Abstract
Breast cancer (BC) is a multifactorial disease caused by an interaction between genetic predisposition and environmental exposures. MicroRNAs are a group of small non-coding RNA molecules, which seem to have a role either as tumor suppressor genes or oncogenes and seem to be related to cancer risk factors. We conducted a systematic review and meta-analysis to identify circulating microRNAs related to BC diagnosis, paying special attention to methodological problems in this research field. A meta-analysis was performed for microRNAs analyzed in at least three independent studies where sufficient data to make analysis were presented. Seventy-five studies were included in the systematic review. A meta-analysis was performed for microRNAs analyzed in at least three independent studies where sufficient data to make analysis were presented. Seven studies were included in the MIR21 and MIR155 meta-analysis, while four studies were included in the MIR10b metanalysis. The pooled sensitivity and specificity of MIR21 for BC diagnosis were 0.86 (95%CI 0.76–0.93) and 0.84 (95%CI 0.71–0.92), 0.83 (95%CI 0.72–0.91) and 0.90 (95%CI 0.69–0.97) for MIR155, and 0.56 (95%CI 0.32–0.71) and 0.95 (95%CI 0.88–0.98) for MIR10b, respectively. Several other microRNAs were found to be dysregulated, distinguishing BC patients from healthy controls. However, there was little consistency between included studies, making it difficult to identify specific microRNAs useful for diagnosis.
Keywords: breast cancer, microRNA, miRNA, serum, plasma, blood
1. Introduction
Breast cancer (BC) is the most frequently diagnosed cancer in Europe, accounting for 13% of all new cancer cases [1].
BC is a multifactorial disease caused by the interaction between genetic predisposition and environmental exposures [2]. The environmental exposures include several modifiable risk factors such as overweight or obesity (post-menopausal), use of menopausal hormone therapy, a low level of physical activity, consumption of alcohol, cigarette smoking, shift work, and some reproductive factors [2]. A genetic predisposition or family history account for about 10%, with some geographical variations [2]. The most common are germline mutations, such as BRCA1, BRCA2, PALB2, ATM, and TP53 genes, among others [3,4].
MicroRNA are a group of short noncoding regulatory RNAs that modulate gene expression at the post transcriptional level [5]. The dysregulation of microRNAs is linked to many human diseases, including cancer. Cell-free circulating microRNAs probably released from cells in lipid vescicles, microvescicles, or exosomes have been detected in peripheral blood circulation [6].
Due to the stability and resistance to the endogenous RNase activity, microRNAs have been investigated as diagnostic biomarkers of BC. Accessing circulating BC biomarkers from peripheral blood (through the so-called liquid biopsy) is a promising non-invasive and cost-effective procedure [7]. In fact, dysregulated microRNAs have both oncogenic and tumour-suppressing actions, depending on their targets [7]. This is a complex matter because some dysregulations of microRNAs seem to be common in most cancers, possibly due to their role in cancer-associated biological processes and not in aetiology targets [7].
Circulating microRNA may reflect the response of the organism to environmental exposures, as well as early signs of disease.
This review aims to report the potential use of altered circulating microRNA levels in the diagnosing of BC, paying special attention to methodological problems in this research field.
2. Materials and Methods
The protocol of this review was registered in the international database of prospective registered systematic reviews (PROSPERO 2022; CRD42022354439). The workflow and methodology were based on the guidelines of Preferred Reporting Items for Systematic Reviews and Meta-Analyses of Diagnostic Test Accuracy (PRISMA-DTA) [8].
2.1. Publication Search
We conducted a comprehensive literature search in PubMed, Cochrane Library, EMBASE, Google Scholar, and NCBI PubMed Central until 31 August 2022 to identify relevant studies. The article search was performed using the following search strategy:
((Circulating) AND (microRNA OR miRNA) AND (breast AND Cancer)) NOT (cells) NOT (tissue) AND ((English[Filter]) AND (Humans[Filter]) AND (“31 August 2022”[Date—Publication]))
Furthermore, other relevant studies were identified by manually searching for references of eligible publications.
2.2. Inclusion and Exclusion Criteria
Studies were considered eligible for the systematic review if they met the following criteria: (1) The study includes patients with BC and healthy controls; (2) The levels of one or more microRNAs were measured in blood, serum, or plasma; (3) They presented sufficient data to collect the number of patients and a measure of diagnostic performance (e.g., sensibility and sensitivity, or fold change) or a measure of association (e.g., Odds Ratio or Relative Risk).
Studies were included in the meta-analysis if there were at least three studies focused on the same microRNA, they met criteria (1), (2), and (3), and the frequencies of true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN) could be directly or indirectly extracted.
Studies were excluded if they were reviews, meta-analysis, letters, commentaries, or abstracts presented in conferences; lacking sufficient data; duplication of previous publications; or languages other than English.
2.3. Data Extraction
After the selection of studies was made, other relevant studies were searched from the references in the articles. According to inclusion criteria, data were extracted by two independent authors (LP and CS). Disagreements were solved through face-to-face discussion and consensus. Extracted data form included: first author’s name and reference, country, sample size, biological sample (plasma, serum, or blood), microRNA, cut-off value, AUC value (95% CI), sensitivity (95% CI), specificity (95% CI), fold change (95% CI), p-value, microRNA source (candidate or discovery if found in a screening phase), and expression (upregulation or downregulation). Diagnostic performance data were extracted or calculated for the studies included in the meta-analysis (FP, FN, TP, TN).
2.4. Quality Assessment
We estimated the quality of each study using the revised Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) by two independent authors (LP and CS) [9].
2.5. Statistical Analysis
The STATA17.0 software was used to realize the statistical analyses.
Descriptive statistics on directions of microRNA expression are displayed in a pyramidal graph by type of specimen. Direction of microRNA expression was defined as the direction of the ratio between the microRNA concentration in breast cancer cases and the microRNA concentration in breast cancer controls.
For each study included in the meta-analysis, we built a contingency table to be used to carry out the meta-analysis. After selecting suitable studies, forest plot, and summary receiver operating characteristic curve (SROC), with the pooled sensitivity and specificity, were built for each microRNA [10,11]. We analyzed the heterogeneity between studies using the I2 statistics. Funnel plots were used to evaluate publication bias [12].
The analyses were repeated on subgroups as sensitivity analysis. Subgroups analyses were based on specimen type, ethnicity, and quality of the study (by QUADAS-2 score).
3. Results
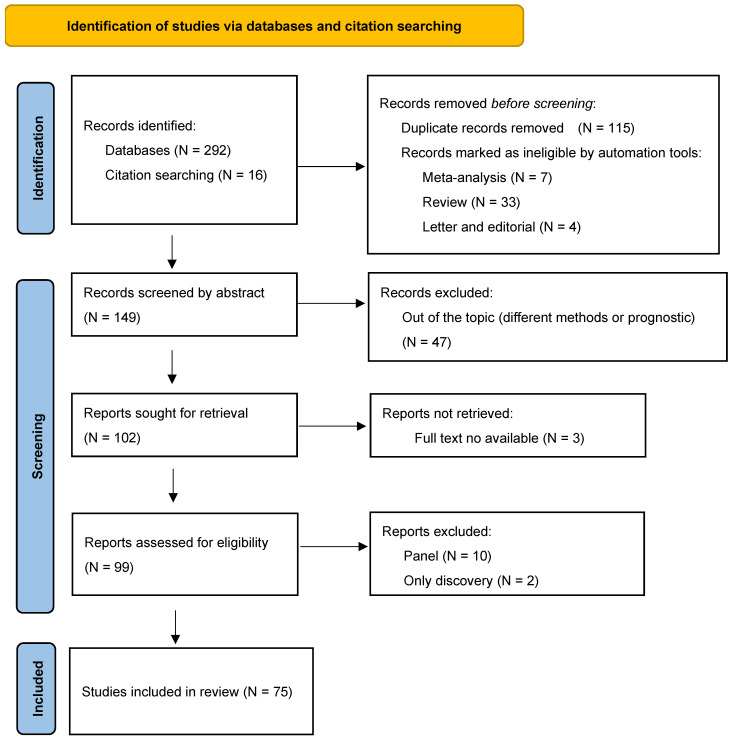
In total, 149 eligible studies were obtained from online database searching after automation screening. After a manual check of titles and abstracts, 47 papers were excluded because of the type of study (review, prognostic studies) or they were out of topic. After screening full texts, 24 publications were excluded because they did not satisfy the inclusion criteria. Finally, 75 publications were considered in this review [13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87]. The flow-chart of the excluded papers is presented in Figure 1. The main characteristics of the studies were summarized in Table 1.
Figure 1.
The flow chart of identification, screening, and eligibility of the included studies [8].
Table 1.
General characteristics of the studies included in systematic review on the role of microRNA in breast cancer diagnosis.
| First Author, Year | Country | Type | Specimen Source | Cases Size | Controls Size | MIR | Internal Reference |
|---|---|---|---|---|---|---|---|
| Zhu W, 2009 [13] | USA | Candidate | Serum | 13 | 8 | 16 | MIR18 |
| 145 | |||||||
| 155 | |||||||
| Heneghan H, 2010 [14] | Ireland | Candidate | Blood | 83 | 44 | 21 | MIR16 |
| 145 | |||||||
| 155 | |||||||
| 195 | |||||||
| 10b | |||||||
| Let-7a | |||||||
| Roth C, 2010 [15] | Germany | Candidate | Serum | 59 | 29 | 141 | MIR16 |
| 155 | |||||||
| 10b | |||||||
| 34a | |||||||
| Wang F, 2010 [16] | China | Candidate | Serum | 68 | 40 | 21 | MIR16 |
| 126 | |||||||
| 155 | |||||||
| 335 | |||||||
| 106a | |||||||
| 199a | |||||||
| Asaga S, 2011 [17] | USA | Validation | Serum | 62 | 10 | 21 | MIR16 |
| Guo LJ, 2012 [18] | China | Candidate | Serum | 152 | 75 | 181a | MIR16 |
| Schrauder MG, 2012 [19] | Germany | Validation | Serum | 24 | 24 | 202 | MIR16 |
| 718 | |||||||
| Schwarzenbach H, 2012 [20] | Germany | Candidate | Serum | 34 | 53 | 21 | MIR16 |
| 214 | |||||||
| 19a | |||||||
| 20a | |||||||
| Sun Y, 2012 [21] | China | Candidate | Serum | 103 | 55 | 155 | MIR39 |
| van Schooneveld E, 2012 [22] | Belgium | Candidate | Serum | 75 | 20 | 215 | |
| 299 | |||||||
| 411 | |||||||
| 452 | |||||||
| Wu Q, 2012 [23] | China | Validation | Serum | 50 | 50 | 222 | |
| Zhao FL, 2012 [24] | China | Candidate | Serum | 122 | 59 | 10b | MIR16 |
| Chan M, 2013 [25] | Singapore | Validation | Serum | 132 | 101 | 1 | MIR103, MIR191 |
| 133a | |||||||
| 133b | |||||||
| 92a | |||||||
| Cuk K, 2013 [26] | Germany | Validation | Plasma | 127 | 80 | 409 | MIR39 |
| 801 | |||||||
| 148b | |||||||
| 376c | |||||||
| Eichelser C, 2013 [27] | Germany | Candidate | Serum | 40 | 40 | 17 | MIR16 |
| 93 | |||||||
| 155 | |||||||
| 373 | |||||||
| 10b | |||||||
| 34a | |||||||
| Godfrey AC, 2013 [28] | USA | Validation | Serum | 5 | 5 | 222 | MIR1825 |
| 181a | |||||||
| 18a | |||||||
| Kumar S, 2013 [29] | India | Candidate | Plasma | 14 | 8 | 21 | MIR16 |
| 146a | |||||||
| Ng EKO, 2013 [30] | China | Validation | Plasma | 170 | 100 | 16 | RNU6B |
| 21 | |||||||
| 145 | |||||||
| 451 | |||||||
| Si H, 2013 [31] | China | Validation | Serum | 100 | 20 | 21 | MIR16 |
| 92a | |||||||
| Wang PY, 2013 [32] | China | Candidate | Serum | 46 | 58 | 182 | 5S rRNA |
| Zeng RC, 2013 [33] | China | Candidate | Plasma | 100 | 64 | 30a | MIR16 |
| Eichelser C, 2014 [34] | Germany | Candidate | Serum | 168 | 28 | 101 | MIR16, MIR 484 |
| 372 | |||||||
| 373 | |||||||
| Hamdi K, 2014 [35] | Tunisia | Candidate | Serum | 20 | 20 | 24 | RNU48 |
| 320 | |||||||
| 335 | |||||||
| 337 | |||||||
| 451 | |||||||
| 486 | |||||||
| 548 | |||||||
| 15a | |||||||
| 29a | |||||||
| 30b | |||||||
| 342-3p | |||||||
| 342-5p | |||||||
| Joosse SA, 2014 [36] | Germany | Candidate | Serum | 102 | 37 | 202 | MIR16 |
| Let-7b | |||||||
| Kodahl AR, 2014 [37] | Denmark | Validation | Serum | 60 | 51 | 107 | |
| 139 | |||||||
| 143 | |||||||
| 145 | |||||||
| 365 | |||||||
| 425 | |||||||
| 133a | |||||||
| 15a | |||||||
| 18a | |||||||
| Mar-Aguilar F, 2014 [38] | Mexico | Validation | Serum | 61 | 10 | 21 | MIR18S |
| 145 | |||||||
| 155 | |||||||
| 191 | |||||||
| 382 | |||||||
| 10b | |||||||
| 125b | |||||||
| McDermott AM, 2014 [39] | Ireland | Validation | Serum | 44 | 46 | 223 | MIR16 |
| 652 | |||||||
| 181a | |||||||
| 29a | |||||||
| Shen J, 2014 [40] | USA | Validation | Plasma | 50 | 50 | 409 | MIR93 |
| 133a | |||||||
| 148b | |||||||
| Sochor M, 2014 [41] | Chez Republic | Candidate | Serum | 63 | 21 | 24 | Let-7a |
| 155 | |||||||
| 181b | |||||||
| 19a | |||||||
| Zearo S, 2014 [42] | Australia | Validation | Serum | 98 | 25 | 484 | |
| Zhao FL, 2014 [43] | China | Candidate | Serum | 210 | 102 | 195 | MIR16 |
| Antolin S, 2015 [44] | Spain | Candidate | Blood, serum and plasma | 57 | 20 | 141 | 5S, U6 sn |
| 200c | |||||||
| Li XX, 2015 [45] | China | Candidate | Serum | 90 | 64 | Let-7c | 5SrRNA |
| Mangolini A (A), 2015 [46] | Italy | Candidate | Serum | 28 | 27 | 145 | MIR39 |
| 425 | |||||||
| 652 | |||||||
| 10b | |||||||
| 148b | |||||||
| Mangolini A (B), 2015 [46] | USA | Candidate | Serum | 59 | 35 | 145 | MIR39 |
| 425 | |||||||
| 652 | |||||||
| 10b | |||||||
| 148b | |||||||
| Matamala N, 2015 [47] | Spain | Validation | Plasma | 114 | 116 | 21 | MIR103a |
| 96 | |||||||
| 505 | |||||||
| 125b | |||||||
| Shaker O, 2015 [48] | Egypt | Candidate | Serum | 100 | 30 | 155 | SNORD |
| 197 | |||||||
| 205 | |||||||
| 29b | |||||||
| Zhang L, 2015 [49] | China | Validation | Serum | 76 | 52 | 424 | MIR132 |
| 199a | |||||||
| 29c | |||||||
| Frères P, 2016 [50] | Belgium | Validation | Plasma | 108 | 88 | 16 | Median of 50 mirna |
| 22 | |||||||
| 103 | |||||||
| 107 | |||||||
| 148a | |||||||
| 19b | |||||||
| Let-7d | |||||||
| Let-7i | |||||||
| Fu L, 2016 [51] | China | Candidate | Serum | 100 | 40 | 184 | |
| 382 | |||||||
| 598 | |||||||
| 1246 | |||||||
| Hamam R, 2016 [52] | Saudi Arabia | Validation | Serum and Plasma | 46 | 50 | 188 | MIR21 |
| 1202 | |||||||
| 1207 | |||||||
| 1225 | |||||||
| 1290 | |||||||
| 3141 | |||||||
| 4270 | |||||||
| 4281 | |||||||
| 642b | |||||||
| Hannafon BN, 2016 [53] | USA | Candidate | Plasma | 16 | 42 | 21 | MIR54 |
| 122 | |||||||
| 1246 | |||||||
| Let-7a | |||||||
| Motawi TM, 2016 [54] | Egypt | Candidate | Serum | 50 | 25 | 21 | REF SNORD 62 |
| 221 | |||||||
| Shimomura A, 2016 [55] | Japan | Validation | Serum | 1206 | 1343 | 1246 | MIR149 |
| 1307 | |||||||
| 4634 | |||||||
| 6861 | |||||||
| 6875 | |||||||
| Thakur S, 2016 [56] | India | Candidate | Serum | 85 | 85 | 21 | Sn U6 |
| 145 | |||||||
| 195 | |||||||
| 210 | |||||||
| 221 | |||||||
| Let-7a | |||||||
| Gao S, 2017 [57] | USA | Validation | Plasma | 75 | 50 | 155 | RNU6B |
| Zhang K, 2017 [58] | China | Validation | Blood | 15 | 13 | 96 | MIR16 |
| 182 | |||||||
| 942 | |||||||
| 30b | |||||||
| 374b | |||||||
| Heydari N, 2018 [59] | Iran | Candidate | Serum | 40 | 40 | 140 | MIR16 |
| Zaleski M, 2018 [60] | Germany | Validation | Plasma | 55 | 28 | 21 | MIR16 |
| 92 | |||||||
| 155 | |||||||
| 222 | |||||||
| 34a | |||||||
| Let-7c | |||||||
| Kaharam M, 2019 [61] | Germany | Validation | Blood | 21 | 21 | 101-3p | RNU48 |
| 126-3p | |||||||
| 126-5p | |||||||
| 144-3p | |||||||
| 144-5p | |||||||
| 301a | |||||||
| 664b | |||||||
| McAnena P, 2019 [62] | Ireland | Validation | Blood | 31 | 34 | 195 | MIR16, MIR425 |
| 331 | |||||||
| 181a | |||||||
| Peña-Cano MI, 2019 [63] | Mexico | Candidate | Serum | 50 | 50 | 17 | MIR26b |
| 195 | |||||||
| 221 | |||||||
| Raheem AR, 2019 [64] | Iraq | Candidate | Serum | 30 | 30 | 34a | MIRU6 |
| Soleimanpour E, 2019 [65] | Iran | Candidate | Plasma | 30 | 25 | 21 | MIR5s |
| 155 | |||||||
| Anwar SL, 2020 [66] | Indonesia | Candidate | Plasma | 102 | 15 | 155 | Sp6 |
| Arabkari V, 2020 [67] | Ireland | Validation | Blood | 38 | 20 | 16 | MIR1, MIR16 |
| 21 | |||||||
| 145 | |||||||
| 155 | |||||||
| 195 | |||||||
| 486 | |||||||
| 181a | |||||||
| 451a | |||||||
| Ashirbekov Y, 2020 [68] | Kazakhstan | Candidate | Plasma | 35 | 33 | 16 | MIR222 |
| 21 | |||||||
| 29 | |||||||
| 145 | |||||||
| 191 | |||||||
| 210 | |||||||
| 222 | |||||||
| Guo H, 2020 [69] | China | Validation | Plasma | 39 | 40 | 21 | cel-39 |
| 1273g | |||||||
| Holubekova V, 2020 [70] | Slovakia | Validation | Plasma | 65 | 34 | 484 | MIR16, MIR103a |
| 1260a | |||||||
| 130a | |||||||
| 99a | |||||||
| Hosseini Mojahed FH, 2020 [71] | Iran | Candidate | Serum | 36 | 36 | 155 | |
| Ibrahim AM, 2020 [72] | Egypt | Candidate | Plasma | 30 | 20 | 21 | MIR16 |
| 145 | |||||||
| 10b | |||||||
| 181a | |||||||
| Let-7 | |||||||
| Jang JY, 2020 [73] | Korea | Validation | Plasma | 80 | 56 | 21 | |
| 24 | |||||||
| 202 | |||||||
| 206 | |||||||
| 223 | |||||||
| 373 | |||||||
| 1246 | |||||||
| 6875 | |||||||
| 219b | |||||||
| Kim J, 2020 [74] | South Korea | Candidate | Plasma | 30 | 30 | 202 | |
| Pastor-Navarro B, 2020 [75] | Spain | Candidate | Serum | 45 | 16 | 21 | MIR16, MIR1228 |
| 155 | |||||||
| 205 | |||||||
| Bakr NM, 2021 [76] | Egypt | Validation | Blood | 196 | 49 | 373 | |
| Diansyah MN, 2021 [77] | Indonesia | Candidate | Plasma | 26 | 16 | 21 | MIR16 |
| Itani MM, 2021 [78] | Lebanon | Candidate | Plasma | 41 | 32 | 21 | |
| 139 | |||||||
| 145 | |||||||
| 155 | |||||||
| 425 | |||||||
| 451 | |||||||
| 130a | |||||||
| 23a | |||||||
| Mohmmed EA, 2021 [79] | Egypt | Candidate | Serum and Plasma | 50 | 30 | 106a | |
| Nashtahosseini Z, 2021 [80] | Iran | Candidate | Serum | 40 | 40 | 210 | MIR16 |
| 660 | |||||||
| Zhang K, 2021 [81] | China | Validation | Blood | 68 | 13 | 185 | |
| 362 | |||||||
| 106b | |||||||
| 142-3p | |||||||
| 142-5p | |||||||
| 26b | |||||||
| Zhao T, 2021 [82] | China | Candidate | Serum | 88 | 40 | 25 | MIR39 |
| Li X, 2022 [83] | China | Candidate | Serum | 49 | 49 | 9 | MIR16 |
| 17 | |||||||
| 148a | |||||||
| Liu H, 2022 [84] | China | Candidate | Serum | 112 | 59 | 103a | U6 sn |
| Mohamed AA, 2022 [85] | Egypt | Candidate | Serum | 99 | 40 | 155 | RNU6 |
| 373 | |||||||
| 10b | |||||||
| 34a | |||||||
| Zavesky L, 2022 [86] | Czech Republic | Validation | Plasma | 52 | 46 | 451a | MIR590, MIR19a, MIR222 |
| 548b | |||||||
| Zou R, 2022 [87] | Mix | Validation | Serum | 177 | 197 | 24 | MIR128, MIR652, MIR106b |
| 324 | |||||||
| 377 | |||||||
| 497 | |||||||
| 125b | |||||||
| 133a | |||||||
| 19b | |||||||
| 374c |
The publications involved a total of 6380 BC cases and 4517 health controls. Studies with two different approaches were included in this review: (I) The validation phase of studies where investigated microRNA were selected on the basis of a previous discovery phase (N = 26); (II) Candidate studies where microRNA were selected based on a priori knowledge (N = 50).
The studies with a number of BC cases ≥ 100 were only 20/75 (≈26%). The studies were conducted in China (N = 19), Germany (N = 9), Egypt (N = 6), USA (N = 6), Iran (N = 4), Ireland (N = 4), Spain (N = 3), Belgium (N = 2), Czech Republic (N = 2), India (N = 2), Indonesia (N = 2), Mexico (N = 2), Singapore (N = 2), and others (N = 12). Notably, the majority of studies were conducted in the Eastern Asian population (N = 29), while the majority of the others were conducted among European or white U.S. populations. Black and Hispanic populations are very little represented in microRNA and breast cancer studies.
Some studies were performed in serum (N = 44), while others were performed in plasma (n = 21), whole blood (N = 7), plasma and serum (N = 2), or blood, serum, and plasma (N = 1).
Among the 75 studies included in the review, 53 studies conducted multiple microRNA assays, while the other 22 studies focused on single microRNA assay (covering in total 141 microRNAs).
In the Supplementary Table S1, the clinical information on cases is presented for each study.
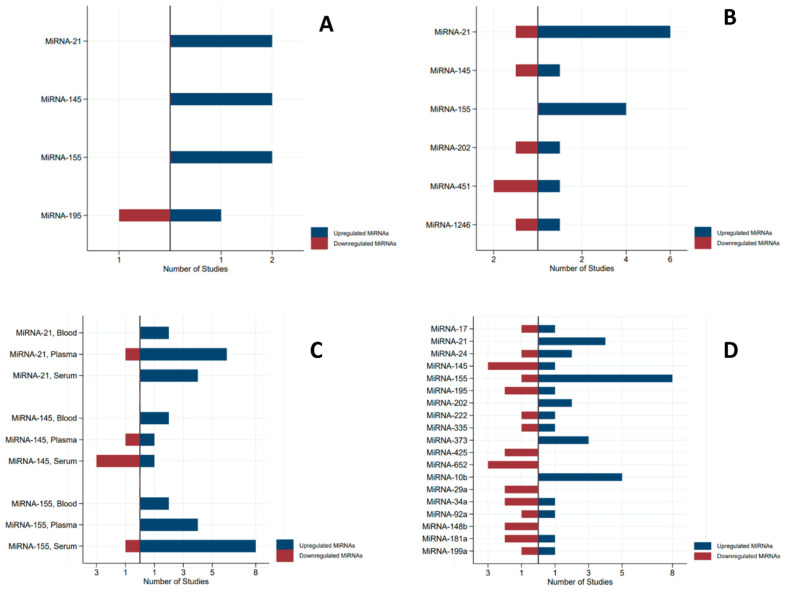
The results of the studies included in the review are presented in Table 2. The microRNAs that were proposed as a biomarker of BC in at least two independent clinical studies in the same biological specimen (serum, plasma, or whole blood) were: MIR17, MIR21, MIR24, MIR145, MIR155, MIR195, MIR202, MIR222, MIR335, MIR373, MIR425, MIR652, MIR10b, MIR29b, MIR34a, MIR92a, MIR148b, MIR181a, MIR199, and MIR1246. The direction of the ratio of microRNA concentrations in breast cancer cases versus controls was generally coherent for MIR21 (12 up versus 1 down), MIR155 (14 up versus 1 down), MIR10b (5 up, only serum), MIR373 (3 up, only serum), MIR652 (3 down, only serum), MIR425 (2 down, only serum), MIR29a (2 up, only serum), and MIR148b (2 down, only serum). The direction was not coherent for MIR145 (5 up versus 4 down), MIR17, MIR24, MIR195, MIR202, MIR222, MIR335, MIR451, MIR1246, MIR34a, MIR92a, MIR181a, and MIR199a (Figure 2).
Table 2.
Summary of the results of the studies included in the systematic review on the role of microRNA in breast cancer diagnosis.
| First Author, Year | Specimen Source | MiR | Direction | Cut_Off (ng/mL) |
AUC | Sens | Spec | Fold Change |
|---|---|---|---|---|---|---|---|---|
| Zhu W, 2009 [13] | Serum | 16 | Up | |||||
| 145 | Up | |||||||
| 155 | Down | |||||||
| Heneghan H, 2010 [14] | Blood | 21 | Up | |||||
| 145 | Up | |||||||
| 155 | Up | |||||||
| 195 | Up | 0.94 (0.91–0.97) |
87.70 | 91.00 | 25.00 | |||
| 10b | Down | |||||||
| Let-7a | Up | |||||||
| Roth C, 2010 [15] | Serum | 141 | ||||||
| 155 | Up | 1.60 | ||||||
| 10b | ||||||||
| 34a | ||||||||
| Wang F, 2010 [16] | Serum | 21 | Up | 2.50 | ||||
| 126 | Down | 2.00 | ||||||
| 155 | Up | 3.50 | ||||||
| 335 | Up | 2.00 | ||||||
| 106a | Up | 1.90 | ||||||
| 199a | Down | 2.00 | ||||||
| Asaga S, 2011 [17] | Serum | 21 | Up | 3.30 | 0.72 | 75.00 | 67.00 | |
| Guo LJ, 2012 [18] | Serum | 181a | Down | 0.74 | 0.67 (0.60–0.74) |
70.70 | 59.90 | 0.36 |
| Schrauder MG, 2012 [19] | Serum | 202 | Up | 0.68 | 19.38 | |||
| 718 | Down | 0.77 | 5.44 | |||||
| Schwarzenbach H, 2012 [20] | Serum | 21 | 0.85 (0.78–0.91) |
|||||
| 214 | 0.92 (0.88–0.97) |
|||||||
| 19a | ||||||||
| 20a | 0.68 (0.59–0.77) |
|||||||
| Sun Y, 2012 [21] | Serum | 155 | Up | 1.91 | 0.80 (0.65–0.82) |
65.00 | 81.80 | 2.94 |
| van Schooneveld E, 2012 [22] | Serum | 215 | Up | |||||
| 299 | Down | |||||||
| 411 | Down | |||||||
| 452 | Down | |||||||
| Wu Q, 2012 [23] | Serum | 222 | Up | 0.01 | 0.67 (0.57–0.78) |
74.00 | 60.00 | |
| Zhao FL, 2012 [24] | Serum | 10b | Up | |||||
| Chan M, 2013 [25] | Serum | 1 | Up | 2.67 | ||||
| 133a | Up | 2.62 | ||||||
| 133b | Up | 2.41 | ||||||
| 92a | Up | 1.32 | ||||||
| Cuk K, 2013 [26] | Plasma | 409 | Up | 0.66 (0.59–0.74) |
||||
| 801 | Up | 0.64 (0.56–0.72) |
||||||
| 148b | Up | 0.65 (0.58–0.73) |
||||||
| 376c | Up | 0.66 (0.59–0.74) |
||||||
| Eichelser C, 2013 [27] | Serum | 17 | Down | 0.68 | 18.80 | 100.00 | ||
| 93 | Up | 0.70 | 44.90 | 100.00 | ||||
| 155 | Up | 0.78 | 70.60 | 42.70 | ||||
| 373 | Up | 0.88 | 76.60 | 100.00 | ||||
| 10b | Up | 21.80 | 92.10 | |||||
| 34a | Up | 0.64 | 59.80 | 76.00 | ||||
| Godfrey AC, 2013 [28] | Serum | 222 | Down | |||||
| 181a | Up | |||||||
| 18a | Up | |||||||
| Kumar S, 2013 [29] | Plasma | 21 | Up | |||||
| 146a | Up | |||||||
| Ng EKO, 2013 [30] | Plasma | 16 | Up | 0.91 (0.87–0.95) |
||||
| 21 | Up | 0.81 (0.74–0.88) |
||||||
| 145 | Down | 0.63 (0.52–0.74) |
||||||
| 451 | Up | 0.94 (0.91–1.00) |
||||||
| Si H, 2013 [31] | Serum | 21 | Up | 0.93 (0.89–0.92) |
||||
| 92a | Down | 0.92 (0.87–0.97) |
||||||
| Wang PY, 2013 [32] | Serum | 182 | Up | |||||
| Zeng RC, 2013 [33] | Plasma | 30a | Down | 0.01 | 0.76 (0.68–0.83) |
74.00 | 65.60 | |
| Eichelser C, 2014 [34] | Serum | 101 | Up | |||||
| 372 | Up | |||||||
| 373 | Up | |||||||
| Hamdi K, 2014 [35] | Serum | 24 | Down | |||||
| 320 | Down | |||||||
| 335 | Down | |||||||
| 337 | Down | |||||||
| 451 | Down | 15.80 | ||||||
| 486 | Down | |||||||
| 548 | Down | |||||||
| 15a | Down | |||||||
| 29a | Down | |||||||
| 30b | Down | |||||||
| 342-3p | Down | |||||||
| 342-5p | Down | |||||||
| Joosse SA, 2014 [36] | Serum | 202 | Up | |||||
| Let-7b | Up | |||||||
| Kodahl AR, 2014 [37] | Serum | 107 | 0.66 | |||||
| 139 | 1.44 | |||||||
| 143 | 1.65 | |||||||
| 145 | 1.56 | |||||||
| 365 | 1.88 | |||||||
| 425 | 0.84 | |||||||
| 133a | 1.68 | |||||||
| 15a | 1.84 | |||||||
| 18a | 0.65 | |||||||
| Mar-Aguilar F, 2014 [38] | Serum | 21 | 6.48 | 0.95 (0.91–0.99) |
94.40 | 80.00 | ||
| 145 | 15.93 | 0.98 (0.95–1.00) |
94.40 | 100.00 | ||||
| 155 | 7.92 | 0.99 (0.99–1.00) |
94.40 | 100.00 | ||||
| 191 | 4.85 | 0.79 (0.71–0.88) |
72.20 | 90.00 | ||||
| 382 | 0.97 (0.94–1.00) |
94.40 | 90.00 | |||||
| 10b | 59.22 | 0.95 (0.91–0.99) |
83.30 | 100.00 | ||||
| 125b | 8.46 | 0.95 (0.91–0.99) |
88.90 | 80.00 | ||||
| McDermott AM, 2014 [39] | Serum | 223 | Down | |||||
| 652 | Down | |||||||
| 181a | Down | |||||||
| 29a | Down | |||||||
| Shen J, 2014 [40] | Plasma | 409 | ||||||
| 133a | 8.30 | |||||||
| 148b | 5.10 | |||||||
| Sochor M, 2014 [41] | Serum | 24 | Up | |||||
| 155 | Up | |||||||
| 181b | Up | |||||||
| 19a | Up | |||||||
| Zearo S, 2014 [42] | Serum | 484 | 1.60 | |||||
| Zhao FL, 2014 [43] | Serum | 195 | Down | 0.50 | 0.86 (0.82–0.90) |
69.00 | 89.20 | 2.38 |
| Antolin S, 2015 [44] | Blood, serum and plasma | 141 | ||||||
| 200c | Down | 0.79 | 90.00 | 70.20 | ||||
| Li XX, 2015 [45] | Serum | Let-7c | Down | 0.85 (0.79–0.91) |
87.50 | 78.90 | ||
| Mangolini A (A), 2015 [46] | Serum | 145 | Down | |||||
| 425 | Down | |||||||
| 652 | Down | 0.83 (0.73–0.93) |
||||||
| 10b | Up | |||||||
| 148b | Down | 0.74 (0.62–0.86) |
||||||
| Mangolini A (B), 2015 [46] | Serum | 145 | Down | |||||
| 425 | Down | |||||||
| 652 | Down | 0.69 (0.58–0.80) |
||||||
| 10b | Up | |||||||
| 148b | Down | 0.66 (0.51–0.80) |
||||||
| Matamala N, 2015 [47] | Plasma | 21 | Up | 0.61 (0.53–0.68) |
||||
| 96 | Up | 0.72 (0.65–0.78) |
73.00 | 66.00 | ||||
| 505 | Up | 0.72 (0.66–0.79) |
75.00 | 60.00 | ||||
| 125b | Up | 0.64 (0.56–0.71) |
||||||
| Shaker O, 2015 [48] | Serum | 155 | Up | 39.57 | 0.99 (0.99–1.00) |
94.10 | 100.00 | 39.57 |
| 197 | Up | 29.80 | 0.98 (0.95–1.00) |
95.30 | 100.00 | 29.80 | ||
| 205 | Up | 27.48 | 0.99 (0.98–1.00) |
98.80 | 100.00 | 27.48 | ||
| 29b | Up | 41.94 | 0.99 (0.98–1.00) |
98.80 | 100.00 | 41.94 | ||
| Zhang L, 2015 [49] | Serum | 424 | Up | 0.75 (0.67–0.84) |
1.77 | |||
| 199a | Up | 0.92 (0.87–0.96) |
2.65 | |||||
| 29c | Up | 0.72 (0.64–0.81) |
1.97 | |||||
| Frères P, 2016 [50] | Plasma | 16 | 1.70 | |||||
| 22 | 1.00 | |||||||
| 103 | 0.80 | |||||||
| 107 | 0.80 | |||||||
| 148a | 1.40 | |||||||
| 19b | 1.20 | |||||||
| Let-7d | 0.90 | |||||||
| Let-7i | 0.70 | |||||||
| Fu L, 2016 [51] | Serum | 184 | Down | 0.48 | 0.74 (0.66–0.82) |
87.50 | 71.00 | |
| 382 | Up | 1.32 | 0.90 (0.85–0.96) |
93.00 | 75.00 | |||
| 598 | Down | 1.61 | 0.74 (0.66–0.82) |
52.00 | 92.50 | |||
| 1246 | Up | 0.55 | 0.94 (0.90–0.98) |
95.00 | 85.00 | |||
| Hamam R, 2016 [52] | Serum and Plasma | 188 | Up | |||||
| 1202 | Up | |||||||
| 1207 | Up | |||||||
| 1225 | Up | |||||||
| 1290 | Up | |||||||
| 3141 | Up | |||||||
| 4270 | Up | |||||||
| 4281 | Up | |||||||
| 642b | Up | |||||||
| Hannafon BN, 2016 [53] | Plasma | 21 | Up | 0.69 (0.49–0.89) |
||||
| 122 | Up | |||||||
| 1246 | Up | 0.69 (0.50–0.88) |
||||||
| Let-7a | Up | |||||||
| Motawi TM, 2016 [54] | Serum | 21 | 1.14 | 0.98 (0.96–1.00) |
96.00 | 94.00 | 2.20 | |
| 221 | 1.21 | 0.97 (0.94–1.00) |
92.00 | 88.00 | 2.09 | |||
| Shimomura A, 2016 [55] | Serum | 1246 | 88.30 | 93.40 | ||||
| 1307 | 100.00 | 53.10 | ||||||
| 4634 | 3.40 | 73.60 | ||||||
| 6861 | 99.80 | 79.40 | ||||||
| 6875 | 14.70 | 76.80 | ||||||
| Thakur S, 2016 [56] | Serum | 21 | Up | 0.79 (0.71–0.86) |
88.00 | 65.00 | ||
| 145 | Down | 0.73 (0.66–0.81) |
74.00 | 56.00 | ||||
| 195 | Down | 0.80 (0.74–0.87) |
77.00 | 71.00 | ||||
| 210 | 0.64 (0.55–0.72) |
78.00 | 61.00 | |||||
| 221 | 0.63 (0.54–0.71) |
65.00 | 57.00 | |||||
| Let-7a | 0.76 (0.69–0.83) |
71.00 | 67.00 | |||||
| Gao S, 2017 [57] | Plasma | 155 | Up | 0.77 (0.68–0.86) |
||||
| Zhang K, 2017 [58] | Blood | 96 | Up | 2.73 | 0.77 | 53.00 | 100.00 | |
| 182 | Up | 1.01 | 0.76 | 53.00 | 92.00 | |||
| 942 | Up | 1.04 | 0.81 | 67.00 | 100.00 | |||
| 30b | Up | 2.04 | 0.93 | 80.00 | 100.00 | |||
| 374b | Up | 1.52 | 0.82 | 87.00 | 69.00 | |||
| Heydari N, 2018 [59] | Serum | 140 | Up | 0.13 | 0.67 (0.55–0.79) |
70.00 | 50.00 | |
| Zaleski M, 2018 [60] | Plasma | 21 | 0.58 (0.46–0.71) |
|||||
| 92 | 0.46 (0.33–0.60) |
|||||||
| 155 | 0.53 (0.36–0.69) |
|||||||
| 222 | 0.53 (0.40–0.67) |
|||||||
| 34a | 0.72 (0.61–0.84) |
|||||||
| Let-7c | 0.51 (0.38–0.64) |
|||||||
| Kaharam M, 2019 [61] | Blood | 101-3p | ||||||
| 126-3p | ||||||||
| 126-5p | ||||||||
| 144-3p | ||||||||
| 144-5p | ||||||||
| 301a | ||||||||
| 664b | ||||||||
| McAnena P, 2019 [62] | Blood | 195 | 0.73 | |||||
| 331 | 2.94 | |||||||
| 181a | 1.19 | |||||||
| Peña-Cano MI, 2019 [63] | Serum | 17 | Up | 0.50 | ||||
| 195 | Up | 0.04 | 0.88 (0.78–0.98) |
83.30 | 78.30 | 4.33 | ||
| 221 | Down | 0.70 | ||||||
| Raheem AR, 2019 [64] | Serum | 34a | Down | 5.05 | 0.67 (0.53–0.81) |
60.00 | 63.00 | |
| Soleimanpour E, 2019 [65] | Plasma | 21 | Up | 0.92 | ||||
| 155 | Up | 0.99 | ||||||
| Anwar SL, 2020 [66] | Plasma | 155 | Up | |||||
| Arabkari V, 2020 [67] | Blood | 16 | Up | 0.61 (0.47–0.76) |
||||
| 21 | Up | 0.65 (0.51–0.79) |
1.35 | |||||
| 145 | Up | 0.83 (0.72–0.94) |
1.61 | |||||
| 155 | Up | 0.76 (0.66–0.89) |
1.63 | |||||
| 195 | Down | 0.81 (0.69–0.92) |
0.14 | |||||
| 486 | Up | 0.90 (0.81–0.97) |
2.25 | |||||
| 181a | 1.52 | |||||||
| 451a | Up | 0.73 (0.61–0.86) |
1.62 | |||||
| Ashirbekov Y, 2020 [68] | Plasma | 16 | 0.69 | |||||
| 21 | 1.35 | |||||||
| 29 | 0.98 | |||||||
| 145 | 2.36 | |||||||
| 191 | 1.87 | |||||||
| 210 | 0.69 | |||||||
| 222 | 0.98 | |||||||
| Guo H, 2020 [69] | Plasma | 21 | 0.66 (0.53–0.78) |
|||||
| 1273g | 0.63 (0.51–0.76) |
|||||||
| Holubekova V, 2020 [70] | Plasma | 484 | Up | 1.10 | ||||
| 1260a | Up | 1.22 | ||||||
| 130a | Up | 1.20 | ||||||
| 99a | Up | 1.33 | ||||||
| Hosseini Mojahed FH, 2020 [71] | Serum | 155 | Up | 1.40 | 0.89 | 77.80 | 88.89 | 1.00 |
| Ibrahim AM, 2020 [72] | Plasma | 21 | 4.94 | 0.78 | 63.30 | 100.00 | ||
| 145 | 0.78 | 0.70 | 45.00 | 83.30 | ||||
| 10b | 2.52 | 0.73 | 53.30 | 100.00 | ||||
| 181a | 1.51 | 0.70 | 50.00 | 80.00 | ||||
| Let-7 | 0.52 | 0.72 | 50.00 | 93.30 | ||||
| Jang JY, 2020 [73] | Plasma | 21 | Down | 0.92 | ||||
| 24 | Down | 0.96 | 65.00 | 96.00 | ||||
| 202 | Down | 0.86 | ||||||
| 206 | Down | 0.94 | 79.00 | 96.00 | ||||
| 223 | Down | 0.81 | ||||||
| 373 | Down | 0.96 | ||||||
| 1246 | Down | 0.93 | 53.00 | 95.00 | ||||
| 6875 | Down | 0.96 | 86.00 | 96.00 | ||||
| 219b | Down | 0.88 | ||||||
| Kim J, 2020 [74] | Plasma | 202 | Up | 2.10 | 0.95 (0.88–1.02) |
90.00 | 93.30 | 9.60 |
| Pastor-Navarro B, 2020 [75] | Serum | 21 | 0.77 (0.68–0.87) |
|||||
| 155 | 0.32 (0.68–0.87) |
|||||||
| 205 | 0.65 (0.68–0.87) |
|||||||
| Bakr NM, 2021 [76] | Blood | 373 | 360.00 | 0.98 (0.95–0.99) |
90.80 | 98.40 | ||
| Diansyah MN, 2021 [77] | Plasma | 21 | 1.66 | 0.92 (0.83–1) |
92.30 | 81.20 | 4.36 | |
| Itani MM, 2021 [78] | Plasma | 21 | Up | 4.46 | 0.76 (0.64–0.88) |
73.00 | 81.00 | |
| 139 | Up | 11.69 | 0.74 (0.62–0.87) |
78.00 | 75.00 | |||
| 145 | Up | 10.18 | 0.78 (0.66–0.90) |
83.00 | 78.00 | |||
| 155 | Up | 8.54 | 0.83 (0.71–0.95) |
76.00 | 96.00 | |||
| 425 | Up | 9.09 | 0.81 (0.69–0.93) |
78.00 | 91.00 | |||
| 451 | Down | 10.54 | 0.70 (0.57–0.83) |
78.00 | 75.00 | |||
| 130a | Up | 7.96 | 0.83 (0.72–0.94) |
70.00 | 100.00 | |||
| 23a | Up | 2.50 | 0.73 (0.61–0.85) |
73.00 | 72.00 | |||
| Mohmmed EA, 2021 [79] | Serum and Plasma | 106a | Up | 0.95 | 83.00 | 100.00 | 3.63 | |
| Nashtahosseini Z, 2021 [80] | Serum | 210 | Up | 0.82 | 0.72 (0.60–0.83) |
68.00 | 51.00 | 2.72 |
| 660 | Up | 0.77 | 0.77 (0.66–0.88) |
79.00 | 61.00 | 2.71 | ||
| Zhang K, 2021 [81] | Blood | 185 | Up | 1.08 | 0.91 (0.83–0.99) |
91.18 | 76.92 | 4.00 |
| 362 | Up | 1.53 | 0.93 (0.88–0.99) |
83.82 | 100.00 | 2.97 | ||
| 106b | Up | 1.26 | 0.82 (0.68–0.95) |
79.41 | 76.92 | 1.89 | ||
| 142-3p | Up | 6.87 | 0.85 (0.76–0.98) |
97.06 | 61.54 | 3.18 | ||
| 142-5p | Up | 1.60 | 0.85 (0.71–0.99) |
85.29 | 76.92 | 2.46 | ||
| 26b | Up | 1.34 | 0.89 (0.81–0.97) |
83.82 | 84.62 | 3.32 | ||
| Zhao T, 2021 [82] | Serum | 25 | Up | 0.75 (0.66–0.84) |
57.10 | 95.00 | ||
| Li X, 2022 [83] | Serum | 9 | Up | |||||
| 17 | ||||||||
| 148a | Up | |||||||
| Liu H, 2022 [84] | Serum | 103a | Up | 3.40 | 0.70 (0.62–0.78) |
78.20 | 74.70 | |
| Mohamed AA, 2022 [85] | Serum | 155 | Up | 7.50 | 0.94 (0.89–0.98) |
86.90 | 90.00 | |
| 373 | Up | 10.00 | 0.95 (0.90–0.98) |
85.00 | 100.00 | |||
| 10b | Up | 9.50 | 0.77 (0.69–0.84) |
60.00 | 93.00 | |||
| 34a | Down | 10.50 | 0.89 (0.82–0.94) |
91.00 | 75.00 | |||
| Zavesky L, 2022 [86] | Plasma | 451a | Down | 1.39 | ||||
| 548b | Up | 3.60 | ||||||
| Zou R, 2022 [87] | Serum | 24 | Up | 0.76 | 0.62 | |||
| 324 | Down | 0.52 | 0.31 | |||||
| 377 | Down | 0.73 | 0.67 | |||||
| 497 | Up | 0.56 | 0.15 | |||||
| 125b | Up | 0.58 | 0.13 | |||||
| 133a | Up | 0.63 | 0.41 | |||||
| 19b | Down | 0.63 | 0.26 | |||||
| 374c | Down | 0.71 | 0.99 |
Figure 2.
Pyramidal graph of the direction of miRNA expression (microRNA concentration in breast cancer cases versus controls) by type of specimens (only microRNAs that were analysed in two or more independent studies). (A) = whole blood, (B) = plasma; (C) = all specimens; (D) = serum.
Not all the studies presented data on AUC and/or sensitivity and specificity or fold change (N = 26 articles did not report sensitivity/specificity or AUC measures; N = 48 not reported fold change measure for single miRNAs).
A meta-analysis was performed only for microRNAs analysed in at least three independent studies where sufficient data to make an analysis were presented.
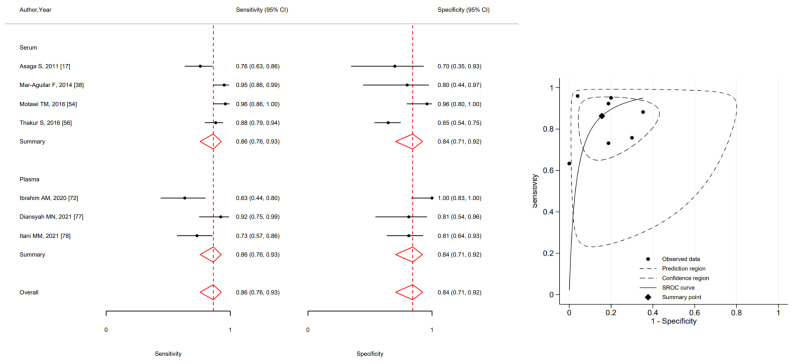
The results of the meta-analysis on MIR21 (upregulated), showed an overall sensitivity of 0.86 (95%CI 0.76–0.93) and a specificity of 0.84 (95%CI 0.71–0.92) (Figure 3).
Figure 3.
Forest plot of included studies assessing the sensitivity and specificity by type of specimen and summary receiver operating characteristic curve (SROC) of MIR21 in breast cancer diagnosis (squares shows sensitivity and specificity, respectively; red diamonds show pooled effect; error bars represents 95% confidence interval) [17,38,54,56,72,77,78].
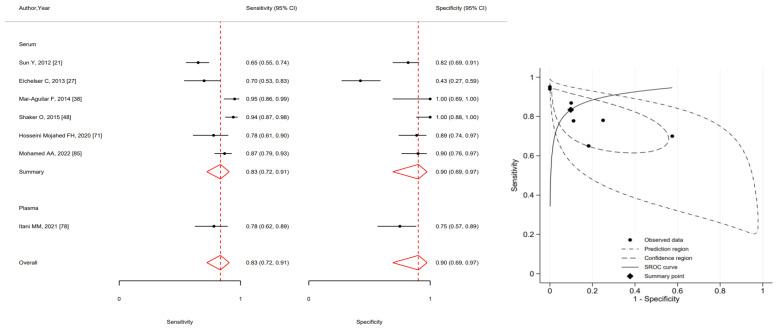
The pooled sensitivity and specificity of MIR155 (upregulated) were respectively 0.83 (95%CI 0.72–0.91) and 0.90 (95%CI 0.69–0.97) (Figure 4).
Figure 4.
Forest plot of included studies assessing the sensitivity and specificity by type of specimen and summary receiver operating characteristic curve (SROC) of MIR155 in breast cancer diagnosis (squares shows sensitivity and specificity, respectively; red diamonds show pooled effect; error bars represents 95% confidence interval) [21,27,38,48,71,78,85].
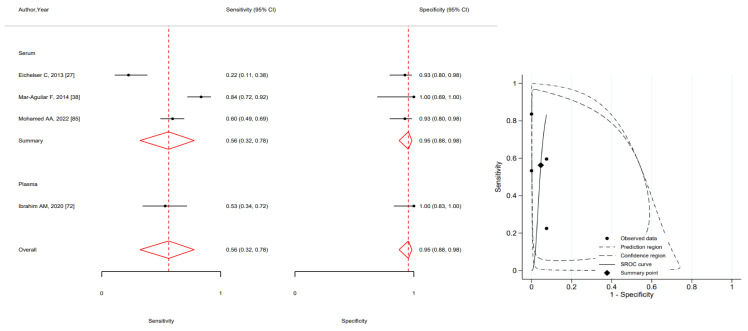
The meta-analysis results on the accuracy of MIR10b demonstrated a very low sensitivity (0.56 95%CI 0.32–0.71) and a high specificity (0.95 95%CI 0.88–0.98) (Figure 5).
Figure 5.
Forest plot of included studies assessing the sensitivity and specificity by type of specimen and summary receiver operating characteristic curve (SROC) of MIR10b in breast cancer diagnosis (squares shows sensitivity and specificity, respectively; red diamonds show pooled effect; error bars represents 95% confidence interval) [27,38,72,85].
Meta-analysis results for MIR34a and MIR195 were presented in the supplementary figure (Supplementary Figures S1 and S2).
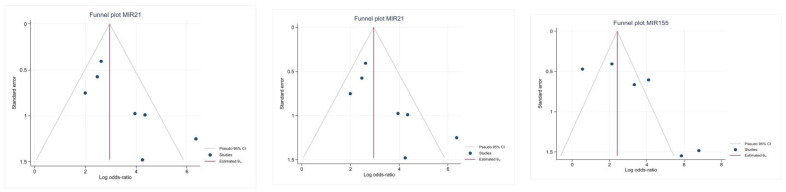
The shape of the funnel plot showed asymmetry in the analyses of MIR21, MIR155, and the overall microRNAs included in the meta-analysis, implying the presence of a publication bias in the analysis of the remaining circulating microRNAs (Figure 6).
Figure 6.
Evaluation of publication bias of all reported microRNAs presented as funnel plots.
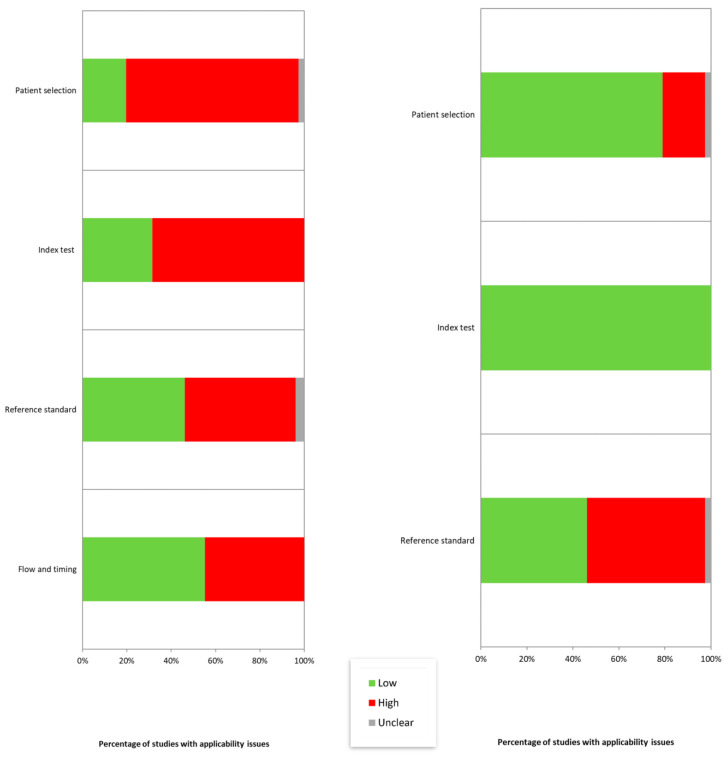
In general, a quality assessment with QUADAS detected a low quality of the studies because of an inadequate sample size and a low attention to the study design, the choice of controls, and the possible confounders (Figure 7 and Supplementary Table S2).
Figure 7.
Quality assessment with the QUADAS-2 tool [9].
4. Discussion
In the present study, we systematically reviewed clinical studies on microRNA for the diagnosis of BC, exploring possible links of most associated microRNA with hallmarks of BC.
Increasing evidence has demonstrated that microRNA may function as either a tumor suppressor or a promoter in a variety of cancers. The association of obesity and inflammation with microRNA has also been proposed [88,89]. The identification of microRNAs that could simultaneously be associated to BC and other hallmarks of cancer could be considered a meet-in-the-middle biomarker, which is useful to disentangle the role of involved factors and to hypothesize a biological pathway from exposures to disease [90].
Only few microRNAs were analyzed in more than two independent studies that presented in the results the essential data to be included in this meta-analysis. Among them, the most interesting microRNAs in terms of coherence among studies in the regulation direction and of results of diagnostic accuracy were two upregulated microRNAs: MIR21 and MIR155.
The MIR21 is an onco-microRNA that inhibits several tumor-suppressor genes (such as PTEN) and promotes cell growth invasion, apoptosis, and immune dysregulation [91]. A significant interaction between obesity and the expression of MIR21 and MIR155 consisting of obesity reducing the expression of these microRNAs in control women was described [89]. MIR155 is another oncogenic microRNA that regulates several signaling pathways related to cell growth [92], and it is also known to target BRCA1 [93]. It has also an important role in reducing inflammation, observed both in vitro and in vivo [94].
We observed little consistency with respect to the circulating microRNAs identified by different studies. This could be due also because of the different method used in the choice of microRNAs, lack of standardization of techniques (different sample retrieval and conservation, laboratory techniques, microRNAs measurements and normalization, cut-off), inconsistent selection of patients, low abundance, small samples size, and inadequate statistical analysis.
The majority of studies analyzed microRNA concentration in serum, but others used plasma or whole blood. Most of the microRNAs in the serum showed higher concentrations than the corresponding plasma samples [95], and some of the discrepancies in the direction of microRNAs presented in Table 1 could be due to the different types of samples.
Furthermore, the most frequent method used to quantify circulating candidate microRNAs, or to validate microRNAs, is the quantitative reverse transcription (PCR RT-qPCR); only two studies have introduced the use of the digital droplet [46,76]. All these techniques have a high sensitivity in detecting a large number of microRNAs at the same time. However, a poor agreement among different microRNA measurement platforms has been reported [96].
For microRNA measurement, data normalization is still a challenge, and this could be another source of variability in the results [95]. Furthermore, the cut-off values of considered microRNAs to calculate sensibility and specificity based on different ROC curves were not uniform, which may contribute to the observed heterogeneity.
The sample sizes in most studies are relatively small, and very few studies included an adequate group of controls that were collected from the same population, rather than cases, and matched at least for age.
In the reviewed articles, two categories of studies on microRNA and BC diagnosis were present: (i) Studies with an agnostic approach based on microRNA profiling (using different array platforms), usually followed by a validation phase in a different population with a more sensitive technique of most promising selected microRNA; (ii) Studies with a Bayesian approach based on microRNAs candidates, selected on the basis of previous biological knowledge, positive results in other studies, or in studies on other cancers.
In the present review, both the categories of studies were included, but only the validation phase of the microRNA profiling studies was described.
Finally, due to genetic heterogeneity, a difference in identified microRNA may be present among different ethnicities.
The result is a high number of microRNA identified as possibly related to BC status, but very few of them were replicated in other studies and other populations.
This review is a very comprehensive collection of studies on circulating microRNA and breast cancer. However, it presents several limitations. In fact, in all the studies, the origin of microRNAs has not been verified, and the contribution of breast cancer tissue has not been verified. There was a high heterogeneity among studies, probably due to different ethnic populations, small sample sizes, different types of sample and laboratory techniques, different statistical analysis, and different cut-offs.
5. Conclusions
The effort of this review has been devoted to exploring the most important microRNA involved in BC pathogenesis.
We found a list of microRNAs possibly involved in the breast cancer pathogenic pathway. Anyway, an effort must be done to try to standardize the microRNA research, with more robust study design, analytical strategies, and a better reporting of results in the published articles. New studies nested in population cohorts are needed to analyze microRNA in pre-diagnostic blood samples in order to strengthen the evidence of the association with breast cancer.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24043910/s1.
Author Contributions
Conceptualization, C.S., L.D.M. and F.R.; methodology, L.D., V.F., P.V., S.C., C.A., F.R. and C.S.; validation, C.S., L.D.M. and F.R.; formal analysis, L.P., L.D., L.M. (Lorenzo Milani) and L.M. (Luca Manfredi); data curation, L.P., L.D., L.M. (Lorenzo Milani), L.M. (Luca Manfredi) and C.S.; writing—original draft preparation, C.S. and L.P.; writing—review and editing, All authors; supervision, All authors; funding acquisition, C.S. and L.D.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Italian Ministry of Health (project n. RF 2018 12366921).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Loibl S., Poortmans P., Morrow M., Denkert C., Curigliano G. Breast cancer. Lancet. 2021;397:1750–1769. doi: 10.1016/S0140-6736(20)32381-3. [DOI] [PubMed] [Google Scholar]
- 3.Chen S., Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniou A.C., Foulkes W.D., Tischkowitz M. Breast-cancer risk in families with mutations in PALB2. N. Engl. J. Med. 2014;371:497–506. doi: 10.1056/NEJMc1410673. [DOI] [PubMed] [Google Scholar]
- 5.Cuk K., Obernosterer G., Leuschner P.J., Alenius M., Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamam R., Hamam D., Alsaleh K.A., Kassem M., Zaher W., Alfayez M., Aldahmash A., Alajez N.M. Circulating microRNAs in breast cancer; novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8:e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shajari E., Mollasalehi H. Ribonucleic-acid-biomarker candidates for early-phase group detection of common cancers. Genomics. 2020;112:163–168. doi: 10.1016/j.ygeno.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 8.McGrath T.A., Alabousi M., Skidmore B., Korevaar D.A., Bossuyt P.M.M., Moher D., Thombs B., McInnes M.D.F. Recommendations for reporting of systematic reviews and meta-analyses of diagnostic test accuracy; a systematic review. Syst. Rev. 2017;6:194. doi: 10.1186/s13643-017-0590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiting P.F., Rutjes A.W., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., Leeflang M.M., Sterne J.A., Bossuyt P.M., QUADAS-2 Group QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y.H. Overview of the process of conducting meta-analyses of the diagnostic test accuracy. J. Rheum. Dis. 2017;25:3–10. doi: 10.4078/jrd.2018.25.1.3. [DOI] [Google Scholar]
- 11.Leeflang M.M., Deeks J.J., Gatsonis C., Bossuyt P.M., Cochrane Diagnostic Test Accuracy Working Group Systematic reviews of diagnostic test accuracy. Ann. Intern. Med. 2008;149:889–897. doi: 10.7326/0003-4819-149-12-200812160-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sterne J.A., Sutton A.J., Ioannidis J.P., Terrin N., Jones D.R., Lau J., Carpenter J., Rücker G., Harbord R.M., Schmid C.H., et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343:d4002. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 13.Zhu W., Qin W., Atasoy U., Sauter E.R. Circulating microRNAs in breast cancer and healthy subjects. BMC Res. Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heneghan H.M., Miller N., Kelly R., Newell J., Kerin M.J. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15:673–682. doi: 10.1634/theoncologist.2010-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth C., Rack B., Müller V., Janni W., Pantel K., Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F., Zheng Z., Guo J., Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol. Oncol. 2010;119:586–593. doi: 10.1016/j.ygyno.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 17.Asaga S., Kuo C., Nguyen T., Terpenning M., Giuliano A.E., Hoon D.S. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin. Chem. 2011;57:84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 18.Guo L.J., Zhang Q.Y. Decreased serum miR-181a is a potential new tool for breast cancer screening. Int. J. Mol. Med. 2012;30:680–686. doi: 10.3892/ijmm.2012.1021. [DOI] [PubMed] [Google Scholar]
- 19.Schrauder M.G., Strick R., Schulz-Wendtland R., Strissel P.L., Kahmann L., Loehberg C.R., Lux M.P., Jud S.M., Hartmann A., Hein A., et al. Circulating micro-RNAs as potential blood-based markers for early stage breast cancer detection. PLoS ONE. 2012;7:e29770. doi: 10.1371/journal.pone.0029770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarzenbach H., Milde-Langosch K., Steinbach B., Müller V., Pantel K. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res. Treat. 2012;134:933–941. doi: 10.1007/s10549-012-1988-6. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y., Wang M., Lin G., Sun S., Li X., Qi J., Li J. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS ONE. 2012;7:e47003. doi: 10.1371/journal.pone.0047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Schooneveld E., Wouters M.C., Van der Auwera I., Peeters D.J., Wildiers H., Van Dam P.A., Vergote I., Vermeulen P.B., Dirix L.Y., Van Laere S.J. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012;14:R34. doi: 10.1186/bcr3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Q., Wang C., Lu Z., Guo L., Ge Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin. Chim. Acta. 2012;413:1058–1065. doi: 10.1016/j.cca.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 24.Zhao F.L., Hu G.D., Wang X.F., Zhang X.H., Zhang Y.K., Yu Z.S. Serum overexpression of microRNA-10b in patients with bone metastatic primary breast cancer. J. Int. Med. Res. 2012;40:859–866. doi: 10.1177/147323001204000304. [DOI] [PubMed] [Google Scholar]
- 25.Chan M., Liaw C.S., Ji S.M., Tan H.H., Wong C.Y., Thike A.A., Tan P.H., Ho G.H., Lee A.S. Identification of circulating microRNA signatures for breast cancer detection. Clin. Cancer Res. 2013;19:4477–4487. doi: 10.1158/1078-0432.CCR-12-3401. [DOI] [PubMed] [Google Scholar]
- 26.Cuk K., Zucknick M., Heil J., Madhavan D., Schott S., Turchinovich A., Arlt D., Rath M., Sohn C., Benner A., et al. Circulating microRNAs in plasma as early detection markers for breast cancer. Int. J. Cancer. 2013;132:1602–1612. doi: 10.1002/ijc.27799. [DOI] [PubMed] [Google Scholar]
- 27.Eichelser C., Flesch-Janys D., Chang-Claude J., Pantel K., Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17; miR-34a; miR-155; and miR-373 in human breast cancer development and progression. Clin. Chem. 2013;59:1489–1496. doi: 10.1373/clinchem.2013.205161. [DOI] [PubMed] [Google Scholar]
- 28.Godfrey A.C., Xu Z., Weinberg C.R., Getts R.C., Wade P.A., DeRoo L.A., Sandler D.P., Taylor J.A. Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. 2013;15:R42. doi: 10.1186/bcr3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S., Keerthana R., Pazhanimuthu A., Perumal P. Overexpression of circulating miRNA-21 and miRNA-146a in plasma samples of breast cancer patients. Indian J. Biochem. Biophys. 2013;50:210–214. [PubMed] [Google Scholar]
- 30.Ng E.K., Li R., Shin V.Y., Jin H.C., Leung C.P., Ma E.S., Pang R., Chua D., Chu K.M., Law W.L., et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS ONE. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Si H., Sun X., Chen Y., Cao Y., Chen S., Wang H., Hu C. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J. Cancer Res. Clin. Oncol. 2013;139:223–229. doi: 10.1007/s00432-012-1315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P.Y., Gong H.T., Li B.F., Lv C.L., Wang H.T., Zhou H.H., Li X.X., Xie S.Y., Jiang B.F. Higher expression of circulating miR-182 as a novel biomarker for breast cancer. Oncol. Lett. 2013;6:1681–1686. doi: 10.3892/ol.2013.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng R.C., Zhang W., Yan X.Q., Ye Z.Q., Chen E.D., Huang D.P., Zhang X.H., Huang G.L. Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med. Oncol. 2013;30:477. doi: 10.1007/s12032-013-0477-z. [DOI] [PubMed] [Google Scholar]
- 34.Eichelser C., Stückrath I., Müller V., Milde-Langosch K., Wikman H., Pantel K., Schwarzenbach H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650–9663. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamdi K., Goerlitz D., Stambouli N., Islam M., Baroudi O., Neili B., Benayed F., Chivi S., Loffredo C., Jillson I.A., et al. miRNAs in Sera of Tunisian patients discriminate between inflammatory breast cancer and non-inflammatory breast cancer. Springerplus. 2014;3:636. doi: 10.1186/2193-1801-3-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joosse S.A., Müller V., Steinbach B., Pantel K., Schwarzenbach H. Circulating cell-free cancer-testis MAGE-A RNA, BORIS RNA, let-7b and miR-202 in the blood of patients with breast cancer and benign breast diseases. Br. J. Cancer. 2014;111:909–917. doi: 10.1038/bjc.2014.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kodahl A.R., Lyng M.B., Binder H., Cold S., Gravgaard K., Knoop A.S., Ditzel H.J. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer; a case control study. Mol. Oncol. 2014;8:874–883. doi: 10.1016/j.molonc.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mar-Aguilar F., Rodríguez-Padilla C., Reséndez-Pérez D. Use of serum-circulating miRNA profiling for the identification of breast cancer biomarkers. Methods Mol. Biol. 2014;1165:71–80. doi: 10.1007/978-1-4939-0856-1_6. [DOI] [PubMed] [Google Scholar]
- 39.McDermott A.M., Miller N., Wall D., Martyn L.M., Ball G., Sweeney K.J., Kerin M.J. Identification and validation of oncologic miRNA biomarkers for luminal A-like breast cancer. PLoS ONE. 2014;9:e87032. doi: 10.1371/journal.pone.0087032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen J., Hu Q., Schrauder M., Yan L., Wang D., Medico L., Guo Y., Yao S., Zhu Q., Liu B., et al. Circulating miR-148b and miR-133a as biomarkers for breast cancer detection. Oncotarget. 2014;5:5284–5294. doi: 10.18632/oncotarget.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sochor M., Basova P., Pesta M., Dusilkova N., Bartos J., Burda P., Pospisil V., Stopka T. Oncogenic microRNAs, miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer. 2014;14:448. doi: 10.1186/1471-2407-14-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zearo S., Kim E., Zhu Y., Zhao J.T., Sidhu S.B., Robinson B.G., Soon P.S. MicroRNA-484 is more highly expressed in serum of early breast cancer patients compared to healthy volunteers. BMC Cancer. 2014;14:200. doi: 10.1186/1471-2407-14-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao F.L., Dou Y.C., Wang X.F., Han D.C., Lv Z.G., Ge S.L., Zhang Y.K. Serum microRNA-195 is down-regulated in breast cancer; a potential marker for the diagnosis of breast cancer. Mol. Biol. Rep. 2014;41:5913–5922. doi: 10.1007/s11033-014-3466-1. [DOI] [PubMed] [Google Scholar]
- 44.Antolín S., Calvo L., Blanco-Calvo M., Santiago M.P., Lorenzo-Patiño M.J., Haz-Conde M., Santamarina I., Figueroa A., Antón-Aparicio L.M., Valladares-Ayerbes M. Circulating miR-200c and miR-141 and outcomes in patients with breast cancer. BMC Cancer. 2015;15:297. doi: 10.1186/s12885-015-1238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X.X., Gao S.Y., Wang P.Y., Zhou X., Li Y.J., Yu Y., Yan Y.F., Zhang H.H., Lv C.J., Zhou H.H., et al. Reduced expression levels of let-7c in human breast cancer patients. Oncol. Lett. 2015;9:1207–1212. doi: 10.3892/ol.2015.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mangolini A., Ferracin M., Zanzi M.V., Saccenti E., Ebnaof S.O., Poma V.V., Sanz J.M., Passaro A., Pedriali M., Frassoldati A., et al. Diagnostic and prognostic microRNAs in the serum of breast cancer patients measured by droplet digital PCR. Biomark. Res. 2015;3:12. doi: 10.1186/s40364-015-0037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matamala N., Vargas M.T., González-Cámpora R., Miñambres R., Arias J.I., Menéndez P., Andrés-León E., Gómez-López G., Yanowsky K., Calvete-Candenas J., et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin. Chem. 2015;61:1098–1106. doi: 10.1373/clinchem.2015.238691. [DOI] [PubMed] [Google Scholar]
- 48.Shaker O., Maher M., Nassar Y., Morcos G., Gad Z. Role of microRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancer females. Gene. 2015;560:77–82. doi: 10.1016/j.gene.2015.01.062. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L., Xu Y., Jin X., Wang Z., Wu Y., Zhao D., Chen G., Li D., Wang X., Cao H., et al. A circulating miRNA signature as a diagnostic biomarker for non-invasive early detection of breast cancer. Breast Cancer Res. Treat. 2015;154:423–434. doi: 10.1007/s10549-015-3591-0. [DOI] [PubMed] [Google Scholar]
- 50.Frères P., Wenric S., Boukerroucha M., Fasquelle C., Thiry J., Bovy N., Struman I., Geurts P., Collignon J., Schroeder H., et al. Circulating microRNA-based screening tool for breast cancer. Oncotarget. 2016;7:5416–5428. doi: 10.18632/oncotarget.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu L., Li Z., Zhu J., Wang P., Fan G., Dai Y., Zheng Z., Liu Y. Serum expression levels of microRNA-382-3p, -598-3p, -1246 and -184 in breast cancer patients. Oncol. Lett. 2016;12:269–274. doi: 10.3892/ol.2016.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamam R., Ali A.M., Alsaleh K.A., Kassem M., Alfayez M., Aldahmash A., Alajez N.M. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci. Rep. 2016;6:25997. doi: 10.1038/srep25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hannafon B.N., Trigoso Y.D., Calloway C.L., Zhao Y.D., Lum D.H., Welm A.L., Zhao Z.J., Blick K.E., Dooley W.C., Ding W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Motawi T.M., Sadik N.A., Shaker O.G., El Masry M.R., Mohareb F. Study of microRNAs-21/221 as potential breast cancer biomarkers in Egyptian women. Gene. 2016;590:210–219. doi: 10.1016/j.gene.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 55.Shimomura A., Shiino S., Kawauchi J., Takizawa S., Sakamoto H., Matsuzaki J., Ono M., Takeshita F., Niida S., Shimizu C., et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107:326–334. doi: 10.1111/cas.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thakur S., Grover R.K., Gupta S., Yadav A.K., Das B.C. Identification of Specific miRNA Signature in Paired Sera and Tissue Samples of Indian Women with Triple Negative Breast Cancer. PLoS ONE. 2016;11:e0158946. doi: 10.1371/journal.pone.0158946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao S., Wang Y., Wang M., Li Z., Zhao Z., Wang R.X., Wu R., Yuan Z., Cui R., Jiao K., et al. MicroRNA-155; induced by FOXP3 through transcriptional repression of BRCA1; is associated with tumor initiation in human breast cancer. Oncotarget. 2017;8:41451–41464. doi: 10.18632/oncotarget.17816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang K., Wang Y.W., Wang Y.Y., Song Y., Zhu J., Si P.C., Ma R. Identification of microRNA biomarkers in the blood of breast cancer patients based on microRNA profiling. Gene. 2017;619:10–20. doi: 10.1016/j.gene.2017.03.038. [DOI] [PubMed] [Google Scholar]
- 59.Heydari N., Nikbakhsh N., Sadeghi F., Farnoush N., Khafri S., Bastami M., Parsian H. Overexpression of serum MicroRNA-140-3p in premenopausal women with newly diagnosed breast cancer. Gene. 2018;655:25–29. doi: 10.1016/j.gene.2018.02.032. [DOI] [PubMed] [Google Scholar]
- 60.Zaleski M., Kobilay M., Schroeder L., Debald M., Semaan A., Hettwer K., Uhlig S., Kuhn W., Hartmann G., Holdenrieder S. Improved sensitivity for detection of breast cancer by combination of miR-34a and tumor markers CA 15-3 or CEA. Oncotarget. 2018;9:22523–22536. doi: 10.18632/oncotarget.25077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kahraman M., Röske A., Laufer T., Fehlmann T., Backes C., Kern F., Kohlhaas J., Schrörs H., Saiz A., Zabler C., et al. MicroRNA in diagnosis and therapy monitoring of early-stage triple-negative breast cancer. Sci. Rep. 2018;8:11584. doi: 10.1038/s41598-018-29917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McAnena P., Tanriverdi K., Curran C., Gilligan K., Freedman J.E., Brown J.A.L., Kerin M.J. Circulating microRNAs miR-331 and miR-195 differentiate local luminal a from metastatic breast cancer. BMC Cancer. 2019;19:436. doi: 10.1186/s12885-019-5636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peña-Cano M.I., Saucedo R., Morales-Avila E., Valencia J., Zavala-Moha J.A., López A. Deregulated microRNAs and Adiponectin in Postmenopausal Women with Breast Cancer. Gynecol. Obstet. Investig. 2019;84:369–377. doi: 10.1159/000496340. [DOI] [PubMed] [Google Scholar]
- 64.Raheem A.R., Abdul-Rasheed O.F., Al-Naqqash M.A. The diagnostic power of circulating micro ribonucleic acid 34a in combination with cancer antigen 15-3 as a potential biomarker of breast cancer. Saudi Med. J. 2019;40:1218–1226. doi: 10.15537/smj.2019.12.24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soleimanpour E., Babaei E., Hosseinpour-Feizi M.A., Montazeri V. Circulating miR-21 and miR-155 as potential noninvasive biomarkers in Iranian Azeri patients with breast carcinoma. J. Cancer Res. Ther. 2019;15:1092–1097. doi: 10.4103/jcrt.JCRT_1227_16. [DOI] [PubMed] [Google Scholar]
- 66.Anwar S.L., Tanjung D.S., Fitria M.S., Kartika A.I., Sari D.N.I., Rakhmina D., Wardana T., Astuti I., Haryana S.M., Aryandono T. Dynamic Changes of Circulating Mir-155 Expression and the Potential Application as a Non-Invasive Biomarker in Breast Cancer. Asian Pac. J. Cancer Prev. 2020;21:491–497. doi: 10.31557/APJCP.2020.21.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arabkari V., Clancy E., Dwyer R.M., Kerin M.J., Kalinina O., Holian E., Newell J., Smith T.J. Relative and Absolute Expression Analysis of MicroRNAs Associated with Luminal A Breast Cancer- A Comparison. Pathol. Oncol. Res. 2020;26:833–844. doi: 10.1007/s12253-019-00627-y. [DOI] [PubMed] [Google Scholar]
- 68.Ashirbekov Y., Abaildayev A., Omarbayeva N., Botbayev D., Belkozhayev A., Askandirova A., Neupokoyeva A., Utegenova G., Sharipov K., Aitkhozhina N. Combination of circulating miR-145-5p/miR-191-5p as biomarker for breast cancer detection. Peer J. 2020;8:e10494. doi: 10.7717/peerj.10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo H., Zeng X., Li H., Guo Y., Wang T., Guo H., Zhu G., Wang L., Zhou H., Liu K., et al. Plasma miR-1273g-3p acts as a potential biomarker for early Breast Ductal Cancer diagnosis. An. Acad Bras. Cienc. 2020;92:e20181203. doi: 10.1590/0001-3765202020181203. [DOI] [PubMed] [Google Scholar]
- 70.Holubekova V., Kolkova Z., Grendar M., Brany D., Dvorska D., Stastny I., Jagelkova M., Zelinova K., Samec M., Liskova A., et al. Pathway Analysis of Selected Circulating miRNAs in Plasma of Breast Cancer Patients; A Preliminary Study. Int. J. Mol. Sci. 2020;21:7288. doi: 10.3390/ijms21197288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hosseini Mojahed F., Aalami A.H., Pouresmaeil V., Amirabadi A., Qasemi Rad M., Sahebkar A. Clinical Evaluation of the Diagnostic Role of MicroRNA-155 in Breast Cancer. Int. J. Genom. 2020;2020:9514831. doi: 10.1155/2020/9514831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ibrahim A.M., Said M.M., Hilal A.M., Medhat A.M., AbdElsalam I.M. Candidate circulating microRNAs as potential diagnostic and predictive biomarkers for the monitoring of locally advanced breast cancer patients. Tumour. Biol. 2020;42:1010428320963811. doi: 10.1177/1010428320963811. [DOI] [PubMed] [Google Scholar]
- 73.Jang J.Y., Kim Y.S., Kang K.N., Kim K.H., Park Y.J., Kim C.W. Multiple microRNAs as biomarkers for early breast cancer diagnosis. Mol. Clin. Oncol. 2020;14:31. doi: 10.3892/mco.2020.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J., Park S., Hwang D., Kim S.I., Lee H. Diagnostic Value of Circulating miR-202 in Early-Stage Breast Cancer in South Korea. Medicina. 2020;56:340. doi: 10.3390/medicina56070340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pastor-Navarro B., García-Flores M., Fernández-Serra A., Blanch-Tormo S., Martínez de Juan F., Martínez-Lapiedra C., Maia de Alcantara F., Peñalver J.C., Cervera-Deval J., Rubio-Briones J., et al. A Tetra-Panel of Serum Circulating miRNAs for the Diagnosis of the Four Most Prevalent Tumor Types. Int. J. Mol. Sci. 2020;21:2783. doi: 10.3390/ijms21082783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bakr N.M., Mahmoud M.S., Nabil R., Boushnak H., Swellam M. Impact of circulating miRNA-373 on breast cancer diagnosis through targeting VEGF and cyclin D1 genes. J. Genet. Eng. Biotechnol. 2021;19:84. doi: 10.1186/s43141-021-00174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diansyah M.N., Prayogo A.A., Sedana M.P., Savitri M., ZakyRomadhon P., NikenAyu Amrita P., Yasmin Wijaya A., May Hendrata W., YudhoBintoro U. Early detection breast cancer; role of circulating plasma miRNA-21 expression as a potential screening biomarker. Turk. J. Med. Sci. 2021;51:562–569. doi: 10.3906/sag-2005-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Itani M.M., Nassar F.J., Tfayli A.H., Talhouk R.S., Chamandi G.K., Itani A.R.S., Makoukji J., Boustany R.N., Hou L., Zgheib N.K., et al. A Signature of Four Circulating microRNAs as Potential Biomarkers for Diagnosing Early-Stage Breast Cancer. Int. J. Mol. Sci. 2021;22:6121. doi: 10.3390/ijms22116121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahmed Mohmmed E., Shousha W.G., El-Saiid A.S., Ramadan S.S. A Clinical Evaluation of Circulating MiR-106a and Raf-1 as Breast Cancer Diagnostic and Prognostic Markers. Asian Pac. J. Cancer Prev. 2021;22:3513–3520. doi: 10.31557/APJCP.2021.22.11.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nashtahosseini Z., Aghamaali M.R., Sadeghi F., Heydari N., Parsian H. Circulating status of microRNAs 660-5p and 210-3p in breast cancer patients. J. Gene Med. 2021;23:e3320. doi: 10.1002/jgm.3320. [DOI] [PubMed] [Google Scholar]
- 81.Zhang K., Wang Y.Y., Xu Y., Zhang L., Zhu J., Si P.C., Wang Y.W., Ma R. A two-miRNA signature of upregulated miR-185-5p and miR-362-5p as a blood biomarker for breast cancer. Pathol. Res. Pract. 2021;222:153458. doi: 10.1016/j.prp.2021.153458. [DOI] [PubMed] [Google Scholar]
- 82.Zhao T., Meng W., Chin Y., Gao L., Yang X., Sun S., Pan X., He L. Identification of miR-25-3p as a tumor biomarker; Regulation of cellular functions via TOB1 in breast cancer. Mol. Med. Rep. 2021;23:406. doi: 10.3892/mmr.2021.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X., Tang X., Li K., Lu L. Evaluation of Serum MicroRNAs (miR-9-5p, miR-17-5p, and miR-148a-3p) as Potential Biomarkers of Breast Cancer. Biomed. Res. Int. 2022;2022:9961412. doi: 10.1155/2022/9961412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu H., Bian Q.Z., Zhang W., Cui H.B. Circulating microRNA-103a-3p could be a diagnostic and prognostic biomarker for breast cancer. Oncol. Lett. 2022;23:38. doi: 10.3892/ol.2021.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohamed A.A., Allam A.E., Aref A.M., Mahmoud M.O., Eldesoky N.A., Fawazy N., Sakr Y., Sobeih M.E., Albogami S., Fayad E., et al. Evaluation of Expressed MicroRNAs as Prospective Biomarkers for Detection of Breast Cancer. Diagnostics. 2022;12:789. doi: 10.3390/diagnostics12040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Záveský L., Jandáková E., Weinberger V., Minář L., Hanzíková V., Dušková D., Faridová A., Turyna R., Slanař O., Hořínek A., et al. Small non-coding RNA profiling in breast cancer: plasma U6 snRNA, miR-451a and miR-548b-5p as novel diagnostic and prognostic biomarkers. Mol. Biol. Rep. 2022;49:1955–1971. doi: 10.1007/s11033-021-07010-8. [DOI] [PubMed] [Google Scholar]
- 87.Zou R., Loke S.Y., Tang Y.C., Too H.P., Zhou L., Lee A.S.G., Hartman M. Development and validation of a circulating microRNA panel for the early detection of breast cancer. Br. J. Cancer. 2022;126:472–481. doi: 10.1038/s41416-021-01593-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ali A.S., Ali S., Ahmad A., Bao B., Philip P.A., Sarkar F.H. Expression of microRNAs; potential molecular link between obesity; diabetes and cancer. Obes. Rev. 2011;12:1050–1062. doi: 10.1111/j.1467-789X.2011.00906.x. [DOI] [PubMed] [Google Scholar]
- 89.Murri M., Insenser M., Fernández-Durán E., San-Millán J.L., Escobar-Morreale H.F. Effects of polycystic ovary syndrome (PCOS); sex hormones; and obesity on circulating miRNA-21; miRNA-27b; miRNA-103; and miRNA-155 expression. J. Clin. Endocrinol. Metab. 2013;98:E1835–E1844. doi: 10.1210/jc.2013-2218. [DOI] [PubMed] [Google Scholar]
- 90.Chadeau-Hyam M., Athersuch T.J., Keun H.C., De Iorio M., Ebbels T.M., Jenab M., Sacerdote C., Bruce S.J., Holmes E., Vineis P. Meeting-in-the-middle using metabolic profiling—A strategy for the identification of intermediate biomarkers in cohort studies. Biomarkers. 2011;16:83–88. doi: 10.3109/1354750X.2010.533285. [DOI] [PubMed] [Google Scholar]
- 91.Frankel L.B., Christoffersen N.R., Jacobsen A., Lindow M., Krogh A., Lund A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 92.Banerjee N., Talcott S., Safe S., Mertens-Talcott S.U. Cytotoxicity of pomegranate polyphenolics in breast cancer cells in vitro and vivo; potential role of miRNA-27a and miRNA-155 in cell survival and inflammation. Breast Cancer Res. Treat. 2012;136:21–34. doi: 10.1007/s10549-012-2224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ali S.A., Abdulrahman Z.F.A., Faraidun H.N. Circulatory miRNA-155; miRNA-21 target PTEN expression and activity as a factor in breast cancer development. Cell Mol. Biol. 2020;66:44–50. doi: 10.14715/cmb/2020.66.7.8. [DOI] [PubMed] [Google Scholar]
- 94.Sonkoly E., Ståhle M., Pivarcsi A. MicroRNAs and immunity; novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 95.Wang K., Yuan Y., Cho J.H., McClarty S., Baxter D., Galas D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE. 2012;7:e41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sato F., Tsuchiya S., Terasawa K., Tsujimoto G. Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PloS ONE. 2009;4:e5540. doi: 10.1371/journal.pone.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.