Abstract
During insect infection Photorhabdus luminescens emits light and expresses virulence factors, including insecticidal toxin complexes (Tcs) and an RTX-like metalloprotease (Prt). Using quantitative PCR and protein assays, we describe the expression patterns of these factors both in culture and during insect infection and compare them to the associated bacterial growth curves. In culture, light and active Prt protease are produced in stationary phase. Tca also appears in stationary phase, whereas Tcd is expressed earlier. These patterns seen in a culture flask are strikingly similar to those observed during insect infection. Thus, in an infected insect, bacteria grow exponentially until the time of insect death at ≈48 h, when both light and the virulence factors Prt protease and Tca are produced. In contrast, Tcd appears much earlier in insect infection. However, at present, the biological significance of this difference in timing of the production of the two toxins in unclear. This is the first documentation of the expression of Tcs and Prt in an insect and highlights the malleability of Photorhabdus as a model system for bacterial infection.
Photorhabdus luminescens is a gram-negative gamma proteobacterium belonging to the family Enterobacteriaceae (7). The bacterium is found in the gut of entomopathogenic nematodes of the family Heterorhabditidae. Upon invasion of an insect host by the nematode, the bacteria are released directly into the open blood system of the insect (the hemocoel), where they multiply and help in killing the insect host. During the course of the insect infection, the carcass becomes visibly luminescent due to the bioluminescence of P. luminescens (14). The role of light production in the biology of this tritrophic interaction is unclear. However, light production does provide a clear and well-documented method for assaying the time course of insect infection. Once the infection is established, the insect cadaver is bioconverted into a source of nutrients for both the bacteria and the nematodes. Infective juvenile nematodes subsequently reacquire the bacteria and leave the insect to infect new hosts (7). P. luminescens has also been found as an opportunistic pathogen in human wounds (4).
Following the observation that there is a positive correlation between the virulence of Pseudomonas aeruginosa mutants in mice and insects (11), we are interested in establishing the invertebrate pathogen P. luminescens as a model system for the study of bacterial virulence. To begin to define this system, this paper compares the expression patterns of several candidate virulence factors both in culture and during insect infection. When P. luminescens is grown in laboratory culture, luminescence is produced and several potential virulence factors can be detected in the culture supernatant. These factors include lipases, proteases, antibiotics, lipopolysaccharides (7), and high-molecular-weight insecticidal toxin complexes (Tcs) (2). Some of the genes encoding these putative virulence factors, such as the toxin complex (tc) genes, are found in other members of the family Enterobacteriaceae (10, 15). Recent sample sequencing of the P. luminescens W14 genome also suggests that as much as 53% of the genome is clearly distinct from that of Escherichia coli K-12, and many sequences show homology to known or putative virulence factors in other species (6). Despite a considerable amount of work on some virulence factors from P. luminescens, such as cloning and gene knockout of the Tc-encoding genes (2), we still have little idea of the function of these factors in vivo. Perhaps more importantly, we have no documentation of their actual expression during insect infection. This leaves open the formal possibility that these genes are expressed at other times during the bacterial life cycle or even within the alternate host (i.e., the nematode).
Several techniques have been developed for the identification of genes expressed during infection (8). There is also growing interest in using invertebrate infections as models for vertebrate pathogenicity (12). With the advent of quantitative PCR, we are interested in quantifying Photorhabdus virulence factor gene expression in an insect infection over time. Here, we describe the patterns of mRNA transcription of the light-producing lux genes in culture and correlate these with the production of three putative virulence factors by two tc genes (tcaB and tcdB) and the gene encoding an RTX-like metalloprotease, prt, using quantitative PCR. We matched their patterns of transcription with production of the associated proteins via measurements of light production, Western blots, and protease zymograms, respectively. Moreover, in each case we tracked virulence factor production during the course of insect infection itself. Our results not only confirm that these factors are actually expressed during infection but also illustrate the potential flexibility of the Photorhabdus-insect system as a model for bacterial infection.
MATERIALS AND METHODS
Bacterial cultures and insect infection.
Cultures of P. luminescens W14 were initiated in 100 ml of 2% PP3 broth, as described previously (3). For insect infection, 10 μl of phosphate-buffered saline (PBS) containing ≈20 washed cells of P. luminescens was injected into third-instar Manduca sexta insects using a 100-μl Hamilton syringe with a disposable 30-gauge needle. Unpublished studies have shown that individual infective juvenile nematodes carry 30 to 250 bacteria (J. Ensign and T. Ciche, personal communication); therefore, 20 is the minimum number of cells released by an individual nematode. The number of cells injected was confirmed by counting the number of bacteria in a fixed volume on a hemacytometer and also by plating the cells on 2% PP3 plates. One hundred twenty third-instar M. sexta insects were injected and incubated at 25°C. Samples consisting of 10 insects were then individually homogenized in 5 ml of PBS at each time point. One milliliter of each homogenate was diluted in PBS and plated on 2% PP3–0.2% arabinose plates, and the number of CFU was assessed after 40 h of incubation at 29°C. We confirmed that the recovered bacteria were P. luminescens by using a strain of W14 marked with green fluorescent protein, inducible with arabinose, and checking for fluorescence of all the counted colonies after plating. For the bacterial cultures, samples were taken at fixed time points, and the optical density (OD) of the culture was measured.
Quantitative RT-PCR.
The abundances of luxA, tcaB, tcdB, and prtA transcripts over time, relative to 16S rRNA, were determined in bacterial cultures by real-time quantitative PCR. RNA was extracted with TRI reagent (Sigma), digested with 1.0 U of DNaseRQ1 (Promega)/μl, and reextracted with TRI reagent. Lack of DNA contamination was confirmed in every RNA sample via 40 cycles of PCR using RNA as the template. Single-step reverse transcription (RT)-PCR was performed with the LightCycler RNA amplification kit SYBR Green I (Roche). RT was performed for 10 min at 55°C, followed by PCR (45 cycles of 0 s at 94°C, 5 s at 62°C, and 20 s at 72°C). The acquisition of fluorescence from the incorporation of SYBR Green I into the double-stranded PCR product was measured at the end of each of 45 PCR cycles to determine the amplification characteristics of each PCR product. Melting analysis was then performed, in which fluorescence is continuously monitored while the sample is heated from 67 to 95°C at a transition rate of 0.1°C/s. The characteristic melting temperature of any product, as highlighted by a peak in fluorescence, is determined from its size and GC content. PCR products were also run on agarose gels to correlate the expected product length with the characteristic melting temperature. For each primer pair (Table 1), a standard curve was constructed using known amounts of W14 genomic DNA (from 100 ng to 10 pg). This was then used to quantify the abundance of each transcript in the RNA samples. As an internal control, the 16S primer set was run concurrently with each set of experimental primers. The 16S primer set is specific for P. luminescens, with one primer designed from within the variable region of the 16S rRNA at positions 440 to 480 (5).
TABLE 1.
Nucleotide sequences of primers used in quantitative PCR
| Gene | Primer | Sequence | Product size (bp) |
|---|---|---|---|
| luxA | Forward | TCGCCAGCTTGAAGAGGTGAATT | 233 |
| Reverse | CCGTCGGCAGCACTTTTACCTTAT | ||
| tcaB | Forward | GCGCTATTGCCACACCGACATTCCTCAG | 227 |
| Reverse | TACCGGCGTGGCATACAGTGGCAGATTTAG | ||
| tcdB | Forward | GGGGCCGGAAGGTGAGTTGGTCGTAG | 183 |
| Reverse | GGGATGCCTGAACTGCCGTGGTAAG | ||
| prtA | Forward | GGGCGGGGAAGGCAAAGATA | 191 |
| Reverse | GCACCGCTTCTCCGACTTCAC | ||
| 16S rRNA | Forward | ACAGAGTTGGATCTTGACGTTACCC | 305 |
| Reverse | AATCTTGTTTGCTCCCCACGCTT |
Assay of virulence-associated proteins.
Western blotting was performed using antibodies raised against specific peptides (Research Genetics) from the predicted amino acid sequences of Tca and Tcd. Zymograms to determine the activity of the 55-kDa Prt protease were performed as described elsewhere (1). Virulence factor expression was correlated with the time course of light production. Light production, either from a sample of bacterial culture or from a homogenate of an infected insect, was measured using a tube luminometer (DYNATECH). Light production is expressed in relative units.
Comparisons of different growth conditions.
In order to compare in vitro (culture flask) with in vivo (insect infection) experiments, we have displayed the relative abundances of RNA, proteins, and light against time. We recognize that in culture, comparisons should be between different ODs, as culture growth rates will differ between experiments. We have therefore compared ODs when contrasting two different culture experiments. Unfortunately, however, it is not readily possible to compare an OD in a flask with the recovery of culturable bacteria from an infected insect. In comparing in vitro and in vivo experiments, therefore, we have confined our discussions to the relative abundance and the relative order of expression of the different factors examined.
RESULTS
Bacterial growth and bioluminescence.
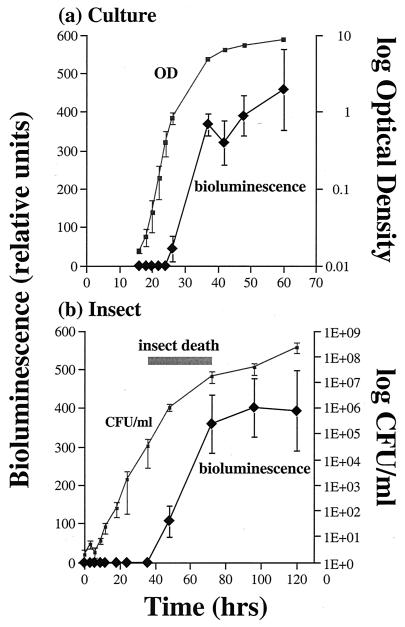
In culture, P. luminescens shows the classic sigmoid growth curve, with stationary phase entered at 30 to 40 h (Fig. 1a). The appearance of bioluminescence follows a similarly shaped curve, with light production increasing dramatically between 30 and 40 h (Fig. 1a). In an insect infection (Fig. 1b), P. luminescens again appears to grow in a rapid exponential phase but at a somewhat lower rate than in culture (note the difference in time scales in Fig. 1a versus 1b). There is rapid bacterial growth until the insect dies (Fig. 1b), at which time both bacterial growth and light production start to plateau.
FIG. 1.
Growth curves for P. luminescens in culture and during an insect infection. (a) Growth curve in culture derived from measurements of OD over time and the associated bioluminescence produced. Note that light production peaks as the cells enter stationary phase. (b) Bacterial growth curve during an insect infection, derived from the mean number of CFU recovered from infected insects at different time points after injection of bacteria. The range of time (shaded bar) during which the study insects died is shown (actual mortality at 36, 48, and 72 h was 4, 60, and 100%, respectively). Note that light production peaks as the bacterial growth rate decreases. All data are shown as the means of three experiments with associated standard errors.
Light and virulence factor expression in culture.
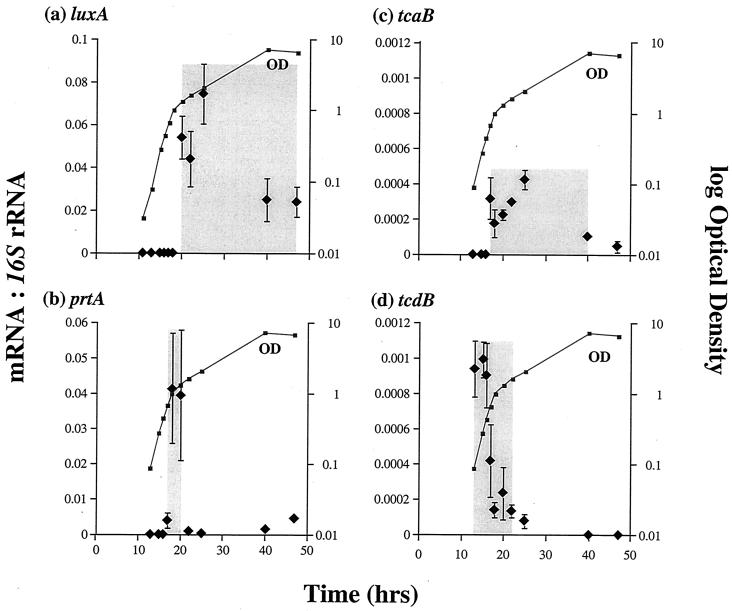
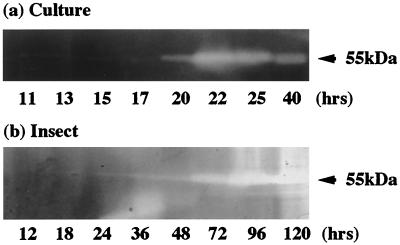
To determine when gene transcription is initiated for luxA and the various virulence factors, we measured their mRNA production over time using quantitative PCR. The luxA transcript is first detected in broth-grown cells at an OD of 1 (at 20 h in this experiment [Fig. 2a]), peaks, and then declines rapidly. This peak of luxA transcript corresponds to maximal light production at an equivalent OD (Fig. 1a) in a different batch culture (with a correspondingly different time course, i.e., reaching an OD of 1 at 27 rather than 20 h) in which light production was monitored. The prtA protease transcript also appears at an OD of 1 (at 17 h in this experiment [Fig. 2b]) and peaks very rapidly (returning to a basal level within 5 h). This peak of transcript abundance correlates with the appearance of active protease in the culture, as detected by zymogram analysis of the same culture broth at an OD of 1 (at 20 h [Fig. 3a]).
FIG. 2.
Relative abundance and times of expression (shaded areas) of luxA (a), prtA (b), tcaB (c), and tcdB (d) mRNAs versus 16S rRNA over time in culture. Note that tcdB message is transcribed at an earlier stage in culture growth than that of tcaB (see the text for discussion). All data are shown as the means of three experiments with their associated standard errors.
FIG. 3.
Detection by zymogram of the active Prt protease in culture supernatants and during an insect infection. Note the zone of protein clearing at ≈55 kDa, corresponding to production of the active Prt protease (see the text for discussion). Note also that the time scales differ for the culture experiment and the insect infection.
Unexpectedly, the overall patterns of tc message abundance differ. Thus, the tcdB transcript is already highly abundant at the initial OD of 0.1 (the first time point: 13 h in this experiment) and message abundance then declines rapidly (Fig. 2d), whereas the tcaB message appears later (OD = 0.5 at 18 h in the same experiment) and is still detectable for the remainder of the observed time course.
Light and virulence factor expression during infection.
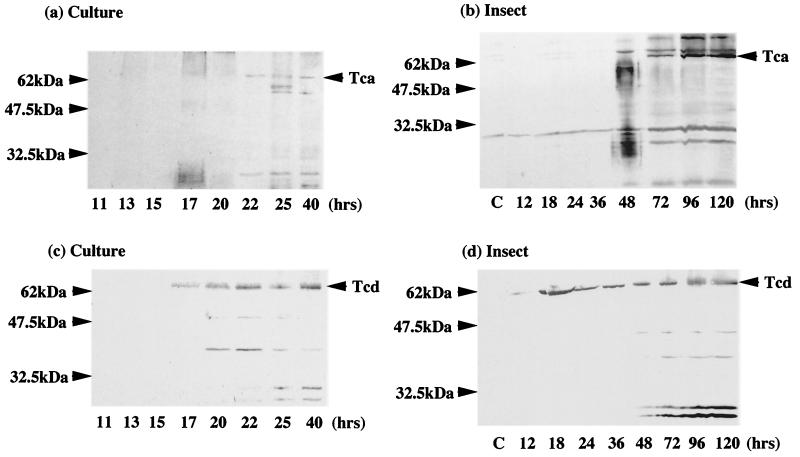
Strikingly, despite the obvious pitfalls of comparing growth in a culture flask with a real infection, the relative order of expression of light and the different virulence factors in an infected insect are similar to that seen in culture. Thus, active Prt protease can be detected in homogenates of infected insects 72 h postinfection (the OD cannot be determined in the context of an in vivo infection) after insect death (Fig. 3). This is also the time postinfection at which the cadaver is most strongly luminescent (Fig. 1b). Again, the relative orders in which the Tc proteins appear are also similar in vivo and in vitro. Thus, the Tcd protein can be detected in Western blots of infected insects at 18 h postinfection, whereas the Tca protein is only detectable 48 h postinfection in the same cohort of infected animals (Fig. 4).
FIG. 4.
Western blot analysis of culture (a) and infected insects (b) with anti-Tca antibody and culture (c) and infected insects (d) with anti-Tcd antibody. Note that species cross-reactive with the anti-Tcd antibody can be detected after 17 h in culture, correlating with the peak of tcd transcription (Fig. 2d). Similarly, in an infected insect, Tcd can be detected after 18 h from the initial infection, before the time of insect death. Tca, in contrast, appears later (72 h postinfection) and in greater relative abundance when all the insects are dead.
DISCUSSION
Light is produced at stationary phase both in vitro and in vivo.
The emission of light by P. luminescens has always been ascribed to the stationary phase of bacterial growth and may be associated with quorum sensing by the bacteria (14). However, as noted above, the biological role of light production during infection is unclear. Our results for light emission in culture support this observation (Fig. 1a), with luxA message being transcribed in late exponential growth phase and light itself subsequently being emitted in late exponential or stationary phase. However, observations of light emission from infected insects are hard to interpret in the absence of an in vivo bacterial growth curve. To document the growth of P. luminescens W14 during an insect infection, therefore, we plated bacteria from samples of infected M. sexta insects at different time points during the course of infection. The curve shows that P. luminescens rapidly enters exponential growth after insect infection and that light is again produced at the equivalent of late exponential-stationary phase, when bacterial growth is beginning to slow. Note that the insect host dies 48 to 72 h postinfection.
The Tcs are expressed at different times.
The appearance of the insecticidal Tcs in culture supernatant has been assumed to occur during stationary phase (3). However, examination of their patterns of transcription in culture shows that tcdB mRNA is present earlier than that of tcaB, which appears during exponential growth (Fig. 2a). Similar differences in the timing of tc message relative abundance may also occur in infected M. sexta insects, as reflected by the earlier appearance of protein species cross-reacting with an anti-Tcd antibody in a Western blot (Fig. 4). Despite earlier implications that these insecticidal complexes were virulence factors (2), these are the first data showing that they are actually expressed during the course of an insect infection. Although Western blot analysis records only the presence of detectable antigen (Tc toxin) with a given antibody (anti-Tc antibody), the Western blot results do support the hypothesis that Tcd is expressed earlier both in a culture flask and in an infected insect. The biological significance of the difference in timing between Tcd and Tca expression is unclear. However, we can speculate that Tcd may play an earlier and perhaps more specific role in killing the insect.
The Prt protease is expressed late in insect infection.
The message for the prtA-encoded RTX-like metalloprotease (Fig. 2a) is found in culture in a dramatic peak at an OD of 1 (18 h after culture initiation in this experiment), and active protease is detectable in the culture shortly thereafter (at 20 h [Fig. 3a]). In an insect infection itself, zymograms can detect active protease 48 h postinfection (Fig. 3b). Although growth curves from a culture flask are not directly comparable with those from an insect infection, we note that active Prt in both cases is detected at a time when straight exponential growth of the bacteria (compare the shapes of growth curves at 20 h in a flask [Fig. 2b] and at 72 h in an insect [Fig. 1b]) is beginning to slow. The precise role of the Prt protease in insect death and/or bioconversion is uncertain, but the documentation of the presence of active enzyme after insect death suggests that it may degrade insect tissues during bioconversion.
Implications for the use of quantitative PCR.
Here, we have shown that the LightCycler is useful for tracking the expression of Photorhabdus transcripts in culture. Mathews et al. have also used this system to document temporal transcription of sigma factors during the infection of cell lines by the human pathogen Chlamydia tracomatis (13). However, our attempts to detect P. luminescens transcripts during the course of an insect infection have met with limited success. This is because the signal (specific bacterial mRNA)-to-noise (combined bacterial and insect RNAs) ratio in RNA isolated from an infected caterpillar is too low to facilitate amplification of a specific bacterial transcript early in the infection process. Thus, we were unable to detect our P. luminescens 16S rRNA standard in an infected Manduca insect until 48 h postinfection. Between 36 and 48 h, the number of bacterial cells per ml of infected caterpillar hemolymph increases dramatically from 1,200 to 4.9 × 105/ml, facilitating the detection of bacterial transcripts. Therefore, at 48 h postinfection we could readily detect transcripts from the 16S rRNA, luxA, tcaB, and prtA genes at levels similar to those detected in culture. However, none of these transcripts could be detected at earlier time points. Further, tcdB transcript could not be detected at any time point during infection, despite the documentation of TcdB protein expression.
Implications for virulence.
Previous workers have tracked the time course of P. luminescens growth in Galleria mellonella and correlated it with the timing of antibiotic production (9). Here, we have looked at the time course of infection in a different insect, M. sexta, and correlated light production with the expression of several putative virulence factors at the levels of both transcript and protein production. Perhaps the most striking initial finding of this study is the remarkable correlation between the pattern of growth of P. luminescens in culture and that in an insect infection. Despite a lower doubling rate in exponential phase in the insect versus in culture, P. luminescens appears to enjoy relatively uninhibited growth during the initial stages of infection. Further, the expression of the candidate virulence factors appears to show the same relative timing and pattern (e.g., Tcd is expressed earlier than Tca). These findings have two major implications. First, they confirm that the candidate virulence factors, the Tc toxins and the Prt protease, are expressed during infection. Second, they begin to correlate patterns of expression of putative virulence factors in a flask (in vitro) with those detectable within an actual insect infection (in vivo).
Implications for Photorhabdus as a model system.
As noted by Jander et al. in their studies of P. aeruginosa infections in insects, bacterium-insect systems have several advantages for the study of infection (11). First, several basic components of the infection process (such as toxin production, cell adhesion, and invasion) are likely to be important in both vertebrates and invertebrates. Second, large numbers of insect hosts can be infected easily and repeatedly. Third, the insect immune system is well understood, having both cellular and humoral responses like the vertebrate immune system. The results presented here add to these advantages by showing how the pattern of expression of individual bacterial virulence factors can also be readily tracked during the course of insect infections. Ultimately, this will help us clarify which aspects of the insect immune response these virulence factors are designed to overcome. Together, these advantages make the Photorhabdus-insect model a rapid and versatile system for studying bacterial virulence.
ACKNOWLEDGMENT
This work was supported by a grant to R.F.-C. from the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Bowen D, Blackburn M, Rocheleau T, Grutzmacher C, ffrench-Constant R H. Secreted proteases from Photorhabdus luminescens: separation of the extracellular proteases from the insecticidal Tc toxin complexes. Insect Biochem Mol Biol. 2000;30:69–74. doi: 10.1016/s0965-1748(99)00098-3. [DOI] [PubMed] [Google Scholar]
- 2.Bowen D, Rocheleau T A, Blackburn M, Andreev O, Golubeva E, Bhartia R, ffrench-Constant R H. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science. 1998;280:2129–2132. doi: 10.1126/science.280.5372.2129. [DOI] [PubMed] [Google Scholar]
- 3.Bowen D J, Ensign J C. Purification and characterization of a high-molecular-weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl Environ Microbiol. 1998;64:3029–3035. doi: 10.1128/aem.64.8.3029-3035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colepicolo P, Cho K W, Poinar G O, Hastings J W. Growth and luminescence of the bacterium Xenorhabdus luminescens from a human wound. Appl Environ Microbiol. 1989;55:2601–2606. doi: 10.1128/aem.55.10.2601-2606.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehlers R U, Niemann I. Molecular identification of Photorhabdus luminescens strains by amplification of specific fragments of the 16S ribosomal DNA. Syst Appl Microbiol. 1998;21:509–519. doi: 10.1016/S0723-2020(98)80063-5. [DOI] [PubMed] [Google Scholar]
- 6.ffrench-Constant R H, Waterfield N, Burland V, Perna N T, Daborn P J, Bowen D, Blattner F R. A genomic sample sequence of the entomopathogenic bacterium Photorhabdus luminescens W14: potential implications for virulence. Appl Environ Microbiol. 2000;66:3310–3329. doi: 10.1128/aem.66.8.3310-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forst S, Dowds B, Boemare N, Stackebrandt E. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol. 1997;51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 8.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu K, Webster J M. Antibiotic production in relation to bacterial growth and nematode development in Photorhabdus-Heterorhabditis infected Galleria mellonella larvae. FEMS Microbiol Lett. 2000;189:219–223. doi: 10.1111/j.1574-6968.2000.tb09234.x. [DOI] [PubMed] [Google Scholar]
- 10.Hurst M R, Glare T R, Jackson T A, Ronson C W. Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J Bacteriol. 2000;182:5127–5138. doi: 10.1128/jb.182.18.5127-5138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jander G, Rahme L G, Ausubel F M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol. 2000;182:3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahajan-Miklos S, Rahme L G, Ausubel F M. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol Microbiol. 2000;37:981–988. doi: 10.1046/j.1365-2958.2000.02056.x. [DOI] [PubMed] [Google Scholar]
- 13.Mathews S A, Volp K M, Timms P. Development of a quantitative gene expression assay for Chlamydia trachomatis identified temporal expression of sigma factors. FEBS Lett. 1999;458:354–358. doi: 10.1016/s0014-5793(99)01182-5. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt T M, Kopecky K, Nealson K H. Bioluminescence of the insect pathogen Xenorhabdus luminescens. Appl Environ Microbiol. 1989;55:2607–2612. doi: 10.1128/aem.55.10.2607-2612.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterfield N R, Bowen D J, Fetherston J D, Perry R D, ffrench-Constant R H. The toxin complex genes of Photorhabdus: a growing gene family. Trends Microbiol. 2001;9:185–191. doi: 10.1016/s0966-842x(01)01978-3. [DOI] [PubMed] [Google Scholar]