Abstract
The crystal structure of Escherichia coli FhuA reveals a β-barrel domain that is closed by a globular cork domain. It has been assumed that the proton motive force of the cytoplasmic membrane through the interaction of the TonB protein with the TonB box of the cork opens the FhuA channel. Yet, deletion of the cork results in an FhuA derivative, FhuAΔ5–160, that still displays TonB-dependent substrate transport and phage receptor activity. To investigate this unexpected finding further, we constructed FhuAΔ5–160 derivatives of FhuA proteins from Salmonella paratyphi B, Salmonella enterica serovar Typhimurium, and Pantoea agglomerans. The FhuAΔ5–160 proteins inserted correctly into the outer membrane, and with the exception of the P. agglomerans protein, transported ferrichrome and albomycin. FhuA hybrids consisting of the β-barrel of one strain and the cork of another strain were active and showed higher TonB-dependent ferrichrome transport rates than the corkless derivatives. Exceptions were the E. coli β-barrel/Salmonella serovar Typhimurium cork hybrid protein and the Salmonella serovar Typhimurium β-barrel/P. agglomerans cork hybrid protein, both of which were less active than the β-barrels alone. Each of the FhuA mutant proteins displayed activity for each of their ligands, except for phage T5, only when coupled to TonB. The hybrid FhuA proteins displayed a similar activity with the E. coli TonB protein as with their cognate TonB proteins. Sensitivity to phages T1, T5, and φ80, rifamycin CGP 4832, and colicin M was determined by the β-barrel, whereas sensitivity to phage ES18 and microcin J25 required both the β-barrel and cork domains. These results demonstrate that the β-barrel domain of FhuA confers activity and specificity and responds to TonB and that the cork domains of various FhuA proteins can be interchanged and contribute to the activities of the FhuA hybrids.
The FhuA outer membrane transport protein of Escherichia coli consists of 22 antiparallel β-sheets that form a β-barrel into which a globular domain is inserted from the periplasmic side. The globular domain seems to close the β-barrel channel and prevent entry of even small molecules and was for this reason designated the “cork” (7) or “plug” (20). Ferrichrome, the natural substrate of FhuA, binds in a cavity located well above the outer membrane lipid bilayer. The cork domain and the β-barrel domain contribute five and six amino acid side chains to the cavity, respectively, which are less than 4 Å away from the ferrichrome (7). It is thought that opening of the FhuA channel requires dislocation of the cork, resulting in a connection between the cavity exposed to the cell surface and the region exposed to the periplasm. Although binding of ferrichrome to FhuA moves the cork about 2 Å towards ferrichrome, this does not open the channel.
Energy provided by the cytoplasmic membrane in the form of the proton motive force (3) and the TonB-ExbB-ExbD protein complex are required for active transport through FhuA. Binding of ferrichrome results in the movement of Glu19 17 Å away from its former α-carbon position, which probably facilitates binding of FhuA to TonB. This hypothesis is supported by the finding that chemical cross-linking of FhuA to TonB is enhanced in vivo upon binding of ferrichrome (25). An N-proximal region of FhuA, residues 7 to 11 (TonB box), interacts with a region around residue 160 of TonB, as shown by mutations in the TonB box that are suppressed by mutations in TonB (9, 30).
A similar suppression analysis revealed the same interacting regions in the BtuB vitamin B12 transport protein and in TonB (11). Moreover, in vivo a segment of the TonB box of BtuB is chemically cross-linked via disulfide bonds with a segment around residue 160 of TonB (6). Cross-linking at several positions is increased when BtuB is loaded with vitamin B12, and the cross-linking pattern changes in mutants containing amino acid substitutions in BtuB that impair TonB-dependent BtuB activity. Site-directed spin labeling and electron paramagnetic resonance assays have suggested that the TonB box of BtuB in the unliganded conformation is located in a helix that forms specific interactions with side chain residues of the periplasmic turns of the β-barrel domain of BtuB (23). Binding of vitamin B12 to BtuB converts this segment into an extended, disordered, and highly dynamic structure that likely extends into the periplasm to interact physically with TonB. A TonB-uncoupled TonB box mutant of BtuB shows a strongly altered electron paramagnetic resonance spectrum and no longer responds to the addition of vitamin B12. These experiments strongly support the interaction of the transporter TonB box with the region around residue 160 of TonB.
In a previous study, we deleted the cork domain, including the TonB box, of E. coli FhuA. To our surprise, the protein FhuAΔ5–160 was found in the outer membrane, although in amounts lower than that of wild-type FhuA; FhuAΔ5–160 could still transport ferrichrome (at 30 to 40% the rate of wild-type FhuA) and albomycin in a TonB-dependent manner and conferred the same or almost the same degree of sensitivity as wild-type FhuA to the TonB-dependent colicin M and the phages T1 and φ80 and to the TonB-independent phage T5 (4). Since FhuAΔ5–160 lacks the TonB box, TonB must interact with other regions of FhuA, and this interaction suffices for TonB-dependent FhuA activities. FhuAΔ5–160 mediates slow diffusion, since sensitivity to larger hydrophilic antibiotics to which the outer membrane normally forms a permeability barrier is only moderately increased and cells remain resistant to sodium dodecyl sulfate (SDS) and EDTA.
In this study, we intended to corroborate our previous results with the E. coli FhuAΔ5–160 protein by constructing FhuAΔ5–160 derivatives of Salmonella paratyphi B, Salmonella enterica serovar Typhimurium, and Pantoea agglomerans; we have previously determined the fhuA nucleotide sequences of these strains (17). Comparison of the E. coli FhuA amino acid sequence with that of S. paratyphi B, Salmonella serovar Typhimurium and P. agglomerans revealed 94, 79, and 60% identity in the cork domain and 92, 74, and 58% identity in the β-barrel domain, respectively. In addition, we exchanged the cork domains to determine whether cork domains insert into heterologous β-barrel domains and whether the resulting FhuA hybrid proteins still respond to TonB and the proton motive force.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli strains and plasmids used are listed in Table 1. Cells were grown in TY medium (10 g of Bacto tryptone [Difco Laboratories]/liter, 5 g of yeast extract/liter, 5 g of NaCl/liter) or NB medium (8 g of nutrient broth/liter, 5 g of NaCl/liter, pH 7) at 37°C. To reduce the available iron of the NB medium, 2,2′-dipyridyl (0.2 mM) was added (NBD medium). The antibiotics ampicillin (40 μg/ml) and chloramphenicol (25 μg/ml) were added when required.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| AB2847 | aroB tsx malT thi | 10 |
| 41/2 | AB2847 cir fepA fhuA | 10 |
| HK99 | AB2847 tonB fhuA | 13 |
| CH1857 | AB2847 ΔfhuACDB tonB | 14 |
| HK97 | F−araD139 lacU169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR aroB thi fhuE::λplacMu53 fhuA | 12 |
| BL21(DE3) omp8 | F−hsdSB(rB− mB−) gal ompT dcm (DE3) ΔlamB ompF::Tn5 ΔompA T7 polymerase under lacUV5 control | 27 |
| CH21 | BL21 omp8 fhuA | This study |
| Plasmids | ||
| pHK763 | pT7-6 fhuA (Ec) wild type | 13 |
| pAB | pT7-6 fhuA (Ec) with BamHI E159D | 16 |
| pBK7 | pT7-6 fhuA (Ec) Δ5–160 E3D | 4 |
| p76Sp | pT7-6 fhuA (Sp) wild type | 17 |
| p76SpB | pT7-6 fhuA (Sp) with BamHI E159D | This study |
| pSpΔ5–160 | pT7-6 fhuA (Sp) Δ5–160 Q3D | This study |
| p76St | pT7-6 fhuA (St) wild type | 17 |
| p76StB | pT7-6 fhuA (St) with BamHI E159D | This study |
| pStΔ5–160 | pT7-6 fhuA (St) Δ5–160 Q3D | This study |
| p76Pa | pT7-6 fhuA (Pa) wild type | 17 |
| p76PaB | pT7-6 fhuA (Pa) with BamHI L159D R160P | This study |
| pPaΔ5–160 | pT7-6 fhuA (Pa) Δ5–160 A3D E4P | This study |
| pEcBSpC | pT7-6 fhuA (Ec) with the first 160 aa of fhuA (Sp) | This study |
| pEcBStC | pT7-6 fhuA (Ec) with the first 160 aa of fhuA (St) | This study |
| pEcBPaC | pT7-6 fhuA (Ec) with the first 160 aa of fhuA (Pa) | This study |
| pSpBEcC | pT7-6 fhuA (Sp) with the first 160 aa of fhuA (Ec) | This study |
| pSpBStC | pT7-6 fhuA (Sp) with the first 160 aa of fhuA (St) | This study |
| pSpBPaC | pT7-6 fhuA (Sp) with the first 160 aa of fhuA (Pa) | This study |
| pStBEcC | pT7-6 fhuA (St) with the first 160 aa of fhuA (Ec) | This study |
| pStBSpC | pT7-6 fhuA (St) with the first 160 aa of fhuA (Sp) | This study |
| pStBPaC | pT7-6 fhuA (St) with the first 160 aa of fhuA (Pa) | This study |
| pPaBEcC | pT7-6 fhuA (Pa) with the first 160 aa of fhuA (Ec) | This study |
| pPaBSpC | pT7-6 fhuA (Pa) with the first 160 aa of fhuA (Sp) | This study |
| pPaBStC | pT7-6 fhuA (Pa) with the first 160 aa of fhuA (St) | This study |
| pAM33 | pHSG576 tonB (Ec) | 21 |
| p576St | pHSG576 tonB (St) | This study |
| p576Pa | pHSG576 tonB (Pa) | This study |
| pBK71 | pT7-6 fhuA Δ25–160 P24D | This study |
| pTO4 | pBR322 cma cmi | 26 |
| pTUC203 | pACYC184 mcjABCD | 31 |
| pT7-6 | Ampr | 32 |
| pHSG576 | Cmr | 33 |
Ec, E. coli; Sp, S. paratyphi; St, Salmonella serovar Typhimurium; Pa, P. agglomerans; B, β-barrel; C, cork.
To construct plasmids p76SpB, pSpΔ5–160, p76StB, pStΔ5–160, p76PaB, and pPaΔ5–160, a BamHI restriction site was introduced into the fhuA gene of p76Sp (S. paratyphi), p76St (Salmonella serovar Typhimurium), and p76Pa (P. agglomerans) using PCR and the following primers (mismatches are underlined): Sp_160for (5′-CCGACGACGGATCCGCTGAAAG-3′), Sp_160rev (5′-CTTTCAGCGGATCCGTCGTCGG-3′), and Sp_BamAnf (5′-CTTCTTTCGGATCCACCGCCGC-3′) (BamHI in S. paratyphi); St_160for (5′-CCGACTACGGATCCGCTGAAAGAAATTC-3′), St_160rev (5′-CTTTCAGCGGATCCGTAGTCGGCCG-3′), and St_BamAnf (5′-GTTTCTTCTTTCGGATCCACCGCCGCCTG-3′) (BamHI in Salmonella serovar Typhimurium); and Pa_160for (5′-CCAGGAAACGGATCCCGAAGTGCAGTTCC-3′), Pa_160rev (5′-CTGCACTTCGGGATCCGTATCCTGGGTCGG-3′), and Pa_BamAnf (5′-GACCATCGTCGGATCCTGCGCGGCGTAAAG-3′) (BamHI in P. agglomerans). The primers of the complementary strands were pT7_ (5′-GCGAGGCCCAGCTGGCTTATCG-3′) and T7_uni (5′-GATTAAGCATTGGTAACTGTCAGACC-3′). All PCR products were purified by agarose gel electrophoresis and recovered from agarose using the EasyPure DNA purification kit (Biozym, Oldendorf, Germany).
Each of the DNA fragments obtained with primers Sp_160rev, St_160rev, and Pa_160rev was digested with HindIII and BamHI and ligated into HindIII/BamHI-cleaved vector pT7-6, resulting in plasmids p76SpBN, p76StBN, and p76PaBN, respectively. The DNA fragments obtained with primers Sp_160for, St_160for, and Pa_160for were digested with EcoRI and BamHI and ligated into EcoRI/BamHI-cleaved plasmids p76SpBN, p76StBN, and p76PaBN, respectively, resulting in plasmids p76SpB′, p76StB′, and p76PaB′. To avoid complete sequencing of the fhuA genes, plasmids p76SpB′, p76StB′, and p76PaB′ were digested with HindIII and Eco47III and ligated into HindIII/Eco47III-cleaved plasmids p76Sp, p76St, and p76Pa, respectively, resulting in plasmids p76SpB, p76StB, and p76PaB. The exchanged HindIII/Eco47III fragments were completely sequenced.
Each of the DNA fragments obtained with primers Sp_BamAnf, St_BamAnf, and Pa_BamAnf was digested with HindIII and BamHI and ligated into HindIII/BamHI-cleaved plasmids p76SpB, p76StB, and p76PaB, resulting in plasmids pSpΔ5–160, pStΔ5–160, and pPaΔ5–160, respectively.
Plasmid pAB was digested with HindIII and BamHI, and the obtained 992-bp fragment was ligated into HindIII/BamHI-cleaved p76SpB, p76StB, and p76PaB, resulting in plasmids pSpBEcC, pStBEcC, and pPaBEcC, respectively. Plasmid p76SpB was digested with HindIII and BamHI, and the obtained 831-bp fragment was ligated into HindIII/BamHI-cleaved pAB, p76StB, and p76PaB, resulting in plasmids pEcBSpC, pStBSpC, and pPaBSpC, respectively. Plasmid p76StB was digested with HindIII and BamHI, and the obtained 828-bp fragment was ligated into HindIII/BamHI-cleaved pAB, p76SpB, and p76PaB, resulting in plasmids pEcBStC, pSpBStC, and pPaBStC, respectively. Plasmid p76PaB was digested with HindIII and BamHI, and the obtained 726-bp fragment was ligated into HindIII/BamHI-cleaved pAB, p76SpB, and p76StB, resulting in plasmids pEcBPaC, pSpBPaC, and pStBPaC, respectively.
To construct plasmid pBK71, a BamHI restriction site was introduced into the fhuA gene on pHK763 (E. coli) using PCR and the primer Bam23_fhuA (5′-CAATAGTTGCAGGATCCCCCCATGCGCTTTC-3′). The primer of the complementary strand was pT7_(5′-GCGAGGCCCAGCTGGCTTATCG-3′). The PCR fragment was digested with HindIII and BamHI and ligated into HindIII/BamHI-cleaved pBK7, resulting in plasmid pBK71.
Plasmid pGB312 was digested with HindIII and EcoRI and ligated into HindIII/EcoRI-cleaved vector pHSG576, resulting in plasmid p576St.
Strain CH21 was constructed by picking a phage T5-resistant clone of strain BL21 (DE3) omp8.
Recombinant DNA techniques.
Isolation of plasmids, use of restriction enzymes, ligation, agarose gel electrophoresis, and transformation were performed according to standard techniques (29). All genetic constructions were examined by DNA sequencing using the dideoxy chain-termination method with fluorescence-labeled or unlabeled nucleotides (Auto Read Sequencing Kit, Pharmacia Biotech, Freiburg, Germany) and the ALF sequencer (Pharmacia).
Protein analytical methods.
E. coli BL21 cells (optical density at 578 nm of 0.5) transformed with one of various plasmids encoding complete FhuA, corkless FhuA, or reconstituted FhuA hybrids were collected by centrifugation and resuspended in 1 ml of M9 salts (24) supplemented with 0.4% glucose, 0.01% methionine assay medium, 0.01% thiamine, and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to induce T7 RNA polymerase synthesis. After shaking the cells for 1 h at 37°C, rifamycin (10 μl of a 5-mg/ml solution in methanol) was added and incubation was continued at 37°C for 30 min. [35S]methionine was added, and the suspension was incubated for 10 min. Cells were then collected by centrifugation and suspended in sample buffer. The radioactively labeled proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (12). In addition, cells transformed with wild-type fhuA and mutant fhuA genes were grown in NB medium, the outer membrane fractions were isolated, and the proteins were separated by SDS-PAGE and stained with Serva blue.
Phenotype assays.
All phenotype assays were carried out with freshly transformed E. coli K-12 strains 41/2 aroB fhuA, HK97 aroB fhuA fhuE, and HK99 aroB fhuA tonB. These strains carry the same four amino acid replacements and an amino acid deletion in fhuA and contain the mutated FhuA protein in the outer membrane (12). The plasmid-encoded fhuA genes in the transformants were transcribed from the fhuA promoter. The sensitivity of cells against the FhuA ligands (phages T1, T5, φ80, and ES18, colicin M, microcin J25, rifamycin CGP 4832, and albomycin) was tested by spotting 10-fold-diluted solutions (4 μl) on TY agar plates overlaid with 3 ml of TY soft agar containing 108 cells of the strain to be tested. The colicin M solution was a crude extract of a strain carrying plasmid pTO4 cma cmi (26). The microcin J25 solution was a supernatant of E. coli MC4100 carrying the plasmid pTUC203 mcjABCD (31) after growth of the transformants in brain heart infusion medium (37 g/liter; Difco Laboratories) at 37°C.
Growth inhibition by SDS and various antibiotics was detected by placing filter paper disks supplemented with 10 μl of the agents in concentrations as indicated on TY agar plates overlaid with 3 ml of TY soft agar containing 108 cells of the strain to be tested. Growth promotion by siderophores was tested by placing filter paper disks containing 10 μl of a siderophore solution concentrated as indicated on NBD agar plates overlaid with 3 ml of NB soft agar containing 108 cells of the strain to be tested. After overnight incubation, the diameter and the growth density around the filter paper disk were determined.
Transport and binding assays.
E. coli K-12 strains 41/2 aroB fhuA, HK97 aroB fhuA fhuE, HK99 aroB fhuA tonB, and CH1857 ΔfhuACDB tonB aroB freshly transformed with the plasmids to be tested were grown overnight on TY plates. Cells were washed and suspended in transport medium (M9 salts [24], 0.4% glucose), and the cell density was then adjusted to an optical density at 578 nm of 0.5. Free iron ions were removed by adding 25 μl of 10 mM nitrilotriacetate, pH 7.0, to 1 ml of cells. After incubation for 5 min at 37°C, transport or binding assays were started by adding 10 μl of 100 μM [55Fe3+]ferrichrome. Only in the case of binding assays, a 150-fold surplus of nonradioactive ferrichrome was added as a chase after 19 min to show the specificity of the ferrichrome binding. Samples of 100 μl were withdrawn, and cells were harvested on cellulose nitrate filters (pore size, 0.45 μm; Sartorius AG, Göttingen, Germany) and washed twice with 5 ml of 0.1 M LiCl. The filters were dried, and the radioactivity was determined by liquid scintillation counting.
Computer-assisted sequence analysis.
Sequences were analyzed using the program package PC.GENE and the BLAST homology search (1).
RESULTS
FhuAΔ5–160 corkless deletion derivatives display TonB-dependent activities.
Precise excision of the cork domain of E. coli FhuA results in a stable barrel that is inserted into the outer membrane and exerts TonB-dependent FhuA activities. Deletions within the cork domain and deletions in the barrel domain, with the exception of the surface-exposed loops, frequently result in unstable FhuA derivatives (4). In this study, we excised the cork domain of FhuA from S. paratyphi B, Salmonella serovar Typhimurium, and P. agglomerans based on the E. coli FhuA crystal structure. The cork domain of all FhuA proteins used in this study have the same length as that of E. coli FhuA, except for FhuA of P. agglomerans, which contains a three-amino-acid insertion and a two-amino-acid deletion (17). In addition, the amino acid sequences of all four FhuA proteins are rather similar, which makes it likely that the cork domains comprise the same or nearly the same segment of the FhuA polypeptide.
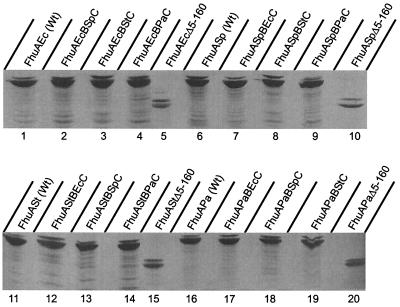
To examine whether the corkless FhuA derivatives were synthesized and to estimate their relative amounts, the E. coli strain CH21 [an fhuA mutant of E. coli BL21 (DE3) omp8] was transformed with plasmids encoding the various fhuA genes which were specifically transcribed by phage T7 RNA polymerase, and the proteins were labeled with [35S]methionine. The proteins of whole cells were separated by SDS-PAGE, and only bands in the region of FhuA were seen on autoradiographs (Fig. 1 shows only the FhuA-containing section of the gel). As observed previously with E. coli (4), less protein of the corkless FhuA derivatives was present (Fig. 1, lanes 5, 10, 15, and 20) than that of the complete FhuA proteins from which they were derived (Fig. 1, lanes 1, 6, 11, and 16). The radioactivity of the major bands of the FhuAΔ5–160 proteins amounted on average to 25% of that of the complete proteins. The faint bands above the major bands probably represent the precursor form with uncleaved signal peptide. The majority of FhuA is processed and presumably inserted into the outer membrane.
FIG. 1.
Comparison of [35S]methionine-labeled FhuA proteins and FhuA barrel-cork hybrids after transformation of E. coli CH21 with plasmid pHK763 (lane 1), pEcBSpC (lane 2), pEcBStC (lane 3), pEcBPaC (lane 4), pBK7 (lane 5), p76Sp (lane 6), pSpBEcC (lane 7), pSpBStC (lane 8), pSpBPaC (lane 9), pSpΔ5–160 (lane 10), p76St (lane 11), pStBEcC (lane 12), pStBSpC (lane 13), pStBPaC (lane 14), pStΔ5–160 (lane 15), p76Pa (lane 16), pPaBEcC (lane 17), pPaBSpC (lane 18), pPaBStC (lane 19), or pPaΔ5–160 (lane 20). No other bands were seen outside the gel section represented here. Nomenclature used: EcBSpC means the β-barrel (B) of E. coli FhuA and cork (C) of S. paratyphi B FhuA; the other designations follow the same rule. Ec, E. coli; Sp, S. paratyphi B; St, Salmonella serovar Typhimurium; Pa, P. agglomerans.
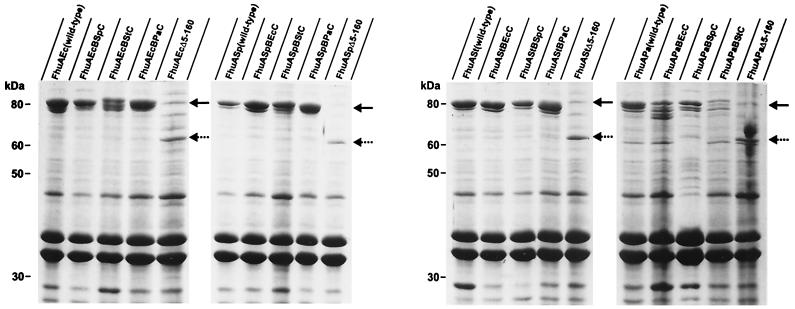
As the corkless FhuA proteins will be used to determine FhuA activities it was important to estimate the amounts of the corkless FhuA proteins in cells that were grown under the same conditions as those under which the activity assays were performed. The fhuA transformants of E. coli HK97 were grown in NB medium. Transcription of the fhuA genes encoded by the same plasmids as those used for T7 transcription proceeded by E. coli RNA polymerase and was controlled by the fhuA promoters. The FhuA proteins in the isolated outer membrane fractions were separated by SDS-PAGE. The FhuA protein of E. coli HK97 carries three amino acid replacements and one amino acid deletion and was not seen after SDS-PAGE when cells were grown in NB medium. Protein bands with electrophoretic mobilities corresponding to the calculated molecular mass of 61 kDa were present in lanes to which outer membranes of corkless fhuA transformants were applied (Fig. 2, marked by dotted arrows) and absent in lanes to which outer membranes of wild-type fhuA transformants were applied (Fig. 2, marked by solid arrows). The amounts of the corkless FhuA proteins were lower than those of the wild-type FhuA proteins, which agrees with the results obtained after transcription of the genes with T7 RNA polymerase (Fig. 1). Since the same relative amounts were obtained with whole cells and outer membrane fractions, the lower amounts of FhuAΔ5–160 may arise from a lower mRNA stability caused by the deletion or by proteolytic degradation of FhuAΔ5–160 in the cytoplasm.
FIG. 2.
Stained proteins after SDS-PAGE of outer membrane fractions of E. coli HK97 fhuA transformed with the plasmids listed in Table 1 that encoded the FhuA proteins indicated in the figure. Solid arrows denote complete FhuA and reconstituted FhuA, and dotted arrows denote corkless FhuA. The molecular masses of standard proteins in kDa are indicated. The nomenclature used is described in the legend for Fig. 1.
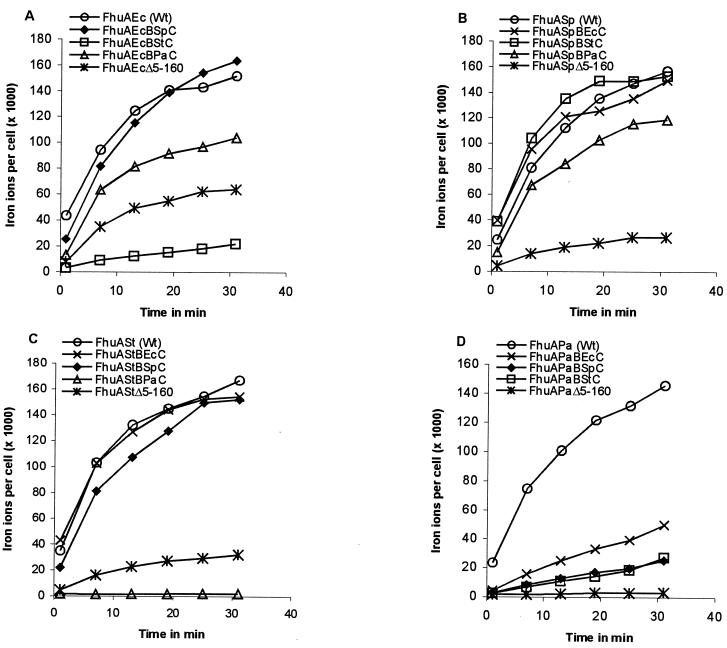
Transport of the corkless derivatives was determined in E. coli HK97 fhuA aroB cells transformed with the plasmids carrying the genes for the corkless FhuA proteins. The fhuA mutations of E. coli HK97 exert no polar effect on the downstream fhuBCD genes required for ferrichrome transport across the cytoplasmic membrane. FhuAΔ5–160 of S. paratyphi B and Salmonella serovar Typhimurium conferred ferrichrome transport (Fig. 3B and C) at rates of 14 and 21% the rate of their respective complete FhuA protein (Fig. 3B and C; Table 2). Each rate was calculated using the value after 31 min of transport minus the value after 1 min. These values were lower than those obtained with E. coli FhuAΔ5–160 (Fig. 3A; Table 2). No transport was observed in E. coli HK99 fhuA tonB transformed with plasmids carrying the genes for the corkless FhuA proteins (data not shown). Transport of these transformants (FhuAΔ5–160 of S. paratyphi B and Salmonella serovar Typhimurium) was restored by transformation with a plasmid encoding a wild-type tonB gene (data not shown). FhuAΔ5–160 of P. agglomerans did not transport ferrichrome (Fig. 3D; Table 2).
FIG. 3.
Time-dependent transport of [55Fe3+]ferrichrome (1 μM) into E. coli HK97 fhuA fhuE aroB expressing the plasmid-encoded FhuA proteins and FhuA barrel-cork hybrids of E. coli (Ec, panel A), S. paratyphi (Sp, panel B), Salmonella serovar Typhimurium (St, panel C), and P. agglomerans (Pa, panel D) as indicated in the figure.
TABLE 2.
Ferrichrome binding and ferrichrome transport rates of complete FhuA, corkless FhuA, and reconstituted FhuA hybrid proteins
| FhuA protein | Ferrichrome binding to CH1857a (iron ions per cell) | Ferrichrome transport rates (% wild type)e into
|
||
|---|---|---|---|---|
| HK97b | HK99 1c | HK99 2d | ||
| FhuAEc (wild type) | 10,322 | 100 | 100 | 100 |
| FhuAEcBSpC | 9,384 | 100 | 100 | 95 |
| FhuAEcBStC | 2,579 | 17 | 7 | 11 |
| FhuAEcBPaC | 84 | 84 | 88 | 86 |
| FhuAEcΔ5–160 | 514 | 35 | 8 | ND |
| FhuASp (wild type) | 12,072 | 100 | 100 | 100 |
| FhuASpBEcC | 11,917 | 71 | 97 | 100 |
| FhuASpBStC | 10,265 | 74 | 86 | 100 |
| FhuASpBPaC | 857 | 67 | 80 | 100 |
| FhuASpΔ5–160 | 629 | 14 | 9 | ND |
| FhuASt (wild type) | 11,188 | 100 | 100 | 100 |
| FhuAStBEcC | 11,327 | 84 | 76 | 89 |
| FhuAStBSpC | 10,225 | 99 | 100 | 100 |
| FhuAStBPaC | 0 | 1 | 1 | 1 |
| FhuAStΔ5–160 | 417 | 21 | 5 | ND |
| FhuAPa (wild type) | 7,925 | 100 | 100 | 100 |
| FhuAPaBEcC | 0 | 37 | 14 | 21 |
| FhuAPaBSpC | 253 | 18 | 4 | 13 |
| FhuAPaBStC | 0 | 20 | 4 | 8 |
| FhuAPaΔ5–160 | 0 | 1 | 5 | ND |
E. coli CH1857 ΔfhuABCD tonB was transformed with the plasmids listed in Table 1 that encoded the FhuA proteins listed in the left panel.
E. coli HK97 fhuA expressing chromosomally encoded tonB was transformed with plasmids that encoded the FhuA proteins listed in the left panel.
E. coli HK99 fhuA tonB was transformed with plasmids that encoded the FhuA proteins listed in the left panel and in addition carried low-copy plasmids (Table 1) that encoded tonB genes of the strains from which the FhuA barrel was derived, i.e., barrel of E. coli (EcB), Salmonella serovar Typhimurium (StB), and P. agglomerans (PaB) combined with tonB of E. coli, Salmonella serovar Typhimurium, and P. agglomerans, respectively. The exception was the barrel S. paratyphi (SpB), which was combined with tonB of Salmonella serovar Typhimurium.
As described above, but the tonB genes were from the strains from which the FhuA cork was derived, e.g., cork of E. coli (EcC) combined with tonB of E. coli. ND, not determined.
The percentage is related to the transport rates of the wild-type strains taken as 100%.
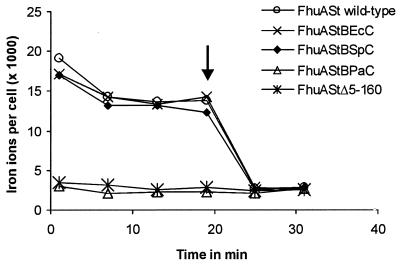
Reduction and lack of ferrichrome transport of the corkless FhuA derivatives could result from impaired ferrichrome binding, translocation, or both. An estimate of ferrichrome binding to E. coli FhuAΔ5–160 was previously derived from the amount of radioactive ferrichrome that was found associated with cells of transport-negative E. coli HK99 fhuA tonB and CH1857ΔfhuABCD tonB in time-dependent transport assays. It amounted to not more than 7% of that of wild-type FhuA (4). In this study we measured the binding of 1 μM radioactive ferrichrome to fhuAΔ5–160 transformants of CH1857 by taking samples after 1, 7, 13, and 19 min, after which the cultures were chased with 150 μM nonradioactive ferrichrome. The amount of ferrichrome that could be chased was taken as the fraction that is bound to FhuA. An example is given in Fig. 4 which shows ferrichrome binding to wild-type FhuA and FhuAΔ5–160 of Salmonella serovar Typhimurium. The curves with a higher value for the 1-min sample than for the following samples are representative for all experiments performed with wild-type and mutant FhuA proteins (data not shown). The data of this and further experiments are listed in Table 2. They show that ferrichrome binding to FhuAΔ5–160 of E. coli amounts to 5% that of wild-type FhuA, to S. paratyphi FhuAΔ5–160 is 5.2% that of the wild type, to Salmonella serovar Typhimurium FhuAΔ5–160 is 3.7% that of the wild type, and to P. agglomerans FhuAΔ5–160 is 0% that of the wild type. The lack of ferrichrome binding to P. agglomerans FhuAΔ5–160 would account for the inability of this corkless derivative to transport ferrichrome. The values have not been quantitatively related to the amounts of the FhuA proteins; however, they reflect the conditions under which transport was measured.
FIG. 4.
Binding of [55Fe3+]ferrichrome to E. coli CH1857 ΔfhuABCD tonB aroB expressing the indicated FhuA proteins. After 19 min a 150-fold surplus of nonradioactive ferrichrome was added (marked by an arrow).
FhuA also transports the structurally related antibiotic albomycin. To examine the albomycin sensitivity of cells that produce the FhuAΔ5–160 proteins, E. coli 41/2 fhuA was transformed with plasmids carrying the genes for the corkless FhuA proteins, and transformants were seeded on nutrient agar plates to which 4 μl of a series of threefold-diluted solutions of the antibiotic were spotted. Transformants carrying the gene for the corkless FhuA were sensitive to albomycin, although to different degrees (Table 3). Only P. agglomerans FhuAΔ5–160 did not confer albomycin sensitivity, which probably results from the lack of binding, as has been observed for ferrichrome. Sensitivity depended on TonB, as shown by the albomycin resistance of TonB-negative fhuAΔ5–160 transformants (data not shown).
TABLE 3.
Sensitivity of E. coli 41/2 fhuA transformed with the indicated plasmids
| Plasmid | Sensitivity to:a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Phage T1 | Phage T5 | Phage φ80 | Coicin M | Microcin J25 | Albomycin | CGP 4832 | Rifamycin | |
| pHK763 (wild type) | 4 | 4 | 4 | 3 | 3 | 3 | 6 | 2 |
| pEcBSpCb | 4 | 4 | 4 | 3 | 3 | 3 | 4 | 2 |
| pEcBStC | − | 3 | 1 | 3 | 2 | 2 | 2 | 2 |
| pEcBPaC | 4 | 4 | 4 | 2 | − | 3 | 2 | 2 |
| pBK7 (EcΔ5–160) | 4 | 4 | 4 | 3 | − | 2 | 4 | 3 |
| p76Sp (wild type) | 4 | 4 | 4 | 3 | 3 | 3 | 4 | 2 |
| pSpBEcC | 4 | 4 | 4 | 3 | 3 | 3 | 5 | 2 |
| pSpBStC | 4 | 4 | 4 | 3 | 2 | 3 | 4 | 2 |
| pSpBPaC | 4 | 4 | 4 | 3 | − | 3 | 2 | 2 |
| pSpΔ5–160 | 3 | 3 | 2 | 2 | − | 1 | 2 | 2 |
| p76St (wild type) | − | − | − | − | − | 3 | 2 | 2 |
| pStBEcC | − | − | − | − | − | 3 | 2 | 2 |
| pStBSpC | − | − | − | − | − | 2 | 1 | 2 |
| pStBPaC | − | − | − | − | − | 1 | 1 | 2 |
| pStΔ5–160 | − | − | − | − | − | 1 | 1 | 2 |
| p76Pa (wild type) | − | − | − | − | − | 3 | 2 | 2 |
| pPaBEcC | − | − | − | − | − | 1 | 2 | 2 |
| pPaBSpC | − | − | − | − | − | 1 | 2 | 2 |
| pPaBStC | − | − | − | − | − | 2 | 2 | 2 |
| pPaΔ5–160 | − | − | − | − | − | − | 2 | 2 |
Sensitivities to the ligands were tested by using E. coli 41/2 aroB fhuA freshly transformed with the plasmids indicated. The sensitivities were tested by spotting 4 μl of 10-fold or 3-fold (for microcin J25, rifamycin CGP 4832, and rifamycin) dilutions onto TY agar plates overlaid with TY top agar containing the strain to be tested. The results are given as the last of a 10-fold or 3-fold dilution series that resulted in a clear zone of growth inhibition. For example, a value of 4 indicates that the phage solution could be diluted 104-fold to yield a clear zone of cell lysis. −, no growth inhibition and no phage plaques.
Ec, E. coli; Sp, S. paratyphi; St, Salmonella serovar Typhimurium; Pa, P. agglomerans; B, β-barrel; C, cork.
Rifamycin CGP 4832, a chemically synthesized derivative of rifamycin, has a much higher activity than rifamycin because it is actively transported across the outer membrane by FhuA (28). However, CGP 4832 is structurally unrelated to either ferrichrome or albomycin. E. coli 41/2 fhuA was found to be equally sensitive to either CGP 4832 or rifamycin (data not shown). However, the sensitivity of E. coli 41/2 fhuA carrying plasmid pHK763 fhuA to CGP 4823 was approximately 100-fold higher than its sensitivity to rifamycin (Table 3). The CGP 4823 sensitivity of E. coli 41/2 fhuA (pBK7) was approximately 10-fold higher, and its sensitivity to rifamycin was threefold higher. The higher sensitivity to CGP 4832 is a result of both active transport, since it depended on active TonB, and passive diffusion through the FhuAΔ5–160 channel (data not shown). An increase in sensitivity by diffusion through FhuAΔ5–160 was evaluated in the E. coli HK99 tonB mutant transformed with pBK7, which for CGP 4832 was as high (threefold) as for rifamycin. Unexpectedly, sensitivity to both antibiotics was not increased in cells that synthesized FhuAΔ5–160 of S. paratyphi B, Salmonella serovar Typhimurium, or P. agglomerans (Table 3).
FhuA of E. coli and S. paratyphi B renders cells sensitive to colicin M and microcin J25 (31). FhuAΔ5–160 of S. paratyphi B conferred sensitivity to colicin M which was 10-fold lower than that of complete FhuA, E. coli FhuA, and E. coli FhuAΔ5–160. Both FhuA deletion derivatives were unable to mediate sensitivity to microcin J25 (Table 3). Cells expressing FhuAΔ5–160 of Salmonella serovar Typhimurium or P. agglomerans were as resistant to colicin M and microcin J25 as cells expressing wild-type FhuA of these strains (Table 3).
FhuA of E. coli and S. paratyphi B serves as a receptor of the phages T1, T5, and φ80. Sensitivity was tested by spotting a series of 10-fold-diluted phage solutions onto a lawn of E. coli 41/2 fhuA transformants that synthesized one of the FhuAΔ5–160 proteins. Cells synthesizing FhuAΔ5–160 of S. paratyphi B were 10-fold less sensitive to phages T1 and T5 and 100-fold less sensitive to phage φ80 than the transformants synthesizing wild-type FhuA of S. paratyphi or E. coli or FhuAΔ5–160 of E. coli (Table 3). Cells that synthesized FhuAΔ5–160 of Salmonella serovar Typhimurium or P. agglomerans were resistant to all the phages (Table 3). E. coli cells that synthesized FhuAΔ5–160 of Salmonella serovar Typhimurium were resistant to phage ES18, which normally infects Salmonella serovar Typhimurium via FhuA. Since E. coli cells that synthesized wild-type FhuA of Salmonella serovar Typhimurium were sensitive to phage ES18, a 103-fold-diluted ES18 stock suspension formed clear plaques, and a 105-fold-diluted suspension formed turbid plaques, we conclude that ES18 infection requires the FhuA cork domain and the β-barrel domain.
FhuAΔ5–160 deletion derivatives display low open channel activities.
For the determination of active transport, 1 μM [55Fe3+]ferrichrome was used. At this ferrichrome concentration, growth on nutrient broth agar plates containing 0.2 mM dipyridyl to suppress low-affinity iron uptake (NBD plates) is not supported. For an estimation of ferrichrome uptake by diffusion across the outer membrane, E. coli HK99 fhuA tonB aroB transformed with plasmids carrying the genes for the corkless FhuA proteins was used. Ferrichrome at concentrations of 0.1, 0.3, 1, 3, and 10 mM was placed on filter paper disks, and growth promotion around the disks on NBD plates seeded with 108 cells of the HK99 transformants was recorded. Slow growth of a small number of cells that synthesized FhuAΔ5–160 of E. coli or P. agglomerans was observed with 0.1 mM ferrichrome. The same result was obtained with cells that synthesized FhuAΔ5–160 of S. paratyphi B when a solution of 0.3 mM ferrichrome was used. At this concentration, cells that synthesized FhuAΔ5–160 of E. coli or P. agglomerans showed a strong growth zone of 10 mm in diameter (6-mm disk diameter not subtracted). At 10 mM ferrichrome, cells synthesizing E. coli FhuAΔ5–160, S. paratyphi B FhuAΔ5–160, Salmonella serovar Typhimurium FhuAΔ5–160, and P. agglomerans FhuAΔ5–160 had growth zones of 18, 13, 18, and 22 mm, respectively.
Another means to measure diffusion through the FhuAΔ5–160 derivatives is provided by antibiotics that are too large to diffuse readily through the porin channels. Growth inhibition around filter paper disks to which these antibiotics had been applied was measured. The sensitivities to erythromycin (734 Da), rifamycin (823 Da), and vancomycin (1,486 Da) of E. coli HK99 synthesizing E. coli FhuAΔ5–160 compared to complete FhuA, measured as zones of growth inhibition, increased from 8 to 14 mm, 15 to 19 mm, and 9 to 12 mm, respectively. The sensitivity of E. coli HK99 tonB fhuA synthesizing P. agglomerans FhuAΔ5–160 to these antibiotics compared to complete FhuA increased from 8 to 13 mm, 15 to 20 mm, and 9 to 11 mm, respectively. The sensitivity of the FhuAΔ5–160 derivatives of S. paratyphi B and Salmonella serovar Typhimurium to antibiotics was not increased significantly. The parental strains AB2847 and 41/2 displayed the same sensitivities to the three antibiotics as the pHK763 (wild-type fhuA) transformants.
Hybrid FhuA proteins consisting of β-barrel domains and unrelated cork domains are active.
The cork and β-barrel domains of the enterobacterial FhuA proteins were mutually exchanged to determine whether complete FhuA can be reconstituted, exported across the cytoplasmic membrane, and inserted correctly into the outer membrane. Moreover, it was of interest to determine whether FhuA hybrids consisting of β-barrel domains and unrelated cork domains display activity with some or all of the ligands and whether the reconstituted FhuA proteins still respond to TonB.
The cork domains of S. paratyphi, Salmonella serovar Typhimurium, and P. agglomerans were each combined with the β-barrel domain of E. coli. The derivatives showed the same electrophoretic mobility (Fig. 1, lanes 2 to 4) as wild-type FhuA of E. coli (Fig. 1, lane 1). The yield of the hybrid FhuA proteins resulting from transcription by T7 RNA polymerase was comparable to the yield of wild-type FhuA cloned in the same vector. Similar results were obtained with each of the unrelated cork domains fused to the β-barrel domain of S. paratyphi B (Fig. 1, lanes 6 to 9), Salmonella serovar Typhimurium (Fig. 1, lanes 11 to 14), and P. agglomerans (Fig. 1, lanes 16 to 19).
To use the same conditions as those under which the FhuA activity assays were performed, FhuA synthesis was examined in transformants in which the fhuA genes were transcribed by E. coli RNA polymerase under the control of the fhuA promoters, and cells were grown under assay conditions. SDS-PAGE analysis revealed a somewhat heterogeneous band pattern at the electrophoretic position of FhuA, especially with FhuAEcBStC and the FhuA hybrids containing the P. agglomerans β-barrel (Fig. 2). However, each of the transformants contained a stronger protein band at the electrophoretic position of FhuA than E. coli contains when it synthesizes chromosomally encoded wild-type FhuA (10, 12). The amount of hybrid FhuA is considered to be sufficient to confer all FhuA activities. The amounts of the hybrid proteins were similar to the amounts of plasmid-encoded wild-type proteins. All FhuA proteins, including E. coli wild-type FhuA, contained a minor band which most likely is a degradation product. The upper band close to FhuA in the samples of the FhuAΔ5–160 proteins is not an FhuA product since it is not contained in Fig. 1 in which the fhuAΔ5–160 genes were specifically transcribed by T7 RNA polymerase (Fig. 1).
The activities of the FhuA hybrids were determined by measuring the ferrichrome transport rate of E. coli HK97 aroB fhuA fhuE transformed with plasmids encoding each of the FhuA hybrids. As shown in Fig. 3 and summarized in Table 2, half of the FhuA hybrids displayed transport activities as high or nearly as high as the wild-type FhuA proteins. For example, the β-barrel of E. coli FhuA fused to the cork of S. paratyphi or P. agglomerans showed 100 and 84% of the transport rate of E. coli wild-type FhuA (Table 2). In contrast, the E. coli β-barrel fused to the Salmonella serovar Typhimurium cork displayed only 17% of the wild-type activity, which may be explained by the lower amounts of the mutant FhuA protein (Fig. 2). The same cork fused to the β-barrel of S. paratyphi showed 74% of the wild-type activity, which agrees with the high amount of the mutant FhuA protein (Fig. 2). The cork of P. agglomerans fused to the β-barrel of E. coli or S. paratyphi B was highly active (84 and 67% of the wild-type activity) but showed no activity when combined with the β-barrel of Salmonella serovar Typhimurium, despite high levels of protein (Fig. 2). The β-barrel of P. agglomerans displayed the lowest tolerance to unrelated cork domains. FhuAPaBPaC, FhuAPaBPaSp, and FhuAPaBStC displayed only 37, 18, and 20% of the FhuAPa wild-type activity, and the amount of unaltered reconstituted FhuA hybrid proteins was the lowest of all the hybrid proteins (Fig. 2).
To see whether the transport rates are related to ferrichrome binding activities, binding of radioactive ferrichrome was measured in cells of CH1857 ΔfhuABCD tonB expressing the FhuA hybrid proteins. As shown in Table 2, FhuAEcBSpC, FhuASpBEcC, FhuASpBStC, FhuAStBEcC, and FhuAStBSpC bound ferrichrome approximately to the same extent as the wild-type FhuA proteins. FhuAEcBStC displayed only 25% of these binding activities, which was correlated with the heterogeneous FhuA protein profile (Fig. 2) and the low ferrichrome transport rate (Fig. 3A; Table 2). Binding of ferrichrome to FhuAPa was lower than to the wild-type FhuA proteins of the other strains, and the hybrid proteins showed low or no binding (Table 2), which was largely correlated with the amounts of FhuA protein and the additional FhuA-derived protein bands (Fig. 2). However, the ferrichrome transport rates were not strictly related to binding since FhuAPaBEcC with no binding transported better than FhuAPaBSpC with residual binding (Table 2). Furthermore, FhuASpBPaC binds poorly (7%) and FhuAEcBPaC binds very poorly (0.8%), but they display high transport activities.
The degree of albomycin sensitivity of E. coli 41/2 fhuA transformed with plasmids encoding the FhuA hybrids is in agreement with the ferrichrome transport rates (Table 3). A few minor deviations may result from the lower transport rate of albomycin than ferrichrome and from the type of assay used, either the transport assay within 30 min or the growth assay within 15 h.
The sensitivity of the FhuA hybrids to phages T1, T5, and φ80 was determined with transformants of E. coli 41/2 fhuA. The FhuA hybrids containing the β-barrel of E. coli or S. paratyphi B FhuA were as phage sensitive as cells synthesizing wild-type FhuA (Table 3). Exceptions were transformants with the β-barrel of E. coli and the cork of Salmonella serovar Typhimurium, which were resistant to phage T1, 1,000-fold less sensitive to phage φ80, and 10-fold less sensitive to phage T5. Cells that synthesized wild-type FhuA of Salmonella serovar Typhimurium or P. agglomerans and FhuA hybrids composed of the β-barrel of Salmonella serovar Typhimurium or P. agglomerans were equally resistant to the phages (Table 3). None of the FhuA hybrids conferred sensitivity to phage ES18, which indicates that infection by phage ES18 requires both the cork and the β-barrel of Salmonella serovar Typhimurium FhuA.
E. coli 41/2 fhuA transformants that synthesized FhuA hybrids containing the E. coli or S. paratyphi B β-barrel were sensitive to colicin M to somewhat variable degrees (Table 3). Transformants that expressed wild-type FhuA of Salmonella serovar Typhimurium or P. agglomerans and FhuA hybrids containing the β-barrel of Salmonella serovar Typhimurium or P. agglomerans were resistant to colicin M. Colicin M sensitivity conferred by the FhuA hybrids was TonB dependent, as transformants of E. coli HK99 tonB were resistant to colicin M (data not shown).
The β-barrel and the cork of E. coli or S. paratyphi B were required to render cells sensitive to microcin J25. However, the cork of P. agglomerans did not reconstitute the activity of FhuAΔ5–160 of E. coli or S. paratyphi B (Table 3).
Of all the FhuA hybrids examined here, only those that synthesized the E. coli β-barrel fused to the S. paratyphi B cork and the β-barrel of S. paratyphi B fused to the cork of E. coli or Salmonella serovar Typhimurium conferred TonB-dependent sensitivity to CGP 4832 that was higher than the sensitivity to rifamycin (Table 3).
There is no preference for the TonB protein related to the FhuA cork or β-barrel.
We first determined the transport activities of all FhuA derivatives in E. coli, which means in combination with the E. coli TonB protein. We then wanted to find out whether it makes a difference in FhuA activity when the FhuA hybrids are combined with the TonB proteins of the same strains from which the FhuA hybrids were derived. In addition, since TonB apparently interacts with the cork and the β-barrel it was of interest to determine whether the cork or the β-barrel should be from the same strain as the TonB protein. We constructed combinations of tonB genes on a low-copy plasmid with the plasmid-encoded wild-type fhuA and mutated fhuA genes in E. coli HK99 fhuA tonB, with the exception of the tonB gene of S. paratyphi B, which was unavailable. All the combinations were active, and the absolute transport rates listed as 100% in Table 2 are similar to the highest transport rates shown in Fig. 3. No alterations of the FhuA activities were observed that could be related to homologous versus heterologous FhuA-TonB combinations or to the cork or the β-barrel (Table 2). The FhuA activities of the E. coli FhuA β-barrel derivatives combined with TonB of E. coli (Table 2, HK97 and HK99 1) differed only slightly from the E. coli FhuA β-barrel derivatives combined with the TonB protein of Salmonella serovar Typhimurium and P. agglomerans (HK99 2). When the β-barrel of S. paratyphi B was combined with TonB of Salmonella serovar Typhimurium, the FhuA activities were somewhat higher (HK99 1) than when combined with the E. coli TonB (HK97). This increase may be a result of the overexpression of plasmid-encoded TonB, although in other cases the FhuA β-barrel derivatives showed lower FhuA activities when combined with plasmid-encoded TonB (HK99) than with chromosomally encoded E. coli TonB (HK97) (Table 2), which has been observed previously (22). There was a tendency of a higher FhuA activity with TonB combined with the related cork domain (HK99 2) than with TonB combined with the related β-barrel domain (HK99 1). However, we doubt that the observed differences are large enough to suggest a stronger impact of the cork than the β-barrel in the interaction of FhuA with TonB.
The β-barrel domain of E. coli FhuA containing the TonB box is less active.
The results reported here and in our previous papers (4, 9, 30) indicated that FhuA activity is mediated by TonB through interaction with the cork and the β-barrel. Therefore, we examined whether the TonB box linked to the β-barrel domain affects the activity of the β-barrel. We constructed FhuAΔ25–160, in which the N-proximal 23 residues of mature FhuA, including the TonB box, were linked to residue 161 of the β-barrel domain. The genetic manipulation replaced Pro24 with Asp. E. coli 41/2 fhuA synthesizing FhuAΔ25–160(P24D) was as sensitive to phage φ80 as E. coli 41/2 fhuA synthesizing FhuAΔ5–160 but was 10-fold less sensitive to phages T1 and T5 and to colicin M and was resistant to albomycin and microcin J25. Since TonB-independent infection by phage T5 was also reduced, the lower activity of FhuAΔ25–160(P24D) cannot be ascribed to an unproductive binding of TonB to the TonB box of FhuAΔ25–160(P24D). This interpretation is supported by the finding that phage T5 sensitivity is also reduced 10-fold in the HK99 tonB mutant. The ferrichrome transport rate was near zero. After 30 min, there were 3,000 ferrichrome molecules per cell compared to 140,000 in cells expressing wild-type FhuA and 45,000 in cells expressing FhuAΔ5–160 in experiments run in parallel. In addition, binding of ferrichrome to FhuAΔ25–160(P24D) was examined. Using E. coli CH21(pBK71), binding of ferrichrome amounted to about 3,000 molecules per cell compared to 20,000 molecules bound to wild-type FhuA of E. coli CH21(pHK763), which after a chase with a 150-fold surplus of unlabeled ferrichrome was reduced to 3,000 molecules per cell. Although the binding site (residue 161) of the N-proximal 24-residue peptide is exposed to the periplasm outside the channel formed by the β-barrel, the N-proximal peptide appears to strongly impair the binding of ferrichrome, which occurs well above the cell surface. In addition to binding, the transport activity must also be impaired since FhuAΔ5–160 also binds ferrichrome poorly but transports ferrichrome rather well. It should be stated that the relative amount of FhuAΔ25–160(P24D) protein observed after SDS-PAGE was comparable to that of wild-type FhuA (data not shown).
DISCUSSION
Our previous finding of high and specific activities of corkless FhuA of E. coli (4) are supported by the results described in this paper with the corkless FhuA proteins of S. paratyphi B and Salmonella serovar Typhimurium. These corkless FhuA derivatives exhibit TonB-dependent ferrichrome transport, although at rates lower than that of the E. coli corkless FhuA. The amounts of the corkless derivatives were also lower (approximately 25% that of wild-type FhuA), which may have reduced the activities. The rates of 14 and 21% in comparison to the rates obtained with the complete FhuA of the same strain decreased to zero in the tonB mutant strain HK99 carrying the same fhuA mutation as that of E. coli HK97 fhuA used for the transport experiments. To rule out complementation of the mutated E. coli HK97 FhuA protein by the E. coli corkless FhuA mutant protein through formation of a functional oligomer, we previously carried out experiments with E. coli H1857 in which the fhuABCD genes are deleted (4). After transformation of E. coli H1857 with fhuAΔ5–160 and the fhuBCD genes for transport across the cytoplasmic membrane, ferrichrome transport is even higher than transport into E. coli HK97 since E. coli H1857 synthesizes greater amounts of plasmid-encoded FhuBCD proteins than E. coli HK97. In addition, X-ray analysis does not support the formation of an FhuA oligomer as the FhuA crystals consisted of a monomer (7, 20).
FhuAΔ5–160 of P. agglomerans was considered inactive, as it did not transport ferrichrome, conferred no sensitivity to albomycin, and showed the same sensitivity to rifamycin CGP 4832 as to rifamycin. Among the FhuA proteins studied, that of P. agglomerans exhibits the least sequence similarity to E. coli FhuA (59%). The construction of the deletion introduced the amino acid replacements A3D and E4P; the latter replacement may not affect FhuAΔ5–160 activity, since similar replacements at the A3 site in FhuAΔ5–160 of E. coli (E3D), S. paratyphi B (Q3D), and Salmonella serovar Typhimurium (Q3D) did not abolish activity.
FhuA of S. paratyphi B is the only non-E. coli FhuA that mediates sensitivities to phages T1, T5, and φ80 and to colicin M and albomycin, and this specificity was retained in the S. paratyphi B corkless FhuA, although at 1 or 2 orders of magnitude lower than the sensitivity conferred by the complete FhuA. Sensitivity to these FhuA ligands was TonB-dependent, except for infection by phage T5, which occurs independent of TonB. FhuAΔ5–160 of S. paratyphi, like that of E. coli, did not mediate sensitivity to microcin J25 and differed from the E. coli FhuAΔ5–160 in that it did not enhance sensitivity to rifamycin CGP 4832.
The β-barrel of E. coli FhuA without the cork mediates all FhuA functions except uptake of microcin J25 (4). Uptake of microcin J25 and infection of Salmonella serovar Typhimurium by phage ES18 may require both the cork and the β-barrel. We have previously shown that the prominent loop of the FhuA β-barrel (18), which is loop 4 in the E. coli FhuA crystal structure (7, 20) and lies above the cell surface, serves as the principal binding site of the phages and colicin M (13, 14, 15). This result implies that TonB, without the help of the cork, can change the conformation of loop 4 such that binding of phages T1 and φ80 triggers DNA release from the phage head. This conformational change is not restricted to loop 4, since release of ferrichrome from its binding sites in the β-barrel (residues Y244, W246, Y313, Y315, F391, and F693) probably also requires a conformational change of the β-barrel, and none of the ferrichrome binding sites are located in loop 4. These binding sites are contained in the four corkless FhuA proteins, with the exception of Y315, which is replaced in Salmonella serovar Typhimurium and P. agglomerans by T and N, respectively, and F693, which is replaced in P. agglomerans by Y. Since aromatic residues play a major role in ferrichrome and albomycin binding, replacement of Y315 by these nonaromatic amino acids may well contribute to the lower transport activity of Salmonella serovar Typhimurium FhuAΔ5–160 and the inactivity of P. agglomerans FhuAΔ5–160. However, this cannot be the only cause since the transport activity of S. paratyphi FhuAΔ5–160 is rather low (after 12 min, 18,000 ions per cell compared to 48,000 per cell with E. coli FhuAΔ5–160), despite the identity of these residues with those of E. coli FhuAΔ5–160.
TonB-dependent conformational changes of the β-barrel may also widen the channel to facilitate diffusion of ferrichrome and albomycin once they are released from their binding sites and/or may properly position the amino acid side chains along which ferrichrome and albomycin diffuse through FhuA. These possibilities should be considered due to the low diffusion rates through the corkless FhuA proteins, as evidenced by the small increase in sensitivity to the antibiotics erythromycin, rifamycin, and vancomycin compared to the same E. coli strain synthesizing plasmid-encoded wild-type FhuA proteins.
If TonB interacts only with β-barrel regions exposed to the periplasm, the conformational change must be transmitted across the entire FhuA molecule up to the cell surface. It is not known whether TonB inserts into the outer membrane. However, the observed shuttling of TonB between the outer membrane and the cytoplasmic membrane (19) excludes a firm integration of TonB in the outer membrane.
Fusions of cork domains with β-barrel domains of different species were constructed to determine whether the corks are inserted into the β-barrels, how they fit into the β-barrels, and whether they restore the activities to those of complete wild-type homologous FhuA proteins. It was conceivable that the corks were not incorporated into the β-barrels, that the hybrid proteins were rapidly degraded in the cytoplasm or the periplasm, that they stayed in the cytoplasm and were not exported across the cytoplasmic membrane, that they remained in the periplasm, or that they were inserted into the outer membrane in an inactive form. The heterologous corks could interact with the β-barrels such that structural transitions in the β-barrels and the corks upon binding of the ligands and TonB were blocked or aberrant. We did not have to investigate all these possibilities since we obtained FhuA hybrids present in processed form in amounts similar to those of the wild-type FhuA proteins. The exceptions were the FhuA hybrids which contained the P. agglomerans β-barrel and heterologous cork domains, which formed several bands of which one was probably the genuine FhuA hybrid. However, the reduced amounts of these hybrids do not fully explain the low activity, as they were present in higher amounts than that observed with chromosomally encoded wild-type FhuA, which confers full FhuA activity (10, 12). FhuAPaΔ5–160 is somewhat unstable, as the band pattern demonstrates, and the hybrid proteins appear to be even less stable. Nevertheless, the hybrid proteins exhibit ferrichrome transport activity, while the corkless mutant does not. Most FhuA hybrids transported ferrichrome with rates higher than those of the corkless FhuA proteins from which they were derived. FhuAEcBStC displayed a low transport rate (17% that of E. coli FhuA wild-type), which may have attributed to the protein's instability (Fig. 2). In contrast, FhuAStBPaC is inactive despite its high amounts in the outer membrane fraction (Fig. 2). In this hybrid protein the cork apparently does not fit into the β-barrel to reconstitute an active FhuA protein. In all mutant FhuA proteins the degree of albomycin sensitivity correlated with the ferrichrome transport rates.
Increased sensitivity to rifamycin CGP 4832, compared to rifamycin and sensitivity to microcin J25 were only mediated by FhuA hybrids containing the β-barrel of E. coli or S. paratyphi B. The binding site of CGP 4832 in FhuA, as derived from the FhuA cocrystal structure (A. D. Ferguson, J. Ködding, G. Walker, C. Bös, J. W. Coulton, K. Diederichs, V. Braun, and W. Welte, unpublished data), largely overlaps with the ferrichrome and albomycin (8) binding site. The same amino acid residues contribute to binding of ferrichrome, albomycin, and CGP 4832 in the E. coli and S. paratyphi B FhuA proteins, except for a single, functionally equivalent E→D exchange in S. paratyphi B. Of the total of 16 residues that bind CGP 4832 to E. coli FhuA, FhuA of Salmonella serovar Typhimurium and P. agglomerans deviate by 4 and 8 residues, respectively. The number of amino acid replacements may explain why the FhuA proteins of Salmonella serovar Typhimurium and P. agglomerans do not show increased sensitivity to CGP 4832. Two out of the 10 residues that in E. coli FhuA bind ferrichrome are different in Salmonella serovar Typhimurium FhuA, and 4 out of 10 differ in P. agglomerans FhuA. These sites also bind albomycin and CGP 4832 in E. coli FhuA.
In addition to the ligand binding sites, the data indicate that other regions are important for the transport activities of the hybrid FhuA proteins. For example, insertion of the Salmonella serovar Typhimurium FhuA cork decreases the activity of the E. coli FhuA β-barrel; however, the E. coli cork strongly increases the transport activity of the Salmonella serovar Typhimurium β-barrel. The Salmonella serovar Typhimurium cork fused to the S. paratyphi B β-barrel results in a highly active transporter. The P. agglomerans cork increases the transport activities when inserted into the E. coli and S. paratyphi B β-barrels but fails to complement the Salmonella serovar Typhimurium β-barrel. These results show that incorporation of a cork into a barrel is not sufficient to restore transport activity; rather, intimate interactions between the cork and the β-barrel must occur in order to form an active transporter. In a previous study, prior to the determination of the FhuA crystal structure, we had replaced the N-proximal 160 amino acids of E. coli FhuA with the first 150 amino acids of FoxA, the ferrioxamine transport protein of Yersinia enterocolitica (16). This FhuA hybrid conferred only low phage T5 sensitivity 4 orders of magnitude below that of wild-type FhuA and even lower phage T1 and φ80 sensitivity and a reduced growth with ferrichrome as the sole iron source. In addition to the presumably impaired fitting of the FoxA cork into the FhuA β-barrel because of the low sequence identity (36%), the approximately 10 residues of the cork that remain on the β-barrel may also have inactivated the FhuA hybrid.
The FhuA hybrid proteins examined here showed nearly wild-type levels of ferrichrome binding, with the exception of those β-barrels that contained the cork of P. agglomerans. However, weak binding did not necessarily result in low transport, as demonstrated by FhuAEcBPaC, FhuASpBPaC, and the corkless FhuA derivatives.
Impairment of the E. coli β-barrel activity by the Salmonella serovar Typhimurium cork (FhuAEcBStC) also abolished the receptor activity for phage T1 and reduced phage φ80 receptor activity 1,000-fold. The sensitivity of FhuAEcBStC to colicin M remained the same as the sensitivity mediated by the β-barrel (and wild-type FhuA). In contrast, cells that synthesized FhuAEcBPaC displayed reduced colicin M sensitivity but full phage sensitivity and low ferrichrome binding but high ferrichrome transport. The data collectively reveal different structural requirements of FhuA for the various FhuA activities.
Our results disclose a rather high tolerance of FhuA β-barrels for different FhuA corks. The β-barrel domains largely determine the activity and specificity of the FhuA proteins. The E. coli cork domain did not confer sensitivity to the E. coli-specific phages, colicin M, rifamycin CGP 4832, and microcin J25 when incorporated into the β-barrels of FhuA proteins of Salmonella serovar Typhimurium and P. agglomerans that did not confer these sensitivities. Conversely, the S. paratyphi and P. agglomerans corks, with the exception of the Salmonella serovar Typhimurium cork, left phage sensitivity of the E. coli β-barrel unaltered and reduced the colicin M sensitivity of FhuAEcBPaC, synthesizing cells only slightly. In this context, it is interesting to note that the E. coli β-barrel fused to the N-terminal 24 FhuA residues bound ferrichrome very poorly and had a low ferrichrome transport rate. It is difficult to predict the effect of the peptide on FhuA β-barrel structure since the fusion site is exposed to the periplasm outside the β-barrel channel. We expected that the peptide might extend into the periplasm and not affect FhuA activity unless TonB binds to it in an unproductive way and is no longer available for activating FhuA. Since the mutant showed a 10-fold reduced sensitivity to phage T5 independent of the presence of TonB, the properties of this FhuA derivative cannot be explained by a locking of the altered FhuA protein in an inactive conformation through binding to TonB. Apparently, the peptide interacts with the β-barrel in a way that affects binding of ferrichrome far away from the peptide binding site, infection by phages, and sensitivity to colicin M, albomycin, and microcin J25. Such long-distance effects have also been observed with the lipoprotein of phage T5 that interacts with periplasmic sites of FhuA but inhibits phage infection, colicin M and albomycin sensitivity, and transport of ferrichrome (5).
We examined whether TonB preferentially activates FhuA of the same strain, and if so, whether interaction with the cork or the β-barrel domain is more important. Therefore, we combined TonB with FhuA and corkless FhuA of the same strain and other strains and with FhuA hybrids of which the cork or the β-barrel was from the same strain as TonB. Only the results collected with HK99 transformed with the various tonB genes on the same low-copy plasmid can be compared, since the level of TonB expression affects TonB-mediated activities (22). We obtained no ferrichrome transport activity pattern that would be consistent with preference for the same strain. The only and nearly consistent tendency found concerns the higher FhuA activities when TonB was derived from the same strain as the cork (Table 2, HK99 2) than when TonB was derived from the same strain as the β-barrel (Table 2, HK99 1). However, the differences in the FhuA activities are not so strong that firm conclusions can be drawn.
It seems to us that FhuA behaves like an allosteric protein, and most of the allosteric transitions induced by binding of TonB, the phages, and ferrichrome take place in the β-barrel, although the B-factors of the crystallographic analysis suggest a rather rigid β-barrel structure and the multiple conformational changes occur independent of the cork. The cork functions in closing the β-barrel channel to avoid uncontrolled diffusion of harmful compounds into the periplasm. It also contributes to ferrichrome binding in that 4 out of 10 ferrichrome binding sites are located in the cork. The strong binding of ferrichrome to FhuA (Kd < 0.1 μM) allows a sufficient iron supply with an iron source present in the culture medium at very low concentrations. For transport, ferrichrome has to be released from the binding sites and the cork has to move to open the channel. This presumably occurs through the concerted action of the cork and the β-barrel, both of which respond to the TonB-mediated energy transfer from the cytoplasmic membrane through direct interaction with TonB. The inactive TonB box mutants of FhuA (30), BtuB (11), and Cir (2) suggest that interaction of TonB with the β-barrel is not sufficient to open the channel of the wild-type proteins, but interaction with the TonB box of the cork has to occur as well. Our studies with the corkless FhuA mutants of E. coli, S. paratyphi B, and Salmonella serovar Typhimurium indicate important TonB-mediated FhuA activities through interaction of TonB with the β-barrel and the cork.
ACKNOWLEDGMENTS
We thank K. A. Brune and M. Ogierman for critically reading the manuscript.
We also thank the Deutsche Forschungsgemeinschaft (BR330/20-1) and the Fonds der Chemischen Industrie for financial support.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bell P E, Nau C D, Brown J T, Konisky J, Kadner R J. Genetic suppression demonstrates interaction of TonB protein with outer membrane transport proteins in Escherichia coli. J Bacteriol. 1990;172:3826–3829. doi: 10.1128/jb.172.7.3826-3829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun M, Killmann H, Braun V. The beta-barrel domain of FhuAΔ5–160 is sufficient for TonB-dependent FhuA activities of Escherichia coli. Mol Microbiol. 1999;33:1037–1049. doi: 10.1046/j.1365-2958.1999.01546.x. [DOI] [PubMed] [Google Scholar]
- 5.Braun V, Killmann H, Herrmann C. Inactivation of FhuA at the cell surface of Escherichia coli K-12 by a phage T5 lipoprotein at the periplasmic face of the outer membrane. J Bacteriol. 1994;176:4710–4717. doi: 10.1128/jb.176.15.4710-4717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadieux N, Kadner R J. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc Natl Acad Sci USA. 1999;96:10673–10678. doi: 10.1073/pnas.96.19.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferguson A D, Hofmann E, Coulton J W, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson A D, Braun V, Fiedler H P, Coulton J W, Diederichs K, Welte W. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 2000;9:956–963. doi: 10.1110/ps.9.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Günter K, Braun V. In vivo evidence for FhuA outer membrane interaction with the TonB inner membrane protein of Escherichia coli. FEMS Microbiol Lett. 1990;274:85–88. doi: 10.1016/0014-5793(90)81335-l. [DOI] [PubMed] [Google Scholar]
- 10.Hantke K, Braun V. Functional interaction of the tonA/tonB receptor system in Escherichia coli. J Bacteriol. 1978;135:190–197. doi: 10.1128/jb.135.1.190-197.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heller, K., R. J. Kadner, and K. Günter. Suppression of the btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene 64:147–153. [DOI] [PubMed]
- 12.Killmann H, Braun V. An aspartate deletion mutation defines a binding site of the multifunctional FhuA outer membrane receptor of Escherichia coli K-12. J Bacteriol. 1992;174:3479–3486. doi: 10.1128/jb.174.11.3479-3486.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Killmann H, Benz R, Braun V. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 1993;12:3007–3016. doi: 10.1002/j.1460-2075.1993.tb05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Killmann H, Videnov G, Jung G, Schwarz H, Braun V. Identification of receptor binding sites by competitive peptide mapping: phages T1, T5, and φ80 and colicin M bind to the gating loop of FhuA. J Bacteriol. 1995;177:694–698. doi: 10.1128/jb.177.3.694-698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Killmann H, Benz R, Braun V. Properties of the FhuA channel in the Escherichia coli outer membrane after deletion of FhuA portions within and outside the predicted gating loop. J Bacteriol. 1996;178:6913–6920. doi: 10.1128/jb.178.23.6913-6920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Killmann H, Braun V. Conversion of the coprogen transport protein FhuE and the ferrioxamine B transport protein FoxA into ferrichrome transport proteins. FEMS Microbiol Lett. 1998;161:59–67. doi: 10.1111/j.1574-6968.1998.tb12929.x. [DOI] [PubMed] [Google Scholar]
- 17.Killmann H, Herrmann C, Wolff H, Braun V. Identification of a new site for ferrichrome transport by comparison of the FhuA proteins of Escherichia coli, Salmonella paratyphi, Salmonella typhimurium, and Pantoea agglomerans. J Bacteriol. 1998;180:3845–3852. doi: 10.1128/jb.180.15.3845-3852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koebnik R, Braun V. Insertion derivatives containing segments of up to 16 amino acids identify surface- and periplasm-exposed regions of the FhuA outer membrane receptor of Escherichia coli K-12. J Bacteriol. 1993;175:826–839. doi: 10.1128/jb.175.3.826-839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letain T E, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 20.Locher K P, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch J P, Moras D. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 21.Mademidis A, Köster W. Transport activity of FhuA, FhuC, FhuD, and FhuB derivatives in a system free of polar effects, and stoichiometry of components involved in ferrichrome uptake. Mol Gen Genet. 1998;258:156–165. doi: 10.1007/s004380050718. [DOI] [PubMed] [Google Scholar]
- 22.Mann B J, Holroyd C D, Bradbeer C, Kadner R J. Reduced activity of TonB-dependent functions in strains of Escherichia coli. FEMS Microbiol Lett. 1986;33:255–260. [Google Scholar]
- 23.Merianos H J, Cadieux N, Lin C H, Kadner R J, Cafiso D S. Substrate-induced exposure of an energy-coupling motif of a membrane transporter. Nat Struct Biol. 2000;7:205–209. doi: 10.1038/73309. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 25.Moeck G S, Coulton J W, Postle K. Cell envelope signaling in Escherichia coli. Ligand binding to the ferrichrome-iron receptor FhuA promotes interaction with the energy-transduction protein TonB. J Biol Chem. 1997;272:28391–28397. doi: 10.1074/jbc.272.45.28391. [DOI] [PubMed] [Google Scholar]
- 26.Ölschläger T, Schramm E, Braun V. Cloning and expression of the activity and immunity genes of colicins B and M on pColBM plasmids. Mol Gen Genet. 1984;196:482–487. doi: 10.1007/BF00436196. [DOI] [PubMed] [Google Scholar]
- 27.Prilipov A, Phale P S, Van Gelder P, Rosenbusch J P, Koebnik R. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol Lett. 1998;163:65–72. doi: 10.1111/j.1574-6968.1998.tb13027.x. [DOI] [PubMed] [Google Scholar]
- 28.Pugsley A P, Zimmerman W, Wehrli W. Highly efficient uptake of a rifamycin derivative via the FhuA-TonB-dependent uptake route in Escherichia coli. J Gen Microbiol. 1987;133:3505–3511. doi: 10.1099/00221287-133-12-3505. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schöffler H, Braun V. Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol Gen Genet. 1989;217:378–383. doi: 10.1007/BF02464907. [DOI] [PubMed] [Google Scholar]
- 31.Solbiati J O, Ciaccio M, Farias R N, Salomon R A. Genetic analysis of plasmid determinants for microcin J25 production and immunity. J Bacteriol. 1996;178:3661–3663. doi: 10.1128/jb.178.12.3661-3663.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ α-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]