Abstract
Glioblastoma multiforme (GBM) is the most common, malignant, poorly promising primary brain tumor. GBM is characterized by an infiltrating growth nature, abundant vascularization, and a rapid and aggressive clinical course. For many years, the standard treatment of gliomas has invariably been surgical treatment supported by radio- and chemotherapy. Due to the location and significant resistance of gliomas to conventional therapies, the prognosis of glioblastoma patients is very poor and the cure rate is low. The search for new therapy targets and effective therapeutic tools for cancer treatment is a current challenge for medicine and science. microRNAs (miRNAs) play a key role in many cellular processes, such as growth, differentiation, cell division, apoptosis, and cell signaling. Their discovery was a breakthrough in the diagnosis and prognosis of many diseases. Understanding the structure of miRNAs may contribute to the understanding of the mechanisms of cellular regulation dependent on miRNA and the pathogenesis of diseases underlying these short non-coding RNAs, including glial brain tumors. This paper provides a detailed review of the latest reports on the relationship between changes in the expression of individual microRNAs and the formation and development of gliomas. The use of miRNAs in the treatment of this cancer is also discussed.
Keywords: glioblastoma multiforme, miRNA
1. miRNA Characteristics
Non-coding RNAs are a type of molecule formed as a result of DNA transcription, which do not encode proteins, but perform a variety of structural, enzymatic, and regulatory functions in the cell. Among such RNAs, we distinguish a particularly important group with regulatory functions—microRNAs. So far, it has been confirmed that miRNA are present in all plants and animals, but of the more than the thousands of identified molecules (over 2000 in human cells), only a very small percentage of them have recognized and described functions [1]. It is suspected that about 30% of human genes can be regulated by appropriate miRNAs [2]. After post-transcriptional treatment, miRNA is a single-stranded, short, ~22 nucleotide long, regulatory molecule found in the cytosol of the cell. Together with the corresponding proteins, miRNA forms RNA-inducing silencing complexes (miRISC). Such a complex, thanks to the complementary sequence of nucleotides in miRNA, inhibits the translation of the target mRNA [3]. In this way, the labeled mRNA is degraded by the Ago protein, which prevents further expression of the protein. This inconspicuous molecule has a large impact on the level of gene expression in the cell, and thus, on the functioning of not only a specific cell or tissue, but the whole organism.
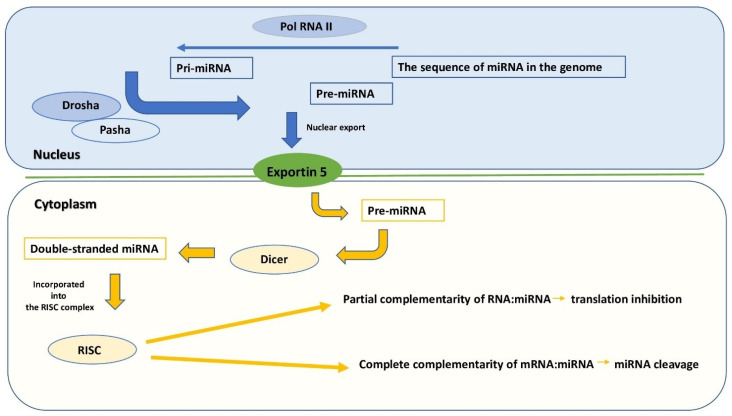
miRNA biogenesis is a very complex and varied process between different types of miRNAs (Figure 1) [4]. The differences between the formation of individual molecules begin at the transcription level, among other things, because the sequences encoding miRNAs can be located in intergenic regions or within genes encoding proteins. The primary product of transcription is double-stranded pri-miRNAs (primary miRNA). These are then shortened to 60–70 nucleotide pre-mirRNAs by RNAse III–Drosha endonuclease (forming a complex in mammalian cells with the DGCR8 protein or its homologues in other animals, e.g., Pasha in Drosophila melanogaster). Pre-miRNAs are exported outside of the cell nucleus through channels formed in the membrane of the cell nucleus by exportin-5. In the cytoplasm, pre-miRNA is bound by the Dicer endoribonuclease and separated into two strands about 22 nucleotides long [4]. Both threads form a duplex, which consists of an antisense strand (guide) and a sense strand (passenger).The antisense strand, responsible for the suppressive activity of miRNA, is built into the miRNA-induced silencing complex (miRISC). On the other hand, the sense strand usually degrades [5]. The suppression mechanism is based on complementary attachment to regulatory mRNA sequences (including the area at the end of 3′-UTR).
Figure 1.
miRNA biogenesis pathway. miRNA genes are transcribed from the sequences encoding them in the genome most often by RNA polymerase II and the resulting pri-miRNA transcripts are subject to modifications of the 5′ (cap) and 3′ (polyadenyl tail) end. They are treated by the Drosha and Pash proteins by cutting out a region with the structure of a hairpin, thanks to which pre-miRNA molecules with a length of about 70 nucleotides are formed. pre-miRNAs are exported from the nucleus by the nuclear transporter exportin 5. In the cytoplasm, pre-miRNA is cut by the Dicer enzyme into double-stranded molecules about 22 nucleotides long. The less thermodynamically stable of the strands is incorporated into the RISC (RNA-induced silencing complex). A high degree of complementarity of the transcript to the RISC-bound miRNA strand results in degradation of the transcript, while partial complementarity inhibits translation.
It has been shown that genes for microRNAs are often located in or near fragile sites [6]. Genome damage, such as translocations, deletions, amplifications, and integrations of foreign DNA, for example HPV (human papillomavirus), affect not only the expression of genes that are tumor suppressors (TS), but also the expression of microRNAs. A change in the level of expression of the latter is observed in various types of tumors [7,8,9].
The diversity of genes regulated by miRNA means that in the context of cancer development, these molecules can behave like oncogenes and as transformation suppressors [10]. Based on the level of miRNA expression, normal tissues can be differentiated from cancerous tissues [11,12,13,14]. microRNA expression profiling can be a good diagnostic tool for assessing disease staging or survival and may also be helpful in choosing treatment tailored to the individual needs of the patient [15]. One of the first reports on the participation of miRNA in the tumorigenesis process concerned the effect of miR-17-92 overexpression on the initiation of the carcinogenesis process in mouse lymphatic cells [16]. Similarly, in the case of cells lining the bile ducts, overexpression of miR-29 led to a decrease in the amount of the antiapoptotic factor Mcl-1 and consequently to the cancer transformation of these cells [17]. Suppressor activity was observed in the case of let-7, which is a negative regulator of the expression of oncogenic proteins Ras and c-Myc in human cancer cells of the large intestine [18]. It is now known that deregulation at the level of miRNA expression affects tumors of various origins, including breast [19], colon [20], lung [21], liver [22], and pancreas [23] cancers, as well brain gliomas [24].
In cancer, miRNAs have been shown to modulate cell proliferation and affect invasiveness, angiogenesis, and recurrence [25,26]. miRNA expression disorders are a direct cause of tumor development and are the result of neoplastic processes involving changes in the activity of transcription factors controlling their expression. One of the arguments supporting the influence of changes in miRNA expression on the initiation of the tumorigenesis process indicates the location of their genes on chromosomes near fragile sites, susceptible to deletions, amplifications, point mutations and DNA methylation disorders [27,28]. An example is miR-15a-16, whose gene (located in the 13q14 region) is subject to frequent deletions in chronic lymphocytic leukemia [28,29]. The result of these changes is inhibition of miR-15a and miR-16-1 expression (acting as negative regulators of the anti-apoptotic factor Bcl-2) and, consequently, impaired activation of apoptosis [28].
The main goal of profiling miRNA expression in glioblastoma cells is to identify specific miRNAs whose changes in the level of expression are correlated with the process of tumorigenesis [30]. Currently, in addition to standard techniques (RT-PCR and Q-PCR), high-resolution techniques such as deep sequencing and microarrays are used for these tests. On the basis of microarrays and deep sequencing, a group of microRNAs were selected that undergo significant overexpression in glioma cancer cells compared to control trials, and miRNAs whose levels in glioma cells are reduced [31,32,33]. Table 1 shows typical miRNAs found in glioblastoma multiforme (GBM).
Table 1.
miRNAs with both suppressor and oncogenic functions in glioblastoma multiforme.
| S. No. | miRNA Name | Expression in Glioblastoma | Role in GBM |
|---|---|---|---|
| 1. | miR-21 | up-regulated [34] | oncomiR |
| 2. | miR-93 | up-regulated [35] | oncomiR |
| 3. | miR-10b | up-regulated [35] | oncomiR |
| 4. | miR-196a | up-regulated [36] | oncomiR |
| 5. | miR-221/222 | up-regulated [37] | oncomiR |
| 6. | miR-182 | up-regulated [38] | oncomiR |
| 7. | miR-7 | down-regulated [39] | tumor suppressor |
| 8. | miR-128 | down-regulated [40] | tumor suppressor |
| 9. | miR-124/137 | down-regulated [41] | tumor suppressor |
| 10. | miR-101 | down-regulated [42] | tumor suppressor |
| 11. | miR-181 | down-regulated [40,43] | tumor suppressor |
| 12. | miR-146a | down-regulated [44] | tumor suppressor |
| 13. | miR-137 | down-regulated [45] | tumor suppressor |
| 14. | miR-34a | down-regulated [43] | tumor suppressor |
2. Glioblastoma Multiforme (GBM)
Gliomas are the most common primary cancers of the central nervous system. They originate from glial cells and account for 40–60% of intracranial tumors. The most common malignant tumor derived from glial tissue is glioblastoma multiforme, accounting for up to 90% of gliomas in adults [46]. Depending on the type of glial cell from which they originate, gliomas are classified as astrocytes (developing from astrocytes), oligodendrocytes (from oligodendrocytes), or linings (from epenymocytes). Due to the degree of biological malignancy, gliomas are divided into low-malignant gliomas with low proliferative potential, slow-growing, minimally invasive gliomas with relatively good prognosis (I and II degree of biological malignancy according to WHO), and high-malignant gliomas characterized by abundant vascularization, intensive proliferation, infiltration of neighboring tissues, and a very poor prognosis (III and IV degree according to WHO) [47]. Glioblastoma multiforme is the most common primary brain tumor, accounts for more than 50% of all gliomas, and has the highest degree of biological malignancy (WHO Grade IV) [47,48,49]. GBM has an infiltrating growth nature, abundant vascularization, and a rapid and aggressive clinical course. GBM is characterized by the occurrence of poorly differentiated neoplastic astrocytic cells, cellular and nuclear atypia, intense mitotic activity, neoangiogenesis, vascular thrombosis, limited apoptosis, and foci of necrosis. Vascular hyperproliferation and necrosis are the basic diagnostic criteria for distinguishing GBM from lower-grade gliomas [50,51]. GBM most often occurs in the supraential part of the cranial cavity—in the frontal, parietal, temporal, and occipital regions—but rarely in the cerebellum [52,53]. Due to the location and significant resistance of gliomas to conventional therapies, the prognosis of patients with glioblastoma is very poor and the cure rate is low [54]. GBM is divided into primary and secondary. The first of these mainly affects elderly patients, while much rarer secondary ones usually affect patients before the age of 45. Primary GBM develops de novo from glial cells, characterized by an aggressive course with a short clinical history of usually less than six months. Secondary multiforme gliomas originate from tumors of the astrocytic series (astrocytics) of the lower degree of WHO malignancy as a result of their transformation and malignancy. Despite differences in their pathogenesis, primary and secondary GBMs are similar in their morphological and clinical terms [55,56]. Neurological symptoms in the course of brain tumors are the consequence of increased intracranial pressure, which is the result of limited possibilities of increasing abnormal mass within the skull, forming a set of symptoms resulting directly from the damage to specific structures in the brain. These symptoms may include headaches, vomiting, disturbances of consciousness, orientation, paresis, changes in personality, mood, and psycho-motor slowdown.
Often, symptoms depend on the location of the tumor rather than on its histological peculiarities. The absence of specific symptoms makes the cancer difficult to diagnose, and therefore it is usually detected only at an advanced stage of the disease, in most cases using magnetic resonance imaging (MRI). New tools and methods of diagnosis, including magnetic resonance spectroscopy, magnetic resonance imaging and cerebral diffusion imaging, and the use of F18-fluorodeoxyglucose in positron emission tomography, allow for the diagnosis of cancer at an early stage of its development [57,58,59]. For many years, the standard in the treatment of gliomas has been surgical treatment supported by radio- and chemotherapy. Maximum cytoreduction in the tumor (>98% of the tumor) prolongs life up to 9–12 months and also improves the patient’s response to radio- and chemotherapy. However, due to the location of the tumor and its infiltrative nature, surgical intervention is often not possible. For high-beech gliomas (WHO III and IV), radiation therapy (RT) is used as the first adjuvant treatment after surgery. In turn, the standard in chemotherapy is temozolomide (TMZ) and carmustine (gliadel) [60,61]. In the case of relapses, a faster and more aggressive tumor growth is observed in the group of patients treated with TMZ and RT, which additionally shows significant resistance to treatment [62,63].
In recent years, many new promising therapeutic targets and potential therapeutics have been identified. The new approaches are based on tools such as small-molecule inhibitors, monoclonal antibodies, and peptide vaccines used to regulate cell pathways crucial for cancer development, angiogenesis, and to abolish the drug resistance of cancer cells. However, clinical trials did not report the expected results in the form of the effective inhibition of glioma cell proliferation [64,65]. The reasons for these failures are seen in the activation of alternative signaling pathways which bypass the factor turned off by the inhibitor [66]. Although the new approaches initially performed very well, most of them were rejected at the clinical trial stage. Therefore, gliomas invariably remain among the most difficult to treat and are the least promising cancers, with an average survival time of less than a year [67]. In the absence of effective treatments for gliomas and their resistance to treatment, the challenge is to research new therapeutic goals and approaches in the treatment of GBM. The first stage of therapy design is the identification of therapeutic targets, the “shutdown” of which provides a chance to stop pathological processes, including a reduction in cell proliferation and the launching of apoptotic processes. The use of high-throughput DNA sequencing techniques, cDNA microarrays, and proteomic methods provided new knowledge on the pathogenesis of gliomas and allowed the identification of potential therapeutic targets [68,69]. Currently, much attention is focused on transcription factors, extracellular matrix proteins, chaperone proteins, and miRNAs as new promising targets for GBM therapy. The advantage of the latter over the others is their ability to regulate expression at almost every stage. miRNAs can regulate the expression of up to 90% of genes, and consequently affect a number of cellular processes, such as growth, cell differentiation, apoptosis, or cell signaling [70,71].
A disturbance in miRNA levels alters the expression of target mRNAs. It is estimated that this may be the cause of over 390 diseases (http://cmbi.bjmu.edu.cn/hmdd, accessed on 1 January 2014), the largest group of these being cancers, including brain tumors.
3. miRNAs Expression in GBM
It has been shown that, just as in a healthy brain during its development, the level of individual miRNAs undergoes dynamic changes in a tumor at various stages of its advancement. The miRNA profile in GBM indicates the stage of the disease and can also facilitate the prognosis and selection of appropriate therapy. Based on the level of individual miRNAs, the miRNAs with the highest prognostic value for GBM were selected [72], indicating that that the diagnosis of GBM is also possible on the basis of the analysis of miRNA from the blood and cerebrospinal fluid of patients [73,74,75]. Functional analysis of individual GBM-specific miRNAs indicates that they can act as both oncogenes and tumor suppressors, are responsible for developing resistance to chemotherapy and radiotherapy, stimulate neo-angiogenesis and cell proliferation, and regulate the cell cycle and apoptosis [76,77,78,79,80] (Table 2).
Table 2.
The role of selected miRNAs in GBM.
| miRNA-Regulated Process/Pathway |
Examples of miRNAs |
|---|---|
| Growth and differentiation of CSCs (cancer stem cells) | miR-7 [81], miR-9/miR-9 [82], miR10a/miR-10b [83], miR-17-92 [84], miR-124a/miR-137 [85], miR-125a/miR-125b [86], miR-302-367 [86], miR-326 [86] |
| Cell cycle | miR-21 [87], miR-15b [88], miR-34a [86], miR-221/miR-222 [86] |
| Proliferation and apoptosis | miR-21, miR-26a [89], miR-101 [90], miR-128 [91], miR-156b-5p, miR-153 [43], miR-181a/miR-181b [92], miR-196a/miR-196b [93], miR-218 [86], miR-381 [86], miR-451 [86], let-7a [86] |
| Neo-angiogenesis | miR-93, miR-296 [94] |
| Cell resistance to radio- and chemotherapy | miR-21 [95], miR-125b-2 [86], miR-195 [96], miR-455-3p [96], miR-10a [96] |
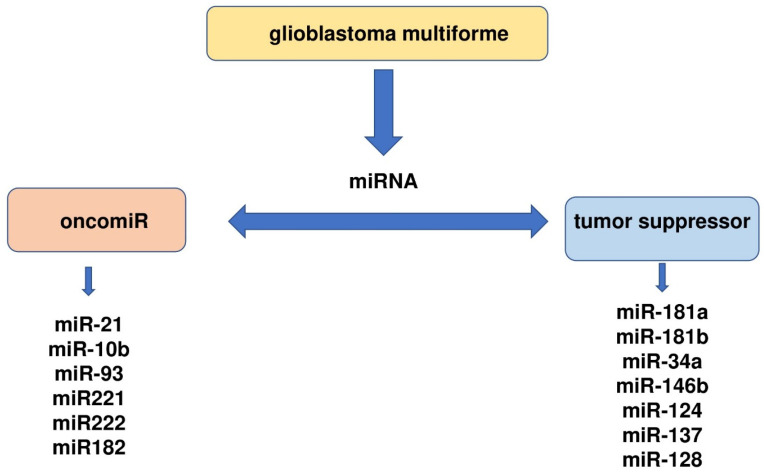
Understanding the GBM-specific miRNA expression profile provides evidence of the involvement of individual miRNAs in the pathogenesis of GBM. This increases the possibility of diagnosing and predicting these cancers. However, we are still far from understanding the mechanisms of cellular regulation involving miRNAs and therefore, also from using miRNAs as potential therapeutic targets. Numerous reports indicate that abnormalities in the expression of selected miRNAs may contribute to the transformation of glial cells and thus to the development of cancer. These include both miRNAs with suppressor and oncogenic functions (Figure 2) [97,98,99,100,101,102,103,104,105,106,107,108,109,110].
Figure 2.
Oncogenic miRNAs and tumor suppressor miRNAs in glioblastoma multiforme.
4. Tumor Suppressor miRNAs
miR-181a and miR-181b are the suppressor molecules whose expression is significantly reduced in glioma cells compared to normal glial cells [111,112]. A decrease in the level of expression of miR-181 genes (miR-181a, 181b, and 181c) was observed in 20–30% of the GBM samples studied [113,114]. It has been shown that the reduction in the pool of miR-181 molecules is proportional to the degree of tumor malignancy (the greatest inhibition of miR-181 expression occurs in stage III and IV tumors). These regulatory factors have been shown to inhibit proliferation, induce apoptosis, and limit cancer cell invasion. Additionally, the induced overexpression of miR-181a and miR-181b in glioma cells results in the loss of the ability of cells to grow independent of contact with the substrate, which is one of the determinants of cell malignancy. Thus, miRNAs from the miR-181 family function as suppressors in glioma cells [115,116].
Reduced expression of miR-34a may also be important in the development of glioma. In glioblastoma cells, the mechanisms that reduce the level of miR-34a probably include epigenetic factors (CpG island methylation disorder) and mutations within the 1p36 locus, which in the case of primary brain tumors, are subject to frequent deletions (70–85%) [117]. This molecule, by regulating the expression of many oncogenes, e.g., C-MET and NOTCH, inhibits the development of cancer [100,118]. It has been proven that transfection of miR-34a cancer cell lines leads to the blockage of cell cycle proliferation and progression and reduces cell survival and invasiveness [119]. One of the direct processes regulated by miR-34a in glioblastoma cells is considered to inhibit the expression of genes of the proteins Notch homolog 1 (Notch1) and Notch homolog 2 (Notch2), which act as transmembrane receptors [120]. With the correct level of miR-34a expression, this mechanism, carried out with the participation of the mesenchymal epithelial transition factor (c-Met), leads to a decrease in signal transduction and subsequently inhibits the process of angiogenesis and proliferation [118]. In GBM cells, elevated levels of Notch1 and 2 receptors stimulate the proliferation and migration of glioma cells by activating the kinases of the AKT-mTOR pathway [121] and also affects the regulation of EGFR receptor expression by the p53 protein, which reduces signal transmission and consequently intensifies the metastasis of cancer cells [30]. Another way to initiate tumor growth with reduced miR-34a expression is to limit the process of inhibition of the transcription of cyclins (E2, D1), kinases (CDK6, CDK4), and Bcl-1, MYC, and E2F3, whose elevated levels in the cell leads to uncontrolled cell cycle progression and the inhibition of apoptosis [122,123]. miR-34a regulates the expression of silent mating type information regulation 2 homolog 1 (SIRT1), the excess of which blocks the process of programmed cell death by binding to the p53 protein [124].
As already stated for cancer stem cells, miR34 plays a visible bimodal role by regulating the Notch and Numb proteins [125]. Numb has been identified as a docking protein involved in the development of Drosophila as an equivalent to Notch, while in various models it works by inducing Notch degradation. In addition, Numb is involved in EGF signaling and internalization of its receptor [126]. Numb also plays a role in stabilizing p53 with a clear implication not only in cancers, but also in stem cells, where p53 has been shown to play a role in stem cell division [127].
miR-34 acts on the regulation of various genes, such as Bcl2, involved in inhibiting the apoptosis pathway, and genes such as NOTCH and NUMB, involved in the development of the nervous system. The effect of miR-34c on NUMB expression can be explained by the interaction on the untranslated 3′ region of NUMB mRNA. This region is conserved among miR-34a and miR-34c. miR34c expression reduces NSC and GSC cell growth and regulates both Bcl2 and NUMB expression. miR-34 clearly inhibits Bcl2, which is involved in resistance to apoptosis, increased cell survival, and response to radiation. Apoptotic resistance can also be affected by NUMB-inducing AKT phosphorylation. miR-34c reduces NUMB expression in both NSC and GSC. miR-34c can inhibit GSC by reducing Bcl2, which could potentially increase the effect of chemotherapy/radiotherapy [128].
miR-34c can be used to treat GBM or other types of cancer. This particular miRNA can be successfully transmitted by viral vectors or extracellular vesicles [129].
The suppressor effect in the development of glioma as well as the migration and invasion of its cells is shown by miR-146b-5p [130,131]. It has been proven that the expression of this miRNA is lower in all types of gliomas than in control astrocytes. miR-146b-5p binds to the 3′UTR region of the EGFR gene transcript, inhibiting its translation. The incorporation of this miRNA into cells results in a reduction in the level of protein kinase phosphorylation (AKT) and inhibition of the Pi3K/AKT pathway. This indicates that restoring the normal expression of this miRNA may be helpful in the treatment of invasive forms of cancer [132,133]. Among the genes regulated by miR-146b in GBM, MMP16 has been identified [134]. The MMP16 protein is responsible for the degradation of extracellular matrix components (ECMs), including collagen III, and is specifically expressed in the central nervous system [135]. In GBM cells with a reduced level of miR-146b, increased expression of MMP affects the expansion of the tumor process by supporting the invasion and migration of cancer cells, as well as by the formation of new blood vessels in the tumor environment [136].
EGFR is also a target gene for miR-7, which is lowered in the expression in glioma cells. It was found that the function of miR-7 associated with the inhibition of the AKT pathway is responsible for limiting the viability and invasiveness of the tumor, which also testifies to the therapeutic potential of this molecule [137,138].
Reduced expression in glioma cancer cells also applies to miR-124, miR-137, and miR-101. miR-124 and miR-137 molecules regulate the expression of the CDK6 gene and contribute to lowering the level of the CDK6 protein, which is involved in the development of a number of malignant tumors [139,140,141]. This results in the blockade of the cell cycle in the G1 phase and the limitation in the proliferation of glioblastoma multiforme cells, which can be extremely valuable in the treatment of this condition [142,143].
The expression of miR-101 varies significantly in glioblastoma cells compared to unchanged cells. Lower levels of this miRNA cause insufficient repression of the enhancer of zeste homolog 2 (EZH2) mRNA translation [73]. This leads to overexpression of methyltransferase EZH2, which induces the proliferation and migration of tumor cells and contributes to the development of tumor vascularization. The level of EZH2 expression correlates with the survival time of patients [142].
In GBM, reduced expression of miR-128 is observed [61]. High levels of miR-128 have been shown to inhibit glioma cell proliferation in vitro and tumor heterograft growth in vivo by directly regulating the Bmi-1 gene [144]. Mechanically, this effect of miR-128 in GBM was associated with self-renewal inhibition of glioma stem cells (GSC) through the Bmi-1 pathway. miR-128 reduces the proliferation of glioma cells by targeting E2F3a [145,146]. miR-128 inhibits the proliferation, invasion, and self-renewal of GBM and glioma stem cells through the BMI1 and E2F3 pathways [147]. miR-128 has been shown to reduce gliogenesis by down-regulating the EGFR and platelet-derived growth factor receptor alpha (PDGFRA). The targets for miR-128 (except EGFR and PDGFRA) for inhibiting GBM cell proliferation are WEE1, MSI1, and E2F3A [148]. In addition, miR-128 regulates angiogenesis by inhibiting P70S6K1 kinase [149]. The upregulation of miR-128 attenuates the effects of cell proliferation, tumor growth, and angiogenesis [149].
5. Onco-miRNA
miR-21 is a miRNA of an oncogenic nature, the overexpression of which is found in many types of cancer, including glioma cells [150,151]. Binding sites for this molecule were found in the 3′UTR regions of transcripts of genes such as programmed cell death 4 (PDCD4), methylthioadenosine phosphorylase (MTAP), and sex-determining region Y box 5 (SOX5). In the development of cancer, PDCD4 is very important, acting as a suppressor gene involved in apoptosis [152]. In the T98G glioma cell line, the level of expression of the PDCD4 gene shows an inverse relationship with the expression of miR-21, and its reduction leads to the inhibition of the process of apoptosis, which is dependent on this gene [153,154].
miR-10b is highly oncogenic in GBM, suggesting that it may regulate oncogenesis and serve as a useful target in GBM therapy. The overexpression of miR-10b has been found in higher-grade gliomas [155,156]. miR-10b has multiple targets such as RhoC, uPAR, and HOXD10 [157]. By influencing these targets, miR-10b is inhibited, resulting in reduced cell growth, invasion, and angiogenesis, as well as increased apoptosis in GBM [126]. In addition, the direct targets of miR-10b associated with cell growth are BCL2L11, TFAP2C, CDKN1A, and CDKN2A [157]. Inhibition of miR-10b can restore target gene expression and reduce glioblastoma cell growth through apoptosis and/or cell cycle arrest.
Many studies have shown that miR-93 is elevated in GBM [30,158,159,160,161]. miR-93 regulates various glioma cell functions such as proliferation, migration, invasion, cell cycle arrest, and chemoresistance by targeting P21 [162]. miR-93 was shown to control autophagic activity in GSC glioblastoma stem cells by inhibiting BECN1/Beclin 1, ATG5, ATG4B, and SQSTM1/p62 [163]. miR-93 regulates GBM cell viability, tumor growth, and vasculogenesis. In particular, miR-93 enhances the formation of blood vessels by targeting integrin-β 8 [163]. These aspects of miR-93 make this it particularly interesting in the treatment of neo-angiogenesis in GBM.
The increased level of expression in glioblastoma with respect to normal glial cells concerns miR-196 [164,165]. It has been proven that significant overexpression of miR-196 in cancer cells is likely associated with a shorter overall survival of glioblastoma patients [166,167].
Other examples of regulatory molecules whose increased expression is observed in glioma cells and in a number of other cancers include miR-221 and miR-222 [168,169]. The key role of these miRNAs is to control the cell cycle and proliferation by regulating the expression of the P27 and P57 proteins [170,171]. miR-221/222 are also involved in the regulation of apoptosis by directly binding to the 3′UTR mRNA region of the PUMA gene (BCL2 binding component 3), which has recently been recognized as the main mediator of apoptosis, in which the transcription factor TP53 is involved [172,173]. Thus, increased expression of these miRNAs may contribute to inhibiting programmed cell death in glioblastoma [174,175]. Increased activity of miR-221/222 in glioma cells may be due to the improper expression of the transcription factors nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) and C-JUN. These factors, binding together to one of the regulatory regions of the miR-221/222 genes, induce their transcription [176,177].
Overexpression of miR-182 increases with the degree of tumor malignancy (a 32-fold increase in miR-182 levels in GBM was observed compared to normal brain tissues) [178,179].
The miR-182 coding sequence was identified in the region of chromosome 7q32.1 within the FRA7H brittle site and the MET gene. MET and FRA7H products have been shown to be frequently amplified in GBM cells [180].
6. The Use of miRNAs in GBM Therapy
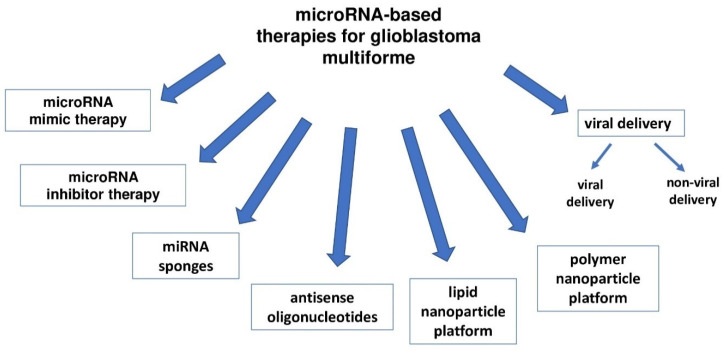
There are high hopes concerning the use of miRNAs in GBM therapy (Figure 3). miRNAs may become a promising therapeutic target due to their ability to target multiple genes.
Figure 3.
The use of miRNA in therapeutic strategies for glioma.
The target of therapy may be microRNAs that undergo both elevated and decreased expression in tumors. According to the latest research, two basic strategies for miRNA-based therapy have been proposed, the first of which is to restore the downward-regulated miRNAs of tumor suppressors using microRNA imitators. The second way is to inhibit oncomiR overexpression with microRNA inhibitors.
Tumor suppressor miRNAs have reduced expression in gliomas. In order to normalize their expression profiles, miRNA-based replacement therapies may be used to increase the expression of a given tumor suppressor molecule.
The inhibition of cancer progression is made possible by exogenous oligonucleotides (also known as miRNA-mimicking), which have the same sequence as the corresponding endogenous miRNAs. These oligonucleotides are synthesized and delivered to GBM cells and strongly inhibit tumor growth. A well-known miRNA with reduced expression suppressor function in GBM is miR-34a [150]. In studies in which cell death was induced using miR-34a mimetics in mutant p53, chemically resistant GBM cells, it was shown that miR-34a mimetics can be used as a novel therapeutic agent [181]. In addition, after transfection into U251 glioma cells, miR-203 mimetics significantly reduce the level of phospholipase D2, which is the target of miR-203. This leads to inhibition of proliferation and invasion of U251 cells [182]. Studies of miR-145 and miR-33a in mouse tumors demonstrated their antitumor activity [183].
miRNA inhibitory therapy is used in GBM to inhibit tumor promoting oncomiR. Recently, many mechanisms have been studied, including the use of antisense oligonucleotides. Antisense oligonucleotides (called antagomiR or antimiR) are synthetically produced oligonucleotides that inhibit levels of upward-regulated miRNAs by blocking the interaction between miRNA and its target mRNAs. In the study, antagomirs coupled to the peptide R3V6 were used to inhibit miR-21. It has been shown that R3V6 peptide can serve as an important tool for the delivery of antisense oligonucleotides [184].
The R3V6 peptide protected oligonucleotides from cleavage by nucleases and also increased their delivery. Conjugate has been found to reduce miR-21 expression and promote apoptosis in GBM cells. Antisense anti-miR-21 oligodeoxynocynucleotides were supplied by R3V6 peptide in vivo. It was noted that apoptosis of cancer cells was strongly promoted, which resulted in effective suppression of tumor growth [185]. In studies using 2′-O-methyl (OMe), the antisense oligonucleotide effectively induced apoptosis in GBM by inhibiting the level of miR-21 expression [186].
miRNA sponges are transcripts containing sites that mimic sequences found in the mRNA complementary to the target miRNA. miRNA sponges are longer nucleic acids such as DNA plasmids or transcribed RNA [187]. They inhibit miRNA function, blocking an entire family of related miRNAs [188]. In GBM, the miR-23b sponge inhibits tumor migration, invasion, and progression in vivo [189]. Natural miRNA sponges include circular RNA (circRNA). ciRS-7 and miR-7 have been found to be overexpressed in the brain [190]. The property of reducing further effects of the target miRNA makes miRNA sponges a tool for studying miRNA function in vitro. However, toxicity and side effects can cause an excess of exogenous nucleic acids, reducing the likelihood that miRNA sponges will be successful as therapeutic agents [191].
Viruses are used to effectively deliver miRNAs to cancer cells [192]. Studies using lentiviral vectors to deliver miR-7-3 to U251 cells showed significant inhibition of proliferation and cell cycle arrest [193]. In vitro and in vivo studies, the crispR/cas9 construct was provided to reduce miR-10 expression using a lentiviral vector [194]. Adenovirus-associated viruses (AAVs) are also candidates for delivering miRNAs. For example, AAV-borne miR-26a was systemically delivered to hepatocellular carcinoma (HCC) cells, resulting in cell cycle arrest, increased apoptosis, and reduced tumor growth [195]. These tests may have potential for other cancers, including glioblastoma. However, side effects such as immunotoxicity, inflammatory responses and tissue degeneration induced by immunogenicity, and mutations caused by the inserted sequence are drawbacks which limit the clinical use of viral miRNA [196]. Therefore, non-viral systems may be more suitable for clinical use.
The most successful delivery systems are polymer and lipid nanoparticles, though magnetic nanoparticles have also been used. In GBM, the widely used miRNA carriers are polymer nanoparticles such as poly (lactic-co-glycolic acid) or PLGA and polyethyleneimine (PEI). In order to deliver antimiR-21 and antimiR-10b to GBM cells, PLGA nanoparticles were used. The result was an increase in the sensitivity to TMZ chemotherapy both in vitro and in vivo [197,198,199]. miRNAs can be successfully delivered by PEI nanoparticles [200]. For example, miR-34a encapsulated in PEI nanoparticles has been delivered across the blood-brain barrier as a treatment for GBM [201]. Lipid nanoparticles are a very useful miRNA carrier for clinical applications due to the stability of miRNAs under physiological conditions [202]. As a result of the simultaneous introduction of the antisense oligonucleotide pemetrexed and miR-21 into glioma cells via cationic solid lipid nanoparticles, high cell uptake efficiency with low toxicity has been demonstrated [203].
Studies in mouse models confirm that lipid-based nanoparticle carriers could become a powerful tool for delivering miRNAs and are likely to find a wide clinical application. In studies on mouse models, stable nucleic acid lipid molecules conjugated with chlorotoxin (CTX-conjugated SNALPs) were used. Systemic delivery of anti-miR-21 resulted in reduced proliferation, tumor growth inhibition, and increased apoptosis in a mouse model of GBM [204]. Lipid nanoparticles containing miR-124 have been found to prolong survival, prevent tumor recurrence, and induce immune memory [53,205,206,207,208,209].
miRNAs can be used as new therapeutic approaches in the treatment of glioblastoma multiforme, Alzheimer’s disease (AD), Parkinson’s disease (PD), and other neurodegenerative diseases [210,211]. The improvement of miRNA changes in GBM and neurodegenerative diseases may be helpful in their early detection. Although glioblastoma multiforme and Alzheimer’s disease share the same molecular pathways, there are significant differences in their modulation. Rapid cell proliferation and cell apoptosis arrest are typical features of GBM. In the case of AD, cell damage and subsequent cell death are common consequences. A set of dysregulated 12 miRNAs in both GBM and AD was identified, demonstrating the existence of an inverse relationship between miRNA expression levels in GBM and AD. Three miRNAs were up-regulated in GBM and down-regulated in AD—hsa-miR-106a, hsa-miR-20b and hsa-miR-424)—and 9 were down-regulated in GBM and up-regulated in AD—hsa-miR-1224, hsa-miR-129, hsa-miR-139, hsa-miR-330, hsa-miR-433, hsa-miR-485, hsa-miR-487b, hsa-miR-584, and hsa-miR-885. In addition, hsa-miR-29c was down-regulated in both GBM and AD, suggesting its involvement in both pathologies [210].
Reverse-expressed miRNAs targeting an identical molecule or modulating the same pathway in both GBM and neurodegenerative diseases may provide attractive entry points to a deeper understanding of the underlying molecular physio-pathological mechanism. For example, miRNA-210 targeting brain-derived neurotrophic factor (BDNF), microglia modulating miRNA-21, and miRNA-27a and -132 modulating Tau would help to explain what pathway triggers a neuron to turn into an undifferentiated and immortal cancer cell or broken dying cell. It is also worth noting that miRNA-10b is not expressed in normal brain tissue, so this would provide an attractive diagnostic approach [211].
Extracellular vesicles (EVs) are a heterogeneous population of vesicles released by cells both in vivo and in vitro. They are an extremely important element of information transfer between different cells without requiring their direct contact. EVs are of a high biological importance and are the subject of intensive research. miRNA-transporting exosomes are one of the key elements of intercellular communication in cancer biology. Exosomes play a key role in GBM, Alzheimer’s disease, Parkinson’s disease, epilepsy, and other brain disorders [212]. An interrelation was observed between GBM exosomes and neuronal damage responsible for neuronal disorders. Exosomal miRNAs are present in the body fluids of patients suffering from malignant gliomas [212].
GBM-derived exosomes can increase oxidative stress in cerebellar neurons by reducing cellular antioxidant defenses and increasing oxidative damage [213].
Attention has recently been drawn to the occurrence of subpopulations of stem cells, called “cancer stem cells” (CSCs), in lesions. miRNAs play an important role in CSCs as important regulators of proliferation and differentiation. Several miRNAs are associated with GBM CSC [214]. The most significant are miR-21 and miR-95, which may affect the molecular profiling of GBM and patient survival. Other miRNAs have been identified as potential regulators of CSC immunogenicity. However, further analysis is needed to elucidate the molecular mechanisms behind GBM CSC.
The difficulty in treating glioblastoma multiforme arises from the fact that many microRNAs, including miR-21, miR-34a, miR-135b, and let-7 are associated with chemotherapy and tumor radiation resistance [215]. A significant increase in miR-24-1 and miR-151-5b expression was demonstrated as a result of irradiation of glioblastoma cell lines with doses commonly used in the treatment of brain tumors—2 Gy [215]. Levels of miR-590-3p were elevated in glioblastoma multiforme tissues and radiation-resistant glioblastoma cells. A potential therapeutic target in increasing the radiosensitivity of cancer cells is miR-221/222. The synthesis of CYP3A4, which metabolizes most chemotherapy drugs, including those used to treat gliomas, is increased in brain tumors, and can be inhibited with the participation of miR-148a, -27b and -125b. These miRNAs are designed to reduce glioblastoma chemoresistance. miR-210 is also a promising diagnostic and prognostic biomarker that can be detected in the peripheral blood of glioblastoma patients. Serum miR-210 levels in glioma are significantly elevated. Studying microRNAs circulating in cerebrospinal fluid can help diagnose brain tumors. This is due to the fact that primary brain tumors with a tendency to spread can secrete microRNAs with oncogenic properties that can be detected in the cerebrospinal fluid. Simultaneous testing of the level of expression of miR-15b and miR-21 in the cerebrospinal fluid allows for the differentiation of glioblastoma patients from healthy individuals and CNS lymphoma patients with 90% sensitivity and 100% specificity [31].
7. Liquid Biopsy
In the diagnosis of brain tumors, a liquid biopsy may be useful [215]. This is a minimally invasive procedure through which information is obtained from body fluids. This information is similar to what is usually obtained from a tissue biopsy sample. A liquid biopsy can analyze circulating tumor cells (CTCs), circulating DNA-free cells (cfDNA), circulating tumor DNA (ctDNA), circulating cell-free tumor RNA (ctDNA), exosomes, proteins, metabolites, and platelets produced by tumors (TEPs). In the case of glioblastoma multiforme, this material may be derived from tumor tissue and may therefore constitute a genuine and representative sample thereof. The best tested of these include ctDNA and ctRNA.
ctDNA can comprehensively represent the glioblastoma genome image. Depending on its histopathological stage, the rate of detection of ctDNA varies [216,217]. Using the NGS technique, the most common gene mutations (TP53, EGFR, MET, PIK3CA, and NOTCH1, TP53, NF1, EGFR1, MET, APC, and PDGFRA, ERBB2, MET, and EGFR) were selected in the patient’s plasma [216,218]. In patients with glioblastoma, methylation of the MGMT gene promoter was observed in tissues and serum ctDNA [214]. In the case of increased methylation, patients had a better response to treatment with alkylating agents.
A potential diagnostic and prognostic target may be the analysis of miRNA in the serum of patients with glioblastoma. It has been shown that the most significant miRNAs are miR-15b, miR-23a, miR-133a, miR-150, miR-197, miR-497, miR-548b, miR-21, miR-128, miR-342, and miR-205. A change in the expression of these miRNAs has been demonstrated in patients with glioblastoma [219,220,221], and their return to normal levels was observed after surgery and chemotherapy, which indicates their use as biomarkers of response to therapy [220,221].
lncRNAs may be potential diagnostic and prognostic biomarkers in glioma multiforme [220]. Several lncRNAs (HOTAIR, GAS5, H19, and MALAT1) with altered expression levels were detected in blood samples of glioblastoma patients compared with healthy subjects [98,221,222,223]. GAS5 has been shown to be associated with patients’ responses to temozolomide (TMZ) therapy. siRNA, circRNA, snRNA, and snoR-NA may also have potential as biomarkers in the diagnosis and prognosis of glioblastoma [224,225].
In non-invasive diagnostics, exosomal vesicles that are present in body fluids (blood, cerebrospinal fluid, urine) can be used. EVs have a diverse molecular composition (nucleic acids, proteins, lipids, metabolites). These components can be transferred to nearby or distant cells through direct EV contact with the cell membrane, fusion, or internalization [226,227]. Analysis of EVs in the blood, cerebrospinal fluid, or other biological fluid of patients with glioblastoma may have diagnostic and prognostic significance [228,229].
EVs can cross anatomical barriers such as BBBs, which increases their value as a potential biomarker for glioblastoma [230,231,232,233]. In glioblastoma-derived EVs, changes in the EGFRvIII, IDH1, PTEN, and PD-L1 genes were detected [231,233,234]. The release of EVs is affected by the treatment of TMZ. EVs derived from TMZ-resistant patients show increased levels of MGMT expression [235,236] and dysregulated levels of proteins associated with cellular adhesion, such as transglutaminase 2 (TGM2), NESTIN, glycoproteins, CD44, and CD133, which are expressed on the surface of EVs [237]. EVs can therefore serve as tumor biomarkers to monitor TMZ treatment.
TEPs are platelets that have received cancer-related molecules from cancer cells [238]. In glioma, TEPs have been shown to capture tumor-derived EVs with mutant EGFRvIII. The EGFRvIII mutation was detected in 80% of glioblastoma multiforme [239]. RNA derived from TEPs could complement currently used biosources and biomolecules used in the diagnosis of a liquid biopsy. This would improve the early detection of cancer and facilitate non-invasive monitoring of the disease [240]. By analyzing TEPs, it is possible to distinguish cancer patients from healthy ones with an accuracy of 84–96%. In addition, TEP profiling can be used to determine the organ origin of the primary tumor with 71% accuracy. TEP profiles can be differentiated between subtypes of molecular tumors based on EGFR and K-RAS [241].
The presence of CTCs in the patient’s bloodstream, resulting from their separation from the primary tumor, can be used for early diagnosis of the disease, as well as for the selection of a therapy and the monitoring of its effectiveness. The representative presence of CTCs for tumors has been detected in patients with glioblastoma of various stages, including glioblastoma multiforme [242,243]. CTCs derived from glioblastoma exhibited EGFR amplification, which was associated with aggressiveness and with the presence of EGFRvIII [49] and increased expression of SERPINE1, TGFB1, TGFBR2, and VIM genes associated with the mesenchymal subtype [244]. CTCs derived from glioblastoma multiforme have been shown to possess stem cell properties, contributing to the formation of local tumors and relapses [245,246]. Additional elements have been detected in the cerebrospinal fluid of glioblastoma patients that may be components of a liquid biopsy, such as circulating miRNAs and miRNAs derived from EVs [247,248]. These may serve as biomarkers of cerebrospinal fluid for diagnosing and monitoring responses to treatment in patients with glioblastoma [248].
The levels of the nine-miRNA panel (miR-21, miR-218, miR-193b, miR-331, miR374a, miR548c, miR520f, miR27b, and miR-30b) were associated with tumor volume and exhibited a 67% sensitivity and 80% specificity [249]. Elevated levels of miR-21, miR-10b, and miR-15b in cerebrospinal fluid have been shown to be associated with glioma stage, prognosis, and response to treatment [250,251,252]. Cerebrospinal fluid miRNAs have a better diagnostic value, with a higher sensitivity (84%) and specificity (92%) than their serum levels [242].
8. Conclusions
Our knowledge of disorders occurring in various types and degrees of glioma malignancy has increased dramatically in recent years [53,207], and many of the changes (confirmed by histopathological analysis) are now complementary to basic diagnostic techniques [207,208].
miRNA studies in cancer processes significantly enrich modern knowledge on the pathogenesis of glioblastoma multiforme cells. miRNAs can be new goals in diagnosis and therapy.
Establishing the miRNA expression profile characteristic of GBM cells is an alternative to obtaining a precise picture of the type and extent of cancerous changes in glioma cells. Experimental verification of high-resolution techniques and in silico analyses provides the chance to obtain a reliable answer to the question of the causes and mechanisms of miRNA disorders in cancer cells. It also creates the possibility of using them as prognostic elements or potential targets in the therapy of brain tumors.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research has received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Ouellet D.L., Perron M.P., Gobeil L.-A., Plante P., Provost P. MicroRNAs in Gene Regulation: When the Smallest Governs It All. J. Biomed. Biotechnol. 2006;2006:69616. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 5.Leonov G., Shah K., Yee D., Timmis J., Sharp T.V., Lagos D. Suppression of AGO2 by miR-132 as a determinant of miRNA-mediated silencing in human primary endothelial cells. Int. J. Biochem. Cell Biol. 2015;69:75–84. doi: 10.1016/j.biocel.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali Syeda Z., Langden S.S.S., Munkhzul C., Lee M., Song S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020;21:1723. doi: 10.3390/ijms21051723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behl T., Kumar C., Makkar R., Gupta A., Sachdeva M. Intercalating the Role of MicroRNAs in Cancer: As Enemy or Protector. Asian Pac. J. Cancer Prev. 2020;21:593–598. doi: 10.31557/APJCP.2020.21.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B., Pan X., Cobb G., Anderson T. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Liang Y., Ridzon D., Wong L., Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira P., Marques J.P., Soares A.R., Carreto L., Santos M.A.S. MicroRNA Expression Variability in Human Cervical Tissues. PLoS ONE. 2010;5:e11780. doi: 10.1371/journal.pone.0011780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leichter A.L., Sullivan M., Eccles M.R., Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol. Cancer. 2017;16:15. doi: 10.1186/s12943-017-0584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sempere L.F., Azmi A.S., Moore A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip. Rev. RNA. 2021;12:e1662. doi: 10.1002/wrna.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mott J.L., Kobayashi S., Bronk S.F., Gores G.J. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akao Y., Nakagawa Y., Naoe T. let-7 MicroRNA Functions as a Potential Growth Suppressor in Human Colon Cancer Cells. Biol. Pharm. Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 19.Abdalla F., Singh B., Bhat H.K. MicroRNAs and gene regulation in breast cancer. J. Biochem. Mol. Toxicol. 2020;34:e22567. doi: 10.1002/jbt.22567. [DOI] [PubMed] [Google Scholar]
- 20.Mei H., Wen Y. MicroRNAs for Diagnosis and Treatment of Colorectal Cancer. Endocr. Metab. Immune Disord. Drug Targets. 2021;21:47–55. doi: 10.2174/1871530320999200818134339. [DOI] [PubMed] [Google Scholar]
- 21.Iqbal M.A., Arora S., Prakasam G., Calin G.A., Syed M.A. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol. Asp. Med. 2019;70:3–20. doi: 10.1016/j.mam.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Sorop A., Constantinescu D., Cojocaru F., Dinischiotu A., Cucu D., Dima S. Exosomal microRNAs as Biomarkers and Therapeutic Targets for Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021;22:4997. doi: 10.3390/ijms22094997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia T., Chen X.-Y., Zhang Y.-N. MicroRNAs as biomarkers and perspectives in the therapy of pancreatic cancer. Mol. Cell. Biochem. 2021;476:4191–4203. doi: 10.1007/s11010-021-04233-y. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Q., Liu J., Quan J., Liu W., Tan H., Li W. MicroRNAs as potential biomarkers for the diagnosis of glioma: A systematic review and meta-analysis. Cancer Sci. 2018;109:2651–2659. doi: 10.1111/cas.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Q., Hou W. Regulation of angiogenesis by microRNAs in cancer. Mol. Med. Rep. 2021;24:583. doi: 10.3892/mmr.2021.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uzuner E., Ulu G.T., Gürler S.B., Baran Y. The Role of MiRNA in Cancer: Pathogenesis, Diagnosis, and Treatment. Methods Mol. Biol. 2022;2257:375–422. doi: 10.1007/978-1-0716-1170-8_18. [DOI] [PubMed] [Google Scholar]
- 27.Hosseini S.A., Horton S., Saldivar J.C., Miuma S., Stampfer M.R., Heerema N.A., Huebner K. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Genes Chromosomes Cancer. 2013;52:1017–1029. doi: 10.1002/gcc.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottoni A., Calin G.A. MicroRNAs as main players in the pathogenesis of chronic lymphocytic leukemia. Microrna. 2014;2:158–164. doi: 10.2174/2211536602666131126002337. [DOI] [PubMed] [Google Scholar]
- 30.Piwecka M., Rolle K., Belter A., Barciszewska A.M., Żywicki M., Michalak M., Nowak S., Naskręt-Barciszewska M.Z., Barciszewski J. Comprehensive analysis of microRNA expression profile in malignant glioma tissues. Mol. Oncol. 2015;9:1324–1340. doi: 10.1016/j.molonc.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sufianov A., Begliarzade S., Ilyasova T., Liang Y., Beylerli O. MicroRNAs as prognostic markers and therapeutic targets in gliomas. Non-Coding RNA Res. 2022;7:171–177. doi: 10.1016/j.ncrna.2022.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S.-W., Ali N.-D., Zhong L., Shi J. MicroRNAs as biomarkers for human glioblastoma: Progress and potential. Acta Pharmacol. Sin. 2018;39:1405–1413. doi: 10.1038/aps.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M., Medarova Z., Moore A. Role of microRNAs in glioblastoma. Oncotarget. 2021;12:1707–1723. doi: 10.18632/oncotarget.28039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monfared H., Jahangard Y., Nikkhah M., Mirnajafi-Zadeh J., Mowla S.J. Potential Therapeutic Effects of Exosomes Packed with a miR-21-Sponge Construct in a Rat Model of Glioblastoma. Front. Oncol. 2019;9:782. doi: 10.3389/fonc.2019.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rezaei O., Honarmand K., Nateghinia S., Taheri M., Ghafouri-Fard S. miRNA signature in glioblastoma: Potential biomarkers and therapeutic targets. Exp. Mol. Pathol. 2020;117:104550. doi: 10.1016/j.yexmp.2020.104550. [DOI] [PubMed] [Google Scholar]
- 36.Yang G., Han D., Chen X., Zhang D., Wang L., Shi C., Zhang W., Li C., Chen X., Liu H., et al. MiR-196a exerts its oncogenic effect in glioblastoma multiforme by inhibition of IκBα both in vitro and in vivo. Neuro-Oncology. 2014;16:652–661. doi: 10.1093/neuonc/not307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C.-Z., Zhang J.-X., Zhang A.-L., Shi Z.-D., Han L., Jia Z.-F., Yang W.-D., Wang G.-X., Jiang T., You Y.-P., et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S., Liu H., Deng M., Wang H. MiR-182 promotes glioma progression by targeting FBXW7. J. Neurol. Sci. 2020;411:116689. doi: 10.1016/j.jns.2020.116689. [DOI] [PubMed] [Google Scholar]
- 39.Alamdari-Palangi V., Amini R., Karami H. MiRNA-7 enhances erlotinib sensitivity of glioblastoma cells by blocking the IRS-1 and IRS-2 expression. J. Pharm. Pharmacol. 2020;72:531–538. doi: 10.1111/jphp.13226. [DOI] [PubMed] [Google Scholar]
- 40.Bahreyni-Toossi M.-T., Dolat E., Khanbabaei H., Zafari N., Azimian H. microRNAs: Potential glioblastoma radiosensitizer by targeting radiation-related molecular pathways. Mutat. Res. Mol. Mech. Mutagen. 2019;816–818:111679. doi: 10.1016/j.mrfmmm.2019.111679. [DOI] [PubMed] [Google Scholar]
- 41.Sabelström H., Petri R., Shchors K., Jandial R., Schmidt C., Sacheva R., Masic S., Yuan E., Fenster T., Martinez M., et al. Driving Neuronal Differentiation through Reversal of an ERK1/2-miR-124-SOX9 Axis Abrogates Glioblastoma Aggressiveness. Cell Rep. 2019;28:2064–2079. doi: 10.1016/j.celrep.2019.07.071. [DOI] [PubMed] [Google Scholar]
- 42.Li L., Shao M.-Y., Zou S.-C., Xiao Z.-F., Chen Z.-C. MiR-101-3p inhibits EMT to attenuate proliferation and metastasis in glioblastoma by targeting TRIM44. J. Neuro-Oncol. 2019;141:19–30. doi: 10.1007/s11060-018-2973-7. [DOI] [PubMed] [Google Scholar]
- 43.Toraih E.A., El-Wazir A., Abdallah H.Y., Tantawy M.A., Fawzy M.S. Deregulated MicroRNA Signature Following Glioblastoma Irradiation. Cancer Control. 2019;26:1073274819847226. doi: 10.1177/1073274819847226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mei J., Bachoo R., Zhang C.-L. MicroRNA-146a Inhibits Glioma Development by Targeting Notch1. Mol. Cell. Biol. 2011;31:3584–3592. doi: 10.1128/MCB.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L., Wang X., Wang H., Li Y., Yan W., Han L., Zhang K., Zhang J., Wang Y., Feng Y., et al. miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur. J. Cancer. 2012;48:3104–3111. doi: 10.1016/j.ejca.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Davis M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016;20((Suppl. S5)):S2–S8. doi: 10.1188/16.CJON.S1.2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K., Burger P.C., Jouvet A., Schithauer B.W., Kleihues P. The 2007 WHO classification of tumors of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Louis D.N., Perry A., Reifenberger G., Von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 49.Mathew E.N., Berry B.C., Yang H.W., Carroll R.S., Johnson M.D. Delivering Therapeutics to Glioblastoma: Overcoming Biological Constraints. Int. J. Mol. Sci. 2022;23:1711. doi: 10.3390/ijms23031711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adamson C., Kanu O., Mehta A., Di C., Lin N., Mattox A.K., Bringer D.D. Glioblastoma multiforme: A review of where we have been and where we are going. Expert Opin. Investig. Drugs. 2009;18:1061–1083. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- 51.Sejda A., Grajkowska W., Trubicka J., Szutowicz E., Wojdacz T., Kloc W., Iżycka-Świeszewska E. WHO CNS5 2021 classification of gliomas: A practical review and road signs for diagnosing pathologists and proper patho-clinical and neuro-oncological cooperation. Folia Neuropathol. 2022;60:137–152. doi: 10.5114/fn.2022.118183. [DOI] [PubMed] [Google Scholar]
- 52.Chamberlain M.C., Kormanik P.A. Practical guidelines for the treatment of malignant gliomas. West. J. Med. 1998;168:114–120. [PMC free article] [PubMed] [Google Scholar]
- 53.Hanif F., Muzaffar K., Perveen K., Malhi S.M., Simjee S.U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. APJCP. 2017;18:3–9. doi: 10.22034/APJCP.2017.18.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bausart M., Préat V., Malfanti A. Immunotherapy for glioblastoma: The promise of combination strategies. J. Exp. Clin. Cancer Res. 2022;41:35. doi: 10.1186/s13046-022-02251-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Furnari F.B., Fenton T., Bachoo R.M., Mukasa A., Stommel J.M., Stegh A., Hahn W.C., Ligon K.L., Louis D.N., Brennan C., et al. Malignant astrocylic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 56.Ohgaki H., Kleihues P. The Definition of Primary and Secondary Glioblastoma. Clin. Cancer Res. 2013;19:764–772. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 57.Kanu O.O., Mehta A., Di C., Lin N., Bortoff K., Bigner D.D., Yan H., Adamson D.C. Glioblastoma multiforme: A review of therapeutic targets. Expert Opin. Ther. Targets. 2009;13:701–718. doi: 10.1517/14728220902942348. [DOI] [PubMed] [Google Scholar]
- 58.Krebs S., Barasch J.G., Young R.J., Grommes C., Schöder H. Positron emission tomography and magnetic resonance imaging in primary central nervous system lymphoma-a narrative review. Ann. Lymphoma. 2021;5:15. doi: 10.21037/aol-20-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hassanzadeh C., Rao Y.J., Chundury A., Rowe J., Ponisio M.R., Sharma A., Miller-Thomas M., Tsien C.I., Ippolito J.E. Multiparametric MRI and [18F]Fluorodeoxyglucose Positron Emission Tomography Imaging Is a Potential Prognostic Imaging Biomarker in Recurrent Glioblastoma. Front. Oncol. 2017;7:178. doi: 10.3389/fonc.2017.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fisher J., Adamson D. Current FDA-Approved Therapies for High-Grade Malignant Gliomas. Biomedicines. 2021;9:324. doi: 10.3390/biomedicines9030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minniti G., Niyazi M., Alongi F., Navarria P., Belka C. Current status and recent advances in reirradiation of glioblastoma. Radiat. Oncol. 2021;16:36. doi: 10.1186/s13014-021-01767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ortiz R., Perazzoli G., Cabeza L., Jiménez-Luna C., Luque R., Prados J., Melguizo C. Temozolomide: An Updated Overview of Resistance Mechanisms, Nanotechnology Advances and Clinical Applications. Curr. Neuropharmacol. 2021;19:513–537. doi: 10.2174/1570159x18666200626204005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu Y., Chen Z., Na Kim S., Gan C., Ryl T., Lesjak M.S., Rodemerk J., De Zhong R., Wrede K., Dammann P., et al. Characterization of Temozolomide Resistance Using a Novel Acquired Resistance Model in Glioblastoma Cell Lines. Cancers. 2022;14:2211. doi: 10.3390/cancers14092211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu H., Qiu W., Sun T., Wang L., Du C., Hu Y., Liu W., Feng F., Chen Y., Sun H. Therapeutic strategies of glioblastoma (GBM): The current advances in the molecular targets and bioactive small molecule compounds. Acta Pharm. Sin. B. 2022;12:1781–1804. doi: 10.1016/j.apsb.2021.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weil R.J. Incorporating Molecular Tools into Early-Stage Clinical Trials. PLoS Med. 2008;5:e21. doi: 10.1371/journal.pmed.0050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen H.-M., Guz-Montgomery K., Lowe D., Saha D. Pathogenetic Features and Current Management of Glioblastoma. Cancers. 2021;13:856. doi: 10.3390/cancers13040856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown N.F., Ottaviani D., Tazare J., Gregson J., Kitchen N., Brandner S., Fersht N., Mulholland P. Survival Outcomes and Prognostic Factors in Glioblastoma. Cancers. 2022;14:3161. doi: 10.3390/cancers14133161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riddick G., Fine H.A. Integration and analysos of genome-scale data form gliomas. Nat. Rev. Neuro. 2011;17:439–450. doi: 10.1038/nrneurol.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramkissoon L.A., Britt N., Guevara A., Whitt E., Severson E., Sathyan P., Gay L., Elvin J., Ross J.S., Brown C., et al. Precision Neuro-oncology: The Role of Genomic Testing in the Management of Adult and Pediatric Gliomas. Curr. Treat. Options Oncol. 2018;19:41. doi: 10.1007/s11864-018-0559-4. [DOI] [PubMed] [Google Scholar]
- 70.Jang J.H., Lee T.-J. The role of microRNAs in cell death pathways. Yeungnam Univ. J. Med. 2021;38:107–117. doi: 10.12701/yujm.2020.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bartel D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Srinivasan S., Patric I.R., Somasundarman K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS ONE. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia C.M., Toms S.A. The Role of Circulating MicroRNA in Glioblastoma Liquid Biopsy. World Neurosurg. 2020;138:425–435. doi: 10.1016/j.wneu.2020.03.128. [DOI] [PubMed] [Google Scholar]
- 74.Mafi A., Rahmati A., BabaeiAghdam Z., Salami R., Salami M., Vakili O., Aghadavod E. Recent insights into the microRNA-dependent modulation of gliomas from pathogenesis to diagnosis and treatment. Cell Mol. Biol. Lett. 2022;27:65. doi: 10.1186/s11658-022-00354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kopkova A., Sana J., Machackova T., Vecera M., Radova L., Trachtova K., Vybihal V., Smrcka M., Kazda T., Slaby O., et al. Cerebrospinal Fluid MicroRNA Signatures as Diagnostic Biomarkers in Brain Tumors. Cancers. 2019;11:1546. doi: 10.3390/cancers11101546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gareev I., Beylerli O., Liang Y., Xiang H., Liu C., Xu X., Yuan C., Ahmad A., Yang G. The Role of MicroRNAs in Therapeutic Resistance of Malignant Primary Brain Tumors. Front. Cell Dev. Biol. 2021;9:740303. doi: 10.3389/fcell.2021.740303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sati I.S.E.E., Parhar I. MicroRNAs Regulate Cell Cycle and Cell Death Pathways in Glioblastoma. Int. J. Mol. Sci. 2021;22:13550. doi: 10.3390/ijms222413550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mahinfar P., Mansoori B., Rostamzadeh D., Baradaran B., Cho W.C., Mansoori B. The Role of microRNAs in Multidrug Resistance of Glioblastoma. Cancers. 2022;14:3217. doi: 10.3390/cancers14133217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahmed S.P., Castresana J.S., Shahi M.H. Role of Circular RNA in Brain Tumor Development. Cells. 2022;11:2130. doi: 10.3390/cells11142130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olivier C., Oliver L., Lalier L., Vallette F.M. Drug Resistance in Glioblastoma: The Two Faces of Oxidative Stress. Front. Mol. Biosci. 2021;7:620677. doi: 10.3389/fmolb.2020.620677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korać P., Antica M., Matulić M. MiR-7 in Cancer Development. Biomedicines. 2021;9:325. doi: 10.3390/biomedicines9030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Geng L., Xu J., Zhu Y., Hu X., Liu Y., Yang K., Xiao H., Zou Y., Liu H., Ji J., et al. Targeting miR-9 in Glioma Stem Cell-Derived Extracellular Vesicles: A Novel Diagnostic and Therapeutic Biomarker. Transl. Oncol. 2022;22:101451. doi: 10.1016/j.tranon.2022.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu A., Zhao H., Sun B., Han X., Zhou D., Cui Z., Ma X., Zhang J., Yuan L. A predictive analysis approach for paediatric and adult high-grade glioma: miRNAs and network insight. Ann. Transl. Med. 2020;8:242. doi: 10.21037/atm.2020.01.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schnabel E., Knoll M., Schwager C., Warta R., Mock A., Campos B., König L., Jungk C., Wick W., Unterberg A., et al. Prognostic Value of microRNA-221/2 and 17-92 Families in Primary Glioblastoma Patients Treated with Postoperative Radiotherapy. Int. J. Mol. Sci. 2021;22:2960. doi: 10.3390/ijms22062960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ryspayeva D., Halytskiy V., Kobyliak N., Dosenko I., Fedosov A., Inomistova M., Drevytska T., Gurianov V., Sulaieva O. Response to neoadjuvant chemotherapy in breast cancer: Do microRNAs matter? Discov. Oncol. 2022;13:43. doi: 10.1007/s12672-022-00507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karsy M., Arslan E., Moy F. Current Progress on Understanding MicroRNAs in Glioblastoma Multiforme. Genes Cancer. 2012;3:3–15. doi: 10.1177/1947601912448068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Masoudi M.S., Mehrabian E., Mirzaei H. MiR-21: A key player in glioblastoma pathogenesis. J. Cell. Biochem. 2018;119:1285–1290. doi: 10.1002/jcb.26300. [DOI] [PubMed] [Google Scholar]
- 88.Gasparello J., Papi C., Zurlo M., Gambari L., Rozzi A., Manicardi A., Corradini R., Gambari R., Finotti A. Current Progress on Understanding MicroRNAs in Glioblastoma Multiforme. Molecules. 2022;27:1299. doi: 10.3390/molecules27041299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.ParvizHamidi M., Haddad G., Ostadrahimi S., Ostadrahimi N., Sadeghi S., Fayaz S., Fard-Esfahani P. Circulating miR-26a and miR-21 as biomarkers for glioblastoma multiform. Biotechnol. Appl. Biochem. 2019;66:261–265. doi: 10.1002/bab.1707. [DOI] [PubMed] [Google Scholar]
- 90.Lombard A., Goffart N., Rogister B. Glioblastoma Circulating Cells: Reality, Trap or Illusion? Stem Cells Int. 2015;2015:182985. doi: 10.1155/2015/182985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jovčevska I. Sequencing the next generation of glioblastomas. Crit. Rev. Clin. Lab. Sci. 2018;55:264–282. doi: 10.1080/10408363.2018.1462759. [DOI] [PubMed] [Google Scholar]
- 92.Witusik-Perkowska M., Zakrzewska M., Jaskolski D.J., Liberski P.P., Szemraj J. Artificial microenvironment of in vitro glioblas-toma cell cultures changes profile of miRNAs related to tumor drug resistance. Onco Targets Ther. 2019;12:3905–3918. doi: 10.2147/OTT.S190601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takkar S., Sharma V., Ghosh S., Suri A., Sarkar C., Kulshreshtha R. Hypoxia-inducible miR-196a modulates glioblastoma cell proliferation and migration through complex regulation of NRAS. Cell Oncol. (Dordr) 2021;44:433–451. doi: 10.1007/s13402-020-00580-y. [DOI] [PubMed] [Google Scholar]
- 94.Wang L., Lee A.Y., Wigg J.P., Peshavariya H., Liu P., Zhang H. miRNA involvement in angiogenesis in age-related macular de-generation. J. Physiol. Biochem. 2016;72:583–592. doi: 10.1007/s13105-016-0496-2. [DOI] [PubMed] [Google Scholar]
- 95.Beylerli O., Gareev I., Sufianov A., Ilyasova T., Zhang F. The role of microRNA in the pathogenesis of glial brain tumors. Non-Coding RNA Res. 2022;7:71–76. doi: 10.1016/j.ncrna.2022.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ujifuku K., Mitsutake N., Takakura S., Matsuse M., Saenko V., Suzuki K., Hayashi K., Matsuo T., Kamada K., Nagata I., et al. miR-195, miR-455-3p and miR-10a are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 2010;296:241–248. doi: 10.1016/j.canlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 97.Shenouda S.K., Alahari S.K. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 98.Ahir B.K., Ozer H., Engelhard H.H., Lakka S.S. MicroRNAs in glioblastoma pathogenesis and therapy: A comprehensive review. Crit. Rev. Oncol. Hematol. 2017;120:22–33. doi: 10.1016/j.critrevonc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 99.Møller H.G., Rasmussen A.P., Andersen H.H., Johnsen K.B., Henriksen M., Duroux M. A systematic review of microRNA in glio-blastoma multiforme: Micro-modulators in the mesenchymal mode of migration and invasion. Mol. Neurobiol. 2013;47:131–144. doi: 10.1007/s12035-012-8349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu B., Mei J., Ji W., Huo Z., Bian Z., Jiao J., Li X., Sun J., Shao J. MicroRNAs involved in the EGFR pathway in glioblastoma. Biomed. Pharm. 2021;134:111115. doi: 10.1016/j.biopha.2020.111115. [DOI] [PubMed] [Google Scholar]
- 101.Chuang H.Y., Su Y.K., Liu H.W., Chen C.H., Chiu S.C., Cho D.Y., Lin S.Z., Chen Y.S., Lin C.M. Preclinical evidence of STAT3 inhibitor pacritinib overcoming temozolomide resistance via downregulating miR-21-enriched exosomes from M2 glioblastoma-associated macrophages. J. Clin. Med. 2019;8:959. doi: 10.3390/jcm8070959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gabriely N.M., Teplyuk A.M. Krichevsky Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy. 2011;7:1384–1386. doi: 10.4161/auto.7.11.17371. [DOI] [PubMed] [Google Scholar]
- 103.Khayamzadeh M., Niazi V., Hussen B.M., Taheri M., Ghafouri-Fard S., Samadian M. Emerging role of extracellular vesicles in the pathogenesis of glioblastoma. Metab. Brain Dis. 2022;38:177–184. doi: 10.1007/s11011-022-01074-6. [DOI] [PubMed] [Google Scholar]
- 104.Di Martino M.T., Arbitrio M., Caracciolo D., Cordua A., Cuomo O., Grillone K., Riillo C., Caridà G., Scionti F., Labanca C., et al. miR-221/222 as biomarkers and targets for therapeutic intervention on cancer and other diseases: A systematic review. Mol. Ther. Nucleic Acids. 2022;27:1191–1224. doi: 10.1016/j.omtn.2022.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gulluoglu S., Tuysuz E.C., Sahin M., Kuskucu A., Yaltirik C.K., Ture U., Kucukkaraduman B., Akbar M.W., Gure A.O., Bayrak O.F., et al. Dalan Simultaneous miRNA and mRNA transcriptome profiling of glioblastoma samples reveals a novel set of OncomiR candidates and their target genes. Brain Res. 2018;1700:199–210. doi: 10.1016/j.brainres.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 106.Schäfer A., Evers L., Meier L., Schlomann U., Bopp M.H.A., Dreizner G.-L., Lassmann O., Ben Bacha A., Benescu A.-C., Pojskic M., et al. The Metalloprotease-Disintegrin ADAM8 Alters the Tumor Suppressor miR-181a-5p Expression Profile in Glioblastoma Thereby Contributing to Its Aggressiveness. Front. Oncol. 2022;12:826273. doi: 10.3389/fonc.2022.826273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jesionek-Kupnicka D., Braun M., Trąbska-Kluch B., Czech J., Szybka M., Szymańska B., Kulczycka-Wojdala D., Bieńkowski M., Kordek R., Zawlik I. MiR-21, miR-34a, miR-125b, miR-181d and miR-648 levels inversely correlate with MGMT and TP53 expression in primary glioblastoma patients. Arch. Med. Sci. 2019;15:504–512. doi: 10.5114/aoms.2017.69374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schneider B., Lamp N., Zimpfer A., Henker C., Erbersdobler A. Comparing tumor microRNA profiles of patients with long and short term surviving glioblastoma. Mol. Med. Rep. 2023;27:8. doi: 10.3892/mmr.2022.12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Silber J., Lim D.A., Petritsch C., Persson A.I., Maunakea A.K., Yu M., Vandenberg S.R., Ginzinger D.G., James C.D., Costello J.F., et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Papagiannakopoulos T., Friedmann-Morvinski D., Neveu P., Dugas J.C., Gill R.M., Huillard E., Liu C., Zong H., Rowitch D.H., Barres B.A., et al. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene. 2011;31:1884–1895. doi: 10.1038/onc.2011.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shea A., Harish V., Afzal Z., Chijioke J., Kedir H., Dusmatova S., Roy A., Ramalinga M., Harris B., Blancato J., et al. MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics. Cancer Med. 2016;5:1917–1946. doi: 10.1002/cam4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu C.-H., Xiao L.-M., Zeng E.-M., Chen L.-K., Zheng S.-Y., Li D.-H., Liu Y. MicroRNA-181 inhibits the proliferation, drug sensitivity and invasion of human glioma cells by targeting Selenoprotein K (SELK) Am. J. Transl. Res. 2019;11:6632–6640. [PMC free article] [PubMed] [Google Scholar]
- 113.Buruiană A., Florian Ș.I., Florian A.I., Timiș T.L., Mihu C.M., Miclăuș M., Oșan S., Hrapșa I., Cataniciu R.C., Farcaș M., et al. The Roles of miRNA in Glioblastoma Tumor Cell Communication: Diplomatic and Aggressive Negotiations. Int. J. Mol. Sci. 2020;21:1950. doi: 10.3390/ijms21061950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Indrieri A., Carrella S., Carotenuto P., Banfi S., Franco B. The Pervasive Role of the miR-181 Family in Development, Neurodegeneration, and Cancer. Int. J. Mol. Sci. 2020;21:2092. doi: 10.3390/ijms21062092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Marisetty A., Wei J., Kong L.-Y., Ott M., Fang D., Sabbagh A., Heimberger A.B. MiR-181 Family Modulates Osteopontin in Glioblastoma Multiforme. Cancers. 2020;12:3813. doi: 10.3390/cancers12123813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wei J., Marisetty A., Schrand B., Gabrusiewicz K., Hashimoto Y., Ott M., Grami Z., Kong L.Y., Ling X., Caruso H., et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J. Clin. Invest. 2019;129:137–149. doi: 10.1172/JCI121266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y., Guo L. MicroRNA-9 regulates the proliferation, migration and invasion of human glioma cells by targeting CDH1. J. BUON. 2020;25:1091–1097. [PubMed] [Google Scholar]
- 118.Ghaemmaghami A.B., Mahjoubin-Tehran M., Movahedpour A., Morshedi K., Sheida A., Taghavi S.P., Mirzaei H., Ham-blin M.R. Role of exosomes in malignant glioma: microRNAs and proteins in pathogenesis and diagnosis. Cell Commun. Signal. 2020;18:120. doi: 10.1186/s12964-020-00623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li W., Wang Y., Liu R., Kasinski A.L., Shen H., Slack F.J., Tang D.G. MicroRNA-34a: Potent Tumor Suppressor, Cancer Stem Cell Inhibitor, and Potential Anticancer Therapeutic. Front. Cell Dev. Biol. 2021;9:640587. doi: 10.3389/fcell.2021.640587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Misso G., Di Martino M.T., De Rosa G., Farooqi A.A., Lombardi A., Campani V., Zarone M.R., Gullà A., Tagliaferri P., Tassone P., et al. Mir-34: A new weapon against cancer? Mol. Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ghafouri-Fard S., Glassy M.C., Abak A., Hussen B.M., Niazi V., Taheri M. The interaction between miRNAs/lncRNAs and Notch pathway in human disorders. Biomed. Pharmacother. 2021;138:111496. doi: 10.1016/j.biopha.2021.111496. [DOI] [PubMed] [Google Scholar]
- 122.Huang Q., Chen L., Liang J., Huang Q., Sun H. Neurotransmitters: Potential Targets in Glioblastoma. Cancers. 2022;14:3970. doi: 10.3390/cancers14163970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kalfert D., Ludvikova M., Pesta M., Ludvik J., Dostalova L., Kholová I. Multifunctional Roles of miR-34a in Cancer: A Review with the Emphasis on Head and Neck Squamous Cell Carcinoma and Thyroid Cancer with Clinical Implications. Diagnostics. 2020;10:563. doi: 10.3390/diagnostics10080563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weiss W., Burns M.J., Hackett C., Aldape K., Hill J.R., Kuriyama H., Kuriyama N., Milshteyn N., Roberts T., Wendland M.F., et al. Genetic determinants of malignancy in a mouse model for oligodendroglioma. Cancer Res. 2003;63:1589–1595. [PubMed] [Google Scholar]
- 125.Iannolo G., Sciuto M.R., Raffa G.M., Pilato M., Conaldi P.G. MiR34 inhibition induces human heart progenitor proliferation. Cell Death Dis. 2018;9:368. doi: 10.1038/s41419-018-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Santolini E., Puri C., Salcini A.E., Gagliani M.C., Pelicci P.G., Tacchetti C., Di Fiore P.P. Numb Is an Endocytic Protein. J. Cell Biol. 2000;151:1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Colaluca I.N., Tosoni D., Nuciforo P., Senic-Matuglia F., Galimberti V., Viale G., Pece S., Di Fiore P.P. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 128.Iannolo G., Sciuto M.R., Cuscino N., Pallini R., Douradinha B., Vitiani L.R., De Maria R., Conaldi P.G. Zika virus infection induces MiR34c expression in glioblastoma stem cells: New perspectives for brain tumor treatments. Cell Death Dis. 2019;10:263. doi: 10.1038/s41419-019-1499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Francipane M.G., Douradinha B., Chinnici C.M., Russelli G., Conaldi P.G., Iannolo G. Zika Virus: A New Therapeutic Candidate for Glioblastoma Treatment. Int. J. Mol. Sci. 2021;22:10996. doi: 10.3390/ijms222010996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yamakuchi M., Ferlito M., Lowenstein C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Qian Z., Zhou S., Zhou Z., Yang X., Que S., Lan J., Qiu Y., Lin Y. miR 146b 5p suppresses glioblastoma cell resistance to te-mozolomide through targeting TRAF6. Oncol. Rep. 2017;38:2941–2950. doi: 10.3892/or.2017.5970. [DOI] [PubMed] [Google Scholar]
- 132.Wang H., Tan L., Dong X., Liu L., Jiang Q., Li H., Shi J., Yang X., Dai X., Qian Z., et al. MiR-146b-5p suppresses the ma-lignancy of GSC/MSC fusion cells by targeting SMARCA5. Aging. 2020;12:13647–13667. doi: 10.18632/aging.103489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ames H., Halushka M.K., Rodriguez F.J. miRNA Regulation in Gliomas: Usual Suspects in Glial Tumorigenesis and Evolving Clinical Applications. J. Neuropathol. Exp. Neurol. 2017;76:246–254. doi: 10.1093/jnen/nlx005. [DOI] [PubMed] [Google Scholar]
- 134.Katakowski M., Zheng X., Jiang F., Rogers T., Szalad A., Chopp M. MiR-146b-5p Suppresses EGFR Expression and Reduces In Vitro Migration and Invasion of Glioma. Cancer Investig. 2010;28:1024–1030. doi: 10.3109/07357907.2010.512596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Khwaja S.S., Cai C., Badiyan S.N., Wang X., Huang J. The immune-related microRNA miR-146b is upregulated in glioblastoma recurrence. Oncotarget. 2018;9:29036–29046. doi: 10.18632/oncotarget.25528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cabral-Pacheco G.A., Garza-Veloz I., La Rosa C.C.-D., Ramirez-Acuña J.M., Perez-Romero B.A., Guerrero-Rodriguez J.F., Martinez-Avila N., Martinez-Fierro M.L. The Roles of Matrix Metalloproteinases and Their Inhibitors in Human Diseases. Int. J. Mol. Sci. 2020;21:9739. doi: 10.3390/ijms21249739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Regazzo G., Terrenato I., Spagnuolo M., Carosi M., Cognetti G., Cicchillitti L., Sperati F., Villani V., Carapella C., Piaggio G., et al. A restricted signature of serum miRNAs distinguishes glioblastoma from lower grade gliomas. J. Exp. Clin. Cancer Res. 2016;35:124. doi: 10.1186/s13046-016-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morales-Martínez M., Vega M.I. Role of MicroRNA-7 (MiR-7) in Cancer Physiopathology. Int. J. Mol. Sci. 2022;23:9091. doi: 10.3390/ijms23169091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou S., Zhu C., Pang Q., Liu H.C. MicroRNA-217: A regulator of human cancer. Biomed. Pharm. 2021;133:110943. doi: 10.1016/j.biopha.2020.110943. [DOI] [PubMed] [Google Scholar]
- 140.Wang Y., Chen R., Zhou X., Guo R., Yin J., Li Y., Ma G. miR-137: A Novel Therapeutic Target for Human Glioma. Mol. Ther. Nucleic Acids. 2020;21:614–622. doi: 10.1016/j.omtn.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sanuki R., Yamamura T. Tumor Suppressive Effects of miR-124 and Its Function in Neuronal Development. Int. J. Mol. Sci. 2021;22:5919. doi: 10.3390/ijms22115919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reséndiz-Castillo L., Minjarez B., Reza-Zaldívar E., Hernández-Sapiéns M., Gutiérrez-Mercado Y., Canales-Aguirre A. The effects of altered neurogenic microRNA levels and their involvement in the aggressiveness of periventricular glioblastoma. Neurología. 2021;19:781–793. doi: 10.1016/j.nrleng.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 143.Papagiannakopoulos T., Kosik K.S. MicroRNAs: Regulators of oncogenesis and stemness. BMC Med. 2008;24:15.:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mirzaei S., Gholami M.H., Hushmandi K., Hashemi F., Zabolian A., Canadas I., Zarrabi A., Nabavi N., Aref A.R., Crea F., et al. The long and shortnon-codingRNAsmodulatingEZH2signaling in cancer. J. Hematol. Oncol. 2022;15:18. doi: 10.1186/s13045-022-01235-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Godlewski J., Nowicki M.O., Bronisz A., Williams S., Otsuki A., Nuovo G., RayChaudhury A., Newton H.B., Chiocca E.A., Lawler S. Targeting of the Bmi-1 Oncogene/Stem Cell Renewal Factor by MicroRNA-128 Inhibits Glioma Proliferation and Self-Renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 146.Zhang Y., Chao T., Li R., Liu W., Chen Y., Yan X., Gong Y., Yin B., Liu W., Qiang B., et al. Mi-croRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J. Mol. Med. 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 147.Cui J.G., Zhao Y., Sethi P., Li Y.Y., Mahta A., Culicchia F., Lukiw W.J. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J. Neuro-Oncol. 2010;98:297–304. doi: 10.1007/s11060-009-0077-0. [DOI] [PubMed] [Google Scholar]
- 148.Shan Z.-N., Tian R., Zhang M., Gui Z.-H., Wu J., Ding M., Zhou X.-F., He J. miR128-1 inhibits the growth of glioblastoma multiforme and glioma stem-like cells via targeting BMI1 and E2F3. Oncotarget. 2016;7:78813–78826. doi: 10.18632/oncotarget.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao B., Bian E.B., Li J., Li J. New advances of microRNAs in glioma stem cells, with special emphasis on aberrant methylation of microRNAs. J. Cell Physiol. 2014;229:1141–1147. doi: 10.1002/jcp.24540. [DOI] [PubMed] [Google Scholar]
- 150.Shi Z.-M., Wang J., Yan Z., You Y.-P., Li C.-Y., Qian X., Yin Y., Zhao P., Wang Y.-Y., Wang X.-F., et al. MiR-128 Inhibits Tumor Growth and Angiogenesis by Targeting p70S6K1. PLoS ONE. 2012;7:e32709. doi: 10.1371/journal.pone.0032709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Feng Y.-H., Tsao C.-J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016;5:395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang C.H., Yue J., Pfeffer S.R., Fan M., Paulus E., Hosni-Ahmed A., Sims M., Qayyum S., Davidoff A.M., Handorf C.R., et al. MicroRNA-21 Promotes Glioblastoma Tumorigenesis by Down-regulating Insulin-like Growth Factor-binding Protein-3 (IGFBP3) J. Biol. Chem. 2014;289:25079–25087. doi: 10.1074/jbc.M114.593863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pin G., Huanting L., Chengzhan Z., Xinjuan K., Yugong F., Wei L., Shifang L., Zhaojian L., Kun H., Weicheng Y., et al. Down-Regulation of PDCD4 Promotes Proliferation, Angiogenesis and Tumorigenesis in Glioma Cells. Front. Cell Dev. Biol. 2020;8:593685. doi: 10.3389/fcell.2020.593685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen Y., Liu W., Chao T., Zhang Y., Yan X., Gong Y., Qiang B., Yuan J., Sun M., Peng X. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 155.Aloizou A.M., Pateraki G., Siokas V., Mentis A.A., Liampas I., Lazopoulos G., Kovatsi L., Mitsias P.D., Bogdanos D.P., Paterakis K., et al. The role of MiRNA-21 in gliomas: Hope for a novel therapeutic intervention? Toxicol. Rep. 2020;7:1514–1530. doi: 10.1016/j.toxrep.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sasayama T., Nishihara M., Kondoh T., Hosoda K., Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int. J. Cancer. 2009;125:1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 157.Sun L., Yan W., Wang Y., Sun G., Luo H., Zhang J., Wang X., You Y., Yang Z., Liu N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 158.Gabriely G., Yi M., Narayan R.S., Niers J.M., Wurdinger T., Imitola J., Ligon K.L., Kesari S., Esau C., Stephens R.M., et al. Human Glioma Growth Is Controlled by MicroRNA-10b. Cancer Res. 2011;71:3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rao S.A., Santosh V., Somasundaram K. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod. Pathol. 2010;23:1404–1417. doi: 10.1038/modpathol.2010.135. [DOI] [PubMed] [Google Scholar]
- 160.Huang T., Wan X., Alvarez A.A., James C.D., Song X., Yang Y., Sastry N., Nakano I., Sulman E.P., Hu B., et al. MIR93 (microRNA-93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy. 2019;15:1100–1111. doi: 10.1080/15548627.2019.1569947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dong H., Siu H., Luo L., Fang X., Jin L., Xiong M. Investigation gene and microRNA expression in glioblastoma. BMC Genom. 2010;11:S16-10. doi: 10.1186/1471-2164-11-S3-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hua D., Mo F., Ding D., Li L., Han X., Zhao N., Foltz G., Lin B., Lan Q., Huang Q. A Catalogue of Glioblastoma and Brain MicroRNAs Identified by Deep Sequencing. OMICS J. Integr. Biol. 2012;16:690–699. doi: 10.1089/omi.2012.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen R., Liu H., Cheng Q., Jiang B., Peng R., Zou Q., Yang W., Yang X., Wu X., Chen Z. MicroRNA-93 promotes the malignant phenotypes of human glioma cells and induces their chemoresistance to temozolomide. Biol. Open. 2016;5:669–677. doi: 10.1242/bio.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fang L., Deng Z., Shatseva T., Yang J., Peng C., Du W.W., Yee A.J., Ang L.C., He C., Shan S.W., et al. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene. 2010;30:806–821. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 165.Guan Y., Mizoguchi M., Yoshimoto K., Hata N., Shono T., Suzuki S.O., Araki Y., Kuga D., Nakamizo A., Amano T., et al. MiRNA-196 Is Upregulated in Glioblastoma But Not in Anaplastic Astrocytoma and Has Prognostic Significance. Clin. Cancer Res. 2010;16:4289–4297. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 166.Qu S., Qiu O., Huang J., Liu J., Wang H. Upregulation of hsa-miR-196a-5p is associated with MIR196A2 methylation and affects the malignant biological behaviors of glioma. Genomics. 2021;113:1001–1010. doi: 10.1016/j.ygeno.2021.02.012. [DOI] [PubMed] [Google Scholar]
- 167.Sana J., Bušek P., Fadrus P., Besse A., Radova L., Vecera M., Reguli S., Stollinova-Sromova L., Hilser M., Lipina R., et al. Identification of microRNAs differentially expressed in glioblastoma stem-like cells and their association with patient survival. Sci. Rep. 2018;8:2836. doi: 10.1038/s41598-018-20929-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu Q., Guan Y., Li Z., Wang Y., Liu Y., Cui R., Wang Y. miR-504 suppresses mesenchymal phenotype of glioblastoma by directly targeting the FZD7-mediated Wnt-β-catenin pathway. J. Exp. Clin. Cancer Res. 2019;38:358. doi: 10.1186/s13046-019-1370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Guo C.M., Liu S.Q., Sun M.Z. miR-429 as biomarker for diagnosis, treatment and prognosis of cancers and its potential action mechanisms: A systematic literature review. Neoplasma. 2020;67:215–228. doi: 10.4149/neo_2019_190401N282. [DOI] [PubMed] [Google Scholar]
- 170.Janaki Ramaiah M., Divyapriya K., Kartik Kumar S., Rajesh Y.B.R.D. Drug-induced modifications and modulations of microRNAs and long non-coding RNAs for future therapy against Glioblastoma Multiforme. Gene. 2020;723:144126. doi: 10.1016/j.gene.2019.144126. [DOI] [PubMed] [Google Scholar]
- 171.Fornari F., Gramantieri L., Ferracin M., Veronese A., Sabbioni S., Calin G.A., Grazi G.L., Giovannini C., Croce C.M., Bolondi L., et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 172.Garofalo M., Quintavalle C., Romano G., Croce C.M., Condorelli G. miR221/222 in cancer: Their role in tumor progression and response to therapy. Curr. Mol. Med. 2012;12:27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr. Pharm. Biotechnol. 2014;15:429. doi: 10.2174/138920101505140828161335. [DOI] [PubMed] [Google Scholar]
- 174.Lai T.C., Lee T.L., Chang Y.C., Chen Y.C., Lin S.R., Lin S.W., Pu C.M., Tsai J.S., Chen Y.L. MicroRNA-221/222 Mediates ADSC-Exosome-Induced Cardioprotection Against Ischemia/Reperfusion by Targeting PUMA and ETS-1. Front. Cell Dev. Biol. 2020;8:569150. doi: 10.3389/fcell.2020.569150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang C., Zhang J., Hao J., Shi Z., Wang Y., Han L., Yu S., You Y., Jiang T., Wang J., et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J. Transl. Med. 2012;10:119. doi: 10.1186/1479-5876-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cai G., Qiao S., Chen K. Suppression of miR-221 inhibits glioma cells proliferation and invasion via targeting SEMA3B. Biol. Res. 2015;48:37. doi: 10.1186/s40659-015-0030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Galardi S., Mercatelli N., Farace M.G., Ciafrè S.A. NF-kB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Res. 2011;39:3892–3902. doi: 10.1093/nar/gkr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vilar J.B., Christmann M., Tomicic M.T. Alterations in Molecular Profiles Affecting Glioblastoma Resistance to Radiochemotherapy: Where Does the Good Go? Cancers. 2022;14:2416. doi: 10.3390/cancers14102416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schneider B., William D., Lamp N., Zimpfer A., Henker C., Classen C.F., Erbersdobler A. The miR-183/96/182 cluster is upregulated in glioblastoma carrying EGFR amplification. Mol. Cell Biochem. 2022;477:2297–2307. doi: 10.1007/s11010-022-04435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jiang L., Mao P., Song L., Wu J., Huang J., Lin C., Yuan J., Qu L., Cheng S.-Y., Li J. miR-182 as a Prognostic Marker for Glioma Progression and Patient Survival. Am. J. Pathol. 2010;177:29–38. doi: 10.2353/ajpath.2010.090812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Beroukhim R., Getz G., Nghiemphu L., Barretina J., Hsueh T., Linhart D., Vivanco I., Lee J.C., Huang J.H., Alexander S., et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc. Natl. Acad. Sci. USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fan Y.N., Meley D., Pizer B., Sée V. Mir-34a Mimics Are Potential Therapeutic Agents for p53-Mutated and Chemo-Resistant Brain Tumour Cells. PLoS ONE. 2014;9:e108514. doi: 10.1371/journal.pone.0108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chen Z., Li D., Cheng Q., Ma Z., Jiang B., Peng R., Chen R., Cao Y., Wan X. MicroRNA-203 inhibits the proliferation and invasion of U251 glioblastoma cells by directly targeting PLD2. Mol. Med. Rep. 2014;9:503–508. doi: 10.3892/mmr.2013.1814. [DOI] [PubMed] [Google Scholar]
- 184.Ibrahim A.F., Weirauch U., Thomas M., Grünweller A., Hartmann R.K., Aigner A. MicroRNA Replacement Therapy for miR-145 and miR-33a Is Efficacious in a Model of Colon Carcinoma. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 185.Song H., Oh B., Choi M., Oh J., Lee M. Delivery of anti-microRNA-21 antisense-oligodeoxynucleotide using amphiphilic peptides for glioblastoma gene therapy. J. DrugTarget. 2015;23:360–370. doi: 10.3109/1061186X.2014.1000336. [DOI] [PubMed] [Google Scholar]
- 186.Oh B., Song H., Lee D., Oh J., Kim G., Ihm S.H., Lee M. Anti-cancer effect of R3V6 peptide-mediated delivery of an anti-microRNA-21 antisense-oligodeoxynucleotide in a glioblastoma animal model. J. Drug Target. 2017;25:132–139. doi: 10.1080/1061186X.2016.1207648. [DOI] [PubMed] [Google Scholar]
- 187.Kang C., Zhou X., Zhang J., Jia Q., Ren Y., Wang Y., Shi L., Liu N., Wang G., Pu P., et al. Reduction of miR-21 induces glioma cell apoptosis via activating caspase 9 and 3. Oncol. Rep. 2010;24:195–201. doi: 10.3892/or_00000846. [DOI] [PubMed] [Google Scholar]
- 188.Janssen H.L.A., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., Van Der Meer A.J., Patick A.K., Chen A., Zhou Y., et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 189.Ebert M.S., Sharp P.A. Emerging Roles for Natural MicroRNA Sponges. Curr. Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chen L., Zhang K., Shi Z., Zhang A., Jia Z., Wang G., Pu P., Kang C., Han L. A lentivirus-mediated miR-23b sponge diminishes the malignant phenotype of glioma cells in vitro and in vivo. Oncol. Rep. 2014;31:1573–1580. doi: 10.3892/or.2014.3012. [DOI] [PubMed] [Google Scholar]
- 191.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 192.Baumann V., Winkler J. miRNA-based therapies: Strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014;6:1967–1984. doi: 10.4155/fmc.14.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen Y., Gao D.-Y., Huang L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dong L., Han C., Zhang H., Gu X., Li J., Wu Y., Wang X. Construction of a recombinant lentivirus containing human microRNA-7-3 and its inhibitory effects on glioma proliferation. Neural Regen. Res. 2012;7:2144–2150. doi: 10.3969/j.issn.1673-5374.2012.27.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.El Fatimy R., Subramanian S., Uhlmann E.J., Krichevsky A.M. Genome Editing Reveals Glioblastoma Addiction to Mi-croRNA-10b. Mol. Ther. 2017;25:368–378. doi: 10.1016/j.ymthe.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.-W., Chang T.-C., Vivekanandan P., Torbenson M., Clark K.R., et al. Therapeutic microRNA Delivery Suppresses Tumorigenesis in a Murine Liver Cancer Model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yang N. An overview of viral and nonviral delivery systems for microRNA. Int. J. Pharm. Investig. 2015;5:179–181. doi: 10.4103/2230-973X.167646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ananta J.S., Paulmurugan R., Massoud T.F. Nanoparticle-Delivered Antisense MicroRNA-21 Enhances the Effects of Te-mozolomide on Glioblastoma Cells. Mol. Pharm. 2015;12:4509–4517. doi: 10.1021/acs.molpharmaceut.5b00694. [DOI] [PubMed] [Google Scholar]
- 199.Ananta J.S., Paulmurugan R., Massoud T.F. Tailored Nanoparticle Codelivery of antimiR-21 and antimiR-10b Augments Glioblastoma Cell Kill by Temozolomide: Toward a “Personalized” Anti-microRNA Therapy. Mol. Pharm. 2016;13:3164–3175. doi: 10.1021/acs.molpharmaceut.6b00388. [DOI] [PubMed] [Google Scholar]
- 200.Malhotra M., Sekar T.V., Ananta J.S., Devulapally R., Afjei R., Babikir H.A., Paulmurugan R., Massoud T.F. Targeted nanoparticle delivery of therapeutic antisense microRNAs presensitizes glioblastoma cells to lower effective doses of te-mozolomide in vitro and in a mouse model. Oncotarget. 2018;9:21478–21494. doi: 10.18632/oncotarget.25135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Park T.G., Jeong J.H., Kim S.W. Current status of polymeric gene delivery systems. Adv. Drug Deliv. Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 202.Vega R.A., Zhang Y., Curley C., Price R.L., Abounader R. 370 Magnetic Resonance-Guided Focused Ultrasound Delivery of Polymeric Brain-Penetrating Nanoparticle MicroRNA Conjugates in Glioblastoma. Neurosurgery. 2016;63:210. doi: 10.1227/01.neu.0000489858.08559.c8. [DOI] [Google Scholar]
- 203.Küçüktürkmen B., Bozkır A. Development and characterization of cationic solid lipid nanoparticles for co-delivery of pemetrexed and miR-21 antisense oligonucleotide to glioblastoma cells. Drug Dev. Ind. Pharm. 2017;44:306–315. doi: 10.1080/03639045.2017.1391835. [DOI] [PubMed] [Google Scholar]
- 204.Küçüktürkmen B., Devrim B., Saka O.M., Yilmaz Ş., Arsoy T., Bozkir A. Co-delivery of pemetrexed and miR-21 antisense oligonucleotide by lipid-polymer hybrid nanoparticles and effects on glioblastoma cells. Drug Dev. Ind. Pharm. 2016;43:12–21. doi: 10.1080/03639045.2016.1200069. [DOI] [PubMed] [Google Scholar]
- 205.Costa P.M., Cardoso A.L., Custódia C., Cunha P., Pereira de Almeida L., Pedroso de Lima M.C. MiRNA-21 silencing mediated by tumor-targeted nanoparticles combined with sunitinib: A new multimodal gene therapy approach for glio-blastoma. J. Control. Release. 2015;207:31–39. doi: 10.1016/j.jconrel.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 206.Yaghi N.K., Wei J., Hashimoto Y., Kong L.-Y., Gabrusiewicz K., Nduom E.K., Ling X., Huang N., Zhou S., Kerrigan B.C.P., et al. Immune modulatory nanoparticle therapeutics for intracerebral glioma. Neuro-Oncology. 2017;19:372–382. doi: 10.1093/neuonc/now198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lowe S., Bhat K.P., Olar A. Current clinical management of patients with glioblastoma. Cancer Rep. 2019;2:e1216. doi: 10.1002/cnr2.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Aftab K., Aamir F.B., Mallick S., Mubarak F., Pope W.B., Mikkelsen T., Rock J.P., Enam S.A. Radiomics for precision medicine in glioblastoma. J. Neuro-Oncol. 2022;156:217–223. doi: 10.1007/s11060-021-03933-1. [DOI] [PubMed] [Google Scholar]
- 209.Mian S.Y., Nambiar A., Kaliaperumal C. Phytotherapy for the Treatment of Glioblastoma: A Review. Front. Surg. 2022;9:844993. doi: 10.3389/fsurg.2022.844993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Candido S., Lupo G., Pennisi M., Basile M.S., Anfuso C.D., Petralia M.C., Gattuso G., Vivarelli S., Spandidos D.A., Libra M., et al. The analysis of miRNA expression profiling datasets reveals inverse microRNA patterns in glioblastoma and Alzheimer’s disease. Oncol. Rep. 2019;42:911–922. doi: 10.3892/or.2019.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Thomas L., Florio T., Perez-Castro C. Extracellular Vesicles Loaded miRNAs as Potential Modulators Shared between Glioblastoma, and Parkinson’s and Alzheimer’s Diseases. Front. Cell. Neurosci. 2020;14:590034. doi: 10.3389/fncel.2020.590034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Quezada C., Torres Á., Niechi I., Uribe D., Contreras-Duarte S., Toledo F., San Martín R., Gutiérrez J., Sobrevia L. Role of extracellular vesicles in glioma progression. Mol. Aspects Med. 2018;60:38–51. doi: 10.1016/j.mam.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 213.Genc S., Pennisi M., Yeni Y., Yildirim S., Gattuso G., Altinoz M.A., Taghizadehghalehjoughi A., Bolat I., Tsatsakis A., Hacımüftüoğlu A., et al. Potential Neurotoxic Effects of Glioblastoma-Derived Exosomes in Primary Cultures of Cere-bellar Neurons via Oxidant Stress and Glutathione Depletion. Antioxidants. 2022;11:1225. doi: 10.3390/antiox11071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tomei S., Volontè A., Ravindran S., Mazzoleni S., Wang E., Galli R., Maccalli C. MicroRNA Expression Profile Distinguishes Glioblastoma Stem Cells from Differentiated Tumor Cells. J. Pers. Med. 2021;11:264. doi: 10.3390/jpm11040264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Saenz-Antoñanzas A., Auzmendi-Iriarte J., Carrasco-Garcia E., Moreno-Cugnon L., Ruiz I., Villanua J., Egaña L., Otaegui D., Samprón N., Matheu A. LiquidBiopsy in Glioblastoma: Opportunities, Applications and Challenges. Cancers. 2019;11:950. doi: 10.3390/cancers11070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Piccioni D.E., Achrol A.S., Kiedrowski L.A., Banks K.C., Boucher N., Barkhoudarian G., Kelly D.F., Juarez T., Lanman R.B., Raymond V.M., et al. Analysis of cell-freecirculating tumor DNA in 419 patients with glioblastoma and otherprimarybrain-tumors. CNS Oncol. 2019;8:CNS34. doi: 10.2217/cns-2018-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zill O.A., Banks K.C., Fairclough S.R., Mortimer S.A., Vowles J.V., Mokhtari R., Gandara D.R., Mack P.C., Odegaard J.I., Nagy R.J., et al. The Landscape of ActionableGenomicAlterations in Cell-FreeCirculating Tumor DNA from 21,807 Advanced Cancer Patients. Clin. Cancer Res. 2018;24:3528–3538. doi: 10.1158/1078-0432.CCR-17-3837. [DOI] [PubMed] [Google Scholar]
- 218.Schwaederle M., Husain H., Fanta P.T., Piccioni D.E., Kesari S., Schwab R.B., Banks K.C., Lanman R.B., Talasaz A., Parker B.A., et al. Detectionrate of actionablemutations in diversecancersusing a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget. 2016;7:9707–9717. doi: 10.18632/oncotarget.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Balana C., Ramirez J.L., Taron M., Roussos Y., Ariza A., Ballester R., Sarries C., Mendez P., Sanchez J.J., Rosell R. O6-methyl-guanine-DNA methyltransferasemethylation in serum and tumor DNA predictsresponse to1,3-bis(2-chloroethyl)-1-nitrosourea but not to temozolamide plus cisplatin in glioblastomamultiforme. Clin. Cancer Res. 2003;9:1461–1468. [PubMed] [Google Scholar]
- 220.Rynkeviciene R., Simiene J., Strainiene E., Stankevicius V., Usinskiene J., MiseikyteKaubriene E., Meskinyte I., Cicenas J., Suziedelis K. Non-CodingRNAs in Glioma. Cancers. 2018;11:17. doi: 10.3390/cancers11010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tan S.K., Pastori C., Penas C., Komotar R.J., Ivan M.E., Wahlestedt C., Ayad N.G. Serum longnoncoding RNA HOTAIR as a noveldiagnostic and prognosticbiomarker in glioblastomamultiforme. Mol. Cancer. 2018;17:74. doi: 10.1186/s12943-018-0822-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Shen J., Hodges T.R., Song R., Gong Y., Calin G.A., Heimberger A.B., Zhao H. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol. Carcinog. 2018;57:137–141. doi: 10.1002/mc.22739. [DOI] [PubMed] [Google Scholar]
- 223.Zhang J.X., Han L., Bao Z.S., Wang Y.Y., Chen L.Y., Yan W., Yu S.Z., Pu P.Y., Liu N., You Y.P., et al. HOTAIR, a cell-cycle-associatedlongnoncoding RNA and a strongpredictor of survival, ispreferentiallyexpressed in classical and mesenchy-malglioma. NeuroOncology. 2013;15:1595–1603. [Google Scholar]
- 224.Zhu J., Ye J., Zhang L., Xia L., Hu H., Jiang H., Wan Z., Sheng F., Ma Y., Li W., et al. DifferentialExpression of CircularRNAs in GlioblastomaMultiforme and ItsCorrelation with Prognosis. Transl. Oncol. 2017;10:271–279. doi: 10.1016/j.tranon.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chen L., Han L., Wei J., Zhang K., Shi Z., Duan R., Li S., Zhou X., Pu P., Zhang J., et al. SNORD76, a box C/D snoRNA, acts as a tumor suppressor in glioblastoma. Sci. Rep. 2015;5:8588. doi: 10.1038/srep08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tkach M., Thery C. Communication by ExtracellularVesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 227.Yanez-Mo M., Siljander P.R., Andreu Z., Zavec A.B., Borras F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J., et al. Biologicalproperties of extracellularvesicles and theirphysiologicalfunctions. J. Extracell. Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Santiago-Dieppa D.R., Steinberg J., Gonda D., Cheung V.J., Carter B.S., Chen C.C. Extracellularvesicles as a platform for ‘liquidbiopsy’ in glioblastomapatients. Expert Rev. Mol. Diagn. 2014;14:819–825. doi: 10.1586/14737159.2014.943193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Osti D., Del Bene M., Rappa G., Santos M., Matafora V., Richichi C., Faletti S., Beznoussenko G.V., Mironov A., Bachi A., et al. ClinicalSignificance of ExtracellularVesicles in Plasma from GlioblastomaPatients. Clin. Cancer Res. 2019;25:266–276. doi: 10.1158/1078-0432.CCR-18-1941. [DOI] [PubMed] [Google Scholar]
- 230.Shao H., Chung J., Balaj L., Charest A., Bigner D.D., Carter B.S., Hochberg F.H., Breakefield X.O., Weissleder R., Lee H. Protein typing of circulatingmicrovesiclesallows real-time monitoring of glioblastomatherapy. Nat. Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Garcia-Romero N., Carrion-Navarro J., Esteban-Rubio S., Lazaro-Ibanez E., Peris-Celda M., Alonso M.M., Guzman-De-Villoria J., Fernandez-Carballal C., de Mendivil A.O., Garcia-Duque S., et al. DNA sequenceswithinglioma-derivedextracellularvesiclescan cross the intactblood-brainbarrier and be detected in peripheralblood of patients. Oncotarget. 2017;8:1416–1428. doi: 10.18632/oncotarget.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Skog J., Wurdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumor growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bible E. Neuro-oncology: Glioblastomadetection and therapy monitoring by microvesiclerelease. Nat. Rev. Neurol. 2013;9:4. doi: 10.1038/nrneurol.2012.247. [DOI] [PubMed] [Google Scholar]
- 234.Ricklefs F.L., Alayo Q., Krenzlin H., Mahmoud A.B., Speranza M.C., Nakashima H., Hayes J.L., Lee K., Balaj L., Passaro C., et al. Immuneevasionmediated by PD-L1 on glioblastoma-derivedextracellularvesicles. Sci. Adv. 2018;4:eaar2766. doi: 10.1126/sciadv.aar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ramakrishnan V., Kushwaha D., Koay D.C., Reddy H., Mao Y., Zhou L., Ng K., Zinn P., Carter B., Chen C.C. Post-transcriptional regulation of O6-methylguanine-DNA methyltransferase MGMT in glioblastomas. Cancer Biomark. 2011;10:185–193. doi: 10.3233/CBM-2012-0245. [DOI] [PubMed] [Google Scholar]
- 236.Garnier D., Meehan B., Kislinger T., Daniel P., Sinha A., Abdulkarim B., Nakano I., Rak J. Divergentevolution of te-mozolomideresistance in glioblastomastemcellsisreflected in extracellularvesicles and coupled with radiosensitization. Neuro-Oncology. 2018;20:236–248. doi: 10.1093/neuonc/nox142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Akers J.C., Ramakrishnan V., Kim R., Skog J., Nakano I., Pingle S., Kalinina J., Hua W., Kesari S., Mao Y., et al. MiR-21 in the extracellularvesicles (EVs) of cerebrospinal fluid (CSF): A platform for glioblastomabiomarker development. PLoS ONE. 2013;8:e78115. doi: 10.1371/journal.pone.0078115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Joosse S.A., Pantel K. Tumor-EducatedPlatelets as Liquid Biopsy in CancerPatients. Cancer Cell. 2015;28:552–554. doi: 10.1016/j.ccell.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 239.Nilsson R.J., Balaj L., Hulleman E., van Rijn S., Pegtel D.M., Walraven M., Widmark A., Gerritsen W.R., Verheul H.M., Vandertop W.P., et al. Blood plateletscontain tumor-derived RNA biomarkers. Blood. 2011;118:3680–3683. doi: 10.1182/blood-2011-03-344408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Best M.G., Wesseling P., Wurdinger T. Tumor-EducatedPlatelets as a NoninvasiveBiomarker Source for CancerDetection and Progression Monitoring. Cancer Res. 2018;78:3407–3412. doi: 10.1158/0008-5472.CAN-18-0887. [DOI] [PubMed] [Google Scholar]
- 241.Best M.G., Sol N., Kooi I., Tannous J., Westerman B.A., Rustenburg F., Schellen P., Verschueren H., Post E., Koster J., et al. RNA-Seq of Tumor-EducatedPlateletsEnables Blood-Based Pan-Cancer, Multiclass, and MolecularPathwayCancer Diagnostics. Cancer Cell. 2015;28:666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gao F., Cui Y., Jiang H., Sui D., Wang Y., Jiang Z., Zhao J., Lin S. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget. 2016;7:71330–71340. doi: 10.18632/oncotarget.11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Muller C., Holtschmidt J., Auer M., Heitzer E., Lamszus K., Schulte A., Matschke J., Langer-Freitag S., Gasch C., Stoupiec M., et al. Hematogenousdissemination of glioblastomamultiforme. Sci. Transl. Med. 2014;6:247ra101. doi: 10.1126/scitranslmed.3009095. [DOI] [PubMed] [Google Scholar]
- 244.Sullivan J.P., Nahed B.V., Madden M.W., Oliveira S.M., Springer S., Bhere D., Chi A.S., Wakimoto H., Rothenberg S.M., Sequist L.V., et al. Brain tumor cells in circulationareenriched for mesenchymalgeneexpression. Cancer Discov. 2014;4:1299–1309. doi: 10.1158/2159-8290.CD-14-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Liu T., Xu H., Huang M., Ma W., Saxena D., Lustig R.A., Alonso-Basanta M., Zhang Z., O’Rourke D.M., Zhang L., et al. Circulating glioma cells exhibit stem cell-like properties. Cancer Res. 2018;78:6632–6642. doi: 10.1158/0008-5472.CAN-18-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Van Schaijik B., Wickremesekera A.C., Mantamadiotis T., Kaye A.H., Tan S.T., Stylli S.S., Itinteang T. Circulating tumor stemcells and glioblastoma: A review. J. Clin. Neurosci. 2019;61:5–9. doi: 10.1016/j.jocn.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 247.Drusco A., Bottoni A., Lagana’ A., Acunzo M., Fassan M., Cascione L., Antenucci A., Kumchala P., Vicentini C., Gardiman M.P., et al. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget. 2015;6:20829–20839. doi: 10.18632/oncotarget.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kopkova A., Sana J., Fadrus P., Slaby O. Cerebrospinal fluid microRNAs as diagnosticbiomarkers in braintumors. Clin. Chem. Lab. Med. 2018;56:869–879. doi: 10.1515/cclm-2017-0958. [DOI] [PubMed] [Google Scholar]
- 249.Akers J.C., Hua W., Li H., Ramakrishnan V., Yang Z., Quan K., Zhu W., Li J., Figueroa J., Hirshman B.R., et al. A cerebrospinal fluid microRNAsignature as biomarker for glioblastoma. Oncotarget. 2017;8:68769–68779. doi: 10.18632/oncotarget.18332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Baraniskin A., Kuhnhenn J., Schlegel U., Maghnouj A., Zollner H., Schmiegel W., Hahn S., Schroers R. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro-Oncology. 2012;14:29–33. doi: 10.1093/neuonc/nor169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Teplyuk N.M., Mollenhauer B., Gabriely G., Giese A., Kim E., Smolsky M., Kim R.Y., Saria M.G., Pastorino S., Kesari S., et al. MicroRNAs in cerebrospinal fluid identifyglioblastoma and metastaticbraincancers and reflectdiseaseactivity. Neuro-Oncology. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Qu K., Lin T., Pang Q., Liu T., Wang Z., Tai M., Meng F., Zhang J., Wan Y., Mao P., et al. Extracellular miRNA-21 as a novelbiomarker in glioma: Evidence from meta-analysis, clinicalvalidation and experimentalinvestigations. Oncotarget. 2016;7:33994–34010. doi: 10.18632/oncotarget.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article.