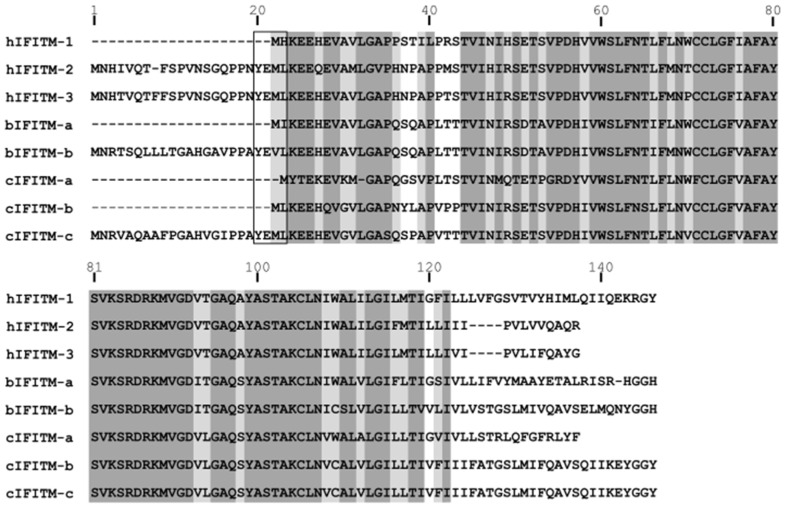
Figure 1.
Sequence alignment of human, bovine, and camel IFITMs. The alignment was performed using MUltiple Sequence Comparison (MUSCLE) software. Aligned amino acids are highlighted in dark or light grey depending on whether the residue is present in at least 80% or 50% of the sequences, respectively. Note that shorter N-terminal domains are characterized by the absence of the YXXφ motif (rectangle) present in hIFITM-3 as well as in other IFITMs. hIFITM-1, human IFITM-1; hIFITM-2, human IFITM-2; hIFITM-3, human IFITM-3; bIFITM-a, bovine IFITM-a; bIFITM-b, bovine IFITM-b; cIFITM-a, camel IFITM-a; cIFITM-b, camel IFITM-b; cIFITM-c, camel IFITM-c.