Abstract
Lagerstroemia indica L. is a well-known ornamental plant with large pyramidal racemes, long flower duration, and diverse colors and cultivars. It has been cultivated for nearly 1600 years and is essential for investigating the germplasm and assessing genetic variation to support international cultivar identification and breeding programs. In this study, 20 common Lagerstroemia indica cultivars from different varietal groups and flower morphologies, as well as multiple wild relative species, were analyzed to investigate the maternal donor of Lagerstroemia indica cultivars and to discover the genetic variation and relationships among cultivars based on plastome and nuclear ribosomal DNA (nrDNA) sequences. A total of 47 single nucleotide polymorphisms (SNPs) and 24 insertion/deletions (indels) were identified in the 20 L. indica cultivars’ plastome and 25 SNPs were identified in the nrDNA. Phylogenetic analysis based on the plastome sequences showed that all the cultivars formed a clade with the species of L. indica, indicating that L. indica was the maternal donor of the cultivars. Population structure and PCA analyses supported two clades of cultivars, which exhibited significant genetic differences according to the plastome dataset. The results of the nrDNA supported that all 20 cultivars were divided into three clades and most of the cultivars had at least two genetic backgrounds and higher gene flow. Our results suggest that the plastome and nrDNA sequences can be used as molecular markers for assessing the genetic variation and relationships of L. indica cultivars.
Keywords: Lagerstroemia indica, crape myrtles, genetic variation, plastome, nuclear ribosomal DNA
1. Introduction
Crape myrtles (Lagerstroemia indica L.), belonging to the genus Lagerstroemia of the family Lythraceae, are important summer-blooming ornamental trees or shrubs. They are excellent woody plants for environmental protection, since they can absorb smoke and dust in the air and are resistant to toxic gases such as sulfur dioxide, hydrogen fluoride, and chlorine released from industrial pollution [1]. Crape myrtles are valuable, with high economic value in city gardening and hill planting. There are more than 200 cultivars of L. indica in the world, and crape myrtles have been cultivated for nearly 1600 years [1,2]. The breeding of new cultivars with various outstanding features, such as a longer flowering time; different floral colors; stronger aroma; seeds with higher oil production; or resistance to drought, coldness, insects, or disease, is an important issue.
Morphological characteristics, such as flower color, length and width of inflorescence, flower diameter, floral fragrance, number of flowerlets per inflorescence, petal claw color, leaf color, and thousand-seed weight, are major elements for understanding phenotypic differentiation in crape myrtle cultivars. The complex genetics and the high phenotypic variation of L. indica cultivars are thought to be the result of multiple cycles of artificial selection and intraspecific hybridization [3,4]. Intraspecific hybridization is mainly used for cultivar improvement, with selection of the parent cultivar a critical step in hybrid breeding. Phenotypic traits are frequently used as the main guide for choosing the germplasm to develop new cultivars with novel appearances and improved stress tolerance. In fact, selecting the germplasm with relatively large genetic differences can help improve breeding efficiency, as hybrid breeding according to phenotype may not produce offspring with the expected characteristics. Understanding the genetic background of L. indica cultivars is essential for choosing germplasm resources and ultimately facilitating genetic improvement and breeding programs.
The origin of cultivated crape myrtles has attracted great attention over the past decades. Previous studies show that the donor of cultivated crape myrtles involved five Lagerstroemia species based on the morphological and genetic data, including L. indica, L. fauriei, L. speciosa, L. subcostata, and L. limii [3,5]. However, the maternal donor of the cultivated crape myrtles is not clear. Morphological characteristics [6], RAPD [7], AFLP [8], and SSRs [2,9,10,11,12] markers were used to assess genetic diversity among the L. indica cultivars. However, few studies have evaluated the genetic variation and diversity of L. indica cultivars using DNA sequence markers. Improved DNA sequence markers need to be developed to facilitate research access to the genetic diversity of crape myrtles.
Plastome and nuclear ribosomal DNA (nrDNA) sequences are popular molecular markers used for plant phylogeny [13,14,15,16,17] and species identification [18,19] because these sequences are conserved across plant species and show variability among interspecies levels. Past studies show that the plastome and nrDNA sequences contain numerous SSRs, indels, and SNPs at the intraspecies levels, which have been used to research the genetic divergence of endangered species [20,21], biogeographical structure [22,23,24], gene flow among subpopulations [25,26], and origins and domestication of cultivars [27,28,29]. For the Lagerstroemia species, more than 17 species have sequenced the whole chloroplast genome, using these data to infer the phylogeny and divergence time of Lagerstroemia [30]. Dong et al. [30] also identified several polymorphism sites in the plastome and nrDNA sequences at the intraspecies level in L. indica, indicating that these molecular data will resolve genetic variation among the L. indica cultivars at the genome level.
In this study, we performed comprehensive sampling in cultivars of L. indica. A total of 20 accessions were collected to represent different varietal groups and flower morphologies [6,31]. All of the plastome and nrDNA sequences were assembled to discover the sequence variation among the cultivars. Phylogenetic analyses combining data of the cultivars and of wild species elucidated the relationships between and maternal origin of crape myrtle cultivars. Genetic diversity and population differentiation analyses evaluated the genetic structure and genetic divergence in the cultivars. This study sheds light on the diversity of crape myrtle cultivars and provides variable genetic resources for the breeding of new cultivars.
2. Results
2.1. Plastome and nrDNA Sequences of Lagerstroemia indica Cultivars
In this study, the plastomes of 20 cultivars of L. indica were assembled (Figure 1 and Table 1). All the plastomes had the typical quadripartite structure of most angiosperm plants. The length of these plastomes varied between 152,174 bp and 152,232 bp, with the LSC (length: 84,006 bp–84,062 bp) and SSC (length: 16,908 bp–16,910 bp) separated by two IRs (length: 25,630) (Table S2). The overall GC content was 37.6%. The L. indica plastome harbored 112 different genes, including 78 protein coding genes, 30 tRNA genes, and 4 rRNA genes. The annotated plastomes were deposited in GenBank (Table 1). The positions of the IR and SC boundaries were conserved among the cultivars. The LSC and IRb boundary was located in the rps19 gene, and the IRb and SSC in the ndhF gene. The boundary between LSC and IRa was located between the rps19 and trnH-GUG. The trnH-GUG gene was located at the beginning of the LSC region. The nrDNA sequences were each assembled into a single contig using the GetOrganelle toolkit. The nrDNA sequences were aligned with 6419 bp.
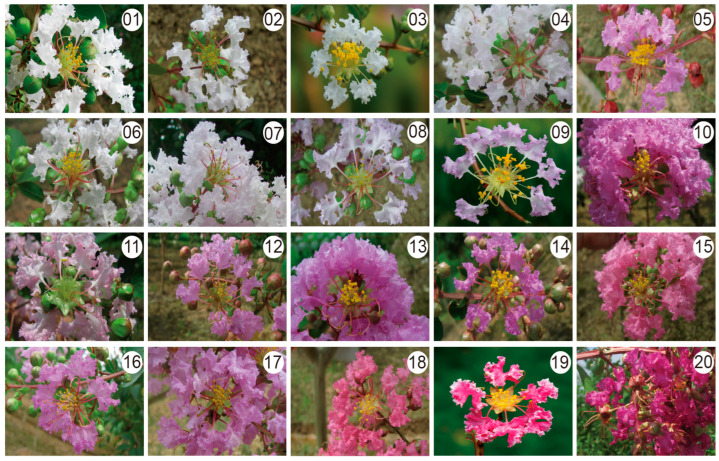
Figure 1.
The floral traits of 20 Lagerstroemia indica cultivars. 01. ‘Baimixiang’; 02. ‘Baiyunyingxia’; 03. ‘Bingqingyudie’; 04. ‘Zizhuainwei’; 05. ‘Dahuazhaolu’; 06. ‘Xiaohuayinwei’; 07. ‘Dahuaziyun’; 08. ‘Zhoubanjinwei’; 09. ‘Dahuacuipanjinwei’; 10. ‘Duohuazi’; 11. ‘Qiaojiaren’; 12. ‘Jinwei’; 13. ‘Lanzi’; 14. ‘Duohuajinxiu’; 15. ‘Fenjing’; 16. ‘Ziyu’; 17. ‘Zixia’; 18. ‘Caixiamantian’; 19. ‘Yinbianhong’; 20. ‘Hongzhuashenzi’.
Table 1.
Samples information and characteristics of the 20 Lagerstroemia indica cultivars.
| Samples | Varietal Group | Flower Number | Flowering Times | Flower Color | Origin | GenBank Accession Number of Plastome |
GenBank Accession Number of nrDNA |
|---|---|---|---|---|---|---|---|
| ‘Dahuazhaolu’ | ‘Bicolor’ Group | 43–98 | Middle period (July and August) | Bicolor | Jishou, Hunan | OP613198 | OP723643 |
| ‘Ziyu’ | ‘Amabilis’ Group | 62–112 | Middle period (July and August) | Purple | Changde, Hunan | OP613199 | OP723644 |
| ‘Duohuazi’ | ‘Amabilis’ Group | 64–172 | Middle period (July and August) | Purple | Liuyang, Hunan | OP613200 | OP723645 |
| ‘Qiaojiaren’ | ‘Bicolor’ Group | 61–138 | Middle period (July and August) | Bicolor | Wuhan, Hubei | OP613201 | OP723646 |
| ‘Zizhuayinwei’ | ‘Alba’ Group | 69–102 | Middle period (July and August) | White | Guiyang, Guizhou | OP613202 | OP723647 |
| ‘Yinbianhong’ | ‘Bicolor’ Group | 86–113 | Early flowering period (before July) | Bicolor | Jishou, Hunan | OP613203 | OP723648 |
| ‘Bingqingyudie’ | ‘Alba’ Group | 76–158 | Middle period (July and August) | White | Guiyang, Guizhou | OP613204 | OP723649 |
| ‘JinWei’ | ‘Amabilis’ Group | 83–185 | Middle period (July and August) | Purple | Changsha, Hunan | OP613205 | OP723650 |
| ‘Zixia’ | ‘Amabilis’ Group | 66–119 | Middle period (July and August) | Purple | Changde, Hunan | OP613206 | OP723651 |
| ‘Zhoubanjinwei’ | ‘Amabilis’ Group | 153–225 | Middle period (July and August) | Purple | Changsha, Hunan | OP613207 | OP723652 |
| ‘Caixiamantian’ | ‘Rubra’ Group | 98–198 | Late flowering (September) | Red | Chengdu, Sichuan | OP613208 | OP723653 |
| ‘Baiyunyingxia’ | ‘Alba’ Group | 57–125 | Middle period (July and August) | White | Changsha, Hunan | OP613209 | OP723654 |
| ‘Baimixiang’ | ‘Alba’ Group | 100–198 | Flowering long | White | Changsha, Hunan | OP613210 | OP723655 |
| ‘Lanzi’ | ‘Amabilis’ Group | 59–106 | Middle period (July and August) | Purple | Changde, Hunan | OP613211 | OP723656 |
| ‘Fenjing’ | ‘Rubra’ Group | 97–198 | Middle period (July and August) | Red | Chengdu, Sichuan | OP613212 | OP723657 |
| ‘Dahuaziyun’ | ‘Bicolor’ Group | 43–122 | Middle period (July and August) | Bicolor | Shaoyang, Hunan | OP613213 | OP723658 |
| ‘Xiaohuayinwei’ | ‘Alba’ Group | 69–145 | Middle period (July and August) | White | Chengdu, Sichuan | OP613214 | OP723659 |
| ‘Duohuajinxiu’ | ‘Amabilis’ Group | 138–222 | Middle period (July and August) | Purple | Shaoyang, Hunan | OP613215 | OP723660 |
| ‘Hongzhuashenzi’ | ‘Amabilis’ Group | 64–164 | Middle period (July and August) | Purple | Shaoyang, Hunan | OP613216 | OP723661 |
| ‘DahuacuipanJinwei’ | ‘Amabilis’ Group | 75–156 | Middle period (July and August) | Purple | Shaoyang, Hunan | OP613217 | OP723662 |
2.2. Plastome Variation in the Lagerstroemia indica Cultivars
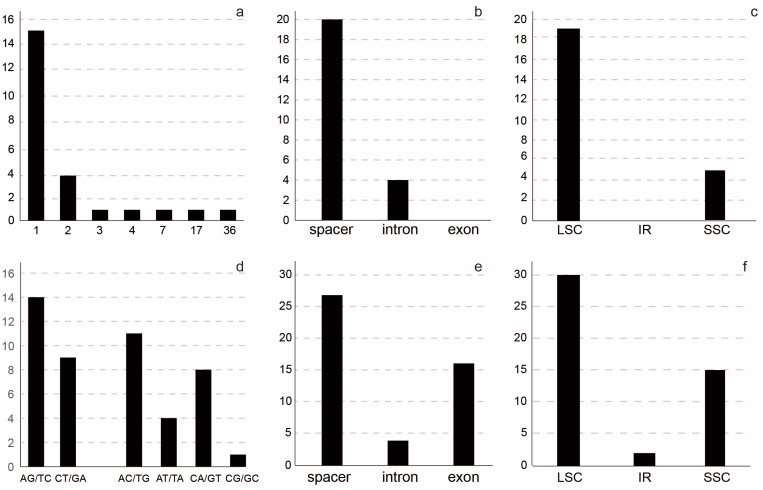
The L. indica cultivars’ plastomes were aligned with 152,250 bp in length. Indels and SNPs were identified in the plastomes, and most of the intraspecific L. indica variable sites and indels were located in the LSC and SSC regions (Figure 2). A total of 24 indels were discovered in the 20 L. indica cultivars’ plastomes, including 14 SSR-related indels, 4 repeat-related indels, and 6 normal indels. All the indels occurred in the noncoding regions, including 4 in introns (ndhA, clpP, atpF, and petB) and 20 in the spacer regions (Figure 2c). The indels’ size ranged from 1 to 36 bp (Figure 2a), with 1 bp indels occurring in the highest frequency (62.5%). The two largest indels were located in the rpl33-rps18 and accD-psaI regions, both of which were repeat-related. Both of these indels were found in an insert in the ‘Qiaojiaren’, ‘JinWei’, ‘Zhoubanjinwei’, ‘Baimixiang’, and ‘Lanzi’ cultivars.
Figure 2.
Plastome variation in the Lagerstroemia indica cultivars. (a) length of indels, X-axis indicated the length of the indels; (b) number of indels in the spacer, intron, and exon regions; (c) number of indels in the LSC, IR, and SSC regions; (d) patterns of SNPs, X-axis indicated the patterns of the SNPs; (e) number of SNPs in the spacer, intron, and exon regions; (f) number of SNPs in the LSC, IR, and SSC regions.
There were 47 SNPs in the 20 L. indica plastomes, and the average number of intraspecific variable sites was 0.31 per kb. Among the 47 SNPs, there were 45 parsimony-informative sites, including 23 transition (Ts) and 24 transversion (Tv) sites. The most frequent SNP mutation types were A to G and T to C, with G to C or C to G mutations occurring far less frequently, and only once in the psbE-petL region. A total of 30, 15, and 2 SNPs occurred in the LSC, SSC, and IR regions, respectively. SNPs were harbored in 38 sequence regions, including 23 spacer regions, 12 coding regions, and 4 intron regions. The trnK-UUU-rps16 spacer region and the ycf1 gene had three SNPs; trnD-GUC-trnY-GUA, rpl32-trnL-UAG, rpoC2, ndhD, and ndhF had two SNPs, and the rest of the regions had one SNP.
2.3. Nuclear Ribosomal DNA Variability
The nrDNA sequences were highly homogeneous among the 20 L. indica cultivars, with an aligned length of 6,419 bp. The GenBank accession numbers of the nrDNA of the cultivars are shown in Table 1. Comparison of the sequences revealed 25 SNPs: 11 in the 18S rRNA region, 11 in the ITS region, and 3 in the 26S region.
2.4. Maternal Origin of the Cultivars
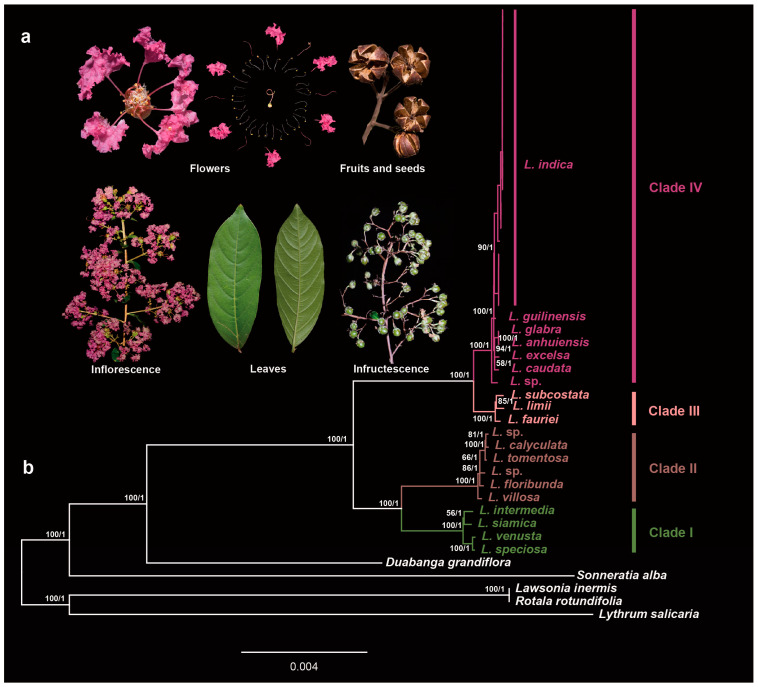
Combining the wild species and cultivars, ML and BI analyses based on the whole plastome dataset produced similar trees (Figure 3). The dataset strongly supported the monophyly and revealed four clades in the genus Lagerstroemia. This result is consistent with recent phylogenetic results [30,32]. All 20 cultivars of L. indica formed a strongly supported group with the wild species of L. indica, with higher supported values (BS/PP = 90/1) in clade IV indicating that the maternal parentage of all 20 cultivars was the L. indica.
Figure 3.
Phylogenetic relationships between Lagerstroemia indica and other related wild species. (a) morphological characteristics of Lagerstroemia indica; (b) ML bootstrap support values/Bayesian posterior probabilities are shown at each node.
2.5. Genetic Variation Based on the Plastome Sequences
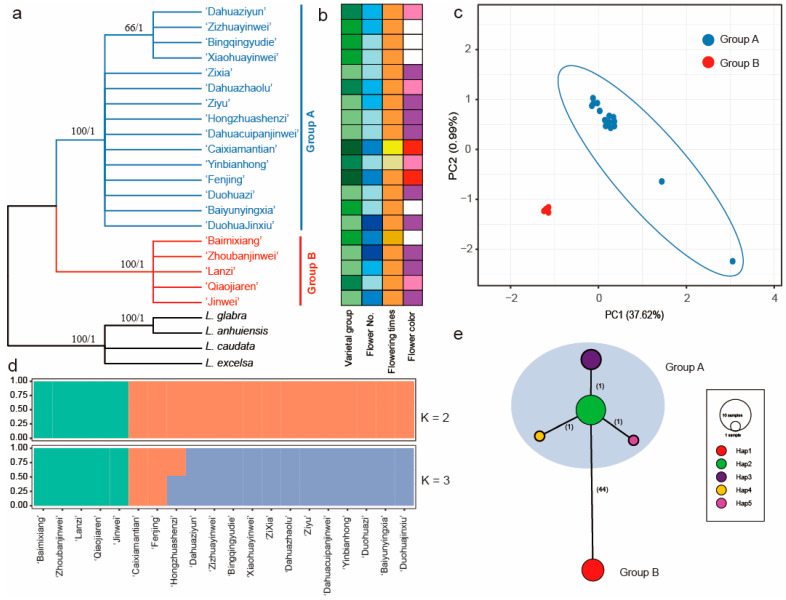
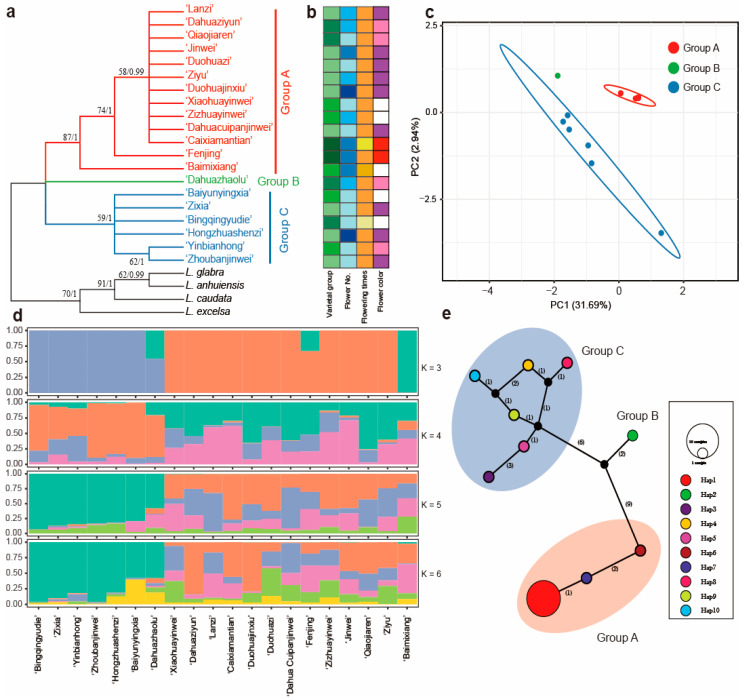
ML and BI tree analyses performed from the whole plastome sequences indicated that all 20 cultivars of L. indica formed two clades (Figure 4a). The five cultivars of ‘Qiaojiaren’, ‘JinWei’, ‘Zhoubanjinwei’, ‘Baimixiang’, and ‘Lanzi’ formed a clade. The PCA scatterplot is presented in Figure 4c. The first two PCA axes account for about 38.61%, revealing a clear clustering in the two groups. Population structure results from ADMIXTURE suggest that there are two clades (Figure 4d and Figure S1).
Figure 4.
Intraspecific diversity and genetic structure of 20 Lagerstroemia indica cultivars based on plastome dataset. (a) phylogenetic tree. ML bootstrap support values/Bayesian posterior probabilities are shown at each node; (b) phenotypic characterization; (c) principal component analysis; (d) population structure analysis with K = 2, 3; (e) TCS network of five haplotypes from the plastome sequences. The number of mutational steps is shown on the lines, and the size of the pie chart represents the number of the accessions. The haplotype for each sample is listed in Table S3.
All the polymorphisms allowed for the identification of five haplotypes (Figure 4e, Table S3). The five haplotypes also formed two clades, exhibiting a significant genetic difference with the number of mutational steps (44 steps). Further evidence of the phylogenetic structure of plastome variation in the L. indica cultivars was provided by the distribution of SNP variation among the phylogenetic clades. Haplotype 1 contained five cultivars of ‘Qiaojiaren’, ‘JinWei’, ‘Zhoubanjinwei’, ‘Baimixiang’, and ‘Lanzi’. Haplotype 2, containing nine cultivars, formed a clade with haplotypes 3, 4, and 5, showing a star-like topology consisting of a central haplotype (hap2) from which the other three haplotypes radiate, separated by one step (Figure 4e).
2.6. Genetic Variation Based on the nrDNA Sequences
Phylogenetic reconstruction using the nrDNA sequences revealed three clades dividing the twenty cultivars of L. indica (Figure 5a). The PCA scatterplot is presented in Figure 5c. The first two PCA axes account for about 34.63%, revealing a clear clustering in the three groups. Population structure was analyzed using K values ranging from 1 to 10, and the cross validation (CV) error was also the lowest with K = 5 (Figure 5d and Figure S1). The results show that most of the cultivars had at least two genetic backgrounds and had higher gene flow.
Figure 5.
Intraspecific diversity and genetic structure of 20 Lagerstroemia indica cultivars based on nrDNA dataset. (a) phylogenetic tree. ML bootstrap support values/Bayesian posterior probabilities are shown at each node; (b) phenotypic characterization; (c) principal component analysis; (d) population structure analysis with K = 3, 4, 5, and 6; (e) TCS network of 10 haplotypes from the nrDNA sequences. The number of mutational steps is shown on the lines, and the size of the pie chart represents the number of the accessions. The black circles are extinct haplotypes, and the haplotype for each sample is listed in Table S4.
Network analysis supported three clades, consistent with the phylogeny result (Figure 5e). Ten haplotypes were identified (Table S4), with only haplotype 1 (hap 1) represented across eleven cultivars. With haplotype 2, haplotypes 1, 6, and 7 formed a clade (Group A in Figure 5a) with nine mutational steps. Haplotype 2 was the ‘Dahuazhaolu’ from the Bicolor group. The population structure shows ‘Dahuazhaolu’ with multiple instances of crossbreeding (Figure 5d). Group C included six haplotypes, containing six cultivars of ‘Yinbianhong’, ‘Bingqingyudie’, ‘Zixia’, ‘Baiyunyingxia’, Hongzhuashenzi’, and ‘Zhoubanjinwei’.
2.7. Phenotypic Characterization
The 20 cultivars of L. indica showed very high morphological variation (Figure 1). The different varietal groups exhibited variation in flower numbers and flowing time, and neither the plastome nor nrDNA data supported a finding that the varietal groups were monophyletic. (Figure 4b and Figure 5b). There were four orders of flower numbers in the inflorescence of the twenty cultivars, showing greater differences. The two cultivars of ‘Duohuajinxiu’ and ‘Zhoubanjinwei’ exhibited a large number of flowers (more than 200) per inflorescence, while the ‘Dahuazhaolu’ cultivar from the ‘Bicolor’ group only had half that amount. Most of the cultivars (17) bloom in July and August. The flowering time of cultivar ‘Yinbianhong’ is relatively early, before July, while ‘Caixiamantian’ flowers relatively late, in September. The ‘Baimixiang’ cultivar from the ‘Alba’ group has a strong fragrance and a long flowering time (from July to September). Finally, flower color was documented in 20 cultivars (Figure 1). The most common colors are white, purple, red, and bicolor (mostly purple and pink). Flower color is the main varietal group-based phenotypic character for L. indica cultivars. However, flower color-based grouping was not supported by molecular data.
3. Discussion
3.1. Maternal Donor of Crape Myrtle Cultivars
Crape myrtles have been cultivated as ornamental trees for more than 1600 years, owing to their long flowering season, high resistance to pollution, and ease of training. Although crape myrtles have more than 300 cultivars [2], the maternal donor has not historically been clear. In this study, we used whole plastome sequences to explore the maternal donor of crape myrtle cultivars and assess their genetic variation.
The five species of L. indica, L. fauriei, L. speciosa, L. subcostata, and L. limii have been introduced in crape myrtle breeding programs, releasing at least 200 varieties with a wide range in plant size, habitat, flower color, and size [3,5]. L. fauriei, from central and southern Japan, has been an important donor of crape myrtle, owing to its strong resistance to mildew, disease, and cold temperatures. L. speciosa, native to Australia, Southern New Guinea, India, and the Philippines, is a woody plant growing 25 m high [33] which is widely cultivated as an ornamental tree in tropical and subtropical areas. L. subcostata and L. limii, native to Southern China, are mostly shrubs or small trees; they bloom earlier [33] and have been used to breed early-flowering cultivars [34].
Cross-breeding is one of the primary strategies for the breeding of crape myrtles, with five main wild species involved in the breeding of its cultivars. Our phylogeny results revealed that all 20 cultivars formed a strongly supported clade within the wild species of L. indica (Figure 3), indicating that L. indica was the maternal donor of these cultivars. Lagerstroemia guilinensis, narrowly distributed in Guangxi Province, is sister to L. indica in Clade IV. Phylogeny results supported that the three species of L. fauriei, L. subcostata, and L. limii form a separate clade (Clade III).
3.2. Phenotypic Diversity and Genetic Variation of Lagerstroemia indica Cultivars
The L. indica cultivars are rich in phenotypic diversity (Figure 1), including quantitative and qualitative trait variations such as flower color, claw color, flower diameter, flower number, length and width of inflorescence, and 1000-seed weight [6,8]. The high degree of phenotypic diversity varies among the different varietal groups; the ‘Rubra’ group had the highest phenotypic diversity, followed by the ‘Amabilis’ group and the ‘Alba’ group [6]. Based on the cluster of phenotypic characteristics, the L. indica cultivars were further divided into five groups, and all the varietal groups did not form a clade except the ‘Bicolor’ group [6]. Meanwhile, genetic evidence did not support the finding of a monophyletic group within the varietal groups from either the plastome or nrDNA sequences (Figure 4a and Figure 5a). Phylogenetic relationships showed there was conflict between plastome and nrDNA datasets (Figure 4a and Figure 5a). Topological cytonuclear discordance is commonly observed in plant phylogenetics [14,35], and incomplete lineage sorting and gene flow can cause cytonuclear discordance within the species or among closely related species [36,37]. For L. indica cultivars, the intraspecific hybridization during the process of cross-breeding may lead to the discordance between the plastome and nrDNA. Plastome and nrDNA sequencing revealed high genetic diversity among crape myrtle cultivars. The plastome sequences unveiled unexplored genetic variation, and 47 variable sites and 24 indels were identified, giving rise to 5 haplotypes and dividing the 20 cultivars into 2 groups (Figure 4). The nrDNA sequence included 25 variable sites and divided the 20 cultivars into 3 clades (Figure 5). The plastome and nrDNA sequences exhibited high variability in crape myrtle cultivars, compared to Chrysanthemum [27], sweet potato (Ipomoea batatas) [38], and Panax ginseng [39] cultivars. Compared to the SSR and AFLP markers, the plastome sequence markers show lower genetic variability; however, as maternal markers, it is essential to identify the maternal parentage.
Further evidence showed that most of the crape myrtle cultivars are of hybrid origin, even interspecific hybrids (for example, some cultivars from America) [5,12]. The structure of the nrDNA also supported the finding that most of the selected cultivars were at least two crosses (Figure 5d). Several studies showed the cultivars mostly tended to group by geographic regions [29]. With wide sampling, the crape myrtle cultivars also showed the same pattern [12]. Interestingly, according to the morphological database, several cultivars exhibited similar traits when forming a clade, such as the same flower color (e.g., ‘Yinbianhong’ and ‘Hongzhuashenzi’). This indicates that they have a similar genetic background (Figure 4 and Figure 5).
3.3. Utility of Plastome and nrDNA for Accessing Genetic Diversity of Cultivars
With the advantage of next-generation sequencing technologies and bioinformatics tools, plastomes and nrDNA can be assembled from genome skimming data, avoiding the high experimental technology requirements of chloroplast isolation and purification [40,41,42]. Plastomes are maternally inherited and structurally conserved in most angiosperm plants, and nrDNA sequences are variable, leading these markers to be widely used for tracking the evolution and species identification of plants at both high and low taxonomic levels. Therefore, the plastome markers rbcL, matK, ycf1, and ITS are typically selected as the DNA barcodes for land plants [43,44]. Some of the regions of the plastomes, including the ndhF, trnH-psbA, and trnL-F, have been identified as mutation hotspot regions [45]. For the wild species of Lagerstroemia, four variable loci, trnD-trnY-trnE, rrn16-trnI, ndhF-rpl32-trnL, and ycf1, were discovered in the Lagerstroemia plastomes [30].
Plastome sequences are widely used to infer phylogenetic relationships at different taxonomic levels. The phylogeny of the species in Lagerstroemia was well resolved based on the whole plastome sequences from this paper and from previous studies [30,32,46], revealing the maternal donor of crape myrtle cultivars. Only a few studies have assessed the intraspecific variation of the whole plastomes and nrDNA sequences [18,20,21,47,48]. The plastid genome markers have less use in analyzing the genetic diversity of cultivars, owing to their limited polymorphic sites. In this study, we sequenced the whole plastome and nrDNA of common and representative crape myrtle cultivars to assess the variations in these sequences. In total, 47 SNPs and 24 indels, and 28 SNPs in plastome and nrDNA sequences, respectively, were identified among these 20 crape myrtle cultivars.
Mutation rate variation among different lineages of plastomes and nrDNA sequences has been examined in various studies [49,50,51]. Most of the cultivars originated from one species, and it is difficult to discover the genetic difference owing to these variations occurring at the intra-specific level or even the intra-group level. However, more variable sites were identified in the crape myrtle cultivars. There are three main factors which have the possibility of introducing more genetic variations. First, at least five wild species (L. indica, L. fauriei, L. speciosa, L. subcostata, and L. limii) are involved in the formation of crape myrtle cultivars [2,3,52], leading to broad genetic variation. Second, most of the cultivars were formed by hybridization, and cross-breeding is one of the primary strategies for the breeding of crape myrtles. Third, longer cultivation and selection based on traits such as flower color and number of flowers per inflorescence have led to the maintenance of more genetic variations.
Additional studies have shown that the plastome has mutational hotspot regions and that the IR region was better conserved than SC regions. In the crape myrtle cultivars’ plastome, intra-specific variable sites and indels were mostly located in the SC regions and the trnK-UUU-rps16 spacer region. The ycf1 gene demonstrated higher variability. The nrDNA internal transcribed spacer (ITS) sequences are highly variable in the kingdom Plantae, with a potentially high resolution of inter- and intra-specific relationships [53]. ITS sequences have been used to authenticate ginseng cultivars [54], assess the genetic variability and relationship of banana cultivars (Musa L.) [55], and conduct molecular identification of Malaysian pineapple cultivars [56].
4. Materials and Methods
4.1. Sampling, DNA Extraction, and Sequencing
Twenty L. indica cultivars representing different varietal groups and flower morphologies were collected (Figure 1 and Table 1), including all four varietal groups (four cultivars in the ‘Bicolor’ group, five cultivars in the ‘Alba’ group, nine cultivars in the ‘Amabilis’ group, and two cultivars in the ‘Rubra’ group) [1,6,31]. The selected cultivars represented different flower numbers and flowering times (Table 1). We also downloaded the plastome sequences of all the published wild relatives of L. indica to elucidate the maternal origin of crape myrtle cultivars.
Total genomic DNA was extracted using the modified CTAB method [57]. DNA quantity and quality were examined by electrophoresis in 1% agarose. Total DNA was sheared by an ultrasonicator to 350 bp fragments, and a paired-end DNA library for Illumina HiSeq X-ten platform sequencing was constructed. Each sample yielded approximately 5 Gb of data.
4.2. Plastome and nrDNA Assembly
Trimmomatic 0.36 [58] was used to conduct a quality control of the raw data within the default parameters. Plastome and nrDNA sequences were assembled using the GetOrganelle toolkit [59] with k-mer lengths of 95. If GetOrganelle failed, we used the following method to assemble it: The SPAdes 3.6.1 program (k-mer = 95) [60] was selected to assemble the contigs using the clean data. Plastome contigs, which were selected using the Blast program [61], were manually assembled using Sequencher 5.4.5 (Gene Codes Corporation, Ann Arbor, MI, USA, http://www.genecodes.com, accessed on 10 July 2022). Gaps and assembly errors were filled and checked using the clean reads that were mapped to the contigs using Geneious Prime (Biomatters Ltd., Auckland, New Zealand) [62]. Plastomes were annotated using the perl scripts Plann.pl [63], with the published genome of L. indica (GenBank accession number: KX263727) as the reference sequence. Annotation errors and missing genes were checked and manually added with Geneious Prime. Our annotation of plastomes and nrDNA sequences was submitted to GenBank.
4.3. Plastome and nrDNA Variation Analyses
All the plastome and nrDNA sequences of the 20 cultivars were aligned using MAFFT 7 [64]. We identified intra-species polymorphism, including SNPs and indel markers. SNPs were identified and calculated using MEGA 7.0 [65], and DnaSP 6 [66] was used to identify the indels. Their number, location, and direction were calculated using the ‘Dahua Zhaolu’ chloroplast genome as the standard reference to determine the mutation direction.
4.4. Phylogenetic Analyses
To infer the maternal origins of the 20 L. indica cultivars, we combined the plastome data of cultivars with 4 wild L. indica samples, 19 other wild Lagerstroemia species, and 5 Lythraceae species used as outgroups (Table S1). The plastome and nrDNA of cultivars were also used to infer phylogenetic relationships, with four Lagerstroemia species as the outgroups according to the phylogenetic relationships of Lagerstroemia. Phylogenetic analyses were performed using the maximum likelihood (ML) and Bayesian inference (BI) methods. For both analyses, the best-fit substitution mode GTR+GAMMA was chosen by ModelFinder [67] under the Bayesian information criterion. ML analysis was conducted in RAxML-NG [68], and the best tree was selected to calculate the node support values using 500 rapid bootstrap replicates.
BI analysis was performed in Mrbayes v3.2 [69]. Markov chain Monte Carlo (MCMC) simulations were run for 10 million generations, with a sampling of 1000 generations. The stationary phase was examined using Tracer 1.6 [70], and the first 25% of the sampled trees were discarded. The majority-rule consensus tree was generated using the remaining trees and estimated posterior probabilities.
4.5. Genetic Variation and Diversity Analyses
Plastome and nrDNA sequences of the 20 cultivars were used for structure and PCA analyses. The population structure used the filtered intraspecific SNPs and an admixture model-based clustering method implemented in Admixture v1.3. The optimal number of clusters was evaluated by running the K-means clustering algorithm (K = 1 to K = 10). The most likely number of clusters was determined based on CV error. Principal component analysis (PCA) was conducted using Plink [71], and the ggplot package [72] in R was used to draw the figure. Both of these data were used to perform network analyses. The haplotype data were exported in DnaSP v6 [66], and the haplotype frequencies were performed in Arlequin v3.5 [73]. PopArt v1.7 was used to build the TCS network [74].
4.6. Phenotypic Analyses
Four phenotypic characters were used for phenotypic analyses. We analyzed the varietal group according to the classification system of Zhang [1]. The 20 cultivars included four varietal groups. The flower numbers in the inflorescence were divided into four classes: less than 100, 100–150, 150–200, and more than 200. Flowering time was divided into four periods: early flowering (before July), middle flowering (July and August), late flowering (September), and flowering long. The flower color was divided into four groups: white, purple, red, and bicolor (mostly purple and pink).
5. Conclusions
In this study, based on plastome and nrDNA, we discovered the genetic variations of crape myrtle (L. indica) cultivars and identified genome-wide variances, which contribute to better understanding the origin and relationships of the cultivars. The phylogenetic tree of the plastome, including wild species and cultivars, reveals the maternal origins of cultivars. The structure results of the nrDNA show that most of the cultivars are of hybrid origins. The haplotype identification and phylogeny provide novel insights into the cultivation history of crape myrtle cultivars.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24043606/s1.
Author Contributions
Conceptualization, project administration, W.D.; data curation, investigation, conceptualization, and resources, C.G., K.L., Z.S., E.L., Y.C., J.H., W.L. and W.D.; funding acquisition, Z.S. and W.L.; writing—review and editing, visualization, and supervision, W.D., C.G., K.L. and Z.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The plastome and nrDNA sequences under this study are deposited in the GenBank database under the following accession numbers: OP613198–OP613217 and OP723643–OP723662. The sequenced raw data are deposited in the SRA database with the accession number PRJNA909618.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was financially supported by the National Natural Science Foundation of China (No. 31770744), the Training Program of Innovation and Entrepreneurship for Undergraduates (202210022242), and the National Forest Genetic Resources Platform (2005DKA21003).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Zhang Q. Studies on cultivars of crape-myrtles (Lagerstroemia indica) and their uses in urban greening. J. Beijing For. Univ. 1991;13:57–66. [Google Scholar]
- 2.Cai M., Pan H.-T., Wang X.-F., He D., Wang X.-Y., Wang X.-J., Zhang Q.-X. Development of novel microsatellites in Lagerstroemia indica and DNA fingerprinting in Chinese Lagerstroemia cultivars. Sci. Hortic. 2011;131:88–94. doi: 10.1016/j.scienta.2011.09.031. [DOI] [Google Scholar]
- 3.Pounders C., Rinehart T., Sakhanokho H. Evaluation of Interspecific Hybrids between Lagerstroemia indica and L. speciosa. HortSci. Horts. 2007;42:1317–1322. doi: 10.21273/HORTSCI.42.6.1317. [DOI] [Google Scholar]
- 4.Ye Y.M., Tong J., Shi X.P., Yuan W., Li G.R. Morphological and cytological studies of diploid and colchicine-induced tetraploid lines of crape myrtle (Lagerstroemia indica L.) Sci. Hortic. 2010;124:95–101. doi: 10.1016/j.scienta.2009.12.016. [DOI] [Google Scholar]
- 5.Rinehart T.A., Pounders C.T. Estimating Diversity among Lagerstroemia Species and Hybrids Using SSR Markers. Acta Hortic. 2010;885:285–290. doi: 10.17660/ActaHortic.2010.885.39. [DOI] [Google Scholar]
- 6.Wang Y.-S., Hou B.-X., Suo Z.-L., Liu T.-H., Chen L.-J., Yang X.-J. Genetic Diversity of Lagerstroemia indica Germplasm Resources in Hunan Based on Morphological Characteristics. J. Plant Genet. Resour. 2015;16:71–79. [Google Scholar]
- 7.Margaret R.P. Molecular Genetic Diversity Among 12 Clones of Lagerstroemia fauriei Revealed by AFLP and RAPD Markers. HortScience. 2003;38:256–259. [Google Scholar]
- 8.Ming C., Miao T., Min W., Pan H.-T., Qi-Xiang Z. Analysis of genetic diversity and relationship of Chinese Lagerstroemia indica cultivars based on AFLP and morphological markers. Acta Hortic. 2012;938:509–516. doi: 10.17660/ActaHortic.2012.938.67. [DOI] [Google Scholar]
- 9.Peng C., Li Z., Ma L., Huang G., Xu H., Yang Y. Identification and genetic analysis in Lagerstroemia interspecific hybrids using SSR markers. North. Hortic. 2020;13:83–90. [Google Scholar]
- 10.Liu Y., He D., Cai M., Tang W., Li X.Y., Pan H.T., Zhang Q.X. Development of microsatellite markers for Lagerstroemia indica (Lythraceae) and related species. Appl. Plant Sci. 2013;1:1200203. doi: 10.3732/apps.1200203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suo Z., Li W., Jin X., Zhang H. A new nuclear DNA marker revealing both microsatellite variations and single nucleotide polymorphic loci: A case study on classification of cultivars in Lagerstroemia indica L. J. Microb. Biochem. Technol. 2016;8:266–271. doi: 10.4172/1948-5948.1000296. [DOI] [Google Scholar]
- 12.He D., Liu Y., Cai M., Pan H., Zhang Q., Wang X., Wang X. Genetic diversity of Lagerstroemia (Lythraceae) species assessed by simple sequence repeat markers. Genet. Mol. Res. 2012;11:3522–3533. doi: 10.4238/2012.September.26.9. [DOI] [PubMed] [Google Scholar]
- 13.Dong W., Liu Y., Xu C., Gao Y., Yuan Q., Suo Z., Zhang Z., Sun J. Chloroplast phylogenomic insights into the evolution of Distylium (Hamamelidaceae) BMC Genom. 2021;22:293. doi: 10.1186/s12864-021-07590-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong W., Li E., Liu Y., Xu C., Wang Y., Liu K., Cui X., Sun J., Suo Z., Zhang Z., et al. Phylogenomic approaches untangle early divergences and complex diversifications of the olive plant family. BMC Biol. 2022;20:92. doi: 10.1186/s12915-022-01297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong W., Liu Y., Li E., Xu C., Sun J., Li W., Zhou S., Zhang Z., Suo Z. Phylogenomics and biogeography of Catalpa (Bignoniaceae) reveal incomplete lineage sorting and three dispersal events. Mol. Phylogenet. Evol. 2022;166:107330. doi: 10.1016/j.ympev.2021.107330. [DOI] [PubMed] [Google Scholar]
- 16.Gao Y., Liu K., Li E., Wang Y., Xu C., Zhao L., Dong W. Dynamic evolution of the plastome in the Elm family (Ulmaceae) Planta. 2023;257:14. doi: 10.1007/s00425-022-04045-4. [DOI] [PubMed] [Google Scholar]
- 17.Li E., Liu K., Deng R., Gao Y., Liu X., Dong W., Zhang Z. Insights into the phylogeny and chloroplast genome evolution of Eriocaulon (Eriocaulaceae) BMC Plant Biol. 2023;23:32. doi: 10.1186/s12870-023-04034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J., Wang S., Wang Y., Wang R., Liu K., Li E., Qiao P., Shi L., Dong W., Huang L., et al. Phylogenomics and Genetic Diversity of Arnebiae Radix and Its Allies (Arnebia, Boraginaceae) in China. Front. Plant Sci. 2022;13:920826. doi: 10.3389/fpls.2022.920826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong W.-P., Sun J.-H., Liu Y.-L., Xu C., Wang Y.-H., Suo Z.-L., Zhou S.-L., Zhang Z.-X., Wen J. Phylogenomic relationships and species identification of the olive genus Olea (Oleaceae) J. Syst. Evol. 2022;60:1263–1280. doi: 10.1111/jse.12802. [DOI] [Google Scholar]
- 20.Shang C., Li E., Yu Z., Lian M., Chen Z., Liu K., Xu L., Tong Z., Wang M., Dong W. Chloroplast Genomic Resources and Genetic Divergence of Endangered Species Bretschneidera sinensis (Bretschneideraceae) Front. Ecol. Evol. 2022;10:873100. doi: 10.3389/fevo.2022.873100. [DOI] [Google Scholar]
- 21.Torre S., Sebastiani F., Burbui G., Pecori F., Pepori A.L., Passeri I., Ghelardini L., Selvaggi A., Santini A. Novel Insights into Refugia at the Southern Margin of the Distribution Range of the Endangered Species Ulmus laevis. Front. Plant Sci. 2022;13:826158. doi: 10.3389/fpls.2022.826158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Sun J., Zhao Z., Xu C., Qiao P., Wang S., Wang M., Xu Z., Yuan Q., Guo L., et al. Multiplexed Massively Parallel Sequencing of Plastomes Provides Insights into the Genetic Diversity, Population Structure, and Phylogeography of Wild and Cultivated Coptis chinensis. Front. Plant Sci. 2022;13:923600. doi: 10.3389/fpls.2022.923600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perdereau A., Klaas M., Barth S., Hodkinson T.R. Plastid genome sequencing reveals biogeographical structure and extensive population genetic variation in wild populations of Phalaris arundinacea L. in north-western Europe. GCB Bioenergy. 2017;9:46–56. doi: 10.1111/gcbb.12362. [DOI] [Google Scholar]
- 24.Xue C., Geng F.D., Li J.J., Zhang D.Q., Gao F., Huang L., Zhang X.H., Kang J.Q., Zhang J.Q., Ren Y. Divergence in the Aquilegia ecalcarata complex is correlated with geography and climate oscillations: Evidence from plastid genome data. Mol. Ecol. 2021;30:5796–5813. doi: 10.1111/mec.16151. [DOI] [PubMed] [Google Scholar]
- 25.Huang D.I., Hefer C.A., Kolosova N., Douglas C.J., Cronk Q.C. Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae) New Phytol. 2014;204:693–703. doi: 10.1111/nph.12956. [DOI] [PubMed] [Google Scholar]
- 26.Cui H., Ding Z., Zhu Q., Wu Y., Gao P. Population structure and genetic diversity of watermelon (Citrullus lanatus) based on SNP of chloroplast genome. 3 Biotech. 2020;10:374. doi: 10.1007/s13205-020-02372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y., Liao B., Ostevik K.L., Zhou H., Wang F., Wang B., Xia H. The Maternal Donor of Chrysanthemum Cultivars Revealed by Comparative Analysis of the Chloroplast Genome. Front. Plant Sci. 2022;13:923442. doi: 10.3389/fpls.2022.923442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nock C.J., Hardner C.M., Montenegro J.D., Ahmad Termizi A.A., Hayashi S., Playford J., Edwards D., Batley J. Wild Origins of Macadamia Domestication Identified Through Intraspecific Chloroplast Genome Sequencing. Front. Plant Sci. 2019;10:334. doi: 10.3389/fpls.2019.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong W., Kim T.-S., Park Y.-J. Rice Chloroplast Genome Variation Architecture and Phylogenetic Dissection in Diverse Oryza Species Assessed by Whole-Genome Resequencing. Rice. 2016;9:57. doi: 10.1186/s12284-016-0129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong W., Xu C., Liu Y., Shi J., Li W., Suo Z. Chloroplast phylogenomics and divergence times of Lagerstroemia (Lythraceae) BMC Genom. 2021;22:434. doi: 10.1186/s12864-021-07769-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Hou B., Yang Q., Zhou H., Chen L., Yang X. Investigation and analysis of application prospects of Lagerstroemia indica germplasm resources in Hunan Province. Acta Prataculturae Sin. 2014;23:77–91. [Google Scholar]
- 32.Wang J., He W., Liao X., Ma J., Gao W., Wang H., Wu D., Tembrock L.R., Wu Z., Gu C. Phylogeny, molecular evolution, and dating of divergences in Lagerstroemia using plastome sequences. Hortic. Plant J. 2022 doi: 10.1016/j.hpj.2022.06.005. in press . [DOI] [Google Scholar]
- 33.Furtado C., Srisuko M. A revision of Lagerstroemia L. (Lythraceae) Gard. Bull. 1969;24:185–334. [Google Scholar]
- 34.Cai M., Meng R., Pan H.-T., Gao Y.-K., Sun M., Song P., Wang X.-F., Zhang Q.-X. Isolation and characterization of microsatellite markers from Lagerstroemia caudata (Lythraceae) and cross-amplification in other related species. Conserv. Genet. Resour. 2010;2:89–91. doi: 10.1007/s12686-010-9197-2. [DOI] [Google Scholar]
- 35.Xu L.L., Yu R.M., Lin X.R., Zhang B.W., Li N., Lin K., Zhang D.Y., Bai W.N. Different rates of pollen and seed gene flow cause branch-length and geographic cytonuclear discordance within Asian butternuts. New Phytol. 2021;232:388–403. doi: 10.1111/nph.17564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose J.P., Toledo C.A.P., Lemmon E.M., Lemmon A.R., Sytsma K.J. Out of sight, out of mind: Widespread nuclear and plastid-nuclear discordance in the flowering plant genus Polemonium (Polemoniaceae) suggests widespread historical gene flow despite limited nuclear signal. Syst. Biol. 2021;70:162–180. doi: 10.1093/sysbio/syaa049. [DOI] [PubMed] [Google Scholar]
- 37.Sarver B.A.J., Herrera N.D., Sneddon D., Hunter S.S., Settles M.L., Kronenberg Z., Demboski J.R., Good J.M., Sullivan J. Diversification, Introgression, and Rampant Cytonuclear Discordance in Rocky Mountains Chipmunks (Sciuridae: Tamias) Syst. Biol. 2021;70:908–921. doi: 10.1093/sysbio/syaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao S., Xu P., Deng Y., Dai X., Zhao L., Heider B., Zhang A., Zhou Z., Cao Q. Comparative analysis of chloroplast genomes of cultivars and wild species of sweetpotato (Ipomoea batatas [L.] Lam) BMC Genom. 2021;22:262. doi: 10.1186/s12864-021-07544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim K., Lee S.-C., Lee J., Lee H.O., Joh H.J., Kim N.-H., Park H.-S., Yang T.-J. Comprehensive Survey of Genetic Diversity in Chloroplast Genomes and 45S nrDNAs within Panax ginseng Species. PLoS ONE. 2015;10:e0117159. doi: 10.1371/journal.pone.0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L., Hu Y., He M., Zhang B., Wu W., Cai P., Huo D., Hong Y. Comparative chloroplast genomes: Insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genom. 2021;22:138. doi: 10.1186/s12864-021-07427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao F., Chen Y.-P., Salmaki Y., Drew B.T., Wilson T.C., Scheen A.-C., Celep F., Bräuchler C., Bendiksby M., Wang Q., et al. An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol. 2021;19:2. doi: 10.1186/s12915-020-00931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L., Abduraimov O., Tojibaev K., Shomurodov K., Zhang Y.-M., Li W.-J. Analysis of complete chloroplast genome sequences and insight into the phylogenetic relationships of Ferula L. BMC Genom. 2022;23:643. doi: 10.1186/s12864-022-08868-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong W., Xu C., Li C., Sun J., Zuo Y., Shi S., Cheng T., Guo J., Zhou S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015;5:8348. doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong W., Cheng T., Li C., Xu C., Long P., Chen C., Zhou S. Discriminating plants using the DNA barcode rbcLb: An appraisal based on a large dataset. Mol. Ecol. Resour. 2014;14:336–343. doi: 10.1111/1755-0998.12185. [DOI] [PubMed] [Google Scholar]
- 45.Dong W., Liu J., Yu J., Wang L., Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng G., Wei L., Ma L., Wu Z., Gu C., Chen K. Comparative analyses of chloroplast genomes from 13 Lagerstroemia (Lythraceae) species: Identification of highly divergent regions and inference of phylogenetic relationships. Plant Mol. Biol. 2020;102:659–676. doi: 10.1007/s11103-020-00972-6. [DOI] [PubMed] [Google Scholar]
- 47.Hu G., Wu Y., Guo C., Lu D., Dong N., Chen B., Qiao Y., Zhang Y., Pan Q. Haplotype Analysis of Chloroplast Genomes for Jujube Breeding. Front. Plant Sci. 2022;13:841767. doi: 10.3389/fpls.2022.841767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H., Zhao W., Hua W., Liu J. A large-scale population based organelle pan-genomes construction and phylogeny analysis reveal the genetic diversity and the evolutionary origins of chloroplast and mitochondrion in Brassica napus L. BMC Genom. 2022;23:339. doi: 10.1186/s12864-022-08573-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith S.A., Donoghue M.J. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- 50.Schwarz E.N., Ruhlman T.A., Weng M.-L., Khiyami M.A., Sabir J.S.M., Hajarah N.H., Alharbi N.S., Rabah S.O., Jansen R.K. Plastome-wide nucleotide substitution rates reveal accelerated rates in Papilionoideae and correlations with genome features across legume subfamilies. J. Mol. Evol. 2017;84:187–203. doi: 10.1007/s00239-017-9792-x. [DOI] [PubMed] [Google Scholar]
- 51.Choi K., Weng M.-L., Ruhlman T.A., Jansen R.K. Extensive variation in nucleotide substitution rate and gene/intron loss in mitochondrial genomes of Pelargonium. Mol. Phylogenet. Evol. 2021;155:106986. doi: 10.1016/j.ympev.2020.106986. [DOI] [PubMed] [Google Scholar]
- 52.Wang X., Wadl P.A., Pounders C., Trigiano R.N., Cabrera R.I., Scheffler B.E., Pooler M., Rinehart T.A. Evaluation of Genetic Diversity and Pedigree within Crapemyrtle Cultivars Using Simple Sequence Repeat Markers. J. Am. Soc. Hort. Sci. 2011;136:116–128. doi: 10.21273/JASHS.136.2.116. [DOI] [Google Scholar]
- 53.Cheng T., Xu C., Lei L., Li C., Zhang Y., Zhou S. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 2016;16:138–149. doi: 10.1111/1755-0998.12438. [DOI] [PubMed] [Google Scholar]
- 54.Kim O.T., Bang K.H., In D.S., Lee J.W., Kim Y.C., Shin Y.S., Hyun D.Y., Lee S.S., Cha S.W., Seong N.S. Molecular authentication of ginseng cultivars by comparison of internal transcribed spacer and 5.8S rDNA sequences. Plant Biotechnol. Rep. 2007;1:163–167. doi: 10.1007/s11816-007-0019-2. [DOI] [Google Scholar]
- 55.Hapsari L., Azrianingsih R., Arumingtyas E.L. Genetic Variability and Relationship of Banana Cultivars (Musa L.) From East Java, Indonesia based on the Internal Transcribed Spacer Region nrDNA Sequences. J. Trop. Biol. Conserv. 2018;15:101–120. doi: 10.51200/jtbc.v15i0.1482. [DOI] [Google Scholar]
- 56.Hidayat T., Abdullah F.I., Kuppusamy C., Samad A.A., Wagiran A. Molecular Identification of Malaysian Pineapple Cultivar based on Internal Transcribed Spacer Region. APCBEE Procedia. 2012;4:146–151. doi: 10.1016/j.apcbee.2012.11.025. [DOI] [Google Scholar]
- 57.Li J., Wang S., Jing Y., Wang L., Zhou S. A modified CTAB protocol for plant DNA extraction. Chin. Bull. Bot. 2013;48:72–78. [Google Scholar]
- 58.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin J.-J., Yu W.-B., Yang J.-B., Song Y., dePamphilis C.W., Yi T.-S., Li D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 62.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang D.I., Cronk Q.C.B. Plann: A command-line application for annotating plastome sequences. Appl. Plant Sci. 2015;3:1500026. doi: 10.3732/apps.1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rozas J., Ferrer-Mata A., Sanchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sanchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 67.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rambaut A., Suchard M., Xie D., Drummond A. Tracer v1. 6. 2014. [(accessed on 1 July 2022)]. Available online: http://beast.community/tracer.
- 71.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wickham H., editor. ggplot2: Elegant Graphics for Data Analysis. Springer International Publishing; Cham, Switzerland: 2016. Data Analysis; pp. 189–201. [Google Scholar]
- 73.Excoffier L., Laval G., Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2005;1:47–50. doi: 10.1177/117693430500100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leigh J.W., Bryant D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plastome and nrDNA sequences under this study are deposited in the GenBank database under the following accession numbers: OP613198–OP613217 and OP723643–OP723662. The sequenced raw data are deposited in the SRA database with the accession number PRJNA909618.