Abstract
Selective disintegration of membrane-enclosed autophagic bodies is a feature of eukaryotic cells not studied in detail. Using a Saccharomyces cerevisiae mutant defective in autophagic-body breakdown, we identified and characterized Aut5p, a glycosylated integral membrane protein. Site-directed mutagenesis demonstrated the relevance of its putative lipase active-site motif for autophagic-body breakdown. aut5Δ cells show reduced protein turnover during starvation and are defective in maturation of proaminopeptidase I. Most recently, by means of the latter phenotype, Aut5p was independently identified as Cvt17p. In this study we additionally checked for effects on vacuolar acidification and detected mature vacuolar proteases, both of which are prerequisites for autophagic-body lysis. Furthermore, biologically active hemagglutinin-tagged Aut5p (Aut5-Ha) localizes to the endoplasmic reticulum (nuclear envelope) and is targeted to the vacuolar lumen independent of autophagy. In pep4Δ cells immunogold electron microscopy located Aut5-Ha at ∼50-nm-diameter intravacuolar vesicles. Characteristic missorting in vps class E and fab1Δ cells, which affects the multivesicular body (MVB) pathway, suggests vacuolar targeting of Aut5-Ha similar to that of the MVB pathway. In agreement with localization of Aut5-Ha at intravacuolar vesicles in pep4Δ cells and the lack of vacuolar Aut5-Ha in wild-type cells, our pulse-chase experiments clearly indicated that Aut5-Ha degradation with 50 to 70 min of half-life is dependent on vacuolar proteinase A.
All eukaryotic cells use autophagy as a transport pathway to deliver part of their own intracellular constituents to the lysosome (vacuole) for degradation. Autophagy helps cells to survive periods of nutrient limitation by recycling amino acids and other cellular building blocks. Autophagy has initially been studied in mammalian cells (7). During recent years our mechanistic and molecular knowledge of autophagy emerged due to work using Saccharomyces cerevisiae as a model of mammalian cells (21, 19, 38, 36). Autophagy starts with the formation of autophagosomes, double-membrane-layered vesicles, which enclose cytosol, and even whole organelles such as mitochondria. After fusion of autophagosomes with the vacuolar membrane, monolayered autophagic bodies are released into the vacuole, where they are broken down together with their cytosolic contents (3, 33). The breakdown of vesicles significantly distinguishes autophagy from classical protein transport pathways. Classical vesicle-mediated protein transport pathways such as secretion utilize monolayered vesicles, which after fusion with the target membrane directly release their contents. Autophagy, however, uses double-membrane-layered autophagosomes, which release still-membrane-enclosed autophagic bodies into the vacuole. Disintegration of their limiting membrane must occur to allow the vacuolar hydrolases access to the cytosolic content.
Lysis of membranes has been recognized as an important step in the pathogenesis of microorganisms such as Listeria spp., which escape from the phagosome into the host cell cytosol using a pore-forming cytolysin and two phospholipases (12). However, as a feature of the normal life cycle of eukaryotic cells, disintegration of membranes, especially of autophagic bodies, has not been studied in detail so far. To break down membranes is a challenging task for the cell, since a failure in selectivity and control implies an enormous risk for cellular life.
The breakdown of autophagic bodies depends on proteinases A and B (33, 37) and on vacuolar acidification (23). How proteinases alone should be able to break down vesicles enclosed by a lipid membrane remains elusive, however. To answer this question, we isolated S. cerevisiae aut4 and aut5 mutants with a defect in the breakdown of autophagic bodies. AUT4 encodes a protein with limited homologies to permeases (32). Here we identify and characterize Aut5p as a glycosylated integral membrane protein. Aut5p contains a lipase active-site motif, which we demonstrate by site-directed mutagenesis to be essential for the breakdown of autophagic bodies. While preparing this paper, AUT5 was independently identified as CVT17 based on its essential function in the maturation of proaminopeptidase I (35). In growing cells proaminopeptidase I reaches the vacuole enclosed in Cvt bodies, which resemble autophagic bodies but are smaller in size and exclude cytosol (2). Consistent with our findings, Aut5p/Cvt17p was found to be required for the lysis of Cvt bodies and the importance of the lipase active-site serine was shown (35). Our study significantly extends this characterization of Aut5/Cvt17p by focusing on autophagic aspects. Vacuolar acidification and the presence of mature vacuolar proteases are prerequisites for the breakdown of autophagic bodies. Here we additionally check aut5Δ cells for these phenotypes. Furthermore, using a biologically active hemagglutinin-tagged version (Aut5-Ha), we augment the results of the Cvt17 study (35) by localizing the protein at the endoplasmic reticulum (ER) (nuclear envelope) and demonstrate its targeting to the vacuolar lumen at ∼50-nm-diameter vesicles independent of autophagy. Localization in 50-nm-diameter vesicles in the vacuolar lumen of pep4Δ cells together with a characteristic missorting in vps class E and fab1Δ cells, which affects the multivesicular body (MVB) pathway (24, 25), suggests that Aut5-Ha reaches the vacuole by a means similar to the MVB pathway.
The MVB pathway has been implicated in the vacuolar targeting of the integral membrane protein procarboxypeptidase S (proCPS) (24, 25, 30). After entry in the ER, proCPS is sorted from the Golgi apparatus to the vacuole via a prevacuolar compartment (prevacuolar endosome). From the prevacuolar compartment, membrane proteins are normally delivered to the limiting vacuolar membrane. ProCPS, however, is sorted by the novel MVB pathway to 50-nm-diameter internal vesicles. The MVB sorting pathway (24) starts at the prevacuolar endosome, where first invaginations into the lumen of the organelle are formed, followed by specific sorting of integral membrane proteins such as proCPS to these invaginations. Next vesicles pinch off from the limiting membrane of the prevacuolar endosome and bud into its lumen. This leads to the formation of an MVB, a prevacuolar endosome filled with internal vesicles carrying specific cargo molecules such as proCPS (24). Then the multivesicular body fuses with the vacuole, thus releasing its internal vesicles into the vacuolar lumen. Finally, in the vacuole the 50-nm-diameter vesicles are thought to be broken down (24). Our localization of Aut5-Ha at 50-nm-diameter intravacuolar vesicles and the lack of vacuolar Aut5-Ha in wild-type cells, together with the results of our pulse-chase experiments, clearly indicate Aut5-Ha degradation dependent on vacuolar proteinase A, with a half-life of 50 to 70 min.
MATERIALS AND METHODS
Strains, media, and materials.
Standard media were prepared according to the methods described in reference 1; SMD medium contains 0.67% Bacto Yeast Nitrogen Base, 2% glucose, and required supplements. If not otherwise mentioned, starvation was done in 1% potassium acetate.
Strains used are listed in Table 1. A PCR fragment consisting of the canamycin resistance gene flanked by AUT5 sequences was generated using oligonucleotides aut5-1 (CATAAAAGCC CTTCAAGAAA GAGATTTGCT TCTCCTTTGC ATCTACAGCT GAAGCTTCGT ACGC) and aut5-2 (ACACTCCAGC CCTTGTCGTT GACCACATCG TAGACACACA CTCTAGCATA GGCCACTAGT GGATCTG) and plasmid pUG6 (13, 39). Chromosomal replacement of AUT5 by this fragment in WCG4a yielded YIS4. Correct gene replacement was confirmed by Southern blotting (not shown). YUE5 was generated by chromosomal integration of a fragment consisting of a triple Ha (Ha3) tag and a Schizosaccharomyces pombe HIS5 marker (6) in WCG4a. The fragment was created by PCR using plasmid p3×HA-HIS5 (S. Munroe, Cambridge, United Kingdom) and primer AUT5his1 (GTAGGCCGCA ATTGGCTTGG CTTCTGCACC AAATACGAGT TGCATCATCA TCATCATCAT GGAGCAGGGG CGGGTGC) and AUT5his2 (GGCCCTAAAA CAACACTAGG GTCATAATAG ATGTATGGGT CGAGGTCGAC GGTATCGATA AG). Positive transformants were selected on plates lacking histidine. Southern blotting confirmed correct gene replacement (not shown). YUE14 and YUE15 are ascospores from a cross of YUE5 and YMTA. Crossing YUE15 and YMS30 and subsequent tetrad dissection yielded YUE41 and YUE43.
TABLE 1.
Yeast strains used in this study
| Strain | Genotypea | Reference or source |
|---|---|---|
| WCG4a | matα his3–11,15 leu2–3,112 ura3 | 37 |
| YMTA | mata WCG4a pep4Δ::HIS3 | 37 |
| YMTAB | matα WCG4a pep4Δ::HIS3 prb1ΔAV | 37 |
| YMS30 | matα WCG4a aut3Δ::KAN | 31 |
| YIS4 | matα WCG4a aut5Δ::KAN | This study |
| YIS8 | matα WCG4a aut5Δ::KAN pep4Δ::HIS3 | This study |
| YIS10 | matα WCG4a aut5Δ::KAN aut1Δ::URA3 | This study |
| YUE5 | matα WCG4a AUT5::Ha3::HIS5 | This study |
| YUE14 | matα WCG4a pep4Δ::HIS3 AUT5::Ha3::HIS5 | This study |
| YUE15 | mata WCG4a pep4Δ::HIS3 AUT5::Ha3::HIS5 | This study |
| YUE41 | matα WCG4a aut3Δ pep4Δ AUT5::Ha3::HIS5 | This study |
| YUE43 | matα WCG4a aut3Δ AUT5::Ha3::HIS5 | This study |
| YR101 | mataade2 his3 leu2 lys2 ura3 | 32 |
| YR393 | mata YR101 tfp1 (vma1)::URA3 | 32 |
| BY4742 | matα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | 4 |
| Y17080 | matα BY4742 fab1Δ::kanMX4 | Euroscarf collection |
| Y15588 | matα BY4742 vps4Δ::kanMX4 | Euroscarf collection |
| Y15381 | matα BY4742 vps27Δ::kanMX4 | Euroscarf collection |
| Y12763 | matα BY4742 vps28Δ::kanMX4 | Euroscarf collection |
| DYY101 | matα leu2–3, 112 ura3–52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 ape1Δ::LEU2 | 20 |
We used antibodies to Ha clone 16B12 (BabCo); Vph1p, Vma2p, and 3-phosphoglycerate kinase (PGK) (Molecular Probes, Leiden, The Netherlands); and CPS (S. D. Emr, La Jolla, Calif.). We also used horseradish peroxidase-conjugated goat-anti rabbit antibodies (Medac, Hamburg, Germany) and horseradish peroxidase-conjugated goat anti-mouse antibodies (Dianova, Hamburg, Germany).
The following chemicals were used: Zymolyase-100T (Seikagaku, Tokyo, Japan), DNA-modifying enzymes (Roche, Basel, Switzerland), and oligonucleotides (MWG-Biotec, Ebersberg, Germany). All other chemicals were of analytical grade and purchased from Sigma (Deisenhofen, Germany) or Merck (Darmstadt, Germany).
For detection of peroxidase-labeled antibodies on immunoblots, an ECL detection kit (Amersham, Braunschweig, Germany) was used.
Plasmids and DNA manipulation.
For isolation of the complementing genomic plasmid p303/I1, we followed a previously described procedure (31).
Digestion of p303/I1 with XbaI yielded a 4.2-kb fragment, which was ligated into the XbaI site of the centromeric plasmid pRS316 to yield pUE05 (pRS316-AUT5). pUE05 was digested with NotI and SalI, and the resulting AUT5-containing fragment (4.3 kb) was ligated into the 2μm plasmid pRS426 (cut with NotI and SalI) to yield pUE06 (pRS426-AUT5). Primers MUTup (AGAGGACGTATCGTAGGGAATTGTGATGCTTG) and MUTdown (TCCAGTTTAAACGAGCTCGAATTCCTATTAGC) were used to amplify by PCR the AUT5::Ha3 cassette with its native promoter by using chromosomal DNA of strain YUE05 as the template. The PCR product was ligated into the SmaI site of pRS426 to yield pUE07 (pRS426-AUT5::HA3). pUE07 was cut with BamHI and HindIII, and the resulting 2.6-kb fragment was ligated into pRS316 cut with BamHI and HindIII to yield pUE13 (pRS316-AUT5::HA3).
Total protein turnover.
We followed an established protocol to measure total protein turnover (31). Cells were grown at 30°C to 5 × 107/ml in 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% proline, 2% glucose, and required auxotrophic nutrients were pulsed with [35S]methionine during the last 14 h of growth. After transfer to 1% K acetate with 10 mM methionine and incubation at 30°C, samples were drawn and precipitated with trichloroacetic acid. After centrifugation, the radioactivity released to the supernatant was measured with a scintillation counter (Wallac 1410; Pharmacia). Protein breakdown was calculated as increase of soluble radioactivity divided by insoluble radioactivity at 0 h.
Quinacrine staining.
Logarithmically growing cells and cells starved for 4 h in 1% potassium acetate were harvested and washed in 10 mM HEPES–2% glucose (pH 7.4). Cells were then incubated in the same buffer for 3 to 5 min with 200 μM quinacrine (27) and washed with buffer.
Glass bead lysis and membrane association.
Cells were grown to stationary phase and starved for 4 h in 1% potassium acetate. Sixty optical density units at 600 nm (OD600) of cells was harvested, resuspended in breaking buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA) containing Complete protease inhibitors (Roche) and 1 mM phenylmethylsulfonyl fluoride, and vortexed with glass beads at 4°C for 10 min. To remove cell debris, the samples were centrifuged for 5 min at 500 × g. One volume of supernatant was added to 1 volume each of (i) breaking buffer, (ii) 2 M K acetate, (iii) 0.2 M Na2CO3, (iv) 2.5 M urea, and (v) 2% Triton X-100 and incubated on ice for 45 min. Membranes were pelleted by centrifugation at 100,000 × g for 45 min. The supernatant was precipitated with trichloroacetic acid. Pellets (membrane fraction) and trichloroacetic acid precipitates were resuspended in sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Deglycosylation.
Thirty OD600 units of cells starved for 4 h was harvested, washed twice with cold TBS (20 mM Tris-HCl [pH 7.6], 200 mM NaCl), resuspended in 400 μl of lysis buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% sodium deoxycholate, 2% Triton X-100, 0.1% SDS) supplemented with protease inhibitors, and lysed with glass beads (see above). After cell debris was removed, the supernatant was divided in two aliquots and immunoprecipitated with anti-Ha antibody for 3 h at 4°C, followed by 1 h of incubation with protein A-Sepharose (Pharmacia). The samples were washed twice with lysis buffer and once with wash buffer (50 mM KH2PO4 [pH 5.5], 0.02% SDS). Twenty-five microliters of wash buffer containing 0.1 M β-mercaptoethanol was added to the protein A-Sepharose pellets. After addition of 15 mU of endoglycosidase H (Roche) or mock treatment, the samples were incubated at 37°C for 1 h. Sample buffer was added, and the samples were analyzed by SDS-PAGE and immunoblotting.
Pulse-chase analysis of Aut5-Ha.
Cells were grown to mid-log phase in SMD, and for each time point 2 OD600 units was collected. Cells resuspended in fresh SMD were pulse-labeled for 20 min (20 μCi of Expre35S35S-label [NEN, Boston, Mass.] per OD600 unit). Labeled cells were then suspended at 2 OD600 units/ml in SMD containing 0.2% yeast extract, 6 mg of methionine per ml, 3 mg of cysteine per ml, and 1 mg of bovine serum albumin per ml and chased at 30°C. At the times indicated in the figures, 2 OD600 units was transferred to a chilled tube and sodium azide was added to a final concentration of 10 mM. Cells were washed with cold 10 mM sodium azide and resuspended in 100 μl of lysis buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% sodium deoxycholate, 2% Triton X-100, 0.1% SDS). Lysis was done by vigorously vortexing the suspension four times for 1 min each time with glass beads and intermittent cooling for 1 min. For solubilization, 900 μl of lysis buffer was added and the solution was vortexed again and incubated on ice for 30 min. After insoluble material was removed by centrifugation, 5 μl of antiserum to Ha was added to the supernatant and incubated at room temperature for 1 h. Then 70 μl of protein A-Sepharose beads (0.5% in lysis buffer) was added, and the mixture was again incubated for 1 h at room temperature on a rotary mixer. Immune complexes bound to the beads were washed three times with lysis buffer, and samples were deglycosylated as described above. After endoglycosidase H treatment, sample buffer was added, the mixture was incubated for 20 min at 37°C, and 25 μl of the immunoprecipitated protein corresponding to 1 OD600 unit of cells was loaded onto a 7% polyacrylamide–SDS gel. After electrophoresis, the dried gel was visualized using a Storm PhosphorImager (Molecular Dynamics).
Indirect immunofluorescence.
Immunofluorescence was performed as described previously (9) with the following modifications. Approximately 10 OD600 units of starved cells was fixed by adding directly to the culture formaldehyde to a final concentration of 3.5% and 1 M potassium phosphate (pH 6.5) to a final concentration of 100 mM. After 2 h of fixation at room temperature, the cells were washed twice with buffer I (1.2 M sorbitol, 0.1 M KH2PO4 [pH 6.5]), resuspended in buffer I containing 20 mM β-mercaptoethanol and 45 μg of Zymolyase-100T per ml, and spheroplasted at 30°C for 30 min. Cells were first labeled with mouse anti-Ha antibody; as a secondary antibody Cy3-conjugated goat-anti-mouse immunoglobulin G (Dianova) was used. Cells were covered with 1 drop of mounting solution containing DAPI (4′,6′-diamidino-2-phenylindole, 0.4 μg/ml) and visualized with a Zeiss Axioskop 2 Plus equipped with an Axiocam digital image system.
Electron microscopy.
For immunogold electron microscopy, cells were fixed in 4% paraformaldehyde in 0.1 M citrate buffer, embedded in 10% gelatin in phosphate-buffered saline, and infiltrated in 2.1 M sucrose and 20% polyvinylpyrrolidone. Small blocks were mounted on specimen holders and frozen in liquid nitrogen. Thin sections were cut with a diamond knife at −100°C, mounted on grids, and immunolabeled with mouse anti-Ha followed by goat F(ab)2 coupled to 10-nm-thick gold. The sections were contrasted and embedded in methyl cellulose-uranyl acetate.
Electron microscopy using permanganate fixation and Epon embedding were done according to previously published procedures (22).
Site-directed mutagenesis.
Aut5(S332A)-Ha was generated by PCR. For the first PCR, a mutagenic primer binding 14 bp upstream and 17 bp downstream of position 994 of the AUT5 coding region with a guanine instead of a thymine in between (GGGTCACAGGCCACgCACTGGGAGGCGCATTG, where “g” is the guanine in question) and a second primer complementary to a region downstream of AUT5-Ha (MUTdown, see above) were used to amplify a megaprimer (736 bp) with chromosomal DNA of strain YUE05 as a template. In a second PCR, with plasmid pUE05 as a template, this megaprimer and a primer binding to a region 844 to 825 bp upstream of AUT5 (MUTup, see above) were used to amplify the entire AUT5(S332A)-Ha gene, including its native promoter. The PCR product was subcloned into the SmaI site of pRS426, resulting in plasmid pUE09 [pRS426-AUT5(S332A)-Ha]. Sequencing confirmed the correct sequence of the mutated AUT5-Ha gene.
RESULTS
Identification of the AUT5 gene.
After EMS mutagenesis of S. cerevisiae cells, we previously isolated a set of putative autophagy mutants based on their defect in starvation-induced protein breakdown (37). We further screened these strains after a starvation period for nitrogen by light microscopy for the accumulation of autophagic bodies in the vacuole and thus identified aut5-1 mutant cells, which exhibit a defect in the breakdown of autophagic bodies (not shown) (14).
Here we now report a significantly reduced sporulation frequency of homozygous aut5-1 diploid cells. After transformation with a YCp50-based genomic library (28), we looked for reversion of this phenotype using a previously described procedure (22, 31) based on the random spore protocol to isolate a plasmid carrying a complementing genomic insert. Partial sequencing localized the genomic insert to chromosome III and pointed to YCR068w as the corresponding open reading frame (Fig. 1A). We generated a chromosomal deletion of YCR068w and confirmed correct gene replacement by Southern blotting (not shown). Crossing and subsequent tetrad dissection of aut5-1 ycr068wΔ cells confirmed the identity of YCR068w with AUT5. aut5-1 ycr068wΔ diploid cells showed a significantly reduced sporulation frequency, but enough asci were formed to allow tetrad dissection. aut5Δ cells grow normally at 30°C on rich medium.
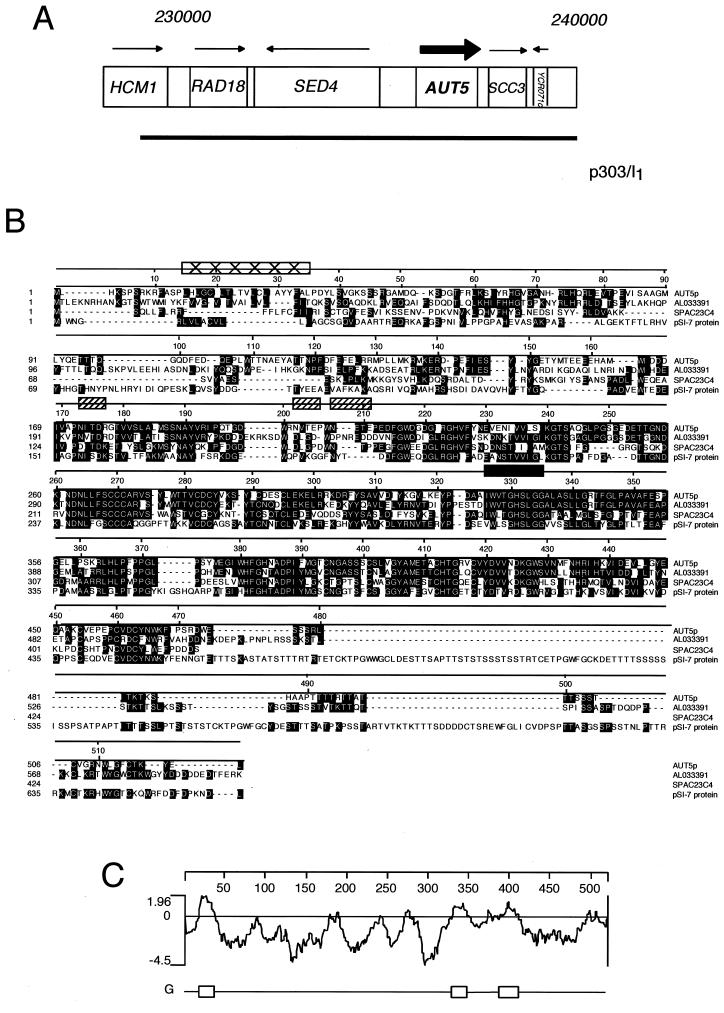
FIG. 1.
Identification and characteristics of AUT5. (A) The AUT5 genetic locus and the isolated complementing genomic fragment are shown. (B) Aut5p shares homologies with AL033391 from C. albicans, SPAC23C4 from S. pombe, and pSI-7 from Cladosporium fulvum. Aut5p contains a putative lipase active-site motif from amino acids 326 to 335 (filled rectangle). Box filled with ×'s, potential transmembrane domain; hatched boxes, potential glycosylation sites. Alignment was created using CLUSTAL, and identical residues are shaded. (C) Hydrophobicity blot of Aut5p generated by using the method of Engelman et al. (10). One potential transmembrane region is predicted between amino acids 15 and 35. Regions with less pronounced hydrophobicity are found from amino acids 324 to 348 and 388 to 408.
Aut5p is a putative membrane protein containing a lipase active-site motif.
Resequencing of AUT5 unraveled a sequencing error in the databases, which was corrected in the latest release. AUT5 encodes a protein of 520 amino acids (Fig. 1B) with a predicted molecular mass of 58 kDa. Aut5p shares 43% identity with AL033391 from C. albicans, 39% identity with SPAC23C4 of S. pombe, and 31% identity with PSI-7, a gene identified in Cladosporium fulvum for its induced expression during starvation and pathogenic growth on tomato plants (5).
Aut5p contains one to three potential transmembrane domains (Fig. 1C). The first ranges from amino acids 15 to 35 and is predicted to be a signal or stop transfer peptide, which might function as an ER import sequence. Two further regions with less pronounced hydrophobicity from amino acids 324 to 348 and 388 to 408 are not identified as putative transmembrane domains by all programs. Most interestingly, a pattern search identified a lipase active-site motif from amino acids 326 to 335 (Prosite accession no. PS00120) (Fig. 1B). This potential lipase active site is located within the second hydrophobic domain. Furthermore, three potential Asn glycosylation sites are predicted.
Aut5p is essential for the breakdown of autophagic bodies in the vacuole.
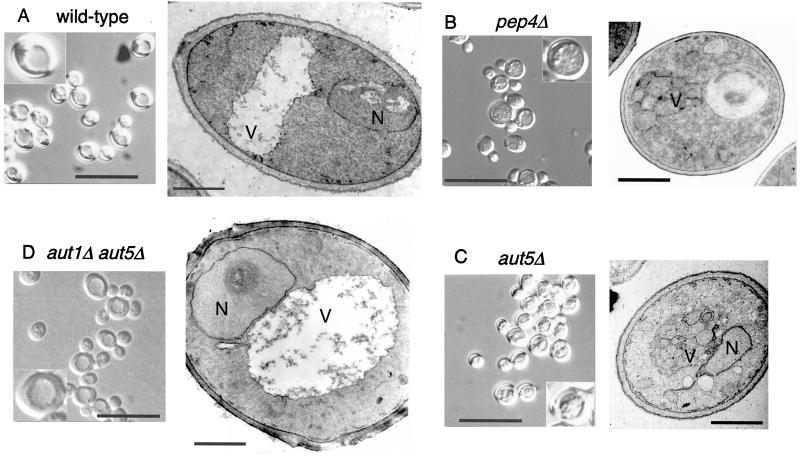
In the vacuoles of wild-type cells autophagic bodies are rapidly broken down dependent on the presence of vacuolar proteinases (Fig. 2A). In contrast, aut5Δ cells grown into late stationary phase or starved for nitrogen accumulate 300- to 400-nm-diameter vesicles in the vacuole (Fig. 2C), similar to cells lacking proteinase A (Fig. 2B). AUT1 is essential for formation of autophagosomes (29) (16). To confirm the autophagic nature of the vesicles accumulating in starved aut5Δ cells, we constructed an aut1Δ aut5Δ double mutant. As expected, these cells lack the starvation-induced accumulation of 300- to 400-nm-diameter vesicles inside the vacuole (Fig. 2D).
FIG. 2.
In starved aut5Δ cells autophagic bodies accumulate in the vacuole. The cells were grown in yeast extract-peptone-dextrose and starved for nitrogen for 4 to 5 h in 1% potassium acetate medium at 30°C. The cells were then checked by light microscopy with Nomarski optics (bar in left panel, 20 μm; insets are enlarged twofold) or prepared for electron microscopy by permanganate fixation and epon embedding (bar in right panel, 1 μm). N, nucleus; V, vacuole.
aut5Δ cells exhibit phenotypes typical for cells defective in autophagy or vacuolar proteolysis.
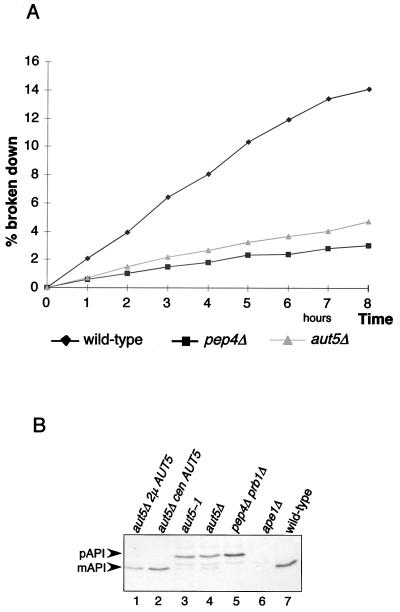
A defect in vacuolar lysis of the membranes of autophagic bodies prevents the access of vacuolar proteinases to the autophagocytosed material and thus should cause a significantly reduced vacuolar protein turnover rate. To check this, growing cells were metabolically labeled with radioactive methionine and then transferred into nitrogen-free starvation medium to induce autophagy. Aliquots were withdrawn, and the amount of acid-soluble small peptides generated by proteolysis was measured. As expected, aut5Δ cells exhibited a significantly reduced total protein breakdown rate (Fig. 3A), similar to that of pep4Δ cells, which due to the lack of vacuolar proteinase A show a severe defect in vacuolar proteolysis (34).
FIG. 3.
aut5Δ cells show phenotypes characteristic of mutants defective in autophagy. (A) Total protein breakdown of aut5Δ cells during starvation is significantly reduced compared to that of wild-type cells. After metabolic pulse-labeling of growing cells with [35S]methionine, cells were shifted to 1% K acetate as a nitrogen-free starvation medium and aliquots were taken at the indicated times. The amount of acid-soluble small peptides generated via proteolysis was quantitated with a liquid scintillation counter and is expressed as a percentage of the acid-insoluble radioactivity of the 0-h sample. (B) Western blot analysis of maturation of proaminopeptidase I (pAPI) using antibodies against aminopeptidase I. mAPI, mature aminopeptidase I. Maturation is impaired in aut5-deficient cells grown to stationary phase, and this defect is complemented by expressing AUT5 from a centromeric (cen) or 2μm (2μ) plasmid.
Proaminopeptidase I, a resident vacuolar protease, is synthesized as a cytosolic proform and proteolytically matured in the vacuole. Proaminopeptidase I is selectively delivered to the vacuole using in large part the autophagic machinery (19, 36). Similar to most autophagy mutants aut5Δ cells grown to stationary phase are further impaired in maturation of proaminopeptidase I (Fig. 3B, lane 4) (14). This finding is consistent with the most recently published analysis of Cvt17p (35).
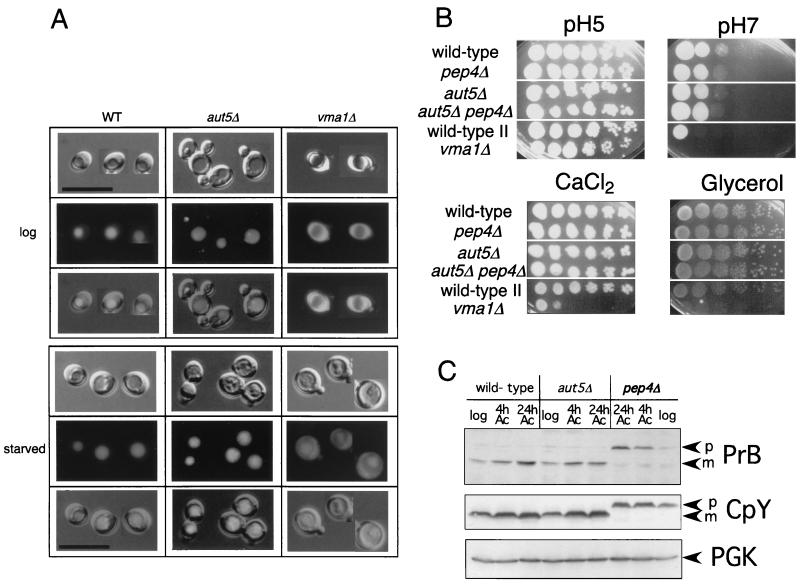
Vacuolar pH appears normal, and mature vacuolar proteinases are detectable in aut5Δ cells.
Vacuolar acidification is a prerequisite for disintegration of autophagic bodies (23). We therefore checked aut5Δ cells for phenotypes of vacuolar acidification mutants (15, 26, 42). Such mutants typically exhibit a defect in accumulation of the fluorescent dye quinacrine in the vacuole (Fig. 4A) or are impaired for growth on media (i) with pH 7.0, (ii) with high concentrations of calcium ions, and (iii) containing a nonfermentable carbon source such as glycerol (Fig. 4B). Growing and starved aut5Δ cells showed normal vacuolar accumulation of quinacrine, and they grew normally in selective media, indicating nonaltered vacuolar acidification. A lack of mature vacuolar proteinases also causes an accumulation of autophagic bodies in the vacuole (33, 37). We therefore confirmed by immunoblotting in growing and starved aut5Δ cells the presence of the mature proteinases B and Y (Fig. 4C).
FIG. 4.
Vacuolar acidification appears normal in aut5Δ cells. (A) The fluorescent dye quinacrine accumulates inside acidic vacuoles. In growing (upper panel) and 4-h-starved (1% K acetate) (lower panel) aut5Δ cells quinacrine accumulation indicates vacuolar acidification. vma1Δ cells, exhibiting a defect in vacuolar acidification, are included as a control. From top to bottom, Nomarski optics, fluorescence, and an overlay of both are shown. Bar, 20 μm. WT, wild type. (B) Growth of aut5Δ cells on media with pHs 5 and 7 and on medium containing 100 mM CaCl2 or glycerol as the carbon source is like that of the wild type. Cells were grown in liquid yeast extract-peptone-dextrose, and from left to right, a dilution series (1:10) of the cultures was dropped on plates and grown at 30°C. (C) Mature carboxypeptidase Y (CpY) and proteinase B (PrB) are detectable in immunoblots of growing and starved aut5Δ cells. Crude extracts of cells of the logarithmic growth phase or starved for 4 or 24 h in 1% K acetate (Ac) were subjected to SDS-electrophoresis, blotted on polyvinylidene difluoride (PVDF) membrane, and analyzed with the indicated antibodies. As a loading control, PGK was detected. p, pro; m, mature.
Biologically active Aut5p-HA is an integral membrane protein.
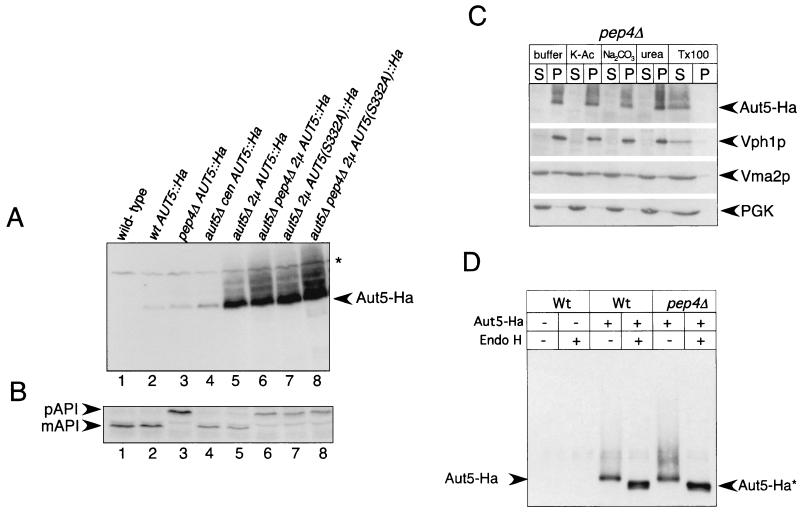
We generated yeast cells carrying a C-terminally Ha3-tagged AUT5 gene under the control of its native promoter on the chromosome. Correct tagging was confirmed by Southern blotting (not shown). In immunoblots of these cells, an antibody against Ha clearly detected a band at 75 kDa (Fig. 5A, lane 2) which was absent in cells lacking the Ha-tagged protein (Fig. 5A, lane 1). Dosage-dependent expression of Aut5-Ha from a centromeric plasmid (Fig. 5A, lane 4) and a 2μm plasmid (Fig. 5A, lanes 5 and 6) further confirmed detection of Aut5-Ha. Chromosome- and plasmid-expressed Aut5-Ha complemented the aut5Δ defect in the breakdown of autophagic bodies during starvation (not shown) and the defect in maturation of proaminopeptidase I in stationary cells (Fig. 5B, lanes 1 to 6), indicating the biological activity of Aut5-Ha.
FIG. 5.
Aut5-Ha is a glycosylated integral membrane protein. (A) Aut5-Ha3 is specifically detected in immunoblots using an antibody to Ha. Crude extracts of stationary-phase cells were subjected to SDS-PAGE, blotted on PVDF membranes, and analyzed with antibodies to Ha. Lane 1, wild type; 2, wild-type (wt) cells expressing Aut5-Ha from the chromosome under the control of the native AUT5 promoter; 3, pep4Δ cells expressing Aut5-Ha from the chromosome; 4, aut5Δ cells carrying a centromeric Aut5-Ha plasmid (cen); 5, aut5Δ cells expressing Aut5-Ha from a 2μm (2μ) plasmid; 6, aut5Δ pep4Δ cells with a 2μm Aut5-Ha plasmid; 7, aut5Δ cells expressing from a 2μm plasmid a mutated version of Aut5-Ha in which serine 332 was replaced by alanine; 8, aut5Δ pep4Δ cells expressing from a 2μm plasmid a mutated version of Aut5-Ha in which serine 332 was replaced by alanine. Aut5-Ha shows a molecular mass of ∼75 kDa; note the smear of the protein to a higher molecular mass. ∗, cross-reacting material. (B) Western blotting of aminopeptidase I. The maturation of aminopeptidase I, which is similar to that of the wild type in strains expressing Aut5-Ha either from the chromosome or from plasmids, indicates the biological activity of the tagged protein (lanes 1, 2, 4, and 5). Replacement of serine 332 with alanine by site-directed mutagenesis impairs the maturation of aminopeptidase I, indicating an impaired biological activity of the mutated protein. The PVDF membrane shown in panel A was stripped and reprobed with antibodies to aminopeptidase I. pAPI, precursor aminopeptidase I; mAPI, mature aminopeptidase I. (C) Aut5-Ha is an integral membrane protein. pep4Δ cells, starved for 4 h in 1% K acetate (K-Ac), were lysed with glass beads, and the homogenate was separated by centrifugation at 100,000 × g for 45 min into pellet (P) and supernatant (S) fractions. Fractions were subjected to SDS-electrophoresis, blotted onto a PVDF membrane, and analyzed using Ha antibodies. As indicated, the cell lysate was incubated with 1 M potassium acetate, 0.1 M Na2CO3, 2.5 M urea, or 1% Triton X-100 (Tx100) prior to centrifugation. As a control, the membrane was stripped and reprobed successively with antibodies against Vph1p (an integral membrane protein), Vma2p (peripheral membrane protein), and PGK as a soluble protein. (D) Aut5-Ha is glycosylated. Cells starved for nitrogen for 4 h in 1% K acetate were lysed with glass beads; Aut5-Ha was immunoprecipitated with Ha antibodies and deglycosylated by treatment with endoglycosidase H (Endo H). After SDS-PAGE and being blotted onto a PVDF membrane, the precipitates were analyzed with Ha antibodies. Aut5-Ha∗ refers to the deglycosylated species.
Since Aut5p contains putative transmembrane domains (Fig. 1C), we checked if Aut5-Ha shows features characteristic of membrane proteins. As shown below, Aut5-Ha is rapidly degraded dependent on vacuolar proteinase A and consistently accumulates to some part in the vacuolar lumen of cells lacking proteinase A. We used pep4Δ cells lacking proteinase A for this experiment to also examine the membrane association status of the part of Aut5-Ha which resides in the vacuolar lumen. In wild-type cells, the vacuolar part of Aut5-Ha is missing due to degradation. Therefore, this experiment extends the study describing Cvt17/Aut5 as an integral membrane protein in wild-type cells (35).
Starved pep4Δ cells expressing Aut5-Ha from the chromosome under the control of its native promoter were homogenized with glass beads and centrifuged to yield a supernatant and pellet fraction. As shown in Fig. 5C, Aut5-Ha was almost completely pelletable. Incubation with 1 M potassium acetate, 0.1 M Na2CO3, or 2.5 M urea did not solubilize significant amounts of Aut5-Ha. Incubation with 1% Triton X-100, however, resulted in detection of most of Aut5-Ha in the supernatant fraction (Fig. 5C). Aut5-Ha in this respect behaves like the integral membrane protein Vph1 and differs from the peripherally membrane-attached Vma2p (Fig. 5C). To exclude the possibility that Aut5-Ha might be trapped inside vesicles, thus mimicking characteristics of an integral membrane protein, we used quite harsh conditions for glass bead lysis of the cells. Therefore, part of the peripheral membrane protein Vma2p was already released into the supernatant with buffer alone. Our findings suggest that in starved pep4Δ cells Aut5-Ha is an integral membrane protein.
Aut5-Ha is glycosylated.
Most integral membrane proteins enter the ER-Golgi sorting pathway, where they undergo posttranslational modifications and are glycosylated. Aut5p contains three putative N-linked glycosylation sites, and Aut5-Ha showed a smear to higher molecular mass in immunoblots (Fig. 5A), which suggests that Aut5p is glycosylated. To test this, we treated cell extracts with endoglycosidase H (Fig. 5D). After endoglycosidase H digestion, Aut5-Ha showed a mobility shift to a lower molecular mass (∼73 kDa) (Fig. 5D), showing glycosylation of the protein.
Aut5-Ha reaches the vacuole via an autophagy-independent pathway.
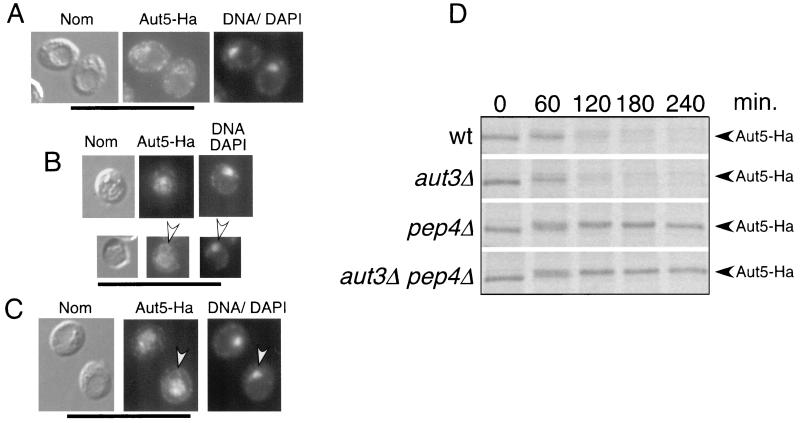
We next performed indirect immunofluorescence microscopy of starved cells, expressing Aut5-Ha from the chromosome under the control of its native promoter, using antibodies to Ha. The ring-like Aut5-Ha immunostaining (Fig. 6A) seen in wild-type cells clearly surrounded the nucleus, which was stained with DAPI (Fig. 6A), suggesting localization of Aut5-Ha at the ER (nuclear envelope). In sucrose density gradient fractionation using starved wild-type cells expressing Aut5-Ha from the chromosome, the ER marker Kar2p sedimented to a position similar to that of Aut5-Ha (not shown), further supporting ER localization.
FIG. 6.
Indirect immunofluorescence microscopy suggests localization of Aut5-Ha at the ER in wild-type cells and in the vacuole and the ER in pep4Δ cells. (A to C) Cells grown to stationary phase were starved for 4 h in 1% K acetate. The cells were then fixed with formaldehyde, followed by spheroplasting with Zymolyase. The spheroplasted cells were then incubated with a primary antibody against Ha and afterwards with a secondary Cy3-coupled antibody. Nuclear DNA was further stained with DAPI. From left to right Nomarski optics (Nom), immunofluorescence microscopy (Aut5-Ha), and nuclear staining with DAPI (DNA/DAPI) are shown. (A) Wild-type cells expressing Aut5-Ha from the chromosome under the control of the endogenous promoter. (B) pep4Δ cells expressing Aut5-Ha chromosomally, which due to the lack of vacuolar endoproteinase A are defective in vacuolar protein breakdown. Arrowheads point to ER staining. (C) Chromosomal expression of Aut5-Ha in aut3Δ pep4Δ cells, which are further defective in autophagy. Bar, 20 μm. (D) Pulse-chase analysis indicates a rapid breakdown of Aut5-Ha dependent on vacuolar proteinase A. The indicated strains were grown to log phase in SMD medium, pulse-labeled for 20 min with [35S]methionine and cysteine, and chased in nonradioactive medium as described in Materials and Methods. At the specified times, aliquots were withdrawn and lysates were immunoprecipitated with antibodies to Ha. After deglycosylation with endoglycosidase H, samples were subjected to SDS-PAGE and labeled proteins were visualized using a PhosphorImager. wt, wild type.
Since autophagic bodies are rapidly broken down in wild-type cells, we next checked pep4Δ cells, which due to the lack of vacuolar proteinase A accumulate autophagic bodies in the vacuole during starvation (33) (37). In these cells, also expressing Aut5-Ha from the chromosome, a significant amount of Aut5-Ha was detected in the vacuole (Fig. 6B). Another part was still seen at a ring-like structure surrounding the nucleus, indicative of the ER (Fig. 6B).
We further tested whether Aut5-Ha reaches the vacuole via autophagy or other pathways such as the ER-Golgi sorting pathway. We localized Aut5-Ha by immunofluorescence in starved aut3Δ pep4Δ double mutant cells expressing Aut5-Ha from the chromosome. The lack of Aut3p, a serine/threonine kinase, blocks early steps of autophagy (31, 17) and thus prevents accumulation of autophagic bodies in the vacuoles of starved aut3Δ pep4Δ cells. Nevertheless, as shown in Fig. 6C, significant amounts of Aut5-Ha were detectable in the vacuole, suggesting a transport of Aut5-Ha independent of the autophagic traffic. No immunofluorescence signal was seen in wild-type and pep4Δ control cells lacking an Ha tag (not shown).
Localizations similar to those in starved cells were seen in wild-type, pep4Δ, and aut3Δ pep4Δ cells grown to log phase in nutrient-rich medium (not shown).
Aut5-Ha is rapidly degraded dependent on vacuolar proteinase A.
The presence of Aut5-Ha in vacuoles of cells defective in vacuolar proteinase A and the absence of the protein in vacuoles of wild-type cells suggests that Aut5-Ha might be rapidly degraded in wild-type vacuoles. To test this hypothesis, we performed a pulse-chase experiment by metabolic labeling and immunoprecipitation. Indeed, chromosomally expressed Aut5-Ha is rapidly degraded in both wild-type and aut3Δ cells, defective in early phases of autophagy (Fig. 6D). Stabilization of the protein is clearly seen in cells defective in vacuolar proteinase A and in aut3Δ pep4Δ cells (Fig. 6D). The stabilization seen in cells lacking vacuolar proteinase A is in contrast to findings in the study of Cvt17p (35), for which degradation of Cvt17p “independent of the major vacuole proteases” was reported. Quantification using a PhosphorImager indicates a half-life of Aut5-Ha of 50 to 70 min in wild-type cells. To get sharper bands, samples were deglycosylated with endoglycosidase H prior to SDS-PAGE. The observed shift to a slightly higher molecular mass during the time course might be due to carbohydrate side chains of the complex type, which are not released by endoglycosidase H.
Immunogold electron microscopy suggests localization of Aut5-Ha at ∼50-nm-diameter intravacuolar vesicles and at the ER (nuclear envelope).
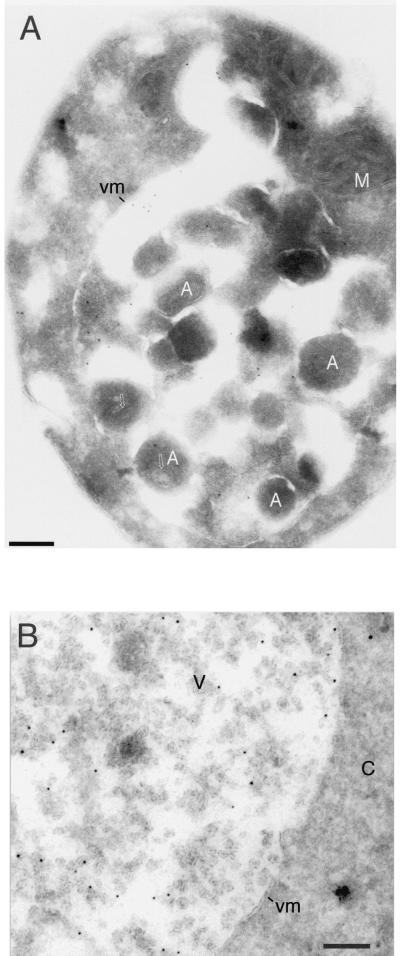
Our immunofluorescence experiments with aut3Δ pep4Δ cells (Fig. 6C) suggest localization of chromosomally expressed Aut5-Ha at the ER and in the vacuolar lumen. Since Aut5-Ha is an integral membrane protein (Fig. 5C), these findings point to the existence of membranous structures in the vacuolar lumen which are formed independent of autophagy. To further specify the nature of these intravacuolar structures, we carried out immunogold electron microscopy using antibodies against Ha. Immunofluorescence also showed that Aut5-Ha expressed from the chromosome and the 2μm plasmid localized to identical cell compartments (not shown). Since higher protein levels are needed to localize epitopes in thin electron microscopical sections, we used the plasmid expression vector for immunoelectron microscopy. Control cells not expressing Aut5-Ha were unlabeled (not shown), indicating the specificity of our labeling. The distribution of gold labeling in different cell compartments was quantitated by systematically screening labeled thin sections cut from cell pellets. In starved pep4Δ cells (Fig. 7A and B) the gold label (about 750 gold particles were counted) was found in the cytoplasm (52% of gold particles) and in the vacuole (48% of gold particles). The cytoplasmic labeling was found in the nuclear envelope and at the cortical ER (42% of total cytoplasmic label) and associated with other unidentified membrane structures. Inside the vacuole, labeling was associated with ∼50-nm-diameter vesicles (78% of total vacuolar label) (Fig. 7B), autophagic bodies (12%), and the vacuolar membrane (10%). Consistent with our immunofluorescence experiments, a block at the early steps of autophagy in aut3 cells did not impair the targeting of Aut5-Ha to ∼50-nm-diameter vesicles in the vacuolar lumen (Fig. 7C and D). Immunogold electron microscopy further confirmed the localization of immunofluorescent Aut5-Ha both at the ER (nuclear envelope) (Fig. 7C and D) and in the vacuolar lumen.
FIG. 7.
Immunogold electron microscopy localizes Aut5-Ha at the ER (nuclear envelope) and at ∼50-nm-diameter vesicles in the vacuolar lumen. Cells expressing Aut5-Ha from a 2μm plasmid and grown to stationary phase were starved for 4 h in 1% K acetate, fixed with paraformaldehyde, and prepared for cryosectioning as detailed in Materials and Methods. The sections were immunolabeled with antibodies to Ha and secondary antibodies coupled to 10-nm-thick gold. (A and B) pep4Δ cells. Arrows in panel A point to material enclosed in autophagic bodies. (C and D) In aut3-1 pep4Δ cells, no autophagic bodies accumulated in the vacuole due to the autophagy defect. In these cells gold particles were detected at the ER (nuclear envelope) (black arrowheads) and at ∼50-nm-diameter vesicles in the vacuolar lumen (white arrowheads). Panel B also shows localization of gold particles at ∼50-nm-diameter vesicles in the vacuolar lumen. A, autophagic body; M, mitochondrion; vm, vacuolar membrane; C, cytoplasm; V, vacuole; N, nucleus. Bar, 200 nm.
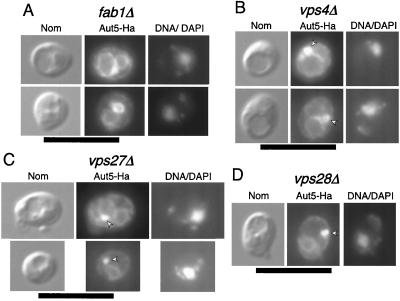
Vacuolar targeting of Aut5-Ha is characteristically affected in vps class E mutants and in mutants defective in the phosphatidylinositol 3-phosphate 5-kinase Fab1p.
The occurrence of 40- to 50-nm-diameter vesicles in the vacuolar lumen has already been described (40) and implicated in the vacuolar targeting of CPS via the MVB pathway (24, 25). CPS, a resident vacuolar hydrolase, is synthesized as an inactive integral membrane proform which reaches the vacuolar lumen attached to 50-nm-long vesicles (24). In the vacuole the prosequence of proCPS, which serves as a membrane anchor, is proteolytically cleaved and mature soluble CPS is released to the vacuolar lumen.
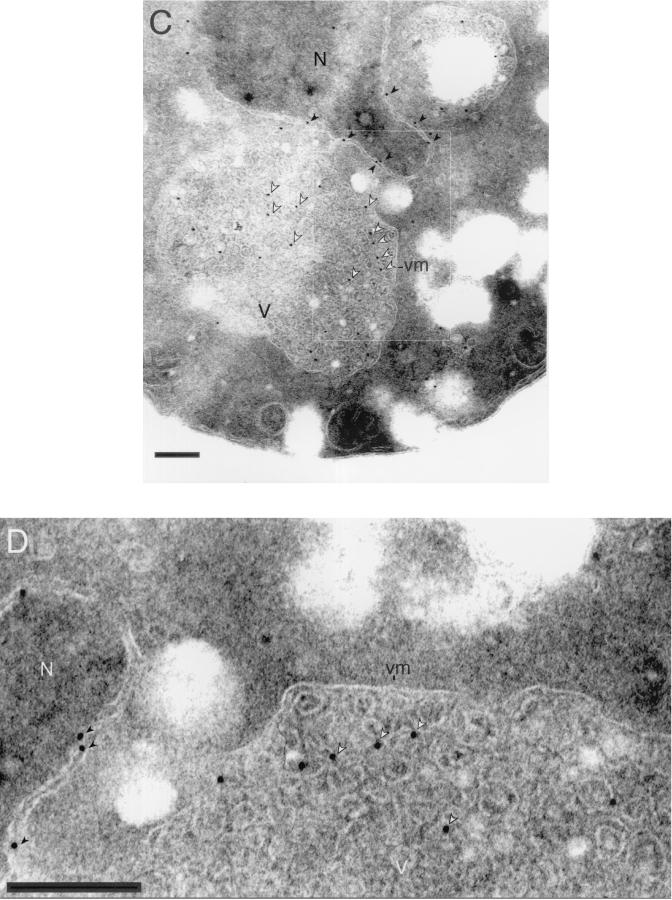
Localization of Aut5-Ha at 50-nm-diameter vesicles in the vacuole suggests that Aut5-Ha might reach the vacuole in a way similar to the MVB pathway. The phosphatidylinositol 3-phosphate 5-kinase Fab1p is specifically required for the sorting of proCPS to the vacuolar lumen via the MVB pathway (24), without affecting the normal biosynthetic vacuolar traffic of carboxypeptidase Y and alkaline phosphatase (41, 11). In the absence of Fab1p, the defect in sorting proCPS to internal vesicles in the prevacuolar endosome causes almost complete mislocalization of the protein to the limiting vacuolar membrane (24). Consistently, indirect immunofluorescence detected mislocalization of a significant part of Aut5-Ha to the vacuolar membrane in fab1Δ cells (Fig. 8A), suggesting vacuolar targeting of Aut5-Ha similar to that of the MVB pathway. In agreement with our previous findings (Fig. 6A to C), also in fab1Δ cells part of Aut5-Ha is detected at the ER (Fig. 8A).
FIG. 8.
The phosphatidylinositol 3-phosphate 5-kinase Fab1p (A) and Vps class E proteins (B to D) are required for targeting of Aut5-Ha. Cells expressing Aut5-Ha from a 2μm plasmid were grown at 25°C to log phase, fixed with formaldehyde, and spheroplasted. After treatment with antibodies to Ha and a secondary Cy3 antibody, the cells were analyzed by indirect immunofluorescence microscopy. From left to right, Nomarski optics (Nom), immunofluorescence microscopy (Aut5-Ha), and nuclear staining with DAPI (DNA/DAPI) are shown. In fab1Δ cells (A) Aut5-Ha is localized to the vacuolar membrane and the ER. In vps class E cells (B to D), Aut5-Ha is seen at the prevacuolar compartment (arrowheads) and to some extent at the vacuolar membrane and the ER. Bar, 10 μm.
Targeting of proCPS to the vacuolar lumen via the MVB pathway further depends on the action of Vps class E proteins (24). A lack of any of these proteins leads to retention of proCPS at the prevacuolar endosome (class E compartment) and to missorting to the limiting vacuolar membrane (24). As for fab1Δ cells, we checked localization of Aut5-Ha in indirect immunofluorescence in vps4Δ (end13Δ), vps27Δ, and vps28Δ mutants, three members of the vps class E family. As expected, all three mutants showed mislocalization of Aut5-Ha (Fig. 8B to D) reminiscent of that described for proCPS, suggesting that, similar to what occurs with proCPS, targeting of Aut5-Ha depends on Vps class E proteins.
Site-directed mutagenesis of the putative lipase active-site serine inhibits the vacuolar breakdown of autophagic bodies.
Aut5p shows the putative lipase active-site motif IWVTGHSLGG from amino acids 326 to 335. To test the relevance of this putative active-site serine for the biological activity of Aut5p, we selectively replaced serine 332 by alanine. Expression of AUT5(S332A)-Ha from a 2μm plasmid in aut5Δ cells led to the detection of a wild-type-like steady-state level of the mutated protein (Fig. 5A, lanes 7 and 8). Also, localization of Aut5(S332A)-Ha in aut5Δ and aut5Δ pep4Δ cells checked by indirect immunofluorescence was like that of the wild type (not shown). However, Aut5(S332A) was unable to complement the defect in maturation of proaminopeptidase I (Fig. 5B, lane7) or the defect in the vacuolar breakdown of autophagic bodies in aut5Δ cells (not shown). These findings suggest an essential function of serine 332 for the biological activity of Aut5p. We further used different commonly used lipase substrates under various conditions but were unable to directly measure a putative lipase activity of Aut5p (not shown). Beside the apparent instability of Aut5p, our inability to measure this activity might be due to the requirement for more specific substrates or the lack of specific cofactors.
DISCUSSION
Yeast cells are able to break down numerous membrane-enclosed autophagic bodies in their vacuoles, leaving the limiting vacuolar membrane intact. Except for our knowledge of the need of vacuolar acidification (23) and vacuolar proteinases (33, 37), components of this inevitably highly specific membrane disintegration machinery remain mysterious. We here identified Aut5p, a component essential for disintegration of autophagic bodies and demonstrated the relevance of its lipase active-site motif via site-directed mutagenesis.
Since autophagic bodies are lysed inside the vacuole, it seems conceivable that Aut5p is targeted to the vacuole, probably simultaneously with autophagic bodies. Indeed, our pulse-chase analysis indicates a rapid (half-life, 50 to 70 min) turnover of Aut5-Ha dependent on the presence of vacuolar proteinase A. Consistently, in cells defective in vacuolar proteolysis, indirect immunofluorescence (Fig. 6B and C) and immunogold electron microscopy (Fig. 7B to D) visualized significant amounts of Aut5-Ha in the vacuole. However, in wild-type cells this vacuolar pool of Aut5-Ha was lacking (Fig. 6A). This finding is in contrast to findings in the study of Cvt17p, for which degradation “independent of the major vacuole proteases” was reported (35). We believe this discrepancy might be due to the use of an antiserum raised against synthetic peptides, since in our hands the protein was hard to detect with two individual antisera against synthetic peptides (not shown), but a reliable strong signal was detectable using the Ha-tagged species.
How does Aut5p reach the vacuole? The breakdown of Aut5-Ha in autophagy-deficient aut3Δ cells (Fig. 6D), together with the vacuolar localization in starved (Fig. 6C) and logarithmically growing (not shown) aut3Δ pep4Δ cells in immunofluorescence, argues against the use of the autophagic pathway. This conclusion is consistent with our statistical analysis of immunogold microscopy. We cannot exclude, however, the possibility that a small portion of Aut5p travels to the vacuole via autophagy at autophagic bodies and then induces their breakdown, but our data clearly indicate that the major part of Aut5-Ha reaches the vacuole independent of autophagy.
The observed glycosylation of Aut5-Ha demonstrates that Aut5-Ha follows the ER-to-Golgi sorting pathway. Immunofluorescence suggests a localization of Aut5-Ha in the vacuolar lumens of pep4Δ cells (Fig. 6B and C). Since Aut5-Ha in these cells behaves as an integral membrane protein (Fig. 5C), we expected a localization at membranous structures inside the vacuole. Immunogold electron microscopy in fact displayed localization at ∼50-nm-diameter vesicles (Fig. 7B to D) in the vacuolar lumen. The appearance of 50-nm-diameter vesicles in the vacuolar lumen is reminiscent of the function of the MVB pathway (24, 25). The MVB pathway at the prevacuolar endosome (compartment) diverges from the biosynthetic ER-to-Golgi vacuolar protein sorting pathway. Integral membrane proteins, which are cargo molecules of the MVB pathway, are specifically directed to invaginations of the membrane of the prevacuolar endosome. Then vesicles carrying these cargo molecules branch off to the interior of the organelle, and a so-called MVB is formed. Fusion of the MVB with the vacuole finally releases the ∼50-nm-diameter vesicles to the vacuolar lumen, where they are thought to be broken down (24).
Vps class E mutants affect both the biosynthetic and MVB traffic to the vacuole. In these mutants proCPS, which is a cargo molecule of the MVB pathway, is retarded at the prevacuolar compartment and, due to a defect in sorting to internal vesicles at the prevacuolar endosome, is mislocalized to the limiting vacuolar membrane. The phosphatidylinositol 3-phosphate 5-kinase Fab1p is specifically required for MVB sorting but not for biosynthetic vacuolar protein sorting, and therefore a lack of this protein causes no retardation at the prevacuolar endosome but only mislocalization to the vacuolar membrane (41, 11, 24). We observed in both mutant types a mislocalization of Aut5-Ha similar to that described for proCPS (Fig. 8), suggesting a sorting similar to that of the MVB pathway. Taken together our findings suggest that Aut5-Ha reaches the vacuolar lumen after transit through the ER and Golgi apparatus in a way similar to that of the MVB pathway.
The lysis of membranes implies a potential high risk for the cell. Very interesting questions therefore are what prevents a premature activation of such a membrane lysis machinery and what impedes lysis of the vacuolar membrane itself? Our observation that two distinct pathways are used for vacuolar targeting of autophagic bodies and Aut5-Ha, respectively, opens up new vistas in answering these questions. Probably Aut5p, a putative lipase, becomes activated only upon direct interaction with a component present exclusively at autophagic bodies. It would be tempting to speculate that this interacting partner acts similarly to a colipase. This idea would explain not only the specificity in disintegrating autophagic bodies without the integrity of the vacuolar membrane being affected but also how the cell prevents untimely activation of Aut5p on its transit to the vacuole by keeping both partners apart by two different transport pathways, which converge at their final destination, the vacuolar lumen. Since Aut5p might remain in its active form even after lysis of the autophagic body, the observed rapid vacuolar degradation of Aut5-Ha would prevent further trouble. This idea implies that Aut5p exhibits its active site and a putative interaction domain towards the vacuolar lumen and not to the interior of the 50-nm-diameter vesicle. The potential glycosylation sites and the lipase active-site motif of Aut5p are located after its predicted transmembrane domain (Fig. 1B). Since Aut5-Ha is glycosylated, this part of the protein including the lipase active-site motif would be estimated to be exposed to the ER lumen. Assuming that the membrane topology is preserved during sorting, one would therefore indeed expect that the lipase active-site motif together with most of Aut5p is exposed to the outside of the 50-nm-diameter vesicle membrane, i.e., to the vacuolar lumen. Another possibility would be that Aut5p selectively attacks lipid molecules present only at the membranes of autophagic bodies and not at the limiting vacuolar membrane.
Interestingly, Aut5-Ha is localized at the ER (nuclear envelope) in wild-type cells and also in pep4Δ cells. This localization is indicated by four lines of evidence: (i) glycosylation, which points to entry in the ER to the Golgi secretory pathway; (ii) indirect immunofluorescence of both starved (Fig. 6) and logarithmic (not shown) cells, which shows a typical ring-like staining around the nucleus; (iii) sucrose density gradient fractionation of starved wild-type cells, in which the ER marker Kar2p cosedimented with Aut5-Ha (not shown); and (iv) immunogold electron microscopy, which showed beside the vacuolar localization a significant localization at the nuclear envelope and ER and at the cortical ER (Fig. 7).
Detection of Aut5-Ha at the ER raises the possibility that instead of the vacuolar lumen Aut5p might alternatively function at the ER by hydrolyzing specific lipids, thus rendering autophagic bodies competent for breakdown. The membrane source of autophagic bodies remains unknown, although there are some hints pointing to the ER (8) or the Golgi apparatus (18). One might therefore speculate that lipids modified at the ER by Aut5p are specifically targeted to autophagosomes. Finally, yet unknown components in the vacuole might catalyze the breakdown of autophagic bodies dependent on the presence of these lipid molecules. In this scenario Aut5p would be catalytically active at the ER and most likely also on its transit to the vacuole. To avoid unspecific hydrolysis of membranes, which surely would lead to cell death, Aut5p must therefore exhibit a very high substrate specificity. When Aut5p functions at the ER, the rapid transport to the vacuolar lumen would reflect only protein degradation. Further extensive studies are necessary to distinguish between these models of Aut5p function.
Aut5p shares significant homologies with several proteins of unknown function (Fig. 1B); the homologies very interestingly include the putative lipase active-site motif. This finding might point to a common function of these proteins. The pSI-7 protein from C. fulvum was identified for its induced expression during starvation and its pathogenic growth on tomato plants (5). Probably our work on Aut5p in yeast therefore might also contribute in the future to the understanding of the pathogenic interaction between C. fulvum and plants.
ACKNOWLEDGMENTS
We are grateful to S. D. Emr for sharing antibodies with us and to M. Bredschneider for doing the transmission electron micrographs.
We thank D. H. Wolf for many helpful discussions and support.
This work was supported by DFG grant Th752/1-1.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1987. [Google Scholar]
- 2.Baba M, Osumi M, Scott S V, Klionsky D J, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Coleman M, Henricot B, Arnau J, Oliver R P. Starvation-induced genes of the tomato pathogen Cladosporium fulvum are also induced during growth in planta. Mol Plant-Microbe Interact. 1997;10:1106–1109. doi: 10.1094/MPMI.1997.10.9.1106. [DOI] [PubMed] [Google Scholar]
- 6.Cottarel G. The Saccharomyces cerevisiae HIS3 and LYS2 genes complement the Schizosaccharomyces pombe his5-303 and lys1-131 mutations, respectively: new selectable markers and new multi-purpose multicopy shuttle vectors, pSP3 and pSP4. Curr Genet. 1995;28:380–383. doi: 10.1007/BF00326437. [DOI] [PubMed] [Google Scholar]
- 7.Dunn W J. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 8.Dunn W J. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egner R, Mahe Y, Pandjaitan R, Kuchler K. Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5879–5887. doi: 10.1128/mcb.15.11.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman D M, Steitz T A, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 11.Gary J D, Wurmser A E, Bonangelino C J, Weisman L S, Emr S D. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goebel W, Kuhn M. Bacterial replication in the host cell cytosol. Curr Opin Microbiol. 2000;3:49–53. doi: 10.1016/s1369-5274(99)00050-8. [DOI] [PubMed] [Google Scholar]
- 13.Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harding T M, Hefner-Gravink A, Thumm M, Klionsky D J. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 15.Hirata R, Umemoto N, Ho M N, Ohya Y, Stevens T H, Anraku Y. VMA12 is essential for assembly of the vacuolar H(+)-ATPase subunits onto the vacuolar membrane in Saccharomyces cerevisiae. J Biol Chem. 1993;268:961–967. [PubMed] [Google Scholar]
- 16.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 17.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Scott S V, Klionsky D J. Alternative protein sorting pathways. Int Rev Cytol. 2000;198:153–201. doi: 10.1016/s0074-7696(00)98005-7. [DOI] [PubMed] [Google Scholar]
- 20.Klionsky D J, Cueva R, Yaver D S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky D J, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf D H, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura N, Matsuura A, Wada Y, Ohsumi Y. Acidification of vacuoles is required for autophagic degradation in the yeast Saccharomyces cerevisiae. J Biochem. 1997;121:338–344. doi: 10.1093/oxfordjournals.jbchem.a021592. [DOI] [PubMed] [Google Scholar]
- 24.Odorizzi G, Babst M, Emr S D. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 25.Odorizzi G, Babst M, Emr S D. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci. 2000;25:229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- 26.Preston R A, Murphy R F, Jones E W. Assay of vacuolar pH in yeast and identification of acidification-defective mutants. Proc Natl Acad Sci USA. 1989;86:7027–7031. doi: 10.1073/pnas.86.18.7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts C J, Raymond C K, Yamahiro C T, Stevens T H. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- 28.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 29.Schlumpberger M, Schaeffeler E, Straub M, Bredschneider M, Wolf D H, Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179:1068–1076. doi: 10.1128/jb.179.4.1068-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spormann D O, Heim J, Wolf D H. Biogenesis of the yeast vacuole (lysosome). The precursor forms of the soluble hydrolase carboxypeptidase yscS are associated with the vacuolar membrane. J Biol Chem. 1992;267:8021–8029. [PubMed] [Google Scholar]
- 31.Straub M, Bredschneider M, Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suriapranata I, Epple U D, Bernreuther D, Bredschneider M, Sovarasteanu K, Thumm M. The breakdown of autophagic vesicles inside the vacuole depends on Aut4p. J Cell Sci. 2000;113:4025–4033. doi: 10.1242/jcs.113.22.4025. [DOI] [PubMed] [Google Scholar]
- 33.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teichert U, Mechler B, Muller H, Wolf D H. Lysosomal (vacuolar) proteinases of yeast are essential catalysts for protein degradation, differentiation, and cell survival. J Biol Chem. 1989;264:16037–16045. [PubMed] [Google Scholar]
- 35.Teter, S. A., K. Eggerton, S. Scott, J. Kim, A. Fischer, and D. Klionsky. 2001. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 2083–2087. [DOI] [PMC free article] [PubMed]
- 36.Thumm M. Structure and function of the yeast vacuole and its role in autophagy. Microsc Res Tech. 2000;51:563–572. doi: 10.1002/1097-0029(20001215)51:6<563::AID-JEMT6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 37.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf D H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 38.Thumm M, Wolf D H. From proteasome to lysosome: studies on yeast demonstrate the principles of protein degradation in the eukaryote cell. Adv Mol Cell Biol. 1998;27:41–67. [Google Scholar]
- 39.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 40.Wurmser A E, Emr S D. Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto A, De W D, Boronenkov I V, Anderson R A, Emr S D, Koshland D. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashiro C T, Kane P M, Wolczyk D F, Preston R A, Stevens T H. Role of vacuolar acidification in protein sorting and zymogen activation: a genetic analysis of the yeast vacuolar proton-translocating ATPase. Mol Cell Biol. 1990;10:3737–3749. doi: 10.1128/mcb.10.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]