Abstract
The Bacillus subtilis inositol operon (iolABCDEFGHIJ) is involved in myo-inositol catabolism. Glucose repression of the iol operon induced by inositol is exerted through catabolite repression mediated by CcpA and the iol induction system mediated by IolR. In this study, we identified two iol catabolite-responsive elements (cre's), to which CcpA complexed with P-Ser-HPr or P-Ser-Crh probably binds. One is located in iolB (cre-iolB, nucleotides +2397 to +2411; +1 is the transcription initiation nucleotide), which was the only cre-iol found in the previous cre search of the B. subtilis genome using a query sequence of WTGNAANCGNWNNCW (W stands for A or T, and N stands for any base). Deletion and base substitution analysis of the iol region indicated that cre-iolB functions even if it is located far downstream of the iol promoter. Further deletion and base substitution analysis revealed another cre located between the iol promoter and the iolA gene (cre-iiolA, nucleotides +86 to +100); the prefix “i” indicates a location in the intergenic region. Both cre-iiolA and cre-iolB appeared to be recognized to almost the same extent by CcpA complexed with either P-Ser-HPr or P-Ser-Crh. Sequence alignment of the six known cre's, including cre-iiolA, which were not revealed in the previous cre search, exhibited another consensus sequence of WTGAAARCGYTTWWN (R stands for A or G, and Y stands for C or T); the right two thymines (TT) were found to be essential for the function of cre-iiolA by means of base substitution analysis. A cre search with this query sequence led to the finding of 14 additional putative cre's.
myo-Inositol is abundant in nature, especially in soil. Various microorganisms, including Bacillus subtilis, are able to grow on myo-inositol as the sole carbon source. The B. subtilis iol divergon consisting of the iolABCDEFGHIJ and iolRS operons is involved in inositol catabolism (29, 31). These two iol operons are induced upon the addition of inositol to the medium (29). This induction is negatively regulated by a repressor (IolR) belonging to the DeoR family through its unique interaction with the extended binding regions close to their promoters (31). Inositol dehydrogenase encoded by iolG catalyzes the first reaction of inositol catabolism by B. subtilis (19, 29). The synthesis of this enzyme induced by inositol was repressed on the addition of glucose to the medium (18, 29). Very recently, DNA microarray analysis implied that not only the expression of iolG but also that of the other 11 iol genes was under glucose repression (30).
The well-characterized mechanisms underlying glucose repression are those of catabolite repression and inducer exclusion. Recently, the mechanism underlying catabolite repression in B. subtilis was extensively investigated. These studies revealed that Bacillus, as well as other low-GC gram-positive bacteria, possesses a negative regulatory mechanism for catabolite repression, which is very different from the positive regulatory mechanisms of enteric bacteria involving cyclic AMP and cyclic AMP receptor protein (20, 25). In low-GC gram-positive bacteria, negative regulation of the transcription of catabolite-repressive genes occurs through the binding of the catabolite control protein (CcpA) (10), which interacts with allosteric effectors such as P-Ser-HPr (6) and P-Ser-Crh (7), to their cis-acting catabolite-responsive elements (cre's) (15).
DNA microarray analysis revealed that the expression of the iolABCDEFGHIJ and iolRS operons was under glucose repression, which was partially CcpA dependent (30). The glucose repression of the synthesis of inositol dehydrogenase (Idh) is dependent on both CcpA and IolR (8, 16, 29, 30), implying that catabolite repression and the induction system mediated by IolR are involved in glucose repression of the iolABCDEFGHIJ operon. Since almost no glucose repression was observed in a doubly mutated strain with respect to ccpA and iolR (30), IolR-independent repression is likely to be exerted through catabolite repression mediated by CcpA. When cre sequences were searched for in the B. subtilis genome using a query sequence of WTGNAANCGNWNNCW (W and N stand for A or T and for any base, respectively), 126 putative and known cre's were found (15). One of them is located within the iolB gene, that is, cre-iolB, which has been found to function as a cre in an in vivo cre test system (15).
In this work, we found on deletion and base substitution analysis of the iol region that cre-iolB, which is located far downstream of the iol promoter (Piol), functioned as a cre. Further deletion and base substitution analysis revealed another functional cre, which is located between Piol and iolA and named cre-iiolA; the prefix “i” indicates the location of cre in the intergenic region. Thus, the CcpA-dependent catabolite repression of the iolABCDEFGHIJ operon was due to the two cre's functioning independently. Interestingly, the sequence of cre-iiolA does not match the 3′ part of the query sequence used for the genome-wide cre search. cre-iiolA was found to belong to a group of cre's exhibiting similar but distinct consensus sequences.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The B. subtilis strains used are listed in Table 1. Strains FU758, FU759, and FU760 carrying iolR::cat were obtained by the transformation of strains 168, QB5223, and QB7096 with DNA of strain YF244 to chloramphenicol (5 μg/ml) resistance on tryptose blood agar base (TBAB) plates (Difco), respectively. Strain FU761 was obtained by the transformation of strain FU759 with DNA of strain QB7096 to kanamycin (10 μg/ml) resistance on TBAB plates, because strains such as strain QB7102 carrying both the ptsH1 and crh::aphA3 mutations were not transformable.
TABLE 1.
Bacterial strains used in this work
| Strain | Genotype | Reference |
|---|---|---|
| GM122 | trpC2 sacB′-′lacZ | 4 |
| SA003 | trpC2 sacB′-′lacZ ptsH1 | 4 |
| 1A250 | trpC2 alsR1 ilvΔ1 | 16 |
| 1A147 | ccpA1 trpC2 alsR1 ilvΔ1 | 16 |
| 168 | trpC2 | 2 |
| YF244 | trpC2 metC7 iolR::cat | 29 |
| QB5223a | trpC2 ptsH1 | 12 |
| QB7096a | trpC2 crh::aphA3 | Not published |
| QB7102a | trpC2 ptsH1 crh::aphA3 | Not published |
| FU704 | trpC2 sacB′-′lacZ iolR::neo | This work |
| FU706 | trpC2 alsR1 ilvΔ1 iolR::neo | This work |
| FU707 | ccpA1 trpC2 alsR1 ilvΔ1 iolR::neo | This work |
| FU709 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol-iolAB′(−107/+2474)-lacZ] | This work |
| FU713 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol-iolAB′(−107/+2270)-lacZ] | This work |
| FU715 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol-iolAB′(−107/+2474,+2405G→T)-lacZ] | This work |
| FU716 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol-iolAB′(−107/+2474,+2410C→T)-lacZ] | This work |
| FU719 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol-iolA′(−107/+1192)-lacZ] | This work |
| FU720 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol-iolA′(−107/+712)-lacZ] | This work |
| FU721 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol-iolA′(−107/+298)-lacZ] | This work |
| FU722 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol(−107/+110)-lacZ] | This work |
| FU723 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol(−107/+82)-lacZ] | This work |
| FU724 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol(−107/+47)-lacZ] | This work |
| FU726 | trpC2 amyE::[cat Pspac-(cre-iiolA)-lacZ] | This work |
| FU727 | trpC2 amyE::[cat Pspac-(cre-iolB)-lacZ] | This work |
| FU728 | trpC2 ptsH1 amyE::[cat Pspac-(cre-iiolA-lacZ)] | This work |
| FU729 | trpC2 ptsH1 amyE::[cat Pspac-(cre-iolB)-lacZ] | This work |
| FU734 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol-iolA′(−107/+298,+91A→G)-lacZ] | This work |
| FU735 | trpC2 sacB′-′lacZ iolR::neo amyE::[cat Piol-iolA′(−107/+298,+94G→T)-lacZ] | This work |
| FU738 | trpC2 sacB′-′lacZ amyE::[cat Pspac-(cre-iiolA)-lacZ] | This work |
| FU742 | trpC2 alsR1 ilvΔ1 amyE::[cat Pspac-(cre-iiolA)-lacZ] | This work |
| FU743 | ccpA1 trpC2 alsR1 ilvΔ1 amyE::[cat Pspac-(cre-iiolA)-lacZ] | This work |
| FU744 | trpC2 alsR1 ilvΔ1 amyE::[cat Pspac-(cre-iolB)-lacZ] | This work |
| FU745 | ccpA1 trpC2 alsR1 ilvΔ1 amyE::[cat Pspac-(cre-iolB)-lacZ] | This work |
| FU748 | trpC2 crh::aphA3 amyE::[cat Pspac-(cre-iiolA)-lacZ] | This work |
| FU749 | trpC2 crh::aphA3 amyE::[cat Pspac-(cre-iolB)-lacZ] | This work |
| FU750 | trpC2 ptsH1 crh::aphA3 amyE::[cat Pspac-(cre-iiolA)-lacZ] | This work |
| FU751 | trpC2 ptsH1 crh::aphA3 amyE::[cat Pspac-(cre-iolB)-lacZ] | This work |
| FU752 | trpC2 sacB′-′lacZ amyE::[cat Pspac-(cre-iiolA,+96T→G)-lacZ] | This work |
| FU753 | trpC2 sacB′-′lacZ amyE::[cat Pspac-(cre-iiolA,+96T→A)-lacZ] | This work |
| FU754 | trpC2 sacB′-′lacZ amyE::[cat Pspac-(cre-iiolA,+97T→G)-lacZ] | This work |
| FU758 | trpC2 iolR::cat | This work |
| FU759 | trpC2 iolR::cat ptsH1 | This work |
| FU760 | trpC2 iolR::cat crh::aphA3 | This work |
| FU761 | trpC2 iolR::cat ptsH1 crh::aphA3 | This work |
Obtained from I. Martin-Verstraete (Institut Pasteur, Paris, France).
Strains FU704, FU706, and FU707 carrying iolR::neo were constructed as follows. Plasmid pIOLR1::neo was first obtained by disruption of iolR encoded in plasmid pIOLR1 (29) with insertion of the neomycin resistance cassette derived from plasmid pBEST513 (11). Plasmid pIOLR1 was digested with EcoRV, ligated with a cassette which had been prepared by EcoRI digestion and subsequent blunt ending of plasmid pBEST513, and was used for the transformation of Escherichia coli strain JM109 (21) to kanamycin resistance (25 μg/ml) on Luria-Bertani (LB) plates (21). The resultant plasmid, pIOLR1::neo, was used for the double-crossover transformation to neomycin (15 μg/ml) resistance of strains GM122, 1A250 and 1A147, resulting in strains FU704, FU706, and FU707, respectively. Construction of the other strains listed in Table 1 is described below.
Construction of strains for deletion analysis of cre's of the iol operon.
To construct strains FU709 and FU713 carrying iolR::neo and transcriptional fusions of iol regions (nucleotides [nt] −107 to +2270 and +2474, respectively; +1 is the transcription initiation nucleotide of the iol operon) to lacZ, we first replaced the EcoRI site of plasmid pCRE-test (15) with an XbaI site, which is absent from the iol region (nt −107 to +2474). This was done through its digestion with EcoRI, blunt ending, attachment of an XbaI linker, digestion with XbaI, and subsequent self-ligation, resulting in plasmid pCRE-test2.
The two iol regions were amplified by PCR using chromosomal DNA of strain 168 and appropriate iol-specific primer pairs, which had been designed to produce 5′ and 3′ flanking XbaI and BamHI sites, respectively (Fig. 1). The PCR products were doubly digested with XbaI and BamHI and then ligated with the XbaI-BamHI arm of plasmid pCRE-test2 from which Pspac had been eliminated. When the iol regions were cloned into plasmid pCRE-test2 in E. coli, unexpected mutations were frequently introduced into the cloned regions, probably due to high expression of iolA from the iol promoter, which is harmful to this bacterium. This gene codes for a protein exhibiting high similarities to methylmalonate-semialdehyde dehydrogenases of various species (29). So, the ligated DNAs were digested with PstI and then used directly for the double-crossover transformation into the amyE locus of B. subtilis strain FU704 to chloramphenicol (5 μg/ml) resistance on TBAB plates, resulting in strains FU709 and FU713 carrying the iol regions (nt −107 to +2474 and +2270) between Piol and lacZ, respectively. Their correct construction was confirmed by sequencing of the inserted iol regions.
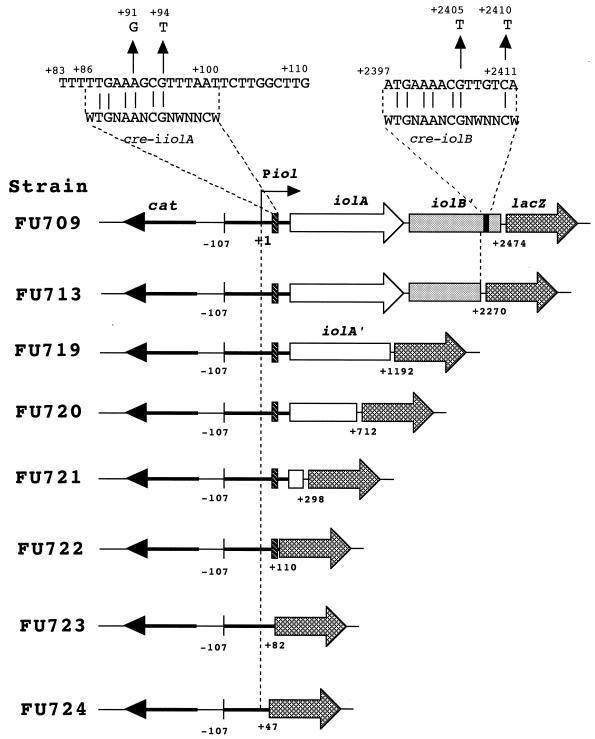
FIG. 1.
Location of cre-iiolA and cre-iolB. The upper part of the figure shows the sequences of cre-iiolA and cre-iolB, which are aligned with a consensus sequence (15). The base substitutions of the indicated bases in the cre sequences which caused the knockout of their function are shown; nt +1 is the iol transcription initiation base. Strains carrying a series of deletion derivatives of Piol (iol promoter)-iolAB′-lacZ in the amyE locus, used for identification of iol-cre's, were constructed as described in the text except for the following. The primers for amplifying the respective iol regions by PCR for construction of strains FU709, FU713, FU719, FU720, FU721, FU722, FU723, and FU724 were a forward one (nt −105 to −85) with an adapter sequence of GTCCTCTAGA and reverse primers (+2448 to +2474, +2250 to +2270, +1173 to +1192, +693 to +712, +279 to +298, +91 to +110, +58 to +82, and +27 to +47) with an adapter sequence of GATAGGATCC; the underlined sequences are XbaI and BamHI sites, respectively.
Strains FU719, FU720, FU721, FU722, FU723, and FU724 carrying a series of further deletions of the iol region (nt −107 to +2270) between Piol and lacZ in amyE of strain FU713 were constructed as follows. Each of the iol regions (nt −107 to +1192, +712, +298, +110, +82, and +47) was amplified by PCR using DNA of strain 168 and its iol-specific primer pair to generate flanking XbaI and BamHI sites (Fig. 1). The PCR products were digested with XbaI and BamHI and then ligated with the XbaI-BamHI arm of plasmid pCRE-test2. The ligated DNAs were used for the transformation of E. coli strain JM109 to ampicillin (50 μg/ml) resistance on Luria-Bertani plates. The correct construction of the Piol-iol-lacZ fusions in the resultant plasmids was confirmed by sequencing. The plasmids were linearized with PstI and then used for the double-crossover transformation of strain FU704 carrying iolR::neo to chloramphenicol resistance, strains FU719, FU720, FU721, FU722, FU723, and FU724 being produced.
Base substitutions in cre sequences within the Piol-iol-lacZ fusions.
Strains FU715 and FU716 carrying base substitutions of +2405G→T and +2410C→T in the cre-iolB sequence (Fig. 1) within the Piol-iolAB′(nt −107 to +2474)-lacZ fusion of strain FU709 were constructed as follows. Introduction of base substitutions was performed by means of recombinant PCR using DNA of strain 168 and the following primers, as described previously (31). The common upstream and downstream primers were 5′-GTCCTCTAGACCTTCTCTTACTTCTCTTACTTG-3′ (the XbaI site is underlined) and 5′-GATAGGATCCTCATTTAATTAGTAGGATGTGTATCCG-3′ (the BamHI site is underlined), respectively. The overlapping primers for +2405G→T were 5′-GGGAAATGAAAACTTTGTCATCG-3′ and 5′-CGATGACAAAGTTTTCATTTCCC-3′, and those for +2410C→T were 5′-AACGTTGTTATCGTTCCTGCGG-3′ and 5′-CCGCAGGAACGATAACAACGTT-3′ (each substituted base is underlined). The resultant recombinant PCR products were digested with XbaI and BamHI and then ligated with the XbaI-BamHI arm of plasmid pCRE-test2. The resultant plasmids were linearized with PstI and then integrated into amyE of strain FU704 through a double-crossover event, resulting in strains FU715 and FU716 being produced.
Strains FU734 and FU735 carrying base substitutions (+91A→G and +94G→T) in the cre-iiolA sequence (Fig. 1) within the iol region (nt −107 to +298) were constructed as follows. Introduction of base substitutions was performed by recombinant PCR using DNA of strain 168 and the following primers, as described above. The common upstream and downstream primers were 5′-GTCCTCTAGACCTTCTCTTACTTCTCTTACTTG-3′ (the XbaI site is underlined) and 5′-GATAGGATCCCATAGCACTTCTTTCGTCGC-3′ (the BamHI site is underlined), respectively. The overlapping primers for +91A→G were 5′-GGTGTTTTTGAAGGCGTTTAATTCTTGGC-3′ and 5′-GCCAAGAATTAAACGCCTTCAAAAACACC-3′, and those for +94G→T were 5′-GGTGTTTTTGAAAGCTTTTAATTCTTGGC-3′ and 5′-GCCAAGAATTAAAAGCTTTCAAAAACACC-3′ (each substituted base is underlined).
Strain construction for further functional analysis of cre-iiolA and cre-iolB.
The respective iol regions containing cre-iiolA and cre-iolB (nt +63 to +121, and +2375 to +2430) were amplified by PCR using DNA of strain 168 and the following two primer pairs to generate flanking BamHI sites. For amplification of the cre-iiolA region, the upstream and downstream primers were 5′-GATAGGATCCCGCCATTTATTTTTTTGGTG-3′ (nt +63 to +82) and 5′-GATAGGATCCCCACTTTTCAGCAAGCCAAG-3′ (nt +102 to +121), and for that of the cre-iolB sequence, they were 5′-GATAGGATCCGACGAGACAATGACTGTGGG-3′ (nt +2375 to +2394) and 5′-GATAGGATCCTGGTATCCCGCAGGAACGAT-3′ (nt +2411 to +2430) (the respective BamHI sites are underlined). The PCR products were digested with BamHI and then cloned into the BamHI site of plasmid pCRE-test (15). The ligated DNA was used for the transformation of E. coli strain JM109 to ampicillin resistance. After confirming the correct orientations and sequences of the Pspac-cre-lacZ fusions in the resulting plasmids pCRE-iiolA and -iolB by sequencing, they were linearized with PstI and used for the double-crossover transformation of strains GM122, 168, QB5223, QB7096, 1A250, and 1A147 to chloramphenicol resistance. The respective resultant strains (FU738, FU726, FU728, FU748, FU742, and FU743) carried the Pspac-(cre-iiolA)-lacZ fusion in their amyE locus, whereas the other strains (FU727 from 168, FU729 from QB5223, FU749 from QB7096, FU744 from 1A250, and FU745 from 1A147) contained the Pspac-(cre-iolB)-lacZ fusion. Strains FU750 and FU751 were obtained by the transformation of strains FU728 and FU729 with DNA of strain QB7096 to kanamycin (10 μg/ml) resistance on TBAB plates, respectively.
Strains FU752, FU753, and FU754 carrying the respective base substitutions of the cre-iiolA sequence (+96T→G, +96T→A, and +97T→A) (Fig. 2) were constructed as follows. Introduction of base substitutions was performed by recombinant PCR using DNA of plasmid pCRE-iiolA as the template and the following primer pairs, as described above. The common upstream and downstream primers were 5′-TGTAAAACGACGGCCAGTTAAAGGATTTGAGCGTAGCG-3′ and 5′-CAGGAAACAGCTATGACCATTACGCCAGCTGGCGAAAG-3′, where the underlined sequences are located in the cat and lacZ genes of plasmid pCRE-iiolA, respectively. The overlapping primers for the +96T→G substitution were 5′-GAAAGCGGTAATTCTTGG-3′ and 5′-CCAAGAATTACACGCTTTC-3′, those for +96T→A were 5′-GAAAGCGTATAATTCTTGG-3′ and 5′-CCAAGAATTATACGCTTTC-3′, and those for +97T→G were 5′-GAAAGCGTTGAATTCTTGG-3′ and 5′-CCAAGAATTCAACGCTTTC-3′ (each substituted base is underlined). The resulting recombinant PCR products were digested with BamHI and then cloned into the BamHI site of plasmid pCRE-test. After linearization of the resulting plasmids with PstI, strain GM122 was transformed, resulting in strains FU752, FU753, and FU754.
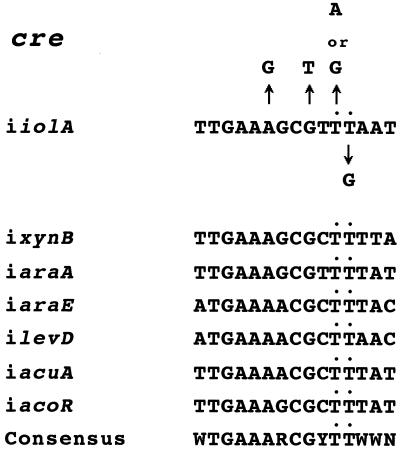
FIG. 2.
Alignment of the cre-iiolA sequence with known cre sequences that were not revealed with a query sequence of WTGNAANCGNWNNCW. The upper part of the figure shows the knockout substitutions of the cre-iiolA sequence. The sequences of five known cre sequences that were not revealed with a query sequence of WTGNAANCGNWNNCW (15) are aligned with that of cre-iiolA; the known cre's are cre-ixynB (7), cre-iaraA (22), cre-iaraE (23), cre-ilevD (12), cre-iacuA (9), and cre-iacoR (1). The consensus sequence carries two thymines (TT) in the consensus sequence, which are indicated with a dot in this alignment. R represents A or G, and Y represents C or T.
Idh and β-Gal assay.
Cells were grown to an absorbance level at 600 nm (A600) of 0.6 in S6 medium (5) containing 0.5% Casamino Acids (Difco) and supplemented with required amino acids (50 μg/ml) with and without a 10 mM concentration of inositol and/or glucose. In addition, neomycin (15 μg/ml), chloramphenicol (5 μg/ml), and kanamycin (5 μg/ml) were added to the media for the growth of strains carrying iolR::neo, iolR::cat or with cat integration into amyE, and crh::aphA3, respectively. The cells (A600 unit = 3.6) were harvested and then lysed by lysozyme treatment and brief sonication (18). Idh activity in crude cell lysates was spectrophotometrically assayed as described previously (18). β-Galactosidase (β-Gal) activity in crude cell extracts was spectrophotometrically assayed as previously described (15). The amounts of protein in cell extracts were determined by the method of Bradford (3) with bovine serum albumin as a standard.
RESULTS
B. subtilis genes involved in glucose repression of Idh synthesis.
Glucose repression of the synthesis of Idh encoded by iolG is known to occur through catabolite repression mediated by CcpA (8, 16, 30) as well as a regulation involving IolR, probably through inducer exclusion (29, 30). We first investigated the effects of the ptsH1 mutation causing the replacement of Ser46 of HPr with alanine and crh::aphA3 disruption on glucose repression of Idh synthesis. As shown in Table 2, Idh synthesis was severely repressed by glucose in strain 168trpC2 (wild type) (repression ratio, >461). Idh synthesis was not relieved from this glucose repression in strain QB7096 (crh::aphA3) at all (repression ratio, >470), but it was partially relieved from catabolite repression in strain QB5223 (ptsH1) (repression ratio = 79). Idh synthesis was more relieved from glucose repression in strain QB7102 carrying the ptsH1 and crh::aphA3 mutations (repression ratio = 7.3) and strain 1A147 carrying the ccpA1 mutation (repression ratio = 4.9). Moreover, this CcpA-independent glucose repression observed in strain 1A147 (ccpA1) was almost completely abolished in strain FU707 carrying the ccpA1 and iolR::neo mutations (repression ratio = 1.1) (Table 2). Part of the results are consistent with those reported by Galinier et al. (8), who found that Crh involvement in catabolite repression of Idh synthesis was solely observed in the ptsH1 background, because HPr alone is likely to be sufficient to cause this catabolite repression. However, it was reported that either the ptsH crh double mutant or the ccpA::spec mutant was completely relieved from glucose repression of Idh synthesis (8). We cannot explain this discrepancy properly, but it might possibly be due to the difference in the media for cell cultivation; we used S6 (this work; 16, 30) or DSM medium (30), whereas they used CSK medium (8).
TABLE 2.
Effects of the iolR, ptsH, crh, and ccpA mutations on catabolite repression of inositol dehydrogenase (Idh) synthesisa
| Strain | Relevant genotype | Idh activity (nmol/min/mg)
|
Repression ratiob | ||
|---|---|---|---|---|---|
| −Iol | +Iol | +Iol and Glc | |||
| 168 | Wild type | 13 | 922 | <2 | >461 |
| QB5223 | ptsH1 | 23 | 1,031 | 13 | 79 |
| QB7096 | crh::aphA3 | 23 | 1,147 | <2 | >470 |
| QB7102 | ptsH1 crh::aphA3 | 19 | 827 | 113 | 7.3 |
| FU758 | iolR::cat | 1,440 | 1,227 | 254 | 4.8 |
| FU759 | ptsH1 iolR::cat | 1,455 | 1,118 | 678 | 1.6 |
| FU760 | crh::aphA3 iolR::cat | 1,575 | 1,226 | 324 | 3.8 |
| 1A147 | ccpA1 | 16 | 760 | 155 | 4.9 |
| FU706 | iolR::neo | 914 | 810 | 133 | 6.1 |
| FU707 | ccpA1 iolR::neo | 963 | 878 | 793 | 1.1 |
Cells of B. subtilis strains were grown with (+) and without (−) myo-inositol (Iol) and with myo-inositol and glucose (Glc), and Idh activities in crude extracts were determined as described in the text. Idh activities are expressed as averages of the values obtained for at least three independent experiments.
Ratio of +Iol to +Iol and Glc.
To more clearly demonstrate the involvement of either HPr or Crh in the CcpA-dependent catabolite repression of Idh synthesis, we constructed ptsH1 and crh::aphA3 isogenic mutants in the iolR::cat background and examined their effect on this catabolite repression (Table 2). Idh synthesis was considerably relieved from glucose repression in strain FU758 carrying iolR::cat (repression ratio = 4.8). The remaining repression is likely to be due to catabolite repression mediated by CcpA, because Idh synthesis was released from glucose repression (repression ratio = 6.1) in strain FU706 carrying iolR::neo and was almost completely released in strain FU707 carrying ccpA1 and iolR::neo (repression ratio = 1.1). This CcpA-dependent catabolite repression, which was slightly decreased in strain FU760 carrying crh::aphA3 and iolR::cat (repression ratio = 3.8), appeared to be still present in strain FU759 carrying ptsH1 and iolR::cat (repression ratio = 1.6), suggesting that Crh might be involved in this repression. However, we could not investigate catabolite repression of Idh synthesis in strain FU761 carrying triple defects of ptsH1, crh::aphA3, and iolR::cat, because this strain could not grow normally in the medium with glucose. The results suggest that Crh as well as HPr is involved in the CcpA-dependent catabolite repression of Idh synthesis.
Actual involvement of cre-iolB in catabolite repression of the iol operon.
Upon a search for cre sequences in B. subtilis, 126 putative and known cre's were revealed (15). Among them, cre-iolB was found to function as a cre in an in vivo cre test system (15). The iol operon is most likely transcribed from only one promoter, Piol (29). Hence, to determine whether or not cre-iolB, which is located approximately 2,400 bp downstream of Piol, is actually involved in the catabolite repression of the iol operon, we constructed a transcriptional fusion, Piol-iolA-iolB′-lacZ, possessing an iol region (nt −107 to +2474) with cre-iolB (nt +2397 to +2411), which expresses lacZ under the direction of Piol (Fig. 1), and integrated it into the chromosomal amyE locus of strain FU704 carrying iolR::neo. The resulting strain, FU709, produced a high level of β-Gal constitutively even on growth without inositol, which was repressed 3.5-fold on the addition of glucose to the medium (Table 3).
TABLE 3.
Deletion and base substitution analyses for cre's of the iol operon to monitor lacZ expression under the control of the iol promoter and cre('s) in the background of iolR::neoa
| Strainb | Fusion | iol region | β-Gal activity (nmol/min/mg)
|
Repression ratioc | |
|---|---|---|---|---|---|
| −Glc | +Glc | ||||
| FU709 | Piol-iolAB′-lacZ | −107 to +2474 | 1,217 | 349 | 3.5 |
| FU715 | Piol-iolAB′-lacZ | −107 to +2474, +2405G→T | 1,123 | 448 | 2.5 |
| FU716 | Piol-iolAB′-lacZ | −107 to +2474, +2410C→T | 942 | 358 | 2.6 |
| FU713 | Piol-iolAB′-lacZ | −107 to +2270 | 2,615 | 1,077 | 2.4 |
| FU719 | Piol-iolA′-lacZ | −107 to +1192 | 1,046 | 449 | 2.3 |
| FU720 | Piol-iolA′-lacZ | −107 to +712 | 4,418 | 1,921 | 2.3 |
| FU721 | Piol-iolA′-lacZ | −107 to +298 | 5,380 | 2,232 | 2.4 |
| FU722 | Piol-lacZ | −107 to +110 | 751 | 307 | 2.4 |
| FU723 | Piol-lacZ | −107 to +82 | 1,813 | 1,927 | 0.9 |
| FU724 | Piol-lacZ | −107 to +47 | 1,649 | 1,753 | 0.9 |
| FU734 | Piol-iolA′-lacZ | −107 to +298, +91A→G | 7,332 | 9,276 | 0.8 |
| FU735 | Piol-iolA′-lacZ | −107 to +298, +94G→T | 6,712 | 7,148 | 0.9 |
Cells of B. subtilis strains were grown with (+) and without (−) glucose (Glc), and β-Gal activities in crude extracts were determined as described in the text. β-Gal activities are expressed as averages of the values obtained for at least three independent experiments.
All the B. subtilis strains assayed carried an iolR disruption (iolR::neo).
Ratio of −Glc to +Glc.
To determine whether or not cre-iolB actually functions in the Piol-iolA-iolB′(nt −107 to +2474)-lacZ transcriptional fusion, we introduced the base substitutions of +2405G→T and +2410C→T in the cre-iolB sequence, which resulted in strains FU715 and FU716 (Fig. 1), with the G and C corresponding to conserved bases (positions 9 and 14 of the query sequence [WTGNAANCGNWNNCW]) for the genome-wide cre search (15). As shown in Table 3, β-Gal synthesis in strains FU715 and FU716 was partially relieved from catabolite repression (repression ratio = 2.5 and 2.6, respectively). We also deleted a cre-iolB region (nt +2271 to 2474) from the Piol-iolA-iolB′-lacZ fusion, which resulted in strain FU713. This deletion decreased the catabolite repression ratio of β-Gal synthesis to 2.4, which is similar to the levels observed for the cases of base substitutions in cre-iolB (Table 3). These results indicate that cre-iolB is actually involved in catabolite repression of the transcription from approximately 2,400 bp downstream of its own promoter (Piol), although it appeared to mediate a low level of catabolite repression. Furthermore, even if cre-iolB had been deleted from or mutated in the Piol-iolA-iolB′-lacZ fusion, β-Gal synthesis was still partially under catabolite repression. This suggests that another cre responsible for this residual catabolite repression exists between nt −107 and +2270 of the iol region, although the genome-wide cre search with the query sequence (WTGNAANCGNWNNCW) failed to reveal any cre candidate (15).
Identification of another cre for catabolite repression of the iol operon.
To find another cre for the iol operon, which might be located between nt −107 and +2270, we further constructed a series of deletion derivatives of strain FU713, from nt +2270 toward the 5′ direction in the Piol-iolA-iolB′-lacZ fusion (Fig. 1), and determined the catabolite repression ratios of β-Gal synthesis (Table 3). The catabolite repression ratios of β-Gal synthesis (2.3 to 2.4) obtained with strains FU719, FU720, FU721, and FU722 carrying iol regions from nt −107 to nt +1192, +712, +298, and +110, respectively, were almost the same as that with strain FU713 carrying one from nt −107 to +2270 (2.4) (Table 3), indicating that no other cre is located in the iol region between nt +111 and +2270 covering iolA and iolB′. However, β-Gal synthesis was completely relieved from catabolite repression in strains FU723 and FU724 carrying iol regions from nt −107 to +82 and +47, respectively (repression ratio = 0.9). These results suggest that another cre for the iol operon is located between nt +83 and +110.
Careful examination of the nucleotide sequence of the iol region (nt +83 to +110) revealed that this region contains a cre-like sequence (nt +86 to +100) exhibiting high similarity to the 5′ side of the consensus sequence (WTGNAANCGNWNNCW) (Fig. 1). In a very recent publication dealing with whole genome analyses (17), this sequence has been also proposed to function as a cre. To determine whether or not the cre-like sequence is another cre for the iol operon, we introduced the base substitutions of +91A→G and +94G→T into this sequence in strain FU721 carrying the Piol-iolA′(nt −107 to +298)-lacZ fusion, which resulted in strains FU734 and FU735, respectively (Fig. 1). As shown in Table 3, these base substitutions completely abolished the catabolite repression of β-Gal synthesis in strains FU734 and FU735 (repression ratios = 0.8 and 0.9, respectively), indicating that the cre-like sequence functioned as a cre for the iol operon, and was designated as cre-iiolA.
Functional analysis of cre-iiolA by means of base substitutions.
Fig. 2 lists several cre's, such as cre-ixynB (7), cre-iaraA (22), cre-iaraE (23), cre-ilevD (12), cre-iacuA (9), and cre-iacoR (1), which have been reported to function or proposed to function as cre's but were not revealed in our previous cre search (15). Interestingly, the last C of the consensus sequence of WTGNAANCGNWNNCW is not conserved in the sequences of these cre's as well as cre-iiolA (Fig. 2), nevertheless the substitution of this C abolished the cre function almost completely, as in the cases of C→T and G in cre-iamyE (27), C→T in cre-gntR (6), C→T in cre-hutP (28), and C→T in cre-iolB (Table 3). Instead, the WN bases underlined in the above sequence are TT in all of these cre sequences (Fig. 2). Thus, we examined whether or not the positioning of the TT bases is essential for cre-iiolA function.
We first constructed a Pspac-(cre-iiolA)-lacZ fusion through cloning of an iol region (nt +63 to +121) containing cre-iiolA into the BamHI site of plasmid pCRE-test (15) and integrated it into the amyE locus of strain GM122, which resulted in strain FU738. As shown in Table 4, β-Gal synthesis in strain FU738, which is directed by a constitutive spac promoter (Pspac), was subjected to catabolite repression (repression ratio = 4.3) due to the presence of cre-iiolA between Pspac and lacZ. Then, we introduced three base substitutions of the TT of cre-iiolA (+96T→G, +96T→A, and +97T→G) into the Pspac-(cre-iiolA)-lacZ fusion of strain FU738, which resulted in strains FU752, FU753, and FU754, respectively. As shown in Table 4, these base substitutions completely abolished the catabolite repression of β-Gal synthesis observed with strain FU738 (repression ratios = 0.9 for strains FU752 and FU754 and 1.0 for strain FU753). The results clearly indicate that the TT of cre-iiolA are essential for its function.
TABLE 4.
Base substitution analysis for cre-iiolA to monitor lacZ expression under the control of the spac promoter and cre-iiolAa
| Strain | Base substitution in cre-iiolA | β-Gal activity (nmol/min/mg)
|
Repression ratiob | |
|---|---|---|---|---|
| −Glc | +Glc | |||
| FU738 | Wild type | 226 | 53 | 4.3 |
| FU752 | +96T→G | 219 | 240 | 0.9 |
| FU753 | +96T→A | 203 | 207 | 1.0 |
| FU754 | +97T→G | 232 | 272 | 0.9 |
Cells of B. subtilis strains were grown with (+) and without (−) glucose (Glc), and β-Gal activities in crude extracts were determined as described in the text. β-Gal activities are expressed as averages of the values obtained for at least three independent experiments.
Ratio of −Glc to +Glc.
Analysis of HPr and Crh involvement in catabolite repression exerted through cre-iiolA and cre-iolB
Not only HPr but also Crh is involved in catabolite repression of Idh synthesis (Table 2) (8), which is likely exerted through cre-iiolA and cre-iolB (Table 2). The sequence of cre-iiolA was found to be distinct from that of cre-iolB (Fig. 2). So, we examined the HPr and Crh involvement in the catabolite repressions exerted through cre-iiolA and cre-iolB. We constructed a series of isogenic strains—FU726 (wild type), FU728 (ptsH1), FU748 (crh::aphA3), and FU750 (ptsH1 crh::aphA3)—carrying the Pspac-(cre-iiolA)-lacZ fusion in the amyE locus and another series of isogenic strains—FU727 (wild type), FU729 (ptsH1), FU749 (crh::aphA3), and FU751 (ptsH1 crh::aphA3)—carrying the Pspac-(cre-iolB)-lacZ fusion in this locus.
As shown in Table 5, β-Gal synthesis in strain FU726 (wild type) was under catabolite repression exerted through cre-iiolA (repression ratio = 3.2). This ratio was reduced to 1.5 in FU728 (ptsH1) but remained almost the same (repression ratio = 3.3) in strain FU748 (crh::aphA3). Also, this synthesis was completely relieved from catabolite repression in strain FU743 (ptsH1 crh::aphA3). In a similar manner, the catabolite repression ratios of β-Gal synthesis exerted through cre-iolB in strains FU727 (wild type), FU729 (ptsH1), FU749 (crh::aphA3), and FU751 (ptsH1 crh::aphA3) were found to be 4.3, 1.3, 4.4, and 0.9, respectively (Table 5). Furthermore, isogenic strains FU742 (wild type) and FU743 (ccpA1) carrying the Pspac-(cre-iiolA)-lacZ fusion exhibited catabolite repression ratios of 3.1 and 0.9, respectively, while isogenic strains carrying the Pspac-(cre-iolB)-lacZ fusion exhibited the ratios of 4.4 and 1.0. These results clearly indicate that the catabolite repression exerted through cre-iiolA and that exerted through cre-iolB occur independently of each other; the latter repression (repression ratios = 4.3 and 4.4 for the wild type) seemed somewhat severer than the former (repression ratios = 3.2 and 3.1). These findings also suggest that HPr is likely to be involved in catabolite repression exerted by both cre-iiolA and cre-iolB to almost the same extents, and if HPr is deficient, Crh can compensate for the HPr function partially.
TABLE 5.
Effects of the ptsH, crh and ccpA mutations on catabolite repression of β-Gal synthesis under the control of the spac promoter and cre-iiolA or cre-iolB
| Strain | cre | Relevant genotype | β-Gal activity (nmol/min/mg)
|
Repression ratiob | |
|---|---|---|---|---|---|
| −Glc | +Glc | ||||
| FU726 | iiolA | Wild type | 212 | 67 | 3.2 |
| FU728 | iiolA | ptsH1 | 221 | 148 | 1.5 |
| FU748 | iiolA | crh::aphA3 | 240 | 73 | 3.3 |
| FU750 | iiolA | ptsH1 crh::aphA3 | 278 | 333 | 0.8 |
| FU742 | iiolA | Wild type | 291 | 95 | 3.1 |
| FU743 | iiolA | ccpA1 | 337 | 370 | 0.9 |
| FU727 | iolB | Wild type | 226 | 53 | 4.3 |
| FU729 | iolB | ptsH1 | 196 | 147 | 1.3 |
| FU749 | iolB | crh::aphA3 | 183 | 42 | 4.4 |
| FU751 | iolB | ptsH1 crh::aphA3 | 259 | 279 | 0.9 |
| FU744 | iolB | Wild type | 307 | 70 | 4.4 |
| FU745 | iolB | ccpA1 | 320 | 328 | 1.0 |
Cells of B. subtilis strains were grown with (+) and without (−) glucose (Glc), and β-Gal activities in crude extracts were determined as described in the text. β-Gal activities are expressed as averages of the values obtained for at least three independent experiments.
Ratio of −Glc to +Glc.
DISCUSSION
Glucose repression of the iol operon is known to be exerted through catabolite repression mediated by CcpA and a regulation system involving IolR, probably through inducer exclusion (30). We investigated the catabolite repression of the iol operon under experimental conditions where the involvement of IolR in its repression was eliminated. Deletion and base substitution analysis allowed us to identify two cre's of the iol operon (cre-iiolA and cre-iolB) (Fig. 1 and Tables 3 and 4). cre-iiolA is located between the iol promoter and the iolA gene (nt +86 to +100), while cre-iolB is in iolB (nt +2397 to +2411). The presence of two cre's has been reported in the gnt (14), ackA (26), ara (22), and rbs (15, 24) operons. Our previous in vivo results (14) implied that catabolite repression exerted by cre-igntR (creup) was probably independent of that exerted by cre-gntR (credown). This study also revealed that cre-iiolA and cre-iolB likely function independently (Table 5). In addition, cre-iolB was found to function at the original location (nt +2397 to +2411), i.e., far downstream of the transcription initiation site (Table 3). Although two cre's of each of the other operons have not been characterized well, it is notable that the relative locations of the two cre's of the ara and rbs operons are very similar to those of iol. These results are well consistent with the idea that the mechanism underlying catabolite repression can be explained by a transcription roadblock if cre is located downstream of the transcription initiation site (6, 13).
Our previous genome-wide cre search using a query sequence of WTGNAANCGNWNNCW (15) failed to reveal cre-iiolA as well as six known cre's (cre-ixynB [7], cre-iaraA [22], cre-iaraE [23], cre-ilevD [12], cre-iacuA [9], and cre-iacoR [1]), although it revealed 126 putative and known cre's, including cre-iolB. Alignment of the sequences of the six cre's not revealed by the search led to another consensus sequence of WTGAAARCGYTTWWN (Fig. 2). The 5′ part of this consensus sequence perfectly coincides with the previous query sequence, but the 3′ one does not match it well. It is notable that the 3′ part of the latter consensus sequence includes conserved TT but is devoid of the last CW of the former consensus sequence. Actually, base substitution analysis of the TT of cre-iiolA indicated that they are indispensable for its function (Fig. 2 and Table 4). Although the sequence of cre-iiolA appeared to be distinct from that of cre-iolB in conserved bases, a protein complex of CcpA with either P-Ser-HPr or P-Ser-Crh likely binds to both cre's to similar extents (Table 5).
Recent analysis of B. subtilis cre sequences led to the following three conclusions (15). (i) Lower mismatching of cre sequences with the query sequence (WTGNAANCGNWNNCW) is required for cre function. (ii) Although cre sequences are partially palindromic, lower mismatching in the same direction as that of transcription of the target genes is more critical for cre function than that in the inverse direction. (iii) Yet, a more palindromic nature of cre sequences is desirable for a better function. Comparison of the above two consensus sequences also implied that the 5′ part of cre sequences should be well conserved for their efficient function and that a protein complex of CcpA with P-Ser-HPr or P-Ser-Crh recognizes this part. The last CW (preferably CA) of the query sequence of WTGNAANCGNWNNCW is likely to be required for pairing with TG, resulting in proper binding of the complex. However, this pairing might be compensated for by another pairing of TT of the second consensus sequence of WTGAAARCGYTTWWN with AA. Of course, both pairings appear to be more desirable for efficient cre function.
We searched for cre sequences in the B. subtilis genome with the currently deduced consensus sequence of WTGAAARCGYTTWW through the DNA pattern search program of the SubtiList Web Server (http://genolist.pasteur.fr/SubtiList/). This search revealed 14 more putative cre's without any mismatch: cre-iybgJ, cre-iycbF, cre-iyceK, cre-iydaA, cre-yebB, cre-iopuE, cre-islp, cre-iyqkI, cre-levR, cre-iysfC, cre-yusL, cre-iyvbQ, cre-iyvfK, and cre-iyycE. Our present study implies that a genome-wide search for certain cis-acting elements with a single query sequence will not reveal most of the elements in question because of additional features due to their secondary structure, such as a palindromic nature (this work) and sequence periodicity (31). In the case of a cre search, a genome-wide search with these two query sequences appears to be able to reveal almost all of them.
ACKNOWLEDGMENTS
We thank K. Adachi, S. Yamada, M. Takatani, M. Abe, and S. Iijima for their help in the experiments.
This work was supported by a grant, JSPS-RFTF96L00105, from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Ali N O, Bignon J, Rapoport G, Debarbouille M. Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis. J Bacteriol. 2001;183:2497–2504. doi: 10.1128/JB.183.8.2497-2504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher J, Reizer J, Fisher C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujita Y, Freese E. Isolation and properties of a Bacillus subtilis mutant unable to produce fructose-bisphosphatase. J Bacteriol. 1981;145:760–767. doi: 10.1128/jb.145.2.760-767.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 7.Galinier A, Deutscher J, Martin-Verstraete I. Phosphorylation of either Crh or HPr mediates binding of CcpA to the Bacillus subtilis xyn cre and catabolite repression of the xyn operon. J Mol Biol. 1999;286:307–314. doi: 10.1006/jmbi.1998.2492. [DOI] [PubMed] [Google Scholar]
- 8.Galinier A, Haiech J, Kilhoffer M-C, Jaquinod M, Stülke J, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundy F J, Turinsky A J, Henkin T M. Catabolite repression of the Bacillus subtilis acetate and acetoin utilization genes by CcpA. J Bacteriol. 1994;176:4527–4533. doi: 10.1128/jb.176.15.4527-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 11.Itaya M, Kondo K, Tanaka T. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 1989;17:4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Verstraete I, Stülke J, Klier A, Rapoport G. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6919–6927. doi: 10.1128/jb.177.23.6919-6927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miwa Y, Fujita Y. Promoter-independent catabolite repression of the Bacillus subtilis gnt operon. J Biochem. 1993;113:665–671. doi: 10.1093/oxfordjournals.jbchem.a124100. [DOI] [PubMed] [Google Scholar]
- 14.Miwa Y, Nagura K, Eguchi S, Fukuda H, Deutscher J, Fujita Y. Catabolite repression of the Bacillus subtilis gnt operon exerted by two catabolite-responsive elements. Mol Microbiol. 1997;23:1203–1213. doi: 10.1046/j.1365-2958.1997.2921662.x. [DOI] [PubMed] [Google Scholar]
- 15.Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 2000;28:1206–1210. doi: 10.1093/nar/28.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miwa Y, Saikawa M, Fujita Y. Possible function and some properties of the CcpA protein of Bacillus subtilis. Microbiology. 1994;140:2567–2575. doi: 10.1099/00221287-140-10-2567. [DOI] [PubMed] [Google Scholar]
- 17.Moreno M S, Schneider B L, Maile R R, Weyler W, Saier M H., Jr Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol Microbiol. 2001;39:1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- 18.Nihashi J, Fujita Y. Catabolite repression of inositol dehydrogenase and gluconate kinase syntheses in Bacillus subtilis. Biochim Biophys Acta. 1984;798:88–95. doi: 10.1016/0304-4165(84)90014-x. [DOI] [PubMed] [Google Scholar]
- 19.Ramaley R, Fujita Y, Freese E. Purification and properties of Bacillus subtilis inositol dehydrogenase. J Biol Chem. 1979;254:7684–7690. [PubMed] [Google Scholar]
- 20.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sá-Nogueira I, Nogueira T V, Soares S, Lencastre H. The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology. 1997;143:957–969. doi: 10.1099/00221287-143-3-957. [DOI] [PubMed] [Google Scholar]
- 23.Sá-Nogueira I, Ramos S S. Cloning, functional analysis, and transcriptional regulation of the Bacillus subtilis araE gene involved in l-arabinose utilization. J Bacteriol. 1997;179:7705–7711. doi: 10.1128/jb.179.24.7705-7711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strauch M A. AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. J Bacteriol. 1995;177:6727–6731. doi: 10.1128/jb.177.23.6727-6731.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stülke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 26.Turinsky A J, Grundy F J, Kim J-H, Chambliss G H, Henkin T M. Transcriptional activation of the Bacillus subtilis ackA gene requires sequences upstream of the promoter. J Bacteriol. 1998;180:5961–5967. doi: 10.1128/jb.180.22.5961-5967.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wray L V, Pettengill P K, Fisher S H. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription initiation site. J Bacteriol. 1994;176:1894–1902. doi: 10.1128/jb.176.7.1894-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida K-I, Aoyama D, Ishio I, Shibayama T, Fujita Y. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J Bacteriol. 1997;179:4591–4598. doi: 10.1128/jb.179.14.4591-4598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida K, Kobayashi K, Miwa Y, Kang C-M, Matsunaga M, Yamaguchi H, Tojo S, Yamamoto M, Nishi R, Ogasawara N, Nakayama T, Fujita Y. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 2001;29:683–692. doi: 10.1093/nar/29.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida K, Shibayama T, Aoyama D, Fujita Y. Interaction of a repressor and its binding sites for regulation of the Bacillus subtilis iol divergon. J Mol Biol. 1999;285:917–929. doi: 10.1006/jmbi.1998.2398. [DOI] [PubMed] [Google Scholar]