Abstract
In the Lyme disease spirochetes, both the ospE and vlsE gene families have been demonstrated to undergo sequence variation during infection. To further investigate the mechanisms associated with the generation of vls variation, single-nucleotide polymorphism and subsequent DNA sequence analyses were performed on the vlsE gene and its paralog, BBJ51, a related gene with a frameshift mutation. These analyses focused on a series of postinfection clonal populations obtained from mice infected with Borrelia burgdorferi B31MIpc or its clonal derivative, B31MIc53. vlsE, but not BBJ51, was found to undergo sequence changes during infection. Consistent with that reported previously (J.-R. Zhang et al., Cell 89:275–285, 1997) many of the sequence changes appear to have arisen through gene conversion events and to be localized to the variable regions of vlsE. However, analysis of the vlsE nucleotide sequences revealed that some sequence changes were the result of point mutations, as these changes did not have potential contributing sources in the vls cassettes. To determine if sequence changes accumulate in vlsE over long-term infection, the vlsE genes of clonal populations recovered after 7 months of infection in mice were analyzed. While new sequence changes developed, a significant number of these changes resulted in the restoration of the vlsE sequence of the original infecting clone. In addition, we noted that some positions within the variable regions (VR) are stable even though the cassettes possess residues that could contribute to sequence variation through gene conversion. These analyses suggest that the total number of amino acid sequence changes that can be maintained by VlsE levels off during infection. In summary, in this report we demonstrate that the development of point mutations serves as a second mechanism by which vlsE sequence variation can be generated and that the capacity for vlsE variation, while still significant, is less than previously postulated.
Lyme disease is a chronic infection caused by certain species of the Borrelia burgdorferi sensu lato complex (2, 17). Even though the Lyme disease spirochetes elicit a rigorous immune response during early infection, the spirochetes survive and persist. The outer surface protein-encoding ospE and vls gene families of B. burgdorferi have been shown to undergo sequence changes during infection, and based on this it has been postulated that OspE and VlsE play a role in immune system evasion (18, 21). The vls system has been extensively characterized in B. burgdorferi B31-5A3 (19–21). In this clone a single expression locus for the vls sequences, vlsE1, resides on a 28-kb linear plasmid (lp28-1). Adjacent to the expression site are a series of partial vls gene cassettes. The vlsE gene has three defined domains: an N-terminus-encoding-invariable domain, an internal variable domain, and a C-terminus-encoding-invariable domain. The variable domain is further divided into six variable regions (VRs), which are separated by invariable regions (IRs) (19). Studies by Zhang and colleagues suggest that many of the sequence changes that arise in vlsE occur from events involving unidirectional gene conversion between the partial vlsE gene cassettes and the expression locus resulting in the specific modification of the VRs of vlsE (19–21).
In this study we have further assessed the kinetics and mechanisms involved in the induction of sequence changes in vlsE in mice infected with B. burgdorferi B31MIpc and its derivative clone, B31MIc53. Analysis of the vlsE variants that arose during infection revealed that many of the sequence changes that developed were likely due to gene conversion events. However, several examples of point mutation were also identified, indicating that a second mechanism for generating vlsE variation exists. Analysis of vlsE variants that developed during long-term infection revealed that within the vlsE VRs some positions undergo sequence change more readily than others. Some positions are either stable or have a tendency to revert back to the original parental vlsE sequence. Collectively, the data suggest that there is a limit to the net accumulation of amino acid changes that can be maintained in VlsE. This important observation suggests that the potential repertoire of VlsE variants, while significant, may not be as great as previously postulated (21).
MATERIALS AND METHODS
Bacterial isolates, cultivation, and experimental infection of mice.
B. burgdorferi B31MI, kindly provided to us by MedImmune Inc. (Gaithersburg, Md.), was used for these analyses since its genome sequence has been determined (5). All clones were cultivated at 33°C in BSK-H complete media (Sigma), and growth was monitored using dark-field microscopy. The isolation and general description of the postinfection clonal populations analyzed in this report have been previously described (18). In brief, a clone of B31MI, designated B31MIpc, was obtained by subsurface plating and used to infect C3H-HeJ mice. Ear punch biopsy samples (1 mm2) were collected at 3 months postinoculation, and spirochetes were cultivated from these samples in BSK-H media (Sigma) containing antibiotics (phosphomycin, 20 μg ml−1; amphotericin B, 2.5 μg ml−1; rifampin, 50 μg ml−1; Sigma). Aliquots of the cultures were then subsurface plated, with colonies becoming evident after 3 weeks. Well-isolated colonies were selected for further analysis. To analyze the stability of vlsE over long-term infection, B31MIc53 was employed. This clone was obtained through the cultivation and subsurface plating of spirochetes from an ear punch biopsy sample obtained from a C3H-HeJ mouse infected with B31MIpc. All methods associated with these analyses were identical to those described above.
Additional B. burgdorferi isolates used in this study were as follows: NY1-86, human erythema migrans isolate from a patient in New York; T2, Ixodes scapularis tick isolate from New York; CA4 and CA9, Ixodes pacificus tick isolates from California; 21343, a mouse isolate from Texas.
PCR analyses.
PCR analyses were performed using isolated DNA (50 ng) or the supernatant from lysed cells as templates. DNA was isolated as previously described (12). To obtain a template for PCR directly from actively growing cultures, a 100-μl culture aliquot was pelleted, washed with phosphate-buffered saline, resuspended in 100 μl of H2O, boiled for 10 min, and centrifuged to pellet the debris and the supernatant was collected. One microliter of the supernatant was used as the amplification template. All primers employed are described in Table 1. PCR was performed with Taq polymerase (Promega) for 30 cycles in an MJ Research PTC100 thermal cycler. Reaction volumes were 30 μl, and final primer set concentrations were 1 pmol of primer pair per μl. Cycling conditions were as follows: 1 cycle of 5 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 50°C, and 1.5 min at 72°C. To analyze the amplicons, 10 μl of each PCR mixture was electrophoresed in a 1.5% GTG-agarose gel using standard Tris-acetate-EDTA running buffer. For single-nucleotide polymorphism (SNP) and sequence analyses some amplicons were further purified using the Wizard system (Promega).
TABLE 1.
Oligonucleotide primers
| Probe | Oligonucleotide sequencea | Target site or function |
|---|---|---|
| vlsF2 | GTTCTTATCAACTGTAAAAGCG | 5′ end of BBJ41 (vlsE allele with an authentic frameshift) |
| vlsR2 | GTCTTGGCTATTTTTGTTAAGC | 3′ end of BBJ41 |
| vlsE292(+) | AGTAGTACGACGGGGAAACCAGA | Binds 5′ of the variable domain of vlsE |
| vlsE960(−) | ACTTTGCGAACTGCAGACTC | Binds 3′ of the variable domain of vlsE |
| LIC70(+) | GACGACGACAAGATGGCTGATAAGGACGACCCAACA | Tailed primer for LIC cloning; binds to the 5′ end of vlsE |
| LIC1080(−) | GAGGAGAAGCCCGGTATTCAAGGCAGGAGGTGTTTC | Tailed primer for LIC cloning; binds to the 3′ end of vlsE |
| LIC292(+) | GACGACGACAAGATGAGTACGACGGGGAAACCAGA | Tailed primer for LIC cloning; binds 5′ of the variable domain of vlsE |
| LIC969(−) | GAGGAGAAGCCCGGTACTTTGCGAACTGCAGAGTC | Tailed primer for LIC cloning; binds 5′ of the variable domain of vlsE |
Underlined sequences indicate the tails for ligase-independent cloning (LIC).
Two-dimensional PFGE.
To separate linear and circular plasmids, genomic DNA was fractionated using the clamped homogenous electric field Mapper system (Bio-Rad). The DNA was separated in the first dimension using pulsed-field gel electrophoresis (PFGE) (field inversion gel electrophoresis) and then in the second dimension by constant-voltage electrophoresis. Bacterial cells were embedded in 1% Incert-agarose (FMC) as previously described (3). To digest the DNA in the agarose plugs with StuI, the plugs were incubated for 1 h in 1× StuI restriction buffer (supplied by the manufacturer) and then the buffer was removed and replaced with 500 μl of fresh buffer containing 20 U of StuI and incubated overnight at 37°C. The buffer-enzyme was removed, and the agarose slices were equilibrated in 0.5× Tris-borate-EDTA at 37°C for 1 h. All plugs were then inserted into a 1.0% GTG-agarose gel and sealed with 1% Incert-agarose (FMC). Digested and intact genomic DNA was fractionated by two-state PFGE using the following parameters: run time, 3 h; voltage, 9 V/cm; angle, 120°; initial switch time, 0.9 s; final switch time, 0.9 s; ramping factor, linear; temperature, 14°C. After electrophoresis in the first dimension the gel was rotated 90° and electrophoresed with constant voltage (80 V) for 2.5 h. The DNA was then visualized by ethidium bromide staining prior to transfer onto membranes for hybridization analysis.
Southern hybridization analyses.
DNA fractionated by two-dimensional PFGE was transferred onto Hybond-N membranes using the VacuGene system as described by the manufacturer (Pharmacia). To assess the vlsE HaeIII restriction fragment length polymorphism patterns, DNA was isolated from B31MIpc as previously described (11). The DNA was digested with HaeIII as instructed by the supplier (New England Biolabs). The digested DNA was fractionated in 0.8% GTG-agarose gels using standard TAE buffer. The DNA was transferred onto Hybond-N membranes by vacuum blotting using the VacuGene system as described by the manufacturer (Pharmacia). PCR-generated probes were labeled by random prime labeling using the Random Primer DNA labeling system (Gibco-BRL) and [α-32P]dATP (3,000 Ci/mmol; NEN-DuPont). Hybridizations were conducted using conditions and buffers previously described (12) in a Hybaid hybridization oven.
Rapid screening for vls sequence changes using SNP analysis.
vlsE and BBJ51 were amplified from postinfection clonal populations using primer sets described in Table 1. The purified amplicons then served as the template for SNP analyses as previously described (18). The SNP approach is essentially a limited sequencing approach in that only one of the four dideoxynucleotide incorporation reactions is performed. Comparison of the resulting ladders serves as a rapid means for screening for sequence changes in the amplified genes. To perform the SNP analyses, the Excel sequencing kit (Epicentre Technologies) and 5′-end 32P-labeled primers were used. The reactions were electrophoresed in a 6% polyacrylamide gel electrophoresis–8M urea gels (17 by 40 cm; 0.4-mm thickness) followed by autoradiography. Select amplicons were chosen for complete sequence analysis.
Cloning and sequence analysis of PCR amplicons.
To determine the complete sequences of select vls amplicons, the amplicons were TA cloned into the pGEM-T Easy vector as described by the manufacturer (Promega). To identify Escherichia coli clones harboring recombinant plasmids, the cells were plated onto Luria-Bertani plates (ampicillin, 50 μg ml−1) and individual colonies were picked with sterile toothpicks and resuspended in 100 μl of distilled H2O. The resuspended cells were boiled for 10 min, and 1 μl of the cell lysate was used as the template in PCR with gene-specific PCR primer sets. The inserts in the recombinant plasmids were sequenced using end-labeled primers and the Excel sequencing kit as described by the manufacturer (Epicentre Technologies) or by automated sequencing methods. The sequence reactions were run on 6% polyacrylamide–8 M urea gels, and autoradiography was performed. The determined sequences were translated using the TRANSLATE program, and both the nucleotide and amino acid sequences were aligned using the PILEUP program (Genetics Computer Group) and manually adjusted.
Nucleotide sequence accession numbers.
All sequences have been deposited in the database, and accession no. AF354775 through AF354793 have been assigned.
RESULTS
PCR analyses of vlsE and BBJ51.
Due to technical problems with the cloning of linear plasmid telomeres and the instability of some B31MI DNA segments upon cloning, limited regions of the B. burgdorferi B31MI genome have not been sequenced (4, 5). Based on data from previous studies (21) it was inferred that vlsE was present in the genome of B31MI on an unsequenced segment of lp28-1 (4). As an essential first step of these analyses, we tested for the presence of vlsE in B31MIpc and in its postinfection clonal derivatives using a PCR approach. The vlsE292-vlsE960 primer set yielded product with B31MIpc and most clones derived from it (data not shown), indicating that a vlsE-related gene is present in B31MIpc. B31MI also carries a vls-related allele called BBJ51 (The Institute of Genomic Research open reading frame designation). BBJ51 is located on lp38 and possesses a frameshift mutation that introduces premature stop codons (5). Because of the homology between vlsE, BBJ51, and the vls cassettes we also sought to determine if BBJ51 undergoes mutation or recombination during infection. As with vlsE the first step toward this goal was to determine if the BBJ51-carrying plasmid, lp38, is maintained in clonal populations after infection in mice. By PCR the presence of BBJ51 in B31MIpc and its postinfection clonal derivatives was confirmed (data not shown). All vlsE and BBJ51 amplicons were of the predicted sizes, indicating that large-scale rearrangements had not occurred in these genes during infection.
Identification of the plasmid carrying vlsE in B. burgdorferi B31MIpc.
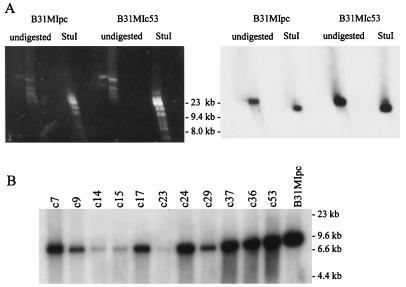
In B31-5A3, vlsE resides on lp28-1. However, in other isolates the size of the plasmid carrying vlsE has been demonstrated to vary (6). To identify the plasmid carrying vlsE in B31MIpc, hybridization analyses were conducted. DNA from B31MIpc and several postinfection clones was fractionated using two-dimensional PFGE and blotted. The membranes were hybridized with a PCR-generated probe that spans positions 292 through 960 of vlsE. The probe bound to an lp28 (Fig. 1A). Additional hybridization analyses were performed to determine if the gene is carried on lp28-1 or on one of the other 28-kb linear plasmids. lp28-1 possesses a single StuI site that upon cleavage should yield a vlsE carrying restriction fragment of 15,151 bp. Consistent with this, the probe bound to a single fragment estimated to be of this size in all clones (Fig. 1A). Hybridization analyses of HaeIII-digested DNA from postinfection clonal populations were also performed. Consistent with the restriction map of lp28-1, all clones yielded an ∼11-kb HaeIII restriction fragment that bound the probe (Fig. 1B). Collectively these analyses confirmed that B31MIpc and all clones derived from it carry vlsE on lp28-1.
FIG. 1.
Southern hybridization analysis of vlsE in B31MIpc and its postinfection clonal derivatives. (A) Two-dimensional PFGE was performed on DNA from B31MIpc and its postinfection clone, B31MIc53, as described in the text. Prior to electrophoresis one set of plugs was digested with StuI while the other was not digested. After electrophoresis the DNA was visualized by ethidium bromide staining (left). The DNA was transferred onto a hybridization membrane and probed with a radiolabeled vlsE PCR-generated probe (right). (B) DNA isolated from 12 different clones (clone designations are indicated above lanes) recovered from a mouse infected with B31MIpc for 3 months was digested with HaeIII. The digested DNA was fractionated in a 0.8% agarose gel and vacuum blotted, and the blot was incubated with a radiolabeled vlsE PCR-generated probe. Molecular weight standards are indicated.
Analysis of the genetic stability of vlsE and BBJ51 during infection.
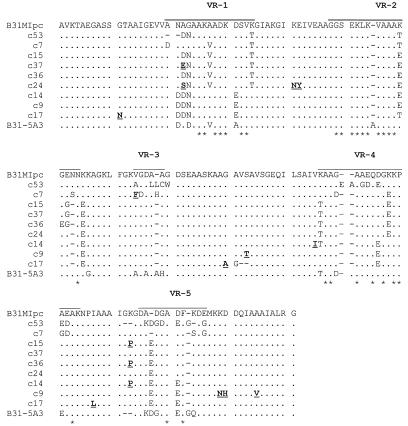
To assess the kinetics of sequence variation in vlsE and BBJ51, SNP analyses were performed on amplicons obtained from postinfection clonal populations recovered after 3 months of infection in mice. These analyses revealed that numerous sequence changes in the vlsE amplicons, but not in BBJ51, had occurred (data not shown). To further analyze the sequence changes, the amplicons were cloned and sequenced. Alignment of the B31MIpc vlsE VR sequence with that from B31-5A3 (21) revealed 21 amino acid (aa) differences. It is interesting to note that while the vlsE sequences of these clones exhibit numerous sequence differences, alignment of their entire vls cassette regions revealed only a single nucleotide difference (data not shown). Alignment of the B31MIpc vlsE sequence with that of its postinfection clones revealed that numerous sequence changes had developed during infection in these clones (Fig. 2). However, extensive rearrangements, deletions, and/or insertions were not observed. With the exception of B31MIc53, which had 28 aa changes, all B31MIpc-derived clones had between 11 and 15 aa changes, with the majority occurring in the VRs of VlsE. Note that the actual total number of sequence changes is probably greater than this since these numbers do not include possible changes in VR6 as this region was not analyzed.
FIG. 2.
Clonal variation in vlsE-encoded sequences after infection in mice. A mouse was infected for 3 months with B31MIpc, and then clonal populations were obtained by cultivation of an ear punch biopsy sample as described in the text. The vlsE gene was amplified from each population, and the amplicons were then sequenced. The determined sequences were translated and aligned as described in the text. VRs are overlined (top sequence). Hyphens, gaps introduced by alignment. Amino acid sequence changes that were identified to have resulted from point mutations are indicated by both bold face and underlining. Asterisks, residues within the VRs that have been found to be conserved in all vlsE-encoded sequences determined in this report.
Analysis of the VlsE sequences in clones recovered from mice infected for 7 months.
To determine if sequence changes continue to accumulate in vlsE over long-term infection, B31MIc53 (a derivative of B31MIpc) was used to infect a mouse and the infection was allowed to persist for 7 months. B31MI53 was one of the clones recovered from the B31MIpc-infected mouse, and its VlsE sequence is known. The vlsE gene was then amplified from the postinfection clones, SNP analyses were performed, and select amplicons were sequenced. The sequences were translated and aligned (Fig. 3). The total number of amino acid differences, relative to the VlsE sequence of the B31MIc53 clone infecting, averaged approximately 28 (Table 2). The number is similar to the number of sequence changes that were detected in some clones of B31MIpc after only 3 months of infection. Interestingly, when the VlsE sequences of the B31MIc53-derived postinfection clones were compared with that of B31MIpc, an average of only 20 aa sequence differences were observed. This observation indicates that a net accumulation of sequence changes did not occur and that restoration of the parental sequence at some positions had taken place. The restoration of the parental sequence or “reversion” could have resulted from one or more gene conversion or point mutation events. The tendency toward reversion and the plateau in the number of sequence changes that are maintained by VlsE suggest that the possible repertoire of VlsE variants that can develop, while significant, is likely to be less than previously postulated (21).
FIG. 3.
Alignment of the variant VlsE proteins carried by postinfection clonal populations derived from B. burgdorferi B31MIc53 after 7 months of infection. A mouse was infected for 7 months with B31MIc53 (a clone obtained from a mouse infected with B31MIpc), and then clonal populations were obtained by cultivation of an ear punch biopsy sample as described in the text. The vlsE gene was amplified from each population and subjected to SNP analyses, and the appropriate amplicons were selected for sequencing. The determined sequences were translated and aligned as described in the text. The VRs are overlined (top sequence). Hyphens, gaps introduced by alignment; asterisks, positions within the VRs that are either absolutely conserved or highly conserved in all of the sequences analyzed in this report.
TABLE 2.
Sequence changes in VlsE
| Infecting clone | Postinfection clone | No. of aa changes relative to:
|
No. of reversions back to the B31MIpc VlsE sequence | |
|---|---|---|---|---|
| Infecting clone | B31MIpc | |||
| B31MIpc | B31MIc7 | 14 | 14 | NA |
| B31MIpc | B31MIc9 | 14 | 14 | NA |
| B31MIpc | B31MIc14 | 13 | 13 | NA |
| B31MIpc | B31MIc15 | 12 | 12 | NA |
| B31MIpc | B31MIc17 | 14 | 14 | NA |
| B31MIpc | B31MIc24 | 14 | 14 | NA |
| B31MIpc | B31MIc36 | 13 | 13 | NA |
| B31MIpc | B31MIc37 | 11 | 11 | NA |
| B31MIpc | B31MIc53 | 25 | 25 | NA |
| B31MIc53 | c53e13 | 28 | 21 | 14 |
| B31MIc53 | c53e53 | 27 | 19 | 16 |
| B31MIc53 | c53e72 | 29 | 23 | 15 |
| B31MIc53 | c53e76 | 28 | 21 | 15 |
| B31MIc53 | c53e79 | 31 | 18 | 19 |
Alignment of the nucleotide sequences of the infection-induced vlsE gene variants with the silent vls cassettes revealed that several of the changes could not be attributed to gene conversion events as there were no potential contributing sources for these sequence changes in the vls cassettes. The corresponding amino acid sequence changes, indicated in Fig. 2, appear to have arisen through point mutation events. From these analyses it is evident that point mutation can serve as a second mechanism by which vlsE sequence variation can be generated.
Sequence analysis of vlsE in divergent Lyme disease isolates.
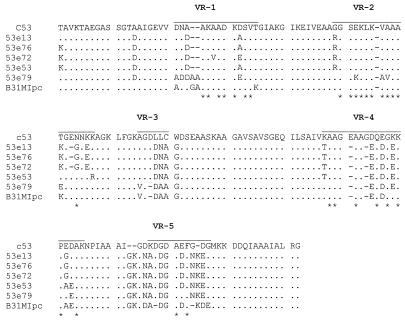
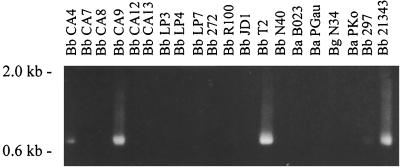
The data presented above suggest that there is apparent limit to the total number of amino acid changes that can be maintained in VlsE. If there were no constraints, then one would expect that the vlsE sequences from divergent B. burgdorferi sensu lato complex isolates would exhibit greater divergence than that found among clonal populations since these isolates would have been subjected to variable immune pressure and selection in the course of their natural enzootic cycles. To test this, we performed PCR and DNA sequence analysis of vlsE from a variety of Lyme disease isolates. Using the vlsE290-vls960 primer set, vlsE could be amplified from only 4 of 26 isolates tested (Fig. 4). This result is consistent with studies by others, who have also reported that a large percentage of isolates tested failed to yield a vlsE PCR amplicon (6), most likely as a result of vlsE sequence variation among isolates. Some of the amplicons were then purified, cloned, and sequenced. Comparison of the vlsE sequences of B31MIpc with those of other isolates revealed similar total numbers of amino acid differences (n = 25) (Fig. 5). Overall the average number of VlsE sequence differences observed among isolates from different geographic regions is equivalent to that observed among postinfection clonal populations that were recovered after only 3 months of infection in mice. These observations support the hypothesis that there are imposed limitations on the degree of variation in the vlsE gene.
FIG. 4.
PCR analysis of the vlsE gene from divergent B. burgdorferi sensu lato complex isolates. Representative PCR data are shown. Isolated DNA from different B. burgdorferi sensu lato isolates (indicated above each lane) served as the template in PCR amplification using the vlsE290-vlsE960 primer set. The amplicons were analyzed in 1.0% GTG-agarose gels. Abbreviations: Bb, B. burgdorferi; Ba, Borrelia afzelii; Bg, Borrelia garinii. Molecular weight standards are indicated.
FIG. 5.
Alignment of the VlsE sequences from divergent B. burgdorferi isolates. vlsE amplicons obtained from each isolate were sequenced, translated, and aligned as described in the text. The VRs are overlined (top sequence). Hyphens, gaps introduced by alignment; asterisks, positions that are either absolutely conserved or highly conserved in all of the sequences analyzed in this report. The isolates from which the vlsE sequences were obtained are indicated to the left. All isolates are B. burgdorferi.
DISCUSSION
In this study we sought to further investigate the mechanisms and kinetics of vlsE variation. While the vls system has been extensively characterized in B31-5A3 (19–21), little is know about the kinetics of vlsE variation in other Lyme disease isolates or clones. We selected B31MIpc for these analyses since the majority of its genome sequence, including its vls cassettes, has been determined (5) and its plasmid content has been verified (13). An essential starting point for these analyses was to verify the presence, map location, and sequence of vlsE in B31MIpc. The vlsE1 locus of B31MI was found to reside on lp28-1 near a telomere, and sequence analysis of the variable domain revealed 21-aa differences with the VlsE variable domain of B31-5A3. Consistent with the observations of Zhang et al. (21) most of the sequence differences were confined to the VRs.
To further characterize the sequence changes that developed in vlsE during infection, vlsE was amplified from postinfection clonal populations after 3- and 7-month time frames of infection and sequenced. Focusing first on the clones recovered after 3 months of infection, (Table 2), a similar number of amino acid sequence changes had occurred in each of the clones obtained 3 months postinfection and these changes were largely localized to the VRs. Many of these changes appear to have their origin in gene conversion events, as they have potential contributing sources in the silent vls cassettes. It is interesting to note that the cassette regions of B31MI, from which B31MIpc was derived, and B31-5A3 have only a single nucleotide difference (occurring within the vls2 cassette). Evidence for large-scale gene conversion events in which the entire sequence located between the direct repeat elements that flank the variable domain of vlsE was not observed (19). The direct repeat elements are thought to be important in mediating the gene conversion events. The largest obvious expanse of sequence transfer that we noted that may have resulted from gene conversion can be seen in the vlsEc53 sequence. It appears that much of VR1 of vlsE of B31MIpc was replaced with VR1 (DNAAKAADKDSVT) from the vls6 cassette. However, it is possible that the sequence of VR1 in vlsEc53 could have resulted from multiple independent gene conversion events. It is striking that most of the apparent gene conversion events appear to have resulted in the transfer of only very limited stretches of sequence.
Analysis of the nucleotide sequences also revealed that point mutations contribute to the generation of vlsE variation. Several examples of sequence changes for which there is no contributing source in the silent vls cassettes were observed. The criterion that we employed in differentiating point mutations from gene conversion was that there should be no possible contributing source in any of the silent cassettes that could have led to the sequence change. It is likely that there are other point mutations that are not discernible due to alignment ambiguities in some regions or that some point mutations are simply masked by possible gene conversion events. In cases where there is a potential contributing source for a sequence change in one of the cassettes there is no way to distinguish between a point mutation and a gene conversion event. Other intuitive considerations also suggest that gene conversion events are not likely to be the only source of vlsE variation. The majority of characterized vlsE variants would have to have undergone an extraordinary number of localized gene conversion events, many of which would have involved only one or a few nucleotides. It can be argued that the probability of rampant gene conversion of this type, over extremely short sequence segments, is low. In any event the data presented above demonstrate that at least some point mutations do develop and that point mutations represent a second mechanism for generating vlsE variation. In view of the remarkable conservation of the cassette region (one nucleotide difference between B31-5A3 and B31MI) it is likely that these point mutations develop post-gene conversion. Precedent for this mechanism can be found in the vmp genes of the relapsing-fever Borrelia sp. (16). In this species, gene conversion events involving extended sequence segments have been demonstrated (14, 15). This process results in the programmed sequential expression of the vmp genes (1). However, in addition to modification by gene conversion, postconversion point mutations have also been shown to further enhance the antigenic diversity of the Vmp proteins (16).
Previous studies suggested that sequence changes in vlsE accumulate over the duration of infection (20). To further assess the nature of the changes that develop and their possible accumulation after an extended period of infection, we infected a mouse with clone B31MIc53. This clone was originally recovered from a B31MIpc-infected mouse. Infection with this clone was allowed to persist for 7 months, and then postinfection clones were recovered. When the VlsE sequences of these clones were compared with that of B31MIc53 (the clone used to infect the mouse), most of these clones were found to possess approximately 28 aa changes. The B31MIc53 infecting clone had been found to possess a total of 28 aa changes when it was recovered from the B31MIpc-infected mouse after 3 months of infection. Hence a more than doubling of the infection time frame did not result in a higher net change in the overall VlsE sequence than that observed after only 3 months of infection with B31MIpc. Consistent with this observation, Zhang and Norris demonstrated that the total number of amino acid sequence changes that were maintained in VlsE did not change significantly in clones collected at either 28 days or 12 months postinfection and remained steady at approximately 25 (20). A comparison of the vlsE sequences from clones derived from the B31MIc53-infected mouse with the B31MIpc vlsE sequence (the vlsE sequence carried by the clone from which all other clones in this study were derived) showed that the total number of amino acid changes in the corresponding products was approximately 20. This indicates that many of the sequence changes that developed during infection with B31MIc53 actually restored the original vlsE sequence of B31MIpc. To further assess the extent to which sequence reversions occur in vlsE, we conducted a retrospective analysis of the vlsE sequences determined by Zhang and Norris (19). Zhang and Norris infected a mouse with B31-5A3, and then after 28 days an ear punch biopsy sample was collected and clone mle4C was obtained by subsurface plating. This clone was then used to infect another mouse, and after 28 days a clone was recovered from this mouse and designated 1396D. Many of the sequence changes that developed in 1396D were also reversions that restored the vlsE sequence of the initial infecting clone, B31-5A3.
Earlier studies demonstrated that vlsE sequence changes occur predominantly within the VRs, and the data presented here are generally consistent with that observation (19–21). In this report, we also noted that sequence changes occur at only a limited number of positions within the VRs. There are a total of 62 aa positions within VRs 1, 2, 3, 4, and 5 (VR6 was not analyzed in this report), and of these 28 (45%) are either absolutely conserved or highly conserved among VlsE variants. The fact that some of these positions do not change is not surprising since some of the nucleotide positions are conserved among the cassette sequences as well. Hence gene conversion events involving these positions would not change the sequence. However, there are several positions that do not change but that do vary in sequence among the cassettes. The reason why these positions are not changed by gene conversion events or point mutations or are not maintained (stable) is unclear. However in some cases it may be due to the absence of flanking repeat elements or sequence variation within some of the repeats. For example, within vls2 there are extensive stretches of unique sequence that do not appear in any of the vlsE variants. vls2 lacks an upstream direct repeat element and thus may be incapable of participating in gene conversion events. The same may hold true for vls16, which does not have a downstream direct repeat. The strict conservation of specific residues that are embedded in regions with high mutational capabilities suggests that these residues may be essential for VlsE function and or structure and thus there is selection against sequence change. It has been estimated that when all the amino acid possibilities at all positions within the VRs are considered, there could be tremendous capacity for VlsE variation, with the number of possible variants being on the order of 1030 (19). However, it is evident from the analyses presented here that, while there is significant potential for variation in vlsE, there are in fact constraints on sequence changes within the vlsE VRs. Further supporting this conclusion is the fact that the extent of vlsE variation among different isolates subjected to different immune pressure histories is less than might be expected if all sequence possibilities were allowed. Iyer et al. have also provided evidence that VlsE variation is less pronounced than expected through their analyses of vlsE sequences from human and tick isolates (6).
While it is clear that vlsE undergoes extensive sequence changes during infection, it could be argued that a direct demonstration for this variation in immune system evasion has not yet been provided. Interestingly, several laboratories have demonstrated that a majority of Lyme disease patients develop anti-VlsE antibodies that recognize recombinant VlsE or VlsE peptides (6–10). In addition, invariable domains and IRs of VlsE have been demonstrated to be highly immunogenic, and it has been suggested that the antibody response to the IRs, specifically IR6, could serve as a useful marker for infection with the Lyme disease spirochetes (10). This observation and the data presented above appear to be at odds with the suggested role of VlsE in immune system evasion. While it is possible that the VRs could be immunodominant and thus play a role in immune system evasion, this seems unlikely in view of the rapid rate at which sequence changes develop and revert in these regions. In addition, there is no evidence that dominant variants emerge during early infection as would be expected for an immune system evasion system.
In closing, these studies have further advanced our understanding of the mechanisms associated with the generation of vlsE variation. Specifically we have demonstrated that post-gene conversion mutational events are involved. The conservation of specific residues within the VRs and the tendency to reversion are important observations that indicate a potential role for these residues in VlsE structure or in its undefined functions. The apparent constraints on the accumulation of sequence changes indicate that the theoretical extent of VlsE variation is less than previously postulated and may indicate a more limited role for VlsE in immune system evasion.
ACKNOWLEDGMENTS
This work was supported in part by grants from the Commonwealth Health Research Board and the Jeffress Trust.
We thank F. T. Mouse for his contributions and our colleagues in the Molecular Pathogenesis Research group at Virginia Commonwealth University for their helpful and insightful discussions.
REFERENCES:
- 1.Barbour A G, Tessier S L, Stoenner H G. Variable major proteins of Borrelia hermsii. J Exp Med. 1982;156:1312–1324. doi: 10.1084/jem.156.5.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 3.Carlyon J A, LaVoie C, Sung S Y, Marconi R T. Analysis of the organization of multicopy linear and circular plasmid-carried open reading frames in Borrelia burgdorferi sensu lato isolates. Infect Immun. 1998;66:1149–1158. doi: 10.1128/iai.66.3.1149-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, Haft D, Hickey E, Gwinn M, White O, Fraser C M. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 5.Fraser C, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischman R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 6.Iyer R, Hardham J M, Wormser G P, Schwartz I, Norris S J. Conservation and heterogeneity of vlsE among human and tick isolates of Borrelia burgdorferi. Infect Immun. 2000;68:1714–1718. doi: 10.1128/iai.68.3.1714-1718.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawabata H, Myouga F, Inagaki Y, Murai N, Watanabe Genetic and immunological analyses of vls (VMP-like sequences) of Borrelia burgdorferi. Microb Pathog. 1998;24:155–166. doi: 10.1006/mpat.1997.0183. [DOI] [PubMed] [Google Scholar]
- 8.Lawrenz M B, Hardham J M, Owens R T, Nowakowski J, Steere A C, Wormser G P, Norris S J. Human antibody response to VlsE antigenic variation protein of Borrelia burgdorferi. J Clin Microbiol. 1999;37:3997–4004. doi: 10.1128/jcm.37.12.3997-4004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang F T, Alvarez A L, Gu Y, Nowling J M, Ramamoorthy R, Philipp M T. An immunodominant conserved region within the variable domain of VlsE, the variable surface antigen of Borrelia burgdorferi. J Immunol. 1999;163:5566–5573. [PubMed] [Google Scholar]
- 10.Liang F T, Steere A C, Marques A R, Johnson B J, Miller J N, Phillip M T. Sensitive and specific serodiagnosis of Lyme disease by enzyme-linked immunosorbent assay with a peptide based on an immunodominant conserved region of Borrelia burgdorferi VlsE. J Clin Microbiol. 1999;37:3990–3996. doi: 10.1128/jcm.37.12.3990-3996.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marconi R T, Samuels D S, Garon C F. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J Bacteriol. 1993;175:926–932. doi: 10.1128/jb.175.4.926-932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marconi R T, Samuels D S, Landry R K, Garon C F. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J Bacteriol. 1994;176:4572–4582. doi: 10.1128/jb.176.15.4572-4582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDowell J V, Skare J T, Sung S-Y, Marconi R T. Analysis of mechanisms associated with the loss of infectivity of clonal populations of Borrelia burgdorferi B31MI. Infect Immun. 2001;69:3670–3677. doi: 10.1128/IAI.69.6.3670-3677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier J T, Simon M I, Barbour A G. Antigenic variation is associated with DNA rearrangements in a relapsing fever Borrelia. Cell. 1985;41:403–409. doi: 10.1016/s0092-8674(85)80013-1. [DOI] [PubMed] [Google Scholar]
- 15.Plasterk R H A, Simon M I, Barbour A G. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium B. hermsii. Nature. 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- 16.Restrepo B I, Barbour A G. Antigen diversity in the bacterium B. hermsii through “somatic” mutations in rearranged vmp genes. Cell. 1994;78:867–876. doi: 10.1016/s0092-8674(94)90642-4. [DOI] [PubMed] [Google Scholar]
- 17.Steere A C, Grodzicki R L, Kornblatt A N, Craft J E, Barbour A G, Burgdorfer W, Schmid G P, Johnson E, Malawista S E. The spirochetal etiology of Lyme disease. N Engl J Med. 1983;308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- 18.Sung S Y, McDowell J, Carlyon J A, Marconi R T. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes result in the development of new antigenic variants during infection. Infect Immun. 2000;68:1319–1327. doi: 10.1128/iai.68.3.1319-1327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J R, Norris S J. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66:3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J R, Norris S J. Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi. Infect Immun. 1998;66:3689–3697. doi: 10.1128/iai.66.8.3689-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J-R, Hardham J M, Barbour A G, Norris A G. Antigenic variation in Lyme disease borreliae by promiscuous recombination of vmp like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]