Abstract
In Synechocystis sp. strain PCC 6803, the genes encoding the proteins involved in nitrate assimilation are organized into two transcription units, nrtABCD-narB and nirA, the expression of which was repressed by ammonium and induced by inhibition of ammonium assimilation, suggesting involvement of NtcA in the transcriptional regulation. Under inducing conditions, expression of the two transcription units was enhanced by nitrite, suggesting regulation by NtcB, the nitrite-responsive transcriptional enhancer we previously identified in Synechococcus sp. strain PCC 7942. The slr0395 gene, which encodes a protein 47% identical to Synechococcus NtcB, was identified as the Synechocystis ntcB gene, on the basis of the inability of an slr0395 mutant to rapidly accumulate the transcripts of the nitrate assimilation genes upon induction and to respond to nitrite. While Synechococcus NtcB strictly requires nitrite for its action, Synechocystis NtcB enhanced transcription significantly even in the absence of nitrite. Whereas the Synechococcus ntcB mutant expresses the nitrate assimilation genes to a significant level in an NtcA-dependent manner, the Synechocystis ntcB mutant showed only low-level expression of the nitrate assimilation genes, indicating that NtcA by itself cannot efficiently promote expression of these genes in Synechocystis. Activities of the nitrate assimilation enzymes in the Synechocystis ntcB mutant were consequently low, being 40 to 50% of the wild-type level, and the cells grew on nitrate at a rate approximately threefold lower than that of the wild-type strain. These results showed that the contribution of NtcB to the expression of nitrate assimilation capability varies considerably among different strains of cyanobacteria.
In cyanobacteria, expression of the genes encoding the proteins involved in uptake and reduction of nitrate, i.e., nrtABCD or nrtP for the nitrate-nitrite transporter (NRT), narB for nitrate reductase (NR), and nirA for nitrite reductase (NiR), is negatively regulated by ammonium (3, 7, 19, 23, 26, 27). These genes are usually clustered on the genome and in Synechococcus sp. strain PCC 7942 and Anabaena sp. strain PCC 7120, organized into a large operon, nirA-nrtABCD-narB (nirA operon) (3, 7, 22, 27). Including the nitrate assimilation genes, cyanobacteria have a number of ammonium-repressible genes related to nitrogen metabolism. Expression of the ammonium-repressible genes commonly requires a Crp-type transcriptional regulator protein, NtcA (28; see reference 11 for a review). Thus, the ammonium-promoted regulation of the nitrate assimilation genes is a part of global nitrogen control in cyanobacteria.
In addition to ammonium-promoted regulation, positive regulation by nitrite of the nitrate assimilation operon has been found in Plectonema boryanum and Synechococcus sp. strain PCC 7942 (14). Studies in Synechococcus sp. strain PCC 7942 showed that the nitrite-promoted regulation is specific to the nirA operon and is mediated by a LysR family protein, NtcB (2). NtcB does not promote transcription by itself but upregulates transcription when transcription is induced by the action of NtcA in the presence of nitrite, either exogenously supplied or endogenously generated by nitrate reduction (14, 16); in other words, NtcB acts as a nitrite-dependent enhancer of nirA operon expression. NtcB is not essential for expression of the nitrate assimilation enzymes and for growth of the Synechococcus strain with nitrate as the nitrogen source (25). Recent studies with Anabaena sp. strain PCC 7120 (6), however, put forward a considerably different view of the role of NtcB in regulation of the nitrate assimilation operon. NtcB appears to be essential for expression of the nitrate assimilation enzymes; NtcB was shown to have nitrite-independent activity in upregulation of nirA operon transcription, which result was taken as evidence for the absence of any specific role of nitrite in regulation of nirA operon expression (6).
In the present study, we identified the ntcB gene of Synechocystis sp. strain PCC 6803, in which cyanobacterium the nitrate assimilation genes constitute two separate loci, and studied its function by construction and characterization of an insertional mutant of the gene. Unlike Synechococcus sp. strain PCC 7942, in which NtcA by itself activates transcription of the nitrate assimilation operon to a significant level, Synechocystis sp. strain PCC 6803 is shown to require NtcB for high-level expression of the nitrate assimilation genes. NtcB is nevertheless shown to be nonessential for expression of the activities of the nitrate assimilation enzymes. It is also shown that Synechocystis NtcB mediates the response to nitrite, although it upregulates transcription of the target genes even in the absence of nitrite. Common features and strain-specific diversities of the NtcB-mediated regulation in cyanobacteria are discussed.
MATERIALS AND METHODS
Strains and growth conditions.
The glucose-tolerant derivative of Synechocystis sp. strain PCC 6803, which was isolated by Williams and has been commonly used for photosynthesis research (31), and the mutant strains derived therefrom (see below) were grown photoautotrophically at 30°C under CO2-sufficient conditions as previously described (26). Continuous illumination was provided by fluorescent lamps at 100 μmol of photons m−2 s−1. The basal medium used was a nitrogen-free medium obtained by modification of BG11 medium (24) as previously described (26). Ammonium-containing medium and nitrate-containing medium were prepared by addition of 3.75 mM (NH4)2SO4 and 15 mM KNO3 to the basal medium, respectively. Both media were buffered with 20 mM HEPES-KOH (pH 8.2). When appropriate, kanamycin and spectinomycin were added to the media at 10 μg ml−1.
Transcription of the nitrate assimilation genes was induced by treatment of ammonium-grown cyanobacterial cells with l-methionine-d,l-sulfoximine (MSX), an inhibitor of glutamine synthetase, or by transfer of the ammonium-grown cells to nitrogen-free medium. MSX was added to cyanobacterial cultures in the mid-logarithmic phase of growth with or without simultaneous addition of NaNO2. The final concentrations of MSX and NaNO2 were 0.1 and 5 mM, respectively. For transfer of the cells to nitrogen-free medium, the ammonium-grown cells were collected by centrifugation at 5,000 × g for 5 min at 25°C, washed twice with the basal medium by resuspension and recentrifugation, and inoculated into the basal medium.
Insertional inactivation of sll1454 and slr0395
For construction of a mutant of sll1454 (narB), a DNA fragment carrying the entire sll1454 coding region (nucleotides −2 to +2145 with respect to the translation start site) was amplified by PCR using the Synechocystis chromosomal DNA as the template and cloned into pT7Blue T-Vector. The plasmid was digested with PacI and MscI to remove a 0.9-kbp internal segment of the cloned sll1454 gene. After blunting of the termini, the linearized plasmid was ligated with a spectinomycin resistance gene cassette excised from plasmid pRL463 (4) with HincII to yield plasmid pNARBS carrying an interrupted copy of sll1454. For construction of a mutant of slr0395 (ntcB), a 1.4-kbp DNA fragment carrying the entire slr0395 coding region (nucleotides −155 to +1201 with respect to the translation start site) was amplified by PCR and cloned into pT7Blue T-Vector. The plasmid was digested with NheI and StyI to remove a 0.5-kbp internal segment of slr0395. The linearized plasmid and a 1.3-kbp kanamycin resistance gene cassette, which had been excised from plasmid pUC4K (29) with BamHI, were mixed and ligated after blunting of the termini of the two fragments. The resulting plasmid, pNTCBK, carried an interrupted slr0395 gene. The plasmids pNARBS and pNTCBK were used to transform the wild-type Synechocystis strain through homologous recombination to spectinomycin and kanamycin resistance, respectively, to obtain an sll1454 insertional mutant (SNAR1) and an slr0395 insertional mutant (SNIC1).
Isolation and analysis of DNA and RNA.
Chromosomal DNA was extracted and purified from Synechocystis sp. strain PCC 6803 cells as described by Williams (31). Total RNA was extracted and purified from the cyanobacterial cells by the method of Aiba et al. (1). For Northern hybridization analysis, RNA samples (10 μg per lane) were denatured by treatment with formamide, fractionated by electrophoresis on 1.2% agarose gels that contained formaldehyde, and transferred to positively charged nylon membranes (Hybond N+; Amersham). For dot hybridization analysis, 1.25- and 2.5-μg aliquots of each of the denatured RNA samples were spotted on the nylon membranes with a dot blot apparatus. The blots were allowed to hybridize as described by Imamura et al. (12) with the following probes: a 1,674-bp DNA fragment carrying the entire nirA gene, extending from nucleotide −13 to 1642 with respect to the translation start site; an 889-bp nrtC fragment corresponding to nucleotides −69 to 820 of the coding region; a 2,149-bp DNA fragment carrying the entire narB gene, extending from nucleotide −2 to 2147 with respect to the translation start site; a 455-bp ntcA fragment corresponding to nucleotides 165 to 619 of the coding region; and a 0.5-kbp NheI-StyI fragment of the ntcB gene. The DNA probes other than the ntcB-specific probe were prepared by amplification by PCR of the respective sequences, with genomic DNA from Synechocystis sp. strain PCC 6803 as the template. The double-stranded DNA probes were labeled with 32P as described by Feinberg and Vogelstein (5). The hybridization signals were detected by autoradiography on X-ray film or by a Bio-Image analyzer (Fuji Photo Film). The radioactivity of the RNA dots was quantified with a Bio-Image analyzer. Primer extension analyses of the transcripts from the nirA gene and the nrt operon were carried out using 10 μg of total RNA samples as the template and 4 pmol of the following oligonucleotides as the primers: a 27-mer oligonucleotide complementary to bases 55 through 81 of the nirA coding region and a 22-mer oligonucleotide complementary to bases 132 through 153 of the nrtA coding region. One-fortieth and one-thirteenth of the reaction products obtained with the nirA- and nrtA-specific primers, respectively, were electrophoresed on a gel containing 6% polyacrylamide and 8.3 M urea to determine the sizes and amounts of the extension products.
Measurements of nitrate uptake.
Nitrate uptake by Synechocystis cells was measured at 30°C in the light by monitoring the changes in concentrations of nitrate in the medium as described previously (17) except that the pH of the assay medium was 8.2. For the assays, cells were collected by centrifugation, washed with nitrogen-free medium, and resuspended in nitrogen-free medium at a chlorophyll concentration of 5 μg ml−1. Nitrate was then added to the cell suspensions to a final concentration of ca. 0.1 mM, and the rate of nitrate uptake was calculated from the linear decrease of nitrate concentration in the medium with respect to time.
Other methods.
NR and NiR activities were determined at 30°C, using toluene-permeabilized cells with dithionite-reduced methylviologen as the electron donor (9, 10). Chlorophyll levels were determined according to the method of Mackinney (15).
RESULTS
Identification of the Synechocystis genes related to nitrate assimilation.
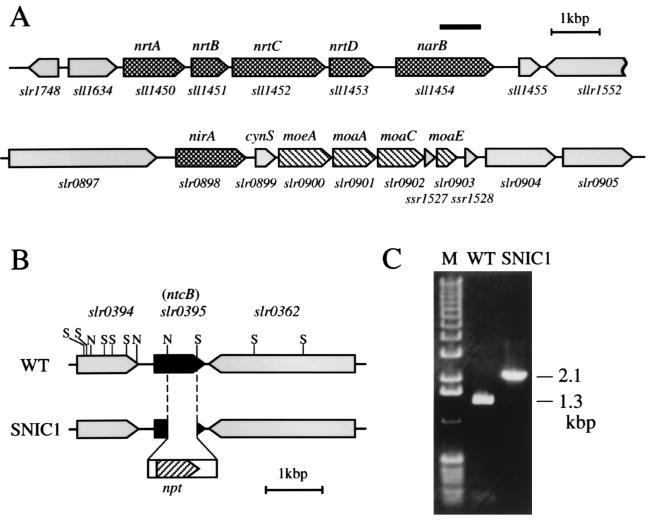
Figure 1A shows the map of the genomic regions carrying the nitrate assimilation genes of Synechocystis sp. strain PCC 6803. The sll1454 gene, encoding a protein 61% identical to NR of Synechococcus sp. strain PCC 7942, was identified as the NR gene (narB) of the Synechocystis strain, because an insertional mutant of this gene, SNAR1, exhibited no NR activity and failed to grow on nitrate (data not shown). The four genes sll1450, sll1451, sll1452, and sll1453, located upstream of the narB gene, were identified as the nitrate transporter genes nrtA, nrtB, nrtC, and nrtD, respectively, because they are the most similar among the genes of Synechocystis sp. strain PCC 6803 (13) to the Synechococcus nrtA, nrtB, nrtC, and nrtD genes (21, 22), respectively, and also because modification of sll1452 abolished ammonium-promoted regulation of nitrate uptake (Kobayashi et al., unpublished results). slr0898 was identified as the NiR gene (nirA) of Synechocystis since it encodes a protein 66 to 71% identical to the nitrite reductase protein (NirA) of various strains of cyanobacteria (3, 20, 26, 27, 30). The Synechocystis nirA gene is located upstream of the cynS gene for cyanase (8), which in turn is located upstream of the putative molybdenum cofactor biosynthesis genes (Fig. 1A).
FIG. 1.
(A) Map of the nrt and nir regions of the genome of Synechocystis sp. strain PCC 6803. The genes encoding NRT, NR, and NiR are indicated by checkered bars. The putative molybdenum cofactor biosynthesis genes are indicated by hatched bars. The bar above the sll1454 gene shows the region replaced by an antibiotic resistance gene cassette to construct the SNR1 mutant. (B) Structure of the slr0395 genomic region of the wild-type strain (WT) and the SNIC1 mutant of Synechocystis sp. strain PCC 6803. The open bar represents the kanamycin resistance gene cassette, with the hatched bar showing the location and orientation of the kanamycin resistance gene (npt). Abbreviations for restriction endonuclease sites: S, StyI; N, NheI. The gene organization in Synechocystis was obtained from CyanoBase (http://www.kazusa.or.jp/cyano/cyano.html). (C) Electrophoretic profiles showing the PCR products amplified from chromosomal DNAs of the wild-type strain and the slr0395 mutant SNIC 1, using a forward primer specific to the slr0394-slr0395 intergenic region and a reverse primer specific to the 3′ region of slr0362. Lane M shows the molecular size markers (1-kbp ladder; BRL).
Nitrogen regulation of the nitrate assimilation genes in Synechocystis sp. strain PCC 6803.
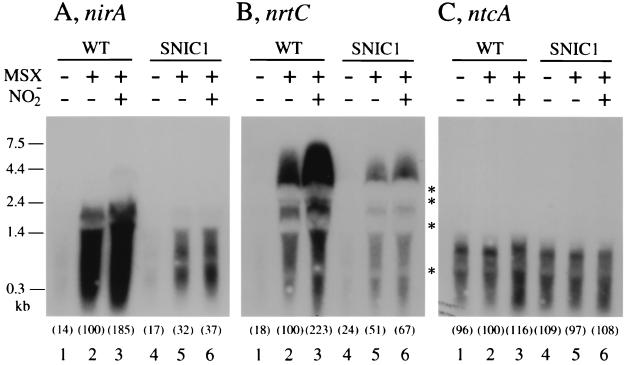
Northern blot analysis, using probes specific to nirA (Fig. 2A, lanes 1 to 3) and nrtC (Fig. 2B, lanes 1 to 3), showed that expression of the two genes is negligible in ammonium-grown cells (lanes 1) and is induced by inhibition of ammonium assimilation with MSX (lanes 2). Expression of these genes was induced also by transfer of the ammonium-grown cells to nitrogen-free medium (data not shown). These results indicated that the nitrate assimilation genes are ammonium-repressible genes which are activated simply by derepression. The abundance of the transcripts was greater when nitrite was added simultaneously with MSX to the cell suspensions (lanes 3) than when MSX alone was added (lanes 2), showing a positive effect of nitrite on transcription. The abundance of the ntcA transcript was, on the other hand, practically unaffected by MSX and nitrite (Fig. 2C, lanes 1 to 3), showing that the gene is transcribed constitutively in Synechocystis sp. strain PCC 6803.
FIG. 2.
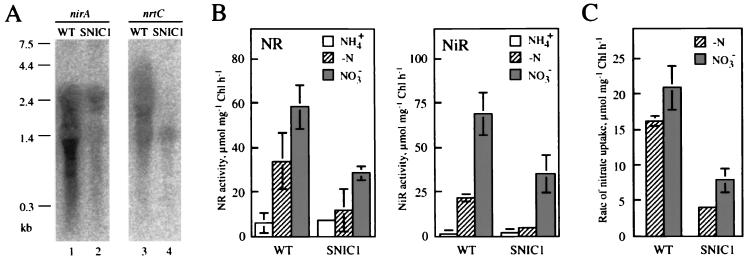
Northern blot analysis of RNA from Synechocystis sp. strain PCC 6803 showing the effects of MSX and nitrite on transcription of nirA (A), nrtC (B), and ntcA (C) in the wild-type (WT) strain (lanes 1 to 3) and the SNIC1 mutant (lanes 4 to 6). Cells were grown with ammonium, the culture was separated into three portions, and total RNA was extracted from the cells before (lanes 1 and 4) and 60 min after the following treatments: addition of MSX (lanes 2 and 5) and addition of MSX plus nitrite (lanes 3 and 6). The numbers in parentheses indicate relative abundances of mRNAs as determined by quantitation of radioactivity using a Bio-Image analyzer (Fuji Photo Film). Asterisks between panels B and C indicate the positions of the rRNA bands as determined by staining of the blots with methylene blue.
In the Northern hybridization analysis, the nrtC-specific probe yielded smeared hybridization signals extending from 0.25 to 7.5 kb, with exclusion of radioactivity in the regions of the rRNA bands (Fig. 2B). The results indicated the presence of a large transcription unit, the transcript from which is rapidly turned over. Since a narB-specific probe yielded essentially the same hybridization profile (data not shown) and since the size of the largest signal was close to the calculated size of the nrtABCD-narB gene cluster, 7.9 kb, we concluded that the nrtABCD-narB genes are cotranscribed as an operon. In the case of the nirA-specific probe, which also yielded smeared hybridization signals (Fig. 2A), estimation of the size of the hybridization signals was difficult because of disturbance of the hybridization profile by the rRNA bands (Fig. 2A, lanes 2 and 3). In most experiments, however, smeared signals of <1.5 kb were observed, suggesting that nirA constitutes a monocistronic transcription unit.
Identification of the ntcB gene of Synechocystis
The positive effect of nitrite on transcription of the nitrate assimilation genes in Synechocystis sp. strain PCC 6803 suggested involvement of NtcB in regulation of the genes. In Synechococcus sp. strain PCC 7942 and in Anabaena sp. strain PCC 7120, the ntcB gene is located in the DNA region upstream of the nitrate assimilation operon (6, 25). In Synechocystis sp. strain PCC 6803, no ntcB-like gene is located around the nirA gene or the nrtABCD-narB operon; however, a gene (slr0395) encoding a protein 47 and 53% identical to NtcB of the Synechococcus and Anabaena strains, respectively, is located between the slr0394 and slr0362 genes (Fig. 1B). Northern hybridization analysis using an slr0395-specific probe showed smeared hybridization signals of <1 kb, suggesting that the gene constitutes a monocistronic transcription unit (data not shown). To determine whether slr0395 represents the ntcB gene of Synechocystis, a mutant (SNIC1) was constructed by replacing a 0.5-kbp internal segment of the slr0395 coding region with a 1.3-kbp kanamycin resistance gene cartridge (Fig. 1B). PCR amplification of the slr0395 genomic region of the SNIC1 mutant, using a set of primers that amplifies a 1.3-kbp DNA fragment from the wild-type DNA, yielded only a 2.1-kbp DNA fragment, showing that complete segregation of the mutant genome was achieved in SNIC1 (Fig. 1C).
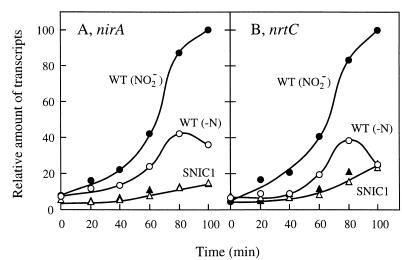
Northern blot analysis showed that the amounts of transcripts from the nirA gene and the nrt operon were much smaller in the slr0395 mutant than in the wild-type strain after induction with MSX treatment (compare lane 2 and lane 5 in Fig. 2A and B). Also, there was no significant effect of nitrite on the level of mRNA (lanes 6). Time course experiments, using dot blots of total RNA samples for quantitative analysis of the transcripts, showed that the wild-type cells rapidly accumulate mRNA from the nirA gene and the nrt operon between 40 and 80 min after addition of MSX to ammonium-utilizing cells (Fig. 3). The maximal levels of mRNA accumulation were much higher in the presence of nitrite than in its absence, confirming the positive effect of nitrite on transcription. The mutant cells, on the other hand, showed only slow, gradual accumulation of mRNA from the two transcription units irrespective of the presence of nitrite (triangles). These results indicated that slr0395 is required for high-level expression of the nitrate assimilation genes and its enhancement in response to nitrite. We therefore identified slr0395 as the ntcB gene of Synechocystis sp. strain PCC 6803. The higher level of the transcripts in the wild-type cells than in the mutant cells, observed in the absence of nitrite, showed that NtcB of Synechocystis sp. strain PCC 6803 is distinct from that of Synechococcus sp. strain PCC 7942 and similar to that of Anabaena sp. strain PCC 7120 in that it is active in upregulation of the nitrate assimilation genes even in the absence of nitrite.
FIG. 3.
Changes in the abundance of the nirA (A) and nrtC (B) transcripts after addition of MSX to the ammonium-grown cultures of the wild-type strain (○ and ●) and the mutant (▵ and ▴), with (● and ▴) and without (○ and ▵) simultaneous addition of nitrite. The amounts of the nirA and nrtC transcripts were quantitated by dot hybridization analysis with 1.25 μg of RNA per dot and are shown relative to the maximum level in the wild-type cells treated with MSX and nitrite.
Nitrate assimilation capacity of the ntcB mutant.
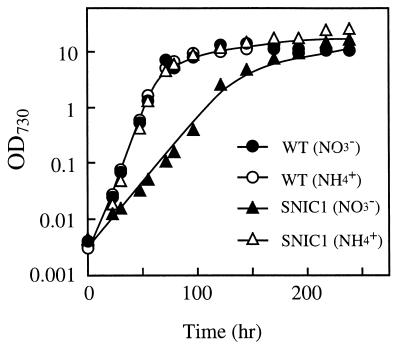
The ntcB mutant grew as fast as the wild-type cells in ammonium-containing medium with a generation time of 5.5 h under the given conditions (Fig. 4). While the wild-type strain grew in nitrate-containing medium with the same generation time as in ammonium-containing medium, the mutant grew very slowly in nitrate-containing medium, with a generation time of 17.4 h. The final cell density in the mutant cultures after prolonged growth in nitrate-containing medium was nevertheless equivalent to that in the cultures of the wild-type strain. These results showed that the mutant has an impaired capacity to assimilate nitrate.
FIG. 4.
Growth curves of the wild-type strain (● and ○) and the SNIC1 mutant (▴ and ▵) of Synechocystis sp. strain PCC 6803 in media containing nitrate (● and ▴) and ammonium (○ and ▵) as the sole source of nitrogen.
When wild-type cyanobacterial cells are transferred from ammonium-containing medium to nitrate-containing medium, the abundance of mRNA of the nitrate assimilation genes increases to a high level and then, as the activities of NR and NiR increase, declines to a low steady-state level within several hours (26, 27). Cells of Synechocystis sp. strain PCC 6803 also showed similar changes in the abundance of mRNA after transfer from ammonium-containing medium to nitrate-containing medium (data not shown). Figure 5A shows that the steady-state levels of mRNA from the nirA gene and the nrt operon of Synechocystis sp. strain PCC 6803 are lower in the ntcB mutant than in the wild-type strain. The NR and NiR activities of the nitrate-utilizing mutant cells were about 50% of the corresponding wild-type levels (Fig. 5B). The rate of nitrate uptake by intact cells was also low in the mutant, being ca. 40% of the wild-type level (Fig. 5C). These results suggested that inactivation of ntcB lowered the steady-state level of expression of the nitrate assimilation genes and hence reduced the capacity of nitrate assimilation.
FIG. 5.
Expression of the nitrate assimilation activities in the wild-type strain and the ntcB-deficient mutant. (A) Northern hybridization analysis of total RNA, comparing the abundance of mRNA from the nitrate assimilation genes in the two strains growing with nitrate. Cells grown with ammonium were transferred to nitrate-containing medium, and total RNA was isolated after 18 h of growth in nitrate-containing medium. (B) NR and NiR activities of wild-type and SNIC1 cells grown under different nitrogen conditions. Cells grown with ammonium (open bars), ammonium-grown cells subsequently grown for 18 h in nitrate containing medium (gray bars), and ammonium-grown cells subjected to 4 h of nitrogen starvation (hatched bars) were used for the assays. (C) Nitrate uptake activity of intact cells of the wild-type strain and the SNIC1 mutant after 18-h incubation in nitrate-containing medium (gray bars) or 4-h incubation in nitrogen-free medium (hatched bars) of ammonium-grown cells. The results shown in panels B and C are the averages and standard deviations (error bars) from three separate experiments with independent cultures.
Figure 5B also shows NR and NiR activities of the cells grown with ammonium and then subjected to a 4-h incubation in nitrogen-free medium. Since Synechocystis sp. strain PCC 6803 has no ability to fix N2, no external source of nitrogen is available for the cells under these conditions. During nitrogen starvation, NR and NiR activities of the wild-type cells increased to levels corresponding to ca. 50 and 30%, respectively, of the cells utilizing nitrate as the nitrogen source. The mutant, on the other hand, showed only a small increase in the NR and NiR activity levels after the same treatment. As a result, the NR and NiR activities in the nitrogen-starved mutant cells were only 35 and 25%, respectively, of the corresponding wild-type levels. The rate of nitrate uptake by the nitrogen-starved mutant cells was also low, corresponding to ca. 25% of that of the wild-type cells (Fig. 5C). Expression of the nitrate assimilation activities was thus enhanced by the presence of ntcB in the absence of an external nitrogen source. This contrasted with the results obtained previously with Synechococcus sp. strain PCC 7942, in which the wild-type strain expressed lower NR and NiR activities than the ntcB mutant under the conditions of nitrogen starvation (2).
Structure of the promoters of the nirA gene and the nrtABCD-narB operon.
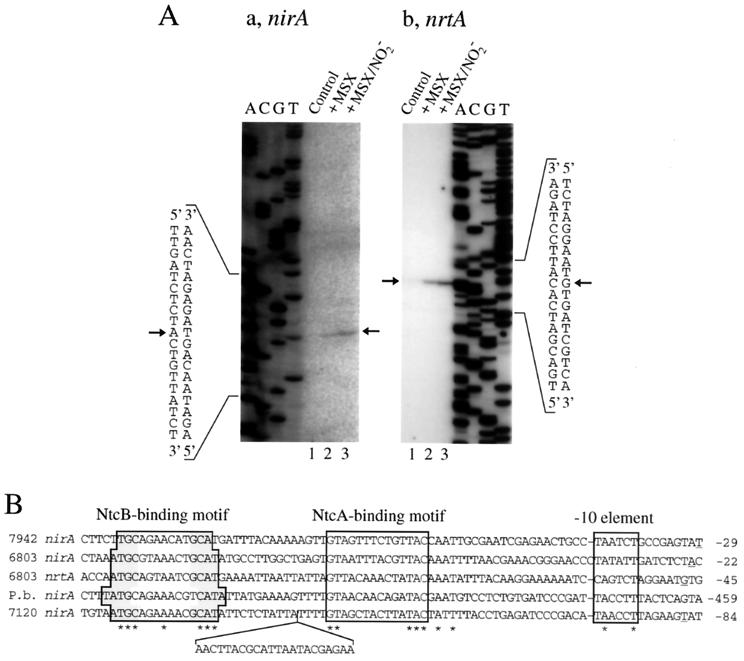
Figure 6A shows the results of primer extension analyses of RNA samples isolated from various nitrogen conditions, obtained using oligonucleotides complementary to the nirA and nrtA coding sequences, respectively. In accordance with the results of Northern hybridization analysis (Fig. 2), no extension product was detected when the RNA samples from ammonium-grown cells were used as templates (Fig. 6A, lanes 1). A single extension product was obtained for each of nirA and nrtA when RNA samples from MSX-treated cells were used as templates (lanes 2). Larger amounts of the extension products were obtained from RNA samples isolated from the cells treated with MSX plus nitrite (lanes 3), confirming the positive effect of nitrite on expression of the nirA and nrtA genes. From the sizes of the extension products, the A residue located 23 bases upstream from the nirA initiation codon and the G residue located 47 bases upstream from the nrtA initiation codon were identified as the transcription start position of the nirA gene and the nrt operon, respectively. The nucleotide sequences upstream of the nirA and nrtA transcription start sites conformed to the consensus sequence of the NtcA-dependent, ammonium-repressible promoters (11), having an NtcA-binding motif (GTAN8TAC) 22 and 20 bases upstream from the −10 promoter element, respectively (Fig. 6B). Also, both of the two Synechocystis promoters contained an inverted repeat carrying a LysR motif (TN11A), centered at position −24 with respect to the NtcA-binding motif, which has been shown to be present in the promoters of the nirA operon of other strains of cyanobacteria (16) and supposed to constitute the binding site for NtcB (6, 16).
FIG. 6.
(A) Primer extension analysis of the expression of the nirA (a) and nrtA (b) genes in the wild-type strain of Synechocystis sp. strain PCC 6803, showing the effects of MSX and nitrite. Cells were grown with ammonium, the culture was separated into three portions, and total RNAs extracted from the cells before (lanes 1) and 60 min after the following treatments were used for the assays: addition of MSX (lanes 2) and addition of MSX plus nitrite (lanes 3). The arrows indicate the extension products and the deduced transcription start sites. (B) Alignment of the promoters of the cyanobacterial nitrate assimilation genes. Only those promoters known to be regulated by NtcB and/or nitrite are included. The regions of the putative NtcB-binding site with a LysR motif (T-N11-A), the NtcA-binding site, and the −10 sequence are boxed. The transcription start position is underlined. The nucleotides forming an inverted repeat with the LysR motif are shaded. Asterisks indicate the nucleotides conserved in the five promoter sequences. Gaps have been introduced into the sequences to maintain optimal alignment. The numbers to the right of the sequences indicate the positions of the rightward-most bases with respect to the translation start site. Strains: 7942, Synechococcus sp. strain PCC 7942; 6803, Synechocystis sp. strain PCC 6803; 7120, Anabaena sp. strain PCC 7120; P.b., Plectonema boryanum.
DISCUSSION
In cyanobacteria, expression of the nitrate assimilation genes is induced simply by inhibition of ammonium assimilation or by withdrawal of combined nitrogen from medium, showing no requirement for nitrate or nitrite (3, 7, 26, 27). Nitrite has nevertheless been shown to enhance transcription of the nitrate assimilation operon in Synechococcus sp. strain PCC 7942 and Plectonema boryanum (14). Since the LysR family protein NtcB (25), which mediates the response to nitrite (2), does not promote transcription by itself and acts as a nitrite-responsive enhancer when transcription is promoted by NtcA (16), and since the activity of NtcA in promotion of transcription is subject to negative feedback by ammonium assimilation (28), the positive effect of NtcB-nitrite is diminished by the ammonium generated internally by nitrate reduction. The action of NtcB-nitrite is hence prominent in cells treated with inhibitors of ammonium assimilation (16). In the present study, using MSX-treated cells, we detected a positive effect of nitrite on expression of the two loci (nirA and nrtABCD-narB) required for nitrate assimilation of Synechocystis sp. strain PCC 6803 (Fig. 2 and 3). Nitrite-responsive, positive regulation of the nitrate assimilation genes has thus been demonstrated in three strains of cyanobacteria and seems to be common to most cyanobacterial strains. Since slr0395 was found to be involved in activation and the nitrite-responsive enhancement of transcription from the two transcription units (Fig. 2 and 3), we have identified the gene as ntcB of Synechocystis sp. strain PCC 6803.
In Synechococcus sp. strain PCC 7942, the positive effect of NtcB on nirA operon transcription is totally dependent on nitrite (2); when incubated in nitrogen-free medium, the levels of nirA operon transcription and of NR and NiR activities were higher in an ntcB deletion mutant than in the wild-type strain, suggesting that NtcB negatively regulates transcription of the nitrate assimilation operon in the absence of nitrite (2). In Synechocystis sp. strain PCC 6803, by contrast, the ntcB mutant SNIC1 showed much lower levels of nirA and the nrt operon expression and of NR and NiR activities than the wild-type strain in nitrogen-free medium (Fig. 2, 3, and 5), indicating that NtcB positively regulates transcription in the absence of nitrite as well as in its presence. Nitrite-independent activity of NtcB in upregulation of expression of the nitrate assimilation operon has been recently reported in Anabaena sp. strain PCC 7120 (6). Thus, in nitrogen-free medium, NtcB has opposite effects on transcription of the nitrate assimilation genes in different strains of cyanobacteria—positive effects in Synechocystis sp. strain PCC 6803 and Anabaena sp. strain PCC 7120 and a negative effect in Synechococcus sp. strain PCC 7942. The molecular basis of this difference is being investigated.
In Anabaena sp. strain PCC 7120, the nitrite-independent activity of NtcB to upregulate nirA operon expression was taken as evidence that NtcB does not mediate effects of nitrate or nitrite on transcription (6). However, the present results obtained with Synechocystis sp. strain PCC 6803 suggest that NtcB can be active in upregulation of transcription in the absence of nitrite and at the same time responsive to nitrite (Fig. 2 and 3). It should be noted that nitrate has been shown to have a positive effect on the activity levels of NR and NiR and the abundance of the nirA operon transcript in Anabaena sp. strain PCC 7120 (7, 18); the effect of nitrate may be due to enhancement of the activity of NtcB by the nitrite generated from nitrate. It has been shown not only in Synechococcus sp. strain PCC 7942 (16) but also in Anabaena sp. strain PCC 7120 (6) that positive regulation by NtcB requires the presence of NtcA. As discussed above, the positive effect of NtcB (and nitrite) would then be diminished by negative feedback through assimilation of internally generated ammonium. To draw a solid conclusion as to the presence or absence of nitrite-responsive regulation and the involvement of NtcB therein in Anabaena sp. stain PCC 7120, effects of nitrate and nitrite on nirA operon transcription need to be examined in cells treated with inhibitors of ammonium assimilation.
The nitrite-independent activity of NtcB in upregulation of transcription of the nitrate assimilation genes in Synechocystis sp. strain PCC 6803 was readily discernible because of the low level of transcription of the nitrate assimilation genes in the ntcB mutant (Fig. 2 and 3). This indicates that NtcA cannot promote high-level expression of the genes by itself and requires NtcB to attain high transcriptional activity. These results contrast with those obtained with Synechococcus sp. strain PCC 7942 (2); when induced with MSX in the absence of nitrite, the level of nirA operon transcription in the Synechococcus ntcB mutant was equivalent to that in the wild-type strain, indicating that NtcA by itself promotes high-level expression of nirA operon transcription. Thus, the contribution of NtcB to expression of the nitrate assimilation genes is much larger in Synechocystis sp. strain PCC 6803 than in Synechococcus sp. strain PCC 7942. It should be noted, however, that despite its large contribution to transcription of the nitrate assimilation genes, NtcB is not essential for expression of the nitrate assimilation activities in the Synechocystis strain (Fig. 5). Since ntcB is reported to be essential for expression of the NR and NiR activities in Anabaena sp. strain PCC 7120 (6), there appears to be a considerable variation among cyanobacteria in dependence on NtcB of expression of the nitrate assimilation activities. The underlying molecular mechanism and the physiological significance of the variation need to be clarified in future studies.
ACKNOWLEDGMENTS
This work was supported by grants-in-aid for Scientific Research in Priority Areas (09274101, 09274103, and 13206027) from the Ministry of Education, Science, Sports, and Culture, Japan.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Aichi M, Omata T. Involvement of NtcB, a LysR family transcription factor, in nitrite activation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1997;179:4671–4675. doi: 10.1128/jb.179.15.4671-4675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai Y, Wolk C P. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J Bacteriol. 1997;179:258–266. doi: 10.1128/jb.179.1.258-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg A, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 6.Frías J E, Flores E, Herrero A. Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors. Mol Microbiol. 2000;38:613–625. doi: 10.1046/j.1365-2958.2000.02156.x. [DOI] [PubMed] [Google Scholar]
- 7.Frías J E, Flores E, Herrero A. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:477–486. doi: 10.1128/jb.179.2.477-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harano Y, Suzuki I, Maeda S, Kaneko T, Tabata S, Omata T. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J Bacteriol. 1997;179:5744–5750. doi: 10.1128/jb.179.18.5744-5750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrero A, Flores E, Guerrero M G. Regulation of nitrate reductase levels in the cyanobacteria Anacystis nidulans, Anabaena sp. strain 7119, and Nostoc sp. strain 6719. J Bacteriol. 1981;145:175–180. doi: 10.1128/jb.145.1.175-180.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrero A, Guerrero M G. Regulation of nitrite reductase in the cyanobacterium Anacystis nidulans. J Gen Microbiol. 1986;132:2463–2468. [Google Scholar]
- 11.Herrero A, Muro-Pastor A M, Flores E. Nitrogen control in cyanobacteria. J Bacteriol. 2001;183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:2691–2696. doi: 10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi H, Aichi M, Suzuki I, Omata T. Positive regulation by nitrite of the nitrate assimilation operon in the cyanobacteria Synechococcus sp. strain PCC 7942 and Plectonema boryanum. J Bacteriol. 1996;178:5822–58525. doi: 10.1128/jb.178.19.5822-5825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackinney G. Absorption of light by chlorophyll solutions. J Biol Chem. 1941;140:315–322. [Google Scholar]
- 16.Maeda S, Kawaguchi Y, Ohe T, Omata T. cis-acting sequences required for NtcB-dependent, nitrite-responsive positive regulation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1998;180:4080–4088. doi: 10.1128/jb.180.16.4080-4088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda S, Omata T. Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in the transport of nitrate and nitrite. J Biol Chem. 1997;272:3036–3041. doi: 10.1074/jbc.272.5.3036. [DOI] [PubMed] [Google Scholar]
- 18.Martín-Nieto J, Herrero A, Flores E. Regulation of nitrate and nitrite reductases in dinitrogen-fixing cyanobacteria and Nif− mutants. Arch Microbiol. 1989;151:475–478. [Google Scholar]
- 19.Merchán F, Kindle K L, Llama M J, Serra J L, Fernández E. Cloning and sequencing of the nitrate transport system from the thermophilic, filamentous cyanobacterium Phormidium laminosum: comparative analysis with the homologous system from Synechococcus sp. PCC 7942. Plant Mol Biol. 1995;28:759–766. doi: 10.1007/BF00021199. [DOI] [PubMed] [Google Scholar]
- 20.Merchán F, Prieto R, Kindle K L, Llama M J, Serra J L, Fernández E. Isolation, sequence and expression in Escherichia coli of the nitrite reductase gene from the filamentous, thermophilic cyanobacterium Phormidium laminosum. Plant Mol Biol. 1995;27:1037–1042. doi: 10.1007/BF00037030. [DOI] [PubMed] [Google Scholar]
- 21.Omata T. Cloning and characterization of the nrtA gene that encodes a 45-kDa protein involved in nitrate transport in the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 1991;32:151–157. [Google Scholar]
- 22.Omata T, Andriesse X, Hirano A. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. PCC 7942. Mol Gen Genet. 1993;236:193–202. doi: 10.1007/BF00277112. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto T, Inoue-Sakamoto K, Bryant D A. A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol. 1999;181:7363–7372. doi: 10.1128/jb.181.23.7363-7372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanier R Y, Kunisawa R, Mandel M, Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki I, Horie N, Sugiyama T, Omata T. Identification and characterization of two nitrogen-regulated genes of the cyanobacterium Synechococcus sp. strain PCC7942 required for maximum efficiency of nitrogen assimilation. J Bacteriol. 1995;177:290–296. doi: 10.1128/jb.177.2.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki I, Kikuchi H, Nakanishi S, Fujita Y, Sugiyama T, Omata T. A novel nitrite reductase gene from the cyanobacterium Plectonema boryanum. J Bacteriol. 1995;177:6137–6143. doi: 10.1128/jb.177.21.6137-6143.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki I, Sugiyama T, Omata T. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 1993;34:1311–1320. [Google Scholar]
- 28.Vega-Palas M A, Flores E, Herrero A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 29.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Li H, Post A F. Nitrate assimilation genes of the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp. strain WH9601. J Bacteriol. 2000;182:1764–1767. doi: 10.1128/jb.182.6.1764-1767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams J G K. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]