Abstract
The trp RNA-binding attenuation protein (TRAP) regulates expression of the Bacillus subtilis trpEDCFBA operon by transcription attenuation and translational control mechanisms. Both mechanisms require binding of tryptophan-activated TRAP to 11 (G/U)AG repeats in the trp leader transcript. trpE translational control involves formation of a TRAP-dependent RNA structure that sequesters the trpE Shine-Dalgarno (SD) sequence (the SD blocking hairpin). By comparing expression levels from trpE′-′lacZ translational fusions controlled by the wild-type leader or by a leader that cannot form the SD blocking hairpin, we found that translational control requires a tryptophan concentration higher than that required for transcription attenuation. We also found that inhibition of trpE translation by the SD blocking hairpin does not alter the stability of the downstream message. Since the coding sequences for trpE and trpD overlap by 29 nucleotides, we examined expression levels from trpED′-′lacZ translational fusions to determine if these two genes are translationally coupled. We found that introduction of a UAA stop codon in trpE resulted in a substantial reduction in expression. Since expression was partially restored in the presence of a tRNA suppressor, our results indicate that trpE and trpD are translationally coupled. We determined that the coupling mechanism is TRAP independent and that formation of the SD blocking hairpin regulates trpD translation via translational coupling. We also constructed a rho mutation to investigate the role of Rho-dependent termination in trp operon expression. We found that TRAP-dependent formation of the SD blocking hairpin allows Rho access to the nascent transcript, causing transcriptional polarity.
Organisms utilize a wide range of regulatory mechanisms to control gene expression. Bacteria have developed several sophisticated regulatory mechanisms that allow the organisms to modulate gene expression after transcription has initiated. In addition, several subtle mechanisms allow the organisms to fine tune the final level of any particular gene product. Expression of the Bacillus subtilis trpEDCFBA operon is regulated in response to changes in the intracellular concentration of tryptophan by the trp RNA-binding attenuation protein (TRAP). TRAP regulates expression of the trp operon by transcription attenuation and translational control mechanisms (reviewed in references 4 and 12). TRAP exists as a complex consisting of 11 identical subunits arranged in a single ring (3). Tryptophan cooperatively activates TRAP by binding between adjacent TRAP subunits (3, 6). When TRAP is activated by tryptophan, 11 KKR motifs that outline the periphery of the TRAP complex bind to 11 (G/U)AG repeats present in the nascent trp leader transcript, thereby wrapping the RNA around the periphery of the TRAP complex (2, 7, 33). In the transcription attenuation mechanism, TRAP binding prevents formation of an antiterminator structure, since six of the (G/U)AG repeats are present within this RNA structure (5, 7). In this case, TRAP binding promotes the formation of an overlapping intrinsic terminator, resulting in transcription termination before RNA polymerase reaches the structural genes. In the absence of TRAP binding, formation of the antiterminator permits transcription of the entire operon (5). A third stem-loop structure that forms at the extreme 5′ end of the trp leader transcript also plays a role in the transcription attenuation mechanism. TRAP-5′ stem-loop interaction increases the affinity of TRAP for trp leader RNA and reduces the number of (G/U)AG repeats that are required for stable TRAP-trp leader RNA association. Thus, TRAP-5′ stem-loop interaction may increase the likelihood that TRAP will bind to the (G/U)AG repeats in time to block antiterminator formation (10, 30).
In addition to regulating transcription of the trp operon, TRAP also regulates translation of trpE (11, 17, 20). RNA structural studies of the trp operon readthrough transcript indicated that the most thermodynamically stable conformation of the leader RNA contains a large secondary structure that includes the last six (G/U)AG repeats in the 5′ half of the stem (Fig. 1). TRAP binding to these repeats prevents or disrupts formation of this large secondary structure, thereby promoting the formation of a structure that sequesters the trpE Shine-Dalgarno (SD) sequence (the trpE SD blocking hairpin) (11, 17, 20). Formation of the trpE SD blocking hairpin inhibits TrpE synthesis by blocking ribosome access to the trpE ribosome binding site (11). In vivo and in vitro studies have established that multiple nucleotide substitutions that destabilize the trpE SD blocking hairpin, without altering the SD sequence itself, reduce the ability of TRAP to regulate TrpE synthesis (11, 20). In this study, we examined the effect of tryptophan concentration on transcription attenuation and translational control and found that translational regulation requires a tryptophan concentration higher than that required for transcription attenuation.
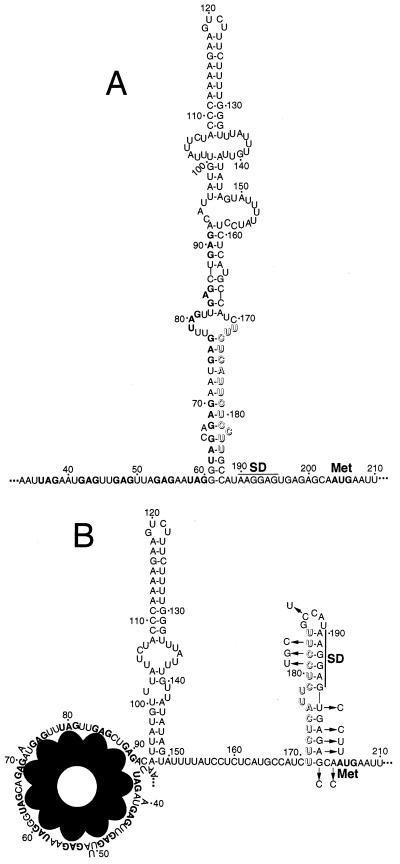
FIG. 1.
Model of the trpE translational control mechanism. (A) Under tryptophan-limiting conditions, TRAP is not activated and is unable to bind to the leader of trp operon readthrough transcripts. In this case the trpE SD sequence is single stranded, allowing efficient translation. (B) Under excess-tryptophan conditions, tryptophan-activated TRAP binds to the (G/U)AG repeats present between nucleotides 36 and 91 in the leader of a trp operon readthrough transcript. In this case, the secondary structure of the downstream RNA is altered such that the trpE SD sequence is sequestered in a stable RNA hairpin (trpE SD blocking hairpin). This RNA secondary structure inhibits TrpE synthesis by preventing 30S ribosomal subunit interaction with the trpE message (11). The nucleotides between 171 and 184 that are responsible for sequestering the trpE SD sequence are shown in outline type in both diagrams. In the absence of bound TRAP, these nucleotides basepair with a segment of the TRAP binding site. The (G/U)AG repeats and the trpE AUG initiation codon are indicated in boldface. The nucleotide substitutions in the SD trpL that prevent formation of the trpE SD blocking hairpin are indicated with arrows. The nucleotides that make up the mutually exclusive antiterminator and terminator structures extend from positions 60 to 111 or 108 to 133, respectively. Periods indicate that sequence information has been omitted. Numbering is from the start of transcription.
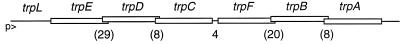
The sequence of the trpEDCFBA operon revealed that the coding sequences of all but two of the genes (trpC and trpF) overlap by several nucleotides (Fig. 2). This gene organization suggested that the trp genes might be regulated by translational coupling (14), a process in which translation of a gene is partially dependent on translation of the gene immediately upstream (19, 24). The first two genes of the trp operon, trpE and trpD, have the most extensive overlap in the operon (29 nucleotides). We found that these two genes are translationally coupled and that formation of the trpE SD blocking hairpin regulates TrpD synthesis via translational coupling. Since inhibition of translation can result in reduced mRNA stability (18) and/or transcriptional polarity (16), we performed experiments to determine if trpE translational control influences trp operon expression by reducing the stability of the message or by allowing Rho access to the nascent trp transcript. Our results establish that formation of the trpE SD blocking hairpin promotes Rho-mediated transcriptional polarity but does not influence the stability of the downstream mRNA.
FIG. 2.
Schematic representation of the B. subtilis trpEDCFBA operon highlighting the overlapping reading frames. With the exception of the 4-nucleotide intercistronic region between trpC and trpF, all of the open reading frames overlap by several residues. The length of each overlap (in nucleotides) is shown in parentheses. The positions of the trp promoter (p>) and leader (trpL) are shown (not drawn to scale).
MATERIALS AND METHODS
Plasmids and bacterial strains.
The genotypes of the B. subtilis strains used in this study are listed in Table 1. Plasmid pPB20 containing the wild-type (WT) B. subtilis trp promoter, the leader, and the first 40 trpE codons was previously described (5). pINT-SDtrpL contains several trp leader point mutations (SDtrpL) that destabilize the trpE SD blocking hairpin without disrupting the trpE SD sequence itself (20). Plasmid pHD15 contains the trp promoter, the leader, the entire trpE coding sequence, and the N-terminal coding sequence of trpD, while pHD22 contains a trpE′-′lacZ translational fusion that is controlled by the trp promoter and leader (11). The integration plasmids ptrpBGI (WT trpL trpE′-′lacZ) (29) and pHD24 (SD trpL trpE′-′lacZ) were previously described (11). The integration vector, ptrpBGI-PLK, used for generation of trpE′-′lacZ translational fusions, was described previously (20). The plasmid constructions used to generate trpED′-′lacZ translational fusions were made in Escherichia coli. Integrative plasmids containing various translational fusions were linearized with PstI and separately integrated into the amyE locus of the B. subtilis chromosome by homologous recombination. For example, to construct strains containing trpE-′lacZ translational fusions in a tryptophan auxotrophic strain, we integrated ptrpBGI or pHD24 into strain 168 (trpC2) to yield strain PLBS176 or PLBS201, respectively. Transformation was by natural competence (1). Selection was for chloramphenicol resistance (5 μg/ml). Integration into amyE was confirmed by screening for the absence of amylase activity on starch plates by using iodine staining (28). In each case, the correct plasmid construction was confirmed by automated DNA sequencing.
TABLE 1.
B. subtilis strains used in this study.
| Strain | Genotypea | Source |
|---|---|---|
| 168 | trpC2 | BGSCb |
| BG4233 | ΔmtrB argC4 | 15 |
| CB312 | his met | C. Stewartc |
| CB313 | sup-3 | C. Stewart |
| PLBS127 | rpoB18 amyE::[trpp(−412 to +203) trpE′-′lacZ Cmr] | 11 |
| PLBS129 | rpoB18 amyE::[trpp(−412 to +203) SDmodtrpE′-′lacZ Cmr] | 11 |
| PLBS138 | his met amyE::[trpp(−412 to +203) trpED′-′lacZ (trpE stop) Cmr] | This study |
| PLBS139 | sup-3 amyE::[trpp(−412 to +203) trpED′-′lacZ (trpE stop) Cmr] | This study |
| PLBS142 | his met amyE::[trpp(−412 to +203) trpED′-′lacZ Cmr] | This study |
| PLBS143 | sup-3 amyE::[trpp(−412 to +203) trpED′-′lacZ Cmr] | This study |
| PLBS144 | his met amyE::[trpp(−412 to +203) SDmodtrpED′-′lacZ (trpE stop) Cmr] | This study |
| PLBS145 | sup-3 amyE::[trpp(−412 to +203) SDmodtrpED′-′lacZ (trpE stop) Cmr] | This study |
| PLBS146 | his met amyE::[trpp(−412 to +203) SDmodtrpED′-′lacZ Cmr] | This study |
| PLBS147 | sup-3 amyE::[trpp(−412 to +203) SDmodtrpED′-′lacZ Cmr] | This study |
| PLBS176 | trpC2 amyE::[trpp(−412 to +203) trpE′-′lacZ Cmr] | This study |
| PLBS201 | trpC2 amyE::[trpp(−412 to +203) SDmodtrpE′-′lacZ Cmr] | This study |
| PLBS283 | ΔmtrB argC4 his met amyE::[trpp(−412 to +203) trpED′-′lacZ (trpE stop) Cmr] | This study |
| PLBS284 | ΔmtrB argC4 sup-3 amyE::[trpp(−412 to +203) trpED′-′lacZ (trpE stop) Cmr] | This study |
| PLBS285 | ΔmtrB argC4 his met amyE::[trpp(−412 to +203) trpED′-′lacZ Cmr] | This study |
| PLBS286 | ΔmtrB argC4 sup-3 amyE::[trpp(−412 to +203) trpED′-′lacZ Cmr] | This study |
| PLBS289 | rpoB18 rho::neo amyE::[trpp(−412 to +203) trpE′-′lacZ Cmr] | This study |
| PLBS291 | rho::neo his met amyE::[trpp(−412 to +203) trpED′-′lacZ (trpE stop) Cmr] | This study |
| PLBS292 | rho::neo sup-3 amyE::[trpp(−412 to +203) trpED′-′lacZ (trpE stop) Cmr] | This study |
| PLBS293 | rho::neo his met amyE::[trpp(−412 to +203) trpED′-′lacZ Cmr] | This study |
| PLBS294 | rho::neo sup-3 amyE::[trpp(−412 to +203) trpED′-′lacZ Cmr] | This study |
| PLBS321 | rho::neo his met | This study |
| PLBS322 | rho::neo trpC2 | This study |
| W168 | rpoB18 | BGSC |
trpp denotes the trp promoter; a prime indicates truncation of the gene; −412 to +203 is the DNA fragment containing trpp and neighboring regions that was incorporated, relative to the transcription start site; (trpE stop) indicates that the strain contains an engineered UAA stop codon in the trpE coding sequence. trpED′-′lacZ contains an in-frame deletion of trpE as well as a translational fusion between trpD and lacZ. sup-3 encodes a UAA tRNA suppressor.
BGSC, Bacillus Genetic Stock Center, Ohio State University, Columbus, Ohio.
These strains were obtained from Charles R. Stewart, Department of Biochemistry and Cell Biology, Rice University, Houston, Tex.
Construction of the trpED′-′lacZ translational fusions used in this study was done in several steps. First, a double-stranded DNA linker was ligated into the HindIII site of pPB20. The resulting plasmid, pYH2, contains four restriction sites (XbaI, BamHI, NheI, and HindIII) derived from the DNA linker. The DNA containing the overlapping trpED region in pHD15 was amplified by PCR. In one case, an in-frame lysine codon (AAA) was retained immediately downstream from an XbaI site. In the other case, we introduced a TAA termination codon in place of the natural lysine codon. The TAA- or AAA-containing PCR products were ligated into pYH2 to produce pYH3 or pYH7, respectively. DNA fragments containing the trp promoter, leader, and portions of trpE and trpD from pYH3 or pYH7 were subcloned into ptrpBGI-PLK to produce pYH5 or pYH10, respectively. Note that pYH10 contains the WT trp promoter and leader followed by an in-frame trpED′-′lacZ translational fusion in which the central region of trpE (codons 41 through 483) was deleted. pYH5 is identical to pYH10 except that the TAA stop codon replaces the lysine codon. Importantly, this stop codon would uncouple translation of trpD from translation of trpE. These translational fusions were separately integrated into the amyE locus of strain CB312 or CB313 as described above. Similar trpED′-′lacZ translational fusions were constructed in which the SD trpL region from pINT-SDtrpL replaced the WT trpL. The SD trpL fusions were separately integrated into strain CB312 or CB313. TRAP-deficient (ΔmtrB) strains were constructed by transforming various strains with chromosomal DNA from strain BG4233 (ΔmtrB) (15). Selection was for 5-fluorotryptophan resistance (200 μg/ml).
A null rho mutation was constructed by replacing nucleotides +155 to +165 relative to the rho initiation codon with the neo (Kmr) gene from plasmid pMK3 (31). The resulting plasmid, pYH14, was linearized with ScaI and subsequently used to replace the WT rho allele in several strains by transformation-mediated homologous recombination. Selection was for kanamycin resistance (10 μg/ml). Proper allelic replacement in each strain was confirmed by PCR amplification of chromosomal DNA.
β-Galactosidase assay.
B. subtilis cultures were grown in minimal-acid casein hydrolysate medium in the presence of 5 μg of chloramphenicol/ml. Tryptophan prototrophs were grown in the presence of 0 or 200 μM tryptophan. Tryptophan auxotrophs were grown in the presence of 20, 50, 100, 200, or 400 μM tryptophan. The cells were harvested during late exponential growth (110 Klett units; filter no. 54; Klett Manufacturing Co., Inc.). Aliquots were then assayed for β-galactosidase activity as previously described (11).
mRNA half-life and steady-state level determinations.
B. subtilis cultures were grown in minimal-acid casein hydrolysate medium in the presence of 20 or 200 μM tryptophan (trpC2 auxotrophic strains) or 0 or 200 μM tryptophan (trp prototrophic strains). The growth medium was supplemented with appropriate antibiotics (5 μg of chloramphenicol/ml or 10 μg of kanamycin/ml). When the cultures reached late exponential phase (110 Klett units; filter no. 54), 100 μg of rifampin/ml was added to inhibit transcription initiation. Six-milliliter aliquots were removed 0, 1, 2, 4, 8, and 16 min after the addition of rifampin and added to an equal volume of frozen killing buffer (8.5 mM Tris-HCl, pH 7.2, 5 mM MgCl2, 25 mM sodium azide, and 500 μg of chloramphenicol/ml). Total RNA was isolated using the RNeasy protocol (Qiagen), and genomic DNA was eliminated using RNase-free DNase I (Promega). RNA was extracted with an equal volume of phenol-chloroform, precipitated with ethanol, and suspended in Tris-EDTA. Quantification of mRNA was performed by slot blot hybridization. Ten micrograms of total RNA from each sample was mixed with an equal volume of denaturing solution (0.55 ml of formamide, 0.2 ml of 37% formaldehyde, 0.2 ml of morpholinepropanesulfonic acid, pH 7.0) and incubated for 15 min at 65°C. Samples were subsequently chilled on ice and spotted onto Hybond-N+ membranes (Amersham Pharmacia Biotech) using a Minifold I dot blot apparatus (Schleicher & Schuel). After filtration, RNA was covalently linked to the membrane using a UV Stratalinker (Strategene). Prehybridization was performed at 65°C for 1 h in a buffer containing 7% sodium dodecyl sulfate (SDS), 0.5 M sodium phosphate buffer, pH 7.2, and 10 mM EDTA (9). Hybridization was carried out at 65°C for 12 h by adding lacZ, trpE, or trpD DNA probes that were radiolabeled using the random-primer DNA-labeling kit (Boehringer Mannheim Biochemical). The filters were washed twice for 30 min each time with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS at 62°C followed by two 30-min washes with 0.1× SSC containing 0.1% SDS at the same temperature. mRNA levels were quantified with a PhosphorImager (Molecular Dynamics, Inc.) and the ImageQuant software package. Determination of the steady-state levels of trpE and trpD mRNAs was performed as described above except that only 0-min time points were analyzed and rifampin was omitted.
RESULTS
The trpE translational control mechanism requires a concentration of tryptophan higher than that required for transcription attenuation.
Expression of the trpEDCFBA operon is regulated by TRAP-mediated transcription attenuation and translational control mechanisms. In the transcription attenuation mechanism, only transient TRAP interaction with the (G/U)AG repeats would be necessary to block formation of the antiterminator. Once transcription has terminated, it would be advantageous for the TRAP-trp leader RNA complex to dissociate to prevent titration of TRAP. However, for translational control it appears that TRAP must remain associated with the triplet repeats to maintain formation of the trpE SD blocking hairpin because the trpE SD sequence is single stranded in the absence of bound TRAP (11). It was also shown that the affinity of TRAP for trp leader RNA increases with increasing tryptophan concentrations (25, 32) and that the dissociation rate of TRAP-trp leader RNA complexes decreases as the concentration of tryptophan increases (8). Because of these findings, we reasoned that the tryptophan concentration required for translational control would be higher than that required for transcription attenuation.
In a previous study, we compared expression levels from trpE′-′lacZ translational fusions containing the WT trpL (transcription attenuation and trpE translational control mechanisms functioning) or the SD trpL (only transcription attenuation mechanism functioning) (11). Although it was not appreciated at the time, our previously published results (11) suggested that TRAP-mediated translational control required an intracellular tryptophan concentration higher than that required for transcription attenuation. To further explore this possibility, we repeated our previously published expression studies. Since the effect of exogenous tryptophan on expression of the trp operon can be assessed from the −Trp/+Trp ratio, we examined β-galactosidase activity in prototrophic strains containing trpE′-′lacZ translational fusions controlled by either the WT trpL (PLBS127) or the SD trpL (PLBS129). The levels of expression in both strains were similar when the cells were grown in the absence of exogenous tryptophan (Table 2). However, when the cells were grown in the presence of 200 μM tryptophan, we observed 50-fold higher expression in the SD trpL strain (Table 2).
TABLE 2.
Effect of various tryptophan concentrations on expression of trpE′-′lacZ fusions
| Strain | Relevant genotype | β-Gal activitya
|
β-Gal ratioe
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 μMb | 20 μM | 50 μM | 100 μM | 200 μM | 400 μM | 0 μM/200 μM | 20 μM/400 μM | ||
| PLBS127 | WT trpL | 42 ± 11 | NDd | ND | ND | 0.1 ± 0.02 | ND | 420 | ND |
| PLBS129 | SD trpL | 39 ± 9 | ND | ND | ND | 5.3 ± 0.2 | ND | 7 | ND |
| PLBS176 | WT trpL trpC2 | NAc | 44 ± 12 | 7.4 ± 2.1 | 2.6 ± 0.2 | 0.8 ± 0.1 | 0.2 ± 0.06 | NA | 220 |
| PLBS201 | SD trpL trpC2 | NA | 43 ± 11 | 12 ± 1.0 | 8.7 ± 1.0 | 5.0 ± 0.4 | 4.7 ± 0.2 | NA | 9 |
The β-galactosidase (β-Gal) activity expressed from the trpE′-′lacZ fusion is given in Miller units (21). The values are averages from at least three independent experiments ± standard deviation.
Concentration of tryptophan in the growth media.
NA, not applicable. Tryptophan auxotrophic strains require tryptophan for growth.
ND, not determined.
Ratio of activities at the indicated concentrations.
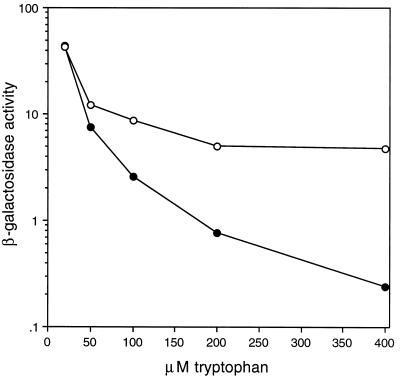
To allow more precise control of the intracellular tryptophan concentrations, we integrated the translational fusions into a trpC2 auxotrophic strain. Expression was examined over a wide range of tryptophan concentrations in the growth media. Since the β-galactosidase activity of the auxotrophic strains grown in the presence of 20 μM tryptophan was similar to the activity of prototrophic strains grown in the absence of tryptophan (Table 2), we used 20 μM as the lower limit for subsequent expression studies. As the tryptophan concentration was gradually increased from 20 to 400 μM, expression from the WT trpL strain (PLBS176) continued to decrease (Fig. 3 and Table 2). However, the decrease in expression in the SD trpL strain (PLBS201) was much more gradual and reached a lower limit at 200 μM tryptophan. Although levels of expression were similar in the two strains when grown in the presence of 20 or 50 μM tryptophan, the difference in expression reached a factor of 3, 6, or 24 when the tryptophan concentration was increased to 100, 200, or 400 μM, respectively. Since these two strains have only the transcription attenuation mechanism in common, the difference in expression at the higher tryptophan concentrations is due to the functional translational control mechanism in the WT trpL strain. These results suggest that the transcription attenuation mechanism is primarily responsible for controlling expression at low tryptophan concentrations and that the translational control mechanism does not play a significant role until the concentration of tryptophan in the growth media is raised above 50 μM.
FIG. 3.
β-Galactosidase activity from trpC2 auxotrophic strains as a function of tryptophan concentration in the growth media. trpE′-′lacZ translational fusions were under control of the WT trpL (solid circles) or SD trpL (open circles). Both the transcription attenuation and trpE translational control mechanisms function in the WT trpL strain (PLBS176), whereas only the transcription attenuation mechanism functions in the SD trpL strain (PLBS201). β-Galactosidase activity is shown in Miller units (21). This figure is derived from the data presented in Table 2.
Inhibition of trpE translation mediated by the SD blocking hairpin does not alter stability of downstream mRNA.
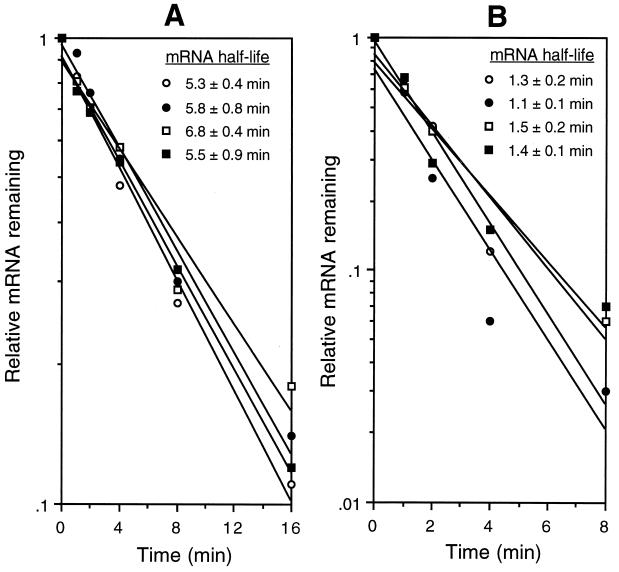
Inhibition of translation has been shown to increase the rate of mRNA decay in some cases (reviewed in reference 18). This presumably results from decreased ribosomal protection of the downstream message from ribonucleases. We tested the possibility that formation of the trpE SD blocking hairpin destabilized the downstream mRNA. We determined the lacZ mRNA half-life in strains containing trpE′-′lacZ fusions controlled by either the WT trpL (PLBS176) or the SD trpL (PLBS201). Since formation of the trpE SD blocking hairpin is dependent on tryptophan-activated TRAP, we expected that the mRNA half-life would be reduced in the WT trpL strain when it was grown in the presence of tryptophan. We were surprised to find that the lacZ mRNA half-lives were essentially identical in both strains when grown under limiting or excess tryptophan conditions (Fig. 4A). To ensure that the lacZ mRNA half-life data accurately reflected trp mRNA stability, we determined the half-life of trpE and trpD mRNAs in the presence of limiting or excess tryptophan. While the decay of trpE and trpD mRNAs was considerably more rapid than that of the lacZ message, formation of the trpE SD blocking hairpin did not influence the stability of the transcripts (Fig. 4B). Thus, trpE translational control does not lead to destabilization of trp mRNA.
FIG. 4.
Time course of lacZ, trpE, and trpD mRNA decay. (A) lacZ mRNA half-life experiments in strains containing trpE′-′lacZ translational fusions that were controlled by either the WT trpL (PLBS176) or the SD trpL (PLBS201) when each strain was grown in the presence of 20 or 200 μM tryptophan. Open circles, WT trpL with 20 μM tryptophan; solid circles, WT trpL with 200 μM tryptophan; open squares, SD trpL with 20 μM tryptophan; solid squares, SD trpL with 200 μM tryptophan. (B) trpE and trpD mRNA half-life experiments from the natural trp operon when strain 168 was grown in the presence of 20 or 200 μM tryptophan. Open circles, trpE with 20 μM tryptophan; solid circles, trpE with 200 μM tryptophan; open squares, trpD with 20 μM tryptophan; solid squares, trpD with 200 μM tryptophan. The relative levels of mRNA remaining at 0, 1, 2, 4, 8, and 16 min after the addition of rifampin were determined by dot blot analysis using the corresponding probe. Each value is an average from at least two independent experiments. The mRNA level corresponding to each 0-min time point was set to 1. The mRNA half-life ± the standard deviation for each strain and growth condition is shown next to the corresponding symbol.
trpE and trpD are translationally coupled.
The coding regions of trpE and trpD overlap by 29 nucleotides (Fig. 2). To determine if this overlap allows translational coupling of these two genes, we constructed two trpED′-′lacZ translational fusions in which the central region of trpE was deleted (Fig. 5). In one of the fusions, we introduced a UAA termination codon in trpE. This stop codon would disrupt translational coupling unless it was translated by a tRNA suppressor. The other fusion contained the natural lysine codon (AAA) instead of the engineered stop codon. In both cases, the reading frame of the internal trpE deletion was maintained (Fig. 5). The translational fusions were integrated into the amyE locus of WT or sup-3 B. subtilis strains. It was previously shown that the sup-3 allele encodes a tRNA suppressor that can place a lysine residue at UAA stop codons approximately 15% of the time (22). Thus, placement of a lysine residue at this position by the tRNA suppressor would result in a trpE polypeptide that is identical in sequence to that encoded by the other fusion. We examined expression from the trpED′-′lacZ fusion and found that the levels of regulation in response to tryptophan for all four strains were comparable (compare the −Trp/+Trp β-galactosidase ratios for the first four strains in Table 3). Introduction of the engineered UAA stop codon reduced expression of the fusion sevenfold in the absence of tryptophan and fourfold in its presence (compare expression levels from PLBS142 and PLBS138 [Table 3]). Importantly, when expression was examined in the sup-3 strain, we found that expression was partially restored in the strain that contained the engineered stop codon; expression increased threefold in the absence of tryptophan and twofold in its presence (compare expression levels from PLBS138 and PLBS139 [Table 3]). The sup-3 allele had little effect on expression when the engineered stop codon was not present (compare expression levels from PLBS142 and PLBS143 [Table 3]). The finding that suppression of the UAA stop codon within the trpE coding sequence partially restored expression of the trpED′-′lacZ translational fusion demonstrates that trpE and trpD are translationally coupled.
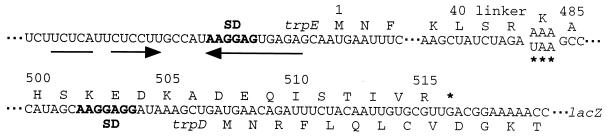
FIG. 5.
Nucleotide and amino acid sequences of the trpED′-′lacZ translational fusion transcripts used in the translational coupling studies. Only the crucial features of the trpE and trpD coding regions are shown. The trpE and trpD amino acid sequences are shown above and below the nucleotide sequence, respectively. The trpE and trpD coding sequences overlap by 29 nucleotides. Two fusions were constructed that contained either an AAA lysine codon or an engineered UAA stop codon (∗∗∗). The tRNA suppressor encoded by the sup-3 allele can place a lysine residue at the UAA stop codon with approximately 15% efficiency (22). The position of the natural trpE UGA termination codon is shown (∗). The trpE codons 41 to 483 were replaced by a short linker. Thus, there are 31 codons between the engineered stop codon and the natural trpE stop codon. The trpE and trpD SD sequences are shown in boldface. The inverted arrows indicate the nucleotides involved in trpE SD blocking hairpin formation. Periods indicate that sequence information has been omitted.
TABLE 3.
Translational coupling of trpE and trpD
| Strain | Relevant genotypea | UAA stop codon in trpEb | β-Gal activityc
|
β-Gal ratio (−Trp/+Trp) | |
|---|---|---|---|---|---|
| −Trp | +Trp | ||||
| PLBS142 | WT trpL | No | 194 ± 25 | 2.3 ± 0.4 | 84 |
| PLBS138 | WT trpL | Yes | 27 ± 4 | 0.6 ± 0.1 | 45 |
| PLBS143 | WT trpL sup-3 | No | 198 ± 28 | 3.2 ± 0.3 | 62 |
| PLBS139 | WT trpL sup-3 | Yes | 79 ± 7 | 1.1 ± 0.1 | 72 |
| PLBS285 | WT trpL ΔmtrB | No | 1,570 ± 90 | 1,660 ± 90 | 1 |
| PLBS283 | WT trpL ΔmtrB | Yes | 230 ± 20 | 240 ± 20 | 1 |
| PLBS286 | WT trpL ΔmtrB sup-3 | No | 1,570 ± 130 | 1,510 ± 60 | 1 |
| PLBS284 | WT trpL ΔmtrB sup-3 | Yes | 460 ± 40 | 460 ± 30 | 1 |
| PLBS146 | SD trpL | No | 169 ± 15 | 18 ± 2 | 9 |
| PLBS144 | SD trpL | Yes | 18 ± 3 | 2.2 ± 0.2 | 8 |
| PLBS147 | SD trpL sup-3 | No | 223 ± 25 | 23 ± 4 | 10 |
| PLBS145 | SD trpL sup-3 | Yes | 50 ± 4 | 4.2 ± 0.3 | 12 |
WT trp leader (WT trpL) or SD trp leader (SD trpL). UAA stop codons are recognized by the tRNA suppressor encoded by the sup-3 allele.
Presence (Yes) or absence (No) of the engineered stop codon within the coding sequence of trpE.
β-Galactosidase (β-Gal) activity expressed from the trpED′-′lacZ fusions is given in Miller units (21). Cells were grown in the absence (−Trp) or presence (+Trp) of tryptophan (200 μM). The values are averages from six independent experiments ± standard deviation.
We also examined expression from TRAP-deficient (ΔmtrB) strains that were otherwise identical to those just described. This allowed us to examine the extent of translational coupling in the absence of TRAP-mediated control of the trp operon. As expected, expression was not regulated in response to tryptophan (compare −Trp/+Trp β-galactosidase ratios for the strains corresponding to lines 5 through 8 in Table 3). However, the same general pattern of expression indicative of translational coupling was retained. We observed a sevenfold reduction in expression from the trpED′-′lacZ fusion when the stop codon was introduced into the trpE coding sequence (compare expression levels from PLBS285 and PLBS283 [Table 3]), while expression was restored twofold when the tRNA suppressor (sup-3) was present (compare expression levels from PLBS283 and PLBS284 [Table 3]). As was observed for the mtrB+ strains, the sup-3 allele had little affect on expression when the engineered stop codon was not present (compare expression levels from PLBS285 and PLBS286 [Table 3]).
The trpE SD blocking hairpin regulates translation of trpD via translational coupling.
Since trpE and trpD are translationally coupled, it was possible that formation of the trpE SD blocking hairpin would also regulate translation of trpD. To test this possibility, we compared expression levels from trpED′-′lacZ fusions containing WT trpL or SD trpL trp leaders. We found that the −Trp/+Trp ratios were similar in all four SD trpL strains and that the SD trpL reduced regulation of the trpED′-′lacZ fusion in response to tryptophan (compare the −Trp/+Trp β-galactosidase ratios for the last four strains in Table 3 with those of the first four strains). This reduction in regulation was primarily due to elevated expression of the SD trpL-containing strains when they were grown in the presence of tryptophan. As expected, we found that the translational coupling expression patterns were retained in the SD trpL strains. In this case, introduction of the engineered stop codon reduced expression eight- to ninefold (compare expression levels from PLBS146 and PLBS144 [Table 3]), while expression was restored two- to threefold when the sup-3 allele was present (compare expression levels from PLBS144 and PLBS145 [Table 3]). Again, the tRNA suppressor (sup-3) had little effect on expression when the UAA stop codon was not present (compare expression levels from PLBS146 and PLBS147 [Table 3]). Taken together, these results demonstrate that the trpE SD blocking hairpin regulates TrpD synthesis via translational coupling of trpE and trpD.
Rho termination factor influences expression of the trp operon.
Polarity in operons can be caused by Rho-dependent termination within transcripts that are not being translated (reference 16 and references therein). Thus, inhibition of trpE translation caused by the SD blocking hairpin could result in Rho-dependent transcriptional polarity. Since we determined that trpE translational control only occurred at relatively high concentrations of tryptophan (see above), we expected Rho to have its greatest effect when cells were grown in the presence of excess tryptophan. To test this possibility, we engineered a Rho-deficient strain by disrupting rho with a kanamycin resistance gene. As was previously reported (16, 26), Rho is not essential for the viability of B. subtilis.
We first compared expression from a trpE′-′lacZ fusion in WT (PLBS127) and rho mutant (PLBS289) strains when the cells were grown in the presence or absence of tryptophan. While expression was only slightly higher in the rho mutant when grown in the absence of tryptophan, we observed threefold-higher expression when the cells were grown in the presence of excess tryptophan (Table 4). We then examined the effect of rho on expression from the trpED′-′lacZ fusion. The rho mutation caused only a slight increase in expression when each strain was grown in the absence of tryptophan (compare expression levels from PLBS142 with those from PLBS293, those from PLBS138 with those from PLBS291, those from PLBS143 with those from PLBS294, and those from PLBS139 with those from PLBS292 [Table 4]). Note that translational coupling was retained in the rho strains when they were grown in the absence of tryptophan. The expression pattern was dramatically different when the same strains were grown in the presence of excess tryptophan. In this case, the expression levels of the four rho mutant strains were similar and did not exhibit the translational coupling pattern. Moreover, expression in all four strains was 7- to 24-fold higher than that observed for their respective isogenic WT strains (Table 4).
TABLE 4.
Effect of Rho termination factor on trp operon expression
| Strain | Relevant genotypea | Translational fusion | UAA stop codon in trpEb | β-Gal activityc
|
β-Gal ratio (−Trp/+Trp) | |
|---|---|---|---|---|---|---|
| −Trp | +Trp | |||||
| PLBS127 | WT trpL | trpE′-′lacZ | NAd | 26 ± 5 | 0.2 ± 0.03 | 130 |
| PLBS289 | WT trpL rho::neo | trpE′-′lacZ | NA | 39 ± 7 | 0.6 ± 0.03 | 65 |
| PLBS142 | WT trpL | trpED′-′lacZ | No | 190 ± 20 | 2.1 ± 0.5 | 90 |
| PLBS138 | WT trpL | trpED′-′lacZ | Yes | 24 ± 6 | 0.5 ± 0.1 | 48 |
| PLBS143 | WT trpL sup-3 | trpED′-′lacZ | No | 240 ± 50 | 2.9 ± 0.5 | 83 |
| PLBS139 | WT trpL sup-3 | trpED′-′lacZ | Yes | 80 ± 8 | 1.0 ± 0.2 | 80 |
| PLBS293 | WT trpL rho::neo | trpED′-′lacZ | No | 210 ± 14 | 14 ± 1 | 15 |
| PLBS291 | WT trpL rho::neo | trpED′-′lacZ | Yes | 27 ± 3 | 12 ± 2 | 2 |
| PLBS294 | WT trpL rho::neo sup-3 | trpED′-′lacZ | No | 316 ± 20 | 21 ± 2 | 15 |
| PLBS292 | WT trpL rho::neo sup-3 | trpED′-′lacZ | Yes | 114 ± 10 | 14 ± 2 | 8 |
WT trp leader (WT trpL). UAA stop codons are recognized by the tRNA suppressor encoded by the sup-3 allele. rho is disrupted by a neo insertion.
Presence (Yes) or absence (No) of the engineered stop codon within the coding sequence of trpE.
β-Galactosidase (β-Gal) activity expressed from the trpE′-′lacZ and the trpED′-′lacZ fusions is given in Miller units (21). Cells were grown in the absence (−Trp) or presence (+Trp) of tryptophan (200 μM). The values are averages from at least four independent experiments ± standard deviation.
NA, not applicable.
To ensure that expression from the translational fusions was indicative of the effect that Rho had on expression from the natural trp operon, we examined trpE and trpD transcript levels in WT and rho strains. We found that both trpE and trpD transcript levels were substantially higher in rho mutants. This was particularly evident when each strain was grown in the presence of excess tryptophan (Table 5). The finding that rho null strains had elevated trp transcript levels is consistent with the translational fusion studies (Table 4). Taken together, our results establish that Rho causes transcriptional polarity of the trp operon under conditions that promote translational control (tryptophan excess).
TABLE 5.
Effect of Rho on trpE and trpD transcript levels
| Strain | Relevant genotype | Tryptophan (μM) | mRNA level (%)a
|
|
|---|---|---|---|---|
| trpE | trpD | |||
| 168 | trpC2 | 20 | 51 ± 6 | 53 ± 6 |
| 168 | trpC2 | 200 | 5.0 ± 0.3 | 4.9 ± 0.4 |
| PLBS322 | rho::neo trpC2 | 20 | 100 | 100 |
| PLBS322 | rho::neo trpC2 | 200 | 43 ± 2 | 37 ± 7 |
| CB312 | WT | 0 | 14 ± 1 | 19 ± 5 |
| CB312 | WT | 200 | 5.0 ± 1.0 | 3.4 ± 0.6 |
| PLBS321 | rho::neo | 0 | 100 | 100 |
| PLBS321 | rho::neo | 200 | 48 ± 1 | 44 ± 4 |
The highest level of both trpE and trpD mRNAs was present in rho mutant strains grown under tryptophan-limiting conditions and was set at 100%. The values are averages of two independent experiments ± standard deviation.
DISCUSSION
Expression of the tryptophan biosynthetic operon (trpEDCFBA) is regulated by TRAP-mediated transcription attenuation and translational control mechanisms. In the transcription attenuation mechanism, only transient TRAP interaction with the (G/U)AG repeats would be necessary to block formation of the antiterminator. However, for translational control it is probably necessary for TRAP to remain associated with the triplet repeats to maintain formation of the trpE SD blocking hairpin. Although the intracellular concentration of free tryptophan was not determined in this study, the results of our expression experiments indicate that the translational control mechanism requires a tryptophan concentration higher than that required for attenuation (Fig. 3 and Table 2). It was recently shown that yhaG encodes a putative transmembrane protein involved in tryptophan transport and that TRAP regulates translation of this gene (27). Since B. subtilis is a soil bacterium, it is reasonable to assume that the organism would encounter environments in which the amounts of available tryptophan differ. Thus, the ability of B. subtilis to utilize different regulatory mechanisms to modulate trp operon expression in response to changes in the environmental supply of tryptophan may provide a growth advantage.
It is generally assumed that inhibition of translation can lead to a decrease in mRNA half-life by increasing the susceptibility of the message to nucleolytic attack. While this has been substantiated in several instances, there are examples where it has been shown that inhibition of translation did not alter the mRNA half-life or even had a stabilizing affect on the message (18). In this study, we found that inhibition of translation by the trpE SD blocking hairpin did not alter the stability of the downstream mRNA (Fig. 4).
The importance of translational coupling in maintaining stoichiometric production of biosynthetic pathway enzymes and ribosomal proteins is well established in E. coli (19, 24). In the majority of cases, the translation termination codon of the upstream cistron overlaps the initiation codon of the downstream cistron. In the case of the B. subtilis trpEDCFBA operon, the lengths of the overlaps are far more extensive (8 to 29 nucleotides) (Fig. 2). Our expression studies establish that trpE and trpD constitute a translationally coupled gene pair (Table 3). In addition, since we still observed substantial translation of trpD when we uncoupled translation by introducing the stop codon in trpE, our results indicate that TrpD synthesis is not dependent on translational coupling. Moreover, our coupling studies demonstrated that the trpE translational control mechanism, relying on TRAP-dependent formation of the trpE SD blocking hairpin, regulates TrpD synthesis via translational coupling (Table 3). However, since we observed the same general pattern of translational coupling in TRAP-deficient (ΔmtrB) strains, our results indicate that the coupling mechanism itself is independent of TRAP (Table 3).
Translational coupling is generally thought to occur by one of two mechanisms (19, 24). In one case, the translating ribosome disrupts an RNA hairpin that can sequester the SD sequence of the downstream cistron. As a consequence, a second ribosome can gain access to the now single-stranded SD sequence and initiate translation. Since our computer predictions have not detected any evidence for a trpD SD blocking hairpin, we do not believe that this mechanism is involved in our case. In other instances, it is thought that the same ribosome that translates the upstream cistron can initiate translation of the downstream cistron because the respective stop and start codons overlap or are close to one another. While it is possible that coupling of trpE and trpD relies on this type of mechanism, it is difficult to envision how such an extensive overlap would be accommodated.
With the exception of trpC and trpF, all of the genes in the B. subtilis trp operon overlap by several nucleotides (Fig. 2). In the case of these two genes, the intercistronic region is only 4 nucleotides. Since it is known that genes with short intercistronic gaps can be coupled (19), it is possible that the entire B. subtilis trpEDCFBA operon is translationally coupled. Thus, it is conceivable that formation of the trpE SD blocking hairpin regulates expression of the entire operon via translational coupling. This possibility remains to be tested. Since overlapping genes are particularly common in B. subtilis, it is likely that translational coupling plays a significant role in the coordinate regulation of several genes in this organism (12).
Transcriptional polarity can occur when Rho is allowed access to a nascent transcript when translation is inhibited. Our results establish that formation of the trpE SD blocking hairpin leads to transcriptional polarity of the trp operon (Tables 4 and 5). Thus, it appears that Rho binds downstream from this RNA structure. It was previously shown that rho is a nonessential gene in B. subtilis (16, 26), in contrast to E. coli, Rhodobacter sphaeroides (13), and Micrococcus luteus (23), in which it is an essential gene. Interestingly, it was determined that B. subtilis Rho regulates expression of its own gene (16). To the best of our knowledge, this is the only other known example of Rho-dependent regulation in B. subtilis.
When cells are growing under conditions of tryptophan excess, TRAP would be activated and most likely bind to the message as it is being synthesized. In most cases this would promote Rho-independent (intrinsic) termination in the trp leader (transcription attenuation). However, in some instances, RNA polymerase will escape termination despite TRAP binding, since transcription termination is never 100% efficient. In other instances, TRAP might bind prior to transcription of the trpE SD sequence but not in time to promote termination. Both of these scenarios would result in a nascent TRAP-bound readthrough transcript that would contain the trpE SD sequence in the SD blocking hairpin. Rho could then bind to the nascent transcript and cause transcriptional polarity by promoting premature transcript release from RNA polymerase.
ACKNOWLEDGMENTS
We thank Charles Stewart for providing us with bacterial strains and Charles Yanofsky for critical reading of the manuscript. We also thank Carol Baker and Igor Morozov for advice during the course of these studies.
This work was supported by grant GM52840 from the National Institutes of Health.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antson A A, Dodson E J, Dodson G, Greaves R B, Chen X-P, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 3.Antson A A, Otridge J, Brzozowski A M, Dodson E J, Dodson G G, Wilson K S, Smith T M, Yang M, Kurecki T, Gollnick P. The structure of trp RNA-binding attenuation protein. Nature. 1995;374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 4.Babitzke P. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol. 1997;26:1–9. doi: 10.1046/j.1365-2958.1997.5541915.x. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke P, Yanofsky C. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci USA. 1993;90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babitzke P, Yanofsky C. Structural features of L-tryptophan required for activation of TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 1995;270:12452–12456. doi: 10.1074/jbc.270.21.12452. [DOI] [PubMed] [Google Scholar]
- 7.Babitzke P, Stults J T, Shire S J, Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem. 1994;269:16597–16604. [PubMed] [Google Scholar]
- 8.Baumann C, Otridge J, Gollnick P. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J Biol Chem. 1996;271:12269–12274. doi: 10.1074/jbc.271.21.12269. [DOI] [PubMed] [Google Scholar]
- 9.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du H, Yakhnin A V, Dharmaraj S, Babitzke P. trp RNA-binding attenuation protein (TRAP)-5′ stem-loop RNA interaction is required for proper transcription attenuation control of the Bacillus subtilis trpEDCFBA operon. J Bacteriol. 2000;182:1819–1827. doi: 10.1128/jb.182.7.1819-1827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du H, Babitzke P. trp RNA-binding attenuation protein-mediated long-distance RNA refolding regulates translation of trpE in Bacillus subtilis. J Biol Chem. 1998;273:20494–20503. doi: 10.1074/jbc.273.32.20494. [DOI] [PubMed] [Google Scholar]
- 12.Gollnick, P., P. Babitzke, E. Merino, and C. Yanofsky. Aromatic amino acid metabolism in Bacillus subtilis. In A. Sonenshein et al. (ed.), Bacillus subtilis and its close relatives: from genes to cells, in press. American Society for Microbiology, Washington, D.C.
- 13.Gomelsky M, Kaplan S. The R. sphaeroides 2.4.1. rho gene: expression and genetic analysis of structure and function. J Bacteriol. 1996;178:1946–1954. doi: 10.1128/jb.178.7.1946-1954.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henner D J, Band L, Shimotsu H. Nucleotide sequence of the Bacillus subtilis tryptophan operon. Gene. 1984;34:169–177. doi: 10.1016/0378-1119(85)90125-8. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman R J, Gollnick P. The mtrB gene of Bacillus pumilus encodes a protein with sequence and functional homology to the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis. J Bacteriol. 1995;177:839–842. doi: 10.1128/jb.177.3.839-842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingham C J, Dennis J, Furneaux P A. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol Microbiol. 1999;31:651–663. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- 17.Kuroda M I, Henner D, Yanofsky C. cis-acting sites in the transcript of the Bacillus subtilis trp operon regulate expression of the operon. J Bacteriol. 1988;170:3080–3088. doi: 10.1128/jb.170.7.3080-3088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kushner S R. mRNA decay. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 849–860. [Google Scholar]
- 19.McCarthy J E G, Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990;6:78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- 20.Merino E, Babitzke P, Yanofsky C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J Bacteriol. 1995;177:6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 22.Mulbry W W, Ambulos N P, Jr, Lovett P S. Bacillus subtilis mutant allele sup-3 causes lysine insertion at ochre codons: use of sup-3 in studies of translational attenuation. J Bacteriol. 1989;171:5322–5324. doi: 10.1128/jb.171.10.5322-5324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nowatzke W L, Keller E, Koch G, Richardson J P. Transcription termination factor Rho is essential for Micrococcus luteus. J Bacteriol. 1997;179:5238–5240. doi: 10.1128/jb.179.16.5238-5240.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oppenheim D S, Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95:785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otridge J, Gollnick P. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan dependent manner. Proc Natl Acad Sci USA. 1993;90:128–132. doi: 10.1073/pnas.90.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quirk P G, Dunkley E A, Jr, Lee P, Krulwich T A. Identification of a putative Bacillus subtilis rho gene. J Bacteriol. 1993;175:647–654. doi: 10.1128/jb.175.3.647-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarsero J P, Merino E, Yanofsky C. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J Bacteriol. 2000;182:2329–2331. doi: 10.1128/jb.182.8.2329-2331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekiguchi J, Takada N, Okada H. Genes affecting the productivity of α-amylase in Bacillus subtilis Marburg. J Bacteriol. 1975;121:688–694. doi: 10.1128/jb.121.2.688-694.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimotsu H, Henner D J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43:85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- 30.Sudershana S, Du H, Mahalanabis M, Babitzke P. A 5′ RNA stem-loop participates in the transcription attenuation mechanism that controls expression of the Bacillus subtilis trpEDCFBA operon. J Bacteriol. 1999;181:5742–5749. doi: 10.1128/jb.181.18.5742-5749.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan M A, Yasbin R E, Young F E. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 32.Yakhnin A V, Trimble J J, Chiaro C R, Babitzke P. Effects of mutations in the L-tryptophan binding pocket of the trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 2000;275:4519–4524. doi: 10.1074/jbc.275.6.4519. [DOI] [PubMed] [Google Scholar]
- 33.Yang M, Chen X-P, Militello K, Hoffman R, Fernandez B, Baumann C, Gollnick P. Alanine-scanning mutagenesis of Bacillus subtilis trp RNA-binding attenuation protein (TRAP) reveals residues involved in tryptophan binding and RNA binding. J Mol Biol. 1997;270:696–710. doi: 10.1006/jmbi.1997.1149. [DOI] [PubMed] [Google Scholar]