Abstract
The amylase-binding protein A (AbpA) of Streptococcus gordonii was found to be undetectable in supernatants of mid-log-phase cultures containing >1% glucose but abundant in supernatants of cultures made with brain heart infusion (BHI), which contains 0.2% glucose. A 10-fold decrease in the level of abpA mRNA in S. gordonii cells cultured in BHI was noted after the addition of glucose to 1%. Analysis of the abpA sequence revealed a potential catabolite responsive element CRE 153 bp downstream of the putative translational start site. A catabolite control protein A gene (ccpA) homolog from S. gordonii, designated regG, was cloned. A regG mutant strain demonstrated moderately less repression of abpA transcription in the presence of 1% glucose. Diauxic growth with glucose and lactose was not affected in the RegG mutant compared to the wild-type parental strain. These results suggest that while RegG plays a role in abpA expression, other mechanisms of catabolite repression are present.
α-Amylase, the predominant salivary enzyme in humans (1), catalyzes the hydrolysis of α-1,4-glucosidic linkages in starch and binds to oral streptococci, a predominant group comprising the oral microflora (9, 10, 25, 28, 30). Amylase binds Streptococcus gordonii through high-affinity protein receptors that cluster around surface cell division sites (31). Bound amylase retains enzymatic activity (9, 29) and may hydrolyze dietary starch to provide fermentable carbohydrates to the bacteria. This hypothesis is supported by growth studies that found that only organisms expressing the amylase-binding phenotype are able to grow in starch-containing medium and only after preincubation of the cells with salivary amylase (9, 29; J. D. Rogers, R. J. Palmer, Jr., P. E. Kolanbrander, and F. A. Scannapieco, submitted for publication).
A 20-kDa amylase-binding protein (AbpA) mediates the binding of amylase to S. gordonii (9, 25, 31). An abpA mutant of S. gordonii did not grow in starch-containing medium despite preincubation with amylase. While AbpA is found in abundance in spent brain heart infusion (BHI) broth, which contains 0.2% glucose, it is absent in supernatants of cultures grown to mid-log phase in defined medium containing 1% glucose. These results suggested that carbohydrates may influence AbpA expression.
Carbon catabolite repression (CCR) is a regulatory mechanism that allows bacteria to use a set of proteins to metabolize a specific carbohydrate source while down-regulating proteins involved in the utilization of other carbohydrates. CCR in gram-negative bacteria is a positive regulatory mechanism that is mediated by cyclic AMP (cAMP)-dependent and -independent mechanisms (5). In cAMP-dependent regulation, the bacterial phosphoenolpyruvate:sugar phosphotransferase system (PTS) promotes a sequence of phosphoryl transfer events that activates transcription of CCR-sensitive operons. cAMP-independent regulation is mediated by a catabolite repressor and activator protein (Cra) that represses proteins involved in sugar metabolism and activates those involved in substrate oxidation. Gram-positive bacteria such as Bacillus and Streptococcus lack detectable levels of cAMP (5) and rely on cAMP-independent mechanisms of catabolite repression (23, 26, 33).
In Bacillus subtilis, catabolite control protein A (CcpA), a DNA-binding protein (14, 18), is thought to negatively regulate transcription by binding to a catabolite-responsive element (CRE) in the promoter region or within the transcribed gene (6, 16). CcpA binding is modulated by the phosphorylation state of HPr, the heat-stable phosphoryl carrier component of the PTS (24). Genes involved in utilization of starch (14), histidine (36), acetate and acetoin (12), gluconate (11), and xylose (19) are all repressed by CcpA. A second protein, CcpB, whose expression depends on growth conditions (7), is involved in catabolite repression of the gluconate and xylose utilization genes. Finally, CcpC regulates the expression of the citB (aconitase) and citZ (citrate synthase) genes (17).
Diauxic growth has been demonstrated in the oral streptococci, and the PTS has a regulatory role in Streptococcus mutans sugar metabolism (8, 20). Recently, RegM has been described as a CcpA homolog in S. mutans. Interestingly, the activity of RegM does not appear to conform to the model of CCR described for B. subtilis (35). For instance, disruption of regM does not affect diauxic growth of S. mutans in a number of sugars in the presence of glucose, and increased glucose repression was noted for α-galactosidase, mannitol-1-P dehydrogenase, and P-β-galactosidase activities in the regM mutant (32).
To investigate the role of carbon catabolite repression on the expression of abpA, culture supernatant levels of AbpA were compared after growth with different carbohydrate sources. Also, a ccpA homolog designated regG was identified in S. gordonii, and the effects of insertional inactivation of this gene on the expression of abpA were determined.
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Streptococci were routinely cultured in a defined medium (FMC) (34) or in BHI for various time periods at 37°C without shaking in a candle jar. Broth media were supplemented to 1% with glucose, maltose, sucrose, lactose, or maltooligosaccharides. Escherichia coli strains were grown under aerobic conditions with shaking for 12 to 16 h at 37°C in Luria-Bertani (LB) broth and maintained on LB agar. Strains containing recombinant clones were plated on LB agar containing erythromycin (300 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80dlacZΔN15Δ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ thi-1 gyrA96 relA1 | Gibco-BRL |
| S. gordonii Challis | Wild type; reference strain; amylase binding | 11 |
| S. gordonii Challis- CE1 | ΔregG::ermAM Emr | This study |
| Plasmids | ||
| pGEM-T | Promega | |
| pTS19E | ermAM Emr Amr | 37 |
| pCRegG-7 | regG Amr Kmr | This study |
| pCReg-E1 | regG::ermAM Amr Kmr Emr | This study |
| pCReg-E2 | regG::ermAM Kmr Emr | This study |
DNA and RNA manipulations.
Standard procedures were used for plasmid extraction from E. coli (3). DNA was prepared from S. gordonii as previously described (25). Total RNA was isolated from S. gordonii cells grown to the mid-logarithmic phase resuspended in 300 μl of diethyl pyrocarbonate-treated distilled H2O (followed by 900 μl of Trizol reagent (Gibco-BRL) using the FastRNA Blue system (Bio 101, Inc, Vista, Calif.). RNA concentration and purity was determined using standard methods (27).
Influence of carbohydrate source on abpA expression.
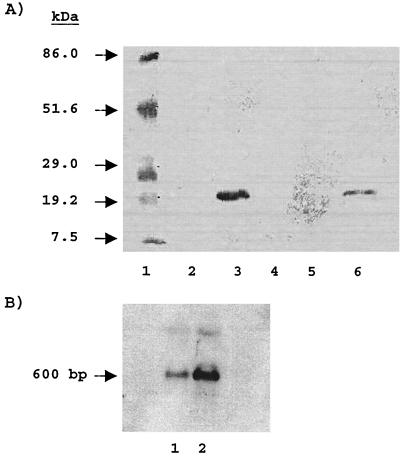
S. gordonii Challis was cultured to mid-log phase (∼8 to 10 h) in FMC supplemented with various sugars. AbpA was detected using a solid-phase amylase ligand-binding assay (9, 13). Relative concentrations of bands were quantitated using a GS 300 scanning densitometer (Hoefer). AbpA was nearly undetectable in the supernatants of bacteria grown to mid-log phase in FMC supplemented with glucose, sucrose, maltose (Fig. 1A, lanes 2, 4, and 5 respectively), or lactose. AbpA was detected when the cells were cultured with maltooligosaccharides (Fig. 1A, lane 6). Growth in unsupplemented BHI resulted in the recovery of ∼50-fold-greater amounts of AbpA than growth in BHI supplemented with 1% glucose. Northern blots probed with biotinylated abpA also demonstrated a large decrease in the abpA transcript when cells were grown in glucose-supplemented BHI (Fig. 1B).
FIG. 1.
(A) Influence of carbohydrate source on AbpA expression. A solid-phase amylase ligand-binding assay was carried out with culture supernatants (100 μg per lane) of S. gordonii Challis grown in defined medium with 1% concentrations of the following sugars: glucose (lane 2), lactose (lane 4), maltose (lane 5), and maltooligosaccharides (lane 6). Lane 1 contains molecular mass standards, and lane 3 is a positive control (BHI with no added sugar). The blot was first probed with purified amylase followed by rabbit antiamylase. Bound antibodies were visualized using goat-anti rabbit immunoglobulin G conjugated to an alkaline phosphatase reporter. (B) Northern blot of cells grown in BHI supplemented with 1% glucose (lane 1) or BHI with no added sugar (positive control) (lane 2). The Northern blot was probed with biotinylated abpA.
Time course of abpA transcription.
BHI (10 ml) was inoculated with 100 μl of an overnight BHI culture of S. gordonii Challis. Cells were incubated (to an optical density of 0.5 at 600 nm), and 500 μl was then removed. Glucose was then added to the cultures to a final concentration of 1%, and 500-μl aliquots were removed at 2, 5, 10, and 15 min. Total RNA was isolated from each sample and analyzed. Biotinylated abpA was used to probe Northern blots (27). Cultures that did not have glucose added were monitored in parallel to control for potential mRNA degradation during processing.
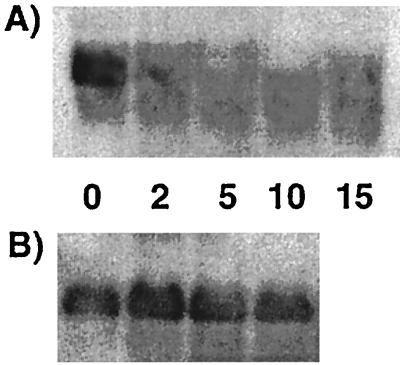
During mid-logarithmic phase in BHI, large amounts of the 600-bp abpA transcript were obtained from cells (Fig. 2A, time zero). The levels of abpA mRNA were decreased 2 min after glucose was added to 1% and remained depressed over the 15-min course of the experiment. No differences in abpA mRNA levels were noted at any of the time points in a control experiment in which BHI was added instead of glucose to the growing cells (Fig. 2B), suggesting that glucose induced the changes seen in the abpA transcript levels.
FIG. 2.
Time course study of abpA mRNA transcript levels. (A) Total RNA (5 μg) was isolated from S. gordonii at mid-logarithmic growth (lane 1) and at 2 min (lane 2), 5 min (lane 3), 10 min (lane 4), and 15 min (lane 5) after the addition of 1% glucose to the culture. (B) Total RNA from parallel cultures grown without additional glucose is included for comparison. Biotinylated abpA was used as the probe.
Analysis of the S. gordonii abpA gene.
cis-acting CREs are nucleotide sequences that mediate CCR for several genes in a variety of bacteria (33). A consensus CRE sequence for the binding of catabolite control proteins was identified by comparing the binding sites for genes known to be regulated by CCR (16). A potential major CRE (CRE 1) 153 bp downstream of the translational start site of the abpA gene was identified (AGTAACCGAATACTT). This is not unusual, since CREs show variability of their positions relative to the transcriptional and translational start sites of regulated genes (15). The abpA CRE shows conservation of bases at 13 of 14 positions with the derived consensus sequence. An additional CRE (CRE 2) at position +109 relative to the translational start site was also identified (CGACGGCGCATACT). The nucleotide sequence of this cre showed conservation at 10 of 14 bases.
Cloning and sequencing of an S. gordonii ccpA homolog.
S. gordonii genomic DNA was used as the template for PCRs using primers based on regions conserved between a hypothetical ccpA homolog from S. mutans (GenBank accession no. AF001316) and a potential ccpA homolog identified from the unfinished Streptococcus pneumoniae genome database (The Institute for Genomic Research website at http://www.tigr.org). The primers ccpA1 (5′-GGAAACAATATGAACACAGACG-3′) and ccpA2 (5′-TTGACGTGTCGTACCACGC-3′) were designed to yield a full-length amplicon of an S. gordonii ccpA homolog. The PCR consisted of a 3-min denaturation at 95°C followed by 35 cycles of 95°C for 1 min, 54°C for 1 min, and 72°C for 1 min. The resulting 1-kb product was cloned into the pGEM-T (Promega) vector, and the nucleotide sequences of both the sense and antisense strands were determined. A search of the incomplete S. gordonii nucleotide sequence database (The Institute for Genomic Research website at http://www.tigr.org) with this sequence revealed a contig containing the 5′ portion of the cloned sequence along with upstream nucleotide sequence data.
The putative ccpA sequence contained an open reading frame 1,002 bp in length preceded by a putative ribosome binding site (AAGGA) 6 bp upstream of a deduced methionine (start) codon. A potential promoter contained −10 (TAATTT) and −35 (GTGTTA) sequences (Fig. 3). This open reading frame, named regG, shared 90% identity with Streptococcus bovis ccpA (GenBank accession number AB28599) and 81% identity with S. mutans regM (32). The deduced amino acid sequence defined a protein of 334 amino acids (molecular weight = 36,606) having 88 and 77% identity with the CcpA of S. bovis and RegM, respectively, and a high degree of similarity to other CcpA proteins and RegM-like regulators (32). The 310-bp partial open reading frame upstream of regG lay on the opposite strand and was preceded by a putative ribosome binding site (GAAGG) located 7 bp upstream. Potential −10 (AAAAAT) and −35 (TTGATT) sequences were also found (Fig. 4). Analysis of the nucleotide and deduced amino acid sequences revealed 84 and 55% identities, respectively, with pepQ and PepQ of S. mutans (32). This gene arrangement is identical to that of the pepQ and ccpA genes of other gram-positive bacteria (21). The nucleotide sequence of regG has been deposited in the GenBank database (accession number AF325223).
FIG. 3.
Sequence analysis of the regG locus. The nucleotide and deduced amino acid sequences are given for regG. The ribosome binding site (RBS) and the −10 and −35 promoter elements for regG are indicated, as is the NdeI restriction site used for insertion of the ermAM gene and inactivation of regG. The nucleotides corresponding to the pepQ partial open reading frame are also indicated, with the putative translational start site underlined.
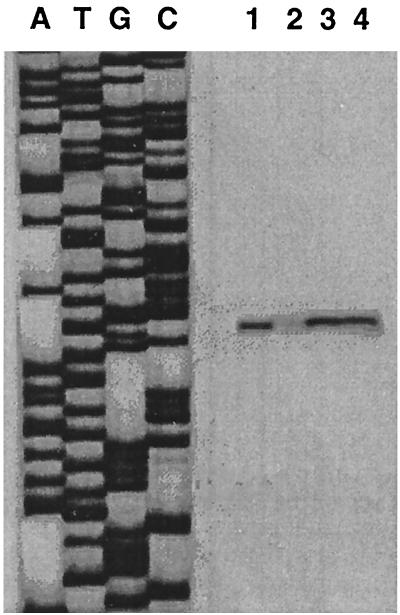
FIG. 4.
Transcriptional regulation of abpA by RegG. Total RNA was extracted from S. gordonii Challis (lanes 1 and 2) and Challis-CE1 (lanes 3 and 4) during exponential growth in BHI (lanes 1 and 3) or in BHI supplemented with 1% glucose (lanes 2 and 4). As a reference, sequencing reactions using the same primer (PE1) were performed (left).
Inactivation of regG.
The ermAM gene from pTS19E (37) was restricted with PvuII, the 2-kb ermAM fragment gel purified on 1% agarose, and the blunt-ended fragment was cloned into the NdeI site of the regG gene in pCRegG. The resulting plasmid was designated pCRegG-E1. A BsaI/ScaI double digest of pCR2KB-E1 was used to cleave a 415-bp portion of the ampicillin gene from the plasmid. The 6.6-kb fragment was purified on 0.8% agarose gels, excised, religated, and designated pCRegG-E2. This vector was constructed to avoid introduction of ampicillin resistance into S. gordonii. pCRegG-E2 was transformed into E. coli DH5α and transformants were initially selected on erythromycin (300 μg/ml) containing LB agar plates. pCRegG-E2, which contained the ermAM gene inserted 200 bp downstream of the translational start site of regG, was linearized by restriction with AspI and gel purified prior to transformation into S. gordonii Challis for 2 h with shaking (120 rpm) at 37°C; aliquots were then plated on BHI agar supplemented with erythromycin (5 μg/ml) and incubated at 37°C overnight in a candle jar (2).
Primers specific for the 5′ (5′-TGATGAAGCTACTGATGC-3′) and 3′ (5′-ATCACTGAACCAATGGCC-3′) ends of regG were used to amplify products from genomic DNA isolated from the wild type and selected erythromycin-resistant mutant strains of S. gordonii. Following an initial denaturation of 4 min at 94°C, 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C were executed, after which an extension time of 10 min at 72°C was added. A 3.0-kb PCR product from genomic DNA from an integrant strain designated Challis-CE1, verified by Southern hybridization using standard methods, confirmed that this mutant was the result of a double-crossover event and contained a disrupted copy of the regG gene. No obvious differences were noted between the wild-type and mutant strains with respect to colony morphology on blood and mitis salivarius agars, and biochemical metabolic profiles using the API Rapid Strep kit (bioMerieux Vitek).
Diauxic growth of S. gordonii Challis-CE1.
Growth of wild-type S. gordonii Challis on sugars in the presence of glucose was compared to that of the regG mutant (data not shown). Cells from overnight cultures in defined medium containing 0.5% glucose were harvested, washed twice in sugar-free medium, and added to fresh medium containing either glucose (6 mM) and/or a secondary carbohydrate (6 mM) to a standard optical density of 0.1 at 600 nm. Cultures were incubated at 37°C, and samples were taken every 30 min in order to monitor growth spectrophotometrically.
The regG mutant showed a slightly reduced growth rate compared to the wild-type strain. Diauxic growth curves were obtained when the wild type was grown in medium containing both lactose and glucose, suggesting catabolite repression of the enzymes responsible for utilization of lactose. The regG mutant displayed identical growth patterns on the same sugars as the wild type (data not shown), indicating that the enzymes responsible for utilization of lactose were still catabolite repressed by glucose even in the absence of RegG. These results are in agreement with similar studies involving RegM of S. mutans (32) and suggest that repressors other than RegG are involved in the regulation of expression of enzymes involved in glucose utilization.
Transcriptional regulation of abpA.
Total RNA from mid-exponential-phase cells was used throughout these studies. Primer extension products were obtained by using oligonucleotide PE1 (5′-GGTAGTATGCGCCGACG-3′) corresponding to the complement of abpA nucleotide positions 102 to 118 or PE2 (5′-CAGCTACTACTGCTGG-3′) corresponding to positions 205 to 220. The oligonucleotides were end labeled with [γ-32P]ATP and T4 polynucleotide kinase, and the reverse transcriptase reaction was performed according to standard protocols (27). The extension products were resolved on polyacrylamide sequencing gels and visualized by autoradiography. As a reference, sequencing reactions were performed by the dideoxy-chain termination method using the same primers as in the primer extension experiments, with pCR2KB serving as the sequence template.
Transcription was initiated 64 bp upstream of the translational start site of abpA. Comparison of transcription levels of abpA for the wild-type organism without and with glucose are shown in Fig. 4 (lanes 1 and 2, respectively). The level of transcription of abpA was nearly undetectable in the presence of the repressing sugar. Examination of transcription of abpA in the regG mutants resulted in a modest reduction in the repression of abpA in the presence of glucose (Fig. 4, lane 4). These results also demonstrate that the position of the transcriptional start site is unaffected by RegG.
Discussion.
CcpA, a LacI/GalR-type trans-acting regulatory protein, mediates CCR in gram-positive bacteria with a low GC content (33). A number of ccpA homologs have been described for Bacillus, Lactobacillus, and Streptococcus spp. (14, 33). RegG of S. gordonii appears to be another CcpA homolog that is closely related to RegM of S. mutans (32). In both cases, a prolidase gene is located upstream of the catabolite control protein gene, and inactivation of regG or regM does not appear to affect glucose-mediated diauxy.
Streptococci are numerically prominent in the oral cavity, and these organisms may be pathogenic in certain hosts (4, 22). High proportions of amylase-binding streptococci are found in supragingival plaque formed immediately after tooth cleaning, and salivary proteins may play an important role in the adhesion of these bacteria to the tooth surface (30). Only host species having measurable salivary amylase activity harbor amylase-binding streptococci on their teeth. These findings provide further evidence that the ability to bind amylase is an important colonization determinant for amylase-binding streptococci.
Amylase may function in streptococcal colonization by supporting bacterial nutrition. Functional amylase on the bacterial surface could hydrolyze dietary starch to oligosaccharides that could be transported into the bacteria and metabolized for energy needs (9, 29). The observation that abpA is regulated by RegG represents the first example of an oral streptococcal extracellular protein that is regulated by a catabolite repression mechanism. This finding further supports a role for AbpA in starch metabolism. AbpA would be expected to be up-regulated under conditions of glucose depletion so that dietary starch would be optimally catabolized by bacterium-bound salivary amylase.
Acknowledgments
This work was supported by grant DE 09838 (F.A.S.) and Institutional Dentist Scientist Award DE 00158 (J.D.R.) from the National Institute of Dental and Craniofacial Research.
We are grateful to Howard K. Kuramitsu and Elaine M. Haase for helpful discussions throughout the course of this work.
REFERENCES
- 1.Aguirre A, Levine M J, Cohen R E, Tabak L A. Immunochemical quantitation of alpha-amylase and secretory IgA in parotid saliva from people of various ages. Arch Oral Biol. 1987;32:297–301. doi: 10.1016/0003-9969(87)90024-0. [DOI] [PubMed] [Google Scholar]
- 2.Behnke D. Plasmid transformation of Streptococcus sanguis (Challis) occurs by circular and linear molecules. Mol Gen Genet. 1981;182:490–497. doi: 10.1007/BF00293940. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochud P Y, Eggiman P, Calandra T, Van Melle G, Saghafi L, Francioli P. Bacteremia due to viridans streptococcus in neutropenic patients with cancer: clinical spectrum and risk factors. Clin Infect Dis. 1994;18:25–31. doi: 10.1093/clinids/18.1.25. [DOI] [PubMed] [Google Scholar]
- 5.Botsford J L, Harman J G. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burne R A, Wen Z T, Chen Y Y, Penders J E. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J Bacteriol. 1999;181:2863–2871. doi: 10.1128/jb.181.9.2863-2871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauvaux S, Paulsen I T, Saier M H., Jr CcpB, a novel transcription factor implicated in catabolite repression in Bacillus subtilis. J Bacteriol. 1998;180:491–497. doi: 10.1128/jb.180.3.491-497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cvitkovitch D G, Boyd D A, Hamilton I R. Regulation of sugar transport via the multiple sugar metabolism operon of Streptococcus mutans by the phosphoenolpyruvate phosphotransferase system. J Bacteriol. 1995;177:5704–5706. doi: 10.1128/jb.177.19.5704-5706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas C W. Characterization of the alpha-amylase receptor of Streptococcus gordonii NCTC 7868. J Dent Res. 1990;69:1746–1752. doi: 10.1177/00220345900690110701. [DOI] [PubMed] [Google Scholar]
- 10.Douglas C W, Pease A A, Whiley R A. Amylase-binding as a discriminator among oral streptococci. FEMS Microbiol Lett. 1990;54:193–197. doi: 10.1016/0378-1097(90)90281-t. [DOI] [PubMed] [Google Scholar]
- 11.Fujita Y, Miwa Y. Catabolite repression of the Bacillus subtilis gnt operon mediated by the CcpA protein. J Bacteriol. 1994;176:511–513. doi: 10.1128/jb.176.2.511-513.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundy F J, Turinsky A J, Henkin T M. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J Bacteriol. 1994;176:4527–4533. doi: 10.1128/jb.176.15.4527-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gwynn J P, Douglas C W. Comparison of amylase-binding proteins in oral streptococci. FEMS Microbiol Lett. 1994;124:373–379. doi: 10.1111/j.1574-6968.1994.tb07311.x. [DOI] [PubMed] [Google Scholar]
- 14.Henkin T M. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 15.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 16.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 17.Jourlin-Castelli C, Mani N, Nakano M M, Sonenshein A L. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J Mol Biol. 2000;295:865–878. doi: 10.1006/jmbi.1999.3420. [DOI] [PubMed] [Google Scholar]
- 18.Kim J H, Voskuil M I, Chambliss G H. NADP, corepressor for the Bacillus catabolite control protein CcpA. Proc Natl Acad Sci USA. 1998;95:9590–9595. doi: 10.1073/pnas.95.16.9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus A, Hueck C, Gartner D, Hillen W. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J Bacteriol. 1994;176:1738–1745. doi: 10.1128/jb.176.6.1738-1745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberman E S, Bleiweis A S. Role of the phosphoenolpyruvate-dependent glucose phosphotransferase system of Streptococcus mutans GS5 in the regulation of lactose uptake. Infect Immun. 1984;43:536–542. doi: 10.1128/iai.43.2.536-542.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahr K, Hillen W, Titgemeyer F. Carbon catabolite repression in Lactobacillus pentosus: analysis of the ccpA region. Appl Environ Microbiol. 2000;66:277–283. doi: 10.1128/aem.66.1.277-283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martino R, Manteiga R, Sanchez I, Brunet S, Sureda A, Badell I, Argiles B, Subira M, Bordes R, Domingo-Albos A. Viridans streptococcal shock syndrome during bone marrow transplantation. Acta Haematol. 1995;94:69–73. doi: 10.1159/000203976. [DOI] [PubMed] [Google Scholar]
- 23.Monedero V, Gosalbes M J, Perez-Martinez G. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J Bacteriol. 1997;179:6657–6664. doi: 10.1128/jb.179.21.6657-6664.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reizer J, Romano A H, Deutscher J. The role of phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, in the regulation of carbon metabolism in gram-positive bacteria. J Cell Biochem. 1993;51:19–24. doi: 10.1002/jcb.240510105. [DOI] [PubMed] [Google Scholar]
- 25.Rogers J D, Haase E M, Brown A E, Douglas C W, Gwynn J P, Scannapieco F A. Identification and analysis of a gene (abpA) encoding a major amylase-binding protein in Streptococcus gordonii. Microbiology. 1998;144:1223–1233. doi: 10.1099/00221287-144-5-1223. [DOI] [PubMed] [Google Scholar]
- 26.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Scannapieco F A, Bergey E J, Reddy M S, Levine M J. Characterization of salivary α-amylase binding to Streptococcus sanguis. Infect Immun. 1989;57:2853–2863. doi: 10.1128/iai.57.9.2853-2863.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scannapieco F A, Bhandary K, Ramasubbu N, Levine M J. Structural relationship between the enzymatic and streptococcal binding sites of human salivary α-amylase. Biochem Biophys Res Commun. 1990;173:1109–1115. doi: 10.1016/s0006-291x(05)80900-3. [DOI] [PubMed] [Google Scholar]
- 30.Scannapieco F A, Solomon L, Wadenya R O. Emergence in human dental plaque and host distribution of amylase-binding streptococci. J Dent Res. 1994;73:1627–1635. doi: 10.1177/00220345940730100701. [DOI] [PubMed] [Google Scholar]
- 31.Scannapieco F A, Haraszthy G G, Cho M I, Levine M J. Characterization of an amylase-binding component of Streptococcus gordonii G9B. Infect Immun. 1992;60:4726–4733. doi: 10.1128/iai.60.11.4726-4733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson C L, Russell R R. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect Immun. 1998;66:2085–2092. doi: 10.1128/iai.66.5.2085-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stulke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 34.Terleckyj B, Willett N P, Shockman G D. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun. 1975;11:649–655. doi: 10.1128/iai.11.4.649-655.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobisch S, Zuhlke D, Bernhardt J, Stulke J, Hecker M. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J Bacteriol. 1999;181:6996–7004. doi: 10.1128/jb.181.22.6996-7004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wray L V, Jr, Fisher S H. Analysis of Bacillus subtilis hut operon expression indicates that histidine-dependent induction is mediated primarily by transcriptional antitermination and that amino acid repression is mediated by two mechanisms: regulation of transcription initiation and inhibition of histidine transport. J Bacteriol. 1994;176:5466–5473. doi: 10.1128/jb.176.17.5466-5473.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita Y, Takehara T, Kuramitsu H K. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J Bacteriol. 1993;175:6220–6228. doi: 10.1128/jb.175.19.6220-6228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]