Abstract
The pH-inducible acid tolerance response (ATR) is believed to play a major role in acid adaptation and virulence of Streptococcus mutans. To study this phenomenon in S. mutans JH1005, differential display PCR was used to identify and clone 13 cDNA products that had increased expression in response to pH 5.0 compared to that of pH 7.5-grown cells. One of these products, confirmed to be pH inducible by RNA dot blot and reverse transcription-PCR analyses, had 67% identity to a uvrA-UV repair excinuclease gene in Bacillus subtilis. Further sequence analysis of the uvrA homologue using the S. mutans genome database revealed that the complete gene was encoded in an open reading frame (ORF) of 2,829 bp (944 amino acids; 104.67 kDa). Immediately 3′ of uvrA was an ORF encoding a putative aminopeptidase gene (pepP). uvrA knockouts were constructed in S. mutans strains JH1005, NG8, and UA159 using allelic-exchange mutagenesis, replacing the entire gene with an erythromycin resistance cassette. As with uvrA mutants in other bacteria, the S. mutans uvrA mutants were extremely sensitive to UV irradiation. The uvrA mutant of S. mutans JH1005 was also more sensitive than the wild type to growth at pH 5.0, showing a 15% reduction in growth rate and a 14% reduction in final resting culture density. Acid-adapted S. mutans JH1005 uvrA mutants were shown to be more resistant to UV irradiation than was the parent but were unable to survive exposure to a killing pH of 3.0. Moreover, agarose gel electrophoretic analysis of chromosomal DNA isolated from uvrA-deficient cells exposed to low pH demonstrated more DNA damage than that for the wild-type strain. Here we suggest that uvrA and the nucleotide excision repair pathway are involved in the repair of acid-induced DNA damage and are associated with successful adaptation of S. mutans to low pH.
The oral bacterium Streptococcus mutans is able to gain a selective advantage over other oral microbes by withstanding extreme fluctuations in plaque pH. In the plaque environment, resident bacteria metabolize dietary carbohydrate, which results in the production of organic acids and a decrease in plaque pH. Telemetric measurements of plaque pH indicate that the pH can drop from 7.0 to values ranging from 4.0 to 3.0 (23). The ability to adapt to moderate pH promotes the survival of S. mutans under lower-pH conditions that would otherwise be lethal (37). This adaptive response in S. mutans is called the acid tolerance response (ATR) (37, 42), and similar mechanisms have been identified in some enteric bacteria (11, 12). Acid adaptation in S. mutans requires de novo protein synthesis (37) of up to 36 acid-regulated proteins (19) presumably encoded by acid-inducible genes.
Aside from the general features of the cellular response to acid pH, relatively little is known about the function of the numerous proteins encoded by the pH-inducible genes that constitute the S. mutans ATR. The genes for the protein repair chaperone, DnaK (22), and the 54-kDa subunit homologue of the eukaryotic signal recognition particle, Ffh (16), have been shown to be acid inducible in S. mutans, and ffh has been linked to the ATR, whereby ffh mutants revealed a lack of adaptive response to acid pH (16). To elucidate the molecular mechanisms of the ATR, we utilized the differential display PCR (dd-PCR) technique adapted from the method of Kwaik and Pederson (25). Here, isolation of total RNA from cells grown at pH 7.5 (unadapted state) and pH 5.0 (adaptive state) was followed by PCR amplification with arbitrary primers and separation by polyacrylamide gel electrophoresis (PAGE) for visualization of differential expression. Our goal was to identify up-regulated genes in S. mutans during acid adaptation. From this analysis, we have identified a gene with homology to the uvrA gene belonging to the nucleotide excision repair (NER) pathway involved in DNA repair in Bacillus subtilis. This pathway primarily consists of the protein complex UvrABC, which functions in locating and excising bulky DNA lesions (34).
It has been proposed elsewhere that the ATR in bacteria can be divided into two main components (13). The first involves mechanisms that maintain internal pH homeostasis. In S. mutans, this primarily involves an increase in H+-ATPase activity and acid end product efflux (3, 9, 18) and a decrease in proton permeability (18) by changes in membrane fatty acid composition (31) and increased synthesis of the cell surface component d-alanyl-lipoteichoic acid (6). The second component of the ATR is thought to involve the repair of cellular components damaged by acidic pH. Previous studies of S. mutans have shown the repair of acid-induced cellular damage to consist of the protein repair chaperone DnaK (22), a DNA repair enzyme exhibiting alkaline phosphatase (AP) endonuclease activity (17), and the DNA damage regulatory-repair protein RecA (30). However, little more is known about other repair mechanisms in S. mutans, specifically those involved in DNA repair.
Several known DNA repair mechanisms in bacteria could potentially be involved in the repair of acid-induced DNA damage, including direct damage reversal repair, recombinational repair (e.g., RecA), mismatch repair, base excision repair (e.g., AP endonuclease), and NER (e.g., UvrA) (2, 15). This picture is further complicated by the existence of specialized, regulated forms of repair such as those potentially found in the SOS, heat shock, and adaptive responses. The NER pathway, however, is thought to be the major system for repairing damaged DNA because of its capacity to repair essentially all types of DNA lesions (28). DNA repair (including NER) has been implicated in the resistance of bacteria to acidic pH (4, 17, 32, 33, 40). In Escherichia coli, mutants defective in the NER constituents uvrA and uvrB (32, 36) were shown previously to be more acid sensitive than the parent strain, suggesting that the NER pathway functions in the repair of acid-induced DNA damage. Whereas the internal pH of E. coli is maintained near neutral during acid challenge (29), S. mutans maintains a pH gradient that is only 0.5 to 1.0 pH units higher than the extracellular pH (9), indicating an increased likelihood of intracellular acidification in S. mutans during low-pH exposure. Therefore, the need for several DNA repair mechanisms in S. mutans, such as the NER pathway, would be paramount in ensuring the integrity of the genome during acid stress and ultimately the survival of the species in its natural habitat.
In this study, we have demonstrated that the S. mutans uvrA gene is up-regulated in response to an acidic environmental pH. We also show that in several strains of S. mutans uvrA mutants were not as resilient as the wild type (WT) in surviving UV irradiation and challenges by pH as low as 3.0.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
S. mutans strains JH1005 (21), NG8 (A. S. Bleiweis, University of Florida), and UA159 (J. Ferretti, Advanced Center for Genome Technology, University of Oklahoma) were grown in Todd-Hewitt (TH) broth containing 0.3% yeast extract (THYE) (BBL; Becton Dickinson, Cockeysville, Md.) at pHs ranging from 7.5 to 3.0, with the pH adjusted by addition of HCl. THYE medium was prepared by autoclaving followed by aseptic titration to the desired pH. Buffering solution (0.1 M morpholinepropanesulfonic acid [MOPS], without sodium acetate) was added to pH 7.5 medium to prevent significant pH drops during incubation for analysis of the ATR. For DNA damage and growth studies, 40 mM K2HPO4-citrate buffer was added to media ranging from pH 3.0 to 7.5. In liquid media, cultures of S. mutans were incubated aerobically in closed screw-cap tubes without agitation at 37°C. Cultivation of S. mutans on solid media was performed in candle jars to provide a CO2-enriched environment. Ten micrograms of erythromycin (Sigma-Aldrich, St. Louis, Mo.) per ml was incorporated into the media when required. Epicurian Coli XL-1 Blue supercompetent cells (Stratagene, La Jolla, Calif.) were used as a cloning host as described by the manufacturer.
Induction of the ATR and phenotypic detection.
An overnight culture of THYE-grown S. mutans JH1005 was diluted 10-fold in fresh THYE, pH 7.5, and incubated at 37°C until the culture reached mid-log phase (optical density at 600 nm [OD600] = 0.4 to 0.5). Cells were harvested by centrifugation and resuspended in THYE, pH 7.5 and 5.0, for unadapted and adapted conditions, respectively. Cells were incubated at 37°C for 1 h for use in dd-PCR and 2 h for phenotypic assessment of the ATR. Phenotypic detection of the ATR was valid only when a proportion of acid-adapted S. mutans JH1005 WT cells were able to withstand a 3-h, 37°C incubation at the killing pH of 3.0 whereas the unadapted cells could not (37). This was quantitatively confirmed by plating cells both before and after incubation at the killing pH on THYE plates using a spiral plater (model D; Spiral System Inc., Cincinnati, Ohio) and incubating them at 37°C for 48 h followed by enumeration. ATR studies performed with S. mutans UA159 used a killing pH of 3.5.
Total RNA isolation.
S. mutans JH1005 cells were disrupted using the FastPrep FP 120 cell disrupter (BIO 101-Savant, Holbrook, N.Y.), and RNA was extracted using the TRIzol reagent (Life Technologies Inc., Gaithersburg, Md.) according to the manufacturer's instructions with the following modification. The isopropanol precipitation step included the addition of a high-salt precipitation solution (Molecular Research Center, Inc., Cincinnati, Ohio) to remove polysaccharide and proteoglycans from the preparation. RNA (∼150 to 200 μg) was dissolved in RNAsecure resuspension solution (Ambion, Austin, Tex.). One-hundred-microgram aliquots of each isolated RNA preparation were treated with 40 U of RQ1 DNase (Promega, Madison, Wis.) for 45 min. at 37°C, extracted with TRIzol reagent and chloroform, and precipitated with ethanol. Washed RNA pellets were then resuspended in diethyl pyrocarbonate (Sigma-Aldrich)-water and stored in aliquots at −80°C.
dd-PCR.
DNA-free RNA samples from S. mutans JH1005 unadapted and adapted cells were subjected to reverse transcription using random hexamers (Pharmacia Biotech, Piscataway, N.J.) and SuperScript II reverse transcriptase (Life Technologies Inc.) following the manufacturer's preamplification protocol. Reaction mixtures containing 5 μg of RNA, 50 ng of random hexamers, 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 500 μM deoxynucleoside triphosphates (dNTPs), 10 mM dithiothreitol, and 200 U of SuperScript II reverse transcriptase were incubated in a volume of 20 μl for 60 min at 42°C. Controls without the addition of reverse transcriptase were also performed. All reaction mixtures were diluted fivefold prior to PCR amplification.
PCRs (20 μl) were performed using a pair of arbitrary primers from the RNAimage mRNA differential display system (GenHunter, Nashville, Tenn.). The reaction mixtures included 1 or 2 μl of the diluted reverse transcription reaction mixture, 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.2 μM (each) arbitrary primer (GenHunter), 2 μM dNTPs, 2.5 μCi of [α-33P]dATP (2,000 to 4,000 Ci/mmol; New England Nuclear, Boston, Mass.), and 1 U of AmpliTaq DNA polymerase (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Primer pairs consisted of H-AP1–H-AP2, H-AP3–H-AP4, H-AP1–H-AP5, H-AP5–H-AP6, and H-AP6–H-AP7 (Table 1). After an initial denaturing step of 94°C for 5 min, 40 PCR cycles were run with the following conditions: 94°C for 15 s, 40°C for 2 min, and 72°C for 30 s, followed by a 10-min, 72°C extension step. An aliquot of each PCR mixture was heated for 3 min at 80°C with DNA sequencing loading dye and separated by electrophoresis on a 5% denaturing Long Ranger (FMC BioProducts, Rockland, Maine) gel. The gel was dried under vacuum at 80°C and exposed to Biomax MR X-ray film (Eastman Kodak Company, Rochester, N.Y.) for 18 to 48 h.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence (5′-3′) | Description |
|---|---|---|
| H-AP1 | AAGCTTGATTGCC | GenHunter arbitrary primers |
| H-AP2 | AAGCTTCGACTGT | GenHunter arbitrary primers |
| H-AP3 | AAGCTTTGGTCAG | GenHunter arbitrary primers |
| H-AP4 | AAGCTTCTCAACG | GenHunter arbitrary primers |
| H-AP5 | AAGCTTAGTAGGC | GenHunter arbitrary primers |
| H-AP6 | AAGCTTGCACCAT | GenHunter arbitrary primers |
| H-AP7 | AAGCTTAACGAGG | GenHunter arbitrary primers |
| uvrAF | GGCTCCTTGGAATCCTATTTCTTC | uvrA probe and RT-PCR |
| uvrAB | GAATGCGTTGGCTTTCACCTC | uvrA probe and RT-PCR |
| ffhF | GGATCCATGGCTTTTGAAAGTTTA | ffh probe and RT-PCR |
| ffhB | GCAACATTTGCGGCTTCT | ffh probe and RT-PCR |
| ldhF | CGGATGCTGACCTTGTTGTTATC | Internal control for RT-PCR |
| ldhB | AAGTGCTTGACGGAAACGAGC | Internal control for RT-PCR |
| ERM CSTP1 | GGCGCGCCCCGGGCCCAAAATTTGTTTGATa | Erm cassette (28) |
| ERM CSTP2 | GCTGGCCGGACTCGGCAGCGACTCATAGAATb | Erm cassette (28) |
| uvrAP1 | TCTGCTGTTGCGTTCTTCTGG | uvrA allelic-exchange mutagenesis |
| uvrAP2 | GGCGCGCCTTTCCTGAACCCGATAAGCCCGa | uvrA allelic-exchange mutagenesis |
| uvrAP3 | GCTGGCCGGAGCTGAGATGACGGAAAGCTATACb | uvrA allelic-exchange mutagenesis |
| uvrAP4 | CAGAACGGTAACCTGATGCAAC | uvrA allelic-exchange mutagenesis |
Underlined bases represent AscI restriction sequence.
Underlined bases represent FseI restriction sequence.
Selected cDNA PCR fragment bands showing differential gene expression at pH 5.0 were recovered and reamplified. Briefly, cDNA PCR products were excised from the dried sequencing gel, eluted in 100 μl of distilled water by boiling, and ethanol precipitated using glycogen as a carrier. An aliquot of each band was reamplified in a reaction volume of 40 μl using the same primer pair and under the same PCR conditions as the differential display reaction, except that the final dNTP concentration was 20 μM and radioisotope was not added. Reamplified PCR products were then run on a 1% agarose gel, excised, purified using the PCR purification kit (Stratagene), and ligated to pCRscript from the PCR-Script Amp SK(+) cloning kit (Stratagene) according to the manufacturer's instructions. Colonies were screened for inserts using a colony screen-PCR procedure (Invitrogen) that involved disrupting the cells by heating them to 94°C for 10 min prior to PCR and using the T7-T3 promoter primers flanking the inserts. Positive colonies were grown overnight in Luria-Bertani broth containing ampicillin, and plasmid DNA was isolated using a commercial plasmid preparation kit (Qiagen Inc., Mississauga, Ontario, Canada). Plasmids harboring inserts were sequenced from both ends using the original amplification primers with either a Pharmacia ALF or a Perkin-Elmer-ABI Prism 377 sequencer, using either dye primer or dye terminator chemistry (DNA Sequencing Facility at The Center for Applied Genomics, The Hospital for Sick Children, Toronto, Ontario, Canada).
DNA sequence analysis.
The cloned dd-PCR products were compared with other sequences in the National Center for Biotechnology Information databases using the programs BLASTX and BLASTP (1) in an attempt to identify the representative genes. Cloned fragments showing high identity to known genes were compared to the partially completed S. mutans genome database using TBLASTN, available from the University of Oklahoma's Advanced Center for Genome Technology (OU-ACGT at http://www.genome.ou.edu/smutans.html). The contigs containing sequence homologous to the gene fragments were analyzed for open reading frames (ORFs) to obtain the complete gene sequences in S. mutans. The ORF analysis and sequence alignments (ClustalW) were performed using MacVector 7.0 (Oxford Molecular, Madison, Wis.).
RNA dot blots.
To confirm that the recovered clones harbored inserts of differentially expressed pH-inducible genes, the DNA fragments were digoxigenin (DIG) labeled by PCR (Table 1) using the PCR DIG probe synthesis kit (Roche Diagnostics, Laval, Quebec, Canada) according to the manufacturer's instructions and as described in reference 16. The PCR probes were then used to screen RNA dot blots of RNA extracted from S. mutans JH1005 cells grown at pH 7.5 and 5.0. Five micrograms of total RNA was serially diluted and fixed onto a nylon membrane (Boehringer Mannheim). A modification of the RNA dot blot procedure described in the DIG System User Guide for Filter Hybridization manual was used (Roche-Boehringer Mannheim) as previously described (16). The ffh gene was incorporated as a positive control for acid-inducible gene expression as previously demonstrated (16).
RT-PCR.
Using the S. mutans genome database, the complete uvrA ORF was retrieved and internal primers to this region were designed and used in the reverse transcription-PCR (RT-PCR) (Table 1). The ldh gene served as an internal control (Table 1). Calypso RT-PCR (DNAmp Ltd.-Bio/Can Scientific, Mississauga, Ontario, Canada), a one-step RT-PCR system, was used as described by the manufacturer. Briefly, 1 μg of DNase-treated total RNA was added to each reaction mixture. Lactate dehydrogenase (ldh) primers along with the primers for the specific target gene were added together at a concentration of 0.3 μg of primer/μl. Standard curves were constructed for each primer set to determine their optimum cycle number. Samples were subjected to RT-PCR as outlined by the manufacturer with an annealing temperature of 52°C with a total of 24 cycles. Controls without the addition of reverse transcriptase were also performed. These reaction mixtures contained 10 mM dNTPs (Life Technologies) and Taq DNA polymerase (MBI Fermentas, Burlington, Ontario, Canada), not provided in the Calypso kit. A Biometra UNOII thermocycler (Biometra, Inc., Tampa, Fla.) was used for all amplification procedures. Ten microliters of each amplified product was analyzed on a 1% agarose gel containing ethidium bromide.
Genomic DNA isolation and qualitative DNA damage analysis.
Genomic DNA was isolated by the following procedure. An overnight culture of S. mutans JH1005 was grown in 10 ml of TH broth at 37°C. Cells were divided into two 2-ml microcentrifuge tubes, centrifuged, washed in 1 ml of Tris-EDTA buffer, resuspended in 545 μl of TE (50 mM Tris and 10 mM EDTA), and incubated at 60°C for 20 min. Mutanolysin (10 μl from 10,000-U/ml stock) and lysozyme (25 μl from 250-mg/ml stock) (Sigma-Aldrich) were added, and cells were further incubated at 37°C with gentle mixing for 1 h. One hundred microliters of 10% sodium dodecyl sulfate (SDS) was added, and the tubes were gently inverted until the cells lysed. This lysate was incubated at 65°C for 15 min and cooled to room temperature, followed by the addition of 50 μg of proteinase K (Roche Biochemicals) and further incubation at 37°C for 30 min. Then, 0.7 M NaCl–10% cetyltrimethylammonium bromide (CTAB) was added to the mixture, incubation continued for an additional 20 min at 65°C, and the mixture was extracted with 900 μl of chloroform. Cells were phenol-chloroform extracted two to four times followed by DNA precipitation and resuspension in double-distilled water. Samples were RNase (Promega) treated with 1 U of enzyme and incubated for 1 h at 37°C.
For qualitative DNA damage analysis, an overnight culture of S. mutans JH1005 was diluted 10-fold into fresh pH 7.5 THYE and grown to mid-log phase. Cells were divided into 10-ml aliquots, harvested, and resuspended into 10 ml of THYE at pH 7.5, 5.0, and 4.0. These cells were incubated for 3 h at 37°C, followed by genomic DNA isolation omitting the RNase treatment to retain the rRNA. The 23S and 16S rRNA bands were used to standardize the amount of total genomic nucleic acid loaded in each well. Each sample was quantitated using the spectrophotometer (OD260), and 5 μg of each sample was visualized on a 1.0% agarose gel containing ethidium bromide for comparison.
Construction of mutants by allelic-exchange mutagenesis.
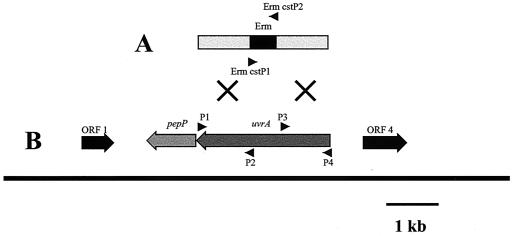
A rapid method for generating mutants in S. mutans JH1005, NG8, and UA159 was employed using PCR. DNA fragments of 1,000 bp which flanked the target gene were ligated to an erythromycin resistance cassette (7, 26). An 860-bp portion of the Erm cassette containing the Ermr marker expressed from a synthetic promoter was amplified for the fusion. The construct was designed so that its integration would not disrupt the original reading frame, minimizing any downstream polar effects. Primers used to amplify the Erm cassette were ERM CSTP1 and ERM CSTP2. uvrAP1-uvrAP2 primers were used to amplify the 3′ flanking region of uvrA, and uvrAP3-uvrAP4 primers were used to amplify the 5′ flanking region (Table 1). Primers P2 and P3 were designed to overlap the target gene within 120 bp of the 5′ and 3′ ends of the ORF sequence, respectively. PCR products for each fragment, generated in triplicate, were purified using the PCR Purification kit (Stratagene); digested with the appropriate restriction enzyme, FseI or AscI (MBI Fermentas); ligated with T4 DNA ligase (Promega); and directly used for natural transformation of S. mutans (Fig. 1).
FIG. 1.
(A) A PCR construct was made by ligating the amplified Erm cassette (Erm CSTP1-CSTP2) and left (P1-P2) and right (P3-P4) flanking regions of uvrA. This was used to transform S. mutans WT strains, resulting in replacement of the uvrA gene with the Erm cassette by double-crossover recombination. Arrowheads mark primer sites used for PCR. The mutagenesis procedure is further described in Materials and Methods. (B) ORF map of S. mutans UA159 uvrA region and neighboring genes and schematic of the mutagenesis of uvrA. ORF 1, potassium channel protein (32% identity, Rattus norvegicus); pepP, aminopeptidase P (50% identity, L. lactis); ORF 4, probable transport protein-membrane protein (33% identity, Deinococcus radiodurans).
Ligated products from the allelic-exchange mutagenesis step (containing the left and right flanking regions and the Erm cassette) were used to transform S. mutans that had been induced to genetic competence by incubation with competence-stimulating peptide (CSP) (27). Briefly, 4 ml of TH broth was inoculated with each strain of S. mutans and incubated overnight at 37°C. A 20-fold dilution of each culture was made into 5 ml of fresh TH broth and incubated to early log phase. Ligated DNA product (10 to 20 μl) along with 500 ng of freshly prepared CSP per ml was added to each tube and incubated a further 1.5 to 2 h. Cells were then centrifuged at 1,000 × g for 10 min, resuspended in 200 μl of TH broth, and plated on THYE plates containing erythromycin. Mutant confirmation was performed by isolating genomic DNA from positive colonies for use as a PCR template with primer sets P1-ERM CSTP2 and P4-ERM CSTP1. The resulting products were analyzed by agarose gel electrophoresis, and their observed sizes were compared to the sizes predicted following successful allelic exchange.
Survival following UV irradiation.
Quantitative assessment of UV-irradiated S. mutans WT and uvrA mutant strains was performed by first inducing the ATR for phenotypic detection (see “Induction of the ATR and phenotypic detection” in Materials and Methods), followed by a 106 dilution of the cells prior to spiral plating on THYE plates. Cells were then incubated at 37°C for 1.5 h and exposed to UV light, as stated above, for 0, 10, 20, and 30 s. Plates were incubated in the dark at 37°C for 24 to 48 h and counted to determine the percent survival.
Growth analysis.
The Bioscreen C (Labsystems, Franklin, Mass.) system was employed to continuously grow cells and measure cell growth for 24 h at 37°C. OD600 was measured every 20 min with shaking every 3 min to prevent cell aggregation. Briefly, overnight S. mutans JH1005 WT and uvrA mutant cells were diluted 10-fold in 1 ml of fresh THYE broth, read at OD600, and adjusted to the same density. Another 10-fold dilution was made into 400 μl of either THYE pH 7.5 or THYE pH 5.0 broth and added to the Bioscreen C wells in triplicate. The growth rates of the cultures were determined by plotting and analyzing their change in OD600 over time using the Bioscreen C BioLink software.
RESULTS
dd-PCR using adapted and unadapted S. mutans JH1005 RNA.
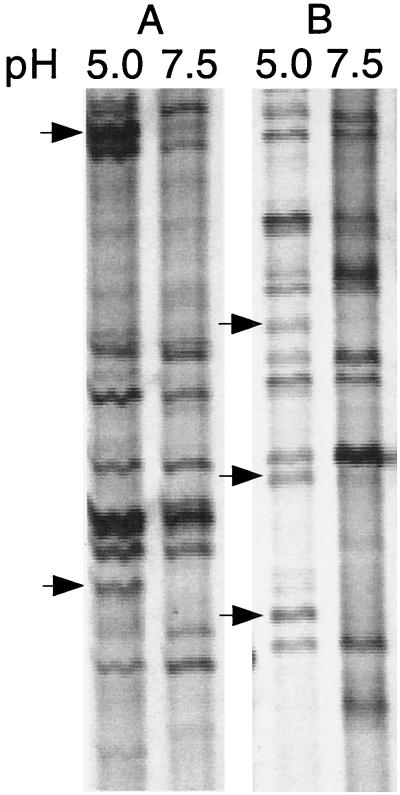
We used dd-PCR to analyze differences in gene expression between unadapted and acid-adapted S. mutans cells. Acid induction was initiated by incubating S. mutans cells for 1 h at pH 7.5 (unadapted) and pH 5.0 (adapted) followed by total RNA isolation to observe early expression of acid-inducible genes. Total RNA from cells incubated at each pH was reverse transcribed into cDNA using random hexamers. Controls in which the addition of reverse transcriptase enzyme had been omitted were used to ensure that the DNA present was newly synthesized cDNA without chromosomal contamination. The two cDNA pools were subjected to PCR using arbitrary primers. Since the entire sequencing gel was difficult to present in a reduced figure, a portion of the entire autoradiograph of the gel resulting from amplification with arbitrary primers H-AP1–H-AP5 and H-AP5–H-AP6 in lanes A and B, respectively, is depicted in Fig. 2. A total of 13 amplicons were observed to be either up-regulated or exclusively present in the pH 5.0 samples, 5 of which are indicated by arrows. Reactions without reverse transcriptase lacked amplification products, indicating that there was no DNA contamination in the RNA preparations (data not shown). Each primer set reaction was performed in triplicate to confirm the reproducibility of the expression patterns observed. DNA in bands representing up-regulation at pH 5.0 was reamplified and cloned into pCRscript. Several ampicillin-resistant colonies were obtained, and plasmid DNA from five or six of these colonies was isolated and sequenced. For each reamplified DNA fragment, multiple products were cloned, since excision of the DNA in a band was not precise, resulting in the presence of products from multiple PCR templates. Three out of the five clones contained DNA representing a partial ORF with 67% identity to a uvrA-DNA repair gene found in B. subtilis. One of the clones harboring a dd-PCR product homologous to the uvrA gene was designated ddPCR4-b and was further characterized in this study. The other 12 clones up-regulated by acid are still under investigation in our laboratory.
FIG. 2.
dd-PCR autoradiograph of amplified S. mutans JH1005 RNA isolated from adapted (pH 5.0) and unadapted (pH 7.5) cells. The cDNAs were amplified with primers H-AP1–H-AP5 (A) and H-AP5–H-AP6 (B) in the portion of the gel shown. The arrows denote pH-inducible gene fragments present at pH 5.0 and absent or greatly reduced at pH 7.5.
Confirmation of the acid inducibility of the uvrA homologue at pH 5.0.
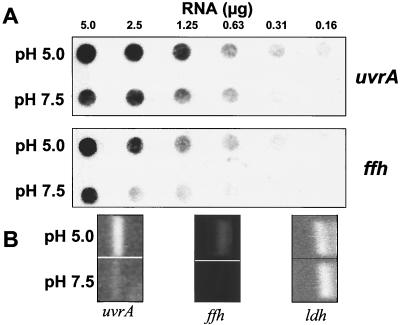
RNA dot blots from S. mutans JH1005 unadapted and adapted cells were probed with the DIG-labeled uvrA homologue cloned insert from the plasmid harbored by clone ddPCR4-b to confirm the acid-inducible expression observed in the dd-PCR experiment. RT-PCR was employed as a second method to confirm up-regulation of the gene at pH 5.0 using uvrA primers that amplified an internal region of the ORF. Parallel experiments were done using ffh as a probe or with ffh-specific primers as a positive control for acid-inducible gene expression. ffh has been shown elsewhere to be up-regulated after incubation of cells at pH 5.0 under the same conditions used in this experiment (16). Acid-adapted cells exhibited an approximately threefold increase in uvrA expression compared to that of unadapted cells (Fig. 3A). As an alternative method to confirm differential expression of the uvrA gene, semiquantitative RT-PCR was performed. Using this method, there appeared to be an even higher difference in relative signal for uvrA and ffh in acid-adapted cells, likely due to the greater sensitivity of the method (Fig. 3B). RT-PCRs included the addition of primers for the internal control gene, lactate dehydrogenase (ldh), known to be equally expressed under the two experimental conditions (16).
FIG. 3.
Dot blot hybridization and RT-PCR of total RNA isolated from S. mutans JH1005 acid-adapted and unadapted cells. The dot blots were probed with the DIG-labeled uvrA cloned fragment and a DNA fragment representing the internal region of ffh (A). RT-PCRs amplified the internal regions of uvrA and ffh (B). The lactate dehydrogenase gene (ldh) served as the internal control for the RT-PCR and was evaluated using the same total RNA preparations for uvrA and ffh. All experiments were repeated more than three times to confirm the acid inducibility of uvrA and ffh.
In silico analysis of the uvrA genetic locus.
Upon confirmation of acid inducibility of the uvrA homologue, we began characterization of the gene in S. mutans. Using the S. mutans genome database, the complete ORF was found to be 2,829 bp in length (contig 51; Feb. 07/01 file). The complete uvrA sequence was translated into protein (∼944 amino acids [104.67 kDa]) and compared with UvrA sequences from other bacteria (Fig. 4). uvrA was highly conserved among both gram-positive and gram-negative bacteria. Analysis of the deduced S. mutans UvrA sequence revealed high structural similarity to common motifs found in other organisms including two zinc finger motif regions and two nucleoprotein-ATP binding sites (2) found at amino acids 252 to 279 and 738 to 764 and amino acids 33 to 40 and 639 to 646, respectively. A search for the remaining NER constituents (uvrB and uvrC) in the S. mutans database revealed that putative uvrB and uvrC genes were located elsewhere in the genome (contigs 53 and 49, respectively; Feb. 07/01 file). Immediately 3′ of uvrA was an ORF showing 50% identity to an aminopeptidase P (pepP) gene from Lactococcus lactis (Fig. 1B). Other neighboring ORFs surrounding uvrA and their putative functions are shown in Fig. 1B.
FIG. 4.
Alignment of the S. mutans UvrA homologue with other UvrA proteins from other bacteria. The sequences were aligned using the program ClustalW. Homologous amino acids are indicated by the shaded areas, and identical amino acids are indicated by the boxed areas. The two zinc finger and nucleoprotein-ATP binding site motifs are located at amino acids 252 to 279 and 738 to 764 and amino acids 33 to 40 and 639 to 646, respectively, in the S. mutans sequence. Abbreviations: S.m, S. mutans; B.s, B. subtilis; M.t, Mycobacterium tuberculosis; E.c, E. coli.
Survival by uvrA mutants of UV exposure.
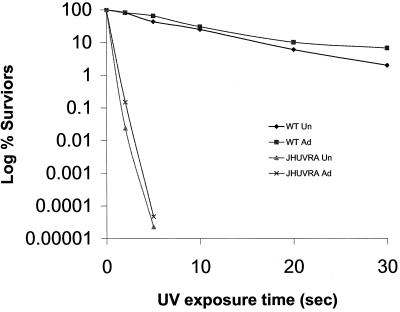
uvrA mutants were constructed in S. mutans strains JH1005, NG8, and UA159 (named JHUVRA, NGUVRA, and UAUVRA, respectively) using allelic-exchange mutagenesis. Here, DNA fragments (approximately 1 kb each) flanking the target gene were amplified (Fig. 1B) and fused to an amplicon containing the erythromycin resistance cassette containing a synthetic promoter (Ermr marker) (Fig. 1A). S. mutans cells were induced to genetic competence by addition of CSP, and the fused construct was then used to transform S. mutans WT strains, resulting in double-crossover recombination and replacement of the target gene with the Erm cassette. Prior to assessment of the uvrA mutant's ability to survive acid challenge, we first determined if a mutant defective in the putative uvrA gene was UV sensitive, since uvrA mutants in other bacteria were extremely sensitive to UV irradiation (8, 10). It has been previously demonstrated that an adaptive response to one stress can often lead to cross-protection against other stresses (20). We sought to determine whether acid-adapted S. mutans JH1005 WT and JHUVRA cells were more resistant to UV irradiation than were unadapted cells. Although it was not immediately apparent from the graph (Fig. 5), analysis by t test revealed that there was a twofold increase (P = 0.02) in survival of adapted cells after 2 s of UV exposure as seen with adapted JHUVRA cells (4.7 × 107 ± 0.1 × 107 CFU/ml) versus unadapted cells (2.3 × 107 ± 0.3 × 107 CFU/ml). We consistently observed more survivors of UV irradiation in the acid-adapted parent strain (6.7 × 106 ± 5.8 × 106 CFU/ml) than in the unadapted parent strain (2.0 × 106 ± 1.2 × 106 CFU/ml) after 30 s; however, this difference was not statistically significant.
FIG. 5.
Survival of unadapted (Un) and adapted (Ad) WT S. mutans JH1005 and uvrA mutant strains after exposure to UV irradiation. Cells were plated at the appropriate dilution after the adaptation period, incubated for 1.5 h, UV irradiated, and incubated for a further 48 h for enumeration. The results are the means of three experiments.
Growth of S. mutans JH1005 WT and JHUVRA during acid challenge.
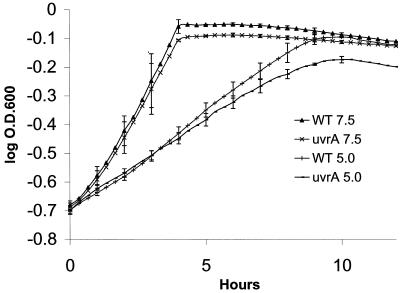
S. mutans JH1005 WT and JHUVRA strains were grown in THYE broth supplemented with 40 mM K2HPO4-citrate buffer adjusted to pH values of 7.5 and 5.0. Using an automated growth reader, we were able to monitor growth of several different cultures simultaneously. This allowed us to perform three independent experiments under each desired condition and to determine the mean for plotting the growth curves. Results showed that both strains grew similarly at pH 7.5, the parent strain JH1005 had a doubling time of 112 min, and JHUVRA doubled every 120 min. Apart from the lower growth rate normally observed at pH 5.0 due to the shift in the cell's energy towards maintaining internal pH homeostasis (9), JH1005 doubled every 269 min while JHUVRA doubled every 312 min (Fig. 6). Similar results where observed with S. mutans strains NG8 and UA159 and their respective mutants (data not shown). Also, the final resting culture density of JHUVRA cells grown at pH 5.0 was shown to be approximately 14% less than that of the WT JH1005. A t test revealed that they were significantly different (P = 0.02). The growth yields of the parent and mutant strains, however, were not statistically different at pH 7.5.
FIG. 6.
Growth curves of S. mutans JH1005 WT and uvrA mutant strains at pH 5.0 and pH 7.5 (control). Each data point is the mean of three independent experiments ± standard deviation.
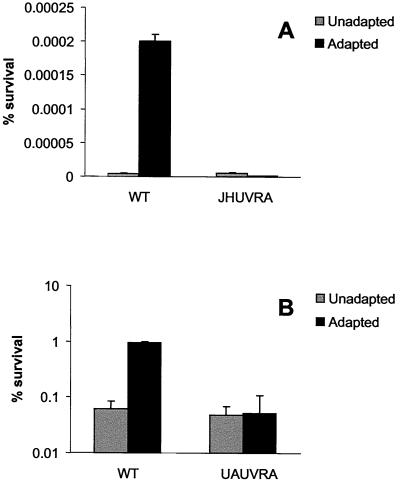
S. mutans JH1005 and UA159 uvrA mutants show deficiency in the ATR.
To determine whether JHUVRA was able to elicit an adaptive response to low pH as observed with the parent, exponential-phase JH1005 WT and JHUVRA cells were incubated at the adaptive pH (5.0) and unadaptive pH (7.5) for 2 h followed by exposure to the killing pH (3.0) for 3 h. Under the conditions outlined previously (37), unadapted JH1005 WT cells showed no survivors after exposure to the killing pH, whereas adapted cells showed a significant number of survivors (Fig. 7A). Unlike the parent, the JHUVRA strain, when exposed to the adaptive pH, did not show enhanced survival after incubation at the killing pH (Fig. 7A). S. mutans JH1005 is, however, known to have a weak adaptive response (ATR) compared to that of other S. mutans strains (37). To strengthen our confidence in the acid-sensitive phenotype of the uvrA mutants, we repeated the ATR experiments with a uvrA mutant that we had constructed with S. mutans UA159. Here, UA159 WT and UAUVRA unadapted and adapted cells were incubated for 3 h at a killing pH of 3.5. The ATR in UA159 displayed a stronger adaptive response, indicated by the significantly larger proportion of survivors found in adapted WT cells. Under the conditions tested, we also observed that UAUVRA demonstrated a reduced capacity in mounting an ATR as demonstrated by a 10-fold decrease in survivors of adapted cells to the killing pH compared to the WT strain (Fig. 7B).
FIG. 7.
ATR of S. mutans JH1005 (A) and UA159 (B) WT and uvrA mutant strains. Mid-log cells were harvested and resuspended in THYE medium, pH 7.5 and 5.0 for unadapted and adapted conditions, respectively. Cells were incubated at 37°C for 2 h followed by a 3-h incubation at the killing pH of 3.0. Each data point represents the mean of three independent experiments ± standard deviation.
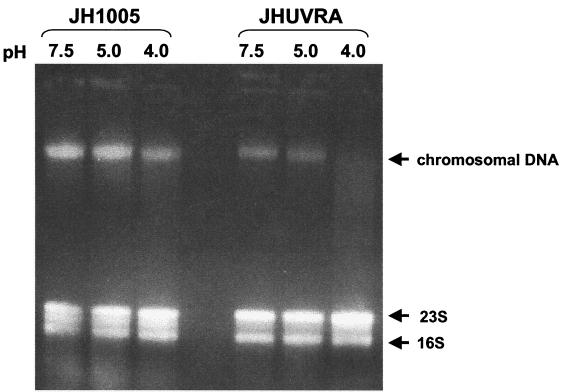
Low external pH (pHo) causes more DNA damage in JHUVRA than in the WT.
To our knowledge, there is no direct evidence that demonstrates that DNA damage occurs in S. mutans during low-pHo exposure. Our observed induction of the DNA repair gene uvrA in S. mutans (by exposure to pH 5.0) suggests that DNA damage may be occurring during low-pHo exposure. To investigate whether uvrA plays a significant role in the repair of acid-induced DNA damage, we grew JH1005 WT and JHUVRA cells to mid-log phase and then exposed them to pH 7.5, 5.0, and 4.0 for 3 h followed by total nucleic acid isolation whereby chromosomal DNA and rRNA could be visualized on an agarose gel. Figure 8 shows JHUVRA chromosomal DNA to be significantly more degraded at pH 4.0 than DNA from the WT. Also, visual assessment of WT DNA indicated that degradation might be occurring at pH 4.0. Slight degradation of JHUVRA DNA is visible in the pH 5.0 sample, while the DNA from the pH 5.0 parent strain appears mostly intact. There appears to be no visible damage to either pH 7.5 sample. There also appears to be less chromosomal DNA in the mutant JHUVRA samples than in the parent strain. This is likely due to a relative increase in the amount of visible rRNA in JHUVRA, since a standardized amount (5 μg) of total nucleic acid was subjected to electrophoresis. Both JH1005 and JHUVRA were not killed (100% survival) after exposure to pH 7.5 and 5.0 for 3 h. After a 3-h exposure to pH 4.0, there was little difference between unadapted parent and mutant cells showing 29.4% (±2.52%) and 19% (±6.49%) survival, respectively (data not shown).
FIG. 8.
Agarose gel electrophoresis of total genomic nucleic acid (consisting of DNA, rRNA, and mRNA) of S. mutans JH1005 and JHUVRA cells exposed to pH 7.5, 5.0, and 4.0 for 3 h. An equal amount (5 μg) of total nucleic acid was added in each well.
DISCUSSION
The dd-PCR technique has been successfully used with prokaryotic cells to evaluate differences in gene expression due to environmental changes (5, 24, 25, 35). We have demonstrated the use of the dd-PCR technique with a gram-positive organism to identify genes differentially expressed due to changes in pH. Further analysis of one gene, uvrA, provided evidence for its role in the ATR. The ATR has been defined as the process of adaptation to acid, whereby exposure to mildly acidic pH (5.5 to 4.0 in S. mutans [37]) affords protection against lower pH values that would otherwise be lethal to the cell. By identifying genes induced at pH 5.0, we hoped to discover genes that encode the proteins necessary for acid adaptation. Using dd-PCR, we visualized increased expression of approximately 13 amplification products in response to exposure to pH 5.0 compared to that of pH 7.5-grown cells (Fig. 1). Each cloned product recovered from the dd-PCR experiment was shown to be heterogeneous, since resolving and excising each band separately was sometimes impossible. The uvrA gene in S. mutans was identified as being acid inducible by dd-PCR and was confirmed by RNA dot blot and reverse transcriptase PCR analysis (Fig. 3).
We retrieved the complete uvrA ORF from the S. mutans genome database and aligned it with sequences of UvrA proteins from other organisms (Fig. 4). The two main DNA binding structural motifs found in UvrA, mainly the two zinc finger and nucleoprotein-ATP binding regions, allow UvrA to preferentially bind to single- and double-stranded DNA breaks which then initiate recruitment of the remaining proteins in the UvrABC complex (2, 39). In bacteria, the UvrABC complex is the principal component of the NER pathway (34), which is the main pathway for the removal of damage caused by UV light. Indeed, in E. coli and other organisms, gene knockouts in any of the NER constituents create mutants that are extremely UV sensitive (8, 10, 40), suggesting that any mutation made in the NER pathway completely obliterates the system. We have shown in S. mutans that uvrA mutants are also extremely sensitive to UV irradiation (Fig. 5), presumably because the entire UV repair system of NER is inoperative. However, mutants with mutations in uvrB and uvrC would need to be constructed and tested for UV sensitivity in S. mutans to make this statement conclusive.
It has been shown previously with S. mutans UR100 (recA mutant) (30) and E. coli K-12 (uvrA and recA mutants) (14) that previous exposure to acidic pH provided cross-protection against UV irradiation. In our present study, we have also shown that acid-adapted S. mutans JHUVRA exhibited a similar phenomenon of increased resistance to UV irradiation. Although the parent strain had consistently higher survival after exposure to UV after acid adaptation, we could not statistically validate these data, suggesting that cross-protection against UV was only partially induced by acid. The extreme UV sensitivity of uvrA mutants suggests that the NER pathway is extremely important in UV damage repair in S. mutans. We can also surmise that another acid-inducible repair system(s) in S. mutans is involved in UV damage repair, since acid-adapted uvrA mutants had a statistically significant resistance to UV irradiation relative to that of unadapted cells (Fig. 5). In addition, other studies with S. mutans have demonstrated that prior adaptation to other stresses, including salt, oxidation, and starvation, increased resistance to acid challenge (38). Also, in L. lactis previous exposure to UV light enhanced resistance to acid challenge (20). These findings collectively suggest that there may be an overlap in DNA repair mechanisms involved in repairing both UV- and acid-induced DNA damage. More data are needed, however, to substantiate this model in S. mutans.
The importance of DNA repair mechanisms for survival of acid shock (pH 4 to 3) has been previously established in S. mutans (30) and other bacteria including E. coli (36) and Helicobacter pylori (40). In these examples, DNA repair-deficient bacteria exposed directly to acid shock have been shown to have poor survival rates compared to the parent strains. Our visual analysis of acid-induced chromosomal DNA damage by gel electrophoresis indicated that there was more DNA damage in JHUVRA than in the DNA repair-proficient JH1005 WT. In the mutant, there was no degradation observable at pH 7.5, a slight amount of degradation observed at pH 5.0, and a substantial amount at pH 4.0 (Fig. 8). The degradation observed with the uvrA-deficient mutant could have resulted from the DNA being less stable and tolerant of the extraction technique than DNA from the parent strain. It is interesting that, although 70% of the parent cells were dead after 3 h of exposure to pH 4.0, the DNA from these samples appears rather intact compared to the DNA of uvrA-deficient cells under the same conditions. These data suggest that uvrA contributes to the repair of acid-induced DNA damage. Additionally, we have shown that disruption of uvrA results in a reduced ability to grow at pH 5.0 (Fig. 6), suggesting that uvrA and possibly other DNA repair systems contribute not only to survival of acid shock but also to growth at moderately acidic pH.
Evidence in E. coli indicates that DNA repair mechanisms are induced during acid adaptation and are responsible for an apparent decrease in DNA damage occurring during growth at pH 5.0 (32, 33). To account for this increase in DNA repair and decrease in DNA damage during acid adaptation, one would envision the necessity of DNA repair mechanisms for maintaining the integrity of the DNA template for successful synthesis of proteins essential for acid adaptation. Without adequate DNA repair from insults such as acidic pH or other damaging agents during the critical adaptation process, the bacteria's ability to successfully adapt and survive at lower pH would be diminished. To evaluate this idea further, we measured the ability of the DNA repair-deficient strain JHUVRA to exhibit an acid-adaptive response (ATR). After incubation at pH 5.0 for 2 h to induce acid adaptation, both WT and uvrA mutant strains were exposed to the killing pH 3.0 for 3 h. Our results showed that adapted JHUVRA cells were unable to survive the killing pH compared to adapted WT cells (Fig. 7A). We also tested the uvrA mutant that we had constructed in UA159 and discovered that the ATR was also impaired (Fig. 7B). Here we observed at a killing pH of 3.5 that adapted UAUVRA mutants had a 10-fold decrease in survival rate compared to that of the WT. These results emphasize the importance of uvrA in survival of acid shock and acid adaptation in S. mutans.
The apparent damage done to DNA during growth at low pH, and the subsequent DNA repair essential for cell survival, suggests that several repair mechanisms may be inducible by acidic pH. The activity of an S. mutans AP endonuclease, involved in repair of damaged or incorrect bases, was also found to be inducible by low pH (17). It is possible that the base repair activity of the AP endonuclease would be responsible for initiating the DNA repair process of UvrABC which is activated by helical distortions, caused by the displacement of bases, rather than by recognition of any particular group (41). Therefore, the role of base excision repair could be to repair minor DNA damage, whereas UvrA and the NER pathway could be responsible for excising larger DNA lesions caused by acid and other DNA-damaging agents. It has been shown with S. mutans that the gene expression and protein levels of the heat shock protein, DnaK, were also up-regulated in response to acid adaptation and acid shock (22). In E. coli, the DnaK protein was also discovered to increase the stability of UvrA during heat stress (43), suggesting that heat shock proteins might indirectly be involved in acid-induced DNA repair by ensuring proper functioning of DNA repair mechanisms in less than optimal conditions. These observations not only support the idea that acid-inducible DNA repair mechanisms exist in S. mutans but also suggest that specialized, regulated forms of DNA repair such as those found in the SOS, heat shock, and adaptive responses potentially exist and likely have a significant overlap with the ATR. These responses could either operate as independently regulated systems or overlap in their activities in response to acid-induced DNA damage. Further evidence that these regulatory networks exist in S. mutans is provided by one- and two-dimensional SDS-PAGE studies that compared protein extracts from acid-induced and uninduced cells and demonstrated that the synthesis of acid-regulated proteins included acid-specific proteins and general stress proteins (e.g., heat shock) (19, 38). This work described up to 36 proteins up-regulated by acid adaptation, while we could observe only 13 clearly defined up-regulated products. This is likely due to the need for further optimization of our primer design for representation of the entire S. mutans genome. Alternately, the two-dimensional studies could have multiple protein spots due to proteolysis of some of the up-regulated products. Obviously, the involvement of these regulatory networks in DNA repair and acid adaptation in S. mutans needs to be investigated further.
This study supports earlier observations made with S. mutans and other acid-tolerant bacteria as to the importance of DNA repair in survival of low pH. We have confirmed that S. mutans mutants defective in uvrA are UV sensitive. We have also confirmed that this acid-inducible gene is involved in growth at moderate pH and the ATR. The dd-PCR technique was also shown to be effective in identifying acid-inducible genes. Future work will involve further characterization of other acid-inducible genes identified in our dd-PCR experiment in combination with two-dimensional SDS-PAGE and microarray techniques to better characterize the genes, proteins, and regulatory networks involved in the process of adaptation to low pH.
ACKNOWLEDGMENTS
We thank J. D. Hillman for providing S. mutans strain JH1005, A. S. Bleiweis for NG8, J. Ferretti for UA159, D. Morrison for the Erm cassette, and also Tammy Flagg from Shands Cancer Center for technical assistance.
We greatly appreciate public release of the Streptococcus mutans Genome Sequencing Project, funded by a USPHS/NIH grant from the National Institute of Dental and Craniofacial Research and B. A. Roe, R. Y. Tian, H. G. Jia, Y. D. Qian, S. P. Linn, L. Song, R. E. McLaughlin, M. McShan, and J. Ferretti from The University of Oklahoma.
Our work was supported by operating grant MT-15431 from the Medical Research Council of Canada and by infrastructure grants from the Canadian Foundation for Innovation and The Ontario Innovation Trust. M. N. Hanna is the recipient of a University of Toronto Open Fellowship and Ontario Graduate Scholarship in Science and Technology. D.G. Cvitkovitch is the recipient of a Canada Research Chair in Microbiology.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Aravind L, Walker D R, Koonin E V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender G R, Sutton S V W, Marquis R E. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect Immun. 1986;53:331–338. doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bijlsma J J E, Lie-A-Ling M, Nootenboom I C, Vandenbroucke-Grauls C M J E, Kusters J G. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J Infect Dis. 2000;182:1566–1569. doi: 10.1086/315855. [DOI] [PubMed] [Google Scholar]
- 5.Bonass W A, Marsh P D, Percival R S, Aduse-Opoku J, Hanley S A, Devine D A, Curtis M A. Identification of ragAB as a temperature-regulated operon of Porphyromonas gingivalis W50 using differential display of randomly primed RNA. Infect Immun. 2000;68:4012–4017. doi: 10.1128/iai.68.7.4012-4017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd D A, Cvitkovitch D G, Bleiweis A S, Kiriukhin M Y, Debabov D V, Neuhaus F C, Hamilton I R. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results [sic] in acid sensitivity. J Bacteriol. 2000;182:6055–6065. doi: 10.1128/jb.182.21.6055-6065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claverys J-P, Dintilhac A, Pestova E V, Martin B, Morrison D A. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- 8.Courcelle J, Crowley D J, Hanawalt P C. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and RecF protein function. J Bacteriol. 1999;181:916–922. doi: 10.1128/jb.181.3.916-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dashper S G, Reynolds E C. PH regulation by Streptococcus mutans. J Dent Res. 1992;71:1159–1165. doi: 10.1177/00220345920710050601. [DOI] [PubMed] [Google Scholar]
- 10.De la Morena M L, Hendrixson D R, St. Geme J W. Isolation and characterization of Haemophilus influenzae uvrA gene. Gene. 1996;177:23–28. doi: 10.1016/0378-1119(96)00264-8. [DOI] [PubMed] [Google Scholar]
- 11.Foster J W. When protons attack: microbial strategies of acid adaptation. Curr Opin Microbiol. 1999;2:170–174. doi: 10.1016/S1369-5274(99)80030-7. [DOI] [PubMed] [Google Scholar]
- 12.Foster J W, Hall H K. Adaptive acidification tolerance response of Salmonella typhimurium. J Bacteriol. 1990;172:771–778. doi: 10.1128/jb.172.2.771-778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster J W, Hall H K. Inducible pH homeostasis and the acid tolerance response of Salmonella typhimurium. J Bacteriol. 1991;173:5129–5135. doi: 10.1128/jb.173.16.5129-5135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodson M, Rowbury R J. RecA-independent resistance to irradiation with U.V. light in acid-habituated Escherichia coli. J Appl Bacteriol. 1991;70:177–180. doi: 10.1111/j.1365-2672.1991.tb04445.x. [DOI] [PubMed] [Google Scholar]
- 15.Grogan D W. The question of DNA repair in hyperthermophilic archaea. Trends Microbiol. 2000;8:180–185. doi: 10.1016/s0966-842x(00)01729-7. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez J A, Crowley P J, Cvitkovitch D G, Brady L J, Hamilton I R, Hillman J D, Bleiweis A S. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology. 1999;145:357–366. doi: 10.1099/13500872-145-2-357. [DOI] [PubMed] [Google Scholar]
- 17.Hahn K, Faustoferri R C, Quivey R G., Jr Induction of an AP endonuclease activity in Streptococcus mutans during growth at low pH. Mol Microbiol. 1999;31:1489–1498. doi: 10.1046/j.1365-2958.1999.01292.x. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton I R, Buckley N D. Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol Immunol. 1991;6:65–71. doi: 10.1111/j.1399-302x.1991.tb00453.x. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton I R, Svensäter G. Acid-regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiol Immunol. 1998;13:292–300. doi: 10.1111/j.1399-302x.1998.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 20.Hartke A, Bouche S, Gansel X, Boutibonnes P, Auffray Y. UV-inducible proteins and UV-induced cross protection against acid, ethanol, H2O2 or heat treatments of Lactobacillus lactis subsp. Lactis Arch Microbiol. 1995;163:329–336. [Google Scholar]
- 21.Hillman J D, Dzuback A L, Andrews S W. Colonization of the human oral cavity by a Streptococcus mutans mutant producing increased bacteriocin. J Dent Res. 1987;66:1092–1094. doi: 10.1177/00220345870660060101. [DOI] [PubMed] [Google Scholar]
- 22.Jayaraman G C, Penders J E, Burne R A. Transcriptional analysis of the Streptococcus mutans hrcA, grpE, and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol. 1997;25:329–341. doi: 10.1046/j.1365-2958.1997.4671835.x. [DOI] [PubMed] [Google Scholar]
- 23.Jensen M E, Polansky P J, Schachtele C F. Plaque sampling and telemetry for monitoring acid production on human buccal tooth surfaces. Arch Oral Biol. 1982;27:21–31. doi: 10.1016/0003-9969(82)90172-8. [DOI] [PubMed] [Google Scholar]
- 24.Kullen M J, Klaenhammer T R. Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus differential display: gene structure, cloning and characterization. Mol Microbiol. 1999;33:1152–1161. doi: 10.1046/j.1365-2958.1999.01557.x. [DOI] [PubMed] [Google Scholar]
- 25.Kwaik Y A, Pederson L L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee M S, Morrison D A. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol. 1999;181:5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y H, Lau P C-Y, Lee J H, Ellen R P, Cvitkovitch D G. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin J J, Sancar A. (A)BC excinuclease: the Escherichia coli nucleotide excision repair enzyme. Mol Microbiol. 1992;6:2219–2224. doi: 10.1111/j.1365-2958.1992.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 29.Paden E, Zilberstein D, Schuldiner S. PH homeostasis in bacteria. Biochim Biophys Acta. 1981;650:151–166. doi: 10.1016/0304-4157(81)90004-6. [DOI] [PubMed] [Google Scholar]
- 30.Quivey R G, Jr, Faustoferri R C, Clancy K A, Marquis R E. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol Lett. 1995;126:257–262. doi: 10.1111/j.1574-6968.1995.tb07427.x. [DOI] [PubMed] [Google Scholar]
- 31.Quivey R G, Jr, Faustoferri R, Monahan K, Marquis R. Shifts in membrane fatty acid profiles associated with acid adaptation of Streptococcus mutans. FEMS Microbiol Lett. 2000;189:89–92. doi: 10.1111/j.1574-6968.2000.tb09211.x. [DOI] [PubMed] [Google Scholar]
- 32.Raja N, Goodson M, Chui W C M, Smith D G, Rowbury R J. Habituation to acid in Escherichia coli: conditions for habituation and its effects on plasmid transfer. J Appl Bacteriol. 1991;70:59–65. doi: 10.1111/j.1365-2672.1991.tb03787.x. [DOI] [PubMed] [Google Scholar]
- 33.Raja N, Goodson M, Smith D G, Rowbury R J. Decreased DNA damage by acid and increased repair of acid-damaged DNA in acid-habituated Escherichia coli. J Appl Bacteriol. 1991;70:507–511. doi: 10.1111/j.1365-2672.1991.tb02748.x. [DOI] [PubMed] [Google Scholar]
- 34.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 35.Shepard B D, Gilmore M S. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl Environ Microbiol. 1999;65:1470–1476. doi: 10.1128/aem.65.4.1470-1476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinha R P. Toxicity of organic acids for repair-deficient strains of Escherichia coli. Appl Environ Microbiol. 1986;51:1364–1366. doi: 10.1128/aem.51.6.1364-1366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svensäter G, Larsson U-B, Greif E C G, Cvitkovitch D G, Hamilton I R. Acid tolerance response and survival by oral bacteria. Oral Microbiol Immunol. 1997;12:266–273. doi: 10.1111/j.1399-302x.1997.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 38.Svensäter G, Sjogreen B, Hamilton I R. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology. 2000;146:107–117. doi: 10.1099/00221287-146-1-107. [DOI] [PubMed] [Google Scholar]
- 39.Thiagalingam S, Grossman L. Both ATPase sites of Escherichia coli UvrA have functional roles in nucleotide excision repair. J Biol Chem. 1991;266:11395–11403. [PubMed] [Google Scholar]
- 40.Thompson S A, Blaser M J. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect Immun. 1995;63:2185–2193. doi: 10.1128/iai.63.6.2185-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voet D, Voet J. Biochemistry. Toronto, Ontario, Canada: John Wiley and Sons, Inc.; 1990. p. 968. [Google Scholar]
- 42.Yousheng M, Curran T M, Marquis R E. Rapid procedure for acid adaptation of oral lactic-acid bacteria and further characterization of the response. Can J Microbiol. 1997;43:143–148. doi: 10.1139/m97-019. [DOI] [PubMed] [Google Scholar]
- 43.Zou Y, Crowley D J, Van Houten B. Involvement of molecular chaperonins in nucleotide excision repair. DnaK leads to increased thermal stability of UvrA, catalytic UvrB loading, enhanced repair, and increased UV resistance. J Biol Chem. 1998;273:12887–12892. doi: 10.1074/jbc.273.21.12887. [DOI] [PubMed] [Google Scholar]