Abstract
More and more clinical trials have explored the role of liquid biopsy in the diagnosis and treatment of EGFR-mutated NSCLC. In certain circumstances, liquid biopsy has unique advantages and offers a new way to detect therapeutic targets, analyze drug resistance mechanisms in advanced patients, and monitor MRD in patients with operable NSCLC. Although its potential cannot be ignored, more evidence is needed to support the transition from the research stage to clinical application. We reviewed the latest progress in research on the efficacy and resistance mechanisms of targeted therapy for advanced NSCLC patients with plasma ctDNA EGFR mutation and the evaluation of MRD based on ctDNA detection in perioperative and follow-up monitoring.
Keywords: liquid biopsy, ctDNA/cfDNA, EGFR, NSCLC
1. Introduction
In recent years, the clinical application of epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) for the targeted therapy of patients with non-small-cell lung cancer (NSCLC) has been fully recognized. Treatment with EGFR-TKIs can significantly improve progression-free survival (PFS) and objective response rate (ORR) in patients with advanced EGFR-mutated NSCLC [1]. With the publication of randomized controlled clinical trials, such as ADJUVANT, ADAURA, EVIDENCE, and EVAN, more optimized targeted treatment regimens have been developed for postoperative adjuvant therapy for patients with EGFR-mutated NSCLC [2]. In addition, preoperative neoadjuvant EGFR-targeted therapy can also lead to higher disease remission rates in patients with NSCLC [3]. The best samples for EGFR testing are fresh tumor tissues and paraffin-embedded specimens, but it is difficult to obtain tumor tissues from some patients with advanced NSCLC. More and more studies have confirmed that the use of liquid biopsy to detect EGFR mutations has great potential for clinical application.
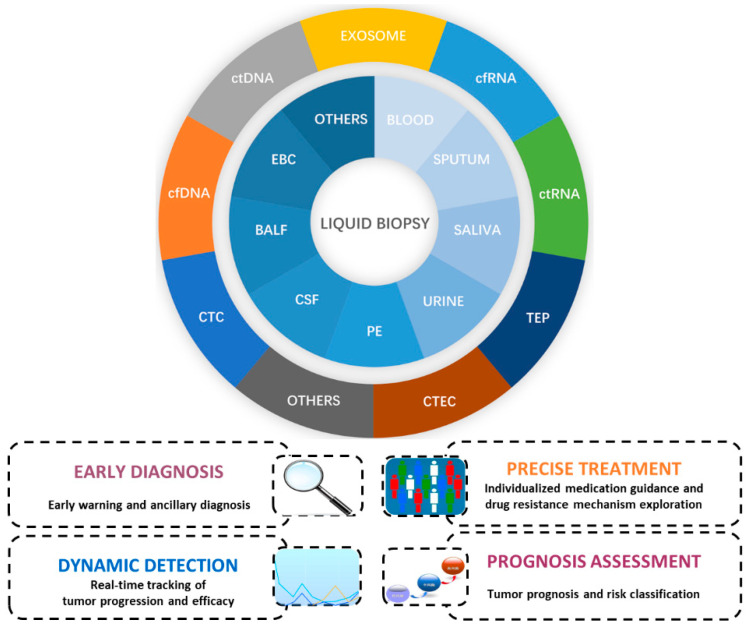
Broadly speaking, liquid biopsy is the process of testing blood or bodily secretions for cancer cells, which helps to further characterize lung cancer by identifying tumor cells or tumor DNA that are released into the blood or bodily secretions following tumor cell growth or apoptosis. As a highly sensitive technology, blood testing can not only diagnose cancer but also reveal key tumor characteristics and more comprehensive tumor genomes and dynamically monitor genetic changes during treatment. Therefore, it has great potential and broad prospects for clinical application in the field of tumor diagnosis and treatment. Currently, liquid biopsy is routinely used to detect circulating tumor cells (CTCs) [4], circulating tumor DNA (ctDNA) [5], circulating free DNA (cfDNA) [6], and exosomes [7], while research on using liquid biopsy to detect peripheral blood circulating RNAs [8], tumor-educated platelets (TEPs) [9], and circulating tumor endothelial cells (CTECs) [10] in the blood and other bodily fluids, including sputum, saliva, urine, pleural effusion (PE) [11], cerebrospinal fluid (CSF) [12], bronchoalveolar lavage fluid (BALF) [11], exhaled breath condensate (EBC) [13], etc., is gradually emerging (Figure 1). Briefly, cfDNA is a genetic material that “runs” in the blood after metabolism of almost all bodily tissues; therefore, healthy tissues, inflammatory tissues, and tumor tissues all release cfDNA into the blood. By extension, ctDNA is a fragment of cfDNA that is released into the bloodstream from primary tumors or metastatic cells. Compared to cfDNA, the amount of ctDNA in the blood is relatively low, making up only 1% of cfDNA (or even less than 0.01%) [14]. Most ctDNA fragments range from 160 to 200 bp in length and are shorter than cfDNA molecules without mutations [15,16,17]. Some studies have shown that ctDNA contains the same molecular characteristics as the tumor DNA of its origin, such as single nucleotide variation (SNV), short insertions and deletions (InDels), copy number variation (CNV) [18,19], and methylation [20,21,22]. Some studies have also found that ctDNA entering the bloodstream has a half-life of about 15 min to 2.5 h [23,24,25]. This feature ensures that ctDNA can be used as a “real-time” tumor biomarker, which can more accurately reflect tumor load than protein markers that may take weeks to present. This review introduces the progress of research on the use of ctDNA/cfDNA in the precision treatment of EGFR-mutated NSCLC, including the screening of therapeutic targets, the analysis of resistance mechanisms, and the monitoring of minimal residual disease (MRD).
Figure 1.
The great potential and broad prospects for the clinical application of liquid biopsy in NSCLC diagnosis and treatment. Abbreviations: PE, pleural effusion; CSF, cerebrospinal fluid; BALF, bronchoalveolar lavage fluid; EBC, exhaled breath condensate; ctDNA, circulating tumor DNA; cfDNA, circulating free DNA; CTC, circulating tumor cells; cfRNA, circulating free RNA; ctRNA, circulating tumor RNA; TEP, tumor-educated platelets; CTEC, circulating tumor endothelial cell.
2. Plasma ctDNA-Based EGFR Mutations Can Guide Targeted Therapy for Advanced NSCLC
Today, the treatment of advanced NSCLC has entered the era of individualized and precise targeted therapy based on molecular typing. Accurate molecular typing is the basis of this precise treatment; however, qualified tumor tissue specimens cannot be obtained from every patient, so liquid biopsy was developed. Many studies have prospectively verified the high specificity (92–100%) and positive predictive value (94.0–98.6%) of ctDNA-based EGFR mutation detection using tissues as a reference [26,27,28,29,30,31,32,33], and these two metrics have also been consistent in guiding the targeted therapy of NSCLC [30,31,34,35]. Recently, research on ctDNA-based next-generation sequencing (NGS) has demonstrated potential clinical utility [36,37,38,39]. For example, one study showed a 12.0% increase in identified actionable alterations when ctDNA-based testing was performed following tissue-based NGS testing, suggesting that ctDNA-based testing could identify additional patients with actionable genomic alterations [40]. In terms of detection timelines, Sehayek et al. compared the results and turnaround times of the molecular analysis of liquid and tissue biopsies, and found that liquid biopsies had consistent analytical results and shorter turnaround times. This means that liquid biopsy is feasible and accurate and can shorten the time required to diagnose NSCLC, especially when tumor tissues are scarce [41]. It is noteworthy that although NGS is widely touted as being more sensitive than qPCR, the available evidence suggests that all commonly used ctDNA detection techniques lack satisfactory sensitivity (e.g., qPCR, ddPCR, and NGS) [42]. A mate-analysis showed that for patients with tissue EGFR-mutated lung cancer, the therapeutic benefit of EGFR-TKIs was similar in the ctDNA+ and ctDNA− subgroups. Approximately 40% of the patients with tumor EGFR mutations that cannot be detected in ctDNA by qPCR assays at baseline could still benefit from the EGFR-TKI therapy. In such patients, repeated EGFR testing using tissue biopsies should be attempted. Of course, the impact of false negatives from the different detection methods also needs to be considered [43]. The limited yield and complex kinetics of ctDNA carry the risk of false negative results, even when using sensitive and well-validated molecular detection methods [44]. When possible, tumor tissues should be tested when ctDNA test results are negative or inconclusive.
Table 1 presents data on the association between plasma ctDNA/cfDNA EGFR mutations and the efficacy and prognosis of targeted therapy in NSCLC patients [28,30,31,32,33,34,35,45,46,47,48,49,50,51,52,53,54,55,56,57,58]. The BENEFIT trial prospectively demonstrated the feasibility of using liquid biopsy to guide the efficacy of first-line EGFR-TKI therapy for the first time. In that study, 188 patients with EGFR mutations in their ctDNA received gefitinib treatment and showed an ORR of 72.1% with a median PFS of 9.2 months, which was basically consistent with the results of IPASS and WJTOG3405 [32]. It has further been confirmed that plasma ctDNA EGFR mutation detection is conducive to the precise targeted therapy of advanced NSCLC. Park et al. evaluated the efficacy of afatinib in treatment-naïve lung cancer patients with ctDNA-based EGFR mutations (exon 19 deletion or exon 21 point mutation). The results showed that afatinib exhibited similar ORR and PFS in lung cancer patients with EGFR mutations in their ctDNA, regardless of the outcome of the tumor EGFR mutations [35]. As a second-line treatment, osimertinib showed a favorable ORR and a median PFS for patients with plasma ctDNA T790M positivity, regardless of their tissue mutation status [45]. However, subgroup analyses of the FLAURA and AURA3 trials showed shorter median PFS for ctDNA-positive vs. ctDNA-negative patients in the osimertinib arm (FLAURA, 15.2 vs. 23.5 months; AURA3, 8.2 months vs. 10.1 months) [59,60]. This suggests that patients with tumor DNA shedding have a worse prognosis than those without tumor DNA shedding, which may, in part, be due to the lower tumor burden in ctDNA-negative patients. Park et al. also evaluated the effect of osimertinib in previously untreated metastatic NSCLC patients (n = 19) harboring activating EGFR mutations in their ctDNA and tumor DNA. The results showed that osimertinib had a favorable effect on such patients, with an ORR of 68% (13/19) and a median PFS of 11.1 months (95% CI = 0.0–26.7). The ORR and median PFS of ex19del (91%; 21.9 months; 95% CI = 5.5–38.3) were significantly better than those of L858R/L861Q (43%; 5.1 months; 95% CI = 2.3–7.9) [54]. Osimertinib has also demonstrated potent activity in T790M-positive NSCLC patients with CNS metastases who previously received EGFR-TKI therapy, with a median PFS of 8.4 months (95% CI = 5.8–10.9) and an ORR of 39.4%. Patients with undetectable EGFR mutations in their plasma at 6 weeks had better PFS compared to patients with detectable mutations (not reached vs. 4.5 months; 95% CI = 0.0–1.1; p < 0.05) [50].
Table 1.
The correlation between plasma ctDNA/cfDNA EGFR mutations and the efficacy and prognosis of EGFR-TKI targeted therapy in patients with NSCLC.
| Reference (Year) | Characteristics of Subjects | n | ctDNA/cfDNA Detection Method | Prognostic Relevance |
|---|---|---|---|---|
| [28] (2015) |
|
97 | TaqMan assay |
L858R mutation in tissues and cfDNA vs. L858R mutation in tissues but not in cfDNA: 13.7 m (95% CI = 7.1–17.7) vs. 27.7 m (95% CI = 16.1–46.2) (HR = 2.22; 95% CI = 1.09–4.52; p = 0.03) |
| [31] (2016) |
Group B: positive for EGFR mutations in blood only (T−/C+) (n = 28) Group C: positive for EGFR mutations in tissue only (T+/C−) (n = 180)
|
472 | ARMS, ddPCR and NGS |
|
| [45] (2018) |
|
19 | Cobas EGFR Mutaion Test v2 or PANA Mutyper |
|
| [46] (2018) |
|
57 | Qualitative (PANAMutyper) and quantitative (PANAGENE-SQI) |
|
| [32] (2018) |
|
183 | ddPCR |
|
| [47] (2019) |
|
22 | dPCR and NGS |
|
| [30] (2019) |
|
71 | ADx-ARMS |
C+EGFR wild type vs. C+EGFR mutant type: 8.7 m vs. 11.0 m (p = 0.001) |
| [48] (2019) |
|
30 | NGS or ARMS |
|
| [49] (2019) |
|
307 | Cobas EGFR Mutation Test v2 |
T790M-positive plasma: 8.3 m with osimertinib (95% CI = 6.8–10.5) ; 4.2 m with platinum–pemetrexed (95% CI = 4.1–5.4) |
| [34] (2019) |
|
66 | ddPCR |
|
| [50] (2020) |
|
12 | NGS |
Patients with undetectable plasma EGFR mutations at week 6 had better overall PFS compared to those with detectable mutations (NR vs. 4.5 m; 95% CI = 0.0–1.1; p < 0.05). No significant changes in PFS were observed based on the absence of detectable EGFR-sensitizing mutations in CSF at week 6 (p = 0.68) |
| [51] (2020) |
|
30 | ddPCR |
|
| [52] (2020) |
|
53 | Cobas EGFR Mutation Test v2 or ddPCR |
|
| [53] (2020) |
|
52 | Cobas EGFR Mutation Test v2, ddPCR and NGS |
|
| [35] (2021) |
Group B: EGFR mutations in ctDNA and tumor DNA (n = 10)
|
21 | PNA-based RT-PCR |
|
| [54] (2021) |
|
19 | Mutyper and Cobas EGFR Mutation Test v2 |
|
| [55] (2021) |
|
54 | cSMART assay |
60.0% for the >0.1% group vs. 21.4% for the ≤0.1% group (p = 0.028)
|
| [56] (2021) |
|
41 | R-superARMS |
ΔCt >4.89 vs. 4.89: NR vs. 11.0 m (p = 0.014) mPFS: mutation clearance vs. incomplete mutation clearance: NR vs. 27.5 m (p = 0.088) |
| [57] (2022) |
|
28 | NGS |
EGFR clearance vs. EGFR non-clearance (week4): 11.4 m vs. 5.67 m (p = 0.011; HR = 0.23; 95% CI = 0.08–0.72) Non-clearance vs. EGFR clearance only vs. total-clearance (week4): 11.4 m vs. 9.2 m vs. 5.07 m
|
| [58] (2022) |
|
116 | SuperARMS ddPCR NGS |
|
| [33] (2022) |
|
158 | Cobas EGFR Mutation Test v2 |
|
Abbreviations: mOS, median overall survival; 95% CI, 95% confidence index; HR, hazard ratio; T+, EGFR mutations in tissues; C+, EGFR mutations in ctDNA; T−, no EGFR mutations in tissues; C−, no EGFR mutations in ctDNA; ddPCR, droplet digital PCR; NGS, next-generation sequencing; ORR, objective response rate; mPFS, median progression-free survival; DoR, duration of response; dPCR, digital PCR; ADx-ARMS, ADx-amplification refractory mutation system; NR, not reached; ctDNA, circulating tumor DNA; cfDNA, circulating free DNA; ARMS, amplification refractory mutation system; CNS, central nervous system; DCR, disease control rate; CSF, cerebrospinal fluid; VEGF, vascular endothelial growth factor. The*ΔCt value (i.e., mutant cycle threshold (Ct) value—internal control Ct value) was calculated to identify the presence of EGFR mutations, and was automatically calculated from PCR amplification fluorescence plots and the corresponding number of cycles.
Previous studies have reported that gene polymorphisms, simultaneous genomic mutations, or tumor size can affect the efficacy of EGFR-TKIs [61,62,63]. Zhou et al. first demonstrated that the abundance of EGFR mutations could predict the efficacy of EGFR-TKIs in the treatment of advanced NSCLC. Patients with low EGFR mutation abundance benefited more from EGFR-TKI treatment than did patients with wild-type EGFR, while patients with high EGFR mutation abundance benefited more from EGFR-TKI treatment than patients with low EGFR mutation abundance [64]. However, it has also been reported that the PFS for patients with low EGFR mutation abundance was similar to that for patients with wild-type EGFR. Therefore, the effect of EGFR mutation abundance on clinical outcomes in patients with advanced NSCLC, especially those with very low EGFR mutation abundance, is still unclear [65,66]. The question of whether untreated NSCLC patients with low EGFR-TKI mutation abundance detected using high-sensitivity techniques should be treated with EGFR-TKIs also remains to be determined. Wang et al. used circulating single-molecule amplification and resequencing technology (cSMART) approach to prospectively assess EGFR mutation status in plasma at the baseline and track dynamic EGFR changes during EGFR-TKI treatment. The results showed that when the cut-off EGFR mutation abundance value in plasma was 0.1%, the ORR of patients with plasma EGFR mutation abundance >0.1% was significantly higher than that of patients with plasma EGFR mutation abundance of ≤0.1% (60.0% vs. 21.4%; p = 0.028). The median PFS was also significantly longer in patients with a plasma EGFR mutation abundance of >0.1% compared to that of patients with a plasma EGFR mutation abundance of 0.01–0.1% (p = 0.0115). Additionally, one Cox multivariate analysis indicated that mutation abundance in plasma was an independent predictor of PFS (HR = 2.41, 95% CI = 1.12–5.20; p = 0.025) [55]. In another study, researchers used a modified semi-quantitative method (reformed-superARMS) based on ΔCt values that were generated during polymerase chain reaction (PCR) assays to investigate the impact of different baseline ctDNA EGFR mutation levels and changes in ctDNA EGFR mutations with reformed-superARMS on the outcomes and prognoses of patients receiving targeted therapy. When the ΔCt cutoff was set at 8.11 at the baseline, a significant difference in median OS was observed between the two groups (EGFR mutation ΔCt ≤ 8.11 vs. >8.11: not reached vs. 11.0 months; log rank p = 0.024). When the cutoff value for ΔCt was set at 4.89 after 1 month of treatment, a significant difference in median OS was again observed between the two groups (change in ΔCt > 4.89 vs. ≤4.89: not reached vs. 11.0 months; log rank p = 0.014) [56]. This indicates that the quantitative stratification of EGFR mutations in ctDNA can not only better select patients to be treated with EGFR-TKIs but can also help to develop better treatment strategies for patients with a low abundance of EGFR mutations.
However, the patients in the above studies had clear pathological diagnoses. The potential role of ctDNA remains unclear in patients with unknown pathological status. Deng et al. conducted a prospective study on 30 patients with suspected advanced lung cancer, in which patients with plasma EGFR-sensitizing mutations were treated with first-generation EGFR-TKIs. The results showed that EGFR-sensitizing mutations were detected in ctDNA of two-thirds of the patients. These patients received EGFR-TKI treatment, resulting in an ORR of 90% and a disease control rate (DCR) of 100%. The median PFS and OS were significantly longer than those in untreated patients (11.0 months vs. 1.0 months, p < 0.001) and not reached vs. 3 months (p < 0.001), respectively. This suggests that patients who have not been pathologically diagnosed have similar clinical outcomes and prognoses to those who have obtained histopathological diagnoses [48]. It is important to note that the amount of ctDNA entering the blood is affected by a variety of biological factors, such as tumor load and the degree of tumor vascularization, which also affect the ctDNA detection rate. For patients without an EGFR mutation in their cfDNA, test samples, test ingredients, and methods may need to be optimized. Thus, patients with poor performance (ECOG 3 to 4) and those who cannot undergo invasive procedures such as bronchoscopy or image-guided needle biopsy due to medical comorbidities or untouchable lesions could also benefit. Another similar study showed that the ORR following first-line treatment with icotinib was 52.6% (95% CI = 43.1–61.9%) for 116 patients with advanced lung cancer of unknown pathological status who had EGFR-sensitizing mutations in their ctDNA. The median PFS and OS were 10.3 months (95% CI = 8.3–12.2) and 23.2 months (95% CI = 17.7–28.0), respectively [58]. The results of these two studies suggest that ctDNA-based EGFR genotyping could help provide viable diagnoses in lung cancer patients for whom tissue biopsy is not possible. Targeted therapy for EGFR based on ctDNA assays could improve patient outcomes, but large-scale real-world exploration is still needed in the future.
3. Evaluation of Resistance to EGFR-TKI Therapy Based on Plasma ctDNA Detection in NSCLC Patients
Targeted therapy has significant clinical effects on lung cancer patients with specific mutated genes, but resistance inevitably develops during treatment. Tumor heterogeneity leads to the development of multiple resistance mechanisms during targeted therapy. Therefore, the dynamic longitudinal detection of gene mutations during the treatment of NSCLC patients has become increasingly important for guiding treatment after disease progression or drug resistance has occurred. EGFR-TKI treatment resistance includes primary resistance and acquired resistance. The primary drug resistance mechanisms that have been reported so far include the presence of drug-resistant mutations in EGFR and EGFR mutations that are also combined with mutations in other related genes, such as KRAS, PIK3CA, BRAF, etc. [67,68,69]. Other non-mutated mechanisms have also been reported; for example, the insulin-like growth factor 1 receptor (IGF1R) induces primary drug resistance by interacting with EGFR [70]. The mechanisms of acquired resistance to EGFR-TKIs are mainly divided into EGFR-dependent and EGFR-independent mechanisms. Among the EGFR-dependent mechanisms, the most common cause of acquired resistance to first- and second-generation EGFR-TKIs is the EGFR T790M mutation [71,72], while the EGFR C797S mutation is considered to be the main EGFR-dependent resistance mechanism to third-generation EGFR-TKIs [73], which accounts for 10–26% of cases of resistance to second-line osimertinib treatment and approximately 7% of cases of resistance to first-line osimertinib treatment [74]. When C797S and T790M mutations are in trans, they cause resistance to third-generation EGFR TKIs but show sensitivity to a combination of first- and third-generation TKIs. On the contrary, when the mutations are in cis, they still cause resistance [75,76,77]. In addition, activation of the PI3K/Akt/mTOR pathway [73,78] and other rare mutations, such as the D761Y, T854A [79], and L747S [80] mutations, are also associated with resistance to EGFR-TKIs. EGFR-independent resistance mechanisms include alternative pathway activation and histological or phenotypic transformation. MET amplification is the most common alternative pathway activation [81,82], but other mechanisms can also lead to acquired resistance to EGFR-TKIs, including the following: HER2 amplification [83]; HER3 overexpression (HER3 activation) [84]; KRAS, BRAF, and MAPK1 mutations [85,86]; ALK fusions; GFR3-TACC3, RET-ERC1, CCDC6-RET, NTRK1-TPM3, NCOA4-RET, GOPC-ROS1, AGKBF and ESYT2-BF fusions [87,88]; the transformation of NSCLC into SCLC [82], adenocarcinoma into squamous cell carcinoma [89], and EMT [90].
In the process of developing drug resistance, the complexity and spatiotemporal diversity of tumor clones determine therapeutic efficacy and prognosis. The composition and competitive evolution of tumor clones have decisive impacts on therapy, which can be modulated using targeted drugs and chemotherapy. These findings could have new implications for the clinical treatment of NSCLC [91]. Liquid biopsy can identify the resistance to targeted therapy in a timely manner, showing a higher coverage of tumor heterogeneity, more complex resistance patterns, and some new resistance mutation sites, such as EGFR p.V769M and KRAS p.A11V [91]. Numerous studies have been conducted to validate the potential of non-invasive strategies for identifying the resistance mechanisms to EGFR-TKIs. Table 2 lists recent literature data on gene alterations in plasma during EGFR-TKI therapy [45,47,52,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109]. More than half of patients with EGFR mutations develop EGFR T790M resistance when using first- and second-generation EGFR-TKIs [110]. The mechanistic patterns of resistance appear to differ between different first-generation TKIs (i.e., gefitinib, erlotinib, and icotinib), reflecting the heterogeneity of TKI resistance [106,111].
Table 2.
Literature data on the genetic alterations detected in plasma during EGFR-TKI treatment.
| References (Year) |
No.of Patients |
Prior Treatment | Detection Method | Genetic Treatment-Resistant Alterations Detected in Plasma (%) |
|---|---|---|---|---|
| [106] (2021) |
50 | First-generation EGFR-TKIs | ddPCR and NGS | T790M: 38% (19/50) |
| [91] (2018) |
53 | First-generation EGFR-TKIs | NGS for 124-genes panel | T790M: 45.28% (24/53) EGFR point mutations: 20.75% (11/53) KRAS/NRAS point mutations: 15.09% (8/53) EGFR amplification: 7.54% (4/53) BRAF amplification: 1.8% (1/53) MET amplification: 3.7% (2/53) ERBB2 amplification: 1.8% (1/53) |
| [101] (2020) |
37 | First-generation EGFR-TKIs | ddPCRand NGS for 223-genes panel | EGFR T790M: 51.35% (19/37) TP53: 67.57% (25/37) KRAS: 8.11% (3/37) c-Met amplification: 5.41% (2/37) STK11: 5.41% (2/37) FANCA: 5.41% (2/37) ERBB2: 5.41% (2/37) PIK3CA: 2.7% (1/37) FGFR1: 2.7% (1/37) BRAF: 2.7% (1/37) |
| [105] (2020) |
147 | First-generation EGFR-TKIs | NGS for 168-genes panel | T790M: 40.13% (59/147) MET and ERBB2 amplification: 2.04% (3/147) TP53: 45.86% (61/133) |
| [96] (2019) |
48 | Icotinib | NGS for 170-genes panel | T790M: 81.2% (39/48) EGFR amplification: 72.9% (35/48) CTNNB1: 2.1% (1/48) PIK3CA: 2.1% (1/48) BRAF: 2.1% (1/48) EML4-ALK: 2.1% (1/48) SLC342-ROS1: 2.1% (1/48) Unknown mutations: 2.1% (1/48) |
| [45] (2018) |
80 | First- and second-generation EGFR-TKIs | Cobas EGFR Mutation Test v2 or PANA Mutyper | T790M: 26.3% (21/80) |
| [95] (2019) |
66 | First- and second-generation EGFR-TKIs | Cobas EGFR Mutation Test v2. |
T790M: 33.3% (22/66) |
| [97] (2019) |
50 | First- and second-generation EGFR-TKIs | NGS | T790M: 71% (30/42) |
| [99] (2019) |
118 | First- and second-generation EGFR-TKIs | ARMS-PCR or super ARMS-PCR | T790M: 41.5% (49/118) |
| [52] (2020) |
276 | First- and second-generation EGFR-TKIs | Cobas EGFR Mutation Test v2 or ddPCR | T790M: 26.8% (74/276) |
| [103] (2020) |
120 | First- and second-generation EGFR-TKIs | Easy EGFR, Therascreen EGFR RGQ PCR and Cobas EGFR Mutation Test v2 |
T790M:25.8% (31/120) |
| [104] (2020) |
104 | First- and second-generation EGFR-TKIs | Cobas EGFR Mutation Test v2 | T790M: 49% (34/104) |
| [94] (2018) |
25 | Afatinib | dPCR and NGS | T790M dPCR: 56.5% (13/23) NGS: 43.5% (10/23) |
| [98] (2019) |
67 | Afatinib | ddPCR | T790M: 73.1% (49/67) |
| [47] (2019) |
22 | First- and second-generation EGFR-TKIs | dPCR and NGS | T790M: 86% (19/22) Progression to osimertinib (16), EGFR C797S (3), A750P (1), S464L (1), amplification (1), PIK3CA E545A (3) and E545K (1) |
| [108] (2021) |
122 | First- and second-generation EGFR-TKIs | NGS for 9-genes panel | T790M: 32% (39/122) EGFR amplification: 6.6% (8/122) PIK3CA: 3.3% (4/122) MET amplification: 3.3% (4/122) HER2 amplification: 4.1% (5/122) |
| [102] (2020) |
11 | Third -generation EGFR-TKIs | Nanowire-based colorimetric cfDNA assay (EGFR mutation and MET amplification) | Plasma cfDNA profiles Drug-sensitive EGFR founder mutations: 36.3% (4/11) De novo EGFR C797S: 18.2% (2/11) MET amplification: 18.2% (2/11) EGFR T790M: 18.2% (2/11) CSF-cfDNA: Drug-sensitive EGFR founder mutations: 45.5% (5/11) De novo EGFR C797S: 36.3% (4/11) MET amplification: 18.2% (2/11) EGFR T790M: 18.2% (2/11) |
| [109] (2022) |
49 | Third-generation EGFR-TKIs | ddPCR (MET copy number gain) | MET CNG: 26.5% (13/49) MET amplification:16.3% (8/49) |
| [100] (2020) |
26 | Osimertinib | ddPCR | EGFR C797S: 20% (3/15) Loss of T790M: 33.3% (4/15) |
| [107] (2021) |
56 | Osimertinib | NGS for 39-genes panel | Second-line osimertinib (n = 41) EGFR C797S: 39% (16/41) Non-C797S EGFR mutations: 12% (5/41) V843I, L718Q, C724S, L792H and one patient with L718V, L718Q, L792H and G796S RB1 and TP53 inactivating mutations: 7% (3/41) EGFR amplification: 10% (4/41) MET amplification: 7% (3/41) CTNNB1 point mutations: 7% (3/41) KRAS mutations: 5% (2/41) PIK3CA activating mutations: 5% (2/41) ERBB2, PTEN, mTOR and RET mutations: 2% (1/41 each) AGK-BRAF, RET-RUFY1, TACC-FGFR3 and DLG1-BRAF fusion: 2% (1/41 each) Loss of T790M: 34% (14/41) First-line osimertinib (n = 7) EGFR alterations (EGFR C797S and EGFR T854A): 28.5% (2/7) MET amplification: 14.2% (1/7) EML4-ALK fusions: 14.2% (1/7) SCC/SCLC switch [RB1(R787*)]: 14.2% (1/7) |
| [93] (2017) |
19 | Osimertinib | NGS for 73-genes panel | MET amplification: 5.3% (n = 1) EGFR and KRAS amplification: 5.3% (n = 1) MEK1, KRAS or PIK3CA mutations: 5.3% (n = 1 each) EGFR C797S: 10.6% (n = 2) JAK2 mutation: 5.3% (n = 1) HER2 exon 20 insertion: 5.3% (n = 1) |
| [92] (2022) |
- | EGFR-TKIs | NGS for 74-genes panel | First- and second-generation EGFR-TKIs (n = 490) EGFR T790M: 48.0% (235/490) MET amplification: 7.1% (35/490) BRAF V600E: 1.0% (5/490) KRAS mutations: 3.6% (20/490) Third-generation EGFR-TKIs (n = 205) MET amplification: 8.9% (16/205) EGFR C797S: 5.6% (10/205) BRAF V600E: 4.5% (8/205) KRAS mutation: 3.4% (7/205) |
Abbreviations: EGFR-TKIs, epidermal growth factor receptor tyrosine kinase inhibitors; NGS, next-generation sequencing; ddPCR, droplet digital PCR; dPCR, digital PCR.
Papadimitrakopoulou et al. evaluated different techniques for the detection of EGFR mutations in the ctDNA of EGFR T790M-positive NSCLC patients in the AURA3 study and found that the positive percent agreement (PPA) between droplet digital polymerase chain reaction (ddPCR) and NGS was similar and that both of these methods were more sensitive than the Cobas EGFR Mutation Test v2 (Cobas plasma) [49]. However, another study showed that the abundance of T790M detected by ddPCR was significantly lower than that detected by NGS [106]. No significant differences have been found between the detection rate of EGFR T790M in tissues and that in ctDNA, and the consistency has been reported to be 50–79.4% [98,99,101,106,112]. The detection of T790M in plasma has been associated with a larger median baseline tumor size and the presence of extrathoracic diseases [49], particularly worsening bone disease [103]. Patients with EGFR ex19del have been found to be more likely to develop EGFR T790M resistance than patients with EGFR L858R (62.1% vs. 19.3%; p = 0.007) [91]. In particular, patients with delE746_A750 have been reported to be more likely to develop T790M mutations than patients with delS752_I759 or L858R [113]. Another study showed that female patients were more likely to develop T790M mutations than male patients [104]. Some investigators have evaluated the sensitivity of EGFR mutation screening to different cfDNA sources and their potential combinations, but unfortunately, no evidence has been reported of any improvements from the use of combinations of alternative sources, such as urine and exhaled breath condensate (EBC) [114].
Based on the results of the AURA3 trial, osimertinib was approved as the standard treatment for NSCLC patients with EGFR T790M mutations who had previously been treated with first- or second-generation EGFR-TKIs [59]. In 2016, the Cobas EGFR mutation Test v2 was approved by the US FDA for the analysis of T790M in plasma. However, for patients who have T790M detected in their plasma using this method, there has been a lack of prospective data on the clinical efficacy of osimertinib. Takahama et al. first evaluated the efficacy of osimertinib for those patients, and the results showed that the ORR of patients with palsma T790M that was detectable by the Cobas test was 55.1% (95% CI = 40.2–69.3%) and that the median PFS was 8.3 months (95% CI = 6.9–12.6 months) [52]. In fact, osimertinib can target not only the T790M mutation in tumors but also EGFR-sensitizing mutations in tumors. One study showed that both EGFR-sensitizing and T790M EGFR mutant fractions (MFs) decreased in ctDNA during osimertinib treatment, while a rebound in EGFR-sensitizing MFs was observed upon PD/treatment cessation. Significant differences were observed in ORR and PFS between the EGFR-sensitizing MF-high and EGFR-sensitizing MF-low groups at treatment cycle 4 [53]. Another study showed that patients with a low T790M relative allele frequency (RAF) in their plasma (<20%), which was calculated as the ratio of the allele frequency (AF) of T790M to the AF of sensitizing mutations, had lower ORR (0 vs. 68.8%; p = 0.03) and DCR (60% vs. 100%; p = 0.048) values when treated with osimertinib compared to patients with a high T790M RAF, suggesting that non-response to osimertinib could be due to alternative resistance mechanisms to T790M, such as MET or ERBB2 amplification and SCLC transformation. Plasma T790M RAF could also become a novel biomarker for prognostic stratification in the future [112]. Although liquid biopsy is a tool for diagnosing the presence of T790M in patients with EGFR-TKI-resistant NSCLC, tissue biopsy should be considered for patients with low T790M/activating mutation ratios in order to rule out the presence of SCLC transformation and/or other concomitant resistance mechanisms [115]. Patients whose conditions progressed in the absence of T790M mutations have been found to have worse PFS following EGFR-TKI therapy and alternative alterations, including SCLC-associated copy number changes and TP53 mutations in their plasma. These alterations are not only beneficial in tracking subsequent therapeutic responses, but they are also especially important in individuals without T790M or other known resistance mechanisms and could justify repeat-biopsies to confirm histological transformation [116]. SCLC transformation occurred in approximately 10% of patients treated with EGFR-TKIs. Such cases have been found to lose RB1 and TP53 while maintaining the original EGFR mutations, and the median OS with platinum-based chemotherapy has been reported to be 10.8 months [117]. The detection of histological transformation using ctDNA was very challenging. At present, only a few cases have been reported in which inactivated mutations in RB1 and TP53 were detected using ctDNA before the transformation of small cell lung cancer was confirmed by tissue biopsy, indicating a certain suggestive role and prompting the use of early tissue biopsy for diagnostic confirmation. In addition, genetic alterations in EGFR detected using liquid biopsy, especially EGFR amplification, have shown marked genomic instability and genome-wide hypomethylation, and these hypomethylation levels have been associated with the duration of the response to EGFR-TKI therapy [108].
For third-generation EGFR-TKIs (e.g., osimertinib), the most common resistance mechanism was the emergence of new C797S mutations in plasma, which have a reported detection rate of 20.0% [100]. Disease progression in osimertinib-treated patients has exhibited distinct molecular patterns, including sensitization+/T790M+/C797S+, sensitization+/T790M+/C797S−, and sensitization+/T790M−/C797S−, with median progression times of 12.27 months, 2.17 months, and 4.87 months, respectively [47]. In another study, the researchers analyzed real-world data from 56 patients with metastatic NSCLC who underwent liquid biopsies as the disease progressed. They found that T790M did not occur in patients receiving first-line osimertinib treatment, while it did occur in about 90% of patients after first- or second-generation EGFR-TKI therapy. After switching to osimertinib for second-line treatment, T790M was “lost” in 34% of these patients, and multiple EGFR and PIK3CA molecular subclones were present in their plasma ctDNA, confirming the complexity of the resistance mechanisms to osimertinib [113]. The second most common resistance mechanism was MET amplification. Patients who were resistant to third-generation EGFR-TKIs have been found to have significantly increased rates of MET amplification [109], occurring in approximately 15% of cases, compared to patients who were resistant to first- and second-generation EGFR-TKIs [118]. Moreover, the patterns of genomic alterations in patients with innate and acquired resistance to osimertinib were significantly different. Researchers have used cancer personalized profiling by deep sequencing (CAPP-seq) to analyze ctDNA and have found that PIK3CA, KRAS, or BRAF mutations and copy number gain in EGFR, ERBB2, or MET were more common in patients with innate drug resistance [119]. Some studies have reported the discovery of some rare acquired resistance mutations through ctDNA testing, such as EGFR C797G [120], TP53 R273C and KRAS G12V [106], which could impair the effectiveness of osimertinib.
Osimertinib has potent activity against EGFR T790M-positive NSCLC with central nervous system (CNS) metastases. However, not much research has been conducted on the accuracy of the molecular analysis of CSF, especially in terms of its relationship to treatment outcomes. The results of a real-world study on NSCLC patients with CNS metastases showed that CSF was superior to plasma for detecting actionable mutations. The maximal somatic allele frequency (MSAF) was a useful bioinformatics tool for estimating the tumor fractions of cfDNA. In terms of consistency between paired CSF and plasma samples, the MSAF of CSF has been reported to be significantly higher than that of paired plasma cfDNA (p < 0.001). It has also been found that CSF was more able to detect changes than plasma, especially changes in copy number variations (CNV) and structural variation (SV). One study confirmed that fluid biopsies using CSF showed great potential for the identification of viable mutations and the exploration of potential resistance mechanisms in NSCLC patients with CNS metastases. However, the study did not analyze the clinical significance of genotyping based on CSF for treatment outcomes [121]. Another study also showed that plasma cfDNA did not reflect leptomeningeal metastases (LM) status very well and that CSF cfDNA was superior to plasma cfDNA for identifying mutations. The lack of effective exposure to first- and second-generation EGFR-TKIs could be one of the reasons that EGFR T790M mutation detection was less common in the CSF cfDNA of patients with meningeal metastases. However, for patients receiving the third-generation EGFR-TKI osimertinib, EGFR C797S mutations were almost evenly distributed in CSF and paired plasma [122]. Zheng et al. explored the clinical significance of paired CSF and plasma genotyping in NSCLC patients with LM for the first time, and the results showed that the identification of EGFR ex19del and T790M in CSF indicated better osimertinib efficacy (the median intracranial PFS and overall PFS were significantly longer in CSF T790M-positive patients than those in T790M-negative patients, while plasma T790M status was not associated with osimertinib efficacy). Patients who tested negative for T790M also benefited from osimertinib, but only those with EGFR ex19del or no detectable FGF3 co-mutations. Changes in the genes involved in cell cycle pathways (e.g., CDK4, CDKN2A, etc.) reduced the efficacy of osimertinib. In addition, CSF could reveal the resistance mechanisms of LM to osimertinib, such as the C797S mutation, MET dysregulation, TP53+RB1 coexistence, etc. [123].
Choi et al. evaluated whether the nanowire-based genotyping of cfDNA in CSF was useful in the treatment of LM in EGFR-mutated NSCLC. The results showed that for patients with EGFR-mutated NSCLC that progressed to LM after treatment with third-generation EGFR-TKIs, EGFR C797S was the most frequently detected mutation in CSF and its level decreased with improvements in radiation or neurological function, whereas T790M mutation levels in plasma were significantly elevated before disease progression, suggesting that nanowire technology-based CSF cfDNA genotyping could be feasible and effective for guiding LM therapy in patients with EGFR-mutated NSCLC [102].
Currently, Guardant360 CDx and FoundationOne Liquid CDx are approved by the FDA for NGS assays for gene fusion detection of DNA extracted from blood samples. However, a number of studies have shown that 5–15% of the samples tested by the DNA-NGS panel as fusion gene negative are still positive by the RNA-NGS test [124,125]. The research results of Li et al. showed that some fusion genes detected by DNA-NGS could not be used as therapeutic targets [126]. In order to solve the limitations of DNA-based fusion detection, clinical practice strategies mainly include “parallel detection” and “sequential detection” [127], but there are still many difficulties in practice. How to make more reasonable and effective use of liquid biopsy technology to explore the mechanism of EGFR-TKI resistance and guide clinical treatment still needs to be further explored.
4. A Marker for the New Era of NSCLC Treatment-Minimal or Molecular Residual Disease (MRD)
MRD refers to the molecular abnormalities of the origin of cancer that cannot be detected using conventional imaging (including PET/CT) or laboratory methods after treatment but can be detected by liquid biopsy and represent the persistence of lung cancer and the possibility of clinical progression. These potential sources of tumor recurrence are strongly associated with poor outcomes for patients. At present, the detection of MRD mainly relies on liquid biopsy. Using non-invasive detection methods, residual tumor lesions can provide tumor progression and specific molecular information, predict the prognosis of patients, and further guide follow-up treatment plans. In lung cancer, MRD-related research is still in the process of accumulating evidence from previous studies. The definitions of and research methods for MRD have varied greatly between different studies, and multiple ongoing trials on ctDNA-determined MRD should reveal more data regarding its value during perioperative and follow-up monitoring.
MRD is used not only to assess the risk of recurrence in patients with early-stage lung cancer but also to inform the selection of adjuvant therapy after radical local treatment (RLT) [128,129]. Since the TRACERx study in 2017 found that the dynamic detection of ctDNA in plasma could predict tumor recurrence in advance and that it had a certain correlation with the efficacy of postoperative adjuvant therapy [130], a number of subsequent studies have been carried out to explore these features further (Table 3) [131,132,133,134,135,136,137,138,139,140]. Postoperative ctDNA detection is more sensitive than imaging and can indicate disease progression or recurrence earlier. It is also helpful for auxiliary diagnosis and the formulation of follow-up treatment plans when the patient tumor burdens are low, thereby playing a key role in improving the prognoses of patients. However, the optimal monitoring time window and the exact lead time still need to be confirmed by extensive research.
Table 3.
The prognostic significance of circulating tumor DNA (ctDNA) detection at different time periods for resectable NSCLC.
| References (Year) |
Sample | Stage | Detection Methods/Study Design | Median Follow-Up Time (Month) | Detection Time | Clinical Relevance | ctDNA Positivity Precedes Radiological Recurrence by a Median Lead Time (Month) |
|---|---|---|---|---|---|---|---|
| [131] (2019) |
26 | I–III | NGS for a 9-gene panel/Pro | 532 days for all patients and 629 days for patients who were free from progression |
B: 5 min; C: 30 min; D: 2 h
|
|
NR |
| [132] (2020) |
20 | IIA–IIIA | NGS for a 197-gene panel/Pro | 12 |
|
|
NR |
| [133] (2020) |
38 | IB–III | NGS for a 425-gene panel/Pro | 15.8 |
|
|
NR |
| [134] (2021) |
174 | I–III | ARMs for EGFR/Pro | NA |
|
|
NR |
| [135] (2021) |
116 | I–IV | NGS for a 139-gene panel/Pro | NA |
|
Proportion of patients who were ctDNA-positive after surgery: 21.2% (18/85); after the completion of ACT: 12.5% (8/64)
|
2.93 |
| [136] (2021) |
77 | I–IV | cSMART for a 127-gene panel/Pro | 46 |
|
|
12.6 |
| [137] (2021) |
119 | I–IIIA | NGS for a 425-gene panel/Pro | 30.7 |
|
|
8.71 |
| [138] (2022) |
21 | IA–IIIB | NGS for an18-gene panel/Pro | 26.2 |
|
|
10.31 |
| [139] (2022) |
330 | I–III | NGS for a 769-gene panel/Pro | 35.6 |
|
|
NR |
| [140] (2022) |
88 | IA–IIIB | RaDaRTMNGS | 36 |
surgery + adjuvant chemotherapy/radiotherapy (n = 8); chemoradiotherapy (n = 19) |
|
7.08 |
Multiple studies have shown that the detection of MRD can have significant clinical relevance for guiding postoperative adjuvant therapy in NSCLC patients. Chen et al. found that ctDNA decayed rapidly after tumor resection in lung cancer patients. In general, 3 days post-surgery can be used as a benchmark for the postoperative monitoring of lung cancer. In patients who were ctDNA-positive 3 days after surgery, the recurrence-free survival (RFS) rate with adjuvant therapy has been found to be 269 days, while the RFS for those who did not receive adjuvant therapy has been found to be 111 days (p = 0.018). Changes in ctDNA are related to therapeutic efficacy, but further research is needed to determine whether there is a beneficial effect on OS from blood-based MRD testing [131]. Qiu et al. evaluated the predictive power of ctDNA for dynamic recurrence risk and adjuvant chemotherapy (ACT) benefit in resectable NSCLC patients. They found that ctDNA-positive patients had a significantly higher risk of recurrence compared to ctDNA-negative patients, whether in the ACT group (p < 0.05) or the non-ACT group (p < 0.05). The risk of recurrence was similar in ctDNA-negative patients, regardless of ACT use (p =0.46). Additionally, ctDNA-positive patients who were treated with ACT had a significantly longer RFS than ctDNA-positive patients who did not receive ACT (p < 0.05) [135]. A study conducted by Xia et al. also showed similar results, in that MRD-positive patients who received adjuvant therapy had improved RFS compared to those who did not receive adjuvant therapy (HR = 0.3; p = 0.008) [139]. Therefore, clinically high-risk NSCLC patients can be divided into two groups based on postoperative ctDNA status: ctDNA-positive patients, who were more likely to benefit from ACT therapy, and ctDNA-negative patients, who did not appear to require ACT but may benefit from slight improvements in reducing the risk of recurrence [135]. Kuang et al. found that, among patients who were ctDNA-positive after surgery, the RFS of patients who were ctDNA-positive after chemotherapy was worse than that of patients who were ctDNA-negative after chemotherapy (HR = 8.68; p = 0.022). For patients who were ctDNA-negative after surgery, those who were ctDNA-negative after chemotherapy had better long-term efficacy than those who were ctDNA-positive after chemotherapy (HR = 4.76; p = 0.047). They also found that post-chemotherapy ctDNA status was closely related to RFS and could serve as a guide for intensive postoperative treatment [133]. Another recent study investigated the association between ctDNA changes and neoadjuvant therapy (NAT) response and postoperative RFS. The results showed that ctDNA kinetics during NAT were highly consistent with pathological responses, with a sensitivity of 100%, a specificity of 83.33%, and an overall accuracy of 91.67%. Preoperatively detectable ctDNA (post-NAT) tended to be associated with worse RFS. The presence of ctDNA 3 months after surgery showed an 83% sensitivity and a 90% specificity for predicting recurrence. Molecular recurrence was better detected using ctDNA than radiography during postoperative disease surveillance, with a median time of 6.83 months. They also found that perioperative ctDNA analysis could evaluate the efficacy of NAT, reflect postoperative MRD, and predict postoperative recurrence [141].
For locally advanced or advanced lung cancer, MRD is mainly used to assess no evidence of disease (NED), i.e., complete response (CR) after treatment, oligometastatic disease (OMD) after surgery, no evidence of active disease based on existing imaging techniques, etc. [142]. CR rates in patients with advanced NSCLC after targeted therapy or immunotherapy have been reported to range from 1% to 7%. In patients with CR, monitoring for disease recurrence is a major clinical concern. The optimal duration of treatment and the availability and duration of “drug holidays” for patients receiving immune checkpoint inhibitors with durable responses remain unknown. At the same time, drug resistance is also a major challenge for patients receiving targeted therapy. In addition, approximately 30–50% of advanced lung cancers are oligometastatic at initial diagnosis [143,144,145]. Whether MRD can guide the next steps of systemic therapy when the metastatic lesions of patients with oligometastases are treated with radical therapy deserves further exploration. However, there have not been many related studies on MRD in patients with advanced NSCLC, and only a few studies have shown that the presence of ctDNA at baseline was an independent marker of poor prognosis in patients with advanced NSCLC who were receiving chemotherapy or targeted therapy. Negative ctDNA detection at the first assessment after treatment initiation has major prognostic impacts on both PFS and OS [146]. Circulating tumor DNA dynamics could predict whether patients with locally advanced NSCLC would benefit from consolidation immunotherapy [147]. For long-term responders (PFS ≥ 12 months) to PD-L1 blockade, ctDNA analysis could differentiate between those who would experience sustained benefits and those at risk of eventual progression [148]. Tang et al. performed dynamic ctDNA detection in 21 patients with oligometastatic lung cancer, 10 of whom were in the local intervention group, and the results suggested that the baseline ctDNA parameters were not associated with the PFS or OS of the patients. In the subsequent follow-up, a significant decrease in ctDNA load was observed in the local intervention group, which suggested that the local intervention could reduce the overall tumor loads of the patients. In addition, an increase in ctDNA burden was also seen in the final five patients with recurrence detected using radiography, with a mean advanced prediction time window of 6.7 months (2.9–17.9 months). Therefore, when patients with oligometastatic diseases undergo local therapy to reduce systemic tumor burden, it would be a feasible scientific hypothesis that MRD could be used to guide systemic therapy [149]. A large number of prospective studies are still needed to explore MRD-based treatment strategies for patients with locally advanced or advanced lung cancer.
Bodily fluids, such as sputum, saliva, urine, pleural effusion, and bronchoalveolar lavage fluid (BALF), could contain more tumor information than blood and reflect tumor heterogeneity in more detail. However, the emerging non-blood-derived fluid biopsy lacks consensus and clinical validation as a pretreatment and analytical method, and there are still many problems that need to be addressed. Thus, although non-blood-derived fluid biopsy could provide more opportunities to improve individualized targeted therapy and prognosis in lung cancer patients with MRD, more research is necessary to determine how best to combine information from tumor biopsy, clinical examinations, and medical imaging with genomic and MRD information from liquid biopsy [150].
5. Conclusions
Liquid biopsy could open up a new era for the diagnosis and treatment of lung cancer, especially when tissue biopsy cannot be performed on patients. However, more advanced molecular biological detection methods are still needed to enhance the reliability and practicality of liquid biopsy. Although liquid biopsy is not yet a substitute for tissue biopsy, with the development of high-throughput detection, early tumor screening, artificial intelligence, and other technologies, it may not merely be a dream. As a detection method for cancer, liquid biopsy would become a powerful tool that is not only consistent with pathological diagnosis but can also provide richer molecular biological information, thus offering greater potential for precise medication, dynamic detection, prognostic evaluation, and even stifling tumors in the cradle.
Author Contributions
Conceptualization and supervision: H.-M.Z.; Data curation and writing—original draft preparation: Y.-Z.L. and S.-N.K.; writing—review and editing: Y.-Z.L., Y.-P.L., and Y.Y. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the Natural Science Basic Research Program of Shaanxi (No. 2021JZ-35) and the National Natural Science Foundation of China (No. 81702554).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Yoneda K., Imanishi N., Ichiki Y., Tanaka F. Treatment of Non-small Cell Lung Cancer with EGFR-mutations. J. UOEH. 2019;41:153–163. doi: 10.7888/juoeh.41.153. [DOI] [PubMed] [Google Scholar]
- 2.Mielgo-Rubio X., Martín M., Remon J., Higuera O., Calvo V., Jarabo J.R., Conde E., Luna J., Provencio M., De, Castro J., et al. Targeted therapy moves to earlier stages of non-small-cell lung cancer: Emerging evidence, controversies and future challenges. Future Oncol. 2021;17:4011–4025. doi: 10.2217/fon-2020-1255. [DOI] [PubMed] [Google Scholar]
- 3.Chaft J.E., Shyr Y., Sepesi B., Forde P.M. Preoperative and Postoperative Systemic Therapy for Operable Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2022;40:546–555. doi: 10.1200/JCO.21.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Matera J., Miller M.C., Reuben J.M., Doyle G.V., Allard W.J., Terstappen L.W., et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 5.Pisapia P., Malapelle U., Troncone G. Liquid Biopsy and Lung Cancer. Acta. Cytol. 2019;63:489–496. doi: 10.1159/000492710. [DOI] [PubMed] [Google Scholar]
- 6.Schwarzenbach H., Hoon D.S., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 7.Cui S., Cheng Z., Qin W., Jiang L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer. 2018;116:46–54. doi: 10.1016/j.lungcan.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Jiang N., Meng X., Mi H., Chi Y., Li S., Jin Z., Tian H., He J., Shen W., Tian H., et al. Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin. Chim. Acta. 2018;486:26–33. doi: 10.1016/j.cca.2018.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Xing S., Zeng T., Xue N., He Y., Lai Y.Z., Li H.L., Huang Q., Chen S.L., Liu W.L. Development and Validation of Tumor-educated Blood Platelets Integrin Alpha 2b (ITGA2B) RNA for Diagnosis and Prognosis of Non-small-cell Lung Cancer through RNA-seq. Int. J. Biol. Sci. 2019;15:1977–1992. doi: 10.7150/ijbs.36284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei Y., Sun N., Zhang G., Liu C., Lu Z., Huang J., Zhang C., Zang R., Che Y., Mao S., et al. Combined detection of aneuploid circulating tumor-derived endothelial cells and circulating tumor cells may improve diagnosis of early stage non-small-cell lung cancer. Clin. Transl. Med. 2020;10:e128. doi: 10.1002/ctm2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y.J., Ji W.J., Lee J.C., Chun S.M., Choi C.M. Assessment of Anti-tumor Efficacy of Osimertinib in Non-Small Cell Lung Cancer Patients by Liquid Biopsy Using Bronchoalveolar Lavage Fluid, Plasma, or Pleural Effusion. Cancer Res. Treat. 2022;54:985–995. doi: 10.4143/crt.2021.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang C.L., Huang H.C., Luo Y.H., Chiu C.H. Cerebrospinal fluid as a medium of liquid biopsy in the management of patients with non-small-cell lung cancer having central nervous system metastasis. Front. Biosci. (Landmark Ed). 2021;26:1679–1688. doi: 10.52586/5060. [DOI] [PubMed] [Google Scholar]
- 13.Ryan D.J., Toomey S., Smyth R., Madden S.F., Workman J., Cummins R., Sheehan K., Fay J., Naidoo J., Breathnach O.S., et al. Exhaled Breath Condensate (EBC) analysis of circulating tumour DNA (ctDNA) using a lung cancer specific UltraSEEK oncogene panel. Lung Cancer. 2022;168:67–73. doi: 10.1016/j.lungcan.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Yong E. Cancer biomarkers: Written in blood. Nature. 2014;511:524–526. doi: 10.1038/511524a. [DOI] [PubMed] [Google Scholar]
- 15.Jiang P., Lo Y.M.D. The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends Genet. 2016;32:360–371. doi: 10.1016/j.tig.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Mouliere F., Robert B., Arnau Peyrotte E., Del Rio M., Ychou M., Molina F., Gongora C., Thierry A.R. High fragmentation characterizes tumour-derived circulating DNA. PLoS ONE. 2011;6:e23418. doi: 10.1371/journal.pone.0023418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Underhill H.R., Kitzman J.O., Hellwig S., Welker N.C., Daza R., Baker D.N., Gligorich K.M., Rostomily R.C., Bronner M.P., Shendure J. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan K.C., Jiang P., Zheng Y.W., Liao G.J., Sun H., Wong J., Siu S.S., Chan W.C., Chan S.L., Chan A.T., et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 19.Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A., Loupakis F., Bardelli A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 20.Wong I.H., Lo Y.M., Zhang J., Liew C.T., Ng M.H., Wong N., Lai P.B., Lau W.Y., Hjelm N.M., Johnson P.J. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59:71–73. [PubMed] [Google Scholar]
- 21.Chan K.C., Lai P.B., Mok T.S., Chan H.L., Ding C., Yeung S.W., Lo Y.M. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin. Chem. 2008;54:1528–1536. doi: 10.1373/clinchem.2008.104653. [DOI] [PubMed] [Google Scholar]
- 22.Chan K.C., Jiang P., Chan C.W., Sun K., Wong J., Hui E.P., Chan S.L., Chan W.C., Hui D.S., Ng S.S., et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA. 2013;110:18761–18768. doi: 10.1073/pnas.1313995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M., Thornton K., Agrawal N., Sokoll L., Szabo S.A., et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.To E.W., Chan K.C., Leung S.F., Chan L.Y., To K.F., Chan A.T., Johnson P.J., Lo Y.M. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin. Cancer Res. 2003;9:3254–3259. [PubMed] [Google Scholar]
- 25.Yao W., Mei C., Nan X., Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene. 2016;590:142–148. doi: 10.1016/j.gene.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 26.Douillard J.Y., Ostoros G., Cobo M., Ciuleanu T., Cole R., McWalter G., Walker J., Dearden S., Webster A., Milenkova T., et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: Circulating-free tumor dna as a surrogate for determination of EGFR status. J. Thorac. Oncol. 2014;9:1345–1353. doi: 10.1097/JTO.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mok T., Wu Y.L., Lee J.S., Yu C.J., Sriuranpong V., Sandoval-Tan J., Ladrera G., Thongprasert S., Srimuninnimit V., Liao M., et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015;21:3196–3203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 28.Karachaliou N., Mayo-de las Casas C., Queralt C., de Aguirre I., Melloni B., Cardenal F., Garcia-Gomez R., Massuti B., Sánchez J.M., Porta R., et al. Spanish Lung Cancer Group. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol. 2015;1:149–157. doi: 10.1001/jamaoncol.2014.257. [DOI] [PubMed] [Google Scholar]
- 29.Mayo-de-Las-Casas C., Jordana-Ariza N., Garzón-Ibañez M., Balada-Bel A., Bertrán-Alamillo J., Viteri-Ramírez S., Reguart N., Muñoz-Quintana M.A., Lianes-Barragan P., Camps C., et al. Large scale, prospective screening of EGFR mutations in the blood of advanced NSCLC patients to guide treatment decisions. Ann. Oncol. 2017;28:2248–2255. doi: 10.1093/annonc/mdx288. [DOI] [PubMed] [Google Scholar]
- 30.Xu H., Baidoo A.A.H., Su S., Ye J., Chen C., Xie Y., Bertolaccini L., Ismail M., Ricciuti B., Ng C.S.H., et al. A comparison of EGFR mutation status in tissue and plasma cell-free DNA detected by ADx-ARMS in advanced lung adenocarcinoma patients. Transl. Lung Cancer Res. 2019;8:135–143. doi: 10.21037/tlcr.2019.03.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan R., Wang Z., Lee J.J., Wang S., Li Q., Tang F., Wang J., Sun Y., Bai H., Wang D., et al. Comprehensive Analysis of the Discordance of EGFR Mutation Status between Tumor Tissues and Matched Circulating Tumor DNA in Advanced Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017;12:1376–1387. doi: 10.1016/j.jtho.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z., Cheng Y., An T., Gao H., Wang K., Zhou Q., Hu Y., Song Y., Ding C., Peng F., et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): A phase 2, single-arm, multicentre clinical trial. Lancet Respir. Med. 2018;6:681–690. doi: 10.1016/S2213-2600(18)30264-9. [DOI] [PubMed] [Google Scholar]
- 33.Behel V., Chougule A., Noronha V., Patil V.M., Menon N., Singh A., Chopade S., Kumar R., Shah S., More S., et al. Clinical Utility of Liquid Biopsy (Cell-free DNA) Based EGFR Mutation Detection Post treatment Initiation as a Disease Monitoring Tool in Patients With Advanced EGFR-mutant NSCLC. Clin. Lung Cancer. 2022;23:410–418. doi: 10.1016/j.cllc.2022.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Chen H.F., Lei L., Wu L.X., Li X.F., Zhang Q.X., Pan W.W., Min Y.H., Zhu Y.C., Du K.Q., Wang M., et al. Effect of icotinib on advanced lung adenocarcinoma patients with sensitive EGFR mutation detected in ctDNA by ddPCR. Transl. Cancer Res. 2019;8:2858–2863. doi: 10.21037/tcr.2019.10.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park C.K., Lee S.Y., Lee J.C., Choi C.M., Lee S.Y., Jang T.W., Oh I.J., Kim Y.C. Phase II open-label multicenter study to assess the antitumor activity of afatinib in lung cancer patients with activating epidermal growth factor receptor mutation from circulating tumor DNA: Liquid-Lung-A. Thorac. Cancer. 2021;12:444–452. doi: 10.1111/1759-7714.13763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leighl N.B., Page R.D., Raymond V.M., Daniel D.B., Divers S.G., Reckamp K.L., Villalona-Calero M.A., Dix D., Odegaard J.I., Lanman R.B., et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin. Cancer Res. 2019;25:4691–4700. doi: 10.1158/1078-0432.CCR-19-0624. [DOI] [PubMed] [Google Scholar]
- 37.Rolfo C., Mack P.C., Scagliotti G.V., Baas P., Barlesi F., Bivona T.G., Herbst R.S., Mok T.S., Peled N., Pirker R., et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018;13:1248–1268. doi: 10.1016/j.jtho.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 38.Thompson J.C., Yee S.S., Troxel A.B., Savitch S.L., Fan R., Balli D., Lieberman D., Morrissette J.D., Evans T.L., Bauml J., et al. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin. Cancer Res. 2016;22:5772–5782. doi: 10.1158/1078-0432.CCR-16-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aggarwal C., Thompson J.C., Black T.A., Katz S.I., Fan R., Yee S.S., Chien A.L., Evans T.L., Bauml J.M., Alley E.W., et al. Clinical Implications of Plasma-Based Genotyping with the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019;5:173–180. doi: 10.1001/jamaoncol.2018.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S., Olsen S., Ku B.M., Lee M.S., Jung H.A., Sun J.M., Lee S.H., Ahn J.S., Park K., Choi Y.L., et al. High concordance of actionable genomic alterations identified between circulating tumor DNA-based and tissue-based next-generation sequencing testing in advanced non-small cell lung cancer: The Korean Lung Liquid Versus Invasive Biopsy Program. Cancer. 2021;127:3019–3028. doi: 10.1002/cncr.33571. [DOI] [PubMed] [Google Scholar]
- 41.Sehayek O., Kian W., Onn A., Stoff R., Sorotsky H.G., Zemel M., Bar J., Dudnik Y., Nechushtan H., Rottenberg Y., et al. Liquid First Is “Solid” in Naïve Non-Small Cell Lung Cancer Patients: Faster Turnaround Time With High Concordance to Solid Next-Generation Sequencing. Front. Oncol. 2022;12:912801. doi: 10.3389/fonc.2022.912801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmero R., Taus A., Viteri S., Majem M., Carcereny E., Garde-Noguera J., Felip E., Nadal E., Malfettone A., Sampayo M., et al. Biomarker Discovery and Outcomes for Comprehensive Cell-Free Circulating Tumor DNA Versus Standard-of-Care Tissue Testing in Advanced Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2021;5:93–102. doi: 10.1200/PO.20.00241. [DOI] [PubMed] [Google Scholar]
- 43.Kok P.S., Lee K., Lord S., Yang J.C., Rosell R., Goto K., John T., Wu Y.L., Mok T.S.K., Lee C.K. Clinical utility of plasma EGFR mutation detection with quantitative PCR in advanced lung cancer: A meta-analysis. Lung Cancer. 2021;154:113–117. doi: 10.1016/j.lungcan.2021.02.027. [DOI] [PubMed] [Google Scholar]
- 44.Selvarajah S., Plante S., Speevak M., Vaags A., Hamelinck D., Butcher M., McCready E., Grafodatskaya D., Blais N., Tran-Thanh D., et al. A Pan-Canadian Validation Study for the Detection of EGFR T790M Mutation Using Circulating Tumor DNA From Peripheral Blood. JTO Clin. Res. Rep. 2021;2:100212. doi: 10.1016/j.jtocrr.2021.100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park C.K., Cho H.J., Choi Y.D., Oh I.J., Kim Y.C. A Phase II Trial of Osimertinib in the Second-Line Treatment of Non-small Cell Lung Cancer with the EGFR T790M Mutation, Detected from Circulating Tumor DNA: LiquidLung-O-Cohort 2. Cancer Res. Treat. 2019;51:777–787. doi: 10.4143/crt.2018.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee Y., Park S., Kim W.S., Lee J.C., Jang S.J., Choi J., Choi C.M. Correlation between progression-free survival, tumor burden, and circulating tumor DNA in the initial diagnosis of advanced-stage EGFR-mutated non-small cell lung cancer. Thorac. Cancer. 2018;9:1104–1110. doi: 10.1111/1759-7714.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romero A., Serna-Blasco R., Alfaro C., Sánchez-Herrero E., Barquín M., Turpin M.C., Chico S., Sanz-Moreno S., Rodrigez-Festa A., Laza-Briviesca R., et al. ctDNA analysis reveals different molecular patterns upon disease progression in patients treated with osimertinib. Transl. Lung Cancer Res. 2020;9:532–540. doi: 10.21037/tlcr.2020.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng Q., Fang Q., Sun H., Singh A.P., Alexander M., Li S., Cheng H., Zhou S. Detection of plasma EGFR mutations for personalized treatment of lung cancer patients without pathologic diagnosis. Cancer Med. 2020;9:2085–2095. doi: 10.1002/cam4.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papadimitrakopoulou V.A., Han J.Y., Ahn M.J., Ramalingam S.S., Delmonte A., Hsia T.C., Laskin J., Kim S.W., He Y., Tsai C.M., et al. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: Osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non-small cell lung cancer. Cancer. 2020;126:373–380. doi: 10.1002/cncr.32503. [DOI] [PubMed] [Google Scholar]
- 50.Xing L., Pan Y., Shi Y., Shu Y., Feng J., Li W., Cao L., Wang L., Gu W., Song Y., et al. Biomarkers of Osimertinib Response in Patients with Refractory, EGFR-T790M-positive Non-Small Cell Lung Cancer and Central Nervous System Metastases: The APOLLO Study. Clin. Cancer Res. 2020;26:6168–6175. doi: 10.1158/1078-0432.CCR-20-2081. [DOI] [PubMed] [Google Scholar]
- 51.Yu H.A., Schoenfeld A.J., Makhnin A., Kim R., Rizvi H., Tsui D., Falcon C., Houck-Loomis B., Meng F., Yang J.L., et al. Effect of Osimertinib and Bevacizumab on Progression-Free Survival for Patients with Metastatic EGFR-Mutant Lung Cancers: A Phase 1/2 Single-Group Open-Label Trial. JAMA Oncol. 2020;6:1048–1054. doi: 10.1001/jamaoncol.2020.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahama T., Azuma K., Shimokawa M., Takeda M., Ishii H., Kato T., Saito H., Daga H., Tsuboguchi Y., Okamoto I., et al. Plasma screening for the T790M mutation of EGFR and phase 2 study of osimertinib efficacy in plasma T790M-positive non-small cell lung cancer: West Japan Oncology Group 8815L/LPS study. Cancer. 2020;126:1940–1948. doi: 10.1002/cncr.32749. [DOI] [PubMed] [Google Scholar]
- 53.Sakai K., Takahama T., Shimokawa M., Azuma K., Takeda M., Kato T., Daga H., Okamoto I., Akamatsu H., Teraoka S., et al. Predicting osimertinib-treatment outcomes through EGFR mutant-fraction monitoring in the circulating tumor DNA of EGFR T790M-positive patients with non-small cell lung cancer (WJOG8815L) Mol. Oncol. 2021;15:126–137. doi: 10.1002/1878-0261.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park C.K., Cho H.J., Choi Y.D., Oh I.J., Kim Y.C. A Phase II Trial of Osimertinib as the First-Line Treatment of Non-Small Cell Lung Cancer Harboring Activating EGFR Mutations in Circulating Tumor DNA: LiquidLung-O-Cohort 1. Cancer Res. Treat. 2021;53:93–103. doi: 10.4143/crt.2020.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., Liu Y., Meng Z., Wu Y., Wang S., Jin G., Qin Y., Wang F., Wang J., Zhou H., et al. Plasma EGFR mutation abundance affects clinical response to first-line EGFR-TKIs in patients with advanced non-small cell lung cancer. Ann. Transl. Med. 2021;9:635. doi: 10.21037/atm-20-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X.L., Bai R.L., Chen X., Zhao Y.G., Wang X., Ma K.W., Tian H.M., Han F.J., Liu Z.L., Yang L., et al. Correlation of circulating tumor DNA EGFR mutation levels with clinical outcomes in patients with advanced lung adenocarcinoma. Chin. Med. J. (Engl.) 2021;134:2430–2437. doi: 10.1097/CM9.0000000000001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y., Xu Z., Wang S., Zhu Y., Ma D., Mu Y., Ying J., Li J., Xing P. Disease monitoring of epidermal growth factor receptor (EGFR)-mutated non-small-cell lung cancer patients treated with tyrosine kinase inhibitors via EGFR status in circulating tumor DNA. Thorac. Cancer. 2022;13:2201–2209. doi: 10.1111/1759-7714.14545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu J., Liu Z., Bai H., Dong G., Zhong J., Wan R., Zang A., Li X., Li Q., Guo J., et al. Evaluation of Clinical Outcomes of Icotinib in Patients With Clinically Diagnosed Advanced Lung Cancer With EGFR-Sensitizing Variants Assessed by Circulating Tumor DNA Testing: A Phase 2 Nonrandomized Clinical Trial. JAMA Oncol. 2022;8:1328–1332. doi: 10.1001/jamaoncol.2022.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mok T.S., Wu Y.L., Ahn M.J., Garassino M.C., Kim H.R., Ramalingam S.S., Shepherd F.A., He Y., Akamatsu H., Theelen W.S., et al. AURA3 Investigators. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gray J.E., Okamoto I., Sriuranpong V., Vansteenkiste J., Imamura F., Lee J.S., Pang Y.K., Cobo M., Kasahara K., Cheng Y., et al. Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: Osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019;25:6644–6652. doi: 10.1158/1078-0432.CCR-19-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morgensztern D., Waqar S., Subramanian J., Gao F., Trinkaus K., Govindan R. Prognostic significance of tumor size in patients with stage III non-small-cell lung cancer: A surveillance, epidemiology, and end results (SEER) survey from 1998 to 2003. J. Thorac. Oncol. 2012;7:1479–1484. doi: 10.1097/JTO.0b013e318267d032. [DOI] [PubMed] [Google Scholar]
- 62.Zhao M., Zhang Y., Cai W., Li J., Zhou F., Cheng N., Ren R., Zhao C., Li X., Ren S., et al. The Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer. 2014;120:2299–2307. doi: 10.1002/cncr.28725. [DOI] [PubMed] [Google Scholar]
- 63.Canale M., Petracci E., Delmonte A., Chiadini E., Dazzi C., Papi M., Capelli L., Casanova C., De Luigi N., Mariotti M., et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin. Cancer Res. 2017;23:2195–2202. doi: 10.1158/1078-0432.CCR-16-0966. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Q., Zhang X.C., Chen Z.H., Yin X.L., Yang J.J., Xu C.R., Yan H.H., Chen H.J., Su J., Zhong W.Z., et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J. Clin. Oncol. 2011;29:3316–3321. doi: 10.1200/JCO.2010.33.3757. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Z.R., Wang J.F., Lin Y.B., Wang F., Fu S., Zhang S.L., Su X.D., Jiang L., Zhang Y.G., Shao J.Y., et al. Mutation abundance affects the efficacy of EGFR tyrosine kinase inhibitor readministration in non-small-cell lung cancer with acquired resistance. Med. Oncol. 2014;31:810. doi: 10.1007/s12032-013-0810-6. [DOI] [PubMed] [Google Scholar]
- 66.Wang H., Zhang M., Tang W., Ma J., Wei B., Niu Y., Zhang G., Li P., Yan X., Ma Z. Mutation abundance affects the therapeutic efficacy of EGFR-TKI in patients with advanced lung adenocarcinoma: A retrospective analysis. Cancer Biol. Ther. 2018;19:687–694. doi: 10.1080/15384047.2018.1450115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linardou H., Dahabreh I.J., Kanaloupiti D., Siannis F., Bafaloukos D., Kosmidis P., Papadimitriou C.A., Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: A systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 68.Poulikakos P.I., Zhang C., Bollag G., Shokat K.M., Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sos M.L., Koker M., Weir B.A., Heynck S., Rabinovsky R., Zander T., Seeger J.M., Weiss J., Fischer F., Frommolt P., et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharma S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A., et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sequist L.V., Waltman B.A., Dias-Santagata D., Digumarthy S., Turke A.B., Fidias P., Bergethon K., Shaw A.T., Gettinger S., Cosper A.K., et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campo M., Gerber D., Gainor J.F., Heist R.S., Temel J.S., Shaw A.T., Fidias P., Muzikansky A., Engelman J.A., Sequist L.V. Acquired Resistance to First-Line Afatinib and the Challenges of Prearranged Progression Biopsies. J. Thorac. Oncol. 2016;11:2022–2026. doi: 10.1016/j.jtho.2016.06.032. [DOI] [PubMed] [Google Scholar]
- 73.Leonetti A., Sharma S., Minari R., Perego P., Giovannetti E., Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer. 2019;121:725–737. doi: 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeng Y., Yu D., Tian W., Wu F. Resistance mechanisms to osimertinib and emerging therapeutic strategies in nonsmall cell lung cancer. Curr. Opin. Oncol. 2022;34:54–65. doi: 10.1097/CCO.0000000000000805. [DOI] [PubMed] [Google Scholar]
- 75.Niederst M.J., Hu H., Mulvey H.E., Lockerman E.L., Garcia A.R., Piotrowska Z., Sequist L.V., Engelman J.A. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin. Cancer Res. 2015;21:3924–3933. doi: 10.1158/1078-0432.CCR-15-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arulananda S., Do H., Musafer A., Mitchell P., Dobrovic A., John T. Combination Osimertinib and Gefitinib in C797S and T790M EGFR-Mutated Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017;12:1728–1732. doi: 10.1016/j.jtho.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 77.Wang Z., Yang J.J., Huang J., Ye J.Y., Zhang X.C., Tu H.Y., Han Z.H., Wu Y.L. Lung Adenocarcinoma Harboring EGFR T790M and In Trans C797S Responds to Combination Therapy of First- and Third-Generation EGFR TKIs and Shifts Allelic Configuration at Resistance. J. Thorac. Oncol. 2017;12:1723–1727. doi: 10.1016/j.jtho.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 78.Eng J., Woo K.M., Sima C.S., Plodkowski A., Hellmann M.D., Chaft J.E., Kris M.G., Arcila M.E., Ladanyi M., Drilon A. Impact of Concurrent PIK3CA Mutations on Response to EGFR Tyrosine Kinase Inhibition in EGFR-Mutant Lung Cancers and on Prognosis in Oncogene-Driven Lung Adenocarcinomas. J. Thorac. Oncol. 2015;10:1713–1719. doi: 10.1097/JTO.0000000000000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bean J., Riely G.J., Balak M., Marks J.L., Ladanyi M., Miller V.A., Pao W. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin. Cancer Res. 2008;14:7519–7525. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Costa D.B., Schumer S.T., Tenen D.G., Kobayashi S. Differential responses to erlotinib in epidermal growth factor receptor (EGFR)-mutated lung cancers with acquired resistance to gefitinib carrying the L747S or T790M secondary mutations. J. Clin. Oncol. 2008;26:1182–1184. doi: 10.1200/JCO.2007.14.9039. [DOI] [PubMed] [Google Scholar]
- 81.Engelman J.A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J.O., Lindeman N., Gale C.M., Zhao X., Christensen J., et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 82.Yu H.A., Arcila M.E., Rekhtman N., Sima C.S., Zakowski M.F., Pao W., Kris M.G., Miller V.A., Ladanyi M., Riely G.J. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patil T., Mushtaq R., Marsh S., Azelby C., Pujara M., Davies K.D., Aisner D.L., Purcell W.T., Schenk E.L., Pacheco J.M., et al. Clinicopathologic Characteristics, Treatment Outcomes, and Acquired Resistance Patterns of Atypical EGFR Mutations and HER2 Alterations in Stage IV Non-Small-Cell Lung Cancer. Clin. Lung Cancer. 2020;21:e191–e204. doi: 10.1016/j.cllc.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yonesaka K., Takegawa N., Watanabe S., Haratani K., Kawakami H., Sakai K., Chiba Y., Maeda N., Kagari T., Hirotani K., et al. An HER3-targeting antibody-drug conjugate incorporating a DNA topoisomerase I inhibitor U3-1402 conquers EGFR tyrosine kinase inhibitor-resistant NSCLC. Oncogene. 2019;38:1398–1409. doi: 10.1038/s41388-018-0517-4. [DOI] [PubMed] [Google Scholar]
- 85.Lin M.W., Su K.Y., Su T.J., Chang C.C., Lin J.W., Lee Y.H., Yu S.L., Chen J.S., Hsieh M.S. Clinicopathological and genomic comparisons between different histologic components in combined small cell lung cancer and non-small cell lung cancer. Lung Cancer. 2018;125:282–290. doi: 10.1016/j.lungcan.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 86.Ho C.C., Liao W.Y., Lin C.A., Shih J.Y., Yu C.J., Yang J.C. Acquired BRAF V600E Mutation as Resistant Mechanism after Treatment with Osimertinib. J. Thorac. Oncol. 2017;12:567–572. doi: 10.1016/j.jtho.2016.11.2231. [DOI] [PubMed] [Google Scholar]
- 87.Offin M., Somwar R., Rekhtman N., Benayed R., Chang J.C., Plodkowski A., Lui A.J.W., Eng J., Rosenblum M., Li B.T., et al. Acquired ALK and RET Gene Fusions as Mechanisms of Resistance to Osimertinib in EGFR-Mutant Lung Cancers. JCO Precis. Oncol. 2018;2:PO.18.00126. doi: 10.1200/PO.18.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schrock A.B., Zhu V.W., Hsieh W.S., Madison R., Creelan B., Silberberg J., Costin D., Bharne A., Bonta I., Bosemani T., et al. Receptor Tyrosine Kinase Fusions and BRAF Kinase Fusions are Rare but Actionable Resistance Mechanisms to EGFR Tyrosine Kinase Inhibitors. J. Thorac. Oncol. 2018;13:1312–1323. doi: 10.1016/j.jtho.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 89.Helissey C., Favre L., Nguyen A.T., Mamou E., Lamboley J.L. What management for epidermal growth factor receptor-mutated non-small-cell lung cancer, with squamous cell transformation and T790M-acquired resistance mechanisms? A Case report and review of literature. Anticancer Drugs. 2022;33:e720–e723. doi: 10.1097/CAD.0000000000001139. [DOI] [PubMed] [Google Scholar]
- 90.Zhu X., Chen L., Liu L., Niu X. EMT-Mediated Acquired EGFR-TKI Resistance in NSCLC: Mechanisms and Strategies. Front. Oncol. 2019;9:1044. doi: 10.3389/fonc.2019.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deng Q., Xie B., Wu L., Ji X., Li C., Feng L., Fang Q., Bao Y., Li J., Jin S., et al. Competitive evolution of NSCLC tumor clones and the drug resistance mechanism of first-generation EGFR-TKIs in Chinese NSCLC patients. Heliyon. 2018;4:e01031. doi: 10.1016/j.heliyon.2018.e01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olsen S., Liao J., Hayashi H. Real-World Clinical Outcomes after Genomic Profiling of Circulating Tumor DNA in Patients with Previously Treated Advanced Non-Small Cell Lung Cancer. Curr. Oncol. 2022;29:382. doi: 10.3390/curroncol29070382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramalingam S.S., Yang J.C., Lee C.K., Kurata T., Kim D.W., John T., Nogami N., Ohe Y., Mann H., Rukazenkov Y., et al. Osimertinib as First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2018;36:841–849. doi: 10.1200/JCO.2017.74.7576. [DOI] [PubMed] [Google Scholar]
- 94.Iwama E., Sakai K., Azuma K., Harada D., Nosaki K., Hotta K., Nishio M., Kurata T., Fukuhara T., Akamatsu H., et al. Exploration of resistance mechanisms for epidermal growth factor receptor-tyrosine kinase inhibitors based on plasma analysis by digital polymerase chain reaction and next-generation sequencing. Cancer Sci. 2018;109:3921–3933. doi: 10.1111/cas.13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Usui K., Yokoyama T., Naka G., Ishida H., Kishi K., Uemura K., Ohashi Y., Kunitoh H. Plasma ctDNA monitoring during epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor treatment in patients with EGFR-mutant non-small cell lung cancer (JP-CLEAR trial) Jpn. J. Clin. Oncol. 2019;49:554–558. doi: 10.1093/jjco/hyz023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lei L., Wang W.X., Zhu Y.C., Li J.L., Fang Y., Wang H., Zhuang W., Zhang Y.B., Wang L.P., Fang M.Y., et al. Real-world efficacy and potential mechanism of resistance of icotinib in Asian advanced non-small cell lung cancer with EGFR uncommon mutations: A multi-center study. Cancer Med. 2020;9:12–18. doi: 10.1002/cam4.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Francaviglia I., Magliacane G., Lazzari C., Grassini G., Brunetto E., Dal Cin E., Girlando S., Medicina D., Smart C.E., Bulotta A., et al. Identification and monitoring of somatic mutations in circulating cell-free tumor DNA in lung cancer patients. Lung Cancer. 2019;134:225–232. doi: 10.1016/j.lungcan.2019.06.010. [DOI] [PubMed] [Google Scholar]
- 98.Hochmair M.J., Buder A., Schwab S., Burghuber O.C., Prosch H., Hilbe W., Cseh A., Fritz R., Filipits M. Liquid-Biopsy-Based Identification of EGFR T790M Mutation-Mediated Resistance to Afatinib Treatment in Patients with Advanced EGFR Mutation-Positive NSCLC, and Subsequent Response to Osimertinib. Target Oncol. 2019;14:75–83. doi: 10.1007/s11523-018-0612-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou J., Zhao C., Zhao J., Wang Q., Chu X., Li J., Zhou F., Ren S., Li X., Su C., et al. Re-biopsy and liquid biopsy for patients with non-small cell lung cancer after EGFR-tyrosine kinase inhibitor failure. Thorac. Cancer. 2019;10:957–965. doi: 10.1111/1759-7714.13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ariyasu R., Uchibori K., Sasaki T., Tsukahara M., Kiyotani K., Yoshida R., Ono Y., Kitazono S., Ninomiya H., Ishikawa Y., et al. Monitoring epidermal growth factor receptor C797S mutation in Japanese non-small cell lung cancer patients with serial cell-free DNA evaluation using digital droplet PCR. Cancer Sci. 2021;112:2371–2380. doi: 10.1111/cas.14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y., Xiong L., Xie F., Zheng X., Li Y., Zhu L., Sun J. Next-generation sequencing of tissue and circulating tumor DNA: Resistance mechanisms to EGFR targeted therapy in a cohort of patients with advanced non-small cell lung cancer. Cancer Med. 2021;10:4697–4709. doi: 10.1002/cam4.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choi W., Cho Y., Park S.Y., Hwang K.H., Han J.Y., Lee Y. A nanowire-based liquid biopsy method using cerebrospinal fluid cell-free DNA for targeted management of leptomeningeal carcinomatosis. J. Cancer Res. Clin. Oncol. 2021;147:213–222. doi: 10.1007/s00432-020-03324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Minari R., Mazzaschi G., Bordi P., Gnetti L., Alberti G., Altimari A., Gruppioni E., Sperandi F., Parisi C., Guaitoli G., et al. DETECTION Study Group. Detection of EGFR-Activating and T790M Mutations Using Liquid Biopsy in Patients With EGFR-Mutated Non-Small-Cell Lung Cancer Whose Disease Has Progressed During Treatment with First- and Second-Generation Tyrosine Kinase Inhibitors: A Multicenter Real-Life Retrospective Study. Clin. Lung Cancer. 2020;21:e464–e473. doi: 10.1016/j.cllc.2020.02.021. [DOI] [PubMed] [Google Scholar]
- 104.Cavic M., Krivokuca A., Pavlovic M., Boljevic I., Rakobradovic J., Mihajlovic M., Tanic M., Damjanovic A., Malisic E., Jankovic R. EGFR mutation testing from liquid biopsy of non-small cell lung cancer at the Institute for Oncology and Radiology of Serbia. J. BUON. 2020;25:2635–2642. [PubMed] [Google Scholar]
- 105.Li Y., Zhang F., Yuan P., Guo L., Jianming Y., He J. High MAF of EGFR mutations and high ratio of T790M sensitizing mutations in ctDNA predict better third-generation TKI outcomes. Thorac. Cancer. 2020;11:1503–1511. doi: 10.1111/1759-7714.13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fu Y., Wang A., Zhou J., Feng W., Shi M., Xu X., Zhao H., Cai L., Feng J., Lv X. Advanced NSCLC Patients with EGFR T790M Harboring TP53 R273C or KRAS G12V Cannot Benefit from Osimertinib Based on a Clinical Multicentre Study by Tissue and Liquid Biopsy. Front. Oncol. 2021;11:621992. doi: 10.3389/fonc.2021.621992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jóri B., Schatz S., Kaller L., Kah B., Roeper J., Ramdani H.O., Diehl L., Hoffknecht P., Grohé C., Griesinger F., et al. Comparison of Resistance Spectra after First and Second Line Osimertinib Treatment Detected by Liquid Biopsy. Cancers. 2021;13:2861. doi: 10.3390/cancers13122861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nguyen H.N., Cao N.T., Van Nguyen T.C., Le K.N.D., Nguyen D.T., Nguyen Q.T., Nguyen T.T., Van Nguyen C., Le H.T., Nguyen M.T., et al. Liquid biopsy uncovers distinct patterns of DNA methylation and copy number changes in NSCLC patients with different EGFR-TKI resistant mutations. Sci. Rep. 2021;11:16436. doi: 10.1038/s41598-021-95985-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fan Y., Sun R., Wang Z., Zhang Y., Xiao X., Liu Y., Xin B., Xiong H., Lu D., Ma J. Detection of MET amplification by droplet digital PCR in peripheral blood samples of non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 2022;18:1–11. doi: 10.1007/s00432-022-04048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gelatti A.C.Z., Drilon A., Santini F.C. Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC) Lung Cancer. 2019;137:113–122. doi: 10.1016/j.lungcan.2019.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lei L., Wang W.X., Zhu Y.C., Li J.L., Fang Y., Wang H., Zhuang W., Zhang Y.B., Wang L.P., Fang M.Y., et al. Potential mechanism of primary resistance to icotinib in patients with advanced non-small cell lung cancer harboring uncommon mutant epidermal growth factor receptor: A multi-center study. Cancer Sci. 2020;111:679–686. doi: 10.1111/cas.14277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Y., He Y., Tian P., Wang W., Wang K., Chuai S., Li Y., Zhao S., Wang Y., Li W. Low T790M relative allele frequency indicates concurrent resistance mechanisms and poor responsiveness to osimertinib. Transl. Lung Cancer Res. 2020;9:1952–1962. doi: 10.21037/tlcr-20-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zou B., Lee V.H.F., Chen L., Ma L., Wang D.D., Yan H. Deciphering mechanisms of acquired T790M mutation after EGFR inhibitors for NSCLC by computational simulations. Sci. Rep. 2017;7:6595. doi: 10.1038/s41598-017-06632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Verzè M., Minari R., Gnetti L., Bordi P., Leonetti A., Cosenza A., Ferri L., Majori M., De Filippo M., Buti S., et al. Monitoring cfDNA in Plasma and in Other Liquid Biopsies of Advanced EGFR Mutated NSCLC Patients: A Pilot Study and a Review of the Literature. Cancers. 2021;13:5403. doi: 10.3390/cancers13215403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Minari R., Bordi P., Del Re M., Facchinetti F., Mazzoni F., Barbieri F., Camerini A., Comin C.E., Gnetti L., Azzoni C., et al. Primary resistance to osimertinib due to SCLC transformation: Issue of T790M determination on liquid re-biopsy. Lung Cancer. 2018;115:21–27. doi: 10.1016/j.lungcan.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 116.Tsui D.W.Y., Murtaza M., Wong A.S.C., Rueda O.M., Smith C.G., Chandrananda D., Soo R.A., Lim H.L., Goh B.C., Caldas C., et al. Dynamics of multiple resistance mechanisms in plasma DNA during EGFR-targeted therapies in non-small cell lung cancer. EMBO. Mol. Med. 2018;10:e7945. doi: 10.15252/emmm.201707945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schmid S., Stewart E.L., Martins-Filho S.N., Cabanero M., Wang A., Bao H., Wu X., Patel D., Chen Z., Law J.H., et al. Early Detection of Multiple Resistance Mechanisms by ctDNA Profiling in a Patient With EGFR-mutant Lung Adenocarcinoma Treated with Osimertinib. Clin. Lung Cancer. 2020;21:e488–e492. doi: 10.1016/j.cllc.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 118.Oxnard G.R., Hu Y., Mileham K.F., Husain H., Costa D.B., Tracy P., Feeney N., Sholl L.M., Dahlberg S.E., Redig A.J., et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients with EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol. 2018;4:1527–1534. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kato R., Hayashi H., Sakai K., Suzuki S., Haratani K., Takahama T., Tanizaki J., Nonagase Y., Tanaka K., Yoshida T., et al. CAPP-seq analysis of circulating tumor DNA from patients with EGFR T790M-positive lung cancer after osimertinib. Int. J. Clin. Oncol. 2021;26:1628–1639. doi: 10.1007/s10147-021-01947-3. [DOI] [PubMed] [Google Scholar]
- 120.De Carlo E., Schiappacassi M., Pelizzari G., Baresic T., Del Conte A., Stanzione B., Da Ros V., Doliana R., Baldassarre G., Bearz A. Acquired EGFR C797G Mutation Detected by Liquid Biopsy as Resistance Mechanism After Treatment with Osimertinib: A Case Report. In Vivo. 2021;35:2941–2945. doi: 10.21873/invivo.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shen F., Liang N., Fan Z., Zhao M., Kang J., Wang X., Hu Q., Mu Y., Wang K., Yuan M., et al. Genomic Alterations Identification and Resistance Mechanisms Exploration of NSCLC With Central Nervous System Metastases Using Liquid Biopsy of Cerebrospinal Fluid: A Real-World Study. Front. Oncol. 2022;12:889591. doi: 10.3389/fonc.2022.889591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu X., Xing P., Shi M., Guo W., Zhao F., Zhu H., Xiao J., Wan J., Li J. Cerebrospinal Fluid Cell-Free DNA-Based Detection of High Level of Genomic Instability Is Associated with Poor Prognosis in NSCLC Patients with Leptomeningeal Metastases. Front. Oncol. 2022;12:664420. doi: 10.3389/fonc.2022.664420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zheng M.M., Li Y.S., Tu H.Y., Jiang B.Y., Yang J.J., Zhou Q., Xu C.R., Yang X.R., Wu Y.L. Genotyping of Cerebrospinal Fluid Associated with Osimertinib Response and Resistance for Leptomeningeal Metastases in EGFR-Mutated NSCLC. J. Thorac. Oncol. 2021;16:250–258. doi: 10.1016/j.jtho.2020.10.008. [DOI] [PubMed] [Google Scholar]
- 124.Benayed R., Offin M., Mullaney K., Sukhadia P., Rios K., Desmeules P., Ptashkin R., Won H., Chang J., Halpenny D., et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin. Cancer Res. 2019;25:4712–4722. doi: 10.1158/1078-0432.CCR-19-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li W., Guo L., Liu Y., Dong L., Yang L., Chen L., Liu K., Shao Y., Ying J. Potential Unreliability of Uncommon ALK, ROS1, and RET Genomic Breakpoints in Predicting the Efficacy of Targeted Therapy in NSCLC. J. Thorac. Oncol. 2021;16:404–418. doi: 10.1016/j.jtho.2020.10.156. [DOI] [PubMed] [Google Scholar]
- 126.Li W., Liu Y., Li W., Chen L., Ying J. Intergenic Breakpoints Identified by DNA Sequencing Confound Targetable Kinase Fusion Detection in NSCLC. J. Thorac. Oncol. 2020;15:1223–1231. doi: 10.1016/j.jtho.2020.02.023. [DOI] [PubMed] [Google Scholar]
- 127.Cohen D., Hondelink L.M., Solleveld-Westerink N., Uljee S.M., Ruano D., Cleton-Jansen A.M., von der Thüsen J.H., Ramai S.R.S., Postmus P.E., Graadt van Roggen J.F., et al. Optimizing Mutation and Fusion Detection in NSCLC by Sequential DNA and RNA Sequencing. J. Thorac. Oncol. 2020;15:1000–1014. doi: 10.1016/j.jtho.2020.01.019. [DOI] [PubMed] [Google Scholar]
- 128.Wu C.Y., Lee C.L., Wu C.F., Fu J.Y., Yang C.T., Wen C.T., Liu Y.H., Liu H.P., Hsieh J.C. Circulating Tumor Cells as a Tool of Minimal Residual Disease Can Predict Lung Cancer Recurrence: A longitudinal, Prospective Trial. Diagnostics. 2020;10:144. doi: 10.3390/diagnostics10030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chaudhuri A.A., Chabon J.J., Lovejoy A.F., Newman A.M., Stehr H., Azad T.D., Khodadoust M.S., Esfahani M.S., Liu C.L., Zhou L., et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7:1394–1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Abbosh C., Birkbak N.J., Wilson G.A., Jamal-Hanjani M., Constantin T., Salari R., Le Quesne J., Moore D.A., Veeriah S., Rosenthal R., et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen K., Zhao H., Shi Y., Yang F., Wang L.T., Kang G., Nie Y., Wang J. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC) Clin. Cancer Res. 2019;25:7058–7067. doi: 10.1158/1078-0432.CCR-19-1213. [DOI] [PubMed] [Google Scholar]
- 132.Ohara S., Suda K., Sakai K., Nishino M., Chiba M., Shimoji M., Takemoto T., Fujino T., Koga T., Hamada A., et al. Prognostic implications of preoperative versus postoperative circulating tumor DNA in surgically resected lung cancer patients: A pilot study. Transl. Lung Cancer Res. 2020;9:1915–1923. doi: 10.21037/tlcr-20-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kuang P.P., Li N., Liu Z., Sun T.Y., Wang S.Q., Hu J., Ou W., Wang S.Y. Circulating Tumor DNA Analyses as a Potential Marker of Recurrence and Effectiveness of Adjuvant Chemotherapy for Resected Non-Small-Cell Lung Cancer. Front. Oncol. 2021;10:595650. doi: 10.3389/fonc.2020.595650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Guo K., Shao C., Han L., Liu H., Ma Z., Yang Y., Feng Y., Pan M., Santarpia M., Carmo-Fonseca M., et al. Detection of epidermal growth factor receptor (EGFR) mutations from preoperative circulating tumor DNA (ctDNA) as a prognostic predictor for stage I-III non-small cell lung cancer (NSCLC) patients with baseline tissue EGFR mutations. Transl. Lung Cancer Res. 2021;10:3213–3225. doi: 10.21037/tlcr-21-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Qiu B., Guo W., Zhang F., Lv F., Ji Y., Peng Y., Chen X., Bao H., Xu Y., Shao Y., et al. Dynamic recurrence risk and adjuvant chemotherapy benefit prediction by ctDNA in resected NSCLC. Nat. Commun. 2021;12:6770. doi: 10.1038/s41467-021-27022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peng M., Huang Q., Yin W., Tan S., Chen C., Liu W., Tang J., Wang X., Zhang B., Zou M., et al. Circulating Tumor DNA as a Prognostic Biomarker in Localized Non-small Cell Lung Cancer. Front. Oncol. 2020;10:561598. doi: 10.3389/fonc.2020.561598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li N., Wang B.X., Li J., Shao Y., Li M.T., Li J.J., Kuang P.P., Liu Z., Sun T.Y., Wu H.Q., et al. Perioperative circulating tumor DNA as a potential prognostic marker for operable stage I to IIIA non-small cell lung cancer. Cancer. 2022;128:708–718. doi: 10.1002/cncr.33985. [DOI] [PubMed] [Google Scholar]
- 138.Waldeck S., Mitschke J., Wiesemann S., Rassner M., Andrieux G., Deuter M., Mutter J., Lüchtenborg A.M., Kottmann D., Titze L., et al. Early assessment of circulating tumor DNA after curative-intent resection predicts tumor recurrence in early-stage and locally advanced non-small-cell lung cancer. Mol. Oncol. 2022;16:527–537. doi: 10.1002/1878-0261.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xia L., Mei J., Kang R., Deng S., Chen Y., Yang Y., Feng G., Deng Y., Gan F., Lin Y., et al. Perioperative ctDNA-Based Molecular Residual Disease Detection for Non-Small Cell Lung Cancer: A Prospective Multicenter Cohort Study (LUNGCA-1) Clin. Cancer Res. 2022;28:3308–3317. doi: 10.1158/1078-0432.CCR-21-3044. [DOI] [PubMed] [Google Scholar]
- 140.Gale D., Heider K., Ruiz-Valdepenas A., Hackinger S., Perry M., Marsico G., Rundell V., Wulff J., Sharma G., Knock H., et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 2022;33:500–510. doi: 10.1016/j.annonc.2022.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yue D., Liu W., Chen C., Zhang T., Ma Y., Cui L., Gu Y., Bei T., Zhao X., Zhang B. Circulating tumor DNA predicts neoadjuvant immunotherapy efficacy and recurrence-free survival in surgical non-small cell lung cancer patients. Transl. Lung Cancer Res. 2022;11:263–276. doi: 10.21037/tlcr-22-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guo H., Li W., Wang B., Chen N., Qian L., Cui J. Coexisting opportunities and challenges: In which scenarios can minimal/measurable residual disease play a role in advanced non-small cell lung cancer? Chin. J. Cancer Res. 2021;33:574–582. doi: 10.21147/j.issn.1000-9604.2021.05.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Miyawaki T., Wakuda K., Kenmotsu H., Miyawaki E., Mamesaya N., Kobayashi H., Omori S., Ono A., Naito T., Murakami H., et al. Proposing synchronous oligometastatic non-small-cell lung cancer based on progression after first-line systemic therapy. Cancer Sci. 2021;112:359–368. doi: 10.1111/cas.14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Spaggiari L., Bertolaccini L., Facciolo F., Gallina F.T., Rea F., Schiavon M., Margaritora S., Congedo M.T., Lucchi M., Ceccarelli I., et al. A risk stratification scheme for synchronous oligometastatic non-small cell lung cancer developed by a multicentre analysis. Lung Cancer. 2021;154:29–35. doi: 10.1016/j.lungcan.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 145.Gauvin C., Krishnan V., Kaci I., Tran-Thanh D., Bédard K., Albadine R., Leduc C., Gaboury L., Blais N., Tehfe M., et al. Survival Impact of Aggressive Treatment and PD-L1 Expression in Oligometastatic NSCLC. Curr. Oncol. 2021;28:59. doi: 10.3390/curroncol28010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pécuchet N., Zonta E., Didelot A., Combe P., Thibault C., Gibault L., Lours C., Rozenholc Y., Taly V., Laurent-Puig P., et al. Base-Position Error Rate Analysis of Next-Generation Sequencing Applied to Circulating Tumor DNA in Non-Small Cell Lung Cancer: A Prospective Study. PLoS. Med. 2016;13:e1002199. doi: 10.1371/journal.pmed.1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moding E.J., Liu Y., Nabet B.Y., Chabon J.J., Chaudhuri A.A., Hui A.B., Bonilla R.F., Ko R.B., Yoo C.H., Gojenola L., et al. Circulating Tumor DNA Dynamics Predict Benefit from Consolidation Immunotherapy in Locally Advanced Non-Small Cell Lung Cancer. Nat. Cancer. 2020;1:176–183. doi: 10.1038/s43018-019-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hellmann M.D., Nabet B.Y., Rizvi H., Chaudhuri A.A., Wells D.K., Dunphy M.P.S., Chabon J.J., Liu C.L., Hui A.B., Arbour K.C., et al. Circulating Tumor DNA Analysis to Assess Risk of Progression after Long-term Response to PD-(L)1 Blockade in NSCLC. Clin. Cancer Res. 2020;26:2849–2858. doi: 10.1158/1078-0432.CCR-19-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tang C., Lee W.C., Reuben A., Chang L., Tran H., Little L., Gumbs C., Wargo J., Futreal A., Liao Z., et al. Immune and Circulating Tumor DNA Profiling After Radiation Treatment for Oligometastatic Non-Small Cell Lung Cancer: Translational Correlatives from a Mature Randomized Phase II Trial. Int. J. Radiat. Oncol. Biol. Phys. 2020;106:349–357. doi: 10.1016/j.ijrobp.2019.10.038. [DOI] [PubMed] [Google Scholar]
- 150.Yan X., Liu C. Application of Non-Blood-Derived Fluid Biopsy in Monitoring Minimal Residual Diseases of Lung Cancer. Front. Surg. 2022;9:865040. doi: 10.3389/fsurg.2022.865040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.