWA254 | wt [pfk] | 0.71 | 0.61 | 52.84 | 950 |
| HWA217 | wt las::CP25 | 0.46 | 2.18 | 39 | |
| HWA256 | wt las::CP25 [pfk] | 0.70 | 0.59 | 33.97 | 611 |
| HWA232 | wt las::CP29 | 0.56 | 3.34 | 60 | |
| HWA258 | wt las::CP29 [pfk] | 0.71 | 0.55 | 50.65 | 911 |
All mutant strains are MG1363 derivatives. The synthetic promoters transcribing the las genes are shown. The strains in which pfk is overexpressed from pMU2916 carrying pfk are indicated. wt, wild type.
Specific growth rates are shown for cultures growing in the absence (−) or presence (+) of erythromycin.
Specific activities (U/OD600), in micromoles per minute per cell density of phosphofructokinase are shown for culture growth in the presence of erythromycin. The activities from each strain were determined in triplicate. The specific activities of phosphofructokinase are percentages of that for MG1363.
Growth was restored by limiting the uptake of glucose.
An explanation for the changed phenotype of the las mutants could be that the low activity of phosphofructokinase resulted in an accumulation of sugar-phosphates, i.e., glucose-6-phosphate or fructose-6-phosphate, which are toxic at elevated levels (9). If this was the case it might be possible to increase the growth rate by limiting the external concentration of glucose. This hypothesis was tested in a chemostat experiment with a limited amount of glucose.
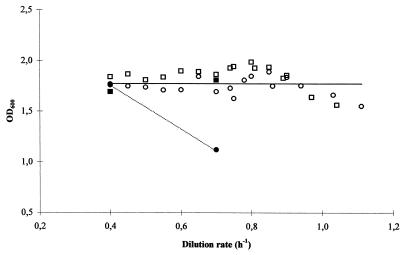
Table 4 shows that the largest difference in growth rate between the las mutants and MG1363 was obtained in M17 medium, and the chemostat experiments were therefore performed with this growth medium. Experiment A was initiated with a dilution rate of 0.4 h−1 and maintained for 5 h. The cell densities of HWA217 and HWA232 are shown in Fig. 4 as functions of the dilution rate. The observation from this experiment was as expected: MG1363 and HWA217 had similar and constant biomass concentrations at a dilution rate of 0.4 h−1, as this rate was below the critical growth rate of HWA217 (0.57 h−1). Subsequently, the dilution rate was changed to 0.7 h−1, and the cell density began to decline rapidly for HWA217, whereas MG1363 was stable, which indicated that the las mutant, in contrast to MG1363, was washed out. Next, experiment B, in which the dilution rate was gradually increased, was performed (Fig. 4). The dilution rate was changed from 0.40 to 1.11 h−1 at small intervals. The biomass concentration of HWA217 remained close to the concentration of MG1363, and thus a washout of the culture was avoided. The data show that by gradually changing the dilution rate it was possible to enhance the growth rate of the las mutant to 195% of its growth rate in batch culture where glucose was abundant.
FIG. 4.
Growth of las mutant HWA217 and MG1363 in chemostat culture at various dilution rates. Biomass concentration of the cultures is shown during the experiments as a function of the dilution rate. Growth was initiated in a batch fermentation. Before the cultures entered the stationary growth phase, the chemostat was inoculated at a dilution rate of 0.40 h−1. The experiments in which the dilution rate was increased rapidly (experiment A) or slowly (experiment B) are shown with closed or open symbols, respectively. The biomass concentrations of MG1363 and HWA217 are shown as squares and circles, respectively. The solid line indicates the biomass concentration of HWA217 when the dilution rate was increased slowly. This line also shows the biomass concentration of MG1363 in both cases. The broken line indicates the biomass concentration of HWA217 when the dilution rate was increased rapidly. The cultures were grown in M17 supplemented with 0.25% glucose, and pH was kept constant at 6.8 by the addition of 2 M NaOH.
To ensure that the phenotype of HWA217 had not changed as a result of acquired mutations, samples from the culture and MG1363 were streaked on plates before, during, and after the experiment to compare the colony sizes. Finally, growth rates of these samples were determined in batch cultures, which showed that the cultures behaved as seen earlier in glucose-abundant medium. This demonstrated that the increased growth rate of the las mutant in the chemostat was not due to selection of mutants during cultivation.
Sugar-phosphates accumulate in las mutants with low phosphofructokinase activity.
Several experiments here indicated that the inhibition of growth in the las mutants is due to the accumulation of sugar-phosphates upstream of phosphofructokinase. We measured the intracellular concentration of these metabolites in cells grown as batch cultures (flasks) (Table 6). Indeed, the concentrations of glucose-6-phosphate were found to be increased to 37 and 55 mM in HWA217 and HWA232, respectively, compared to 16 mM in the wild-type strain MG1363. Fructose-6-phosphate was also increased to 3.9 and 6.2 mM in strains HWA217 and HWA232, respectively, compared to 1.5 mM in MG1363.
TABLE 6.
Intracellular concentrations of sugar-phosphates in las mutants with reduced phosphofructokinase activity
| Strain | Genotypea | Relative phosphofructokinase activity (%) | Intracellular sugar-phosphate concn (mM)b
|
|
|---|---|---|---|---|
| Glucose-6-phosphate | Fructose-6-phosphate | |||
| MG1363 | wt | 100 | 16.4 (2.5) | 1.5 (0.09) |
| HWA217 | wt las::CP25 | 39 | 36.7 (2.1) | 3.9 (1.7) |
| HWA232 | wt las::CP29 | 60 | 55.1 (8.2) | 6.2 (1.4) |
wt, wild type.
Values are averages of 2 to 6 individual samples, and standard deviations are given in parentheses.
Glycolytic flux is unaffected in nongrowing cells.
If the accumulation of sugar-phosphates is responsible for the inhibition of growth rate and glycolytic flux in the mutants with low phosphofructokinase activity, then the glycolytic flux might be less affected in resting cells where glycolysis is uncoupled from growth. We therefore measured the glycolytic flux of the wild-type and mutant cells resuspended in buffer (Table 7). Homolactic fermentation continued in the cells, and the rates of glucose consumption and lactate production were almost identical to the rates observed for the wild-type strain.
TABLE 7.
Glycolytic flux in resting cells of las mutants with reduced phosphofructokinase activity
| Strain | Genotypea | Relative phosphofructokinase activity (%) | Rate of glucose consumption (mmol · g [dry wt]−1 · h−1)b | Rate of lactate production (mmol · g [dry wt]−1 · h−1)b |
|---|---|---|---|---|
| MG1363 | wt | 100 | 15.1 | 27.1 |
| HWA217 | wt las::CP25 | 39 | 14.5 | 25.9 |
| HWA232 | wt las::CP29 | 60 | 14.3 | 24.5 |
wt, wild type.
Rates of glucose consumption and lactate production were determined from the slopes of glucose and lacate concentration versus time and normalized by the cell dry weight. The correlation coefficients for all experiments ranged from 0.980 to 0.986.
DISCUSSION
The original aim of this study was to construct strains with modulated expression of the las operon and to estimate the control on the glycolytic flux by the corresponding three enzymes, using the approach of metabolic control analysis (13, 22). Unfortunately, the relative changes in gene expression in the obtained las mutants were not the same for the individual las genes pfk, pyk, and ldh, and the mutants are therefore not suited for such an analysis. Instead, the strains allowed us to investigate the importance of changing the expression of the pfk gene. We found that moderately reduced phosphofructokinase activity has a strong impact on L. lactis physiology; these changes appear to be due to changes in the concentration of upstream sugar-phosphates.
In both las mutants, the phosphofructokinase activity was reduced approximately twofold. Sequencing the pfk region in the las mutants excluded the possibility that mutations had been introduced in the pfk gene. Instead it is likely that the relative functional half-life of the mRNA for each of the three genes had been altered. The original mRNA of the las operon was not conserved in the mutants because 24 bp were missing in the leader region upstream of pfk. This may have affected the stability of mRNA in this region and lowered the functional half-life of the pfk message compared to the other two genes. The slightly altered pyruvate kinase and lactate dehydrogenase activities in one of the two mutant strains can also be explained from small variations in the leader mRNA sequences conferred by the two different synthetic promoter fragments employed.
It is unlikely that the slightly elevated expression of ldh could be the cause of the altered physiology since much larger variations in lactate dehydrogenase activity were found to have no influence on L. lactis growth rate or glycolytic flux (1; H. W. Andersen, M. B. Pedersen, K. Hammer, and P. R. Jensen, unpublished data). In both mutant strains the activity of pyruvate kinase was quite close to the wild-type level, and it is therefore unlikely that this enzyme was the cause of the observed growth defects. Instead, three lines of evidence indicated that the altered phosphofructokinase activity was the direct cause of the changed physiology: (i) the mutants grew like the wild-type strain in medium containing fructose, which bypasses the use of phosphofructokinase; and (ii) overexpressing the phosphofructokinase activity in trans restored the growth rate to the wild-type rate in the las mutants.
When the glycolytic flux in Lactococcus is reduced, the pattern of fermentation shifts to mixed-acid production, which in turn results in an increased supply of ATP (6, 8, 34). In our las mutants, phosphofructokinase appears to limit the glycolytic flux to a similar extent and the mutants might therefore be expected to show a shift to mixed-acid fermentation. However, it was found that the las mutants were mainly homolactic and probably not energy limited. What causes the lower growth rate of these mutants? In principle, the decrease in phosphofructokinase activity may be limiting for anabolic reactions. However, in complex medium the growth rates of the las mutants were even further reduced relative to the wild-type strain.
Another consequence of reduced phosphofructokinase activity could be an accumulation of upstream metabolites, i.e., glucose-6-phosphate. Glucose-6-phosphate has been reported to be toxic at enhanced levels (9). The concentration of glucose-6-phosphate was found to be increased from 2.2- to 3.4-fold in the two las mutants, and the fructose-6-phosphate concentration was increased 2.6- to 4.1-fold. Curiously, we find that the concentration of both sugar-phosphates is higher in the strain which has intermediary phosphofructokinase activity (60%) than in the strain with the lowest phosphofructokinase activity (39%). We suspect that this may be a consequence of slightly higher expression of pyruvate kinase and lactate dehydrogenase in the strain with the lowest phosphofructokinase activity. Both enzymes affect the phosphoenolpyruvate pool: the higher pyruvate kinase activity will directly lead to a lower phosphoenolpyruvate pool and the higher lactate dehydrogenase activity will lead to a lower pyruvate pool and therefore higher pyruvatekinase activity and lower phosphoenolpyruvate pool. A lower phosphoenolpyruvate pool will in turn affect the activity of the phosphoenolpyruvate:phosphotransferase system and the accumulation of the sugar-phosphates.
In the present study it was found that a sudden change in the dilution rate of mutant HWA217 in a chemostat culture resulted in culture washout. In contrast, when the dilution rate was increased slowly, washout of the las mutant was avoided. Under these conditions, the growth rate could be almost doubled compared to the growth rate in batch cultures. An explanation for this phenomenon is that an acceptable pool of sugar-phosphates in the las mutants was achieved by starving the cells for glucose and thereby eliminating a toxic effect. In Streptococcus mutans a toxic effect, which probably arose from some unidentified glycolytic intermediates, was also avoided by limiting the supply of sugar (14). The likely reason that the fast shift from low to high dilution rate did not result in fast growth was that the cells became trapped with high sugar-phosphate pools.
In cells resuspended in buffer we observed almost the same glycolytic flux of the two las mutants as of the wild-type strain. Glycolysis is uncoupled from growth under these conditions, and this result supports the hypothesis that the accumulation of sugar-phosphates inhibits the growth rate and thereby the glycolytic flux in the las mutants. This result also shows that under these specific conditions in nongrowing cells, phosphofructokinase does not control glycolysis.
The fact that a 40% reduction in the phosphofructokinase activity results in a three- to fourfold-higher concentration of the upstream metabolites indicates that the control exerted by phosphofructokinase on these metabolite concentrations is high. It also suggests that it is important for these cells to accurately balance the activity of the glycolytic enzymes, and the placement of the three genes together in the las operon is likely to be a reflection of this. By coregulating several glycolytic genes, the cells may gain an advantage with respect to regulating the glycolytic flux, because the concomitant changes in metabolite concentrations will tend to be smaller, in contrast to a situation in which only a single enzyme activity is altered. Indeed, computer modeling of the consequences of altering phosphofructokinase activity indicated a poor homeostasis of the glycolytic intermediates (41), which is consistent with the observations in the present study.
Does phosphofructokinase control growth rate and glycolytic flux in L. lactis?
One of the main observations in this study is that the growth rate, the glycolytic flux, and the lactate flux were decreased proportionally by a twofold reduction of the activity of phosphofructokinase, which indicates that phosphofructokinase is not present in large excess in wild-type L. lactis cells. This result is surprising because enzymes are usually present in large excess. In E. coli, it was found that the H+-ATPase, which is essential for growth on succinate minimal medium, has no control over the growth rate of E. coli, and reducing this enzyme activity by one-half has almost no effect on the growth rate (20). In diploid organisms, mutations that lower the cellular enzyme activities by one-half are often found to be recessive with virtually no effect on cell physiology.
The correlation between relative growth rate and relative activity of phosphofructokinase, obtained with the limited data points from Tables 2 and 4, was linear with a slope of 0.6 in defined medium, and this slope is even higher for experiments performed in complex medium. From these data it is tempting to draw conclusions about phosphofructokinase being a flux-controlling enzyme, which is certainly true at reduced phosphofructokinase levels. However, conclusions about flux control by phosphofructokinase can only be drawn if the strong effect of phosphofructokinase is also present at the wild-type phosphofructokinase level and preferably also slightly above this level. The complementation studies showed that eightfold overproduction of phosphofructokinase did not result in an enhanced growth rate or glycolytic flux. This could however be a consequence of excessive overproduction of phosphofructokinase or a protein burden effect, as was observed in Zymomonas mobilis (38). Therefore, whether phosphofructokinase exerts control over the growth rate and the glycolytic flux at the wild-type level of the enzyme in L. lactis remains to be investigated.
ACKNOWLEDGMENTS
We sincerely appreciate the expert technical assistance of Katrine Madsen, and we thank Martin Willemoës for providing the plasmid pMW40. Martin B. Pedersen, Martin Willemoës, Hans V. Westerhoff, and David Fell are acknowledged for suggestions and discussions. We acknowledge A. Hillier for donating plasmid pMU2916.
This work is part of the FO/TEK program supported by the Danish Dairy Research Foundation (Danish Dairy Board) and the Center of Advanced Food Studies (LMC).
REFERENCES
- 1.Andersen H W. Ph.D. thesis. Regulation and control of the glycolytic enzymes phosphofructokinase and lactate dehydrogenase in Lactococcus lactis. Technical University of DenmarkLyngby, Denmark: Department of Microbiology; 2000. [Google Scholar]
- 2.Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- 3.Crow V L, Pritchard G G. Fructose 1,6-diphosphate-activated l-lactate dehydrogenase from Streptococcus lactis: kinetic properties and factors affecting activation. J Bacteriol. 1977;131:82–91. doi: 10.1128/jb.131.1.82-91.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crow V L, Pritchard G G. Pyruvate kinase from Streptococcus lactis. Methods Enzymol. 1982;90:165–170. doi: 10.1016/s0076-6879(82)90122-7. [DOI] [PubMed] [Google Scholar]
- 5.Davidson B E, Llanos R M, Cancilla M R, Redman N C, Hillier A J. Current research on the genetics of lactic acid production in lactic acid bacteria. Int Dairy. 1995;5:763–784. [Google Scholar]
- 6.Demko G M, Blanton S J B, Benoit R E. Heterofermentative carbohydrate metabolism of lactose-impaired mutants of Streptococcus lactis. J Bacteriol. 1972;112:1335–1345. doi: 10.1128/jb.112.3.1335-1345.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fordyce A M, Moore C H, Pritchard G G. Phosphofructokinase from Streptococcus lactis. Methods Enzymol. 1982;90:77–82. doi: 10.1016/s0076-6879(82)90109-4. [DOI] [PubMed] [Google Scholar]
- 8.Fordyce A M, Crow V L, Thomas T D. Regulation of product formation during glucose or lactose limitation in nongrowing cells of Streptococcus lactis. Appl Environ Microbiol. 1984;48:332–337. doi: 10.1128/aem.48.2.332-337.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraenkel D G. The accumulation of glucose 6-phosphate from glucose and its effect in a Escherichia coli mutant lacking phosphoglucose isomerase and glucose 6-phosphate dehydrogenase. J Biol Chem. 1968;243:6451–6457. [PubMed] [Google Scholar]
- 10.Garrigues C, Loubiere P, Lindley N D, Cocaign-Bousquet M. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J Bacteriol. 1997;179:5282–5287. doi: 10.1128/jb.179.17.5282-5287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes F, Law J, Daly C, Fitzgerald G F. Integration and excision of plasmid DNA in Lactococcus lactis subsp. lactis. Plasmid. 1990;24:81–89. doi: 10.1016/0147-619x(90)90010-a. [DOI] [PubMed] [Google Scholar]
- 13.Heinrich R, Rapoport S M, Rapoport T A. Metabolic regulation and mathematical models. Prog Biophys Mol Biol. 1977;32:1–82. [PubMed] [Google Scholar]
- 14.Hillman J D, Chen A, Snoep J L. Genetic and physiological analysis of the lethal effect of l-(+)-lactate dehydrogenase deficiency in Streptococcus mutants: complementation by alcohol dehydrogenase from Zymomonas mobilis. Infect Immun. 1996;64:4319–4323. doi: 10.1128/iai.64.10.4319-4323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen N B. Ph.D. thesis. Influence of oxygen on growth and product formation in lactic acid bacteria. Technical University of DenmarkLyngby, Denmark: Department of Biotechnology; 1999. [Google Scholar]
- 17.Jensen P R, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen P R, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen P R, Michelsen O. Carbon and energy metabolism of atp mutants of Escherichia coli. J Bacteriol. 1992;174:7635–7641. doi: 10.1128/jb.174.23.7635-7641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen P R, Westerhoff H V, Michelsen O. Excess capacity of H+-ATPase and inverse respiratory control in Escherichia coli. EMBO J. 1993;12:1277–1282. doi: 10.1002/j.1460-2075.1993.tb05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansen E, Kibenich A. Characterization of Leuconostoc isolates from commercial mixed strain mesophilic starter cultures. J Dairy Sci. 1992;75:1186–1191. [Google Scholar]
- 22.Kacser H, Burns J A. The control of flux. Symp Soc Exp Biol. 1973;27:65–104. [PubMed] [Google Scholar]
- 23.Le Bourgeois P, Lautier M, Mata M, Ritzenthaler P. New tools for the physical and genetic mapping of Lactococcus strains. Gene. 1992;111:109–114. doi: 10.1016/0378-1119(92)90610-2. [DOI] [PubMed] [Google Scholar]
- 24.Liberman E S, Bleiweis A S. Transport of glucose and mannose by a common phosphoenolpyruvate-dependent phosphotransferase system in Streptococcus mutans GS5. Infect Immum. 1984;43:1106–1109. doi: 10.1128/iai.43.3.1106-1109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llanos R M, Harris C J, Hillier A J, Davidson B E. Identification of a novel operon in Lactococcus lactis encoding three enzymes for lactic acid synthesis: phosphofructokinase, pyruvate kinase, and lactate dehydrogenase. J Bacteriol. 1993;175:2541–2551. doi: 10.1128/jb.175.9.2541-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luesink E J, van Herpen R E, Grossiord B P, Kuipers O P, de Vos W M. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 27.Luesink E J, Beumer C M A, Kuipers O P, De Vos W M. Molecular characterization of the Lactococcus lactis ptsHI operon and analysis of the regulatory role of HPr. J Bacteriol. 1999;181:764–771. doi: 10.1128/jb.181.3.764-771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mason P W, Carbone D P, Cushman R A, Waggoner A S. The importance of inorganic phosphate in regulation of energy metabolism of Streptococcus lactis. J Biol Chem. 1981;256:1861–1866. [PubMed] [Google Scholar]
- 29.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- 30.Novák L, Loubiere P. The metabolic network of Lactococcus lactis: distribution of 14C-labeled substrates between catabolic and anabolic pathways. J Bacteriol. 2000;182:1136–1143. doi: 10.1128/jb.182.4.1136-1143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamato T, Fujita Y, Irie R. Protoplast formation and regeneration of Streptococcus lactis cells. Agric Biol Chem. 1983;47:259–263. [Google Scholar]
- 32.Pedersen M B. Master's thesis. Metabolic control analysis on glycolysis in Lactococcus lactis. Technical University of DenmarkLyngby, Denmark: Department of Microbiology; 1996. [Google Scholar]
- 33.Peri K G, Goldie H, Waygood E B. Cloning and characterization of the N-acetylglucosamine operon of Escherichia coli. Biochem Cell Biol. 1990;68:123–137. doi: 10.1139/o90-017. [DOI] [PubMed] [Google Scholar]
- 34.Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–147. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 35.Poolman B, Bosman B, Kiers J, Konings W N. Control of glycolysis by glyceraldehyde-3-phosphate dehydrogenase in Streptococcus cremoris and Streptococcus lactis. J Bacteriol. 1987;169:5887–5890. doi: 10.1128/jb.169.12.5887-5890.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sjöberg A, Persson I, Quednau M, Hahn-Hägerdal B. The influence of limiting and non-limiting growth conditions on glucose and maltose metabolism in Lactococcus lactis ssp. lactis strains. Appl Microbiol Biotechnol. 1995;42:931–938. [Google Scholar]
- 38.Snoep J L, Yomano L P, Westerhoff H V, Ingram L. Protein burden in Zymomonas mobilis: negative flux and growth control due to overproduction of glycolytic enzymes. Microbiology. 1995;141:2329–2337. [Google Scholar]
- 39.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 40.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas S, Fell D A. A control analysis exploration of the role of ATP utilisation in glycolytic-flux control and glycolytic-metabolite-concentration regulation. Eur J Biochem. 1998;258:956–967. doi: 10.1046/j.1432-1327.1998.2580956.x. [DOI] [PubMed] [Google Scholar]
- 42.Thomas T D. Regulation of lactose fermentation in group N streptococci. Appl Environ Microbiol. 1976;32:474–478. doi: 10.1128/aem.32.4.474-478.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas T D, Ellwood D C, Longyear V M C. Change from homo- to heterolactic fermentation by Streptococcus lactis resulting from glucose limitation in anaerobic chemostat cultures. J Bacteriol. 1979;138:109–117. doi: 10.1128/jb.138.1.109-117.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson J. Characteristics and energy requirements of an α-aminoisobutyric acid transport system in Streptococcus lactis. J Bacteriol. 1976;127:719–730. doi: 10.1128/jb.127.2.719-730.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson J. In vivo regulation of glycolysis and characterization of sugar:phosphotransferase systems in Streptococcus lactis. J Bacteriol. 1978;136:465–476. doi: 10.1128/jb.136.2.465-476.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson J. Sugar transport in lactic acid bacteria. In: Reizer J, Peterkofsky A, editors. Sugar transport and metabolism in gram-positive bacteria. Chichester, England: Ellis Horwood Limited; 1987. pp. 13–38. [Google Scholar]
- 47.Thompson J, Chassy B M. Intracellular phosphorylation of glucose analogs via the phosphoenolpyruvate: mannose-phosphotransferase system in Streptococcus lactis. J Bacteriol. 1985;162:224–234. doi: 10.1128/jb.162.1.224-234.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson J, Thomas T D. Phosphoenolpyruvate and 2-phosphoglycerate: endogenous energy source(s) for sugar accumulation by starved cells of Streptococcus lactis. J Bacteriol. 1977;130:583–595. doi: 10.1128/jb.130.2.583-595.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson J, Torchia D A. Use of 31P nuclear magnetic resonance spectroscopy and 14C fluorography in studies of glycolysis and regulation of pyruvate kinase in Streptococcus lactis. J Bacteriol. 1984;158:791–800. doi: 10.1128/jb.158.3.791-800.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]