Abstract
Current inflammatory bowel disease (IBD) treatments including non-biological, biological, and nutritional therapies aim to achieve remission and mucosal healing. Treatment efficacy, however, is highly variable, and there is growing evidence that the gut microbiota influences therapeutic efficacy. The aim of this study was to conduct a systematic review and meta-analysis to define changes in the gut microbiota following IBD treatment and to identify microbial predictors of treatment response. A systematic search using MEDLINE/Embase and PubMed was performed in July 2022. The review was conducted based on the Preferred Reporting Items for Systematic reviews and Meta-Analyses guidelines. Studies were included if they reported longitudinal microbiota analysis (>2 weeks) using next-generation sequencing or high-throughput sequencing of faecal/mucosal samples from IBD patients commencing treatment. Meta-analysis on alpha-diversity changes following infliximab treatment was conducted. Thirty-nine studies met the inclusion criteria, and four studies were included in the meta-analysis. An increase in alpha diversity was observed following treatment with 5-aminosalicylates, corticosteroids, and biological therapies in most studies. Characteristic signatures involving the enrichment of short-chain-fatty-acid-producing bacteria including Faecalibacterium prausnitzii and a reduction of pathogenic bacteria including various Proteobacteria were demonstrated following treatment with specific signatures identified based on treatment outcome. The meta-analysis demonstrated a statistically significant increase in bacterial richness following infliximab treatment (standardised mean difference −1.16 (−1.50, −0.83), p < 0.00001). Conclusion: Distinct microbial signatures are seen following treatment and are associated with treatment response. The interrogation of large longitudinal studies is needed to establish the link between the gut microbiota and IBD therapeutic outcomes.
Keywords: inflammatory bowel disease, Crohn’s disease, ulcerative colitis, therapeutics, gut micro-biota
1. Introduction
Inflammatory bowel diseases (IBDs), of which the main subtypes are ulcerative colitis (UC) and Crohn’s disease (CD), are chronic immune-mediated inflammatory conditions that require lifelong treatment [1]. The natural history of IBD is that of quiescent disease interspersed with flares in disease activity with significant socio-economic implications [2,3].
Current drug treatment strategies, which include 5-aminosalicylates (5-ASA), corticosteroids, and immunomodulators, aim to achieve remission and mucosal healing through reducing the inflammatory burden during active disease [4]. However, they do not specifically target the cause of the inflammation. Growing understanding of the complex multifactorial aetiopathogenesis of IBD has led to the development and use of more targeted therapies, specifically biological agents (anti-TNF-alpha antibodies, anti-cytokine monoclonal antibodies, and anti-integrin monoclonal antibodies) to treat moderate to severe disease [5,6,7,8].
Anti-TNF-alpha agents, including infliximab, are often first-line in the majority of biologic naïve IBD patients [1]; however, the efficacy and tolerability of these therapies are highly variable among patients. It is estimated that 33–50% of patients, especially in the elderly population, discontinue anti-TNF therapy, with a further 13–20% of them experiencing yearly loss of response [9,10,11]. Nutritional therapies including exclusive enteral nutrition (EEN), probiotics, prebiotics, and a specific carbohydrate diet (SCD) aim to induce and maintain disease remission, but mechanistic insight into the mode of action of nutritional therapy remains in its infancy.
IBD is thought to arise from defective immune regulation, producing an abnormal response to luminal antigens, which enter the intestinal wall via a leaky epithelium, thus triggering inflammation in susceptible individuals [12,13]. IBD risk variants often implicate pathways for microbial recognition or handling/clearance strategies. However, the rapid increase in disease burden in the last 100 years refutes the significance of genetic risk and elevates the importance of other environmental factors, which have changed more dramatically in line with disease prevalence.
The healthy human gut microbiota consists of four major bacteria phyla (Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria) with the gut microbiota composition shaped by genetics, ethnicity, environmental exposures, diet, and lifestyle factors [14,15,16]. Changes or imbalances in the gut microbiota, termed dysbiosis, have been consistently associated with IBD [17,18,19]. Studies have shown that dysbiosis imparts both community and functional changes, which drive a proinflammatory state [16].
The most-consistent finding in IBD, in terms of gut microbiota changes, is decreased bacterial diversity/richness, also referred to as alpha diversity. From a taxonomic perspective, IBD is associated with a reduction in members of the Firmicutes phyla, including beneficial bacterial species such as Faecalibacterium prausnitzii, and an increase in pathogenic proinflammatory species, including members of the Enterobacteriaceae including certain species of Escherichia coli and Bacteroidetes [20,21]. In CD specifically, reductions in Roseburia and Faecalibacterium genera and an increase in Ruminococcus gnavus have been consistently reported [22,23]. Specific features of UC-associated dysbiosis are not so consistent, although increases in sulphate-reducing bacteria have been observed [24,25]. Faecal microbiota transplant (FMT) as a successful UC treatment and FMT colitis mouse models highlight and support the central role of the gut microbiota in IBD pathogenesis [26,27].
Recent studies have established the ability of gut microbes to alter the efficacy of various therapeutics including cardiac drugs (digoxin), L-dopa treatment for Parkinson’s disease [28,29], and cancer treatments [30,31]. For example, a distinct baseline gut microbiota enriched with F. prausnitzii and other Firmicutes was associated with a beneficial clinical response to immune checkpoint inhibitor, ipilimumab, in the treatment of metastatic melanoma [31]. This key finding was corroborated by Gopalakrishnan and colleagues, who analysed the gut microbiome of melanoma patients undergoing PD-1 immunotherapy in which characteristic differences were observed in terms of the diversity and composition of the gut microbiome in responders versus non-responders [32]. These studies demonstrated the relationship between gut microbes and the efficacy of pharmacotherapies and highlighted the promising potential of the gut microbiota as a predictor or modulator of treatment response.
Similarly, in IBD, the potential role of the gut microbiota as a predictive biomarker for treatment response has been an area of intense research. It is currently unclear whether microbiota analysis is superior to existing parameters including C-reactive protein, erythrocyte sedimentation rate, the endoscopic disease activity index, and pathologic findings. Several recent longitudinal studies have analysed the impact of 5-ASA, corticosteroids, biological therapies, and nutritional therapies on the gut microbiota in IBD patients and their association with clinical outcomes [33,34,35,36,37,38,39,40,41,42]. Most studies have identified associations between unique microbial changes during treatments and favourable disease outcomes. However, whether these changes are sustained or treatment-specific remains unknown. Despite the growing number of studies on this topic, there has been no systematic review undertaken to date that has comprehensively summarised the impact of IBD therapeutics including nutritional therapies on the gut microbiota and, conversely, how the microbiota is related to treatment outcomes. The ability to predict treatment responses using personalised gut microbiota signatures would enable targeted, effective, and highly personalised treatment for optimal health outcomes for IBD patients. Therefore, the aims of this systematic review and meta-analysis were: (i) to define changes in the gut microbiota following IBD treatment and (ii) to identify microbial predictors of treatment response.
2. Materials and Methods
2.1. Search Strategy
This systematic review was registered at inception on PROSPERO, an international prospective register of systematic reviews (registration ID: CRD42021244046; 22 April 2021). The study was conducted based on the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) Guidelines [43]. A search of the online bibliographic databases PubMed (includes MEDLINE) and Embase was performed by two independent researchers (C.M. and G.L.) in April 2021 and updated in July 2022 (C.M. and T.J.). No restrictions were placed on the publication period. The following Medical Subject Heading terms were used, which included both the root terms and text words: inflammatory bowel disease, Crohn’s disease, ulcerative colitis, gastrointestinal microbiome, intestinal flora, dysbiosis, biological therapy, tumour necrosis factor antibody, infliximab, adalimumab, golimumab, certolizumab pegol, vedolizumab, ustekinumab, natalizumab, 5-aminosalicylic acid, mesalamine, sulfasalazine, corticosteroids, prednisone, budesonide, immunomodulation, immunomodulator, azathioprine, methotrexate, mercaptopurine, anti-bacterial agents, antibiotics, faecal microbiota transplantation, enteral nutrition, probiotic agent, prebiotic agent, specific carbohydrate diet. Synonyms and word variations were combined using the AND and OR functions. Manual searches of the reference list from potentially relevant review articles, published editorials, and retrieved original studies were performed to identify additional studies that may have been missed using the computer-assisted search strategy. The Covidence systematic review software was used during the screening process to identify the most-relevant papers.
2.2. Study Selection and Eligibility Criteria
Randomised controlled trials (RCTs), cohort studies, and observational studies reporting on microbiota changes in patient faecal samples and/or mucosal biopsies were evaluated. Eligible studies were included in this systematic review if they met all the inclusion criteria. Studies that reported duplicated results or where data could not be extracted were excluded.
Studies involving adults or children (>5 years old) previously diagnosed with IBD using clinical, endoscopic, and/or pathologic features and using a commonly accepted method to assess disease progression and outcomes were eligible for inclusion. Studies were included if subjects on IBD therapy were followed up for at least two weeks with microbial analysis of faecal or biopsy samples performed at baseline and the endpoint using next-generation or high-throughput sequencing. Microbial analysis reporting was required to provide information on the presence or abundance of microbial taxa.
Studies were excluded if they performed solely culture-based analysis, did not report on patterns of individual bacterial taxa differences, or if the microbial analysis did not include information on the baseline status prior to therapy. Additionally, case reports, studies of fewer than ten patients, studies focusing on children <5 years old, conference articles, review articles, animal studies, studies that did not provide information on antibiotic usage, or studies reported in languages other than English were excluded.
2.3. Outcome Assessment
The primary outcomes were (i) changes in the gut microbiome of IBD patients following commencement of treatment with biological therapies, 5-ASA, corticosteroids, antibiotics, or nutritional therapies and (ii) the identification of microbial predictors of treatment response.
2.4. Data Extraction and Analysis
The Newcastle and Ottawa scale (NOS) was used to assess the quality of the studies [44] Cohort studies were scored across 3 categories: selection (4 questions) and comparability (1 question) of study groups and ascertainment of the outcome of interest (3 questions) with all questions with a score of 1, except for the comparability of study groups, in which separate points were awarded for adjusting for confounders (maximum of 2 points). An aggregate score of 8 or greater was suggestive of a high-quality study. Different scales were used to assess cohort studies and RCTs.
2.5. Data Synthesis and Statistical Methods
Information from the included studies was extracted into a Microsoft Excel spreadsheet. Where available, the following clinical data were extracted for each trial: study type, country of origin, diagnosis, age, duration of disease, intervention and its dosage, duration of intervention, sample types, length of follow-up, method of microbiota composition analysis, and microbiota changes. Studies were eligible for the meta-analysis if more than two studies shared the same outcome and had reported usable data in a compatible metric. Revman5 (Review Manager (RevMan) (Computer program), Version 5.4.1, The Cochrane Collaboration, 2020) was used to conduct the meta-analysis. A meta-analysis of four studies involving 82 patients looking at the mean differences in alpha diversity measured by the Shannon index at baseline and after exposure to biologic treatment with anti-TNF-alpha (infliximab) was undertaken. Shannon index values were analysed using an inverse variance model with a 95% confidence interval. Values are reported as standardised mean differences (SMDs). p-values were two-tailed, and statistical significance was set at p < 0.05. The I2 test was used to quantify heterogeneity estimates, i.e., <30%, 30–59%, 60–75%, and >75% defined the thresholds for low, moderate, substantial, and considerable heterogeneity, respectively. When statistically significant heterogeneity was not present, pooled estimates (95% CI) were calculated using the fixed-effects model. If significant heterogeneity was present, pooled estimates (95% CI) were calculated using the random-effects model [45,46,47].
3. Results
3.1. Search Results
The search strategy identified 10,008 references that were imported for screening, and 1266 duplicates were removed. Three reviewers (C.M., G.L., and T.S.) independently performed an initial screen of the titles and abstracts based on the eligibility criteria, with 8479 articles excluded at this stage. Then, 263 studies were assessed for full-text eligibility. Of these, 205 were excluded, and of these, 95 were conference proceedings, 19 full texts that were not retrievable, 16 were review articles/study protocols, 20 reported on the microbiota without using high-throughput sequencing, 12 had insufficient microbial data, 11 lacked longitudinal data, 16 had insufficient data on intervention, 3 had the wrong intervention, 2 had the wrong outcomes, 1 included fewer than six patients, 1 did not specify treatment, 3 had the wrong patient population, and 1 was found to be a duplicate. Disagreements between reviewers were resolved by consensus with third-party experts (G.H. and S.L.). Twenty-eight studies on faecal microbiota transplantation were out of the scope of this systematic review and, therefore, excluded. Thirty-nine studies met the eligibility criteria and were included in the analysis (Figure 1).
Figure 1.
PRISMA diagram showing the flow chart of the included studies.
3.2. Description of Included Studies
The characteristics of the studies involving biological therapies, non-biological therapies, and nutritional therapies stratified by disease subtypes (studies with CD patients, UC patients, or both) are summarised in Table 1. All thirty-nine studies were prospective in nature, with thirty-four cohort studies and five RCTs. All studies reported the impact of IBD therapeutics on alpha diversity and/or taxonomic changes by conducting microbial assessment at baseline prior to therapy initiation and at least two weeks post therapy exposure. Some studies also reported longer-term outcomes of more than a year [42,48,49]. Nineteen studies involved only CD patients; five studies involved only UC patients; fifteen studies included both UC and CD patients. Twenty-one studies focused on adult subjects, while eighteen studies evaluated paediatric cohorts. Twenty studies reported on biological therapies: anti-TNF-alpha therapies (infliximab, adalimumab, etanercept, golimumab; n = 18), anti-integrin antibody therapy (vedolizumab; n = 2), and anti-IL-12/23 monoclonal antibody therapy (ustekinumab; n = 1) (Table 1). One study assessed the impact of 5-ASAs, two corticosteroids, two the impact of antibiotics, and one study the impact of azathioprine (Table 1). There were fourteen studies reporting on nutritional therapies, specifically exclusive enteral nutrition (EEN; n = 8), specific carbohydrate diet (SCD; n = 2), prebiotics (n = 2), adsorptive granulomonocytapheresis (GMA; n = 1), and dietary intervention (n = 1).
Quality assessment using the NOS for observational cohort studies and RCTs was undertaken (Supplementary Tables S1 and S2). Scores of 0–3, 4–6, and 7–9 were considered as low, moderate, and high quality, respectively. Of the cohort studies, fifteen scored the maximum 9 points, twelve scored 8 points, four scored 7 points, and three scored 6 points. All except three studies were deemed to be of high quality. Olbjorn et al. did not include information confirming that the use of the IBD treatment of interest was not present at the start of the study [50]. Valcheva et al. and Suskind et al. did not include information on the selection of the non-exposed cohort [51,52]. Similarly, no study control factors involving the IBD subjects were documented in the three moderate-quality studies. Of the RCTs, two studies attained 9 points, two studies scored 7, and one study scored 6 points, meaning four articles were deemed to be of high quality, while the remaining one was of moderate quality. The study of moderate quality did not have information on the representativeness of the controls and was not blinded, and the patients were recruited in a hospital setting [34].
Table 1.
Study characteristics of the studies that included CD patients only, UC patients only, or both, stratified by biological therapies, non-biological therapies, and nutritional therapies.
| Disease Subtype | Study | Study Type | Country, Single-(SC) /Multi-Centre (MC) |
Diagnosis (# Subjects) |
Age Group | Age Group | Duration of Disease (in Years) (Mean SD) | Intervention (# of Subjects); Route, Dose |
Length of Intervention (Sampling Points (Weeks)) | Sample Type | Length of Follow-up (Weeks + Sampling Points) | Method of Microbial Composition Analysis | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (Years) Mean (SD) | Age (Years) Median/IQR | ||||||||||||
| Biological Therapies | |||||||||||||
| CD CD |
Wang et al. [39] | Prospective cohort study (PCS) | China, SC |
CD (n = 11) HC (n = 16) |
Paediatric | CD: ND (range 4–17) | ND | ND | Infliximab (IFX; n = 11); IV, 5mg/kg | 0, 2, 6 + every 8 weeks | Faeces | ND | 16S rRNA sequencing analysis (16S SA) |
| Doherty et al. [41] | Randomised controlled trial (RCT) | U.S., MC | CD (n = 306) | Adult | CD: 39 (SEM 13) | ND | CD: 12 (SEM 8.8) | Ustekinumab (UST, n = 232); IV, ND (induction therapy (IT)) UST; SC, 270 mg or 90 mg SC (maintenance therapy (MT)) |
8 weeks (ND; IT) 8 weeks (8, 16; MT) |
Faeces | 0, 4, 6, 22 | 16S SA | |
| Ribaldone et al. [53] | PCS | Italy, SC |
CD (n = 20) | Adult | ND | CD: 52.5 (range 26–69) | CD: 14.5, ND | Adalimumab (ADA, n = 20); ND | 6 months (ND) | Faeces | 0, 26 | Metagenomic (MGS) and 16S SA | |
| Kowalska-Duplaga et al. [54] | PCS | Poland, ND | CD (n = 18) HC (n = 18) |
Paediatric | ND | ND | ND | IFX (n = 18); 5 mg/kg, IT | 0, 2, 6 | Faeces | 0, 14 and 6–8 weeks post drug) | 16S SA | |
| Zhuang et al. [40] | PCS | China, SC |
CD (n = 49) | Adult | CD: 25 (range 18–48) | ND | CD: 2 (median range 0.5–17) | IFX (n = 49); IV, ND | 0, 2, 6, 14, 22, 30 | Faeces | 0, 6, 30 | 16S SA | |
| Salamon et al. [55] | PCS | Poland, SC | CD (n = 61) HC (n = 17) |
Paediatric | CD on IFX: 13.09 (3.76) HC: 11.73 (2.88) |
ND | IFX (n = 13); IV 5 mg/kg | 0, 2, 6 | Faeces | 10 weeks (0, 10); 4 weeks after 3rd dose | qPCR analysis | ||
| Wang et al. [38] | PCS | China, MC | CD (n = 18) HC (n = 12) |
Paediatric | ND | CD: 12 (11,14) HC: 12 (11,13) |
ND | IFX (n = 18); IV 5–10 mg/kg | 0, 2, 6 + every 8 weeks | Faeces | >6 weeks (0, 6 or 30 weeks; after 3rd or 6th dose) | 16S SA | |
| Ventin-Holmberg et al. [56] | PCS | Finland, SC | CD-R (n = 12) CD-NR (n = 18) |
Paediatric | ND | CD-R: 13 CD-NR: 14 (ND) |
Median CD-R: 1.4 CD-NR: 0.3 |
IFX (n = 30), IV, ND | 0, 2, 6 | Faeces | 0, 2, 6 | 16S SA | |
| IBD IBD |
Kolho et al. [43] | PCS | Finland, SC | UC (n = 26) CD (n = 36) IBDU (n = 6) HC (n = 26) |
Paediatric | ND | IBD: 15.5 (range 9.6–18.3) HC: 13.4 (range 9.7–18.3) |
UC, CD, IBDU: 3.5 (median range 0–10.7) | IFX (n = 31); ND ADA (n = 1); ND |
ND | Faeces | >6 weeks (0, 2, 6 + 1 during MT) | Phylogenetic microarray and qPCR analysis |
| Ananthakrishnan et al. [42] | PCS | U.S., SC |
UC (n = 43) CD (n = 42) |
Adult | ND | ND | Remission (n = 31): 9.2 (7.2) No remission (n = 54):15.7 (12.7) |
Vedolizumab (VEDO, n = 85); IV, 300mg infusion | 0, 2, 6 + every 8 weeks | Faeces | 0, 6, 14, 30, 54 | MGS | |
| Zhou et al. [57] | PCS | China, SC |
UC (n = 51) CD (n = 16) HC (n = 73) |
Adult | UC: 41.75 (14.36) CD: 31.81 (12.67) HC: 30.07 (6.36) |
ND | ND | IFX (n = 16); 5 mg/kg | 0, 2, 6, 14, 22, 30 | Faeces | 0, 30 | 16S SA | |
| Aden et al. [58] | PCS | Germany, SC | Discovery Cohort (DC): UC (n = 4), CD (n = 8), HC (n = 19) Validation Cohort (VC): UC (n = 13), CD (n = 10), HC (n = 99) |
Adult | DC- IBD: 45.4 (4.5) HC: 26.37 (0.5) VC; ND |
ND | ND | IFX (n = 10); ND Etanercept (n = 12); ND VEDO (n = 13); ND |
ND | Faeces | 0, 2, 6, 30 0, 2, 6, 14 |
16S SA | |
| Olbjorn et al. [50] | PCS | Norway, MC | HC (n = 70) non-IBD (n = 50) CD (n = 80) UC (n = 27) IBDU (n = 3) |
Paediatric | ND | ND | ND | Immunomodulators (n = 98); ND Anti-TNF therapy (n =6 4); ND |
ND | Faeces | 0, 78 | 16S SA | |
| Dovrolis et al. [59] | PCS | Greece, SC | UC (n = 6) CD (n = 14) HC (n = 9) |
Adult | ND | ND | ND | IFX (n = 14); IV, 5 mg/kg | 0, 2, 6 + every 8 weeks | Mucosal Biopsies | 12–20 weeks (0, 12–20) | qPCR and 16S SA | |
| Ding et al. [48] | PCS | ND | CD (n = 76) UC (n = 10) HC (n = 13) |
Adult | ND | CD: 38.49 14.61 (19–82) UC: 25.52 17.34 (19–70) HC: 30.5 2.94(27–35) |
ND | IFX (n = 66); ND (IT) ADA (n = 10); ND (IT) |
ND | Faeces | 16 months (3 monthly) | 16S SA | |
| Schierova et al. [60] | PCS | Czech Republic, SC | CD (n = 34, with 17 at endpoint (EP)) UC (n = 18, with 10 at EP) HC (n=37) |
Adult | CD: 35.0 (26.5, 44.0) UC: 31.0 (26.0, 41.3) HC: 36.5 (28.8, 41.3) |
ND | CD: 26 (22.5, 35) UC: 27 (21.5, 31) |
Anti-TNF therapy (CD (n = 17), ND UC (n = 10), ND) |
38 | Faeces | 0, 2, 8, 14, 20, 26, 32, 38) | 16S SA | |
| Sanchis-Artero et al. [61] | Prospective observational study | Spain, MC |
CD (n = 27) HC (n = 16) |
Adult | CD: 41.4 (17.4) HC: 29.3 (7.2) |
ND | ND | Anti-TNF therapy (n = 27), ND | 26 | Faeces | 0, 13, 26 | 16S SA | |
| Ventin-Holmberg et al. [49] | PCS | Finland, SC | CD (n = 25, with 19 at EP) UC (n = 47 with 33 at EP) |
Adult | ND | 31 (24–45) | Median 2 (0–7) | IFX (n = 72, ND, ND) | 52 | Faeces | 0, 2, 6, 12, 52 | 16S SA | |
| Effenberger et al. [62] | Prospective comparative study | Austria, SC | CD (n = 24) UC (n = 12) |
Adult | CD: 33.9 (+/−12.9) UC: 44.1 (+/−14.5) |
ND | ND | anti-TNF therapy | 30 | Faeces | 0, 12, 30 | 16S SA | |
| Park et al. [63] | PCS | South Korea, MC | CD (n-10) UC (n = 9) HC (n = 19) |
Adult | ND | IBD: 33 (23–52) HC: 31 (28–34) |
ND | IFX (CD n = 7; UC n = 4) ADA (CD n = 3; UCn = 2) Golimumab (UC n = 3) |
13 | Faeces, saliva, serum, urine | 0, 13 | 16S SA | |
| Non-Biological Therapies | |||||||||||||
| CD | Pigneur et al. [34] | RCT | France, ND | CD (n = 19) | Paediatric | CD on corticosteroids: 13.7 (1.8) HC: ND |
ND | ND | Corticosteroids (n = 6); ND, 1 mg/kg/ day (up to max of 60 mg) | 4 daily IT + tapered over 3 months | Faeces | 0, 8 | 16S SA |
| Sprockett et al. [36] | RCT | Europe Canada, Israel, MC | CD (n = 67) | Paediatric | Metronidazole (MET): 13.5 (3.1) Metronidazole + Azithromycin (MET+AZ): 14.2 (3.1) |
ND | MET: 0.7 (1) MET+AZ: 1.1 (1.1) |
MET; n = 36); ND, 20 mg/kg twice daily (max 1000 mg/day) MET+AZ; n = 31 of which AZ, 7.5 mg/kg once a day (max 500 mg/day) |
8 weeks (daily) + 4 weeks if lacked response | Faeces | 0, 4, 8, 12 | 16S SA | |
| UC | Ishikawa et al. [37] | PCS | Japan, SC | UC (n = 41) | Adult | UC on AFM: 44.7 (14.9) | ND | UC on FMT: 7.0 (8.0) | AFM monotherapy (n = 20); | 2 weeks (daily) | Faeces | 0, 4, 8 | 16S SA |
| Schierova et al. [33] | RCT | Czech Republic, SC | UC (n = 16) | Adult | UC on 5-ASA: 40 (range 31–66) | ND | ND | Mesalazine (n = 8); enema, 4 g | 6 weeks (daily for 2 weeks, every alternate day till week 6) | Faeces | 0, 2, 4, 6, 12 | 16S SA | |
| IBD | Hart et al. [35] | PCS | Canada, SC | UC (n = 10) CD (n = 20) |
Paediatric | ND except CS ≤12 years (n = 7) CS ≥13 years (n = 7) |
ND | ND | Methyprednisolone (n = 14); IV, 1 mg/kg/day (max 40 mg/day) Transition to oral corticosteroids 1 mg/kg/day PO x 2 weeks followed by progressive wean by 5 mg/wk |
8 weeks (daily) | Faeces | 0, 0.5, 1, 2, 4, 6, 8, 12 | 16S SA |
| Effenberger et al. [62] | Prospective comparative study | Austria, SC | CD (n = 19) UC (n = 10) |
Adult | CD: 49.2 (+/- 15.5) UC: 45.0 (+/−10.7) |
ND | ND | Azathioprine | 20 | Faeces | 0, 12, 30 | 16S SA | |
| Nutritional therapies | |||||||||||||
| CD | Kaakoush et al. [64] | PCS | Australia SC | CD (n = 5) HC (n = 5) |
Paediatric | CD: 9.88 (ND) HC: ND |
9.7 | de novo presenting | EEN (OSMOLITE) | 8–12 | Faecal | 0, 12 weeks post EEN | 16S SA |
| Quince et al. [65] | PCS | Scotland SC | CD (n = 23) HC (n = 21) |
Paediatric | CD: ND (range 6.9–14.7) HC: ND (range 4.6–16.9) |
NR | NR | EEN (Modulen) | 8 | Faecal | −1, 16, 32, 54 | 16S SA | |
| Tang et al. [66] | PCS | China | CD (n = 31) HC (n = 12) |
Paediatric | HC n = 12; 10.45 ± 2.39; CD RE n = 17; 12.02 ± 1.88; CD NRE n = 8; 9.84 ± 4.51 | NR | de novo presenting | EEN (oral) | 8 | Faecal | 8 weeks | 16S SA | |
| Dunn et al. [67] | PCS | Canada | CD (n = 10) HC (n = 5) |
Paediatric | CD: 12.27 | CD: 12 | mainly de novo presenting | EEN; (nasogastric/gastric) | 12 | Faecal | 12 weeks | 16S SA | |
| Costa-Santos et al. [68] | PCS | Portugal | CD patients receive pre-operative EEN = 10; CD patients receive immediate surgery = 5 |
Adult | CD: 45.4 ± 19.1 | NR | 9 years (0–33 years) | Pre-operative EEN (oral) | 15–70 days | Faecal | 24 weeks | 16S SA | |
| Diederen et al. [69] | PCS | Netherlands | CD (n = 27) HC (n = 18) |
Paediatric | NR | CD: 14 (12–15) HC: 13 (11–16) |
de novo presenting | EEN + concomitant thiopurines | 6 | Faecal | 6 weeks | 16S SA | |
| Tang et al. [66] | Prospective comparative study | China, SC |
CD (n = 31) HC (n = 12) |
Paediatric | CD:11.3 (3.5) HC:10.5 (2.4) |
ND | ND | EEN (n = 31); oral, ND | 0, 2, 8 | Faeces | 0, 2, 6, 8 | 16S SA | |
| Jiang et al. [70] | Prospective study | China, SC |
CD (n = 7) | Adult | 27.9 (12.4) | ND | ND | Enteral nutritional powder (n = 7); oral, 250 mL | 0, 8 | Faeces | 0, 8 | 16S SA | |
| Suskind et al. [71] | RCT | U.S. | CD (n = 10) | Paediatric | 14.3 ±2.9 (7–18) | NR | NR | Specific carbohydrate diet (SCD) | 12 | Faecal | NR | 16S SA | |
| UC | Wilson et al. [72] | Prospective study | U.K., SC |
UC (n = 17) | Adult | 35 (10) | ND | ND | GOS supplement (n = 17); oral, 2.8 g | 0, 6 | Faeces | 0, 6 | 16S SA |
| Chen et al. [73] | Prospective comparative study | China, SC |
UC (n = 14) HC (n = 14) |
Adult | UC and HC: 42.4 (15.0) | ND | ND | GMA (n = 28); intravenous, ND | 5 “sessions”; (baseline and endpoint) (n = 5) 10 “sessions”; (baseline and endpoint) (n = 8) |
Faeces | baseline and endpoint | 16S SA | |
| Valcheva et al. [51] | PCS | Canada | UC (n = 25) Inulin 7.5g/d n = 12. Inulin 15g/d n = 13 |
Adult | Inulin 7.5g/d = 36 Inulin 15g/d = 39 |
NR | NR | Prebiotic (inulin) | 9 | Faecal | NR | 16S SA | |
| IBD | Suskind et al. [52] | Observational study | U.S., MC | CD = 9, UC = 3 | Paediatric | 12.8 ± 2.2.(10–17) | NR | 1.3 ± 1.6 years (0–5) | Specific carbohydrate diet (SCD) | 12 | Faecal | NR | 16S SA |
| Olendzki et al. [74] | Prospective comparative study | U.S., SC |
CD (n = 14) UC (n = 7) |
Adult | 40.5 (12.8) | ND | ND | Dietary intervention (n = 25); oral |
8 | Faeces | 0, 8 | MGS | |
Abbreviations: UC, ulcerative colitis; CD, Crohn’s disease; IBDU, inflammatory bowel disease unclassified; HC, healthy controls; qPCR, quantitative polymerase chain reaction; 16S SA, 16S rRNA sequence analysis; IV, intravenous, ulcerative colitis; CD, Crohn’s disease; IBDU, inflammatory bowel disease unclassified; HC, healthy controls; qPCR, quantitative polymerase chain reaction; 16S rRNA, 16S ribosomal RNA; MGS, metagenomic sequencing; IV, intravenous; EEN, exclusive enteral nutrition; GOS, galactooligosaccharide; GMA, granulomonocytapheresis; ND, not disclosed; NR, not reported, SC, single-centre; MC, multi-centre; IT, induction therapy; MT, maintenance therapy; RCT, randomised controlled study; PCS, prospective cohort study, IFX, Infliximab; ADA, Adalimumab, VEDO, Vedolizumab, UST, Ustekinumab; EP, endpoint; AFM, Amoxicilin; oral, 1500 mg/day; Fosfomycin; oral, 3000 mg/day, Metronidazole; oral 750 mg/day).
3.3. Impact of IBD Treatment on the Gut Microbiota
3.3.1. Biologics
Of the twenty studies on biological therapies, eight studies included CD patients only and twelve studies enrolled both CD and UC patients. Four studies were conducted in China; three studies were from Finland; two studies were from Poland and the United States, respectively. Single studies originated from the United Kingdom, Greece, Italy, Czech Republic, Austria, Germany, South Korea, Spain, and Norway. Eighteen studies evaluated the impact of anti-TNF-alpha agents including infliximab, adalimumab, golimumab, and etanercept; two studies investigated the role of anti-integrin antibody (vedolizumab) and interleukin antibody (ustekinumab), respectively (Table 1).
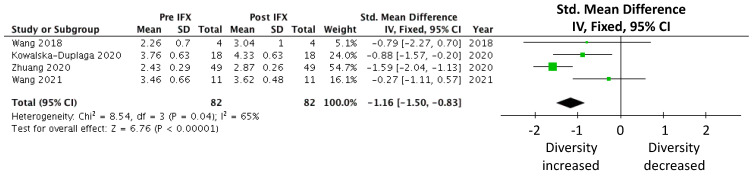
A meta-analysis was carried out on the prospective cohort studies that examined the effects of infliximab on the gut microbiota in CD patients, measured by the Shannon index [38,39,40,54]. Eighty-two subjects were included in the meta-analysis that demonstrated a statistically significant increase in alpha diversity following commencement of infliximab treatment (SMD −1.16 (−1.50, −0.83), p < 0.00001; Figure 2). The degree of variance resulting from between-study heterogeneity, reported as I2, was 65%.
Figure 2.
Meta-analysis of alpha diversity (measured by the Shannon index) in CD cohorts following infliximab treatment [38,39,40,54].
Microbial Diversity
Microbial species diversity in terms of alpha diversity was reported in 16 studies (Table 2). Measures of alpha diversity among studies varied with the use of the following: Shannon, Simpson, inverse Simpson, observed operational taxonomic units (OTUs)/species, observed number of phylotypes, Chao estimated richness, phylodiversity, Good’s coverage, Faith’s phylogenetic diversity, Pielou’s measure of species evenness indexes, ACE index, and Jackknife diversity. Of the 14 alpha diversity measures, the Shannon index was used in 10 studies, the inverse Simpson index in 5 studies, Chao1 index in 5 studies, and observed species in 4 studies. Raw alpha diversity data were only reported for eleven studies, and data from the remaining studies were obtained through manual extrapolation of published graphs.
Sixteen studies reported the impacts of biological therapies on alpha diversity with fifteen studies focusing on anti-TNF-alpha agents and one on interleukin inhibitor, Ustekinumab. Eight studies included only CD patients; one study included only UC patients, while seven studies included both UC and CD patients. Two studies demonstrated a statistically significant increase in alpha diversity [40,58]. Six studies involving CD patients and one study assessing an IBD cohort observed non-statistically significant changes in all measures of alpha diversity measured [43,59]. Dovrolis et al., who analysed both CD and UC patients, respectively, found reduced alpha diversity (Pielou index) in CD patients (regardless of treatment outcome) and UC responders on infliximab [59]. Similarly, Kolho and colleagues reported the same trend in non-responders on infliximab/adalimumab when measured with the inverse Simpson index [43]; however, they did not stratify their findings based on IBD subtypes. The statistical significance of alpha diversity changes compared to baseline were not reported in four studies [38,43,54,58]. One study evaluating the impacts of ustekinumab reported an increase in alpha diversity (measured by the inverse Simpson index) in both responder and non-responder groups [41]. A statistically significant increase compared to the baseline was only achieved at Week 22 in the responder group (Table 2).
In summary, increased alpha diversity following treatment was associated with response to infliximab, adalimumab, and ustekinumab [41,43,57]. Based on the included studies, the study period varied between 6 and 30 weeks. Despite the variability in the study periods, both the short- and longer-term data support the notion that an increase in alpha diversity is characteristic of patients commencing biologics.
Table 2.
Alpha diversity changes following biological therapies, non-biological therapies, and nutritional therapies in studies with CD patients only, UC patients only, or both.
| Disease Subtype | Reference/No. of Subjects | Alpha Diversity Changes | Pre- Treatment (Week) |
Post-Treatment (Week) |
|||
|---|---|---|---|---|---|---|---|
| Biological therapies | |||||||
| CD | Wang et al. [38] CD (n = 4) HC (n = 16) |
Week | 0 * | 14 * | 22 * | 30 * | 38 * |
| IFX Shannon#○ | 2.26 | 3.04 | 3.63 | 3.27 | 2.99 | ||
| IFX Observed Species#◊ | 148 | 219 | 321.5 | 281 | 202 | ||
| IFX Simpson#○ | 0.824 | 0.912 | 0.940 | 0.918 | 0.907 | ||
| IFX Inverse Simpson#○ | 5.68 | 11.4 | 18.3 | 12.1 | 10.8 | ||
| HC Shannon#○ | 3.49 | ||||||
| HC Observed Species#◊ | 393 | ||||||
| HC Simpson#○ | 0.898 | ||||||
| HC Inverse Simpson#○ | 10.00 | ||||||
| Kowalska-Duplaga et al. [54] CD (n = 18) HC (n = 18) |
Week | 0 | 14 | ||||
| IFX Shannon#○ | 3.762 | 4.326 | |||||
| IFX Observed Species#◊ | 46.5 | 64 | |||||
| IFX Faith PD#□ | 6.5723 | 7.65354 | |||||
| IFX Pielou#○ | 0.70198 | 0.6995 | |||||
| HC Shannon#○ | 4.664 | ||||||
| HC Observed Species#◊ | 64 | ||||||
| HC Faith PD#□ | 8.1631 | ||||||
| HC Pielou#○ | 0.7739 | ||||||
| Zhuang et al. [40] CD (n = 49) |
Week | 0 | 6 | 30 | |||
| IFX Shannon#○ | 2.43 | 2.61 | 2.87 | ||||
| IFX Observed Species#◊ | 113 | 145 | 166.5 | ||||
| IFX Chao1#◊ | 148.1 | 183.1 | 209.7 | ||||
| IFX Good’s Coverage#◊ | 0.999 | 0.999 | 0.999 | ||||
| Wang et al. [39] CD (n = 11) HC (n = 12) |
Week | 0 | T1^ | ||||
| IFX Shannon#○ | 3.46 | 3.62 | |||||
| IFX Chao1#◊ | 489.97 | 506.2 | |||||
| IFX Simpson#○ | 0.891 | 0.924 | |||||
| IFX Inverse Simpson#○ | 9.19 | 13.17 | |||||
| HC Shannon○ | 3.7 | - | |||||
| HC Chao1◊ | 475 | - | |||||
| HC Simpson○ | 0.94 | - | |||||
| HC Inverse Simpson○ | 14.5 | - | |||||
| Zhou et al. [57] CD R (n = 9) CD Relapse (n = 7) HC (n = 11) |
Week | 0 | 30 | ||||
| IFX R Shannon○ | 5.5 | 6.25 | |||||
| IFX R PD□ | 15 | 19 | |||||
| IFX Relapse Shannon○ | 5.5 | 5.6 | |||||
| IFX Relapse PD□ | 15 | 16 | |||||
| HC Shannon○ | 5.6 | - | |||||
| HC PD□ | 16 | - | |||||
| Doherty et al. [41] CD R (n = 18) CD NR (n = 30) |
Week | 0 | 4 | 6 | 22 | ||
| UST R Inverse Simpson#○ | 6.65 | 9.44 | 8.42 | 10.7 | |||
| UST NR Inverse Simpson○ | 5.25 | 5.0 | 5.5 | 6.5 | |||
| Dovrolis et al. [59] CD R (n = 5) CD NR (n = 5) |
Week | 0 | 12–20 | ||||
| CD IFX R Pielou○ | 0.74 | 0.73 | |||||
| CD IFX NR Pielou○ | 0.74 | 0.725 | |||||
| Sanchis-Artero et al. [61] CD R (n = 13) CD NR (n = 14) HC (n = 16) |
Week | 0 | 24 | ||||
| CD R anti-TNF Shannon○ | 5.65 | 6.15 | |||||
| CD NR anti-TNF Shannon○ | 5.65 | 5.2 | |||||
| CD R anti-TNF Chao1◊ | 92.5 | 112.5 | |||||
| CD NR anti-TNF Chao1◊ | 92.5 | 68.8 | |||||
| HC Shannon○ | 6.33 | ND | |||||
| HC Chao1◊ | 116.3 | ND | |||||
| UC | Dovrolis et al. [59] UC R (n = 2) UC NR (n = 2) |
Week | 0 | 12–20 | |||
| UC IFX R Pielou○ | 0.74 | 0.69 | |||||
| UC IFX NR Pielou○ | 0.74 | 0.78 | |||||
| IBD | Kolho et al. [43] IBD R (n = 6) IBD NR (n = 5) |
Week | 0 | 2 | 6 | ||
| IFX/ADA Responder Inverse Simpson○ | 173 | 188 | 175 | ||||
| IFX/ADA Non-Responder Inverse Simpson○ | 133 | 113 | 128 | ||||
| Aden et al. [58] IBD (n = 12) HC (n = 19) |
Week | 0 | 2 | 6 | 30 | ||
| Etanercept Shannon○ | 2.4 | 2.5 | 2.7 | 2.7 | |||
| Etanercept Observed Species◊ | 64 | 64 | 61 | 70 | |||
| Etanercept Chao1◊ | 75 | 76 | 80 | 86 | |||
| Etanercept PD□ | 6 | 6.5 | 6 | 7.5 | |||
| HC Shannon○ | 3.2 | ||||||
| HC Observed Species◊ | 72 | ||||||
| HC Chao1◊ | 86 | ||||||
| HC PD□ | 7.5 | ||||||
| Schierova et al. [60] CD (n = 26) UC (n = 18) HC (n = 37) |
Week | 0 | 20 | 38 | |||
| CD Anti-TNF Shannon○ | 5.65 | 5.7 | 5.9 | ||||
| UC Anti-TNF Shannon○ | 5.2 | 5.85 | 5.85 | ||||
| HC Shannon○ | 6.15 | ND | ND | ||||
| Park et al. [63] IBD (n = 19) HC (n = 19) |
Week | 0 | 12 | ||||
| IBD Anti-TNF Shannon○ | 3.28 | 3.35 | |||||
| IBD Anti-TNF Simpson○ | 0.068 | 0.068 | |||||
| IBD Anti-TNF Chao1◊ | 290 | 315 | |||||
| IBD Anti-TNF ACE◊ | 305 | 325 | |||||
| IBD Anti-TNF Jackknife | 295 | 330 | |||||
| IBD Anti-TNF NPShannon○ | 3.29 | 3.36 | |||||
| HC Shannon○ | 3.73 | ND | |||||
| HC Simpson○ | 0.05 | ND | |||||
| HC Chao1◊ | 465 | ND | |||||
| HC ACE◊ | 485 | ND | |||||
| HC Jackknife | 503 | ND | |||||
| HC NPShannon○ | 3.73 | ND | |||||
| Ventin-Holmberg et al. [49] CD R (n = 13) CD PR (n = 4) CD NR (n = 6) UC R (n = 31) UC PR (n = 8) UC NR (n = 8) |
Week | 0 | 2 | 6 | 12 | 52 | |
| CD R IFX Simpson○ | 9 | 9 | 12 | 8.75 | 10.5 | ||
| CD NR IFX Simpson○ | 7 | 8.5 | 8.3 | 9 | ND | ||
| CD PR IFX Simpson○ | 5.5 | 5.5 | 10.3 | 8 | 8 | ||
| CD R IFX OTU◊ | 97.5 | 105 | 107.5 | 107.5 | 110 | ||
| CD NR IFX OTU◊ | 90 | 95 | 97.5 | 95 | ND | ||
| CD PR IFX OTU◊ | 55 | 77.5 | 107.5 | 95 | 107.5 | ||
| UC R IFX Simpson○ | 11.25 | 11 | 10.5 | 10.5 | 11 | ||
| UC NR IFX Simpson○ | 7 | 10.75 | 8 | 10.25 | 4.75 | ||
| UC PR IFX Simpson○ | 9.75 | 8.5 | 10 | 10 | 10.5 | ||
| UC R IFX OTU◊ | 105 | 115 | 107.5 | 105 | 105 | ||
| UC NR IFX OTU◊ | 90 | 105 | 97.5 | 105 | 102.5 | ||
| UC PR IFX OTU◊ | 107.5 | 112.5 | 110 | 105 | 105 | ||
| Ventin-Holmberg et al. [56] IBD R (n = 10) IBD NR (n-16) |
Week | 0 | 2 | 6 | |||
| IBD R IFX Simpson○ | 8.5 | 7 | 6.5 | ||||
| IBD NR IFX Inverse Simpson○ | 6 | 6.5 | 7 | ||||
| IBD R IFX OTU◊ | 75 | 72.5 | 72.5 | ||||
| IBD NR IFX OTU◊ | 65 | 66 | 70 | ||||
| Effenberger et al. [62] IBD R (n = 18) IBD NR (n = 18) |
Week | 0 | 98 days | ||||
| R Anti-TNF Shannon○ | 0.4 | 0.2 | |||||
| NR Anti-TNF Shannon○ | −0.2 | 0.175 | |||||
| Non-biological therapies | |||||||
| CD | Sprockett et al. [36] ^1 MET (n = 36) MET+AZ (n = 31) MET/MET+AZ (n = 11) |
Week | 0 | 4 | 8 | 12 | |
| CD MET Faith’s PD□ | 18.5 | 11.5 | 12 | 16 | |||
| CD MET + AZ Faith’s PD□ | 17.5 | 11 | 10 | 13 | |||
| CD MET/MET+AZ Faith’s PD□ | 18.5 | 15.5 | 6.5 | 9.5 | |||
| UC | Schierova et al. [33] UC R (n = 4) UC NR (n = 4) |
Week | 0 | 2 | |||
| UC R 5-ASA Shannon○ | 6.6 | 6.9 | |||||
| UC NR 5-ASA Shannon○ | 6.65 | 6.85 | |||||
| IBD | Hart et al. [35] ^2 UC (n = 10) CD (n = 4) |
Week | 0 | 12 | |||
| Corticosteroids Shannon○ | 2.85 | 3.6 | |||||
| Effenberger et al. [62] IBD R (n = 15) IBD NR (n = 14) |
Week | 0 | 14 | ||||
| R AZA Shannon○ | −0.2 | −0.2 | |||||
| NR AZA Shannon○ | −0.05 | −0.85 | |||||
| Nutritional therapies | |||||||
| CD | Kaakoush et al. [64] CD (n = 5) HC (n = 5) |
Week | 0 | 8 | 26 | ||
| EEN CD Shannon#○ | 2.25 ± 0.24 | ND | ND | ||||
| HC Shannon#○ | 2.75 ± 0.14 | ND | ND | ||||
| Quince et al. [65] CD (n = 23) HC (n = 21) |
Week | 0 | 2 | 4 | 8 | Free diet | |
| EEN CD Shannon#○ | 2.88 ± 0.597 | 2.82 ± 0.492 | 2.49 ± 0.731 | 2.84 ± 0.513 | 3.04 ± 0.686 | ||
| HC Shannon○ | ND | ||||||
| Tang et al. [66] HC (n = 12) CD Remission (n = 17) CD Non-remission (n = 8) |
Week | 0 | 8 | ||||
| EEN CD-R Shannon#○ |
1.84 ± 0.16 | 2.45 ± 0.1 | |||||
| EEN CD-NR Shannon#○ |
1.33 ± 0.23 | 1.76 ± 0.23 | |||||
| HC Shannon#○ | 3.1 ± 0.2 | ND | |||||
| Dunn et al. [67] CD (n = 10 at baseline, n = 9 at 12 weeks) HC (n = 5) |
Week | 0 | 12 | ||||
| EEN CD Sustained Remission EEN Chao |
~1300 | ~1250 | |||||
| EEN CD Non-Sustained Remission Chao◊ | ~1000 | ~ 700 | |||||
| HC Chao◊ | ~2000 | ND | |||||
| Costa-Santos et al. [68] CD (n = 15) |
Week | 0 | 8 | ||||
| CD Faith’s PD#□ | 8 ± 2.3 | 5.2 ± 1.5 | |||||
| Diederen et al. [69] CD (n = 27) HC (n = 18) |
Week | 0 | 1–5 | 6 | 9 | ||
| EEN CD Inv Simpsons#○ | 19.40 | 11.57 | 12.85 | 16.87 | |||
| HC Inv Simpsons#○ | 20.37 | ||||||
| Suskind et al. [52] CD (n = 8) |
Week | 0 | 12 | ||||
| SCD CD Shannon#○ | 2.62(2.4–3.1) | 2.7(2.3–3.1) | |||||
| Suskind et al. [71] CD (n = 5) |
Week | 0 | 2 | 12 | |||
| SCD CD Inverse Simpson#○ | 103 (89–132) | 115 (95–130) | 112 (107–147) | ||||
| Tang et al. [66] CD (n = 31) HC (n = 12) |
Week | 0 | 2 | 8 | |||
| HC Shannon#○ | 3.1 | ND | ND | ||||
| EEN NR CD Shannon#○ | 1.33 | ND | 1.76 | ||||
| EEN R CD Shannon#○ | 1.84 | ND | 2.45 | ||||
| Observed Species#◊ | 172 | ND | ND | ||||
| Jiang et al. [70] CD (n = 7) |
Week | 0 | 8 | ||||
| Shannon#○ | ND | ||||||
| Observed Species#◊ | 292 | 390 | |||||
| UC | Wilson et al. [72] UC (n = 17) |
Week | 0 | 6 | |||
| Shannon#○ | 2.5 | 2.2 | |||||
| Chao1#◊ | 50.4 | 35.6 | |||||
| Observed Species#◊ | 178 | ND | |||||
| Chen et al. [73] UC (n = 14) HC (n = 14) |
“Sessions” | 0 | 5 | 10 | |||
| UC Shannon#○ | ND | ||||||
| HC Shannon#○ | ND | ||||||
| UC Observed Species#◊ | 523 | 751 | 521 | ||||
| IBD | Olendzki et al. [74] CD (n = 14) UC (n = 7) |
Week | 0 | 6 | |||
| Shannon#○ | CD = 2.1 UC = 2.3 |
||||||
# Reflects accurate alpha diversity values; * [38]: Time points are estimated based on IFX dosing schedule: Weeks 0, 2, and 6 followed by maintenance every 8 weeks. Patients give a baseline sample and one at various time points after IFX infusion; ^ [39]: T1 represents sampling post-treatment (3rd or 6th IFX infusion). IFX is given at Weeks 0, 2, and 6 followed by maintenance every 8 weeks; Bold: statistically significant compared to baseline (p-value < 0.05); # Reflects accurate alpha diversity values; ^1: In Sprockett et al., treatment ended at Week 8, and microbial analysis was investigated till Week 12 (4 weeks post-treatment); ^2 [35]: Treatment ended at Week 8, and microbial analysis was evaluated till Week 12 (day 84); ◊ Richness alpha diversity indices; observed species, Chao1, Good’s coverage, abundance-based coverage estimator (ACE), operational taxonomical units (OTU); ○ Abundance alpha diversity indices; Simpson, inverse (Inv) Simpson, Shannon, Pielou; □ Phylogenetic distance alpha diversity index; Faith’s phylogenetic distance (PD); Abbreviations: UC, ulcerative colitis; CD, Crohn’s disease; HC, healthy controls; EEN, exclusive enteral nutrition; SCD, specific carbohydrate diet; NR, non-responders; R, responders; PR, partial-responders; 5-ASA, 5-aminosalicyclic acid; MET, Metronidazole; AZ, Azithromycin; IFX, infliximab; ADA, adalimumab; UST, ustekinumab; anti-TNF, anti-tumour necrosis factor.
Microbial Composition Changes
Overall, a consistent reduction in the relative abundance of Proteobacteria including Escherichia coli and Enterococcus species and enrichment of more beneficial genera including short-chain fatty acid (SCFA) producers such as Blautia and Faecalibacterium species were observed following biological therapy in the CD, UC, and IBD cohorts [38,39,41,42,43,53,58,59]. Most studies investigated microbial changes in faecal samples of CD patients treated with infliximab [38,39,40,53,54,57,59]. Wang and colleagues conducted two similarly designed studies analysing the effects of infliximab in paediatric CD patients. In the earlier study (2018), the authors reported a sustained increase of SCFA-producing genera (Blautia, Faecalibacterium, Odoribacter, and Sutterella) and decreased abundance of Enterobacteriaceae, Enterococcaceae, Planococcaceae, and Streptococcaceae post-treatment [38]. Similar increases in the abundance of Blautia were also reported in their later study; however, Clostridium Cluster IV, Collinsella, Eubacterium, and Ruminococcus species were also increased. The study also observed a decreased abundance of Abiotrophia and Lactococcus species [39]. The findings by Zhuang and colleagues, in an adult CD cohort, corroborated the results reported by Wang et al., with a sustained enrichment of SCFA-producing taxa (Lachnospira, Roseburia and Blautia), as well as a reduction in pathogenic bacteria such as Enterobacteriaceae including Escherichia-Shigella and Fusobacterium [40]. Interestingly, in the only study evaluating mucosal biopsies of adult CD patients following infliximab treatment, Dovrolis and colleagues observed an increased abundance of Proteobacteria and Rubrobacter, which is in disagreement with other studies, thus highlighting the different microbial composition of stool versus mucosal samples [59]. In a small study involving both CD and UC patients, the restoration of normal levels of 14 phylotypes including several SCFA-producing bacteria following etanercept treatment was noted [58].
Doherty et al. evaluated microbiota changes following the use of ustekinumab on CD patients [41]. The study reported an increase in Bacteroidetes species, as well as F. prausnitzii, Blautia, Ruminococcaceae, and Roseburia species, which agrees with the anti-TNF-a studies. Following vedolizumab treatment, a reduction in Bifidobacterium longum, Eggerthella, Ruminococcus gnavus, Roseburia inulinivorans, and Veillonella parvula species’ abundance were observed between baseline and Week 14 post-treatment in CD patients. These changes were sustained over the 54-week study period [42].
Microbial Predictors of Treatment Outcomes
The association between microbiota changes following biological therapies and treatment outcomes was investigated in seven studies (Table 3). Five studies exploring associations between faecal microbiota profiles and infliximab therapeutic response identified microbial features that distinguished treatment responders from non-responders. Paediatric CD patients with sustained response had a higher abundance of SCFA-producing taxa, Blautia, Faecalibacterium, Lachnospira, and Roseburia compared to non-sustained responders [38]. In a more recent study by the same authors, an increased abundance of Actinomyces, Atopobium, and Parabacteroides genera were observed in patients with sustained treatment response [39]. Interestingly, the authors also demonstrated an association between baseline microbial composition and sustained treatment response. A higher abundance of Methylobacterium, Sphingomonas, Staphylococcus, and Streptococcus genera at baseline was associated with sustained response, with an increased abundance of Clostridium XI and Clostridium XVIII members, Eggerthella, Lachnospiraceae incertae sedis, Parabacteroides, and Peptococcus genera at baseline associated with a loss of response [39]. Increased proportions of Lachnospiraceae and Blautia genera at Week 6 following infliximab treatment were associated with clinical and endoscopic response in an adult CD cohort, whilst CD patients who had a relatively higher abundance of Clostridiales at baseline responded better to infliximab compared to those with lower abundance [49,57]. Clostridiales levels were also restored to almost healthy levels in those achieving clinical remission [61].
Table 3.
Microbiota changes following biological therapies, non-biological therapies, and nutritional therapies in studies with CD patients only, UC patients only, or both.
| Disease Subtype | Reference | Population | Microbiota Changes due to Treatment |
|---|---|---|---|
| Biological therapies | |||
| CD CD CD |
Wang et al. [38] | Paediatric (P) CD (n = 4) |
Following IFX treatment:
|
| Zhou et al. [57] | Adult (A) CD (n = 16) |
Following IFX treatment:
|
|
| Dovrolis et al. [59] | A -CD (n = 10) | Following IFX treatment, CD patients:
|
|
| Kowalska-Duplaga et al. [54] | P -CD (n = 18) | Following IFX treatment:
|
|
| Salamon et al. [55] | P -CD (n = 13) | Following IFX treatment:
|
|
| Zhuang et al. [40] | A -CD (n = 49) | Following IFX treatment:
|
|
| Wang et al. [39] | P -CD (n = 11) | Following IFX treatment:
|
|
| Ribaldone et al. [53] | A -CD (n = 20) | Following ADA treatment:
|
|
| Ananthakrishnan et al. [42] | A -CD (n = 42) | Following Vedolizumab treatment: In CD remission patients (n = 10):
|
|
| Doherty et al. [41] | A-CD(n = 306) | Following UST treatment, in subjects in remission 6 weeks after induction compared to those with active disease:
|
|
| Sanchis-Artero et al. [61] | CD R (n = 13) CD NR(n = 14) |
Following IFX/ADA treatment:
|
|
| CD | Ventin-Holmberg et al. [56] | CD R (n = 12) CD NR(n = 18) |
Following IFX treatment:
|
| UC | Dovrolis et al. [59] | A -UC (n = 4) | Following IFX treatment, in UC patients:
|
| IBD | Kolho et al. [43] | P -IBD (n = 11) | Following IFX/ADA treatment:
|
| Aden et al. [58] | A-IBD (n = 12) | Following anti-TNF Etanercept treatment:
|
|
| Ding et al. [48] | Adult CD (n = 76) UC (n = 10) |
Following anti-TNF-alpha treatment:
|
|
| Olbjorn et al. [50] | P -CD (n = 22) UC (n = 9) |
Following anti-TNF-alpha treatment:
|
|
| Schierova et al. [60] | CD (n = 17) UC (n= 10) |
Following IFX/ADA treatment:
|
|
| Ventin-Holmberg et al. [49] | CD R (n = 13) CD PR (n = 4) CD NR (n = 6) UC R (n = 31) UC PR (n = 8) UC NR (n = 8) |
Following IFX treatment:
|
|
| Effenberger et al. [62] | CD R (n = 13) CD NR(n = 11) UC R (n = 7) UC NR (n = 5) |
Following anti-TNF treatment:
|
|
| IBD | Park et al. [63] | UC (n = 10) CD (n = 9) |
Following anti-TNF treatment:
|
| Non-biological therapies | |||
| CD | Pigneur et al. [34] | P -CD (n = 4) | Following Corticosteroid treatment:
|
| Sprockett et al. [36] | P -CD (n = 74) | Following antibiotic treatment: Metronidazole:
|
|
| UC | Hart et al. [35] | P -UC (n = 10) | Following corticosteroid treatment:
|
| Ishikawa et al. [37] | A -UC (n = 19) | Following amoxicillin, fosfomycin and metronidazole treatment:
|
|
| Schierova et al. [33] | UC (n = 8) | Following 5-ASA treatment, no changes in faecal microbiome observed. | |
| IBD | Hart et al. [35] | Paediatric CD (n = 4) UC (n = 10) |
Following corticosteroid treatment:
|
| Effenberger et al. [62] | CD R (n = 10) CD NR (n = 9) UC R (n = 5) UC NR (n = 5) |
Following AZA treatment:
|
|
| Nutritional therapies | |||
| CD | Kaakoush et al. [64] | Paediatric CD (n = 5) HC (n = 5) |
Following EEN:
|
| Quince et al. [65] | Paediatric CD (n = 23) HC (n = 21) |
Following EEN:
|
|
| Tang et al. [66] | Paediatric CD remission (n = 17) CD non-remission (n = 8) HC (n = 12) |
Following EEN:
|
|
| Dunn et al. [67] | Paediatric CD (n = 10) HC (n = 5) |
Following EEN:
|
|
| Costa-Santos et al. [68] | A -CD (n = 10) | Following EEN:
|
|
| Diederen et al. [69] | P -CD (n = 43) | Following EEN:
|
|
| Suskind et al. [71] | P -CD (n = 5) | Following SCD:
|
|
| Tang et al. [66] | CD (n = 31) HC (n = 12) |
Following EEN treatment:
|
|
| Jiang et al. [70] | CD (n = 7) | Following EEN treatment:
|
|
| UC | Valcheva et al. [51] | A -UC (n = 25) | Following prebiotics (inulin-type fructans):
|
| Wilson et al. [72] | UC (n = 17) | Following GOS prebiotic treatment:
|
|
| Chen et al. [73] | UC (n = 14) HC (n = 14) |
Following GMA treatment:
|
|
| IBD | Olendzki et al. [74] | CD (n = 14) UC (n = 7) |
After dietary intervention:
|
Abbreviations: UC, ulcerative colitis; CD, Crohn’s disease; IBD, inflammatory bowel disease; HC, healthy controls; EEN, exclusive enteral; GMA, granulomonocytapheresis; GOS, galactooligosaccharide; R, responders; NR, non-responders; PR, partial responders; SCFA, short-chain fatty acid; SCD, specific carbohydrate diet; 5-ASA, 5-aminosalicylic acid; anti-TNF, anti-tumour necrosis factor; IFX, infliximab; ADA, adalimumab; UST, ustekinumab; LEfSE, linear discriminant analysis effect size.
In contrast, in the study that analysed mucosal biopsies, the authors identified enterotypes distinct for responders and non-responders, which was inconsistent with the findings of faecal-based studies [59]. A high abundance of Hungatella, Ruminococcus gnavus, and Parvimonas genera at baseline were associated with clinical and endoscopic response, while a higher abundance of Blautia, Faecalibacterium, Roseburia, and Negativibacillus genera in CD patients at baseline was associated with non-response determined by clinical and endoscopic parameters. Increased prevalence of Chloroflexi, Ruminococcus, Eubacterium hallii, Eubacterium eligens, Escherichia, Shigella, and Butyricicoccus genera and decreased abundance of Fusobacteria were noted in CD patients who responded to treatment. In UC, responders had increased Bacteroidetes populations, Veillonella, Tyzzerella, Ruminococcus torques, Parabacteroides, Erisipela, Clostridium, and Bilophila genera, and loss of Spirochaetes and Plantomycetes. Non-responders had an increased abundance of Actinobacteria, Porphyromonas, Granulicatella, and Corynebacterium genera and a reduction in Anaerostipes abundance after 3 months of infliximab [59].
Higher abundance of Bifidobacterium, Clostridium colinum, Eubacterium rectale, an uncultured Clostridiales, and Vibrio species, as well as decreased abundance of Streptococcus mitis at baseline were associated with response to infliximab or adalimumab treatment compared to non-responders [43]. Similar findings were seen in adult CD patients on ustekinumab, with Bacteroides, Faecalibacterium, Blautia, Ruminococcaceae, and Roseburia species significantly more abundant in subjects in remission compared to those with active disease at 6 weeks post-treatment initiation [41]. In a study involving infliximab refractory CD patients switched onto anti-integrin, vedolizumab, a higher abundance of Roseburia inulinivorans and Burkholderiales genera at baseline was associated with remission at Week 14 compared to non-remitters [42]. Further analysis at 1 year post-treatment revealed that these specific microbial changes observed in patients that achieved remission at Week 14 were sustained and the treatment effect was durable.
3.3.2. Non-Biological Therapies
Microbial Diversity
Seven studies evaluated the impact of non-biologic therapeutics on alpha diversity, and adequate data were available for extraction from four studies (Table 2). Schierova et al. reported that 5-ASA use in UC patients was associated with an increase in alpha diversity after 14 days of treatment [33]. The extent of alpha diversity increase was greater in treatment responders than non-responders; however, the results did not reach statistical significance. Hart et al. demonstrated that corticosteroid treatment was associated with a statistically significant increase in alpha diversity (Shannon index) in a cohort of CD and UC patients [35]. The increase in alpha diversity measured from baseline continued throughout 8 weeks of therapy and persisted at Week 12. Sprockett and colleagues evaluated the therapeutic impact of three antibiotic regimens: (1) Metronidazole (MET), (2) MET, and Azithromycin (AZ) or (3) initial MET and the combination of MET+AZ over a 12-week period in a CD cohort [41]. Alpha diversity, measured by Faith’s PD index, decreased over 8 weeks of treatment, rebounding slightly at Week 12 in all groups. No significant changes in the distance between baseline and Weeks 12 or 30 in either patients in remission or without remission were noted following azathioprine treatment [62].
Microbial Composition
There were six studies on non-biological therapies. Pigneur et al. observed an enrichment of SCFA-producing genera including Roseburia intestinalis, Eubacterium species, Ruminococcus, and Bifidobacterium bifidum, along with reduced numbers of Blautia species at Week 8 following corticosteroid treatment in a small paediatric CD study [34]. A similar increase in uncharacterised Ruminococcaceae was observed in paediatric UC and CD patients, which also reported an increased abundance of Blautia and Sellimonas species and a decreased abundance of Granulicatella, Haemophilus, and Streptococcus genera at Week 12 compared to baseline [35].
Antibiotic treatment is known to induce an ecosystem-wide disturbance and decrease microbial diversity. A Japanese study evaluated the impact of combination antibiotic regimen (amoxicillin, fosfomycin, and metronidazole) in nineteen adult patients with active UC [37]. The treatment resulted in an increased abundance of Proteobacteria, particularly Enterobacteriaceae, and a near complete depletion of Bacteroidetes species. Some recovery in the proportion of Bacteroidetes species was observed at both 4 and 8 weeks post-treatment [37]. A large multinational longitudinal study involving CD paediatric patients investigated the microbial impacts of two specific antibiotic regimens, MET and MET+AZ [36]. The finding of increased abundances of potentially pathogenic Enterococcus in the MET group concurred with the Japanese study’s findings, which may indicate the presence of antibiotic resistant microbes and their enrichment predisposing patients to invasive infections. Interestingly, the opposite effect was observed in the MET+ AZ treatment, which might be reflective of the effects of AZ against adherent and invasive E. coli strains [36]. The 5-ASA study failed to observe specific microbial changes over the 12-week study period [33]. CD patients not benefiting from azathioprine treatment had increased abundance of Lactobacillus and Klebsiella genera [62]. In contrast, AZA responders had a significant decrease in Proteobacteria and an associated increase in Bacteroidetes (Table 3).
3.3.3. Nutritional Therapies
Of the fourteen studies on nutritional therapies, five studies were conducted in North America, four studies in China, three studies in Europe, and one study in Australia and Canada, respectively. Nine studies involved solely CD patients; three studies enrolled UC patients; two involved both UC and CD patients. Eight of the studies evaluated the impact of EEN (6 paediatric and 2 adult cohorts); two paediatric studies assessed the impact of SCD; two adult studies assessed the impact of the prebiotic inulin and galactooligosaccharide (GOS) supplement, respectively; one study assessed adsorptive (GMA); another studied dietary intervention on adult cohorts (Table 1).
Microbial Diversity
The results of eight studies evaluating the impact of EEN-specific microbial changes on CD cohorts were essentially study specific. EEN therapy was seen to reduce the abundance of several bacterial genera including Blautia, Subdoligranulum, Halomonas, Ruminococcus, Faecalibacterium, Bifidobacteriaceae, Lachnospiraceae, Bacteroidales, and Enterobacterales. Concomitant increases in Clostridiales, Nesterenkonia, Rhizobium, Gemella, and Ruminococcus gnavus were also reported [64,65,66,67,68,69]. When microbial changes were evaluated based on clinical response vs. non-sustained response, again, specific signatures were study-dependent, but generally, responders had increased levels of Firmicutes including Faecalibacterium, Roseburia, Anaerostipes genera, and Ruminococcus bromii, as well as Bacteroidetes including Parabacteroides and Flavonifractor species. Non-sustained responders had higher abundance of Proteobacteria including E. coli, Blautia, Streptococcus, Granulicatella, and Rothia genera (Table 3).
When the impact of inulin-type fructan supplementation was assessed, Valcheva et al. showed an increase in Bifidobacteriaceae and Lachnospiraceae abundance along with an increase in colonic butyrate levels in UC patients, but this did not correlate with disease activity scores [51]. GOS prebiotic treatment was associated with a reduction in Oscillospira and Dialister abundance and an increase in Alistipes (Table 3).
4. Discussion
The concept of altering the gut microbiota to improve health is now a well-established concept in medicine. Microbiome-based therapies include dietary interventions, prebiotics, probiotics, antibiotics, phage therapy, FMT, live biotherapeutics, and microbiome mimetics. In the context of IBD, understanding the interaction between gut microbes, IBD therapeutics, as well as environmental factors and disease progression is essential if we want to achieve symptom resolution and/or avoid adverse reactions. Several prospective studies have investigated the role of gut microbiota as a potential biomarker for treatment response; yet, a systematic evaluation of microbiota changes following IBD treatment has not been undertaken. Therefore, the aims of this systematic review and meta-analysis were to define the microbial changes following pharmacological and nutritional IBD treatments and identify microbial predictors of treatment response.
The review demonstrated a consistent increase in alpha diversity indices following biological therapies, 5-ASA, and corticosteroids. Additionally, higher microbial diversity at baseline and/or following treatment was shown to be predictive of treatment response in studies on infliximab, adalimumab, and ustekinumab [41,43,57,59]. Increased microbial richness in the gut has been strongly associated with good health, and a less diverse microbiome is consistently implicated in IBD [19,75]. Therefore, a more diverse gut microbiome at baseline might indicate a greater abundance of microbial taxa with anti-inflammatory properties (directly or metabolite mediated). A subsequent reduction in colonic inflammation and preservation of mucosal barrier function/integrity may lead to stronger treatment response. However, the exact mechanisms by which these therapeutics impact microbial diversity remains to be elucidated. The studies included in this review tracked alpha diversity changes for varying periods ranging from 2–30 weeks. Considering that IBD is a life-long disease that is progressive in nature, it would be of significant interest to evaluate if the increase in microbial diversity can be sustained longitudinally and its association with long-term health outcomes. By generating this insight through extended interrogation of large longitudinal IBD cohorts, which comprise a range of disease presentations, as well as ethnic diversity, will contribute to the improvement of IBD treatment results and also IBD management.
Comparison of the studies looking at biologic therapies demonstrated considerable heterogeneity in microbial changes in response to treatment; however, an enrichment of putative SCFA-producing bacteria was consistently noted. SCFAs such as acetate, butyrate, and propionate exert immunomodulatory and anti-inflammatory effects through mechanisms such as the modulation of intestinal wall permeability and reducing oxidative stress and decreasing inflammation through inhibition of mediators such as NF-κB and IL-8 and are primary sources of energy for colonic epithelial cells [76,77,78]. Therefore, the expansion of SCFA-producing bacteria, following certain treatments, highlights that those alterations in microbial metabolic functions could be more important than identifying individual taxonomic changes in the context of IBD. This concept accords with a recent study by Anathakrishnan et al., which observed that there were significantly greater metagenomic alterations in microbial functions than there were changes in microbial composition in both CD and UC cohorts following treatment with the anti-integrin agent, vedolizumab [42]. This highlights the need for future studies to characterise both compositional and functional alterations in the gut microbiota.
Evidence supporting the role of the gut microbiota as a predictor of treatment response is accumulating, with most studies in the systematic review identifying microbial predictors associated with treatment outcomes. An important consideration is understanding that IBD therapeutics can alter microbial profiles, and conversely, the gut microbiota can influence therapeutic outcomes. Interestingly, the observation in three studies on infliximab and vedolizumab that certain baseline signatures are predictive of treatment response or non-response is strongly suggestive that specific gut microbial factors can influence treatment efficacy [39,42,43]. However, the findings were strikingly heterogenous between studies; thus, it was not possible to perform a meta-analysis to evaluate the potential of microbial predictors of treatment response in IBD reported in these studies. Therefore, the microbiota results have to be interpreted with caution, especially since significant variation between studies in terms of subject age, disease subtype and severity, clinical outcome assessments, geography, medications, the length of the study period, and analysis techniques were noted.
The modulation of the gut microbiota by faecal microbiota transplantation (FMT) is now being used to induce remission in UC. Published randomised clinical trials have highlighted that clinical response relies on the enrichment of certain beneficial bacteria including SCFA-producing strains. Interestingly, recent evidence suggests that baseline microbiome profiles are a good predictor of response, with patients achieving a successful outcome having significantly higher microbial richness prior to, during, and after FMT [79]. This further reinforces the potential of baseline or pretreatment microbiome assessment in defining therapeutic outcomes.
There are several limitations in this review. First, the majority of the studies had small subject numbers and, notably, the patients that were included in the detailed longitudinal microbial analysis often represented a significantly smaller subset of the actual cohort. Another significant limitation of the studies included in this systematic review is that most did not account for confounders that are known to impact gut microbiota such as dietary intake and polypharmacy effects. The lack of studies focusing on certain treatments (vedolizumab (n = 1), ustekinumab (n = 1), etanercept (n = 1), 5-ASA (n = 1), antibiotics (n = 2), corticosteroid (n = 2), prebiotic (n = 1), and SCD (n = 2) also limited the comparisons. Additionally, the lack of standardisation in disease phenotyping, microbial analysis methods, defining outcome assessments (clinical/endoscopic response/remission), study periods, and sampling points remain major barriers to understanding the intricate association between the gut microbiota and therapeutics. Overall, the heterogeneity of the gut microbiota findings between different patient populations is challenging to interpret. These limitations must be considered within the design of future studies. This includes the need to focus on well-phenotyped prospective longitudinal patient cohorts, the appreciation of inflammatory and treatment confounders, as well as considering differences due to geography, age, and diet. Finally, harmonisation across studies in terms of establishing a robust standardised scientific methodology is fundamental to validate, reproduce, and ensure the quality of findings in future research.
5. Conclusions
In conclusion, this systematic review contributes to the growing appreciation of the relationship between gut microbes and IBD therapeutics; however, defining optimal treatment strategies remains a challenge. The identification of microbial predictors of treatment response is fundamental to achieving precision medicine in IBD; however, there is still a long way to go before these findings can be translated into clinical practice. Further validation in large longitudinal prospective cohorts is needed.
Supplementary Materials
The following Supporting Information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens12020262/s1, Table S1: Summary of Newcastle-Ottawa Scale of each included cohort study; Table S2: Summary of Newcastle-Ottawa Scale of each included RCT.
Author Contributions
G.L.H. conceptualised the study. C.M., T.J. and G.L. performed a systematic search of the literature. C.M., G.L.H., S.T.L., T.J. and G.L. conducted the analysis and interpretation of the results. C.M., S.K. and G.L.H. wrote the manuscript. G.L.H. and T.R. provided statistical support for the meta-analysis. G.L.H., M.C.G. and S.T.L. supervised the project. S.K., M.C.G., D.L., T.J., S.J.C., S.T.L. and G.L.H. were involved in the reviewing and editing of manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.McQuaid K.R. Inflammatory Bowel Disease. In: Papadakis M.A., McPhee S.J., Rabow M.W., McQuaid K.R., editors. Current Medical Diagnosis & Treatment 2022. McGraw-Hill Education; New York, NY, USA: 2022. [Google Scholar]
- 2.Alatab S., Sepanlou S.G., Ikuta K., Vahedi H., Bisignano C., Safiri S., Sadeghi A. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020;5:17–30. doi: 10.1016/S2468-1253(19)30333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lichtenstein G.R., Shahabi A., Seabury S.A., Lakdawalla D.N., Espinosa O.D., Green S., Brauer M., Baldassano R.N. Lifetime Economic Burden of Crohn’s Disease and Ulcerative Colitis by Age at Diagnosis. Clin. Gastroenterol. Hepatol. 2020;18:889–897.e810. doi: 10.1016/j.cgh.2019.07.022. [DOI] [PubMed] [Google Scholar]
- 4.Peyrin-Biroulet L., Sandborn W., Sands B.E., Reinisch W., Bemelman W., Bryant R.V., D’Haens G., Dotan I., Dubinsky M., Feagan B., et al. Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE): Determining Therapeutic Goals for Treat-to-Target. Am. J. Gastroenterol. 2015;110:1324–1338. doi: 10.1038/ajg.2015.233. [DOI] [PubMed] [Google Scholar]
- 5.Privitera G., Pugliese D., Lopetuso L.R., Scaldaferri F., Neri M., Guidi L., Gasbarrini A., Armuzzi A. Novel trends with biologics in inflammatory bowel disease: Sequential and combined approaches. Ther. Adv. Gastroenterol. 2021;14:17562848211006669. doi: 10.1177/17562848211006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colombel J.F., Narula N., Peyrin-Biroulet L. Management Strategies to Improve Outcomes of Patients with Inflammatory Bowel Diseases. Gastroenterology. 2017;152:351–361.e355. doi: 10.1053/j.gastro.2016.09.046. [DOI] [PubMed] [Google Scholar]
- 7.Berg D.R., Colombel J.F., Ungaro R. The Role of Early Biologic Therapy in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2019;25:1896–1905. doi: 10.1093/ibd/izz059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khanna R., Bressler B., Levesque B.G., Zou G., Stitt L.W., Greenberg G.R., Panaccione R., Bitton A., Paré P., Vermeire S., et al. Early combined immunosuppression for the management of Crohn’s disease (REACT): A cluster randomised controlled trial. Lancet. 2015;386:1825–1834. doi: 10.1016/S0140-6736(15)00068-9. [DOI] [PubMed] [Google Scholar]
- 9.de Jong M.E., Smits L.J.T., van Ruijven B., den Broeder N., Russel M., Römkens T.E.H., West R.L., Jansen J.M., Hoentjen F. Increased Discontinuation Rates of Anti-TNF Therapy in Elderly Inflammatory Bowel Disease Patients. J. Crohns Colitis. 2020;14:888–895. doi: 10.1093/ecco-jcc/jjaa012. [DOI] [PubMed] [Google Scholar]
- 10.Qiu Y., Chen B.L., Mao R., Zhang S.H., He Y., Zeng Z.R., Ben-Horin S., Chen M.H. Systematic review with meta-analysis: Loss of response and requirement of anti-TNFα dose intensification in Crohn’s disease. J. Gastroenterol. 2017;52:535–554. doi: 10.1007/s00535-017-1324-3. [DOI] [PubMed] [Google Scholar]
- 11.Ben-Horin S., Chowers Y. Tailoring anti-TNF therapy in IBD: Drug levels and disease activity. Nat. Rev. Gastroenterol. Hepatol. 2014;11:243–255. doi: 10.1038/nrgastro.2013.253. [DOI] [PubMed] [Google Scholar]
- 12.Hold G.L., Smith M., Grange C., Watt E.R., El-Omar E.M., Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J. Gastroenterol. 2014;20:1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue B., Luo X., Yu Z., Mani S., Wang Z., Dou W. Inflammatory Bowel Disease: A Potential Result from the Collusion between Gut Microbiota and Mucosal Immune System. Microorganisms. 2019;7:440. doi: 10.3390/microorganisms7100440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashiro Y. Gut Microbiota in Health and Disease. Ann. Nutr. Metab. 2017;71:242–246. doi: 10.1159/000481627. [DOI] [PubMed] [Google Scholar]
- 15.Huttenhower C., Gevers D., Knight R., Abubucker S., Badger J.H., Chinwalla A.T., Creasy H.H., Earl A.M. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W., Andrews E., Ajami N.J., Bonham K.S., Brislawn C.J., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pascal V., Pozuelo M., Borruel N., Casellas F., Campos D., Santiago A., Martinez X., Varela E., Sarrabayrouse G., Machiels K., et al. A microbial signature for Crohn’s disease. Gut. 2017;66:813–822. doi: 10.1136/gutjnl-2016-313235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halfvarson J., Brislawn C.J., Lamendella R., Vázquez-Baeza Y., Walters W.A., Bramer L.M., D’Amato M., Bonfiglio F., McDonald D., Gonzalez A., et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2017;2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokol H., Leducq V., Aschard H., Pham H.P., Jegou S., Landman C., Cohen D., Liguori G., Bourrier A., Nion-Larmurier I., et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokol H., Seksik P. The intestinal microbiota in inflammatory bowel diseases: Time to connect with the host. Curr. Opin. Gastroenterol. 2010;26:327–331. doi: 10.1097/MOG.0b013e328339536b. [DOI] [PubMed] [Google Scholar]
- 22.Joossens M., Huys G., Cnockaert M., De Preter V., Verbeke K., Rutgeerts P., Vandamme P., Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 23.Sokol H., Seksik P., Furet J.P., Firmesse O., Nion-Larmurier I., Beaugerie L., Cosnes J., Corthier G., Marteau P., Doré J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 24.Kushkevych I., Castro Sangrador J., Dordević D., Rozehnalová M., Černý M., Fafula R., Vítězová M., Rittmann S.K.R. Evaluation of Physiological Parameters of Intestinal Sulfate-Reducing Bacteria Isolated from Patients Suffering from IBD and Healthy People. J. Clin. Med. 2020;9:1920. doi: 10.3390/jcm9061920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kushkevych I., Dordević D., Vítězová M. Possible synergy effect of hydrogen sulfide and acetate produced by sulfate-reducing bacteria on inflammatory bowel disease development. J. Adv. Res. 2021;27:71–78. doi: 10.1016/j.jare.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Britton G.J., Contijoch E.J., Mogno I., Vennaro O.H., Llewellyn S.R., Ng R., Li Z., Mortha A., Merad M., Das A., et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity. 2019;50:212–224.e214. doi: 10.1016/j.immuni.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sokol H., Landman C., Seksik P., Berard L., Montil M., Nion-Larmurier I., Bourrier A., Le Gall G., Lalande V., De Rougemont A., et al. Fecal microbiota transplantation to maintain remission in Crohn’s disease: A pilot randomized controlled study. Microbiome. 2020;8:12. doi: 10.1186/s40168-020-0792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haiser H.J., Seim K.L., Balskus E.P., Turnbaugh P.J. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes. 2014;5:233–238. doi: 10.4161/gmic.27915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rekdal V.M., Bess E.N., Bisanz J.E., Turnbaugh P.J., Balskus E.P. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019;364:eaau6323. doi: 10.1126/science.aau6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gopalakrishnan V., Weiner B., Ford C.B., Sellman B.R., Hammond S.A., Freeman D.J., Dennis P., Soria J.C., Wortman J.R., Henn M.R. Intervention strategies for microbial therapeutics in cancer immunotherapy. Immunooncol. Technol. 2020;6:9–17. doi: 10.1016/j.iotech.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaput N., Lepage P., Coutzac C., Soularue E., Le Roux K., Monot C., Boselli L., Routier E., Cassard L., Collins M., et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 32.Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schierová D., Březina J., Mrázek J., Fliegerová K.O., Kvasnová S., Bajer L., Drastich P. Gut Microbiome Changes in Patients with Active Left-Sided Ulcerative Colitis after Fecal Microbiome Transplantation and Topical 5-aminosalicylic Acid Therapy. Cells. 2020;9:2283. doi: 10.3390/cells9102283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pigneur B., Lepage P., Mondot S., Schmitz J., Goulet O., Doré J., Ruemmele F.M. Mucosal Healing and Bacterial Composition in Response to Enteral Nutrition Vs Steroid-based Induction Therapy-A Randomised Prospective Clinical Trial in Children With Crohn’s Disease. J. Crohns Colitis. 2019;13:846–855. doi: 10.1093/ecco-jcc/jjy207. [DOI] [PubMed] [Google Scholar]
- 35.Hart L., Farbod Y., Szamosi J.C., Yamamoto M., Britz-McKibbin P., Halgren C., Zachos M., Pai N. Effect of Exclusive Enteral Nutrition and Corticosteroid Induction Therapy on the Gut Microbiota of Pediatric Patients with Inflammatory Bowel Disease. Nutrients. 2020;12:1691. doi: 10.3390/nu12061691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sprockett D., Fischer N., Boneh R.S., Turner D., Kierkus J., Sladek M., Escher J.C., Wine E., Yerushalmi B., Dias J.A., et al. Treatment-Specific Composition of the Gut Microbiota Is Associated with Disease Remission in a Pediatric Crohn’s Disease Cohort. Inflamm. Bowel Dis. 2019;25:1927–1938. doi: 10.1093/ibd/izz130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishikawa D., Sasaki T., Osada T., Kuwahara-Arai K., Haga K., Shibuya T., Hiramatsu K., Watanabe S. Changes in Intestinal Microbiota Following Combination Therapy with Fecal Microbial Transplantation and Antibiotics for Ulcerative Colitis. Inflamm. Bowel Dis. 2017;23:116–125. doi: 10.1097/MIB.0000000000000975. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y., Gao X., Ghozlane A., Hu H., Li X., Xiao Y., Li D., Yu G., Zhang T. Characteristics of Faecal Microbiota in Paediatric Crohn’s Disease and Their Dynamic Changes During Infliximab Therapy. J. Crohns Colitis. 2018;12:337–346. doi: 10.1093/ecco-jcc/jjx153. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Gao X., Zhang X., Xiao F., Hu H., Li X., Dong F., Sun M., Xiao Y., Ge T., et al. Microbial and metabolic features associated with outcome of infliximab therapy in pediatric Crohn’s disease. Gut Microbes. 2021;13:1865708. doi: 10.1080/19490976.2021.1900996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhuang X., Tian Z., Feng R., Li M., Li T., Zhou G., Qiu Y., Chen B., He Y., Chen M., et al. Fecal Microbiota Alterations Associated with Clinical and Endoscopic Response to Infliximab Therapy in Crohn’s Disease. Inflamm. Bowel Dis. 2020;26:1636–1647. doi: 10.1093/ibd/izaa253. [DOI] [PubMed] [Google Scholar]
- 41.Doherty M.K., Ding T., Koumpouras C., Telesco S.E., Monast C., Das A., Brodmerkel C., Schloss P.D. Fecal Microbiota Signatures Are Associated with Response to Ustekinumab Therapy among Crohn’s Disease Patients. mBio. 2018;9:e02120-17. doi: 10.1128/mBio.02120-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ananthakrishnan A.N., Luo C., Yajnik V., Khalili H., Garber J.J., Stevens B.W., Cleland T., Xavier R.J. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe. 2017;21:603–610.e603. doi: 10.1016/j.chom.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolho K.L., Korpela K., Jaakkola T., Pichai M.V., Zoetendal E.G., Salonen A., de Vos W.M. Fecal Microbiota in Pediatric Inflammatory Bowel Disease and Its Relation to Inflammation. Am. J. Gastroenterol. 2015;110:921–930. doi: 10.1038/ajg.2015.149. [DOI] [PubMed] [Google Scholar]
- 44.Wells G.A., Shea B., O'Connell D., Peterson J., Welcon V., Losos M., Tugwell P. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. [(accessed on 19 October 2022)]. Available online: https://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- 45.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 46.Sutton A.J., Abrams K.R., Jones D.R., Sheldon T.A., Song F. Methods for Meta-Analysis in Medical Research. John Wiley & Sons, Ltd.; New York, NY, USA: 2000. [Google Scholar]
- 47.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding N.S., McDonald J.A.K., Perdones-Montero A., Rees D.N., Adegbola S.O., Misra R., Hendy P., Penez L., Marchesi J.R., Holmes E., et al. Metabonomics and the Gut Microbiome Associated with Primary Response to Anti-TNF Therapy in Crohn’s Disease. J. Crohns Colitis. 2020;14:1090–1102. doi: 10.1093/ecco-jcc/jjaa039. [DOI] [PubMed] [Google Scholar]
- 49.Ventin-Holmberg R., Eberl A., Saqib S., Korpela K., Virtanen S., Sipponen T., Salonen A., Saavalainen P., Nissilä E. Bacterial and Fungal Profiles as Markers of Infliximab Drug Response in Inflammatory Bowel Disease. J. Crohns Colitis. 2021;15:1019–1031. doi: 10.1093/ecco-jcc/jjaa252. [DOI] [PubMed] [Google Scholar]
- 50.Olbjørn C., Cvancarova Småstuen M., Thiis-Evensen E., Nakstad B., Vatn M.H., Jahnsen J., Ricanek P., Vatn S., Moen A.E.F., Tannæs T.M., et al. Fecal microbiota profiles in treatment-naïve pediatric inflammatory bowel disease–associations with disease phenotype, treatment, and outcome. Clin. Exp. Gastroenterol. 2019;12:37–49. doi: 10.2147/CEG.S186235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valcheva R., Koleva P., Martínez I., Walter J., Gänzle M.G., Dieleman L.A. Inulin-type fructans improve active ulcerative colitis associated with microbiota changes and increased short-chain fatty acids levels. Gut Microbes. 2019;10:334–357. doi: 10.1080/19490976.2018.1526583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suskind D.L., Cohen S.A., Brittnacher M.J., Wahbeh G., Lee D., Shaffer M.L., Braly K., Hayden H.S., Klein J., Gold B., et al. Clinical and Fecal Microbial Changes With Diet Therapy in Active Inflammatory Bowel Disease. J. Clin. Gastroenterol. 2018;52:155–163. doi: 10.1097/MCG.0000000000000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ribaldone D.G., Caviglia G.P., Abdulle A., Pellicano R., Ditto M.C., Morino M., Fusaro E., Saracco G.M., Bugianesi E., Astegiano M. Adalimumab Therapy Improves Intestinal Dysbiosis in Crohn’s Disease. J. Clin. Med. 2019;8:1646. doi: 10.3390/jcm8101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kowalska-Duplaga K., Kapusta P., Gosiewski T., Sroka-Oleksiak A., Ludwig-Słomczyńska A.H., Wołkow P.P., Fyderek K. Changes in the Intestinal Microbiota Are Seen Following Treatment with Infliximab in Children with Crohn’s Disease. J. Clin. Med. 2020;9:687. doi: 10.3390/jcm9030687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salamon D., Gosiewski T., Krawczyk A., Sroka-Oleksiak A., Duplaga M., Fyderek K., Kowalska-Duplaga K. Quantitative changes in selected bacteria in the stool during the treatment of Crohn’s disease. Adv. Med. Sci. 2020;65:348–353. doi: 10.1016/j.advms.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Ventin-Holmberg R., Höyhtyä M., Saqib S., Korpela K., Nikkonen A., Salonen A., de Vos W.M., Kolho K.L. The gut fungal and bacterial microbiota in pediatric patients with inflammatory bowel disease introduced to treatment with anti-tumor necrosis factor-α. Sci. Rep. 2022;12:6654. doi: 10.1038/s41598-022-10548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Y., Xu Z.Z., He Y., Yang Y., Liu L., Lin Q., Nie Y., Li M., Zhi F., Liu S., et al. Gut Microbiota Offers Universal Biomarkers across Ethnicity in Inflammatory Bowel Disease Diagnosis and Infliximab Response Prediction. mSystems. 2018;3:e00188-17. doi: 10.1128/mSystems.00188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aden K., Rehman A., Waschina S., Pan W.H., Walker A., Lucio M., Nunez A.M., Bharti R., Zimmerman J., Bethge J., et al. Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology. 2019;157:1279–1292.e1211. doi: 10.1053/j.gastro.2019.07.025. [DOI] [PubMed] [Google Scholar]
- 59.Dovrolis N., Michalopoulos G., Theodoropoulos G.E., Arvanitidis K., Kolios G., Sechi L.A., Eliopoulos A.G., Gazouli M. The Interplay between Mucosal Microbiota Composition and Host Gene-Expression is Linked with Infliximab Response in Inflammatory Bowel Diseases. Microorganisms. 2020;8:438. doi: 10.3390/microorganisms8030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schierova D., Roubalova R., Kolar M., Stehlikova Z., Rob F., Jackova Z., Coufal S., Thon T., Mihula M., Modrak M., et al. Fecal Microbiome Changes and Specific Anti-Bacterial Response in Patients with IBD during Anti-TNF Therapy. Cells. 2021;10:3188. doi: 10.3390/cells10113188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchis-Artero L., Martínez-Blanch J.F., Manresa-Vera S., Cortés-Castell E., Valls-Gandia M., Iborra M., Paredes-Arquiola J.M., Boscá-Watts M., Huguet J.M., Gil-Borrás R., et al. Evaluation of changes in intestinal microbiota in Crohn’s disease patients after anti-TNF alpha treatment. Sci. Rep. 2021;11:10016. doi: 10.1038/s41598-021-88823-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Effenberger M., Reider S., Waschina S., Bronowski C., Enrich B., Adolph T.E., Koch R., Moschen A.R., Rosenstiel P., Aden K., et al. Microbial Butyrate Synthesis Indicates Therapeutic Efficacy of Azathioprine in IBD Patients. J. Crohns Colitis. 2021;15:88–98. doi: 10.1093/ecco-jcc/jjaa152. [DOI] [PubMed] [Google Scholar]
- 63.Park Y.E., Moon H.S., Yong D., Seo H., Yang J., Shin T.S., Kim Y.K., Kim J.R., Lee Y.N., Kim Y.H., et al. Microbial changes in stool, saliva, serum, and urine before and after anti-TNF-α therapy in patients with inflammatory bowel diseases. Sci. Rep. 2022;12:6359. doi: 10.1038/s41598-022-10450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaakoush N.O., Day A.S., Leach S.T., Lemberg D.A., Nielsen S., Mitchell H.M. Effect of exclusive enteral nutrition on the microbiota of children with newly diagnosed Crohn’s disease. Clin. Transl. Gastroenterol. 2015;6:e71. doi: 10.1038/ctg.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quince C., Ijaz U.Z., Loman N., Eren A.M., Saulnier D., Russell J., Haig S.J., Calus S.T., Quick J., Barclay A., et al. Extensive Modulation of the Fecal Metagenome in Children with Crohn’s Disease During Exclusive Enteral Nutrition. Am. J. Gastroenterol. 2015;110:1718–1729; quiz 1730. doi: 10.1038/ajg.2015.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang W., Huang Y., Shi P., Wang Y., Zhang Y., Xue A., Tang Z., Hu W., Sun H., Zhang P., et al. Effect of Exclusive Enteral Nutrition on the Disease Process, Nutrition Status, and Gastrointestinal Microbiota for Chinese Children with Crohn’s Disease. J. Parenter. Enter. Nutr. 2021;45:826–838. doi: 10.1002/jpen.1938. [DOI] [PubMed] [Google Scholar]
- 67.Dunn K.A., Moore-Connors J., MacIntyre B., Stadnyk A.W., Thomas N.A., Noble A., Mahdi G., Rashid M., Otley A.R., Bielawski J.P., et al. Early Changes in Microbial Community Structure Are Associated with Sustained Remission after Nutritional Treatment of Pediatric Crohn’s Disease. Inflamm. Bowel Dis. 2016;22:2853–2862. doi: 10.1097/MIB.0000000000000956. [DOI] [PubMed] [Google Scholar]
- 68.Costa-Santos M.P., Palmela C., Torres J., Ferreira A., Velho S., Ourô S., Glória L., Gordo I., Maio R., Cravo M. Preoperative enteral nutrition in adults with complicated Crohn’s disease: Effect on disease outcomes and gut microbiota. Nutrition. 2020;70s:100009. doi: 10.1016/j.nutx.2020.100009. [DOI] [PubMed] [Google Scholar]
- 69.Diederen K., Li J.V., Donachie G.E., de Meij T.G., de Waart D.R., Hakvoort T.B.M., Kindermann A., Wagner J., Auyeung V., Te Velde A.A., et al. Exclusive enteral nutrition mediates gut microbial and metabolic changes that are associated with remission in children with Crohn’s disease. Sci. Rep. 2020;10:18879. doi: 10.1038/s41598-020-75306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang J., Chen L., Chen Y., Chen H. Exclusive enteral nutrition remodels the intestinal flora in patients with active Crohn’s disease. BMC Gastroenterol. 2022;22:212. doi: 10.1186/s12876-022-02293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suskind D.L., Lee D., Kim Y.M., Wahbeh G., Singh N., Braly K., Nuding M., Nicora C.D., Purvine S.O., Lipton M.S., et al. The Specific Carbohydrate Diet and Diet Modification as Induction Therapy for Pediatric Crohn’s Disease: A Randomized Diet Controlled Trial. Nutrients. 2020;12:3749. doi: 10.3390/nu12123749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson B., Eyice Ö., Koumoutsos I., Lomer M.C., Irving P.M., Lindsay J.O., Whelan K. Prebiotic Galactooligosaccharide Supplementation in Adults with Ulcerative Colitis: Exploring the Impact on Peripheral Blood Gene Expression, Gut Microbiota, and Clinical Symptoms. Nutrients. 2021;13:3598. doi: 10.3390/nu13103598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X., Lou L., Tang H., Tan X., Bi J., Wu H., Li N., Wang Y., Mao J. Adsorptive granulomonocytapheresis alters the gut bacterial microbiota in patients with active ulcerative colitis. J. Clin. Apher. 2021;36:454–464. doi: 10.1002/jca.21887. [DOI] [PubMed] [Google Scholar]
- 74.Olendzki B., Bucci V., Cawley C., Maserati R., McManus M., Olednzki E., Madziar C., Chiang D., Ward D.V., Pellish R., et al. Dietary manipulation of the gut microbiome in inflammatory bowel disease patients: Pilot study. Gut Microbes. 2022;14:2046244. doi: 10.1080/19490976.2022.2046244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Forbes J.D., Van Domselaar G., Bernstein C.N. The Gut Microbiota in Immune-Mediated Inflammatory Diseases. Front. Microbiol. 2016;7:1081. doi: 10.3389/fmicb.2016.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 78.Martin-Gallausiaux C., Béguet-Crespel F., Marinelli L., Jamet A., Ledue F., Blottière H.M., Lapaque N. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci. Rep. 2018;8:9742. doi: 10.1038/s41598-018-28048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paramsothy S., Nielsen S., Kamm M.A., Deshpande N.P., Faith J.J., Clemente J.C., Paramsothy R., Walsh A.J., van den Bogaerde J., Samuel D., et al. Specific Bacteria and Metabolites Associated with Response to Fecal Microbiota Transplantation in Patients with Ulcerative Colitis. Gastroenterology. 2019;156:1440–1454.e1442. doi: 10.1053/j.gastro.2018.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.