Abstract
The VirB11 ATPase is a subunit of the Agrobacterium tumefaciens transfer DNA (T-DNA) transfer system, a type IV secretion pathway required for delivery of T-DNA and effector proteins to plant cells during infection. In this study, we examined the effects of virB11 mutations on VirB protein accumulation, T-pilus production, and substrate translocation. Strains synthesizing VirB11 derivatives with mutations in the nucleoside triphosphate binding site (Walker A motif) accumulated wild-type levels of VirB proteins but failed to produce the T-pilus or export substrates at detectable levels, establishing the importance of nucleoside triphosphate binding or hydrolysis for T-pilus biogenesis. Similar findings were obtained for VirB4, a second ATPase of this transfer system. Analyses of strains expressing virB11 dominant alleles in general showed that T-pilus production is correlated with substrate translocation. Notably, strains expressing dominant alleles previously designated class II (dominant and nonfunctional) neither transferred T-DNA nor elaborated detectable levels of the T-pilus. By contrast, strains expressing most dominant alleles designated class III (dominant and functional) efficiently translocated T-DNA and synthesized abundant levels of T pilus. We did, however, identify four types of virB11 mutations or strain genotypes that selectively disrupted substrate translocation or T-pilus production: (i) virB11/virB11∗ merodiploid strains expressing all class II and III dominant alleles were strongly suppressed for T-DNA translocation but efficiently mobilized an IncQ plasmid to agrobacterial recipients and also elaborated abundant levels of T pilus; (ii) strains synthesizing two class III mutant proteins, VirB11, V258G and VirB11.I265T, efficiently transferred both DNA substrates but produced low and undetectable levels of T pilus, respectively; (iii) a strain synthesizing the class II mutant protein VirB11.I103T/M301L efficiently exported VirE2 but produced undetectable levels of T pilus; (iv) strains synthesizing three VirB11 derivatives with a four-residue (HMVD) insertion (L75.i4, C168.i4, and L302.i4) neither transferred T-DNA nor produced detectable levels of T pilus but efficiently transferred VirE2 to plants and the IncQ plasmid to agrobacterial recipient cells. Together, our findings support a model in which the VirB11 ATPase contributes at two levels to type IV secretion, T-pilus morphogenesis, and substrate selection. Furthermore, the contributions of VirB11 to machine assembly and substrate transfer can be uncoupled by mutagenesis.
Agrobacterium tumefaciens VirB11 is a member of a family of ATPases widely distributed among members of the domains Bacteria and Archaea (38, 48, 67). Mutational studies have established the importance of VirB11 homologs for translocation of macromolecules across the cell envelope in association with type IV secretion (17, 29, 32, 44, 52) and competence (1) systems and type II protein secretion and pilus biogenesis systems (54). These proteins hydrolyze ATP, as demonstrated for VirB11 (17), TrbB of the IncP plasmid RP4 (38), TrwD of the IncW plasmid R388 (38, 52), and HP0525 of the cag pathogenicity island of Helicobacter pylori (38). Interestingly, recent electron microscopy studies determined that the last three proteins assemble as hexameric rings in solution (37, 38), and a crystallography study identified a binary complex of HP0525 bound to ADP as a two-tiered hexameric ring with a central cavity of 50 nm (67). This structural information and the demonstrated importance of the VirB11 ATPases for assembly or function of supramolecular surface organelles has prompted the proposal that the VirB11 ATPases function as chaperones for trafficking of unfolded substrates across the cytoplasmic membrane.
VirB11 is one of 11 VirB proteins required for efficient assembly of the A. tumefaciens transfer DNA (T-DNA) transfer system (7). This type IV secretion system translocates oncogenic T-DNA and effector proteins to susceptible plant cells during the course of A. tumefaciens infection (15). The T-DNA transfer system is composed of a transenvelope channel for substrate translocation and the T pilus for establishment of A. tumefaciens contacts with susceptible plant cells (15, 35). Like HP0525, VirB11 self-assembles as a higher-order homomultimeric complex via domains in its N- and C-terminal halves (49, 50, 69). VirB11 localizes at the inner face of the cytoplasmic membrane independently of interactions with other VirB proteins, and studies of mutant proteins with defects in the nucleoside triphosphate binding pocket (Walker A motif) suggest that this membrane interaction is modulated by ATP binding or hydrolysis (51). No heterologous protein contacts have yet been reported, but VirB11 is stabilized by the production of other VirB proteins, most notably the outer membrane VirB7 lipoprotein-VirB9 protein complex (28, 51). In addition, dominant virB11 mutations are suppressed by overproduction of VirB proteins, including the outer membrane protein VirB9, the bitopic cytoplasmic membrane protein VirB10, and VirB11 itself (7, 28, 69). The current data, therefore, support a model in which the VirB11 homooligomer, probably configured as a homohexameric ring, is situated at the cytoplasmic membrane in direct contact both with the lipid bilayer and with subunits of the translocation channel.
Analyses of virB11 null mutants have established the importance of VirB11 for the transfer of several secretion substrates (7, 17, 29, 62). These substrates include (i) the T-DNA transfer intermediate, which minimally consists of the VirD2 endonuclease attached covalently to the 5′ end of a single strand of T-DNA (T-strand) (60, 71); (ii) the VirE2 single-stranded-DNA-binding protein (SSB) and another virulence factor termed VirF (16, 62); and (iii) the mobilizable IncQ plasmid RSF1010 (9, 11, 29). Interestingly, wild-type A. tumefaciens harboring an RSF1010 derivative inefficiently transfers the T-DNA to plants, whereas the inhibitory effect of this IncQ plasmid on T-DNA transfer is suppressed by overexpression of virB9, virB10, and virB11 (64). These early findings led to the suggestion that VirB9, VirB10, and VirB11 complex formation is rate limiting for assembly of functional T-DNA transfer machines in a cell (64). More recent studies showed that the IncQ plasmid preferentially blocks VirE2 export but is also inhibitory for T-strand–VirD2 export, suggesting that the secretion substrates compete for available transfer machines (9, 60).
VirB11 also is thought to contribute to assembly of the T-DNA transfer system. The principal finding supporting a morphogenetic function is that virB11 null mutants fail to elaborate T pili (41, 53). However, nonpolar mutations of all 11 virB genes abolish T-pilus formation (41, 53), and no studies have distinguished direct from indirect contributions of the VirB proteins to pilus morphogenesis.
In this study, we characterized the effects of various virB11 dominant or recessive alleles on T-pilus formation and substrate transfer. Our findings support a model whereby ATP-dependent activities of VirB11 are required at two distinct stages for translocation, biogenesis of the T pilus, and selection of secretion substrates. The functional importance of the T pilus is discussed in the context of the finding that certain virB11 mutations uncouple T-pilus production from substrate translocation.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
Table 1 lists the bacterial strains and plasmids used in this study. Conditions and media for growth of A. tumefaciens and Escherichia coli and for induction of A. tumefaciens vir genes in induction medium (IM) containing acetosyringone (AS) have been described previously (69). Plasmids were maintained in E. coli and A. tumefaciens by addition of carbenicillin (100 μg/ml), kanamycin (100 μg/ml), tetracycline (5 μg/ml), gentamycin (50 μg/ml), or spectinomycin (500 μg/ml) to the growth medium. ColE1 plasmids were introduced into A. tumefaciens by ligation to the IncP plasmid pSW172 or pXZ151; these cointegrate plasmids are given the ColE1 name plus a B to denote ligation to a broad-host-range replicon.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | λ− φ80d/lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1 | GIBCO-BRL |
| CJ236 | dut ung thi relA; pCJ105 (Camr) | Bio-Rad |
| S17-1 | Tra genes from pRP4 integrated into chromosome for plasmid mobilization | 58 |
| A. tumefaciens | ||
| A136 | Strain C58 cured of pTi plasmid | 65 |
| A348 | A136 containing pTiA6NC | 31 |
| A348Spcr | A348 with Spcr by spontaneous mutagenesis | This study |
| PC1002 | A348 with virB2 from pTiA6NC deleted | 7 |
| PC1004 | A348 with virB4 from pTiA6NC deleted | 6 |
| PC1011 | A348 with virB11 from pTiA6NC deleted | 7 |
| Plasmid vectors | ||
| pBSIISK+NdeI | Crbr; cloning vector | 7 |
| pBSIIKS+NdeI | Crbr; cloning vector | 7 |
| pSW172 | Tetr; broad-host-range IncP plasmid containing Plac with downstream polylinker sequence | 13 |
| pXZ151 | pSW172 with Kanr gene | 70 |
| pXZ1000K | pSW172 ligated at KpnI site to ColE1 pBSKSK+ | 69 |
| pML122ΔKm | Genr; IncQ RSF1010 plasmid derivative with gentamycin resistance | 30 |
| Expression plasmids | ||
| pSR1 | Crbr; pBKS+NdeI with virB11 expressed from PvirB | 51 |
| pJC902 | Crbr; pSR1 digested with SalI-XhoI and religated | This study |
| pJC903 | Crbr; pJC902 with XhoI site introduced after stop codon by site-directed mutagenesis | This study |
| pJC8xxx series | pJC903 derivatives with 12-bp insertions corresponding to tandem NdeI-SalI sites immediately after codons indicated by xxx | This study |
| pXZB100 | Crbr Kanr; pXZ151 with PvirB::virB11 | 69 |
| pXZB1xx series | Crbr Kanr; pXZB100 with wild-type virB11 gene replaced with PCR-mutagenized virB11 alleles 1 through 12 | 69 |
| pSRB9114 | Crbr Tetr; pSW172 with PvirB::virB11 | 49 |
| pPCB7111 | Crbr Tetr; pSW172 with Plac::virB11 | 51 |
| pPCB7112 | Crbr Tetr; pSW172 with Plac::virB11ΔGKT174–176 | 51 |
| pPCB7113 | Crbr Tetr; pSW172 with Plac::virB11K175Q | 51 |
| pZDH10 | Crbr Tetr; pSW172 with Plac::virB4 | 6 |
| pBBB15 | Crbr Tetr; pSW172 with Plac::virB4K439Q | 6 |
| pBBB17 | Crbr Tetr; pSW172 with Plac::virB4ΔGKT438–440 | 6 |
| pVSB10 | Crbr Tetr; pSW172 with PvirB::virB2C64S | 53 |
Insertion mutagenesis of virB11.
In-frame insertions of a four-residue sequence (HMVD) were introduced at ∼20-residue intervals along the length of VirB11 as follows. Plasmid pSR1, a pBSIIKS+ derivative with PvirB::virB11, was digested with XhoI and SalI and religated to destroy these restriction sites, yielding pJC902. Single-stranded pJC902 served as a template for introduction of an XhoI site immediately following the virB11 TAG stop codon with the oligonucleotide CCTAAATCAATAGCTCGAGTAGCTGTAACC (the XhoI site is underlined; stop codons are in boldface) according to the method of Kunkel (40). The resulting plasmid, pJC903, served as the template for introduction of tandem, in-frame NdeI-SalI restriction sites (CATATGGTCGAC) at ∼60-bp intervals along the virB11 gene by oligonucleotide-directed mutagenesis. Mutant alleles were identified by restriction enzyme digestion, and the insertion mutations were confirmed by sequencing across the entire virB11 gene. Plasmids expressing the mutant alleles were designated pJC8xxx, where xxx is the position of the residue relative to the beginning of the protein that immediately precedes the four-residue insertion.
Protein analysis, immunoblotting, and cell fractionation.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or a Tricine–SDS-PAGE system as previously described (50). Vir proteins were visualized by SDS-PAGE, protein transfer to nitrocellulose membranes, and immunoblot development with goat anti-rabbit antibodies conjugated to alkaline phosphatase and histochemical substrates. For enhanced sensitivity, blots were alternatively developed with anti-rabbit antibodies conjugated to horseradish peroxidase, and antibody-antigen interactions were visualized by chemiluminescence (Amersham, Arlington Heights, Ill.). Proteins were loaded on a per-cell equivalent basis to compare VirB protein abundance in different strains. Molecular size markers were from GIBCO-BRL (Grand Island, N.Y.). Antibody specificities were previously documented for the VirB1, VirB2, VirB4, VirB5, VirB7 through VirB11, and VirE2 proteins (20, 50, 53, 68).
T-pilus isolation and sucrose fractionation.
T pili were isolated as previously described (53). Briefly, A. tumefaciens strains were grown to an optical density at 600 nm OD600 of 0.5 in MG/L medium (27) at 28°C. The cells were pelleted, diluted fivefold in IM, and incubated for 6 h at 22°C. Two hundred microliters of AS-induced culture was spread on IM agar plates, and the plates were incubated for 3 days at 18°C. The cells were then gently scraped off the plates in 50 mM KH2PO4 buffer, pH 5.5 (buffer A), and pelleted by centrifugation at 14,000 × g for 15 min at room temperature. The supernatant was removed, and the cell pellet was resuspended in 50 mM phosphate buffer. This suspension was passed through a 25-gauge needle 10 times to collect flagella, pili, and surface proteins. The sheared bacterial cells were pelleted by centrifugation at 14,000 × g for 30 min at 4°C. The remaining supernatant was filtered through a 0.22-μm-pore-size cellulose acetate membrane to remove unpelleted cells. When necessary, the culture supernatants and sheared materials were concentrated with trichloroacetic acid (53).
T pili were harvested by centrifugation of filtered exocellular material at 100,000 × g for 1 h at 4°C. The pelleted material was analyzed by SDS-PAGE and immunostaining. The material was also solubilized in buffer A and loaded onto a 20 to 70% linear sucrose density gradient (5 ml) prepared with buffer A. The T pili were then fractionated by ultracentrifugation in an SW55 Beckman rotor at 80,000 × g for 20 h at 4°C. Fractions (0.5 ml) were collected from the bottoms of the centrifugation tubes, analyzed for the presence of Vir proteins by immunoblotting, and analyzed for other proteins by silver staining (53).
Conjugation assay.
The RSF1010 derivative pML122ΔKm was introduced into various A. tumefaciens donor strains by diparental mating with E. coli strain S17-1(pML122Δ Km) (29, 58). A. tumefaciens strains carrying pML122ΔKm were mated with an Spcr derivative of A348 by use of a protocol described previously (10). Briefly, mid-log-phase (OD600 = 0.5) cells were harvested and incubated in liquid IM containing AS (200 μM) at 22°C for 6 h to induce expression of the vir genes. Five microliters each of preinduced donor and recipient cells was mixed on a cellulose acetate filter on an IM agar plate containing AS (200 μM), and the plate was incubated at 18°C for 3 days. Mating mixtures were recovered from filters in IM medium and directly plated onto MG/L medium selective for transconjugants, or cultures were serially diluted and plated for determination of transconjugant and donor cell numbers. Frequencies of transfer were estimated as transconjugants recovered per donor cell. Each assay was performed in triplicate, and three or more independent experiments were performed.
Virulence assay.
A. tumefaciens strains were tested for virulence by inoculating wound sites of Kalanchoe daigremontiana leaves (69). Controls for the tumorigenesis assay included coinoculating the same leaf with wild-type A348 (virulent) and PC1011 (avirulent) strains. Each experiment was repeated at least three times for each strain on separate leaves.
RESULTS
Effects of Walker A mutations on T-pilus production.
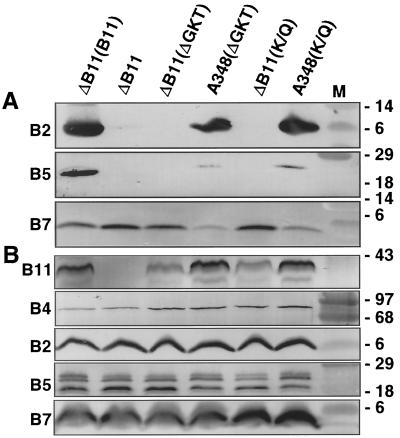
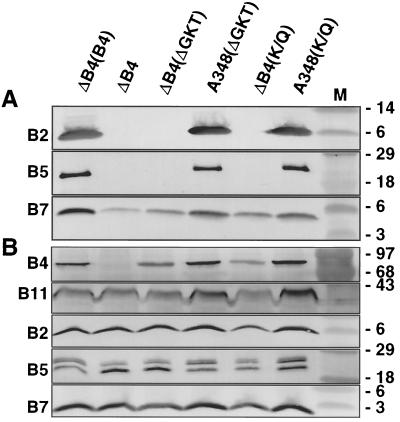
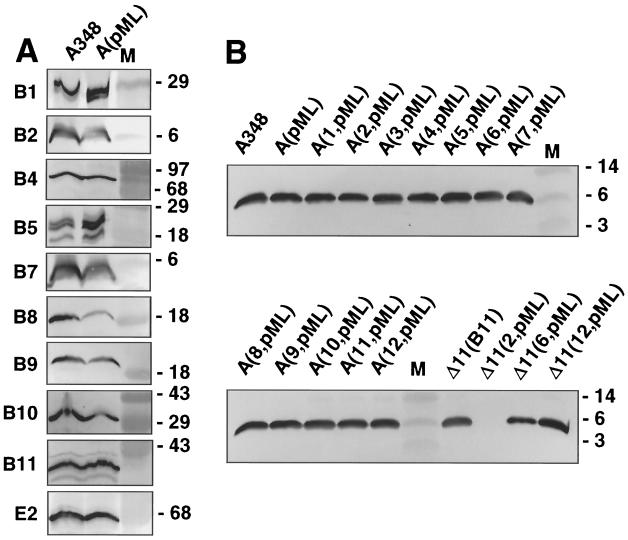
A. tumefaciens strains synthesizing mutant forms of the VirB4 or VirB11 ATPases with defects in the conserved nucleoside triphosphate binding sites (Walker A domain) fail to export T-DNA to plant cells (6, 30, 51, 61). To determine whether strains synthesizing these mutant proteins elaborate T pili, exocellular material was examined for the presence of VirB2 pilin (42) and the T-pilus-associated proteins VirB5 (53, 55) and VirB7 lipoprotein (53). As shown in Fig. 1A and 2A, mutant strains with nonpolar deletions of virB4 (strain PC1004) or virB11 (strain PC1011) engineered to express wild-type virB4 or virB11 from an IncP replicon possess abundant amounts of exocellular VirB2 and VirB5 in the exocellular fractions. These strains translocate substrates at wild-type frequencies (6, 7). By contrast, the isogenic strains expressing alleles for the Walker A mutant proteins possessed undetectable levels of these proteins, indicative of defects in T-pilus production. virB11∗ alleles encoding the Walker A derivatives are recessive to wild-type VirB11 (51, 61), whereas the corresponding virB4∗ alleles are dominant with respect to substrate transfer (8, 30). Interestingly, strain A348 expressing the mutated virB11 or virB4 allele, hereafter designated virB11/virB11∗ and virB4/virB4∗ merodiploid strains, respectively, possessed abundant levels of exocellular VirB2 and VirB5 (Fig. 1A and 2A). These findings suggest that the dominance of virB4∗ alleles encoding the Walker A derivatives most probably is not due to a disruption in T-pilus production.
FIG. 1.
Effects of VirB11 Walker A mutations on accumulation of T-pilus proteins and other VirB proteins in exocellular (A) and cellular (B) fractions obtained as described in the text. Strains: ΔB11(B11), strain PC1011(pSRB1) expressing virB11 from an IncP replicon; ΔB11, strain PC1011; ΔB11(ΔGKT), PC1011(pPCB7112) expressing virB11ΔGKT174-176; A348(ΔGKT), A348(pPCB7112) coexpressing virB11 and virB11ΔGKT174-176; ΔB11(K/Q), PC1011(pPCB7113) expressing virB11K175Q; A348(K/Q), A348(pPCB7113) coexpressing virB11 and virB11K175Q. M, molecular mass markers, with sizes in kilodaltons indicated on the right. The blots were developed with antisera to the VirB proteins listed on the left.
FIG. 2.
Effects of VirB4 Walker A mutations on accumulation of T-pilus-associated proteins and other VirB proteins in exocellular (A) and cellular (B) fractions obtained as described in the text. Strains: ΔB4(B4), strain PC1004(pZDH10) expressing virB4 from an IncP replicon; ΔB4, strain PC1004; ΔB4(ΔGKT), PC1004(pBB17) expressing virB4ΔGKT174–176; A348(ΔGKT), A348(pBB17) coexpressing virB4 and virB4ΔGKT174–176; ΔB4(K/Q), PC1004(pBB15) expressing virB4K175Q; A348(K/Q), A348(pBB15) coexpressing virB4 and virB4K175Q. M, molecular mass markers, with sizes in kilodaltons indicated on the right. The blots were developed with antisera to the VirB proteins listed on the left.
Figures 1A and 2A show that the presence of VirB5 in exocellular fractions correlates well with that of VirB2 pilin. By contrast, all strains examined possessed detectable levels of exocellular VirB7, although the virB4 and virB11 mutations did influence accumulation of the exocellular lipoprotein in different ways. PC1004 engineered to produce native VirB4 accumulated abundant levels of exocellular VirB7, whereas PC1004 itself or PC1004 engineered to produce Walker A mutant proteins accumulated the lipoprotein at appreciably lower levels (Fig. 2A). By contrast, PC1011 itself or PC1011 engineered to produce either native VirB11 or the Walker A mutant proteins accumulated abundant levels of exocellular VirB7, yet the virB11/virB11∗ merodiploid strains accumulated exocellular lipoprotein at low levels (Fig. 1A). These findings suggest that the VirB4 and VirB11 ATPases influence the sorting of the VirB7 lipoprotein across the outer membrane by mechanisms influenced by their oligomeric structures and by the capacity to bind or hydrolyze ATP.
All PC1011 and PC1004 strains accumulated abundant levels of cellular forms of VirB2, VirB5, and VirB7 (Fig. 1B and 2B), as well as other VirB proteins (data not shown). Of further note, the virB4 and virB11 mutations did not affect the migration of either the cellular or exocellular forms of the T-pilus-associated proteins in SDS-polyacrylamide gels. Each of these proteins possesses cleavable signal sequences, and VirB2 and VirB7 are further processed in ways that affect their migration in SDS-polyacrylamide gels (25, 27). Therefore, VirB4 and VirB11 probably do not participate in the maturation of T-pilus proteins. Our anti-VirB5 antiserum reacts against three VirB5 species present in cell extracts (Fig. 1B and 2B). The two smaller species presumably correspond to degradation products that fail to interact productively with the T pilus, as deduced by their absence in exocellular fractions (Fig. 1A and 2A). Although VirB4 and VirB11 clearly are required for VirB5 sorting to the exocellular fraction, neither protein seems to influence the formation of the presumed VirB5 degradation products (Fig. 1B and 2B).
Effects of virB11 dominant mutations on T-pilus production.
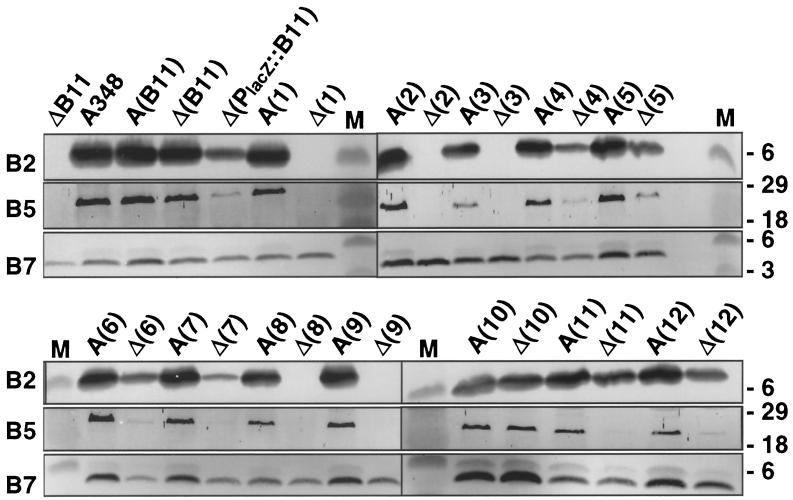
Next, we determined whether other VirB11 mutations also disrupt both T-DNA transfer and T-pilus production. Of special interest was a set of dominant alleles shown in a previous study to strongly suppress T-DNA transfer when expressed in wild-type A348 cells (69). These dominant alleles fell into two classes. Alleles designated class II encode nonfunctional mutant proteins, whereas the class III alleles encode functional VirB11 proteins, as judged by the capacity of these alleles to restore the virulence of strain PC1011.
All merodiploid strains expressing wild-type virB11 and a dominant allele accumulated exocellular VirB2 and VirB5 at levels comparable to those in isogenic wild-type A348 (Fig. 3; Table 2). By contrast, most of the PC1011 strains expressing the class III alleles accumulated exocellular VirB2 and VirB5, whereas none of the corresponding strains expressing the class II alleles accumulated these pilus proteins at detectable levels. As shown below, PC1011 strains expressing the class III alleles elaborate wild-type T pili. Thus, in general, the class II and III mutations are distinguished by their capacity to support T-pilus production in the PC1011 genetic background.
FIG. 3.
Effects of virB11 dominant mutations on T-pilus production. Exocellular fractions from PC1011 (designated ΔB11 or Δ) and A348 (designated A348 or A) strains expressing wild-type virB11 and alleles 1 to 12 were analyzed for accumulation of T-pilus-associated proteins, VirB2, VirB5, and VirB7, as listed on the left of each immunoblot. Strain Δ(B11) is pC1011(pSRB1) expressing PvirB::virB11, and strain Δ(PlacZ::B11) is PC1011(pPCB117) expressing PlacZ::virB11; alleles 1 to 12 expressed from PvirB are carried on plasmids pXZB101 to pXZB112. Plasmid pXZB104 was shown previously to encode a wild-type virB11 gene (69); however, we identified a mutation in the PvirB promoter of pXZ104 that results in a reduction in VirB11 production levels. M, molecular mass markers, with sizes in kilodaltons indicated on the right.
TABLE 2.
Phenotypes of virB11 dominant alleles
| Allele or strain type | pXZB1xxa plasmids or strain | Mutation | PC1011(pXZB1xx) strains
|
A348(pXZB1xx) merodiploid strains
|
||||
|---|---|---|---|---|---|---|---|---|
| T-DNA transferb | pML122ΔKm mobilization (Tcs/donor)c | T-pilus productiond | T-DNA transferb | pML122ΔKm mobilization (Tcs/donor)c | T-pilus productiond | |||
| Class III (dominant | pXZB102 | I265T | +++ | 8.5 × 10−4 | − | ± | 6 × 10−3 | +++ |
| functional) | pXZB105 | L75F | +++ | 1.6 × 10−3 | ++ | + | 3.6 × 10−3 | +++ |
| pXZB106 | Q135E | +++ | 1 × 10−3 | ++ | ± | 1.6 × 10−4 | +++ | |
| pXZB107 | V258G | +++ | 2.5 × 10−5 | + | + | 6 × 10−3 | +++ | |
| pXZB110 | V116I | +++ | 1.4 × 10−3 | +++ | + | 2.5 × 10−3 | +++ | |
| pXZB111 | V49A/E256G | +++ | 2 × 10−4 | ++ | + | 4.5 × 10−3 | +++ | |
| pXZB112 | N73Y/T98I | +++ | 3.3 × 10−4 | ++ | + | 3.5 × 10−3 | +++ | |
| Class II (dominant | pXZB101 | I88T/I103T | − | 2.4 × 10−7 | − | + | 5 × 10−3 | +++ |
| nonfunctional) | pXZB103 | H269R | − | <10−7 | − | + | 2.5 × 10−4 | +++ |
| pXZB108 | L11P/D56G/E335G | − | 5 × 10−7 | − | + | 4.2 × 10−3 | +++ | |
| pXZB109 | I103T/M301L | −e | <10−7 | − | + | 8 × 10−3 | +++ | |
| Control | A348 | WTg | NAf | NA | NA | +++ | 3.7 × 10−3 | +++ |
| PC1011 | ΔvirB11 | − | <10−7 | − | NA | NA | NA | |
| PC1011(pXZB100) | PvirB::virB11 | +++ | 4.5 × 10−3 | +++ | NA | NA | NA | |
| PC1011(pPCB7111) | Plac::virB11 | +++ | 4.1 × 10−4 | ++ | NA | NA | NA | |
Underlined number denotes allele, as originally named in Zhou et al. (69).
Monitored by virulence assays on Kalanchoe wound sites. +++, wild-type transfer; ±, barely detectable transfer; −, no transfer
Data are the means of three trials from a single experiment. Three independent experiments yielded similar results.
+++ abundant pilus production, − no detectable pilus proteins in exocellular fraction.
PC1011(pXZB109) exported VirE2 but not DNA substrates, as determined by mixed-infection assays.
NA, not applicable.
WT, wild type.
Several PC1011 strains expressing class III alleles (e.g., alleles 2, 5, 6, and 7) accumulated exocellular VirB2 and VirB5 at reduced levels (Fig. 3). In the most extreme case, PC1011(pXZB102) expressing virB11.I265T did not accumulate any detectable exocellular VirB2 or VirB5 (Fig. 3). These results are of considerable interest, because all PC1011 strains expressing class III alleles, including PC1011(pXZB102), transfer T-DNA at wild-type frequencies (Table 2) (69). Thus, certain class III mutations, most notably I265T, are permissive for T-DNA transfer but disrupt or prevent T-pilus production when synthesized in the absence of native VirB11.
All merodiploid and PC1011 strains produced exocellular VirB7 (Fig. 3). Interestingly, however, there was some allele-specific variation in levels of exocellular VirB7; for example, compare strains expressing alleles 10, 11, and 12 (Fig. 3). These findings further support the notion that VirB11 influences the release of VirB7 lipoprotein to the extracellular milieu.
Effects of virB11 mutations on VirB2 pilin distribution in sucrose gradients.
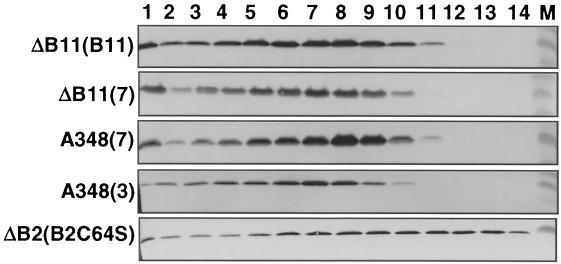
The fractionation of VirB2 through sucrose gradients can be used to monitor the production and structural integrity of the T pilus (41, 42, 53, 55). Figure 4 shows the distribution profiles for VirB2 from strains expressing wild-type virB11, a class II allele (V258G), and a class III allele (H269R). Exocellular VirB2 from each of these strains displayed similar distribution profiles in sucrose gradients. These findings are representative of results obtained for all strains expressing class II and III alleles that were shown to accumulate exocellular T-pilus proteins (data not shown). At this level of resolution, therefore, the virB11/virB11∗ merodiploid strains expressing class II and III alleles and the PC1011 strains expressing class III alleles produce wild-type T pili. For comparison, we previously determined that PC1002(pVSB10) engineered to produce the VirB2C64S mutant pilin elaborates a T pilus with a distinct distribution profile in sucrose gradients (53) (Fig. 4). Based on its distribution pattern, we suspect VirB2C64S polymerizes as a pilus that is shorter or less stable than the native T pilus (53).
FIG. 4.
Sucrose density gradient distribution profiles of T pili from various virB11 mutant strains. Exocellular proteins from the strains listed on the left were centrifuged through identically prepared sucrose density gradients, and the fractions were analyzed for the presence of VirB2 pilin. Strains: ΔB11(B11), PC1011(pXZB100); ΔB11(7), PC1011(pXZB107); A348(7), A348(pXZB107); A348(3), A348(pXZB103); ΔB2(B2C64S), PC1002(pVSB10) that synthesizes mutant pilin (53).
Further evidence for uncoupling of substrate transfer and T-pilus production.
The pilus− Tra+ phenotype of PC1011(pXZB102) and the pilus+ Tra-deficient phenotypes of virB11/virB11∗ merodiploid strains expressing both classes of dominant alleles suggest that pilus production and substrate translocation can be uncoupled by mutation of VirB11. To further test this model, we assayed strains expressing the dominant virB11 alleles for translocation of substrates other than T-DNA. Wild-type A348 harboring an IncQ plasmid, pML122ΔKm, efficiently mobilizes the IncQ plasmid to agrobacterial recipient cells by a VirB-dependent mechanism (10, 29). Of considerable interest, all virB11/virB11∗ merodiploid strains mobilized the IncQ plasmid to agrobacterial recipients at frequencies comparable to that of wild-type A348 (Table 2). These findings show that the dominant mutations preferentially block T-DNA transfer without disrupting T-pilus production or IncQ plasmid mobilization.
Most PC1011 strains expressing class III alleles transferred the IncQ plasmid at frequencies comparable to that of wild-type A348, whereas strains expressing the class II alleles transferred the IncQ plasmid at low or undetectable frequencies (Table 2). Thus, strains synthesizing these mutant proteins without coproduction of native VirB11 transferred T-DNA and IncQ plasmid substrates at frequencies that correlated with the level of T-pilus production. Again, strain PC1011(pXZB102) expressing the class III allele virB11.I265T was an exception in that it efficiently transferred both DNA substrates even in the absence of detectable T-pilus production.
Otten et al. (47) discovered that two avirulent strains, a T-DNA+ virE2 mutant and a T-DNA− virE2+ mutant, can incite tumor formation when coinoculated on a plant wound site. To explain this phenomenon, it was proposed that these strains translocate the T-strand–VirD2 transfer intermediate and the VirE2 SSB, respectively, to the same plant cell. Once inside the plant cell, these molecules assemble as a T-strand–VirD2–VirE2 complex for delivery of the T-DNA to the plant nucleus (18). The export of VirE2 independently of T-DNA has now been shown unequivocally (62). We used a mixed-infection assay to test whether any virB11 mutants can separately export VirE2 and the T-strand–VirD2 transfer intermediate. Because this assay requires the coinoculation of avirulent strains, our studies were restricted to the analysis of the class II (dominant and nonfunctional) mutations. PC1011 strains expressing these alleles were coinoculated on plant wound sites with the virE2 mutant A348mx358 to test for the capacity to export VirE2 and were coinoculated with the T-DNA deletion mutant LBA4404 to test for the capacity to export the T-strand–VirD2 intermediate. No strain expressing a class II allele transferred the T-strand–VirD2 intermediate at detectable frequencies. However, PC1011(pXZB109) producing the VirB11.I103T/M301I mutant protein efficiently exported VirE2 (Table 2). This strain therefore translocates VirE2 in the absence of detectable T-pilus production.
Effects of i4 insertion mutations on VirB11 function.
The virB11 dominant mutations generally consist of conservative substitution mutations that map to two regions in the N- and C-terminal halves of VirB11 (see Discussion). To expand our structure-function studies, we constructed a set of four-residue (HMVD) insertion mutations at ∼20- to 30-residue intervals across the entire length of VirB11. A348 and PC1011 strains synthesizing the VirB11.i4 derivatives were assayed for VirB protein and T-pilus production and for substrate translocation.
Only 3 of the 14 alleles coding for the mutant proteins exerted dominant effects. Of these, only the allele encoding the E196.i4 mutant protein displayed strong dominance (Table 3). These i4 mutations probably more strongly perturb VirB11's tertiary structure, and hence, its partner protein interactions, than the substitution mutations described above. Interestingly, all of the virB11/virB11.i4 merodiploid strains produced abundant levels of cellular VirB proteins and exocellular VirB2, VirB5, and VirB7, suggesting that the VirB11.i4 mutant proteins do not interfere with the capacity of otherwise-wild-type cells to assemble this transfer system.
TABLE 3.
Phenotypes of the virB11.i4 alleles
| PC1011(pJC8xxx) or control strain | i4 insertion sitea | Dominanceb | T-DNA transferc | Transfer by mixed infectionc
|
pML122ΔKm mobilization (Tcs/input donor)cd | T-pilus productiona | |
|---|---|---|---|---|---|---|---|
| VirE2 | T-strand/D2 | ||||||
| pJC8018 | L18 | − | + | − | − | <10−7 | − |
| pJC8056 | D56 | − | − | − | − | <10−7 | − |
| pJC8075 | L75 | + | − | ++ | − | 9.5 × 10−6 | − |
| pJC8088 | L88 | − | − | − | − | 5 × 10−7 | − |
| pJC8115 | E115 | − | − | − | − | 2 × 10−7 | − |
| pJC8135 | Q135 | − | +++ | NA | NA | 2 × 10−3 | +++ |
| pJC8168 | C168 | − | − | ++ | − | 1.5 × 10−5 | − |
| pJC8196 | E196 | +++ | − | − | − | <10−7 | − |
| pJC8217 | G217 | − | +++ | NA | NA | 5.5 × 10−4 | +++ |
| pJC8237 | P237 | − | +++ | NA | NA | 1.2 × 10−3 | +++ |
| pJC8260 | G260 | − | − | − | − | 1 × 10−7 | − |
| pJC8281 | F281 | − | − | − | − | 1.3 × 10−7 | − |
| pJC8302 | L302 | − | − | ++ | − | 9.3 × 10−6 | − |
| pJC8323 | E323 | + | − | − | − | 1.5 × 10−7 | − |
| A348 | WT | +++ | NA | NA | 3 × 10−3 | +++ | |
| PC1011 | ΔvirB11 | − | − | − | − | <10−7 | − |
| PC1011(pXZB100) | (PvirB::virB11) | − | +++ | NA | NA | 3 × 10−3 | +++ |
Indicated residue immediately precedes i4 insertion.
Virulence of the virB11/virB11.i4 merodiploid strain is strongly (+++) or weakly (+) attenuated or not attenuated (−).
See footnotes to Table 1 for meaning of symbols.
Data are the means of three trials from a single experiments. Three independent experiments yielded similar results.
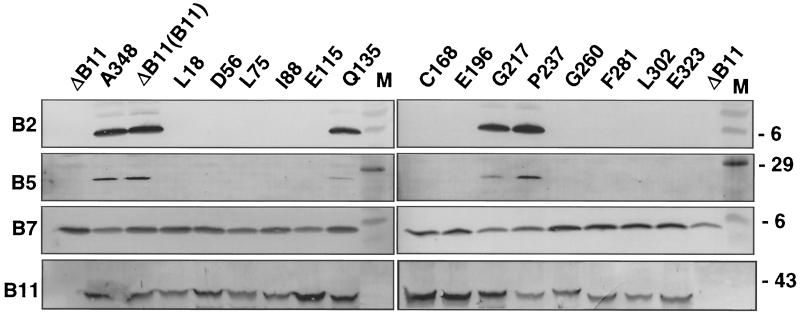
All PC1011 strains expressing the virB11.i4 alleles accumulated abundant levels of the VirB11.i4 mutant proteins as well as other VirB proteins (Fig. 5 and data not shown). However, only three strains, encoding the Q135.i4, G217.i4, and P237.i4 mutant proteins, accumulated detectable levels of exocellular VirB2 pilin and VirB5. These strains elaborated wild-type T pili, as judged by VirB2 distribution profiles in sucrose gradients (Fig. 5 and data not shown).
FIG. 5.
Effects of virB11.i4 mutations on T-pilus production. Exocellular fractions from PC1011 (designated ΔB11) expressing wild-type virB11 (designated B11; from plasmid pJCB903) and alleles for the i4 mutant proteins indicated were analyzed for accumulation of T-pilus proteins, VirB2, VirB5, and VirB7. Corresponding cellular levels of native and mutant forms of VirB11 are shown at the bottom. M, molecular mass markers with sizes in kilodaltons indicated on the right.
PC1011 strains producing the Q135.i4, G217.i4, and P237.i4 mutant proteins also transferred T-DNA at wild-type levels (Table 3). Mutants defective in T-pilus production did not translocate T-DNA, yet three derivatives, encoding the L75.i4, C168.i4, and L302.i4 mutant proteins, transferred the IncQ plasmid to agrobacterial recipient cells. The transfer frequencies were 1 to 2 orders of magnitude lower than those of the isogenic PC1011 producing native VirB11 but were still appreciably higher (>2 orders of magnitude) than background (Table 3). In addition, these three strains translocated VirE2 protein, as determined by the mixed-infection assay (Table 3). Thus, three i4 mutant proteins support the translocation of selected substrates but interfere with production of the T pilus.
Effect of an IncQ plasmid on T-pilus production and T-DNA transfer.
Previous work has shown that RSF1010 derivatives suppress the capacity of A. tumefaciens to transfer T-DNA and VirE2 substrates to plants (9, 60, 64). To determine whether cells carrying an IncQ plasmid show defects in assembly of this transfer system, we assayed for production of cellular and exocellular VirB proteins by wild-type A348 and an isogenic strain carrying pML122ΔKm. As shown in Fig. 6A, A348 with and without the IncQ plasmid accumulated most VirB proteins at comparable levels; the only reproducible effects of the IncQ plasmid were slightly diminished levels of VirB8, VirB9, and VirB10. Both A348 and A348(pML122ΔKm) cells also accumulated comparable levels of exocellular VirB2, VirB5, and VirB7 (Fig. 6B). Moreover, exocellular VirB2 from both strains fractionated similarly in sucrose gradients, suggesting that both strains elaborate abundant levels of wild-type T pili (data not shown). Further studies showed that the presence of an IncQ plasmid also does not influence the T-pilus production of A348 merodiploid and PC1011 strains expressing the dominant virB11 alleles (Fig. 6B).
FIG. 6.
Effects of IncQ plasmid on accumulation of VirB proteins and T-pilus proteins. (A) VirB and VirE2 protein levels in total-cell extracts of A348 and A348(pML122), with VirB proteins listed on the left of each immunoblot. M, molecular mass markers, with sizes in kilodaltons indicated on the right. (B) VirB2 pilin levels in exocellular fractions from A348 and merodiploid strains expressing the dominant alleles 1 through 12 and pML122ΔKm (designated pML). Also shown are the pilin levels in PC1011 (designated Δ11) expressing virB11 (B11; from plasmid pXZB100) and class III alleles.
Overexpression of virB9, virB10, and virB11 suppresses IncQ plasmid-mediated inhibition of T-DNA transfer, prompting the proposal that the IncQ plasmid competes with the T-DNA transfer intermediate and/or VirE2 for available transfer machines (9, 60, 64). To determine whether any virB11 mutations counterract the suppressive effect of the IncQ plasmid, we compared the relative efficiencies with which the virB11 mutant strains with and without the IncQ plasmid transfer T-DNA to plants. We found that the presence of the IncQ plasmid suppressed transfer of T-DNA and/or VirE2 by all virB11 mutant strains capable of translocating these substrates in the absence of the IncQ plasmid (data not shown). Therefore, none of the virB11 mutations appears to selectively block the interaction of the IncQ transfer intermediate with the transfer machine.
DISCUSSION
In this report, we showed that VirB11 regulates in an ATP binding-dependent manner both the assembly of the T pilus and the selection or translocation of secretion substrates. Although most mutations exerted similar effects on pilus production and substrate translocation, a few selectively impaired one or the other of these functions. Moreover, some mutations disrupted the transfer of one or more substrates without affecting the transfer of other substrates. We (50, 69) and others (37, 38, 67) have postulated that the VirB11-type ATPases function as chaperones to facilitate the movement of unfolded proteins and DNA substrates across the cytoplasmic membrane. As discussed in more detail below, if this chaperone model is correct, it must satisfactorily explain the dual role of VirB11 in pilus biogenesis and substrate transfer and also the finding that VirB11's contributions to these processes can be uncoupled by mutagenesis. In addition, the chaperone model must also be considered in the context of other biochemical reactions that are known to be required for assembly of a functional T-DNA transfer system. For reference during this discussion, Fig. 7 shows the positions and phenotypes of VirB11 mutations characterized in this study along with an alignment of VirB11 and its conserved motifs with the HP0525 secondary structure (67).
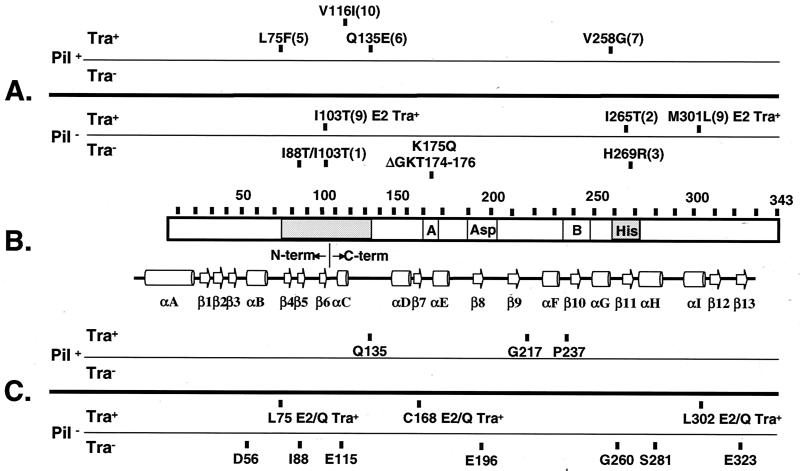
FIG. 7.
Positions of VirB11 mutations grouped according to their effects on pilus production and substrate transfer. (A) Substitution mutations with allele numbers in parantheses as defined by Zhou et al. (69). Phenotypic descriptions are provided for mutations of special interest. Note that all virB11/virB11∗ merodiploid strains expressing the dominant alleles exhibit a T-pilus+ IncQ plasmid Tra+ T-DNA Tra-deficient phenotype. (B) VirB11 (343 residues) with conserved Walker A and B domains and the Asp and His boxes denoted. The shading identifies the two regions of VirB11 in which dominant mutations were predominantly located. Below the VirB11 representation is the HP0525 secondary structure with β-sheets and α-helices as presented by Yeo et al. (67). The junction between the N- and C-terminal domains, shown by the HP0525 crystal structure to assemble as independent hexameric rings, is indicated. (C) i4 mutations with insertion sites indicated.
Effects of ATP-binding mutations on T-pilus production and substrate transfer.
Defining the structural and biological consequences of ATP binding or hydrolysis is central to developing a detailed mechanistic understanding of VirB11's biological activity. In a previous study, we showed that mutations in the Walker A motif disrupted the capacity of the C-terminal domain of VirB11 to self-assemble (50). In addition, the electron microscopy studies of VirB11 homologs (36, 37) together with the HP0525 crystal structure (67) establish that nucleotide binding is critical for coordination of the N- and C-terminal domain rings. ATP binding therefore is likely required for the formation of both homo- and heteromultimeric complexes. Here, we demonstrated that mutations of VirB11 residues predicted to coordinate nucleotide binding disrupted or abolished T-pilus production, with corresponding effects on substrate translocation. Walker A residues are required for the binding of phosphate groups (63, 67), and a substitution (K175Q) and a deletion (ΔGKT174–176) of these residues of VirB11 abolished both T-pilus assembly and substrate transfer. One mutation near the Walker A motif, C168.i4, also abolished both processes, whereas a second, P170L, led to diminished T-pilus production and T-DNA transfer (reference 51 and data not shown). The structural studies further identified several additional contacts between N- and C-terminal residues of HP0525 and the adenine and ribose moieties (67). Two residues in the N-terminal domain, T46 and N61, coordinate the ribose moiety of ADP, whereas Y140 and F145, located at the beginning of the C-terminal domain, contact the adenine moiety (67). For VirB11, the E25G and E115.i4 mutations are at sites aligning near T46 and F145 of HP0525, respectively, and both mutations abolished T-DNA transfer as well as T-pilus production (reference 51 and data not shown).
Reactive Glu residues in two other highly-conserved motifs of HP0525, the Asp box and the Walker B domain (37, 52, 63), are proposed to coordinate Mg2+ binding and ATP hydrolysis (67). For VirB11, the Asp box is located between residues 198 and 210, and the E196.i4 mutation near this box abolished T-pilus production and substrate transfer (Fig. 7). Interestingly, however, the Walker B domain is located between residues 234 and 245, and the P237.i4 mutation is completely permissive both for pilus production and substrate transfer. Consistent with this finding, a substitution mutation (R217T) within the Walker B motif of the VirB11 homolog, TrbB of plasmid RP4, was found to abolish ATPase activity without affecting RP4 plasmid transfer, although effects on ATP binding or pilus production were not reported (38).
Our findings firmly establish that an intact ATP binding site is important for T-pilus production and substrate transfer. At this time, however, we cannot distinguish between the relative contributions of ATP binding and ATP hydrolysis to the assembly of a functional system. The structural findings indicating that nucleotide binding is critical for VirB11 oligomerization raise the possibility that ATP binding suffices at least for a subset of VirB11 activities. Walker B mutations might be found to disrupt ATP hydrolysis without affecting ATP binding, and further studies of such mutations should help to resolve this question.
Walker A mutations of the VirB4 ATPase also abolished T-pilus biogenesis, consistent with previous work showing that these mutations disrupt substrate export (6, 30, 57). Although there is no structural information about VirB4, we previously supplied evidence for a transmembrane topology (21), and we further demonstrated that an N-terminal membrane-spanning domain mediates VirB4 self-assembly (20). The ATP binding pocket is located in a central domain of this large (87-kDa) protein, and VirB4 Walker A mutations do not abolish self-interaction of the N-terminal domain (20). Of further interest, an independent study showed that the production of VirB4 and a subset of other VirB proteins in agrobacterial recipient cells stimulates the acquisition of an IncQ plasmid in matings with agrobacterial donor cells (10). However, production of VirB4 Walker A mutant proteins in agrobacterial recipients stimulates IncQ plasmid acquisition to the same extent as production of native VirB4, suggesting that ATP binding is not a prerequisite for assembly of a VirB protein substructure with a discernible biological activity (20). Taken together, the data support a working model in which VirB4 establishes initial contacts with other VirB proteins independent of ATP binding. Then, by an ATP binding-dependent mechanism, VirB4 promotes T-pilus production and configures the transfer apparatus as a dedicated export machine. This model will be refined by studies examining the contributions of ATP binding and hydrolysis activities to the VirB4 structure and partner protein interactions.
Uncoupling of T-pilus production and substrate transfer.
The T-DNA transfer machine is usually depicted as a supramolecular complex composed of a translocation channel physically connected to the T pilus (19, 43, 66). Interestingly, however, our mutational analyses supplied strong evidence that T-pilus production is not obligatorily coupled to substrate transfer. The Pil+ Tra-deficient phenotype of the virB11/virB11∗ merodiploid strains expressing dominant alleles indicates that VirB11 can support pilus production while simultaneously blocking substrate transfer. More intriguingly, this block is substrate specific, preventing T-DNA transfer to plants without affecting IncQ plasmid transfer to agrobacterial recipients. We suggest below that this class of VirB11 mutations might disrupt recognition of the T-DNA transfer intermediate at the cytoplasmic face of the transfer channel.
It is intriguing to note that the Pil+ Tra-defective phenotype is also observed when the IncQ plasmid is introduced into wild-type cells. Previous studies led to a prediction that the IncQ plasmid inhibits the export of T-DNA and VirE2 transfer intermediates (9, 64). In support of this prediction, we found that the IncQ plasmid does not interfere with production of VirB proteins or the T pilus. Apparently, both conditions, expression of the virB11 dominant alleles and the presence of the IncQ plasmid in a wild-type background, permit assembly of a functional transfer system, but this system is somehow configured for selective substrate translocation, e.g., IncQ plasmid mobilization. We have reported that synthesis of VirE2::GFP in an otherwise wild-type strain also interferes with T-DNA transfer without disrupting IncQ plasmid transfer (70). None of the virB11 mutations examined in the present study counterracted the suppressive effects of the IncQ plasmid (this study) or VirE2::GFP (data not shown) on T-DNA transfer. Further studies might identify such mutations, although it is also possible that the IncQ plasmid and VirE2::GFP transfer intermediates selectively block T-DNA transfer by a mechanism(s) that bypasses VirB11.
The Pil− Tra+ phenotypes of PC1011 strains expressing substitution (I265T) or i4 insertion (L75.i4, C168.i4, and L302.i4) mutants supplied further evidence for uncoupling of pilus production and substrate transfer. Clearly, the 1265T substitution mutation is the best example of an uncoupling mutation in that it completely abolished T-pilus formation without disrupting translocation of any substrates tested. Of possible significance, this mutation is in the highly conserved His box (37, 52), and other His box mutations also led to a reduction or loss of pilus production and substrate transfer. As noted above, besides virB11 mutations, other elements have been shown to selectively block substrate transfer without impairing pilus production. By contrast, the isolation of mutations conferring a Pil− Tra+ phenotype is completely novel, strongly indicating that production of a wild-type T pilus is dispensible for substrate transfer. Because each A. tumefaciens cell elaborates only a few pili (41), we consider it unlikely that a reduction in the number of T pili on a per-cell basis would account for the Pil− phenotype. However, we cannot exclude the possibility that these cells elaborate morphologically aberrant pili, e.g., stubby pili that do not protrude beyond the cell surface and thus are refractive to isolation by shearing.
The dominant mutations generally map to two regions of VirB11, one located between residues 75 and 135 and the second in the His box between residues 258 and 270 (69) (Fig. 7). The corresponding regions of HP0525 were found to establish intra- and intersubunit contacts, and as noted above, several residues in the N-terminal region also contact nucleotide moieties (67). While these observations suggest that the dominant mutations might exert their effects by perturbing the overall oligomeric structure of VirB11, we have also shown that all VirB11 mutant proteins exerting dominant effects can self-assemble and also interact with native VirB11 (50, 69). With these considerations in mind, we propose that the dominant mutant proteins retain the capacity to self-assemble, forming mixed multimers with native VirB11 in merodiploid strains and homomultimers in PC1011 strains. Depending on their overall structures, these multimers may or may not productively enter a pilus morphogenetic pathway described below or direct the transfer of substrates through the translocation channel.
Entry point into the morphogenetic pathway and models for VirB11 function.
If VirB11 indeed functions as a chaperone to drive assembly of the T-DNA transfer system, we should be able to assign its entry point into the morphogenetic pathway. The available data suggest that an initial series of reactions occurs independently of VirB11: (i) VirB7 undergoes maturation as a lipoprotein, (ii) VirB7 forms a disulfide-cross-linked complex with VirB9, (iii) the VirB7-VirB9 heteromultimer is sorted to the outer membrane, (iv) the VirB7-VirB9 complex interacts with the bitopic inner membrane proteins, VirB8 and VirB10, and (v) the VirB7-VirB9 heteromultimer directs assembly of a VirB10 homooligomer (2, 3, 4, 22, 23, 27, 39, 59). These reactions are proposed to lead to the assembly of a VirB protein substructure that spans the cytoplasmic and outer membranes (14). This core complex, composed of VirB7 through VirB10 and probably also the polytopic proteins VirB4 ATPase and VirB6, is stable and can act independently of VirB11 to stimulate IncQ plasmid acquisition by recipient cells during matings with agrobacterial donors (10). VirB11 does seem to influence the sorting of VirB7 monomers or homomultimers to the extracellular milieu, but further studies are needed to determine whether this is an on- or off-pathway reaction with respect to biogenesis of the T-DNA transfer system.
We suggest that VirB11 contributes to T-pilus assembly subsequent to its formation of a homooligomeric complex and interaction of the homooligomer with the core structure. One possibility is that a VirB11 hexameric chaperone configures VirB2 pilin for translocation across the cytoplasmic membrane through a preassembled VirB channel (43). Although it is intriguing, we do not favor this mechanism of action because VirB2 possesses a signal sequence and is exported to the periplasm in various A. tumefaciens virB mutants and in heterologous E. coli, as determined by reporter protein fusion studies (5, 24, 34, 42). Another argument against a role for VirB11 in the translocation of pilin across the cytoplasmic membrane is that VirB2 and its homolog, TrbC of plasmid RP4, are processed to their mature forms via reactions occurring in the periplasm independent of their cognate type IV components (25, 26, 42). The mature form of VirB2, which, intriguingly, is a cyclic polypeptide (25), embeds in the cytoplasmic membrane, forming a reservoir of pilin available for T-pilus polymerization (34, 56). Thus, an alternative possibility is that a VirB11 chaperone acts to catalyze the release of pilin monomers from the cytoplasmic membrane. Such a chaperone-pilin interaction might be dynamic, driven by cycles of ATP binding and hydrolysis and ADP release, with the net effect that pilin monomers are successively delivered to the site of pilus polymerization. Although there is no precedent for this type of chaperone activity, an appealing aspect of this model is that it offers a solution to a long-standing problem of how pilin proteins that are integrated into the cytoplasmic membrane can be recruited to assemble as conjugative pili.
It should be noted that VirB11's contribution to machine assembly alternatively might be entirely structural. For example, the ADP- or ATP-bound forms of VirB11 might interact with the VirB core structure in a way that induces a conformational change required for the structure to serve as a platform for T-pilus assembly. As noted above, studies examining the relative importance of ATP or ADP binding versus ATP hydrolysis should help distinguish structural from catalytic contributions to pilus biogenesis.
Finally, we propose that once VirB11 assembles with the base of the core structure and induces the production of the T pilus, presumably on top of the core structure, it can then participate in substrate translocation. While substrate selection or translocation might temporally follow the elaboration of the supramolecular channel or pilus structure, the isolation of uncoupling mutations suggests the assembly pathway can be blocked at some stage without disrupting substrate transfer. With respect to VirB11's role in substrate transfer, again a chaperone model is enticing, in which VirB11 situated at the base of the secretion machine configures secretion substrates for translocation (67). Of considerable interest, however, another putative ATPase termed VirD4 has been shown to be essential for substrate translocation but dispensible for pilus biogenesis (16, 41, 45). There is also genetic evidence based on construction of chimeric transfer systems that VirD4 and its homologs participate in substrate selection (12, 33, 46). An intriguing question for future studies is how VirB11 functions independently of VirD4 to direct T-pilus biogenesis and also coordinates its activities with VirD4 to direct substrate transfer.
ACKNOWLEDGMENTS
We thank Simon Jakubowski and Zhiyong Ding for helpful discussions and Juan Fernandez for excellent technical assistance. We thank Gabriel Waksman and Thierre Rose for helpful discussions and critiques of the manuscript.
Work in this laboratory is supported by NIH grant GM48746.
REFERENCES
- 1.Albano M, Breitling R, Dubnau D A. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J Bacteriol. 1989;171:5386–5404. doi: 10.1128/jb.171.10.5386-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson L B, Hertzel A V, Das A. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc Natl Acad Sci USA. 1996;93:8889–8894. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron C, Thorstenson Y R, Zambryski P C. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J Bacteriol. 1997;179:1211–1218. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaupré C E, Bohne J, Dale E M, Binns A N. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beijersbergen A, Smith S J, Hooykas P J J. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid. 1994;32:212–218. doi: 10.1006/plas.1994.1057. [DOI] [PubMed] [Google Scholar]
- 6.Berger B R, Christie P J. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993;175:1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger K H, Isberg R R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 9.Binns A, Beaupre C, Dale E. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J Bacteriol. 1995;177:4890–4899. doi: 10.1128/jb.177.17.4890-4899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohne J, Yim A, Binns A N. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc Natl Acad Sci USA. 1998;95:7057–7062. doi: 10.1073/pnas.95.12.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchanan-Wollaston V, Passiatore J E, Cannon F. The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature. 1987;328:172–175. [Google Scholar]
- 12.Cabezon E, Sastre J I, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 13.Chen C-Y, Winas S C. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J Bacteriol. 1991;173:1139–1144. doi: 10.1128/jb.173.3.1139-1144.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christie P J. The Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie P J. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally-related to conjugation machines. Mol Microbiol. 2001;40:294–305. doi: 10.1046/j.1365-2958.2001.02302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie P J, Ward J E, Winans S C, Nester E W. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christie P J, Ward J E, Winans S C, Nester E W. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc Natl Acad Sci USA. 1989;86:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Citovsky V, Zambryski P. Transport of nucleic acids through membrane channels: snaking through small holes. Annu Rev Microbiol. 1993;47:167–197. doi: 10.1146/annurev.mi.47.100193.001123. [DOI] [PubMed] [Google Scholar]
- 19.Covacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 20.Dang T A, Zhou X-R, Graf B, Christie P J. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on assembly and function of the T-DNA transporter. Mol Microbiol. 1999;32:1239–1253. doi: 10.1046/j.1365-2958.1999.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang T A T, Christie P J. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J Bacteriol. 1997;179:453–462. doi: 10.1128/jb.179.2.453-462.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das A, Anderson L B, Xie Y H. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J Bacteriol. 1997;179:3404–3409. doi: 10.1128/jb.179.11.3404-3409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das A, Xie Y-H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol. 2000;182:758–763. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das A, Xie Y H. Construction of transposon Tn3PhoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol Microbiol. 1998;27:405–414. doi: 10.1046/j.1365-2958.1998.00688.x. [DOI] [PubMed] [Google Scholar]
- 25.Eisenbrandt R, Kalkum M, Lai E M, Lurz R, Kado C I, Lanka E. Conjugative pili of IncP plasmids and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 26.Eisenbrandt R, Kalkum M, Lurz R, Lanka E. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J Bacteriol. 2000;182:6751–6761. doi: 10.1128/jb.182.23.6751-6761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez D, Dang T A T, Spudich G M, Zhou X-R, Berger B R, Christie P J. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J Bacteriol. 1996;178:3156–3167. doi: 10.1128/jb.178.11.3156-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez D, Spudich G M, Zhou X-R, Christie P J. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fullner K J. Role of Agrobacterium virB genes in transfer of T complexes and RSF1010. J Bacteriol. 1998;180:430–434. doi: 10.1128/jb.180.2.430-434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fullner K J, Stephens K M, Nester E W. An essential virulence protein of Agrobacterium tumefaciens, VirB4, requires an intact mononucleotide binding domain to function in transfer of T-DNA. Mol Gen Genet. 1994;245:704–715. doi: 10.1007/BF00297277. [DOI] [PubMed] [Google Scholar]
- 31.Garfinkel D J, Simpson R B, Ream L W, White F F, Gordon M P, Nester E W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981;27:143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 32.Haase J, Lurz R, Grahn A M, Bamford D H, Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage production, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol. 1995;177:4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton C M, Lee H, Li P-L, Cook D M, Piper K R, Beck von Bodman S, Lanka E, Ream W, Farrand S K. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol. 2000;182:1541–1548. doi: 10.1128/jb.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones A L, Lai E-M, Shirasu K, Kado C I. VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J Bacteriol. 1996;178:5706–5711. doi: 10.1128/jb.178.19.5706-5711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kado C I. The role of the T-pilus in horizontal gene transfer and tumorigenesis. Curr Opin Microbiol. 2000;3:643–648. doi: 10.1016/s1369-5274(00)00154-5. [DOI] [PubMed] [Google Scholar]
- 36.Krasan G P, Sauer F G, Cutter D, Farley M M, Gilsdorf J R, Hultgren S J, St. Geme J W., III Evidence for donor strand complementation in the biogenesis of Haemophilus influenzae haemagglutinating pili. Mol Microbiol. 2000;35:1335–1347. doi: 10.1046/j.1365-2958.2000.01816.x. [DOI] [PubMed] [Google Scholar]
- 37.Krause S, Barcena M, Panseqrau W, Lurz R, Carazo J, Lanka E. Sequence related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc Natl Acad Sci USA. 2000;97:3067–3072. doi: 10.1073/pnas.050578697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krause S, Pansegrau W, Lurz R, de la Cruz F, Lanka E. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar R B, Xie Y H, Das A. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol Microbiol. 2000;36:608–617. doi: 10.1046/j.1365-2958.2000.01876.x. [DOI] [PubMed] [Google Scholar]
- 40.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 41.Lai E M, Chesnokova O, Banta L M, Kado C I. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J Bacteriol. 2000;182:3705–3716. doi: 10.1128/jb.182.13.3705-3716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai E M, Kado C I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai E M, Kado C I. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 2000;8:361–369. doi: 10.1016/s0966-842x(00)01802-3. [DOI] [PubMed] [Google Scholar]
- 44.Li P, Hwang I, Miyagi H, True H, Farrand S. Essential components of the Ti plasmid trb system, a type IV macromolecular transporter. J Bacteriol. 1999;181:5033–5041. doi: 10.1128/jb.181.16.5033-5041.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin T S, Kado C I. The virD4 gene is required for virulence while virD3 and orf5 are not required for virulence of Agrobacterium tumefaciens. Mol Microbiol. 1993;9:803–812. doi: 10.1111/j.1365-2958.1993.tb01739.x. [DOI] [PubMed] [Google Scholar]
- 46.Moncalian G, Cabezon E, Alkorta I, Valle M, Moro F, Valpuesta J M, Goni F M, de La Cruz F. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J Biol Chem. 1999;274:36117–36124. doi: 10.1074/jbc.274.51.36117. [DOI] [PubMed] [Google Scholar]
- 47.Otten L, De Greve H, Leemans J, Hain R, Hooykaas P, Schell J. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol Gen Genet. 1984;195:159–163. [Google Scholar]
- 48.Planet P J, Kachlany S C, DeSalle R, Figurski D H. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci USA. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rashkova S. Studies of subcellular localization and complex formation of the Agrobacterium tumefaciens transport ATPase VirB11. Ph.D. thesis. Houston: University of Texas—Houston Medical School; 1998. [Google Scholar]
- 50.Rashkova S, Zhou X-R, Christie P J. Self-assembly of the Agrobacterium tumefaciens VirB11 traffic ATPase. J Bacteriol. 2000;182:4137–4145. doi: 10.1128/jb.182.15.4137-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rashkova S, Spudich G M, Christie P J. Mutational analysis of the Agrobacterium tumefaciens VirB11 ATPase: identification of functional domains and evidence for multimerization. J Bacteriol. 1997;179:583–589. doi: 10.1128/jb.179.3.583-591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rivas S, Bolland S, Cabezon E, Goni F M, de la Cruz F. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J Biol Chem. 1997;272:25583–25590. doi: 10.1074/jbc.272.41.25583. [DOI] [PubMed] [Google Scholar]
- 53.Sagulenko V, Sagulenko E, Jakubowski S, Spudich E, Christie P J. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T-pilus. J Bacteriol. 2001;183:3642–3651. doi: 10.1128/JB.183.12.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandkvist M. Biology of type II secretion. Mol Microbiol. 2001;40:271–283. doi: 10.1046/j.1365-2958.2001.02403.x. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt-Eisenlohr H, Domke N, Augerer C, Wanner G, Zambryski P C, Baron C. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J Bacteriol. 1999;181:7485–7492. doi: 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shirasu K, Kado C. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol Lett. 1993;111:287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- 57.Shirasu K, Koukolikova N Z, Hohn B, Kado C I. An inner-membrane-associated virulence protein essential for T-DNA transfer from Agrobacterium tumefaciens to plants exhibits ATPase activity and similarities to conjugative transfer genes. Mol Microbiol. 1994;11:581–588. doi: 10.1111/j.1365-2958.1994.tb00338.x. [DOI] [PubMed] [Google Scholar]
- 58.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 59.Spudich G M, Fernandez D, Zhou X-R, Christie P J. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci USA. 1996;93:7512–7517. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stahl L E, Jacobs A, Binns A N. The conjugal intermediate of plasmid RSF1010 inhibits Agrobacterium tumefaciens virulence and VirB-dependent export of VirE2. J Bacteriol. 1998;180:3933–3939. doi: 10.1128/jb.180.15.3933-3939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens K M, Roush C, Nester E. Agrobacterium tumefaciens VirB11 protein requires a consensus nucleotide-binding site for function in virulence. J Bacteriol. 1995;177:27–36. doi: 10.1128/jb.177.1.27-36.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vergunst A C, Schrammeijer B, den Dulk-Ras A, de Vlaam C M, Regensburg-Tuink T J, Hooykaas P J. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 63.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ward J E, Dale E M, Binns A N. Activity of the Agrobacterium T-DNA transfer machinery is affected by virB gene products. Proc Natl Acad Sci USA. 1991;88:9350–9354. doi: 10.1073/pnas.88.20.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watson B, Currier T C, Gordon M P, Chilton M D, Nester E W. Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1975;123:255–264. doi: 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winans S C, Burns D L, Christie P J. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yeo H-J, Savvides S N, Herr A B, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV system. Mol Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 68.Zhao Z, Sagulenko E, Ding Z, Christie P. Activities of virE1 and the VirE1 secretion chaperone for export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J Bacteriol. 2001;183:3855–3865. doi: 10.1128/JB.183.13.3855-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou X-R, Christie P J. Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J Bacteriol. 1997;179:5835–5842. doi: 10.1128/jb.179.18.5835-5842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou X-R, Christie P J. Mutagenesis of Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J Bacteriol. 1999;181:4342–4352. doi: 10.1128/jb.181.14.4342-4352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu J, Oger P M, Schrammeijer B, Hooykaas P J, Farrand S K, Winans S C. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]