Abstract
The genome of Bacillus subtilis contains two genes that code for membrane proteins that belong to the 2-hydroxycarboxylate transporter family. Here we report the functional characterization of one of the two, yxkJ, which codes for a transporter protein named CimHbs. The gene was cloned and expressed in Escherichia coli and complemented the citrate-negative phenotype of wild-type E. coli and the malate-negative phenotype of the E. coli strain JRG4008, which is defective in malate uptake. Subsequent uptake studies in whole cells expressing CimHbs clearly demonstrated the citrate and malate transport activity of the protein. Immunoblot analysis showed that CimHbs is a 48-kDa protein that is well expressed in E. coli. Studies with right-side-out membrane vesicles demonstrated that CimHbs is an electroneutral proton-solute symporter. No indications were found for the involvement of Na+ ions in the transport process. Inhibition of the uptake catalyzed by CimHbs by divalent metal ions, together with the lack of effect on transport by the chelator EDTA, showed that CimHbs translocates the free citrate and malate anions. Among a large set of substrates tested, only malate, citramalate, and citrate competitively inhibited citrate transport catalyzed by CimHbs. The transporter is strictly stereoselective, recognizing only the S enantiomers of malate and citramalate. Remarkably, though citramalate binds to the transporter, it is not translocated.
Di- and tricarboxylate uptake in the gram-positive bacterium Bacillus subtilis has not been studied extensively. The scarce reports stem from the 1970s and early 1980s. Two uptake systems have been described for C4-dicarboxylates. Willecke and Lange (37) reported a malate uptake system that was induced by malate and responsible for growth on malate minimal medium. The second system was less specific and was claimed to transport malate, succinate, and fumarate. It was described in cells grown in yeast extract (15) and in the presence of citrate (14) or was claimed to be expressed constitutively (9). The last study showed, by studies with membrane vesicles, that the transporter was of the secondary type. Similarly, it was demonstrated that the uptake of the tricarboxylate citrate in B. subtilis was catalyzed by a secondary transporter (8). Remarkably, uptake of citrate was dependent on the availability of divalent metal ions (8, 27).
The publication of the complete genome sequence of B. subtilis (20) provoked the molecular characterization of di- and tricarboxylate transport in the organism. A screen using deletion strains identified an operon containing two open reading frames (ydbE and ydbH) that was responsible for growth on fumarate and succinate but not malate (1). The gene product YdbH is a homologue of the dicarboxylate transporters (Dct) found in gram-negative bacteria. In the same screen, no transporter for malate was identified. A malate transporter was identified by heterologous expression of the yufR gene of B. subtilis in Escherichia coli (36). The gene product, termed MaeNbs, catalyzed Na+-coupled malate transport, but neither the physiological function nor the expression was studied in B. subtilis. MaeNbs is a member of the 2-hydroxycarboxylate transporter (2HCT) family, which contains secondary transporters for citrate and/or malate (see below). In the same study, a second malate transporter with a more complicated mode of action was described. YqkI catalyzes antiport of H+-malate against Na+-lactate. It was proposed that the transporter plays a role in malate uptake and Na+-H+ exchange under low-oxygen or low proton motive force (PMF) conditions (36).
For citrate, the situation is at least as complicated. Two secondary transporters, CitMbs and CitHbs, have been studied in some detail. They were characterized upon expression in E. coli (10) and shown to transport citrate in complex with divalent metal ions, but with complementary specificity for the metal ions (19). CitMbs transports citrate in complex with Mg2+, Mn2+, Ni2+, Co2+, and Zn2+, while CitHbs transports citrate in complex with Ca2+, Sr2+, and Ba2+. CitMbs is the transporter responsible for growth of B. subtilis on minimal medium containing citrate as a sole carbon source. Its expression is subject to carbon catabolite repression (35, 38). The physiological function of CitHbs is unknown. It was also demonstrated that B. subtilis takes up the free citrate anion; however, neither CitMbs nor CitHbs accounted for this activity (19). CitMbs and CitHbs are in a novel family of secondary transporters that contains a third gene from B. subtilis, yraO, of unknown function.
The 2HCT family mentioned above contains another potential transporter for carboxylate uptake in B. subtilis coded by the open reading frame yxkJ. This transporter, termed here CimHbs (for citrate, malate, H+ symporter of B. subtilis), is the subject of the present study. The 2HCT family contains transporters for malate and citrate, some of which have been studied extensively. The family contains Na+ and H+ symporters (CitSkp of Klebsiella pneumoniae, MaeNbs of B. subtilis, and MaePsb of Streptococcus bovis [17, 24, 34, 36]) but also citrate-lactate and malate-lactate exchangers found in lactic acid bacteria (CitPlcm of Leuconostoc mesenteroides and MlePll of Lactococcus lactis [2, 25]). The last two transporters are involved in secondary metabolic energy generation (23, 26, 28). Substrate specificity studies revealed that the symporters in the family are very specific, transporting only citrate or malate, while the exchangers catalyze a wide range of 2-hydroxycarboxylates (2). Here we show that CimHbs of B. subtilis is a proton symporter that transports both citrate and malate.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. E. coli strains were routinely grown in flasks in Luria-Bertani broth (LB) at 37°C under continuous shaking at 150 rpm. When appropriate, carbenicillin and spectinomycin were added at final concentrations of 50 μg/ml. Expression of CimHbs from the plasmid pETCimH (see below) was induced by adding 100 μM isopropylthiogalactopyranoside (IPTG) when the optical density of the culture measured at 660 nm was 0.6. The cells were harvested by centrifugation 1 h after induction.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| B. subtilis 168 | trpC2 | |
| E. coli DH5α | Δ(argF-lac)U169 (φ80DlacZM15) recA | Life Technologies, Inc. |
| E. coli BL21(DE3) | hsdS gal (Δclts857 ind 1 sam7 nin5 lacUV5-T7 gene 1) | Novagen |
| E. coli TOP10 | (φ80DlacZM15) ΔlacX74 recA | Invitrogen |
| E. coli SF100 | F ΔlacX74 galE galK thi rpsL (strA) ΔphoA(PvuII) ΔompT | 7 |
| E. coli JRG4008 | AN387 dctA::spc | 12 |
Complementation of the Mal− phenotype of E. coli strain JRG4008 was assayed on a minimal malate broth (11) supplemented with 0.1% LB. The medium was inocculated with a single colony of JRG4008 carrying pET302 or pETCimH from an LB plate and incubated for 14 h at 37°C with vigorous shaking.
Cloning of CimHbs.
The yxkJ gene was amplified from chromosomal DNA isolated from B. subtilis 168 by PCR. The forward primer (5′-GGGAGGTTTTTCCATGGGAGAGC-3′) was centered around the beginning of the gene and contained an NcoI site (italicized) around the ATG start codon. The reverse primer (5′-CGTTTGACGAAGATCTAGATTGCGGGGATCA-3′) was centered around the end of the gene and contained an XbaI site (italicized) just downstream of the stop codon. The PCR product was digested with NcoI and XbaI and ligated into the vector pET302, a pTRC99 derivative (32) that was digested with the same two restriction enzymes. The resulting plasmid, pETCimH, codes for CimHbs extended with six histidines (His tag) at the N terminus. Expression is under the control of the IPTG-inducible lac promoter. The nucleotide sequence of the insert was checked by sequencing (Biomedical Technology Centre, Groningen, The Netherlands); no unexpected mutations had occurred.
Immunoblot analysis.
Crude membranes were prepared by sonication of the cells followed by removal of debris by centrifugation for 5 min in an Eppendorf tabletop centrifuge operated at 6,000 rpm. Subsequently, the supernatant was centrifuged for 5 min at 14,000 rpm in an Eppendorf tabletop centrifuge to collect the membranes. The protein concentrations in the samples were determined with the DC protein assay kit (Bio-Rad Laboratories, Richmond, Calif.). Samples containing 10 μg of total membrane protein were loaded onto a sodium dodecyl sulfate–12.5% polyacrylamide gel. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes (Boehringer Mannheim) by semidry electroblotting. The blots were analyzed using monoclonal antibodies directed against a six-His tag (Dianova, Hamburg, Germany). The antibodies were visualized using the Western-light chemiluminescence detection kit (Tropix, Bedford, Mass.).
Preparation of RSO membrane vesicles.
Right-side-out (RSO) membrane vesicles were prepared by the osmotic lysis procedure as described previously (16). Membrane vesicles resuspended in 50 mM potassium phosphate (KPi) buffer( pH 6.1) were rapidly frozen in liquid nitrogen and stored at −80°C. Protein concentrations were determined with the DC protein assay kit.
Uptake in whole cells.
Citrate and malate uptake in whole cells was measured by rapid filtration as described previously (21). Briefly, cells were harvested by centrifugation and washed once with ice-cold 50 mM KPi (pH 6.1), after which they were resuspended to an optical density measured at 660 nm of 10. The cells were diluted 10-fold in 50 mM KPi (pH 6.1), and 100-μl samples were incubated for 8 min at 30°C with magnetic stirring. At time zero, [1,5-14C]citrate (114 mCi/mmol) or l-[1,4(2,3)-14C]malate (25 mCi/mmol) was added to yield final concentrations of 4.4 and 19.6 μM, respectively. Uptake was stopped by the addition of 2 ml of ice-cold 0.1 M LiCl solution immediately followed by filtering through a 0.45-μm-pore-size nitrocellulose filter. The filter was washed once with 2 ml of ice-cold 0.1 M LiCl, after which the filters were submerged in scintillation fluid and the retained radioactivity was counted in a liquid scintillation counter. The background was estimated by adding the radiolabeled substrate to the cell suspension after the addition of 2 ml of ice-cold LiCl, immediately followed by filtering.
Uptake in RSO membranes.
Uptake in RSO vesicles was essentially performed like uptake in whole cells with the following differences. Before the addition of the radiolabeled substrates, the membranes (final protein concentration, 0.5 mg/ml) in 50 mM KPi (pH 6.1) were incubated for 2 min at 30°C with 200 μM phenazine methosulfate (PMS) and 10 mM potassium ascorbate under a flow of water-saturated air with magnetic stirring. To determine the influence of Na+ on citrate transport, membranes containing CimHbs of B. subtilis or CitSkp of K. pneumoniae were washed four times in a special low-Na+ KPi buffer (pH 6.1; Merck, Darmstadt, Germany) and resuspended to 0.5 mg of membrane protein/ml in the same buffer. Initial rates of uptake were determined in duplicate from the uptake during the first 10 s. The reported kinetic parameters are the averages of two or three independent measurements.
Homologous and heterologous exchange.
RSO membrane vesicles were allowed to accumulate radiolabeled [1,5-14C]citrate driven by the electron donor system ascorbate-PMS for 1 min as described above. The PMF was dissipated by the addition of the uncoupler carbonylcyanide p-trifluormethoxy-phenylhydrazone (FCCP; 10 μM). When indicated, at the same time, citrate, l-malate, or l-citramalate was added to a final concentration of 1 mM. The release of label from the membranes was followed during 1 min by rapid filtration at various time points.
Transport driven by artificial gradients.
Artificial ion gradients across RSO membranes were imposed essentially as described previously (31). Briefly, RSO vesicles were resuspended to a final protein concentration of 5 mg/ml in a buffer containing 25 mM KPi (pH 6.1), 100 mM K-acetate, and 100 μM valinomycin. A PMF consisting of a pH gradient (ΔpH) and a membrane potential (ΔΨ) was generated by rapid 100-fold dilution of the concentrated membranes in a buffer containing 25 mM sodium phosphate (NaPi) (pH 6.1) and 100 mM NaCl. To generate a ΔpH alone, valinomycin was omitted from the membrane suspension, and to generate a ΔΨ alone, NaCl in the dilution buffer was replaced with Na-acetate. The dilution buffer contained 4.5 μM radiolabeled [1,5-14C]citrate, and the internal labeled substrate was measured after 10 and 15 s by rapid filtration as described above.
RESULTS
The product of the open reading frame yxkJ on the genome of B. subtilis, termed CimHbs, belongs to the 2HCT family. Members of the 2HCT family transport citrate and/or malate (see the introduction).
E. coli does not grow on citrate under aerobic conditions because it lacks an uptake system for citrate (18). E. coli strain DH5α was transformed with pETCimH, harboring the yxkJ gene of B. subtilis under the control of the trc promoter, and plated on Simmons citrate agar. The cells grew as large colonies surrounded by blue halos, indicative of citrate metabolism. DH5α, or DH5α transformed with the vector alone, showed poor growth (not shown), strongly suggesting that the insert was responsible for the Cit+ phenotype.
E. coli strain JRG4008 is defective in dctA, a gene coding for a malate transporter, and is unable to grow on malate as the sole carbon source under aerobic conditions (12). When JRG4008 was grown in Christensen indicator broth containing malate (11), the color of the culture changed only slightly, while growth of the wild-type E. coli strain DH5α in the same medium shifted the color to purple. The low residual malate metabolism in JRG4008 is probably due to low-level expression of one of the other malate transport systems (13). JRG4008 expressing the yxkJ gene product CimHbs showed the same color change that was observed with the wild-type DH5α strain, indicating an improved malate uptake capacity (not shown).
The complementation of E. coli DH5α for growth on citrate and of strain JRG4008 for growth on malate suggests that CimHbs, the product of the B. subtilis yxkJ gene, transports both citrate and malate.
Transport activity and expression of CimHbs.
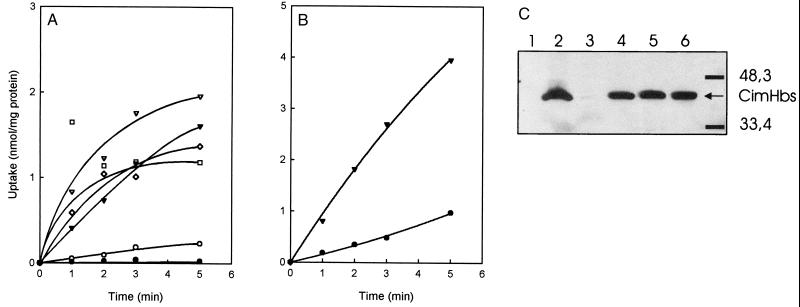
Citrate and malate transport activity in E. coli catalyzed by CimHbs was measured in whole cells using radiolabeled substrates. The lack of citrate uptake in wild-type E. coli is well documented in the literature (10, 18, 19, 33). E. coli DH5α carrying the empty plasmid pET302 did not transport [1,5-14C]citrate, while DH5α expressing CimHbs showed a low but significant uptake (Fig. 1A). A number of other E. coli strains, including JRG4008, BL21(DE3), SF100, and TOP10 transformed with the pETCimH plasmid, were also tested for citrate uptake with much better results (Fig. 1A). The expression levels of CimHbs in the cells were analyzed by Western blotting using an antibody directed against the N-terminal His tag (Fig. 1C). A clear band running with an apparent molecular mass of 48 kDa was observed in cells expressing CimHbs. In line with the observed uptake activities, expression was very low in DH5α.
FIG. 1.
Citrate and malate uptake in whole cells expressing CimHbs. (A) Uptake of [1,5-14C]citrate was measured in whole cells of E. coli strain JRG4008 carrying the empty plasmid pET302 (●) and strains JRG4008 (▾), DH5α (○), BL21(DE3) (□), TOP10 (⋄), and SF100 (▿) expressing CimHbs. (B) Malate transport activity was measured in whole cells of E. coli JRG4008 carrying the empty pET302 vector (●) or expressing CimHbs (▾). (C) Expression levels of CimHbs in the cells used for the uptake experiments shown in panel A. The immunoblot of membrane extracts of strains JRG4008 carrying plasmid pET302 (lane 1) and JRG4008 (lane 2), DH5α (lane 3), BL21(DE3) (lane 4), TOP10 (lane 5), and SF100 (lane 6) expressing CimHbs were analyzed using antibodies directed against the N-terminal His tag. Molecular mass markers are indicted on the right.
A low level of l-[1,4(2,3)-14C]malate accumulation was observed in the dctA-deficient strain JRG4008, which was in agreement with the low level of malate metabolism on Christensen medium observed for this strain (Fig. 1B). Expression of CimHbs in JRG4008 resulted in a rate of uptake of l-[1,4(2,3)-14C]malate that was four to five times faster. The initial rate of uptake of malate in the same batch of JRG4008 cells expressing CimHbs was about twice the initial rate of uptake of citrate (compare Fig. 1A and B). Kinetic analysis of malate transport in whole cells is hampered by the residual uptake activity in the JRG4008 strain and by the increasing PMF across the cell membrane in the presence of l-malate, as evidenced by the stimulation of l-[U-14C]proline uptake in the presence of l-malate (not shown). Therefore, the kinetics of citrate and malate transport catalyzed by CimHbs was analyzed in RSO membrane vesicles (see below).
Mode of energy coupling.
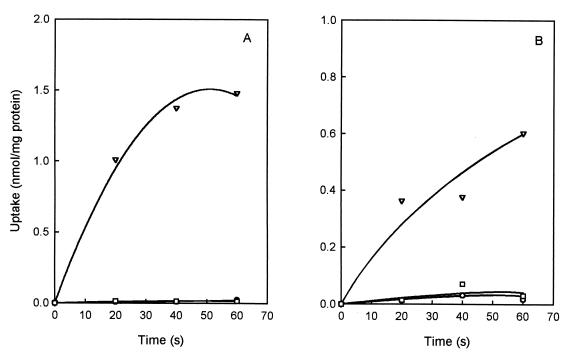
RSO membrane vesicles were prepared from E. coli TOP10 cells expressing CimHbs. Upon the generation of a PMF by the artificial electron donor system ascorbate-PMS, the vesicles took up citrate at an initial rate of 3 nmol/min · mg. In the absence of the electron donor system, or in the presence of the uncoupler FCCP, no uptake was observed (Fig. 2A). Similar results were obtained for malate transport in RSO membrane vesicles prepared from the malate-negative strain JRG4008 expressing CimHbs, which took up malate at a rate of 1.2 nmol/min · mg in the presence of ascorbate-PMS (Fig. 2B). Importantly, in contrast to what was observed in whole cells (Fig. 1B), no background malate uptake activity was observed in membrane vesicles prepared from the host cells. Probably, binding-protein-dependent systems are involved in the background activity. It was consistently observed that in membrane vesicles the rate of malate transport was lower than the rate of citrate transport, in contrast to the results in whole cells. We have no explanation for this phenomenon.
FIG. 2.
Citrate and malate uptake catalyzed by CimHbs in RSO membranes. [1,5-14C]citrate uptake was measured in RSO vesicles prepared from E. coli TOP10 expressing CimHbs (A), and l-[1,4(2,3)-14C]malate uptake was measured in RSO vesicles prepared from E. coli JRG4008 expressing CimHbs (B). The membranes were either not energized (○) or energized with Asc-PMS to generate a PMF (▿), or the PMF was dissipated with 10 μM FCCP (□).
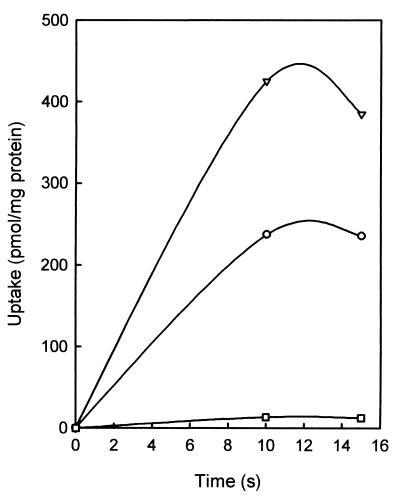
The PMF consists of a ΔpH and the ΔΨ. The contribution of ΔpH and ΔΨ to the driving force on citrate transport catalyzed by CimHbs was investigated by applying artificial gradients (31). Dilution of membrane vesicles equilibrated with 100 mM potassium acetate 100-fold in a buffer without potassium acetate and in the presence of the potassium ionophore valinomycin results in a PMF consisting of both ΔpH and ΔΨ generated by the diffusion of K+ ions and undissociated acetic acid across the membrane. The result was a rapid uptake of citrate into the lumen of the vesicles (Fig. 3). When the membranes were diluted in the absence of valinomycin, where only a ΔpH develops, uptake of citrate was even higher. In contrast, dilution in a buffer containing 100 mM sodium acetate, where only a ΔΨ is imposed across the membrane, does not result in detectable uptake of citrate. The experiments suggest that CimHbs is an electroneutral proton symporter. The number of protons transported is equal to the valence of the transported anions.
FIG. 3.
Citrate uptake driven by artificial gradients. Artificial gradients were imposed across RSO membranes prepared from E. coli TOP10 expressing CimHbs as described in Materials and Methods. [1,5-14C]citrate uptake was measured at pH 6.0 in the presence of both the ΔpH and ΔΨ components (○), only ΔpH (▿), and only ΔΨ (□).
Cation specificity of CimHbs.
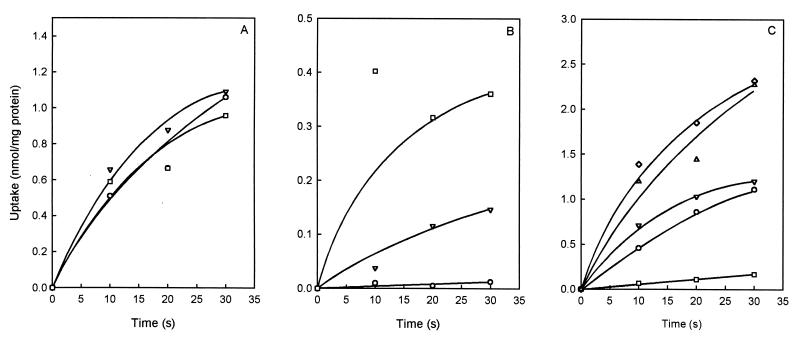
The 2HCT family of secondary transporters contains transporters that use H+ as well as Na+ as the coupling ion. For instance, citrate transport by CitSkp of K. pneumoniae is strictly coupled to transport of Na+ (21, 33), while the exchangers CitPlcm and MlePll from lactic acid bacteria cotransport protons. Vesicles of E. coli TOP10 expressing CimHbs were prepared in low-Na+ potassium phosphate buffer. Uptake of citrate by the vesicles with and without added NaCl revealed no influence of Na+ (Fig. 4A), indicating that sodium ions are not required for activity. In a control experiment, E. coli vesicles containing CitSkp of K. pneumoniae resuspended in the same buffer showed citrate uptake only in the presence of NaCl (Fig. 4B).
FIG. 4.
Cation dependency of the activity of CimHbs. (A and B) Effect of an Na+ ion gradient on the citrate transport activity of CimHbs and CitSkp of K. pneumoniae. RSO membranes prepared from E. coli expressing CimHbs (A) and CitSkp (B) were washed four times in low-Na+ KPi buffer. [1,5-14C]citrate transport was measured in the absence (○) and presence of 1 (▿) and 10 (□) mM NaCl. (C) Effect of divalent cations on citrate transport by CimHbs. [1,5-14C]citrate transport was measured in RSO vesicles prepared from E. coli TOP10 containing CimHbs in the absence (▵) or presence (⋄) of 1 mM EDTA and in the presence of 10 mM MgCl2 (○), 10 mM CaCl2 (▿), and 1 mM NiCl2 (□).
The effect of divalent metal ions on citrate transport by CimHbs was tested in membrane vesicles. Ca2+ and Mg2+ were added at 10 mM concentration, which resulted in clear inhibition of citrate transport (Fig. 4C). The presence of 1 mM Ni2+ resulted in almost complete abolition of citrate uptake. The different degrees of inhibition are caused by differences in complexation efficiency of the Ca2+, Mg2+, and Ni2+ ions (19). The addition of the chelator EDTA (1 mM) had no significant effect on citrate uptake (Fig. 4C), indicating that the free citrate anion is the preferred substrate for CimHbs.
Substrate specificity.
A range of carboxylates was tested for the ability to competitively inhibit citrate transport catalyzed by CimHbs in RSO membranes. At 1 mM concentrations, propionate, succinate, fumarate, and tricarbalate showed no inhibition. A similar result was observed for the 2-oxo-carboxylates, α-ketoglutarate and pyruvate. Of the 2-hydroxy-carboxylates, lactate, glycolate, citramalate, malate, and citrate, only citramalate caused significant inhibition, in addition to citrate and malate (not shown).
The exchangers CitPlcm and MlePll in the 2HCT family are strictly stereoselective for the S enantiomers of dicarboxylates (4). A 1 mM concentration of the S enantiomers of malate and citramalate inhibited citrate uptake by CimH by 60 and 90%, respectively (Fig. 5). The R enantiomers did not result in significant inhibition even at a five-times-higher concentration, indicating that CimHbs is also stereoselective.
FIG. 5.
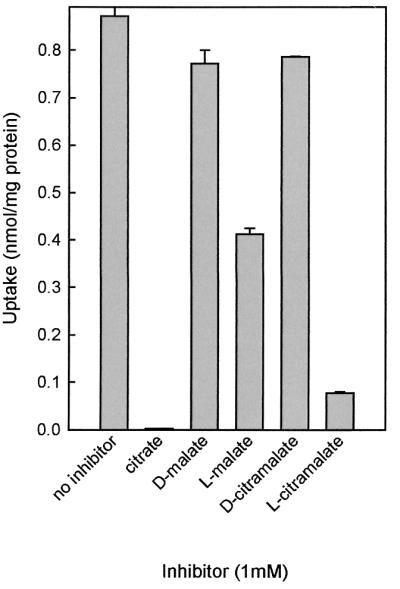
Competitive inhibition of citrate transport in RSO vesicles. Initial rates of [1,5-14C]citrate uptake were determined in the presence of 1 mM concentrations of the competing substrates. The bars represent the initial rates of uptake in the absence and presence of competing substrates. Shown are the averages and standard deviations (error bars) of the results of two independent experiments.
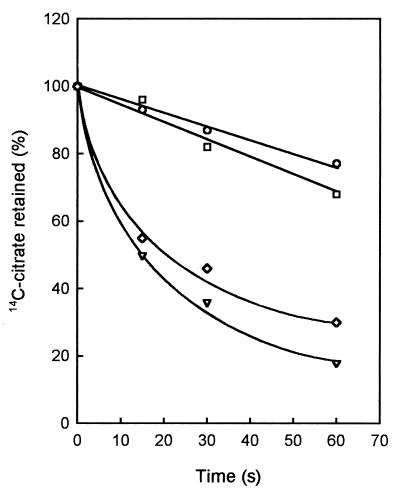
The inhibition assay does not discriminate between a substrate that is transported and a substrate that only binds to the transporter. The ability of CimHbs to transport l-citramalate was determined by heterologous exchange (2). RSO membranes containing CimHbs were allowed to accumulate [1,5-14C]citrate for 1 min driven by the artificial electron donor system ascorbate-PMS (Fig. 2). Addition of the uncoupler FCCP (time zero [Fig. 6]) resulted in a slow release of the label from the membranes during the first minute. Addition of 1 mM citrate or S-malate together with FCCP resulted in a much faster release of label, indicative of homologous citrate-citrate and heterologous citrate–S-malate exchange catalyzed by CimHbs, respectively. However, addition of S-citramalate at a concentration of 1 mM, which resulted in 90% inhibition of PMF-dependent citrate uptake (Fig. 5), together with the uncoupler FCCP did not increase the rate of release of label. Apparently, S-citramalate binds to CimHbs, but is not or is very poorly translocated.
FIG. 6.
Homologous and heterologous exchange catalyzed by CimHbs. RSO membranes prepared from E. coli TOP10 expressing CimHbs were energized with Asc-PMS and allowed to accumulate [1,5-14C]citrate for 1 min. At time zero, 10 μM FCCP was added alone (○) or together with 1 mM concentrations of citrate (▿), l-malate (◊), and l-citramalate (□). Indicated is the fraction of label retained in the membranes relative to that at the zero time point.
Kinetic parameters.
The kinetic parameters of CimHbs were determined in RSO membrane vesicles in 50 mM KPi (pH 6.1) and at 30°C. The data are summarized in Table 2. CimHbs catalyzes citrate transport with a Km of 10 μM and a maximal rate of transport of 37 nmol/min · mg of protein. Because of the lower transport rates for malate, it was difficult to measure the Km for malate accurately. Rather, the affinity of CimHbs for S-malate was inferred from the inhibition of citrate uptake by unlabeled S-malate. The affinity for S-malate was 2 orders of magnitude lower than that observed for citrate (Ki = 1.5 mM). On the other hand, the maximal rate of transport was fourfold higher (167 nmol/min · mg). The affinity of CimHbs for S-citramalate was likewise determined from the inhibition of citrate uptake and was between the affinities for citrate and S-malate (Ki = 100 μM).
TABLE 2.
Kinetic parameters of CimHbs for uptake activity in RSO membrane vesicles
| Substrate | Km (μM) | Kia (μM) | Vmaxd (nmol/min · mg of protein) |
|---|---|---|---|
| Citrate | 12 ± 2 | 37 ± 5 | |
| S-citramalate | 100 ± 20 | −b | |
| S-malate | 1,500 ± 100 | 167 ± 20c |
Inhibition constant for the inhibition of citrate uptake.
No significant turnover was observed.
Calculated using the inhibitor constant and the rate of uptake of l-malate at 19.8 μM.
Vmax, maximal rate of transport.
DISCUSSION
The molecular basis of di- and tricarboxylate uptake in B. subtilis is poorly understood. Three genes involved in C4-dicarboxylate transport have been identified. A mutant strain defective in the ydbH gene showed a clear growth defect on the dicarboxylates fumarate and succinate but not malate, suggesting that the gene product is involved in the uptake of these substrates (1). YdbH is a member of the glutamate transporter family that contains a cluster of C4-dicarboxylate transporter proteins (30). The YdbH protein was not further characterized. yufR of B. subtilis was expressed in E. coli and shown to be a sodium-dependent malate transporter (36). The gene product, termed MaeNbs, is a member of the 2HCT family, which contains both citrate and malate transporters (2). The third gene, yqkI, codes for a transporter that combines Na+-H+ antiporter activity and malate-lactate antiporter activity (36). The transporter is homologous to other Na+-H+ antiporters in the NhaC family (29). The roles of the last two gene products in the physiology of B. subtilis are unclear. Two genes products, CitMbs and CitHbs, have been shown to be involved in the uptake of the tricarboxylate citrate in complex with divalent metal ions.
In this study, we characterized another transporter involved in carboxylate uptake in B. subtilis. CimHbs, the product of the yxkJ gene, is a member of the 2HCT family. CimHbs is a PMF-driven secondary transporter for citrate and malate. Transport catalyzed by CimHbs appears to be electroneutral, i.e., driven by the ΔpH across the membrane. Furthermore, the Na+ ion motive force is not involved, and citrate is transported in its free ionic form. The apparent affinity of CimHbs for citrate is 2 orders of magnitude higher than for malate (10 μM and 1.5 mM, respectively). On the other hand, the maximal rate of transport was fourfold higher for l-malate than for citrate (167 and 37 nmol/min · mg protein, respectively). Therefore, CimHbs is a high-affinity, low-capacity citrate transporter and a low-affinity, high-capacity l-malate transporter that could be involved in the uptake of both substrates under physiological conditions.
In the 2HCT family, two groups of transporters can be discriminated, with distinct physiological functions. One group consists of the “classical” secondary transporters involved in the uptake of substrates, a process that consumes metabolic energy. The second group consists of exchangers that are involved in the generation of metabolic energy by a secondary mechanism (22). The transporters in the latter group, CitPlcm and MlePll, catalyze the uptake of a divalent anion, citrate and malate, respectively, coupled to the excretion of monovalent lactate, which is a metabolic end product in lactic acid bacteria. Electrogenic exchange of the divalent precursor and the monovalent product in combination with scalar proton consumption in the cytoplasmic conversion of precursor into product result in the generation of a PMF (22). The exchangers have three characteristics that distinguish them from the other transporters in the family: (i) exchange is catalyzed much faster than unidirectional transport (4), (ii) an inverse membrane potential drives unidirectional transport (26), and (iii) a wide range of 2-hydroxycarboxylates is transported (2). The characteristics of CimHbs classify the transporter as a “traditional” one functioning in the uptake of citrate and/or malate. CimHbs catalyzes exchange faster than efflux (Fig. 6), but the rate is still much lower than that observed for CitPlcm and MlePll. Attempts to drive citrate uptake catalyzed by an SCN− diffusion potential failed (not shown). Finally, though CimHbs transports both citrate and malate, it fails to translocate one of the monocarboxylic 2-hydroxycarboxylates, such as lactate, which is essential for precursor-product exchange.
The 2HCT family contains transporters with very strict substrate specificities (e.g., CitSkp) as well as transporters with very broad substrate specificities (the exchangers). Substrate specificity and mutational studies have resulted in a model for the binding site of the transporter in the latter group (3, 5, 6). In the model, two interactions between protein and substrate are essential for binding: one with the hydroxyl and one with the carboxylate group of the 2-hydroxycarboxylate motif present in all substrates. A third interaction with a “second” carboxylate group is responsible for high-affinity binding of di- and tricarboxylates and determines the preference of the transporter for the S enantiomers of the substrates. The residue involved in this interaction in CitPlcm is Arg425. Mutation of this residue to Cys resulted in a dramatic loss of affinity for S-malate, while the affinity for the monocarboxylate 2-hydroxyisobutyrate increased somewhat (6). As observed for the exchangers, CimHbs translocates only 2-hydroxycarboxylates, indicating that interactions with the hydroxyl and the carboxyl group are essential. CimHbs is strictly stereospecific and interacts only with the S enantiomers of malate and citramalate. The Arg residue responsible for the interaction in CitPlcm is conserved in the family and corresponds to Arg432 in CimHbs. The data suggest, as may be expected for homologous proteins, that the global structure of the binding site in CitPlcm and CimHbs is the same. Nevertheless, there are important differences. In CitPlcm, the interaction of the Arg residue with the second carboxylate is not essential, while it is in CimHbs. CimHbs does not accept substrates that lack the second carboxylate, i.e., monocarboxylates and the R enantiomers of dicarboxylates. In the future, we plan to study this modulation of substrate specificity within one family of transporters by mutational studies.
ACKNOWLEDGMENTS
We acknowledge J. R. Guest for the gift of E. coli strain JRG4008 and I. Sobczak for the RSO membrane vesicles containing CitSkp of K. pneumoniae.
This research was supported by the Ministry of Economic Affairs in the framework of the IOP Milieutechnologie/Zware metalen IZW97404.
REFERENCES
- 1.Asai K, Baik S H, Kasahara Y, Moriya S, Ogasawara N. Regulation of the transport system for C4-dicarboxylic acids in Bacillus subtilis. Microbiology. 2000;146:263–271. doi: 10.1099/00221287-146-2-263. [DOI] [PubMed] [Google Scholar]
- 2.Bandell M, Ansanay V, Rachidi N, Dequin S, Lolkema J S. Membrane potential-generating malate (MleP) and citrate (CitP) transporters of lactic acid bacteria are homologous proteins. Substrate specificity of the 2-hydroxycarboxylate transporter family. J Biol Chem. 1997;272:18140–18146. doi: 10.1074/jbc.272.29.18140. [DOI] [PubMed] [Google Scholar]
- 3.Bandell M, Lhotte M E, Marty-Teysset C, Veyrat A, Prevost H, Dartois V, Divies C, Konings W N, Lolkema J S. Mechanism of the citrate transporters in carbohydrate and citrate cometabolism in Lactococcus and Leuconostoc species. Appl Environ Microbiol. 1998;64:1594–1600. doi: 10.1128/aem.64.5.1594-1600.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandell M, Lolkema J S. Stereoselectivity of the membrane potential-generating citrate and malate transporters of lactic acid bacteria. Biochemistry. 1999;38:10352–10360. doi: 10.1021/bi9907577. [DOI] [PubMed] [Google Scholar]
- 5.Bandell M, Lolkema J S. Arg425 of the citrate transporter CitP is responsible for high affinity binding of di- and tricarboxylates. J Biol Chem. 2000;275:39130–39136. doi: 10.1074/jbc.M005940200. [DOI] [PubMed] [Google Scholar]
- 6.Bandell M, Lolkema J S. The conserved C-terminus of the citrate (CitP) and malate (MleP) transporters of lactic acid bacteria is involved in substrate recognition. Biochemistry. 2000;39:13059–13067. doi: 10.1021/bi0011882. [DOI] [PubMed] [Google Scholar]
- 7.Baneyx F, Georgiou G. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J Bacteriol. 1990;172:491–494. doi: 10.1128/jb.172.1.491-494.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergsma J, Konings W N. The properties of citrate transport in membrane vesicles from Bacillus subtilis. Eur J Biochem. 1983;134:151–156. doi: 10.1111/j.1432-1033.1983.tb07545.x. [DOI] [PubMed] [Google Scholar]
- 9.Bisschop A, Doddema H, Konings W N. Dicarboxylic acid transport in membrane vesicles from Bacillus subtilis. J Bacteriol. 1975;124:613–622. doi: 10.1128/jb.124.2.613-622.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boorsma A, van der Rest M E, Lolkema J S, Konings W N. Secondary transporters for citrate and the Mg(2+)-citrate complex in Bacillus subtilis are homologous proteins. J Bacteriol. 1996;178:6216–6222. doi: 10.1128/jb.178.21.6216-6222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen W B. Hydrogen sulfide production and citrate utilization in the differentiation of enteric pathogens and coliform bacteria. Res Bull Weld County Health Dep, Greeley, Colorado. 1949;1:3–16. [Google Scholar]
- 12.Davies S J, Golby P, Omrani D, Broad S A, Harrington V L, Guest J R, Kelly D J, Andrews S C. Inactivation and regulation of the aerobic C(4)-dicarboxylate transport (dctA) gene of Escherichia coli. J Bacteriol. 1999;181:5624–5635. doi: 10.1128/jb.181.18.5624-5635.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engel P, Kramer R, Unden G. Anaerobic fumarate transport in Escherichia coli by an fnr-dependent dicarboxylate uptake system which is different from the aerobic dicarboxylate uptake system. J Bacteriol. 1992;174:5533–5539. doi: 10.1128/jb.174.17.5533-5539.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghei O K, Kay W W. A dicarboxylic acid transporter system in Bacillus subtilis. FEBS Lett. 1972;20:140. doi: 10.1016/0014-5793(72)80777-4. [DOI] [PubMed] [Google Scholar]
- 15.Ghei O K, Kay W W. Properties of an inducible C4-dicarboxylic acid transport system in Bacillus subtilis. J Bacteriol. 1973;114:65–79. doi: 10.1128/jb.114.1.65-79.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaback H R. The lac carrier protein in Escherichia coli. J Membr Biol. 1983;76:95–112. doi: 10.1007/BF02000610. [DOI] [PubMed] [Google Scholar]
- 17.Kawai S, Suzuki H, Yamamoto K, Kumagai H. Characterization of the l-malate permease gene (maeP) of Streptococcus bovis ATCC 15352. J Bacteriol. 1997;179:4056–4060. doi: 10.1128/jb.179.12.4056-4060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kay W W, Sweet G D, Widenhorn K, Somers J M. Transport of organic acids in prokaryotes. In: Rosen B P, Silver S, editors. Ion transport in prokaryotes. San Diego, Calif: Academic Press; 1987. pp. 269–302. [Google Scholar]
- 19.Krom B P, Warner J B, Konings W N, Lolkema J S. Complementary metal ion specificity of the metal-citrate transporters CitM and CitH of Bacillus subtilis. J Bacteriol. 2000;182:6374–6381. doi: 10.1128/jb.182.22.6374-6381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Lolkema J S, Enequist H, van der Rest M E. Transport of citrate catalyzed by the sodium-dependent citrate carrier of Klebsiella pneumoniae is obligatorily coupled to the transport of two sodium ions. Eur J Biochem. 1994;220:469–475. doi: 10.1111/j.1432-1033.1994.tb18645.x. [DOI] [PubMed] [Google Scholar]
- 22.Lolkema J S, Poolman B, Konings W N. Role of scalar protons in metabolic energy generation in lactic acid bacteria. J Bioenerg Biomembr. 1995;27:467–473. doi: 10.1007/BF02110009. [DOI] [PubMed] [Google Scholar]
- 23.Lolkema J S, Poolman B, Konings W N. Secondary transporters and metabolic energy generation in bacteria. In: Konings W N, Kaback H R, Lolkema J S, editors. Transport processes in eukaryotic and procaryotic organisms. Amsterdam, The Netherlands: Elsevier; 1996. pp. 229–260. [Google Scholar]
- 24.Magni C, Lopez P, de Mendoza D. The properties of citrate transport catalyzed by CitP of Lactococcus lactis ssp. lactis biovar diacetylactis. FEMS Microbiol Lett. 1996;142:265–269. [Google Scholar]
- 25.Marty-Teysset C, Lolkema J S, Schmitt P, Divies C, Konings W N. Membrane potential-generating transport of citrate and malate catalyzed by CitP of Leuconostoc mesenteroides. J Biol Chem. 1995;270:25370–25376. doi: 10.1074/jbc.270.43.25370. [DOI] [PubMed] [Google Scholar]
- 26.Marty-Teysset C, Posthuma C, Lolkema J S, Schmitt P, Divies C, Konings W N. Proton motive force generation by citrolactic fermentation in Leuconostoc mesenteroides. J Bacteriol. 1996;178:2178–2185. doi: 10.1128/jb.178.8.2178-2185.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oehr P, Willecke K. Citrate-Mg2+ transport in Bacillus subtilis. Studies with 2-fluoro-l-erythro-citrate as a substrate. J Biol Chem. 1974;249:2037–2042. [PubMed] [Google Scholar]
- 28.Poolman B, Molenaar D, Smid E J, Ubbink T, Abee T, Renault P P, Konings W N. Malolactic fermentation: electrogenic malate uptake and malate/lactate antiport generate metabolic energy. J Bacteriol. 1991;173:6030–6037. doi: 10.1128/jb.173.19.6030-6037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saier M H. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slotboom D J, Konings W N, Lolkema J S. Structural features of the glutamate transporter family. Microbiol Mol Biol Rev. 1999;63:293–307. doi: 10.1128/mmbr.63.2.293-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolner B, Ubbink-Kok T, Poolman B, Konings W N. Characterization of the proton/glutamate symport protein of Bacillus subtilis and its functional expression in Escherichia coli. J Bacteriol. 1995;177:2863–2869. doi: 10.1128/jb.177.10.2863-2869.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Does C, Manting E H, Kaufmann A, Lutz M, Driessen A J. Interaction between SecA and SecYEG in micellar solution and formation of the membrane-inserted state. Biochemistry. 1998;37:201–210. doi: 10.1021/bi972105t. [DOI] [PubMed] [Google Scholar]
- 33.van der Rest M E, Molenaar D, Konings W N. Mechanism of Na+-dependent citrate transport in Klebsiella pneumoniae. J Bacteriol. 1992;174:4893–4898. doi: 10.1128/jb.174.15.4893-4898.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Rest M E, Siewe R M, Abee T, Schwarz E, Oesterhelt D, Konings W N. Nucleotide sequence and functional properties of a sodium-dependent citrate transport system from Klebsiella pneumoniae. J Biol Chem. 1992;267:8971–8976. [PubMed] [Google Scholar]
- 35.Warner J B, Krom B P, Magni C, Konings W N, Lolkema J S. Catabolite repression and induction of the Mg2+-citrate transporter CitM of Bacillus subtilis. J Bacteriol. 2000;182:6099–6105. doi: 10.1128/jb.182.21.6099-6105.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y, Guffanti A A, Ito M, Krulwich T A. Bacillus subtilis YqkI is a novel malic/Na+-lactate antiporter that enhances growth on malate at low protonmotive force. J Biol Chem. 2000;275:30287–30292. doi: 10.1074/jbc.M001112200. [DOI] [PubMed] [Google Scholar]
- 37.Willecke K, Lange R. C4-dicarboxylate transport in Bacillus subtilis studied with 3-fluoro-l-erythro-malate as a substrate. J Bacteriol. 1974;117:373–378. doi: 10.1128/jb.117.2.373-378.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto H, Murata M, Sekiguchi J. The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol Microbiol. 2000;37:898–912. doi: 10.1046/j.1365-2958.2000.02055.x. [DOI] [PubMed] [Google Scholar]