Abstract
The Streptomyces venezuelae pikD gene from the pikromycin biosynthetic cluster was analyzed, and its deduced product (PikD) was found to have amino acid sequence homology with a small family of bacterial regulatory proteins. Database comparisons revealed two hypothetical domains, including an N-terminal triphosphate-binding domain and a C-terminal helix-turn-helix DNA-binding motif. Analysis of PikD was initiated by deletion of the corresponding gene (pikD) from the chromosome of S. venezuelae, resulting in complete loss of antibiotic production. Complementation by a plasmid carrying pikD restored macrolide biosynthesis, demonstrating that PikD is a positive regulator. Mutations were made in the predicted nucleotide triphosphate-binding domain, confirming the active-site amino acid residues of the Walker A and B motifs. Feeding of macrolide intermediates was carried out to gauge the points of operon control by PikD. Although the pikD mutant strain was unable to convert macrolactones (10-deoxymethynolide and narbonolide) to glycosylated products, macrolide intermediates (YC-17 and narbomycin) were hydroxylated with high efficiency. To study further the control of biosynthesis, presumed promoter regions from pik cluster loci were linked to the xylE reporter and placed in S. venezuelae wild-type and pikD mutant strains. This analysis demonstrated that PikD-mediated transcriptional regulation occurs at promoters controlling expression of pikRII, pikAI, and desI but not those controlling pikRI or pikC.
The soil bacteria belonging to the genus Streptomyces have been of great interest due to their well-known capacity to produce a diverse range of antibiotics and other secondary metabolites (4, 24). Production typically occurs according to a growth phase-dependent profile (11, 12) and is often accompanied by the development of spore-bearing aerial mycelia (9).
The regulatory elements involved in generating antibiotics are of significant interest. An entire family of regulatory genes called SARPs (Streptomyces antibiotic regulatory proteins) has been identified based on sequence and motif homology, as well as by complementation studies (2, 8). Sequence analysis links the SARP family together by the presence of OmpR-like DNA-binding domains (25). Members of this family include the positive regulators ActII-ORF4 of the actinorhodin biosynthetic cluster (2), RedD of the undecylprodigiosin gene cluster (26, 36), DnrI of the daunorubicin biosynthetic system (33, 36), and CcaR, which regulates both the cephamycin and clavulanic acid pathways (28, 36). Other genes have been identified that encode transcriptional activators of specific SARPs. These include redZ and dnrN, which activate redD and dnrI, respectively (15, 35).
A general network or system of regulatory elements involved in the control of secondary metabolite pathways has not yet emerged. For example, the srmR gene of Streptomyces ambofaciens, which regulates spiramycin production, shows no sequence homology to other regulatory proteins (16). The tylosin biosynthetic pathway of Streptomyces fradiae contains putative regulatory genes from several different families (3). These genes include tylR, whose product is a global regulator of tylosin production, and the putative regulators encoded by tylT, tylS (both belonging to the SARP family), and tylP, whose product is a γ-butyrolactone receptor. In other notable cases, as with the polyketide-producing erythromycin pathway, no regulatory genes have been found in the region of the biosynthetic cluster (14).
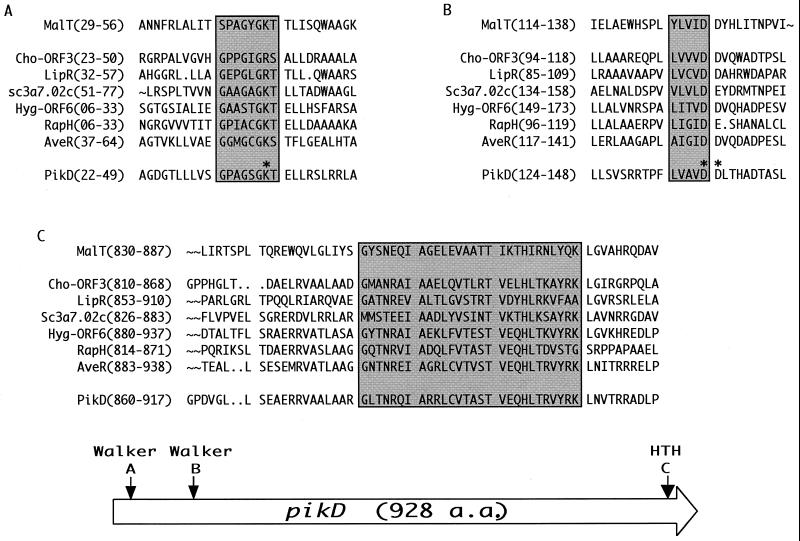
Significantly, recent characterization of several other macrolide antibiotic pathways has led to the identification of a set of novel transcriptional regulators. This new family is based on recent observations (13) involving a unique group of putative regulators with several shared traits, including a relatively large size (872 to 1,159 amino acids [a.a.]) compared to the SARP family (typically between 277 and 665 a.a.), an N-terminal ATP-binding domain represented by discernable Walker A and B motifs (34), and a C-terminal LuxR type DNA-binding domain (17) (Fig. 1). The name given to this group is large ATP-binding regulators of the LuxR family (LAL), and the prototype member is the Escherichia coli MalT protein, involved in the uptake and catabolism of maltodextrins (29). Currently, three regulators from modular polyketide synthase (PKS) biosynthetic pathways appear to fall into this group, including RapH from the rapamycin cluster of Streptomyces hygroscopicus (1, 32), Orf6 from a second PKS cluster (whose product remains unknown) found in S. hygroscopicus (30), and PikD from the multi-drug producing pikromycin pathway (39, 41). The role of these LALs in regulation of polyketide biosynthesis has not been reported previously, and providing insight into their function motivated the current study.
FIG. 1.
General structure and amino acid alignment of hypothetical LAL proteins, including PikD. Shaded regions encompass putative and confirmed NTP- and DNA-binding motifs, corresponding to arrows on the schematic drawing of PikD. Asterisks denote amino acid residues in PikD that were mutated in this study.
In this report we provide a detailed characterization of the PikD regulator from the pikromycin biosynthetic system. Through chromosomal gene deletion, plasmid complementation, and site-directed mutagenesis, its role as a pathway-specific positive regulator and the location of its nucleotide triphosphate-binding site residues have been verified. In addition, we describe the level of control exerted by PikD through conversion of pathway intermediates and promoter probing using the xylE reporter system.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
E. coli DH5α was used as a cloning host and grown on Luria-Bertani (LB) liquid or solid medium. S. venezuelae ATCC 15439 is the pikromycin-producing strain used throughout this study. S. venezuelae mutants AX906 (40), AX916 (Y. Xue, unpublished), and LZ3001 (41) were used for precursor feeding experiments. Liquid SGGP (22) was employed for vegetative growth. Solid R2YE (21) was used for transformation and selection of S. venezuelae, while liquid SCM (22) was used for antibiotic production. pUC119 (42) was used for general DNA cloning. The pikD gene disruption construct pDHS2062 was derived from pKC1139 (6) (see Table 2), which is an E. coli-Streptomyces conjugative shuttle plasmid. pDHS618 (37), also an E. coli-Streptomyces shuttle plasmid containing the pikA promoter, was the vector used to create the pikD complementation construct pDHS2063. pDHS702 (38), a shuttle vector containing the pikA promoter coupled with pikAV, was used to create the promoter-probing plasmids pDHS2100 to pDHS2112. For a complete list of strains and plasmids, see Tables 1 and 2.
TABLE 2.
Strains and plasmids used
| Strain or plasmids | Relevant characteristicsa | Source or reference | |
|---|---|---|---|
| S. venezuelae | |||
| ATCC 15439 | Wild-type pikromycin, methymycin, and neomethymycin producer | ATCCb | |
| AX906 | pikC insertion mutant of ATCC 15439 | 34 | |
| AX916 | In-frame fusion of pikAI module 1 and pikAIII module 5 | Y. Xue, unpublished | |
| DW900 | pikD replacement by aphII in ATCC 15439 | This study | |
| LZ3001 | desVI replacement by aphII in ATCC 15439 | 41 | |
| Plasmids | |||
| pUC119 | Ap lacZα MCS, E. coli cloning vector | 36 | |
| pKC1139 | Am lacZα MCS oriT repts | 4 | |
| pDHS618 | Am PpikA, containing pikAV and pikC | 42 | |
| pDHS702 | Ap Ts PpikA, containing pikAV | 29 | |
| pDHS2100–pDHS2112 | pDHS702 containing a T7 terminator upstream of various S. venezuelae putative promoter sequences fused to the reporter xy1E | This study | |
| pDHS2061 | pUC119 containing 1.5 kb upstream and 1.2 kb downstream of pikD flanking a Kan gene | This study | |
| pDHS2062 | pKC1139 containing replacement deletion cassette excised from pDHS2061 | This study | |
| pDHS2063 | pDHS618 containing wild-type pikD | This study | |
| pDHS2070 | pDHS618 containing pikD K38A mutation | This study | |
| pDHS2071 | pDHS618 containing pikD K38R mutation | This study | |
| pDHS2072 | pDHS618 containing pikD D138N mutation | This study | |
| pDHS2073 | pDHS618 containing pikD D139N mutation | This study |
Am, apramycin resistance; Ap, ampicillin resistance; Kan, kanamycin resistance; Ts, thiostrepton resistance. pikAV was removed from all constructs using pDHS702 prior to use. All constructs using pDHS618 were digested with EcoRI and XbaI to remove pikAV and pikC before further manipulation.
ATCC, American Type Culture Collection.
TABLE 1.
Plasmidsa
| Plasmid | Promoter region amplified | Forward primer | Reverse primer |
|---|---|---|---|
| pDHS2100 | None (control) | AAATTAATTAAGAGGAATTCATCGAGGGG | AAAAGATCTCACTTGCCCGCCCCCTCGATG |
| pDHS2101 | pikAI | AAATTAATTAACCCCTCCCTGGATGCCGTGGTCA | AAAAGATCTCACTTGCCCGCCCCCTCGATG |
| pDHS2102 | pikAIV | AAATTAATTAACGACCTGGACGCCGAGGCCCTGAT | AAAAGATCTCATCGGGTGTGGTCTTCCG |
| pDHS2103 | desI-I | AAATTAATTAACACGGAGAACTCCAGACCG | AAAAGATCTCACGGATGTTCCCTCCGGGCC |
| pDHS2104 | pikC | AAATTAATTAACACGGATGTTCCCTCCGGG | AAAAGATCTCACGGAGAACTCCAGACCGG |
| pDHS2105 | pikRI | AAATTAATTAACCGGCTCCGTCTCCGGAAG | AAAAGATCTCATGAACGATCCCCTCCCTG |
| pDHS2106 | pikRII | AAATTAATTAATGACCTCGGCCGATGCCCC | AAAAGATCTCATGAGTCTGCTCCGCGAAA |
| pDHS2107 | pikD | AAATTAATTAATGAACCCGCACGTCACCCA | AAAAGATCTCATCGCGGAAGTCCCCCCTCG |
| pDHS2108 | None (control) | AAATTAATTAACGCCGGCCAGGGCCATCAC | AAAGAATTCGCACAGCCTCGGCGCCTCCG |
| pDHS2109 | None (control) | AAATTAATTAAGTCCAAGAGTGAGTCCGAG | AAAGAATTCGGCGCCGGGCACCCGGCAGG |
| pDHS2110 | desI-II | AAATTAATTAATCGCGGAAGTCCCCCCTCG | AAAAGATCTCACGGATGTTCCCTCCGGGCC |
| pDHS2111 | desVIII (full length) | AAATTAATTAAGATCCGGCGCTTCCACCCC | AAAGGATCCCACCGTGGGTTCTGCCATCTC |
| pDHS2112 | desVIII (100 bp) | AAATTAATTAATGCCTCCGGGCGTACTCCG | AAAGGATCCCACCGTGGGTTCTGCCATCTC |
The primers used to amplify the desired promoter region are shown with the engineered restriction sites in bold type.
DNA sequencing and analysis.
pikD was sequenced as part of the pikCD operon as previously described (39, 40). DNA and deduced protein sequence analyses were performed using MacVector or GCG sequence analysis packages (Oxford Molecular Group) employing the program default-specific parameters.
Gene disruption.
Deletion of pikD was accomplished by gene replacement via homologous recombination. For this purpose, two regions were amplified by PCR. The first region began 1.5 kb upstream of pikD and terminated short of its ribosome-binding site (RBS). HindIII and BamHI sites were engineered into the upstream and downstream primers, respectively (primers GGTCTGGAGTTCAAGCTTCGCCGTACCCAGCAGGGAACG and CTGGATCCCTCTAACCAGGTCTTGTTACGGCG). This first fragment was cloned into HindIII- and BamHI-digested pUC-NEO, a version of pUC119 containing a neomycin-kanamycin resistance gene and promoter cloned into the SmaI site of the multiple cloning site (MCS). The second PCR-amplified region extended 1.2 kb downstream of the pikD stop codon. The upstream and downstream primers contained KpnI and EcoRI, respectively (primers TGAGGTACCCCCGGTGTCCCCGTGCGACGACC and GCGAATTCGACGAGGACGGCAATCAGCTCGCC). Cloning the second fragment into KpnI- and EcoRI-digested pUC-NEO containing the first fragment created pDHS2061, an intermediate containing the necessary insert for deletion of pikD. pDHS2061 was subsequently digested with HindIII and EcoRI, and the 3.7-kb insert was ligated into pKC1139, creating pDHS2062.
pDHS2062 was transformed into S. venezuelae by previously described methods (21). Taking advantage of the instability of pKC1139 in the absence of thiostrepton, transformants were plated alternately on medium containing thiostrepton and kanamycin and medium containing only kanamycin. The cells were inoculated into liquid SGGP, and serial dilutions were plated onto R2YE containing kanamycin. These plates yielded over 90% kanamycin-resistant, thiostrepton-sensitive colonies. The genotype was checked in all colonies selected by Southern blot analysis (data not shown), which confirmed construction of the pikD replacement strain DW900.
Complementation of pikD deletion mutant.
Beginning with its RBS, pikD was PCR amplified using primers designed to incorporate EcoRI and XbaI sites into the upstream and downstream regions, respectively (primers AAAGAATTCGAGGGGGGACTTCCGCGATGA and TTTTCTAGAGGTGGCTCAGGCCGTGACGG). The gene was ligated into the EcoRI and XbaI sites of pDHS618, an E. coli-Streptomyces shuttle vector carrying the PpikA region, which drives transcription of the PKS genes. Genes placed under PpikA have previously been shown to be expressed in S. venezuelae (10, 38). Incorporation of pikD into pDHS618 created the complementation vector pDHS2063 (Table 2), which was transformed into DW900, and colonies were selected for resistance to kanamycin and apramycin. The presence of the plasmid was confirmed by plasmid extraction before metabolite fermentation analysis was performed.
Site-directed mutagenesis of pikD.
The QuickChange site-directed mutagenesis kit (Stratagene) was used to create point mutations in the active-site residues of the Walker A and B motifs (Fig. 1). To this end pikD was cloned into pUC119 to reduce the overall size needed for amplification. Four pairs of complementary primers were designed to mutate three residues. K38 was changed to either an R or A to confirm the position of the Walker A motif (primers CCGGCAGCGGGGCGACGGAGCTGC and GCAGCTCCGTCGCCCCGCTGCCGG and primers CCGGCAGCGGGAGGACGGAGCTGC and GCAGCTCCGTCCTCCCGCTGCCGG, respectively). Both aspartic acid residues 138 and 139 were mutated to asparagine residues to elucidate which was the terminal amino acid of the Walker B motif (primers CTCGTCGCCGTCAACGACCTGACCC and GGGTCAGGTCGTTGACGGCGACGAG and primers GTCGCCGTCGACAACCTGACCCACG and CGTGGGTCAGGTTGTCGACGGCGAC, respectively). The mutations were confirmed through sequencing and cloned back into pDHS618 to give pDHS2070 (K38R), pDHS2071 (K38A), pDHS2072 (D138N), and pDHS2073 (D139N) (Table 2). The plasmids were transformed into DW900 and tested for antibiotic production.
Antibiotic production, isolation, and analysis.
The antibiotics methymycin and neomethymycin were considered representative of the ability of S. venezuelae to produce all pik cluster antibiotics (37). Therefore, production of antibiotics was performed using solely SCM medium, incubated at 30°C for 72 h. Methymycin and neomethymycin were extracted following published procedures (7). Thin-layer chromatography (TLC) comparison with wild-type S. venezuelae was used to confirm the presence or absence of antibiotics. The solvent system employed for TLC was chloroform–methanol–25% ammonium hydroxide (90:10:1) followed by vanillin staining (0.75% vanillin and 1.5% H2SO4 in methanol) and development by heating.
Biosynthetic intermediate feeding experiments.
Macrolactone and macrolide intermediates suspended in ethanol were added to fresh SCM medium in the ratio of 150 μl per 50 ml. This inoculum contained half of the extract from one 50-ml culture from either AX906 or LZ3001. The pikD deletion mutant was added to the broth containing either nonhydroxylated macrolides (YC-17 or narbonolide) or 10-deoxymethonolide. AX916, an in-frame fusion mutant between pikAI module 1 and pikAIII module 5, which contains intact des and pikCD loci, was used as a control.
Promoter probe assays.
Each plasmid (Table 1) contained a specific hypothetical promoter region followed by the original start codon of the downstream gene coupled to the xylE start codon by a BglII site. The pikAI promoter was an exception, as it was fused to pikAV as previously described (38). The fusion was made at the native EcoRI site 216 bp into the coding region of pikAI and an engineered EcoRI site 23 bp upstream of the pikAV start codon. Promoter regions were PCR amplified with a PacI site in the upstream primer and a BglII site in the downstream primer directly following the native start codon. The xylE gene was amplified to include a BglII site in the upstream primer directly preceding its start codon and an NsiI site in the downstream primer.
Three control plasmids were created to test the stringency of the XylE assay. The first control (pDHS2100) was amplified from pDHS2101. The control region placed a PacI site 3 bp upstream of the native EcoRI site and a BglII site directly following the pikAV start codon, which was ligated into PacI-BglII-digested pDHS2101. The other two controls (pDHS2108 and pDHS2109) were created by amplifying regions of pik DNA (within known coding regions) that contained no promoter. These regions were amplified with an upstream PacI site and a downstream EcoRI site. The fragments were ligated into pDHS2101 digested with PacI and EcoRI, which resulted in a plasmid that contained the control DNA sequence while retaining the active RBS and start codon of pikAV. pDHS2108 includes a 300-bp fragment starting 236 bp upstream of the pikAV stop codon and ending at the serine active-site residue. The fragment is in the opposite orientation relative to pikAV. pDHS2109 contains the 20-bp region starting at the second hypothetical pikAI start site and ending at the native EcoRI site. A T7 terminator was incorporated upstream of the promoter region of each plasmid inserted between the NdeI and PacI sites.
For promoter-probe analysis, each vector was transformed into either the S. venezuelae wild-type strain or DW900. To determine XylE activity, transformants were inoculated into 50 ml of SCM medium in 250-ml baffled flasks. Cultures were harvested after 67 h and processed as previously described (20). Cells were washed in 5 ml of 20 mM potassium phosphate (pH 7.5) and suspended in a final volume of 3 ml of sample buffer (100 mM potassium phosphate [pH 7.5], 20 mM EDTA, 10% acetone [vol/vol]). Cells were lysed by sonication for 1 min, and 30 μl of 10% Triton X-100 was added. Cell debris was removed by centrifugation for 30 min at 4°C in a Beckman JA-10 rotor at 17,000 rpm. Catechol dioxygenase activities were determined using a spectrophotometer as previously described (20). One milliliter of reaction buffer (100 mM potassium phosphate [pH 7.5], 0.2 mM catechol) was incubated for 1 min at 37°C prior to the addition of 20 μg of total protein. The optical density at 375 nm was measured over 1 min. Catechol dioxygenase activity was calculated as the rate of change in optical density per minute per milligram of protein used and converted to milliunits per milligram (31). Protein concentration was determined using the Bradford protein assay (Bio-Rad).
RESULTS AND DISCUSSION
Sequence characteristics of pikD.
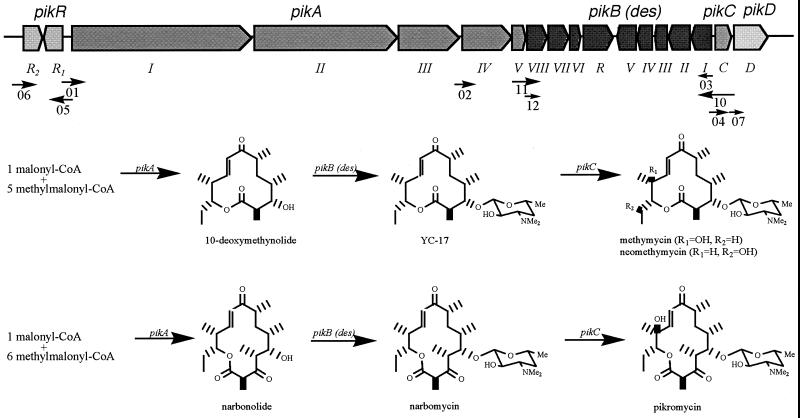
The entire PKS cluster responsible for methymycin, neomethymycin, narbomycin, and pikromycin biosynthesis (pik) in S. venezuelae was characterized by nucleotide sequence analysis (39, 41). It begins with a resistance locus that includes the divergently transcribed genes pikRI and pikRII. It continues through three biosynthetic loci: pikA, PKS genes responsible for the macrolide backbone; des, the desosamine biosynthesis and transfer genes; and pikC, the sole P450 hydroxylase gene. Finally, at the presumed terminus of the pik cluster, upstream of several primary metabolite genes, lies pikD (Fig. 2), whose protein product (PikD) is a deduced polypeptide of 929 a.a. (predicted molecular mass, 101 kDa).
FIG. 2.
Organization of the pikromycin cluster and locations of putative promoter regions. Numbered arrows correspond to the promoter regions cloned into promoter-probe constructs, with the numbers representing the last two digits in the name of the corresponding plasmid (see also Table 1). The length of the promoter region is denoted by the tail of the arrow, and the arrow point reflects the direction of transcription. CoA, coenzyme A.
Initial analysis of PikD was performed using SwissProt protein motif identification software. Query returns revealed a hypothetical N terminus nucleotide triphosphate (NTP)-binding domain or Walker A motif GXXXGKT (34) (a.a. 32-39), as well as a possible C-terminus helix-turn-helix (HTH) DNA-binding domain (a.a. 880 to 907) belonging to the LuxR family (17) (Fig. 1). A BlastP sequence homology search identified several regulatory and putative regulatory proteins with significant similarity profiles. A number of the proteins were relatively large (800 to 1,200 a.a.) and contained an N-terminal NTP-binding motif in addition to a C-terminal HTH DNA-binding domain. The proteins showing the greatest homology to PikD were putative positive regulators, such as ORF6 of an uncharacterized type I PKS found in the rapamycin-producing strain S. hygroscopicus (30), and the well-defined maltotriose and ATP-requiring activator MalT of the maltose system in E. coli (29). This group was previously assigned to the LALs (11). Members of this family were proposed to operate by a similar biochemical mechanism. The less conserved middle region of the protein is proposed to be the possible site for inducer interaction, such as those found in MalT and AcoK, which require the coinducers maltotriose and acetoin, respectively (27, 29).
Along with PikD, RapH, and S. hygroscopicus ORF6, sequence homology searches showed that AveR of the avermectin cluster from Streptomyces avermitilis (18, 19) could be added to the list of hypothetical LAL proteins. Currently, of these four regulatory genes, no direct analysis has been reported other than the transposon mutagenesis of aveR, which eliminated avermectin production (19).
Functional characterization of PikD.
In order to begin probing its function, the pikD gene was deleted from the chromosome using a kanamycin resistance gene (aphII) replacement cassette. After transformation of S. venezuelae by pDHS2062, clones selected for the kanamycin resistance phenotype were genotypically confirmed by Southern blotting (data not shown). The pikD deletion mutant strain DW900 had growth and morphological characteristics identical to those of wild-type S. venezuelae when grown on solid media, indicating that pikD plays no role in bacterial growth and differentiation. Moreover, no trace of methymycin, pikromycin, or pathway intermediates was observed in the DW900 mutant, indicating that PikD plays a key role as a positive regulator in antibiotic production. The absence of YC-17 and narbomycin indicates that PikD exhibits control in the early part of polyketide biosynthesis, supporting the hypothesis that it acts on the initiation of macrolide production by positively regulating expression of at least the pikA-encoded PKS (e.g., some or all of pikAI to pikAV) genes.
To confirm that deletion of pikD was the sole reason for loss of antibiotic production in S. venezuelae DW900, the gene was reintroduced into the mutant strain on the pDHS618-based expression plasmid (37) pDHS2063. The recombinant strain S. venezuelae DW900/pDHS2063 was cultured under macrolide production conditions and found to produce antibiotics at levels comparable to those of wild-type S. venezuelae (data not shown). This result was not surprising, since pikD was expressed under the control of a strong heterologous promoter (PpikAI) that would result in more PikD in the recombinant strain than in wild-type S. venezuelae.
Dissection of PikD through mutagenesis.
In an effort to confirm the presence of functional domains within PikD, mutations were made in the putative Walker A and B motifs (Fig. 1). Point mutations were introduced into pikD (borne on a plasmid), and subsequent analysis was performed in the DW900 (pikD deletion mutant) strain. The binding-site lysine residue (K38) of the Walker A motif was mutated to both alanine (pDHS2070) and arginine (pDHS2071). The binding-site residues of the Walker B motif include one of two different aspartic acid residues (D138 and D139). To elucidate which one serves as the active-site residue, each was individually mutated to asparagine (pDHS2072 and pDHS2073). DW900 was transformed with each plasmid and checked for production of macrolide antibiotics (Table 3). Both pDHS2070 (K38A) and pDHS2071 (K38R) failed to complement DW900, indicating that the binding-site lysine had been correctly recognized. Likewise, pDHS2072 (D138N) failed to complement DW900. In contrast, the plasmid carrying the D139N mutation (pDHS2073) fully complemented the pikD deletion strain (Table 3). These results provide strong evidence that PikD contains an active NTP-binding domain. MalT requires ATP both to form the open complex and to bind DNA (29) and thus, perhaps PikD utilizes an NTP in the same manner.
TABLE 3.
Antibiotic productiona
| Construct | Walker A | Walker B | Antibiotic production |
|---|---|---|---|
| Wild type | + | ||
| pDHS2070 | GPAGSGAT | LVAVDD | − |
| pDHS2071 | GPAGSGRT | LVAVDD | − |
| pDHS2072 | GPAGSGKT | LVAVND | − |
| pDHS2073 | GPAGSGKT | LVAVDN | + |
Antibiotic production phenotype was determined in S. venezuelae strains that contain mutations in the Walker A and B sequence motifs. Asterisks denote amino acid residues that were mutated in this study. The complete Walker A motif is shown, while the Walker B motif contains an extra amino acid corresponding to two possible aspartic acid residues at the C terminus of the domain.
Mapping PikD site of action by biosynthetic intermediate feeding experiments.
The lack of macrolactone production in DW900 indicates that PikD is necessary for transcription of the pikA-encoded modular PKS genes. In order to assess which other pikromycin biosynthetic genes were transcribed in the absence of pikD, a precursor feeding regimen was devised. S. venezuelae mutants (41) blocked in either pikC (strain AX906, accumulates YC-17) or des (LZ3001, accumulates 10-deoxymethynolide) were cultured, and their products were extracted. The extracts derived from the corresponding mutant strains containing the desired precursor metabolites were then used for biosynthetic intermediate feeding studies with DW900 or AX916, a mutant containing deleted pikA genes but possessing functional des and pikC genes.
The results demonstrate that 10-deoxymethynolide and the nonhydroxylated macrolides YC-17 and narbomycin freely cross the S. venezuelae cell membrane, where their cognate biosynthesis enzymes efficiently convert them to final products. Thus, as positive control AX916 (containing a nonfunctional Pik PKS but functional Des and PikCD) converted 10-deoxymethynolide to methymycin and neomethymycin and narbonolide to narbomycin and pikromycin. Similarly, AX916 converted YC-17 to methymycin and neomethymycin and narbomycin to pikromycin. In contrast, recovery of metabolites from the pikD deletion mutant (DW900) revealed no detectable conversion of the aglycones (10-deoxymethynolide and narbonolide), but quantitative conversion of YC-17 and narbomycin to their hydroxylated products (Fig. 2). The lack of conversion of 10-deoxymethynolide by DW900 shows that the des genes are also directly or indirectly under pikD control and not regulated solely by substrate concentration. At the same time, these results suggest that pikC is regulated independently of pikD.
Quantitative analysis of PikD activity by promoter probing.
To examine points of PikD control in greater detail, a series of promoter-probe constructs linked to the xylE reporter gene were engineered (Table 1). The sites were initially chosen at intergenic regions between divergently transcribed genes. This was the basis for selection of the promoter regions within pikRI, pikRII (resistance genes), pikAI (PKS genes), desI (desosamine genes), and pikC (P450 gene) (Fig. 2). Recently, a small region between pikAIII and pikAIV showed possible promoter activity, so this region was also examined (Y. Xue, unpublished). In addition, a sequence within pikAV has recently been reported to have promoter-like activity (10). In the latter report, two constructs of different lengths, both containing the C-terminal region of pikAV, were engineered. Promoter-probe plasmids that include both fragments were cloned, with the full-length version comprising the entire pikAV gene. The intergenic region between pikC and pikD was also included for experimental completeness.
The regions used to construct the promoter-probe plasmids are depicted in Fig. 2. Each putative promoter region followed by the original start codon of the downstream gene was coupled to the xylE start codon by a BglII site (see Materials and Methods). This maintained the wild-type configuration of each promoter with its native RBS. The pikA promoter (PpikAI) was an exception and was derived from pDHS702 (38), which uses the pikAV start site in place of its native site.
Three control promoter-probe plasmids devoid of promoter sequences were engineered to determine background levels of XylE. The first one (pDHS2100) included 18 bp between the T7 terminator region and the RBS. pDHS2108 added 317 bp to this region (from pikAV) in the opposite orientation relative to the native gene sequence. pDHS2109 added 19 bp to give a total of 37 bp between the T7 terminator and the RBS. The sequence cloned for construction of pDHS2109 was taken from within the coding region of pikAI. For subsequent analysis, the data derived from the controls were averaged and subtracted from subsequent pik promoter-probe experiments to give the corresponding weighted data.
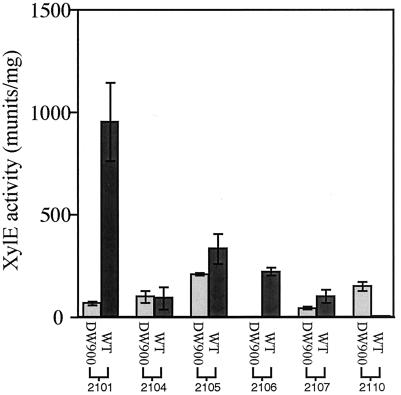
pDHS2102, pDHS2103, pDHS2111, and pDHS2112 (Table 1) containing putative promoters PpikAIV, PdesI (the first construct restricted to the intergenic region between pikC and desI), and PdesVIII (the full-length construct and 100 bp constructs), respectively, failed to produce XylE above background levels and were judged to be devoid of significant promoter activity. The remaining constructs and their activities are displayed in Fig. 3.
FIG. 3.
Comparison of XylE activities obtained from promoter-probe analysis in wild-type (WT) and DW900 strains of S. venezuelae.
Promoter-probe analysis of PpikAI, PpikC, and PpikD.
Consistent with the deletion and complementation experiments, PpikAI is about 14 times more active in wild-type S. venezuelae than in the DW900 pikD deletion mutant strain. There is still a low level of promoter activity in DW900, which is probably insufficient to drive transcription of the pikA operon. In accordance with the biosynthetic intermediate feeding experiments, pDHS2104 (PpikC) showed equivalent promoter activity in the wild-type and DW900 strains, providing further corroboration that PikD does not regulate PpikC. Interestingly, the 135-bp intergenic region separating pikC and pikD containing a presumed PpikD (pDHS2107) also drove xylE expression in both DW900 and wild-type S. venezuelae (Fig. 3). Although both promoters showed the same activity in wild-type S. venezuelae, PpikD expressed xylE at lower levels than PpikC in DW900 (47% less). These results could indicate that the pikC promoter drives expression of both genes initially and that the pikD promoter (upon accumulation of PikD) increases production of the regulator to promote high-level pikAI expression. Alternatively, pikD expression might be completely independent of PpikC and expressed at low levels until enough gene product is accumulated to increase its own expression and subsequently induce expression of pikAI.
Analysis of desosamine subcluster promoter PdesI.
Two desI promoter constructs were designed to evaluate their dependence on the PikD regulator. The first, pDHS2103, is limited to the intergenic region between pikC and desI (Fig. 2). The second, pDHS2110, begins at the pikD start codon and proceeds to the start codon of desI. pDHS2110 was constructed after initial studies showed that pDHS2103 had almost no promoter activity in either wild-type S. venezuelae or DW900. In contrast, pDHS2110 shows a 12-fold increase in activity in DW900 compared to the wild-type strain. This result implies that PikD directly or indirectly represses pPikB in the wild-type S. venezuelae host. In concert with this observation, growth of wild-type S. venezuelae often produces significant levels of aglycone intermediates (e.g., 10-deoxymethynolide and narbonolide) along with glycosylated products (Y. Xue, unpublished). It seems reasonable that PikD (or some other regulatory factor dependent on PikD) represses the des genes, causing macrolactone intermediates to remain unglycosylated. It has also been observed that ermE* upregulation of the desVIII to desR genes does not alter the levels of macrolactone glycosylation (10). This is consistent with desI to desV being the primary point of control for sugar pathway gene expression.
Analysis of PpikRI and PpikRII resistance gene promoters.
An important question regarding antibiotic production in microorganisms is the mechanism of cellular self-protection. In S. venezuelae, colocalized with the pikromycin biosynthetic genes lay pikRI and pikRII, which code for two putative N-methyltransferases of the macrolide-lincosamide-streptogramin B family, as determined by BlastP sequence analysis. To explore the role of PikD in regulating resistance, the pikRI and pikRII intergenic regions were cloned to give pDHS2105 and pDHS2106, respectively. Both constructs showed higher levels of XylE activity in wild-type S. venezuelae than in DW900. Interestingly, pDHS2105 displayed a high level of promoter activity in DW900, which increased 1.6-fold when expressed in the wild-type strain. In contrast, PpikRII (pDHS2106) expressed high levels of XylE in wild-type S. venezuelae, but no activity was observed in the DW900 mutant (Fig. 3). These results are consistent with the notion that the presence of two resistance genes provides an additional level of cellular self-protection. Specifically, pikRI is constitutively expressed to maintain a constant level of antibiotic resistance, whereas pikRII is not expressed until antibiotic production occurs with onset of pikD expression. These results demonstrate that in wild-type S. venezuelae, when PikD and macrolide antibiotics are present, the expression of pikRI is elevated and pikRII is strongly activated.
As a pathway-specific activator, PikD is the likely regulator for the additional level of control of pikRI/pikRII gene expression. Interestingly, no regulatory gene has been identified in the erythromycin biosynthetic gene cluster from Saccharopolyspora erythraea (14). In accordance with the finding that erythromycin is produced throughout the S. erythraea growth cycle (albeit at low levels) (23), the erythromycin resistance protein ErmE (which shows significant homology to PikRI and PikRII [30 and 28%, respectively, using Clustal W]) is constitutively expressed (5).
The discovery of PikD as a novel regulatory element for macrolide production has significant implications. Relatively few regulators have been identified or analyzed in detail from type I polyketide pathways, and little is known about their precise mechanisms of genetic control. The pik cluster represents an attractive system for the study of gene regulation in a complex antibiotic pathway. The architecture of the pik cluster is linear and includes resistance genes followed by four PKS genes, a subcluster of sugar biosynthetic genes (including two divergent transcripts), a unique P450 hydroxylase gene, and finally a single positive regulatory gene.
PikD is currently one of only four proteins found within polyketide biosynthetic pathways that share the three characteristics of size, NTP-binding domain, and HTH motif. As such, it may offer insights into a new method for macrolide biosynthetic pathway control. PikD is also one of only a few proteins overall in prokaryotes identified with these traits and assigned to a possible subfamily. If the recent classification of LAL proteins (13) is correct, the study of PikD might be greatly aided by comparisons with similar, well-studied proteins such as MalT. At the same time, further analysis of PikD might help refine this new classification of regulators found within the eubacteria.
The characterization of PikD may also prove useful for future applications to control and improve expression of genes involved in secondary metabolism. For example, it may be possible to upregulate the entire pikromycin antibiotic pathway to increase the production of known metabolites. Moreover, the ability to increase levels of metabolic flux could facilitate the detection of new compounds resulting from engineered pathways in which antibiotic biosynthesis may be inherently less efficient.
ACKNOWLEDGMENTS
We thank Brian Beck and Shuo Chen for helpful discussions.
This work was supported by NIH grant GM48562 to D.H.S.
REFERENCES
- 1.Aparicio J F, Molnar I, Schwecke T, Konig A, Haydock S F, Khaw L E, Staunton J, Leadlay P F. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene. 1996;169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 2.Arias P, Fernandez-Moreno M A, Malpartida F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol. 1999;181:6958–6968. doi: 10.1128/jb.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bate N, Butler A R, Gandecha A R, Cundliffe E. Multiple regulatory genes in the tylosin biosynthetic cluster of Streptomyces fradiae. Chem Biol. 1999;6:617–624. doi: 10.1016/s1074-5521(99)80113-6. [DOI] [PubMed] [Google Scholar]
- 4.Bérdy J. New ways to obtain antibiotics. Chin J Antibiot. 1984;7:272–290. [Google Scholar]
- 5.Bibb M, Janssen G, Ward J M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythaeus. Gene. 1985;38:215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 6.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 7.Cane D, Lambalot R, Prabhakaran P C. Macrolide biosynthesis. 7. Incorporation of polyketide chain elongation intermediates into methymycin. J Am Chem Soc. 1993;115:522–526. [Google Scholar]
- 8.Champness W. Cloning and analysis of regulatory genes involved in Streptomyces secondary metabolite biosynthesis. In: Davies J E, Demain A, Sherman D H, editors. Manual of industrial microbiology and biotechnology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 725–739. [Google Scholar]
- 9.Chater K F. Multilevel regulation of Streptomyces differentiation. Trends Genet. 1989;5:372–377. doi: 10.1016/0168-9525(89)90172-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Roberts J B, Sherman D H, Reynolds K A. The Streptomyces venezuelae pikAV gene contains a transcription unit essential for expression of enzymes involved in glycosylation of narbonolide and 10-deoxymethynolide. Gene. 2001;263:255–263. doi: 10.1016/s0378-1119(00)00560-6. [DOI] [PubMed] [Google Scholar]
- 11.Demain A L, Fang A. Emerging concepts of secondary metabolism in actinomycetes. Actinomycetologica. 1995;9:98–117. [Google Scholar]
- 12.Demain A L. Microbial secondary metabolism: a new theoretical frontier for academia, a new opportunity for industry. CIBA Found Symp. 1992;171:3–16. doi: 10.1002/9780470514344.ch2. [DOI] [PubMed] [Google Scholar]
- 13.De Schrijver A, De Mot R. A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology. 1999;145:1287–1288. doi: 10.1099/13500872-145-6-1287. [DOI] [PubMed] [Google Scholar]
- 14.Donadio S, Stassi D, McAlpine J B, Staver M J, Sheldon P J, Jackson M, Swanson S J, Wendt-Pienkowski E, Yi-Guang W, Jarvis B, Hutchinson C R, Katz L. Recent developments in the genetics of erythromycin formation. In: Hegeman G D, Baltz R H, Skatrud P L, editors. Industrial microorganisms: basic and applied molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 257–265. [Google Scholar]
- 15.Furuya K, Hutchinson C R. The DnrN protein of Streptomyces peucetius, a pseudo-response regulator, is a DNA-binding protein involved in the regulation of daunorubicin biosynthesis. J Bacteriol. 1996;178:6310–6318. doi: 10.1128/jb.178.21.6310-6318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geistlich M, Losick R, Turner J R, Rao R N. Characterization of a novel regulatory gene governing the expression of a polyketide synthase gene in Streptomyces ambofaciens. Mol Microbiol. 1992;6:2019–2029. doi: 10.1111/j.1365-2958.1992.tb01374.x. [DOI] [PubMed] [Google Scholar]
- 17.Henikoff S, Wallace J C, Brown J P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc Natl Acad Sci USA. 1999;96:9509–9514. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda H, Takada Y, Pang C H, Tanaka H, Omura S. Transposon mutagenesis by Tn4560 and applications with avermectin-producing Streptomyces avermitilis. J Bacteriol. 1993;175:2077–2082. doi: 10.1128/jb.175.7.2077-2082.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingram C, Brawner M, Youngman P, Westpheling J. XylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J Bacteriol. 1989;171:6617–6624. doi: 10.1128/jb.171.12.6617-6624.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieser T, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical Streptomyces genetics. Norwich, England: Crowes; 2000. [Google Scholar]
- 22.Lambalot R H, Cane D E. Isolation and characterization of 10-deoxymethynolide produced by Streptomyces venezuelae. J Antibiot. 1992;45:1981–1982. doi: 10.7164/antibiotics.45.1981. [DOI] [PubMed] [Google Scholar]
- 23.Marini F, Teatini L. Erythromycin production kinetics. Biotechnol Bioeng Symp. 1973;4:209–215. [PubMed] [Google Scholar]
- 24.Miyadoh S. Research on antibiotic screening in Japan over the last decade: a producing microorganism approach. Actinomycetologica. 1993;7:100–106. [Google Scholar]
- 25.Mizuno T, Tanaka I. Structure of the DNA-binding domain of the OmpR family of response regulators. Mol Microbiol. 1997;24:665–667. doi: 10.1046/j.1365-2958.1997.3571723.x. [DOI] [PubMed] [Google Scholar]
- 26.Narva K E, Feitelson J S. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:326–333. doi: 10.1128/jb.172.1.326-333.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng H L, Yang Y H, Deng W L, Chang H Y. Identification and characterization of acoK, a regulatory gene of the Klebsiella pneumoniae acoABCD operon. J Bacteriol. 1997;179:1497–1504. doi: 10.1128/jb.179.5.1497-1504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Llarena F J, Liras P, Rodriguez-Garcia A, Martin J F. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both β-lactam compounds. J Bacteriol. 1997;179:2053–2059. doi: 10.1128/jb.179.6.2053-2059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richet E, Raibaud O. MalT, the regulatory protein of the Escherichia coli maltose system, is an ATP-dependent transcriptional activator. EMBO J. 1989;8:981–987. doi: 10.1002/j.1460-2075.1989.tb03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan X, Stassi D, Lax S A, Katz L. A second type-I PKS gene cluster isolated from Streptomyces hygroscopicus ATCC 29253, a rapamycin-producing strain. Gene. 1997;203:1–9. doi: 10.1016/s0378-1119(97)00450-2. [DOI] [PubMed] [Google Scholar]
- 31.Sala-Trepat J M, Evans W C. The meta cleavage of catechol by Azotobacter species: 4-oxalocrotonate pathway. Eur J Biochem. 1971;20:400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 32.Schwecke T, Aparicio J F, Molnar I, Konig A, Khaw L E, Haydock S F, Oliynyk M, Caffrey P, Cortes J, Lester J B, Bohm G A, Staunton J, Leadlay P F. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stutzman-Engwall K J, Otten S L, Hutchinson C R. Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J Bacteriol. 1992;174:144–154. doi: 10.1128/jb.174.1.144-154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White J, Bibb M. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J Bacteriol. 1997;179:627–633. doi: 10.1128/jb.179.3.627-633.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wietzorrek A, Bibb M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol. 1997;25:1181–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 37.Xue Y, Sherman D H. Biosynthesis and combinatorial biosynthesis of pikromycin related macrolides in Streptomyces venezuelae. Metab Eng. 2001;3:15–26. doi: 10.1006/mben.2000.0167. [DOI] [PubMed] [Google Scholar]
- 38.Xue Y, Sherman D H. Alternative modular polyketide synthase expression controls macrolactone structure. Nature. 2000;403:571–575. doi: 10.1038/35000624. [DOI] [PubMed] [Google Scholar]
- 39.Xue Y, Wilson D, Sherman D H. Genetic architecture of the polyketide synthases for methymycin and pikromycin series macrolides. Gene. 2000;245:203–211. doi: 10.1016/s0378-1119(00)00003-2. [DOI] [PubMed] [Google Scholar]
- 40.Xue Y, Wilson D, Zhao L, Liu H, Sherman D H. Hydroxylation of macrolactones YC-17 and narbomycin is mediated by the pikC-encoded cytochrome P450 in Streptomyces venezuelae. Chem Biol. 1998;5:661–667. doi: 10.1016/s1074-5521(98)90293-9. [DOI] [PubMed] [Google Scholar]
- 41.Xue Y, Zhao L, Liu H-W, Sherman D H. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc Natl Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]