Abstract
Glutamine synthetase (GS), EC 6.3.1.2, is a central enzyme in the assimilation of nitrogen and the biosynthesis of glutamine. We have isolated the Aspergillus nidulans glnA gene encoding GS and have shown that glnA encodes a highly expressed but not highly regulated mRNA. Inactivation of glnA results in an absolute glutamine requirement, indicating that GS is responsible for the synthesis of this essential amino acid. Even when supplemented with high levels of glutamine, strains lacking a functional glnA gene have an inhibited morphology, and a wide range of compounds have been shown to interfere with repair of the glutamine auxotrophy. Heterologous expression of the prokaryotic Anabaena glnA gene from the A. nidulans alcA promoter allowed full complementation of the A. nidulans glnAΔ mutation. However, the A. nidulans fluG gene, which encodes a protein with similarity to prokaryotic GS, did not replace A. nidulans glnA function when similarly expressed. Our studies with the glnAΔ mutant confirm that glutamine, and not GS, is the key effector of nitrogen metabolite repression. Additionally, ammonium and its immediate product glutamate may also act directly to signal nitrogen sufficiency.
The major route of ammonium assimilation in Aspergillus nidulans is through the action of NADP-linked glutamate dehydrogenase (NADP-GDH), encoded by the gdhA gene, to yield glutamate (4, 22, 23). In addition, ammonium can be assimilated to glutamate by the combined action of glutamine synthetase (GS) and glutamate synthase (GOGAT). First, GS, encoded by the glnA gene, synthesizes glutamine from glutamate and ammonium, and then GOGAT, encoded by the gltA gene, converts glutamine and 2-oxoglutarate to two molecules of glutamate (10, 29). Mutants lacking NADP-GDH show leaky growth on ammonium, whereas mutants lacking both NADP-GDH and GOGAT are strict glutamate auxotrophs (29). As mutants lacking gltA function alone grow well on ammonium, this cycle is not critical for ammonium assimilation or glutamate production in cells able to synthesize an active NADP-GDH enzyme. In contrast, previously described A. nidulans glnA mutants require glutamine supplementation for growth (10).
Glutamine, as well as being an essential amino acid, has been implicated as a key effector for nitrogen metabolite repression. Nitrogen metabolite repression is a global regulatory control mechanism that ensures the preferential utilization of simple nitrogen sources over more complex sources (30). When wild-type cells are grown on nitrogen-sufficient media containing ammonium or glutamine, the expression of a wide range of catabolic enzymes required for the utilization of secondary nitrogen sources, such as nitrate or acetamide, is repressed. Nitrogen catabolic enzyme expression under nitrogen-limiting conditions requires a transcriptional activator encoded by the areA gene (3). AreA binds via a single GATA zinc finger to the promoters of nitrogen-responsive genes (25, 41). The synthesis and activity of AreA are modulated through changes in areA mRNA transcription and stability, as well as interaction with the negatively acting NmrA protein in response to changes in the nitrogen status of the cell (2, 26, 36). The phenotypes of mutants affected in ammonium assimilation indicate that ammonium alone is not the key effector for the modulation of AreA function. gdhA mutants are partially derepressed on ammonium, suggesting that ammonium must be metabolized, possibly to glutamine, to generate the signal(s) for full nitrogen metabolite repression (4, 21, 22). Previous studies of glnA mutants in A. nidulans and gln-1 mutants in Neurospora crassa have presented conflicting evidence about the role of glutamine and have even led to the suggestion that the glutamine synthetase protein itself may have a regulatory role (10, 15, 16, 17, 28, 37, 38). To further investigate the role of this enzyme in the generation of the signal for nitrogen metabolite repression, we have isolated and characterized the A. nidulans glnA gene. By creating a glnAΔ strain, our studies have revealed that glutamate, in addition to glutamine, may be involved in signaling nitrogen status.
MATERIALS AND METHODS
Molecular Methods.
Molecular methods were essentially as described previously (40). Escherichia coli strain NM522 {supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5 (rK− mK+) [F′ proAB lacIqZΔM15]} was used for all bacterial work. DNA was prepared using the High Pure plasmid isolation kit (Boehringer Mannheim) and sequenced using the PRISM Dye Primer Cycle Sequencing Ready Reaction kit (Applied Biosystems Inc.) or by the Australian Genome Research Facility.
A. nidulans strains media, and growth conditions.
A. nidulans strains used in this study were MH1 (biA1), MH50 (yA1 adE20 suA-adE20 areA102 pyroA4 riboB2), MH3018 (yA1 pabA1 argB2), MH5699 (yA1 adE20 suA-adE20 areA102 pyroA4 riboB2 areA::riboB), MH 8694 (tamAΔ riboB2), and and MH9962 (areA102 glnAΔ pyroA4). Gene symbols and genetic manipulations have been described previously (9).
Aspergillus media and growth conditions were as described by Cove (11). Nitrogen sources were added at a final concentration of 10 mM, carbon sources were at 1% (wt/vol), and nitrogen source analogues were used at concentrations described previously (36). Growth tests on solid media were scored after 2 days of incubation at 37°C. Liquid cultures for enzyme assays and RNA extractions were grown for 16 h with shaking at 37°C prior to being harvested or transferred to the appropriate media for 4 h with shaking at 37°C.
Aspergillus transformation and enzyme assays.
A. nidulans strains were transformed as described previously (1). In cotransformation experiments, transformants were selected using either the riboB+ selectable marker plasmid pPL3 (33) on media lacking riboflavin or the pyroA+ selectable marker plasmid pI4 (34) on media lacking pyridoxine. Southern blot analysis was used to confirm that cotransformants contained the plasmids of interest. β-Galactosidase assays of A. nidulans protein extracts were performed as described previously (12).
Cloning of the A. nidulans glnA gene.
Degenerate oligonucleotides (Tom1, 5′ GARCARGARTAYACNCT 3′, and Tom2, 5′ RCANCCNGCNCCRTTCCA 3′) were designed based on conserved regions of eukaryotic GS enzymes. These primers were used to amplify a 300-bp product from A. nidulans genomic DNA. The gel-purified product was labeled and used to probe a cosmid library of A. nidulans genomic DNA. Two cosmids, W22G7 and W3D7, were identified, and a 3-kb XbaI fragment from W3D7 was subcloned into pBluescript SK(−) to yield pCD5.
Construction and integration of a glnA::lacZ fusion plasmid.
A glnA::lacZ fusion construct was created by subcloning a 1-kb KpnI-PstI fragment from pCD5 into pMH3863, which contains a nonfunctional argB gene, allowing selection for targeted integration of the construct at the argB gene in A. nidulans. The resulting plasmid, pMH4627, contains 0.8 kb of glnA promoter sequences and 0.2 kb of glnA coding region specifying residues 1 to 30 of GlnA fused in frame with the lacZ gene of E. coli. Following transformation of the argB2 mutant strain MH3018 by pMH4627, ArgB+ transformants were selected. Correct integration of a single copy of pMH4627 at the argB locus was confirmed by Southern blot analysis.
Inactivation of the A. nidulans glnA gene.
A glnA inactivation construct was created by replacing an internal 0.8-kb PstI-Bg/II fragment of the glnA (−3 to +827) with a 2.2-kb PstI-Bg/II fragment containing the A. nidulans riboB gene from pPL1 (33). The resulting plasmid, pSM4638, was cut with KpnI and transformed into the recipient strain, MH50. Ribo+ transformants were selected on medium lacking riboflavin, with 10 mM glutamine as the sole nitrogen source, and screened on medium lacking added glutamine (with 10 mM ammonium as the sole nitrogen source).
Anabaena sp. strain 7120 glnA and A. nidulans fluG constructs.
For heterologous complementation of A. nidulans glnAΔ, the Anabaena glnA gene was PCR amplified using pAN503 (20, 46) as the template and the oligonucleotides TOM4 (5′-CTT CGA TGA GCT CAA GTT TTA CTC-3′) and GLN1 (5′-GTA ACA ATG AGA TCT CCA CAA GAA G-3′). A 1.6-kb BglII-SacI fragment was inserted into pMF16 (19) to yield pCD7. This results in fusion of Anabaena GlnA to the N terminus of A. nidulans AlcA and places glnA under the control of the alcA promoter. Plasmids pBN57.8, pBN68, and pBN72 encode amino acids 1 to 865, 387 to 865, and 1 to 402, respectively, of the A. nidulans FluG protein expressed from the alcA promoter (C. D'Souza, unpublished data).
RESULTS
A. nidulans GlnA is solely responsible for glutamine synthesis.
The glnA gene of A. nidulans was isolated from a genomic cosmid library (see Materials and Methods), subcloned as a 3-kb XbaI fragment into pBluescript SK(−), and sequenced. The glnA gene is predicted to contain a single intron of 167 bp and to encode a 345-amino-acid protein that is highly conserved with other GS type II enzymes from a variety of fungi and other eukaryotes (Fig. 1). The GlnA protein of A. nidulans has the highest similarity to GlnA of Schizosaccharomyces pombe (74% identity; 84% similarity).
FIG. 1.
Sequence alignment of the predicted GS enzymes of A. nidulans (accession no. AF333968), S. pombe (accession no. Z98977), Colletotrichum gloesosporoides (accession no. Q12613), Nectria haematococca (accession no. AF175498.1), S. cerevisiae (accession no. M65157), and Anabaena sp. (accession no. X00147). The sequences were aligned using CLUSTAL W (45), and identical amino acids (solid boxes) and conservative substitutions (shaded boxes) are indicated.
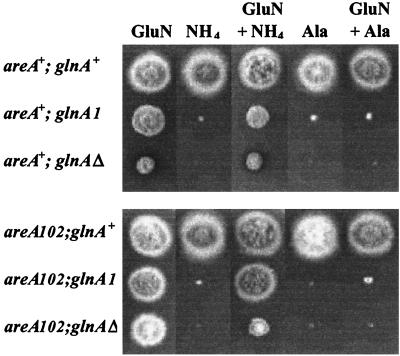
A glnAΔ construct, pSM4638, was made by replacing an internal fragment of glnA with the selectable marker riboB (see Materials and Methods). The riboB2 strain MH50 was transformed with pSM4638, and RiboB+ transformants were selected on medium containing 10 mM glutamine, followed by screening for glutamine auxotrophy on glucose-minimal medium with 10 mM ammonium as the sole nitrogen source (see Materials and Methods). One transformant was identified as unable to grow in the absence of added glutamine. Southern blot analysis of this strain revealed that homologous integration had resulted in two tandem copies of the glnA::riboB fragment replacing the glnA gene (data not shown). Comparison of the glnAΔ strain with the previously isolated glnA1 strain (10) indicated that the glutamine auxotrophy of the glnAΔ strain was more severe (Fig. 2). After extended (3- to 4-day) incubation, some growth of the glnA1 mutant could be seen on ammonium medium, whereas the glnAΔ strain remained unable to grow. The glnA1 mutant has been shown to retain residual levels of GS activity, sufficient to allow very limited glutamine synthesis, whereas inactivation of the glnA gene resulted in a strict glutamine auxotrophy, indicating that no alternative pathways of glutamine synthesis exist in A. nidulans.
FIG. 2.
Growth tests of glnA mutants. Conidia of the relevant strain were inoculated onto 1% glucose-minimal medium containing 10 mM glutamine (GluN), ammonium tartrate (NH4), or alanine (Ala) as nitrogen sources as indicated. The growth of the glnA1 and glnAΔ strains was compared to that of the glnA+ controls after 2 days of growth at 37°C.
glnA expression is not highly regulated.
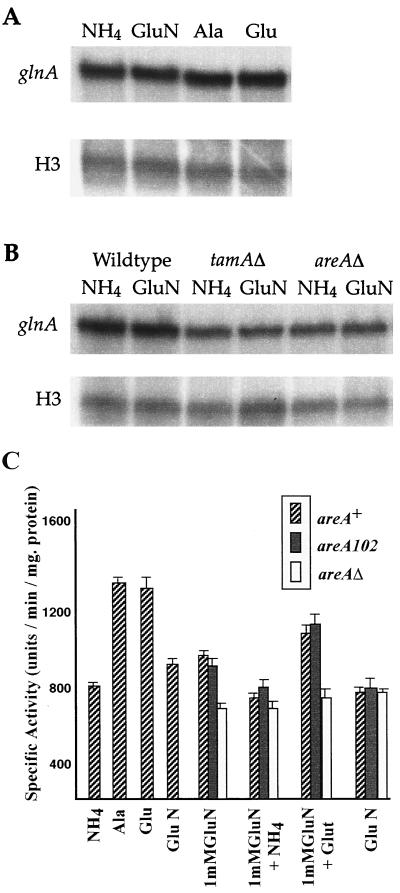
Northern blot analysis revealed that the glnA gene encodes an mRNA of approximately 1.5 kb, consistent with the transcript size predicted from sequence analysis. The glnA transcript was readily detected under all growth conditions tested. Levels in ammonium- and glutamine-grown mycelia of the wild-type strain were similar, and the levels increased approximately two-fold during growth on alanine or glutamate as the sole nitrogen source (Fig. 3). glnA mRNA levels were lower in strains carrying areAΔ or tamAΔ mutations under all conditions tested (Fig. 3). TamA is thought to function as a coactivator of AreA, and mutations in tamA lead to reduced expression of a variety of enzymes involved in nitrogen metabolism (14, 24, 42).
FIG. 3.
Expression of the glnA gene. (A) Mycelia of the wild-type strain, MH1, were grown for 16 h on glucose-minimal medium containing 10 mM ammonium (NH4), glutamine (GluN), alanine (Ala), or glutamate (Glu) as the sole nitrogen source. Total RNAs made from these mycelia were probed with a 1.1-kb HincII fragment from the glnA gene and a 1.3-kb EcoRI fragment from the histone H3 gene (18) as loading controls. (B) Mycelia of MH1, MH8694 (tamAΔ), and MH5699 (areAΔ) were grown, and the RNAs were probed as described above. (C) Mycelia were grown overnight on 1% glucose-minimal medium containing the appropriate nitrogen source at a concentration of 10 mM unless otherwise indicated. The β-galactosidase activities are expressed as units of activity per minute per milligram of soluble protein (+ standard errors).
These results were confirmed using a glnA::lacZ construct integrated in single copy at the argB locus (see Materials and Methods). The glnA::lacZ fusion strain expressed high levels of β-galactosidase, consistent with the strong mRNA signals detected by Northern blot analysis. In an areA+ background, there was less than two-fold variation in levels of β-galactosidase under the different nitrogen source conditions used (Fig. 3). The pattern of expression was similar to that found by Northern blot analysis, suggesting weak areA control. The levels of glnA::lacZ expression were determined in strains carrying either an areAΔ mutation or the altered-function areA102 mutation, which results in an amino acid substitution within the AreA GATA zinc finger, leading to altered DNA binding affinities (25, 39). While the areA102 mutation had little effect on the levels of glnA::lacZ expression, the levels were lower in an areAΔ mutant than in the wild type under all conditions. The variation in glnA transcript and glnA-directed β-galactosidase levels under different growth conditions was less extreme than the variation in GS enzyme activity (10, 35). Thus, it is likely that GS activity itself is subject to posttranslational regulation in response to different nutritional conditions.
Glutamine supplementation of the glnAΔ growth defect.
Besides glutamine, no other amino acid was able to allow growth of the glnAΔ strains. Glutamine levels of 0.5 mM or above were required to supplement the glutamine auxotrophy of the glnAΔ mutant on minimal media. However, even in the presence of 10 mM glutamine, the glnAΔ mutant exhibited a slightly restricted colony size. When glutamine repair of the glnAΔ mutant was tested on complete, rather than minimal, medium, it was found that the glnAΔ mutant produced a more compact, pigmented colony with reduced conidiation. This phenotype could be due to the effects of the areA102 mutation on glutamine uptake or metabolism, because glnA was mutated in an areA102 genetic background. However, when the areA102 glnAΔ strain was crossed with a wild-type strain, the areA+ glnAΔ progeny still produced compact, pigmented colonies with poorer conidiation than those of the wild type. The inhibited growth morphology of areA+ glnAΔ strains on glutamine is more extreme than that of the areA102 glnΔ strain. In an areA+ background, this phenotype was evident even on minimal medium and was even more pronounced on complete medium supplemented with 10 mM glutamine (Fig. 2).
This observation led us to test the effects of ammonium and individual amino acids on the growth of these strains on minimal medium with 10 mM glutamine. In the presence of ammonium, the glnA mutant strains were further restricted in growth. All l-amino acids (10 mM), with the exception of arginine, impaired the growth of the glnAΔ strains to some extent. There appeared to be two different, but not necessarily mutually exclusive, effects on growth. The amino acids valine, proline, glutamate, phenylalanine, tyrosine, isoleucine, lysine, and aspartate, in decreasing order of effectiveness, increased the compactness and pigmentation of the glnAΔ strains. In all cases, the areA102 glnAΔ strain was less severely affected than the areA+ glnAΔ strain. The remaining amino acids had a different effect in that they appeared to interfere with glutamine supplementation. The amino acids with the most extreme effects were alanine (Fig. 2), methionine, leucine, and histidine. In the presence of these amino acids, the glnAΔ strains were completely unable to grow, despite the presence of 10 mM glutamine. It is possible that the metabolism of the former group of amino acids resulted in increased levels of a growth-inhibitory compound, possibly glutamate. The latter class of amino acids may effect glutamine supplementation through competition or inhibition of glutamine uptake. The basis of these effects is not known.
The A. nidulans glnA gene and the cyanobacterium Anabaena glnA gene can complement the glnAΔ mutation.
The phenotypes of glutamine auxotrophy and growth inhibition of strains carrying the glnAΔ mutation were fully reversed by reintroduction of the A. nidulans glnA gene, indicating that the glutamine auxotrophy was indeed conferred by the glnAΔ mutation. This was achieved by transforming the recipient strain, MH9962, to glutamine prototrophy with pCD5 or by cotransformation with pCD5 and the pyroA vector pI4 (34). Transformants were directly selected for complementation of glutamine auxotrophy on medium containing 10 mM ammonium as the sole nitrogen source. Pyro+ transformants were selected on media containing 10 mM glutamine and lacking pyridoxine. Complementing transformants were indistinguishable from glnA+ strains. Reintroduction of the glnA gene on pCD5 was also able to reverse the phenotypes of the glnAΔ mutation in an areA+ background.
In addition, it was found that the glnA gene of the cyanobacterium Anabaena sp. strain 7120 (20, 46) was able to completely restore the Gln+ phenotype when it was expressed in A. nidulans. The Anabaena glnA gene was fused to the A. nidulans alcA promoter (see Materials and Methods), and the plasmid was transformed into MH9962. Full complementation of the glnAΔ mutation was observed in direct selection or cotransformation experiments using pI4 as the selectable marker. In contrast, the Anabaena glnA gene expressed from its native prokaryotic promoter sequences failed to complement the glnAΔ mutation. Clearly, the prokaryotic enzyme has GS activity in A. nidulans provided it is expressed from a promoter recognized by the host.
Previous studies had identified a region in the C terminus of the FluG protein that has similarity to prokaryotic GS type 1 enzymes (27). The fluG gene is required for the initiation of the developmental cycle in A. nidulans that leads to the production of asexual spores (conidia). The entire coding region of fluG (encoding amino acid residues 1 to 865) and sequences encoding the N-terminal and C-terminal portions of FluG (residues 1 to 402 and 387 to 865, respectively) were fused to the alcA promoter (see Materials and Methods). These constructs were introduced by cotransformation with pI4 into MH9962. All fluG constructs failed to complement the glnAΔ mutation, indicating that the FluG protein lacks GS catalytic function.
GS is an effector of nitrogen metabolite repression.
Given the proposed role of glutamine as the key effector of nitrogen metabolite repression and the possibility that GS itself may have a regulatory role, the response of the glnAΔ strains to repression was of considerable interest. A number of standard plate assays were used to assess nitrogen regulation (3, 36). Chlorate, aspartylhydroxamate, and thiourea are analogues of nitrate, asparagine, and urea, respectively. Both ammonium and glutamine are known to protect the wild-type strain from the toxicities of these analogues by repressing the synthesis of permeases and other enzymes required for their assimilation. In both areA+ and areA102 backgrounds, the glnAΔ mutation led to an apparent slight sensitivity to chlorate and hypersensitivity to thiourea and aspartate hydroxymate, even in the presence of glutamine, compared to glnA+ controls. Similar results have been observed previously and have led to the suggestion that glutamine alone is not sufficient to signal repression of catabolic enzyme synthesis and that the GS enzyme itself may have a complex role in nitrogen regulation (28). However, interpretation of these plate tests is complicated by the finding that the growth of glnA mutants on glutamine may be affected by other compounds in the medium. Indeed, further growth tests revealed that the presence of asparagine and, to a lesser extent, nitrate or urea interfered with the growth and/or glutamine supplementation of glnAΔ strains. Therefore, it is difficult to distinguish between direct sensitivity to toxic nitrogen source analogues, indicating derepression, and indirect effects on glutamine supplementation of the glnAΔ mutant strains.
Therefore, the effect of the glnAΔ mutation on nitrogen metabolite repression was assessed in strains carrying an amdS::lacZ fusion reporter gene integrated at the amdS locus (Table 1). Expression of the amdS gene is highly regulated by areA and is not dependent on induction (13). The glnA+ strain had low β-galactosidase levels after overnight growth on glutamine. Subsequent transfer to nitrogen-free conditions led to a substantial increase in enzyme levels, whereas transfer to ammonium prevented this response. Interestingly, transfer to glutamate-containing medium resulted in an increase in β-galactosidase activities to levels only approximately 50% of those of the nitrogen-free culture. This is consistent with assays of a range of different catabolic enzymes (21), where glutamate-grown levels were half those of nitrogen-starved levels. The glnAΔ mutant grown overnight on glutamine exhibited very low levels of β-galactosidase, similar to levels in a glnA+ background. Therefore, the addition of glutamine was sufficient to bring about nitrogen metabolite repression in the absence of the glnA gene product. This suggests that while the GS enzyme is required for synthesizing glutamine, GS itself does not mediate nitrogen metabolite repression. This conclusion was also supported by amdS-directed β-galactosidase assays of glnAΔ transformants expressing the Anabaena glnA gene, in which regulation of amdS::lacZ was equivalent to that of the glnA+ strain (data not shown).
TABLE 1.
Effects of mutations in the glnA and gdhA genes on amdS::lacZ expression
| Relevant genotypeb | β-Galactosidase activitya
|
|||
|---|---|---|---|---|
| Glutamine | Nitrogen-free | Ammonium | Glutamate | |
| glnA+gdhA+ | 8.0 (1.7) | 234.0 (21.6) | 11.4 (2.8) | 93.7 (8.7) |
| glnAΔ gdhA+ | 14.7 (3.1) | 170.0 (19.5) | 23.5 (2.2) | 68.7 (7.6) |
| glnA+gdhA10 | 30.0 (3.8) | 219.6 (24.5) | 76.5 (8.8) | 104.4 (18.2) |
| glnAΔ gdhA10 | 33.2 (4.8) | 148.2 (23.4) | 126.0 (24.5) | 110.8 (15.8) |
Mycelia were grown overnight on 1% glucose medium containing 10 mM glutamine and transferred to medium containing either 10 mM ammonium, 10 mM glutamate, or no added nitrogen source (nitrogen-free) for 4 h. The assay results presented are the average of six independent experiments, with standard errors in parentheses, and β-galactosidase activities are expressed as units per minute per milligram of soluble protein.
All strains contained the areA102 mutation and an amdS::lacZ reporter gene replaced at the amdS locus.
Glutamate as well as glutamine signals nitrogen metabolite repression.
If glutamine alone were the signal for repression, it would be predicted that a glnAΔ strain would be fully derepressed on ammonium or glutamate, as it would be unable to metabolize these compounds to glutamine. Surprisingly, it was found that the glnAΔ mutant showed only partial derepression on ammonium (Table 1). Therefore, even in a strain lacking GS activity, ammonium was able to bring about significant repression. In addition, the effect of glutamate in the glnAΔ mutant was similar to the effect in the glnA+ strain. Therefore, glutamate led to a 50% reduction in levels even though it could not metabolized to glutamine.
These results suggest that ammonium and/or glutamate may generate signals for repression independently of glutamine. Growth of the glnAΔ stain on ammonium may result in an internal build-up of glutamate through the action of NADP-GDH. To investigate the contribution of glutamate synthesized from ammonium, the gdhA10 mutation was introduced into a glnAΔ background by genetic crosses. The double mutant was unable to use any nitrogen source other than glutamine due to the glnAΔ mutation. The growth of the double mutant on glutamine as a sole nitrogen source was poorer than that of glnAΔ or gdhA single mutants. The gdhA10 glnAΔ double mutant is unable to assimilate the ammonium derived from the catabolism of glutamine through either NADP-GDH or GS, leaving only the derived glutamate as a usable nitrogen source. Interestingly, the double mutant did not exhibit the inhibited growth morphology of the glnAΔ single mutant. Therefore, by preventing the conversion of ammonium to glutamate, the gdhA mutation reduced the accumulation of glutamate-derived inhibitory metabolites that occurs with the glnAΔ single mutant.
The gdhA10 mutation alone resulted in elevated levels of amdS::lacZ expression on ammonium, consistent with previous studies of gdhA mutants (4, 21, 22). This mutation did not alter the response to glutamate, with activities again half those of the nitrogen-free culture. While the glnAΔ mutant had low levels of amdS::lacZ expression on ammonium, levels in the gdhA10 glnAΔ double mutant were elevated. Therefore, glutamate derived from ammonium rather than ammonium itself was responsible for the low expression levels in the glnAΔ strain. Comparison of amdS::lacZ expression in glutamate and nitrogen-free cultures in the double mutant indicated that growth on glutamate resulted in only slightly reduced enzyme levels in this genetic background. Internal glutamate levels are likely to be reduced in the double mutant relative to the glnAΔ single mutant, as ammonium derived from glutamate catabolism via NAD-GDH cannot be reconverted to glutamate (see Discussion).
DISCUSSION
In A. nidulans, GS is absolutely required for the biosynthesis of glutamine. Previously isolated alleles have shown, to various degrees, a slight leakiness (10). Lee and Adams (27) have investigated whether the GS-like domain of FluG has sufficient activity to account for the leaky phenotype of glnA− mutants. They found that a fluG glnA− double mutant was no more extreme than the glnA− single mutant, indicating that FluG is not the source of the residual activity. We have now shown that the leakiness is a reflection of residual GS enzymatic activity in these strains, as a complete null mutation is a strict glutamine auxotroph. We have also shown directly that FluG lacks GS activity by virtue of its inability to restore any growth to the glnAΔ strain. Amino acid sequence comparison indicates that the GS-like region of FluG is most similar to the type I GS of prokaryotes. Type I GS enzymes form a 12-subunit complex structure and are unique to prokaryotes, while the type II GSs found in eukaryotes are octamers of a smaller polypeptide which is only distantly similar to the type I form. The presence of a type II GS in certain bacteria is thought to have arisen by lateral gene transfer (8). It is interesting that A. nidulans FluG has greater similarity to the prokaryotic type I enzyme. The lack of GS activity of this region of FluG is not simply a function of its type I similarity, as we have found that the Anabaena enzyme can function effectively in a eukaryotic context provided that the promoter is functional.
A noticeable feature of the glnAΔ strains is their aberrant morphology on glutamine-containing media. This was particularly evident in an areA+ background and was partially suppressed by the areA102 mutation. MacDonald (28) noted that strains carrying the glnA1 mutation also had a slightly restricted morphology on 10 mM glutamine. The glnA1 mutant retains very low transferase and synthetase activities (10) and exhibits residual growth in the absence of glutamine, unlike the glnAΔ strain. Therefore, the altered-morphology phenotype has been observed even with mutants that retain low levels of GS function. As high glutamine concentrations (in the millimolar range) are required to fully supplement the glutamine requirement of glnA mutants, MacDonald (28) has suggested that glutamine uptake may be limiting for A. nidulans. However, glutamine concentrations up to 20 mM were not effective in reversing the phenotype and in fact increased its severity in the glnAΔ strains. As glnA+ (areA+ or areA102) strains grow strongly on 10 mM glutamine, glutamine uptake is not limiting for growth, and externally supplied glutamine is a potent source of nitrogen metabolite repression for both wild-type and areA102 strains.
In a glnAΔ strain, the breakdown of glutamine to glutamate and ammonium may lead to the accumulation of glutamate within the cells. In N. crassa, a leaky gln-1 mutant grown on glutamine as a nitrogen source accumulated more glutamate and alanine than a wild-type strain, and wild-type cells treated with an inhibitor of GS have been shown to accumulate glutamate when grown on glutamine (5, 31). It is known that glutamine synthesis occurs even in cells growing on glutamine and that nitrogen is efficiently cycled between glutamine and glutamate via the GS-GOGAT cycle (31, 47). Loss of the ability to recycle glutamate into glutamine in glnAΔ mutants may result in excessive levels of glutamate or a derivative, leading to toxic effects within the cells. This is supported by the observation that the growth inhibition associated with the glnAΔ mutants is abolished by a gdhA mutation. In N. crassa, a significant fraction of the ammonium derived from glutamine degradation is normally assimilated by NADP-GDH into glutamate (5). As the gdhA mutant lacks NADP-GDH, the ammonium derived from glutamine breakdown would not add to the pool of glutamate within a glnAΔ mutant. Furthermore, areA102 glnAΔ strains were found to be less inhibited on glutamine than glnAΔ single mutants. As areA102 strains grow more strongly than the wild-type strain on glutamate as a sole nitrogen source, the areA102 mutation may reduce the buildup of glutamate by increased glutamate catabolism via NAD-GDH (22).
The growth-restricted morphology, without associated pigmentation, has also been observed on very low glutamine concentrations. At these levels, toxic metabolite accumulation is unlikely. Therefore, the endogenous synthesis of glutamine may be required for hyphal extension, a requirement not met by exogenous glutamine, possibly because young tips do not contain active glutamine permease(s). Cardenas and Hansberg (6, 7) found that in N. crassa, the transition from mycelial mat to aerial hyphae required high glutamine pools, and they proposed that internal glutamine serves as a nitrogen carrier to the growing aerial mycelium.
The availability of a glnAΔ strain has allowed us to investigate the generation of the signaling metabolites for nitrogen metabolite repression. Since the mutant lacks the GS enzyme, we were able to establish that the addition of glutamine is sufficient to bring about repression of the nitrogen-regulated amdS gene. The GS enzyme itself is unlikely to have other than a biosynthetic role in generating the signal. This is also supported by the finding that expression of the Anabaena sp. strain 7120 GS simultaneously restores glutamine prototrophy and normal regulation. The finding that a wide range of compounds can interfere with the growth of glnAΔ strains on glutamine may explain previous data leading to the suggestion that glutamine alone was not effective in glnA mutant strains. These earlier findings were repeated here with plate tests, indicating that glnAΔ mutants were sensitive to chlorate (10, 28). The fact that nitrate could partially mimic these results suggests that they are in fact an indirect consequence of the glnA mutant phenotype rather than an indication of derepression.
The use of an amdS::lacZ reporter construct allowed us to test the response to glutamine without the need for simultaneously inducing enzyme synthesis and hence avoiding the complications of inducer exclusion. From these data it is clear that glutamine is an effective trigger for repression in the glnAΔ strains. If glutamine alone is the required signaling molecule, it would be predicted that glnAΔ mutants unable to synthesize glutamine would be fully derepressed on ammonium. The data clearly indicate that this is not the case and that ammonium and/or glutamate may also act as signaling molecules. By introducing the gdhA10 mutation into the glnAΔ background, we were able to show that ammonium was effective in the glnAΔ mutant by virtue of its conversion to glutamate. While it may be expected that growth on glutamate would result in repression in the double mutant, the data indicate that this is not so. In this mutant background, glutamate levels are not replenished from the ammonium released by glutamate catabolism by either NADP-GDH or the GS-GOGAT cycle, as described above. This correlates with the phenotype of the double mutant, which grows more poorly on glutamine than the single glnAΔ mutant but lacks the growth inhibition phenotype that accompanies the single mutant. It is well established that NAD-GDH is responsible for breaking down glutamate (22). In the gdhA10 glnAΔ double mutant, the inability to convert the derived ammonium back to glutamate prevents intracellular glutamate from accumulating to bring about repression. Glutamate repression has been proposed as a separate mechanism for the control of NADP-GDH synthesis (35). Interestingly, studies of nitrogen regulation in Saccharomyces cerevisiae have suggested that the two GATA factor activators GLN3 and NIL1 may respond to different signals, with GLN3, but not NIL1, active in glutamate-grown cultures (43, 44). Whether these multiple forms of repression operate via a single regulatory pathway involving AreA or whether there are distinct pathways remains to be determined. However, recent evidence suggests that glutamine and glutamate may generate distinct signals that each influence the stability of the areA mRNA through interactions in the 3′ untranslated region of the areA gene (32)
ACKNOWLEDGMENTS
We acknowledge the support of the Australian Research Council (Grant A10020013 to M.A.D. and M.J.H.) and the National Institutes of Health (Grant GM45252 to T.H.A.).
REFERENCES
- 1.Andrianopoulos A, Hynes M J. Cloning and analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol. Cell. Biol. 1988;8:3532–3541. doi: 10.1128/mcb.8.8.3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrianopoulos A, Kourambas S, Sharp J A, Davis M A, Hynes M J. Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J Bacteriol. 1998;180:1973–1977. doi: 10.1128/jb.180.7.1973-1977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arst H N, Jr, Cove D J. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973;126:111–141. doi: 10.1007/BF00330988. [DOI] [PubMed] [Google Scholar]
- 4.Arst H N, Jr, MacDonald D W. A mutant of Aspergillus nidulans lacking NADP-linked glutamate dehydrogenase. Mol Gen Genet. 1973;122:261–265. doi: 10.1007/BF00278601. [DOI] [PubMed] [Google Scholar]
- 5.Calderon J, Mora J. Glutamine cycling in Neurospora crassa. J Gen Microbiol. 1985;131:3237–3242. [Google Scholar]
- 6.Cardenas M E, Hansberg W. Glutamine requirement for aerial mycelium growth in Neurospora crassa. J Gen Microbiol. 1984;130:1723–1732. doi: 10.1099/00221287-130-7-1723. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas M E, Hansberg W. Glutamine metabolism during aerial mycelium growth in Neurospora crassa. J Gen Microbiol. 1984;130:1733–1741. doi: 10.1099/00221287-130-7-1733. [DOI] [PubMed] [Google Scholar]
- 8.Carlson T A, Chelm B K. Apparent eukaryotic origin of glutamine synthetase II from the bacterium Bradyrhizobium japonicum. Nature. 1986;322:568–570. [Google Scholar]
- 9.Clutterbuck A J. Aspergillus nidulans genetics. In: King R C, editor. Handbook of genetics. New York, N.Y: Plenum Press; 1974. pp. 447–510. [Google Scholar]
- 10.Cornwell E V, MacDonald D W. glnA mutations define the structural gene for glutamine synthetase in Aspergillus. Curr Genet. 1984;8:33–36. doi: 10.1007/BF00405429. [DOI] [PubMed] [Google Scholar]
- 11.Cove D J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;133:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 12.Davis M A, Cobbett C S, Hynes M J. An amdS-lacZ fusion for studying gene regulation in Aspergillus nidulans. Gene. 1988;63:199–212. doi: 10.1016/0378-1119(88)90525-2. [DOI] [PubMed] [Google Scholar]
- 13.Davis M A, Kelly J M, Hynes M J. Fungal catabolic gene regulation: molecular genetic analysis of the amdS gene of Aspergillus nidulans. Genetica. 1993;90:133–145. doi: 10.1007/BF01435035. [DOI] [PubMed] [Google Scholar]
- 14.Davis M A, Small A J, Kourambas S, Hynes M J. The tamA gene of Aspergillus nidulans contains a putative zinc cluster motif which is not required for gene function. J Bacteriol. 1996;178:3406–3409. doi: 10.1128/jb.178.11.3406-3409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn-Coleman N S, Tomsett A B, Garrett R H. Nitrogen metabolite repression of nitrate reductase in Neurospora crassa: effect of the gln-la locus. J Bacteriol. 1979;139:697–700. doi: 10.1128/jb.139.2.697-700.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn-Coleman N S, Garrett R H. The role of glutamine synthetase and glutamine metabolism in nitrogen metabolite repression, a regulatory phenomenon in the lower eukaryote Neurospora crassa. Mol Gen Genet. 1980;179:25–32. doi: 10.1007/BF00268442. [DOI] [PubMed] [Google Scholar]
- 17.Dunn-Coleman N S, Garrett R H. Effect of the gln-1b mutation on nitrogen metabolite repression in Neurospora crassa. J Bacteriol. 1981;145:884–888. doi: 10.1128/jb.145.2.884-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehinger A, Denison S H, May G S. Sequence, organization and expression of the core histone genes of Aspergillus nidulans. Mol Gen Genet. 1990;222:416–424. doi: 10.1007/BF00633848. [DOI] [PubMed] [Google Scholar]
- 19.Fernandes M, Keller N P, Adams T H. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol Microbiol. 1998;28:1355–1365. doi: 10.1046/j.1365-2958.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- 20.Fisher R, Tuli R, Haselkorn R. A cloned cyanobacterial gene for glutamine synthetase functions in Escherichia coli, but the enzyme is not adenylated. Proc Natl Acad Sci USA. 1981;78:3393–3397. doi: 10.1073/pnas.78.6.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hynes M J. Effects of ammonium, l-glutamate, and l-glutamine on nitrogen catabolism in Aspergillus nidulans. J Bacteriol. 1974;120:1116–1123. doi: 10.1128/jb.120.3.1116-1123.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinghorn J R, Pateman J A. NAD and NADP L-glutamate dehydrogenase activity and ammonium regulation. J Gen Microbiol. 1973;78:39–46. doi: 10.1099/00221287-78-1-39. [DOI] [PubMed] [Google Scholar]
- 23.Kinghorn J R, Pateman J A. The structural gene for NADP-L-glutamate dehydrogenase in Aspergillus nidulans. J Gen Microbiol. 1975;86:294–300. doi: 10.1099/00221287-86-2-294. [DOI] [PubMed] [Google Scholar]
- 24.Kinghorn J R, Pateman J A. Studies of partially repressed mutants at the tamA and areA loci in Aspergillus nidulans. Mol Gen Genet. 1975;140:137–147. doi: 10.1007/BF00329781. [DOI] [PubMed] [Google Scholar]
- 25.Kudla B, Caddick M X, Langdon T, Martinez-Rossi N, Bennett C F, Sibley S, Davies R W, Arst H N., Jr The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9:1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langdon T, Sheerins A, Ravagnami A, Gielkens M, Caddick M X, Arst H N., Jr Mutational analysis reveals dispensability of the N-terminal region of the Aspergillus transcription factor mediating nitrogen metabolite repression. Mol Microbiol. 1995;17:877–888. doi: 10.1111/j.1365-2958.1995.mmi_17050877.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee B N, Adams T H. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 1994;8:641–651. doi: 10.1101/gad.8.6.641. [DOI] [PubMed] [Google Scholar]
- 28.MacDonald D W. A single mutation leads to loss of glutamine synthetase and relief of ammonium repression in Aspergillus. Curr Genet. 1982;6:203–208. doi: 10.1007/BF00390339. [DOI] [PubMed] [Google Scholar]
- 29.Macheda M L, Hynes M J, Davis M A. The Aspergillus nidulans gltA gene encoding glutamate synthase is required for ammonium assimilation in the absence of NADP-glutamate dehydrogenase. Curr Genet. 1999;34:467–471. doi: 10.1007/s002940050421. [DOI] [PubMed] [Google Scholar]
- 30.Marzluf G A. Genetic regulation of nitrogen metabolism in fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mora J. Glutamine metabolism and cycling in Neurospora crassa. Microbiol Rev. 1990;54:293–304. doi: 10.1128/mr.54.3.293-304.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morozov I Y, Martinez M G, Jones M G, Caddick M A. A defined sequence within the 3′UTR of the areA transcript is sufficient to mediate nitrogen metabolite signalling via accelerated deadenylation. Mol Microbiol. 2000;37:1248–1257. doi: 10.1046/j.1365-2958.2000.02085.x. [DOI] [PubMed] [Google Scholar]
- 33.Oakley C E, Weil C F, Kretz P L, Oakley B R. Cloning of the riboB locus of Aspergillus nidulans. Gene. 1987;53:293–298. doi: 10.1016/0378-1119(87)90019-9. [DOI] [PubMed] [Google Scholar]
- 34.Osmani A H, May G S, Osmani S A. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J Biol Chem. 1999;274:23565–23569. doi: 10.1074/jbc.274.33.23565. [DOI] [PubMed] [Google Scholar]
- 35.Pateman J A. Regulation of synthesis of glutamate dehydrogenase and glutamine synthetase in micro-organisms. Biochem J. 1969;115:769–775. doi: 10.1042/bj1150769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platt A, Langdon T, Arst H N, Jr, Kirk D, Tollervey D, Mates Sanchez J M, Caddick M X. Nitrogen metabolite signalling involves the C-terminus and the GATA domain of the Aspergillus transcription factor AREA and the 3′ untranslated region of its mRNA. EMBO J. 1996;15:2791–2801. [PMC free article] [PubMed] [Google Scholar]
- 37.Premakumar R, Sorger G J, Gooden D. Nitrogen metabolite repression of nitrate reductase in Neurospora crassa. J Bacteriol. 1979;137:1119–1126. doi: 10.1128/jb.137.3.1119-1126.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Premakumar R, Sorger G J, Gooden D. Repression of nitrate reductase in Neurospora studied by using l-methionine-dl-sulphoximine and glutamine auxotroph gln-1b. J Bacteriol. 1980;143:411–415. doi: 10.1128/jb.143.1.411-415.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravagnani A, Gorfinkiel L, Langdon T, Diallinas G, Adjadj E, Demais S, Gorton D, Arst H N, Jr, Scazzocchio C. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 1997;16:3974–3986. doi: 10.1093/emboj/16.13.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Scazzocchio C. The fungal GATA factors. Curr Opin Microbiol. 2000;3:126–131. doi: 10.1016/s1369-5274(00)00063-1. [DOI] [PubMed] [Google Scholar]
- 42.Small A J, Hynes M J, Davis M A. The TamA protein fused to a DNA-binding domain can recruit AreA, the major nitrogen regulatory protein, to activate gene expression in Aspergillus nidulans. Genetics. 1999;153:95–105. doi: 10.1093/genetics/153.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanborough M, Rowen D W, Magasanik B. Role of GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc Natl Acad Sci USA. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanborough M, Magasanik B. Two transcription factors, Gln3p and Nil1p, use the same GATAAG sites to activate expression of GAP1 of Saccharomyces cerevisiae. J Bacteriol. 1996;178:2465–2468. doi: 10.1128/jb.178.8.2465-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tumer N E, Robinson S J, Haselkorn R. Different promoters for the Anabaena glutamine synthetase gene during growth using molecular or fixed nitrogen. Nature. 1983;306:337–342. [Google Scholar]
- 47.Vichido I, Mora Y, Quinto C, Palacios R, Mora J. Nitrogen regulation of glutamine synthetase in Neurospora crassa. J Gen Microbiol. 1978;106:251–259. doi: 10.1099/00221287-106-2-251. [DOI] [PubMed] [Google Scholar]