Abstract
The stationary-phase response exhibited by Escherichia coli upon nutrient starvation is mainly induced by a decrease of the ClpXP-dependent degradation of the alternate primary ς factor RpoS. Although it is known that the specific regulation of this proteolysis is exercised by the orphan response regulator SprE, it remains unclear how SprE's activity is regulated in vivo. Previous studies have demonstrated that the cellular content of SprE itself is paradoxically increased in stationary-phase cells in an RpoS-dependent fashion. We show here that this RpoS-dependent upregulation of SprE levels is due to increased transcription. Furthermore, we demonstrate that sprE is part of the two-gene rssA-sprE operon, but it can also be transcribed from an additional RpoS-dependent promoter located in the rssA-sprE intergenic region. In addition, by using an in-frame deletion in rssA we found that RssA does not regulate either SprE or RpoS under the conditions tested.
Bacteria are constantly sampling their surroundings and regulating gene expression accordingly. Since many of the environments they encounter often have hazardous conditions (for example, limiting nutrients, high osmolarity, extreme pH, or extreme temperature), bacteria have evolved to survive in such hostile habitats. In particular, the gram-negative bacterium Escherichia coli enters a state in its life cycle known as the stationary phase, which renders it highly resistant to unfavorable environmental conditions.
When cells enter stationary phase, they undergo dramatic changes in their morphology and physiology that increase their chance for survival in a wide variety of stresses. This cross-protection results from the global control system regulated by RpoS. RpoS, encoded by the rpoS gene, is the second primary ς factor of E. coli, and it is required for the transcription of stationary-phase-specific genes. Due to the drastic consequences (i.e., slowed metabolism) of entering stationary phase, RpoS is tightly regulated. In fact, RpoS is regulated at all levels: transcription, translation, protein stability, and activity (for a recent review, see reference 13).
Among the possible stresses that can induce RpoS (the stationary-phase response), starvation of an essential nutrient is perhaps the most widely studied. When nutrients are readily available, the levels of RpoS are very low, mainly due to its efficient degradation by the ATP-dependent ClpXP serine protease. Conversely, when nutrients become limiting for growth, this ClpXP-dependent proteolysis stops and, consequently, RpoS levels increase significantly (26). This mode of RpoS regulation has been shown to occur in response to carbon starvation as well as during growth in Luria-Bertani (LB) medium, although the specific signals sensed in the latter medium remain to be determined (25, 32).
The regulation of RpoS proteolysis is not mediated by controlling either the levels of the ClpXP protease itself or its activity (33). Instead, it is orchestrated by the response regulator SprE (named RssB in E. coli, MviA in Salmonella enterica serovar Typhimurium, and ExpM in Erwinia carotovora) (1, 2, 22, 25). In a recent report, Zhou et al. demonstrated in vitro that SprE plays a catalytic role in the delivery of RpoS to ClpX, the regulatory component of ClpXP that is believed to unfold RpoS and eventually feed it to ClpP, the proteolytic component (34). ClpP then degrades RpoS and SprE is released from the proteolytic complex. Furthermore, Zhou et al. also showed that this in vitro degradation is greatly enhanced upon SprE phosphorylation (34).
To date, it remains unknown how SprE is phosphorylated in vivo; therefore, SprE is an orphan response regulator. Moreover, unphosphorylated SprE can still promote, although less efficiently, RpoS degradation (reference 6 and unpublished results cited in reference 34). This raises the possibility that SprE might be regulated by a mechanism(s) other than phosphorylation.
A possible mechanism for regulating SprE-mediated degradation of RpoS is to control the levels of SprE itself. Paradoxically, the levels of SprE (and MviA) have been shown to increase when cells enter stationary phase (11, 21). Specifically, it has been shown that the translation of sprE increases in an RpoS-dependent manner (11). In addition, it was reported that sprE (and mviA) transcription is also upregulated during the stationary phase (11, 21). Although those studies did not prove the exact location of the sprE promoter, their reporter fusion data showed that sprE transcription can occur independently from that of its upstream gene, rssA (11). Prior to these studies it was assumed, based on DNA sequence analysis, that rssA and sprE constitute an operon (5, 22). In addition, RssA itself was implicated in the SprE pathway regulating RpoS, although its function has never been clearly demonstrated (unpublished results cited in references 13 and 22).
Since it is unclear how sprE transcription is regulated and what role RssA plays in RpoS regulation, we have constructed a series of reporter fusions and rssA null alleles that allow us to address these issues. In this report we present data demonstrating that although rssA and sprE constitute an operon, sprE can also be transcribed from an RpoS-regulated promoter located in the rssA-sprE intergenic region. In addition, while RpoS controls RssA levels, we found no role for RssA in the regulation of either SprE or RpoS under the conditions tested.
MATERIALS AND METHODS
Bacterial strains and bacteriophages.
E. coli DH5α (Invitrogen Life Technologies) was used as the host strain for all plasmid constructions. All other bacterial strains used (Table 1) are derivatives of MC4100 (7). Standard microbial techniques were used for strain construction (27). All fusions were recombined with λRZ5 (23).
TABLE 1.
Bacterial strains
| Strain | Genotype | Referencea |
|---|---|---|
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 7 |
| AF633 | MC4100 λφ(uspB'-lacZ+) | 9 |
| NR247 | AF633 rpoS::kan | |
| NR252 | AF633 sprE19::cam | |
| NR253 | AF633 sprE::tet | |
| NR260 | AF633 rssA1::kan | |
| NR270 | AF633 rssA1::kan sprE19::cam | |
| NR286 | AF633 rssA1::kan sprE::tet | |
| NR289 | MC4100 nadA::Tn10 Δ(gal-att-bio) | |
| NR501 | MC4100 λφ(rssA'-'lacZ) | |
| NR502 | NR501 rpoS::kan | |
| NR507 | MC4100 λφ(rssA-sprE'-lacZ+) | |
| NR508 | NR507 rpoS::kan | |
| NR509 | NR289 λφ(rssA-sprE'-lacZ+) | |
| NR510 | NR509 rpoS::kan | |
| NR511 | MC4100 λφ(‘rssA-sprE’-lacZ+) | |
| NR512 | NR511 rpoS::kan |
Strains without a given reference were constructed in this study.
Media and growth conditions.
LB medium was prepared as described previously (27). Unless indicated, all bacterial strains were grown under aeration at 37°C and their growth was monitored by measuring the optical density at 600 nm (OD600).
DNA manipulations.
Plasmid DNA was purified by standard techniques and was introduced into the appropriate strains by the method of Kushner (17). All restriction endonucleases (New England Biolabs), Taq and Pfu DNA polymerases (Roche and Stratagene, respectively), T4 polynucleotide kinase (T4 PNK) (New England Biolabs), and T4 DNA ligase (New England Biolabs) were used according to the recommendations of their respective manufacturers. Primers were synthesized by the Princeton University Department of Molecular Biology Synthesis and Sequencing Facility.
Plasmid construction.
The plasmids used in complementation studies were constructed as follows. A 3.06-kbp fragment containing the ychJ-rssA-sprE region was amplified from MC4100 chromosomal DNA using the primers RSSAUP (5′-CGGGATCCCGCCAGAGAACGTAAAGTATCG) and 3SPREMBP2 (5′-CCACCAGCCAAGCTTAGCAGG), which introduce BamHI and HindIII restrictions sites (underlined) at the 5′ and 3′ ends of the PCR products, respectively. After digestion with these restriction enzymes, the PCR fragment was inserted between the BamHI and HindIII sites of pBluescript KS(+) (Stratagene) and pBBR1MCS (15) to yield pRSSA5 and pRSSA6, respectively.
We also constructed a plasmid (pRSSA9) that contains the ychJ-rssA region but not sprE. The 2,332-bp BamHI-BglII fragment from pRSSA5 containing the ychJ-rssA region (and the first 332 bp of sprE) was introduced into BamHI-digested pBBR1MCS, resulting in pRSSA9.
Insertional inactivation of rssA.
A null rssA allele was constructed by insertion of a kanamycin cassette into the PstI site in the RssA-coding region (580 bp after the first guanine of the GTG start codon). The RssA open reading frame was amplified from MC4100 chromosomal DNA by PCR using primers 5RSSAKPN1 (5′-GCGATATGGGTACCATTGCATTCC) and 3RSSAHIND3 (5′-TTCTGGTCAAGCTTCGGTGCGTACC), which introduce KpnI and HindIII restriction sites (underlined), respectively. The resulting 0.94-kbp fragment was digested with KpnI and HindIII and ligated with KpnI-HindIII-digested pRSETB (Invitrogen Life Sciences), and the new plasmid was named pRSSA2. Then, the 794-bp EcoRV-HindIII fragment containing the 3′ end of rssA was introduced into SmaI-HindIII-digested pBluescript KS(+) to generate pRSSA3. The PstI fragment containing the kanamycin cassette from pUC4K (30) was then inserted into the PstI site of pRSSA3 to create pRSSA3::kan. The kanamycin cassette was inserted in an orientation opposite to that of rssA transcription (confirmed by DNA sequencing). The XbaI-KpnI fragment from pRSSA3::kan, containing rssA::kan, was ligated with XbaI-KpnI-digested pAMPTS (24), and the resulting plasmid, pRSSA::kanTs, was used for the allelic exchange in MC4100 as previously described (12).
Introduction of an in-frame deletion into rssA.
A defined in-frame deletion in rssA was generated in pRSSA5 (which carries the ychJ-rssA-sprE region; see above) by using the method of inside-out PCR previously described (14). Briefly, pRSSA5 served as the template for an inside-out PCR with primers RTRSSA1 (5′-CCTCTCGCCGCGCCAGATCCC) and RTRSSA2 (5′-GCACGATGCTCATTTGATGC), which were designed to create an in-frame deletion of amino acids 30 to 194 of RssA. The 5.47-kbp PCR product was phosphorylated using T4 PNK and self-ligated to yield pRSSA5Δ, which contains the 492-bp in-frame deletion allele of rssA named rssAΔ1. The pRSSA5Δ plasmid was then digested with the BamHI and HindIII restriction endonucleases and the fragment containing the ychJ-rssAΔ1-sprE region was inserted between the BamHI and HindIII sites of pBBR1MCS to generate pRSSA6Δ.
Construction of rssA-lacZ and sprE-lacZ fusions.
An rssA'-‘lacZ translational fusion containing the ychJ-rssA’ region was constructed as described below. Two sprE'-lacZ+ transcriptional fusions were also made: one containing the ychJ-rssA-sprE' region (rssA-sprE'-lacZ+) and the other containing only an ‘rssA-sprE’ fragment (‘rssA-sprE’-lacZ+). The appropriate regions (see below) were inserted into either pRS414 (for the rssA'-‘lacZ fusion) or pRS415 (for both sprE’-lacZ+ fusions) (28). All fusion-containing plasmids were recombined into the phage λRZ5 (23) as described by Simons et al. (28). For integration of the fusions at the λ attachment (att) site, MC4100 was infected with the appropriate recombinant λ phage. For integration of the fusions at the chromosomal sprE locus, att deletion MC4100 derivative strains (Table 1) were infected with the appropriate recombinant λ phage.
The pRS414 derivative plasmid containing the rssA'-‘lacZ protein fusion was constructed as follows. A PCR product was amplified from MC4100 chromosomal DNA using primers 5RSSAECOR1 (5′-GGAATTCGCCGCGATTTCGACATCC), which introduces an EcoRI site (underlined), and 3RSSAHIND3 (see above). The resulting PCR product was digested with EcoRI and EcoRV to generate a 1.07-kbp fragment containing the ychJ-rssA’ region (from 395 bp downstream of ychJ to the first 141 bp of rssA), which was then inserted between the EcoRI and SmaI sites of pRS414, resulting in pRSSA11.
The pRS415 derivative plasmid containing the rssA-sprE'-lacZ+ fusion (pCNP1) was created by introducing into the SmaI site of pRS415 a 2.11-kbp fragment that had been generated by PCR amplification from MC4100 DNA by using primers 5RSSAECOR1 (see above) and 3SPREBAMH1 (5′-CGGGATCCGCCAGTACCGTTGTCGCTC). The amplified region extends from 395 bp downstream of ychJ to 112 bp into the sprE coding region. To construct the pRS415 derivative plasmid containing the ‘rssA-sprE’-lacZ+ fusion (pRSSA10), the 10,660-bp EcoRV fragment from pRSSA9 (see above) containing the region from the last 798 bp of rssA to the first 174 bp of sprE was introduced into the SmaI site of pRS415.
β-Galactosidase assays.
After growing overnight in LB broth, cells were diluted 1:100 into fresh LB broth and grown to an OD600 of ∼0.3 to 0.4 for logarithmic-phase samples or to an OD600 of ∼3.0 for stationary-phase samples. β-Galactosidase assays were performed using a microtiter plate assay as described previously (29). The β-galactosidase activities were expressed as ΔOD420/(OD600 × volume), where volume refers to the amount (in milliliters) of cell lysate used. For each experiment, every sample was assayed three times and the average activity and standard deviation (SD) were obtained. The data shown resulted from a single experiment representative of at least three other independent experiments.
Western blot analysis.
Cells were grown as indicated above for the β-galactosidase assays to obtain both logarithmic- and stationary-phase samples. Once cells reached the indicated OD600, 1-ml samples were pelleted. To standardize samples, the pellets were resuspended in a volume of sodium dodecyl sulfate (SDS) sample buffer (18) equal to the OD600/6. Samples were boiled for 5 min and equal volumes were subjected to SDS–12% polyacrylamide gel electrophoresis as described by Laemmli (18). The proteins were transferred to nitrocellulose membranes (Schleicher & Schuell), and Western blot analyses were performed as previously described (10). When appropriate, polyclonal sera against RpoS or SprE were used as primary antibodies at a dilution of 1:6,000 and 1:4,000, respectively. Donkey anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (Amersham Pharmacia Biotech) was used as secondary antibody at a 1:6,000 dilution. For visualization of bands, the ECL antibody detection kit (Amersham Pharmacia Biotech) and X-Omat film (Kodak) were used.
Primer extension analysis.
AF633 cells were grown to stationary phase as described above for the β-galactosidase assays. Total RNA was extracted using Trizol (Invitrogen Life Sciences). Primer RTSPRE1 (5′-AGCCGCCAGTACCGTTGTCGC) was labeled with [γ-33P]ATP (ICN) using T4 PNK prior to primer extension. For the primer extension reaction, 5 μg of total RNA, labeled primer, and 100 U of Moloney murine leukemia virus reverse transcriptase (U.S. Biochemicals) were used as directed by the manufacturer. For the sequencing reaction, 3 μg of pRSSA5 plasmid was digested with HindIII for 15 min. After the digestion, pRSSA5 was mixed with 0.5 pmol of unlabeled RTSPRE1 primer and denatured by boiling for 3 min. After cooling on ice for 20 min, the sequences were determined by using a Sequenase DNA sequencing kit (version 2; U.S. Biochemicals) with [α-33P]dATP (ICN) according to the directions of the manufacturer. All reactions were subjected to electrophoresis in an 8.3 M urea–6% polyacrylamide gel. The reaction products were visualized on X-Omat film (Kodak).
RESULTS
Disruption of rssA increases RpoS-mediated transcription.
Sequence homology suggests that RssA belongs to a family of serine esterases/proteases found from bacteria to humans (20). Unfortunately, the only members of this family with a characterized function are those found in Drosophila and humans; their function pertains to neuronal development and they contain additional domains not found in their bacterial counterparts (16, 20).
To clarify RssA's role in RpoS regulation, we constructed a null allele (rssA1::kan) by insertionally inactivating rssA with a kanamycin resistance (kan) cassette after nucleotide 576 (where 1 corresponds to the first guanine of the GTG start codon). This rssA1::kan allele was introduced into strain AF633, which carries the RpoS-dependent uspB'-lacZ+ fusion (Table 1) (9), and β-galactosidase activity was measured to monitor RpoS-mediated transcriptional activity. After growing in LB medium, stationary-phase cultures carrying the rssA1::kan allele had a 2.4-fold increase in the levels of β-galactosidase activity above those of the wild-type parent strain. Similar results were found using other known RpoS-dependent LacZ transcriptional fusions (data not shown). Although the increase in RpoS activity caused by the rssA1::kan allele was significant, it must be pointed out that it was not as high as that caused by a sprE null allele tested under the same conditions (a threefold increase with respect to the wild type). This difference in RpoS-dependent activities between the sprE and rssA null strains was also detectable on lactose MacConkey indicator media, and it was further confirmed by the fact that while the sprE null strain cannot grow in minimal succinate medium (due to its high levels of RpoS [see reference 24]), the rssA null strain can. Moreover, since a strain carrying both rssA and sprE null alleles has levels of RpoS activity equivalent to those of the sprE null strain, the rssA and sprE null alleles do not function in additive fashion. Thus, these results show that disruption of rssA increases RpoS-dependent transcription and that a mutation in sprE is epistatic to rssA.
Disruption of rssA increases RpoS levels by altering SprE levels.
Since SprE regulates RpoS at the level of protein stability, it is likely that the increase in RpoS-mediated transcription caused by the rssA1::kan allele reflects increased RpoS levels rather than increased specific activity of RpoS. To test this, we determined the relative levels of RpoS by Western blot analysis in both logarithmic- and stationary-phase cultures of various strains grown in LB medium.
In wild-type cells, the levels of RpoS increased as cells entered stationary phase (Fig. 1A and B, compare wt lanes). This increase in RpoS levels is largely dependent on SprE-mediated regulation of its proteolysis by ClpXP. Therefore, altering SprE levels affects the content of RpoS in the cell. As shown previously (25), cells carrying the sprE19::cam allele contain higher levels of SprE than do wild-type cells (Fig. 1C, compare wt and sprE19::cam lanes), and this results in lower levels of RpoS (Fig. 1B, compare wt and sprE19::cam lanes) and RpoS activity (a ca. threefold decrease in uspB'-lacZ+ activity). Accordingly, depleting cells of SprE (Fig. 1C, sprE::tet lane) increased RpoS levels throughout the entire life cycle (Fig. 1A and B, sprE::tet lanes).
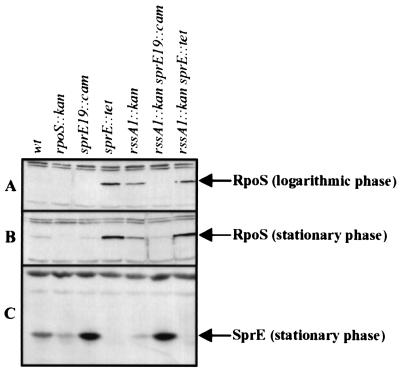
FIG. 1.
Disruption of rssA increases the levels of RpoS by reducing those of SprE, owing to polarity. (A and B) Western blot analysis was used to monitor the levels of RpoS in total cell lysates recovered during the logarithmic (A) and stationary (B) phases of growth in LB medium. (C) The same stationary-phase samples were used to determine the levels of SprE. The bands corresponding to RpoS and SprE are marked with labeled arrows. Lanes in each panel: wt, strain AF633; rpoS::kan, strain NR247; sprE19::cam, strain NR252; sprE::tet, strain NR253; rssA1::kan, strain NR260; rssA1::kan sprE19::cam, strain NR270; rssA1::kan sprE::tet, strain NR286. The sample orders are the same in all panels.
As shown in Fig. 1A and B, both logarithmic- and stationary-phase cells carrying the rssA1::kan allele contained increased levels of RpoS above that of the wild-type parent strain. This increase in RpoS levels correlated with the increase in RpoS-dependent transcription discussed above. Also in line with the results found for RpoS-mediated transcription, the levels of RpoS in the rssA1::kan strain were not as high as those found in the sprE::tet strain (Fig. 1A and B). In addition, RpoS levels did not further increase in a strain carrying both rssA1::kan and sprE::tet alleles with respect to those found in one carrying only sprE::tet (Fig. 1A and B, compare rssA1::kan sprE::tet and sprE::tet lanes). More importantly, the increased levels of RpoS detected in the rssA null strain correlated with reduced levels of SprE (Fig. 1C, compare wt and rssA1::kan lanes). In summary, the results presented here show that disruption of rssA decreases, but does not eliminate, SprE levels in the cell. This most likely accounts for the increase in the cellular RpoS content seen in the rssA1::kan strain.
The decreased levels of SprE in the rssA1::kan null strain are the result of polarity.
The decreased levels of SprE reported above that occurred upon disruption of rssA can be explained as follows. If rssA and sprE are cotranscribed, disruption of rssA would result in lowered sprE expression. Alternatively, RssA could be a positive regulator of SprE levels. In addition, both of these scenarios could be true.
In order to address these issues, we first tested whether the effects of rssA1::kan on sprE expression are the result of polarity. To accomplish this, we uncoupled sprE transcription from that of rssA by using the sprE19::cam allele, which carries a mini-Tncam cassette 27 bp upstream from the adenine of the ATG start codon of sprE (25). As stated in the previous section, strains carrying the sprE19::cam allele contain high levels of SprE and, therefore, low levels of RpoS (Fig. 1). Interestingly, when the sprE19::cam allele was present in cis with the rssA1::kan allele, the levels of SprE, RpoS, and RpoS-dependent transcription (as assessed by measuring the β-galactosidase activity of uspB'-lacZ+) did not change from those found in the strain carrying the sprE19::cam allele alone (Fig. 1A to C, compare sprE19::cam and rssA1::kan sprE19::cam lanes). Thus, when the transcription of sprE is uncoupled from that of rssA, disruption of rssA has no effect on either SprE or RpoS, suggesting that RssA has no role in the posttranscriptional regulation of these proteins under the conditions tested.
The notion that RssA does not regulate either SprE or RpoS was further supported by the following complementation studies. We introduced into a low-copy-number plasmid (pBBR1MCS) either the entire region encompassing ychJ-rssA-sprE (pRSSA6) or just ychJ-rssA (pRSSA9). The presence of the plasmid carrying the entire ychJ-rssA-sprE region (Fig. 2A, ychJ rssA sprE lanes) caused a significant decrease of RpoS levels in both the wild-type and rssA1::kan strains from those found in the same strains carrying the control pBBR1MCS vector (Fig. 2A, vector lanes). On the contrary, the presence of the plasmid-encoded ychJ-rssA region (Fig. 2A, ychJ rssA lanes) did not alter RpoS levels in either the wild-type or rssA1::kan strain with respect to the pBBR1MCS vector (Fig. 2A, vector lanes). Therefore, the presence of rssA in multicopy does not affect RpoS levels.
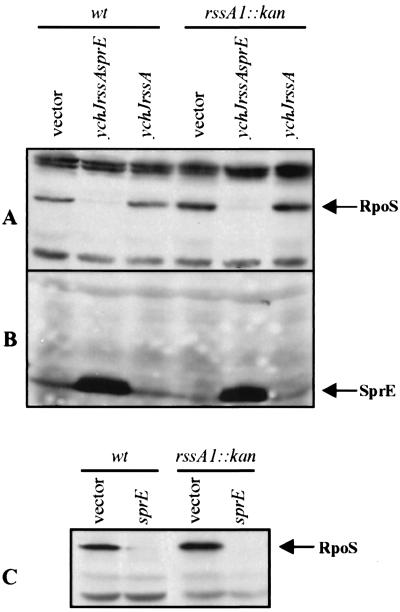
FIG. 2.
RssA does not regulate SprE or RpoS. Complementation studies were done in stationary-phase cells grown in LB medium. RpoS and SprE levels (corresponding bands marked with labeled arrows) were monitored by Western blot analysis. (A) RpoS levels present in wild-type (wt) AF633 and rssA1::kan NR260 strains carrying the vector plasmid (pBBR1MCS), a plasmid encoding the ychJ-rssA-sprE region (pRSSA6), or one carrying the ychJ-rssA region (pRSSA9) (as marked above the lanes). (B) SprE levels present in wild-type AF633 and rssA1::kan NR260 strains carrying either pBBR1MCS, pRSSA6, or pRSSA9. The sample order is the same as for panel A. (C) RpoS levels in wild-type (wt) AF633 and rssA1::kan NR260 strains carrying either the vector plasmid pMAL-c2 (vector) or a plasmid encoding the MBP-SprE hybrid protein pMBPSprE (SprE), respectively.
We also examined the effects on SprE levels caused by the presence of these plasmids and found that they are changed only in cells carrying sprE in multicopy. Specifically, the presence of the plasmid carrying the entire ychJ-rssA-sprE region caused a significant increase in the levels of SprE in both wild-type and rssA1::kan backgrounds (Fig. 2B, ychJ rssA sprE lanes) with respect to the vector control (Fig. 2B, vector lanes), while no changes were detected in those strains carrying the plasmid containing only the ychJ-rssA region (Fig. 2B, ychJ rssA lanes). Furthermore, overexpression of an MBP-SprE hybrid protein (composed of the maltose binding protein lacking the signal sequence and SprE from a high-copy-number plasmid (pMBPSprE) alone is sufficient to increase RpoS degradation to the same extent in both the wild-type and rssA1::kan strains compared to that in the respective strains carrying the control vector pMALc-2 (Fig. 2C, compare sprE and vector lanes). Thus, under the conditions tested, RssA does not appear to participate in the regulation of either RpoS or SprE.
rssA and sprE constitute an operon.
To further support that polarity alone is responsible for the decreased levels of SprE in the rssA1::kan strain, we examined the levels of SprE in a cell depleted of RssA by an in-frame deletion in rssA. We introduced an allele (rssAΔ1) that carries an internal in-frame deletion (encompassing amino acids 30 to 194) in rssA into a low-copy-number plasmid (pRSSA6Δ) and determined its effects on SprE levels by Western blot analysis.
The presence of the plasmid carrying the wild-type ychJ-rssA-sprE region (Fig. 3, wt lanes) increased the levels of SprE in an rssA1::kan null strain in both the logarithmic and stationary phases of growth compared to cells containing the vector control (Fig. 3, vector lanes). In addition, depleting cells of RssA by an in-frame deletion did not have an effect on SprE levels (Fig. 3, compare wt and rssAΔ1 lanes). The same was observed when these plasmids were introduced into our wild-type strain, AF633: there was no detectable difference in the levels of SprE, RpoS, or RpoS activity between strains carrying the wild-type gene versus those carrying the in-frame-deletion rssAΔ1 allele (data not shown). Together with the results presented above, these data further demonstrate that rssA and sprE constitute an operon and that, under the conditions tested, RssA does not function in the regulation of either SprE or RpoS.
FIG. 3.
In-frame deletion in rssA has no effect on SprE levels. A plasmid carrying the in-frame deletion allele rssAΔ1 was introduced into the rssA1::kan strain NR260 and the levels of SprE in logarithmic (L) and stationary (S) phase cells were monitored by Western blot analysis. The relevant genotype from the plasmid is shown above the lanes. Vector, samples from strain NR260(pBBR1MCS); wt, NR260 (pRSSA6); rssAΔ1, samples from NR260(pRSSA6Δ).
Interestingly, rssA1::kan does not result in total depletion of SprE (Fig. 1C), suggesting that under certain conditions sprE can be transcribed independently of rssA. The fact that there is still some SprE made in the strain carrying the rssA1::kan allele indicates that there is an additional promoter in the region between the kan cassette insertion and the translational start of sprE, as has been previously suggested (11). Furthermore, the residual levels of SprE cannot be attributed to an artifact created by the insertion of the kan cassette at the PstI site of rssA, since another independently isolated insertion (rssA3::kan) located upstream of rssA1::kan behaves similarly (C. W. Bowers, unpublished results).
The levels of SprE are growth phase regulated at the level of transcription.
Previous reports have shown that in both E. coli and S. enterica serovar Typhimurium, SprE (MviA) is growth phase regulated (11, 21). Consistent with this, we find that there is a significant increase in SprE levels between samples prepared from logarithmic- versus stationary-phase cells (Fig. 3). Paradoxically, this increase in SprE is RpoS dependent (11). This explains why an rpoS::kan null strain contains less SprE than its parent wild-type strain (Fig. 1C, compare wt and rpoS::kan lanes).
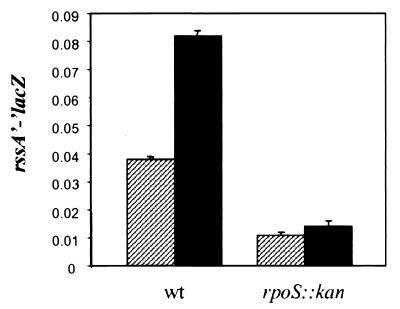
To better understand the growth phase regulation of sprE, we constructed lacZ fusions to both rssA and sprE. First, we constructed a rssA'-‘lacZ fusion that included 929 bp of sequence upstream of the translational start of rssA to ensure that all regulatory sites were present (see below). Interestingly, when this rssA’-‘lacZ fusion is recombined onto a λ phage and integrated at the λ attachment site (att site), it produces such low levels of β-galactosidase activity that this lysogen is unable to grow in lactose minimal medium. Possibly, the low activity of this fusion is the result of rssA having a GTG start codon instead of the more efficiently translated ATG codon. However, the activity of the fusion is still much higher than that derived from a promoterless fusion located at the λ att site (which yields values comparable to background levels [data not shown]). As shown in Fig. 4, the activity of the rssA’-‘lacZ fusion increases about twofold when cells enter the stationary phase. Furthermore, the levels of β-galactosidase in samples obtained from stationary-phase cells decreased about fivefold in an rpoS::kan null strain (Fig. 4), indicating that rssA is growth phase regulated in an RpoS-dependent fashion.
FIG. 4.
RssA levels are growth phase regulated by RpoS. The β-galactosidase levels of an rssA'-‘lacZ fusion were measured in a wild-type (NR501) and an rpoS::kan (NR502) strain during logarithmic (hatched bars) and stationary (black bars) phases of growth in LB medium. The relative levels of β-galactosidase activity are shown as the average and SD of three measurements per sample and they are representative of at least three independent experiments.
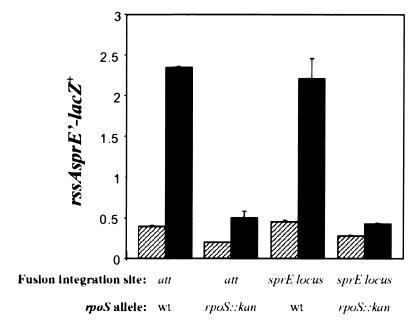
We also constructed a lacZ transcriptional fusion to sprE that included all of rssA (and 929 bp upstream) fused to the first 112 bp of sprE. This fusion was either recombined into the native rssA-sprE chromosomal locus or integrated at the λ att site, and the β-galactosidase levels from both strains were compared. Both strains contained equivalent levels of β-galactosidase under all conditions tested (Fig. 5), suggesting that all of the regulatory sites necessary for the transcriptional regulation of sprE are present in the region used to construct the fusion. In addition, a promoterless fusion did not generate any significant amount of β-galactosidase activity (data not shown). As also shown in Fig. 5, transcription of sprE (when the fusion was either at the sprE locus or at the att site) increased about sixfold in stationary-phase cells compared to that in logarithmically growing cells in an RpoS-dependent fashion (Fig. 5, compare LacZ activity in wild-type and rpoS::kan strains). Thus, RpoS regulates sprE transcription in a positive manner.
FIG. 5.
RpoS regulates SprE at the transcriptional level. The β-galactosidase levels of an rssA-sprE’-lacZ+ fusion were measured in wild-type strains (NR507 carries the fusion at the att site and NR509 carries the fusion at the sprE chromosomal locus) and rpoS::kan strains (NR508 carries the fusion at the att site and NR510 carries the fusion at the sprE chromosomal locus) during logarithmic (hatched bars) and stationary (black bars) phases of growth in LB medium. The location where the fusion was integrated (att site or sprE locus) and the rpoS genotype are indicated. The relative levels of β-galactosidase activity are shown as the average and SD of three measurements per sample, and they are representative of at least three independent experiments.
As predicted, recombination of the sprE’-lacZ+ fusion into the rssA-sprE locus in cis with the rssA1::kan allele yielded two LacZ phenotypes (as determined using lactose MacConkey agar) (data not shown), since the fusion could have recombined before or after the kan cassette (confirmed by PCR analysis). β-Galactosidase assays showed that the presence of the kan cassette upstream of lacZ did not abolish activity of the fusion but significantly reduced it (ca. 50%) with respect to those strains in which the kan cassette was either absent or located downstream of lacZ. This correlates with the results reported above showing that disruption of rssA with the kan cassette decreased but did not eliminate SprE levels.
sprE is transcribed from an RpoS-dependent promoter located in the rssA-sprE intergenic region.
The last result described above confirms that although rssA and sprE constitute an operon, there is an additional promoter(s) from which sprE can be transcribed. This promoter must lie between the location of the kan cassette insertion and the translational start of sprE, since SprE is still made in the presence of the rssA1::kan allele. We have also observed that the already-decreased levels of SprE present in an rssA1::kan strain are growth phase regulated, since they are undetectable in the logarithmic growth phase by Western blot analysis (data not shown), suggesting that this second sprE promoter is also growth phase regulated. To better understand the nature of this promoter, we constructed an additional sprE transcriptional fusion and conducted primer extension analysis.
The new sprE'-lacZ+ fusion differs from the one described in the previous section in two ways. First, the fusion junction (i.e., the 3′ end of the cloned fragment) is 62 bp downstream from that of the previously described fusion. Second, the region before the translational start of sprE that is contained in this new fusion is only 893 bp long (i.e., it contains the last 798 bp of rssA). After analyzing the levels of β-galactosidase produced by this fusion throughout the growth curve, we conclude that it is growth phase regulated, because stationary-phase cells carrying this fusion contained about eightfold more β-galactosidase than their logarithmic counterparts (Fig. 6). Furthermore, transcription from this fusion is considered RpoS dependent, because introducing the rpoS::kan allele reduced the levels of β-galactosidase activity present in stationary-phase cells to that found in wild-type logarithmic-phase cells (Fig. 6). In addition, introduction of other mutations known to alter the levels of RpoS (either increasing or decreasing the levels) caused directly proportional changes in expression from this sprE-lacZ+ fusion.
FIG. 6.
The intergenic rssA-sprE region contains an RpoS-dependent promoter for sprE. The β-galactosidase levels of an ‘rssA-sprE’-lacZ+ fusion were measured in a wild-type strain (NR511) and an rpoS::kan strain (NR512) during logarithmic (hatched bars) and stationary (black bars) phases of growth in LB medium. The relative levels of β-galactosidase activity are shown as the average and SD of three measurements per sample, and they are representative of at least three independent experiments.
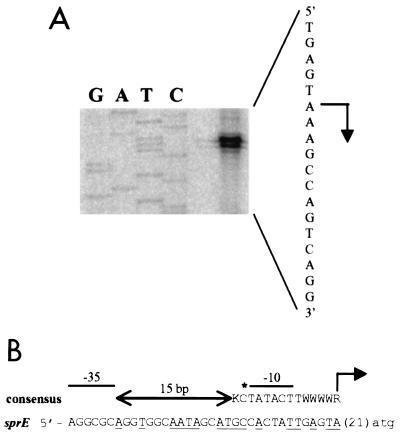
To determine the location of this additional RpoS-dependent sprE promoter, primer extension analysis was performed using RNA isolated from stationary-phase wild-type cells. As shown in Fig. 7A, we determined that there is a transcriptional start site for sprE expression 21 bp upstream of the adenine of its ATG start codon. Sequence analysis revealed that the region immediately upstream of this transcriptional start site has features found in the consensus for promoters recognized by RpoS recently proposed by Becker and Hengge-Aronis (3). Therefore, we propose that this additional promoter, located as shown in Fig. 7B, be named P2.
FIG. 7.
Location of the sprE promoter in the rssA-sprE intergenic region. (A) Primer extension analysis on RNA prepared from stationary-phase wild-type cells (AF633) showed that there is a transcriptional start site located 21 bp upstream of the adenine of the ATG start codon of sprE. The primer extension reaction (shown on the right) was performed using the RTSPRE1 primer as described in Materials and Methods. A sequencing reaction was also performed using pRSSA5 as the template and RTSPRE1 as the primer. The section of the gel corresponding to the sequence of the noncoding strand for the relevant region is shown. The order of loading in the sequencing gel is marked above the lanes and the corresponding sequence of the coding strand appears on the right, with the transcriptional start site marked with an arrow pointing to the direction of transcription. (B) Sequence of the putative promoter (coding strand alone) and comparison with the consensus sequence (similarities appear underlined) of RpoS-dependent promoters (top line) as described by Becker and Hengge-Aronis (3). The location of the transcriptional start is marked with an arrow and it corresponds to +1. The sprE start codon is shown in lower case. The locations of the putative −10 and −35 regions are marked, but because RpoS-dependent promoters do not have a recognizable −35 consensus region, none is shown. The conserved −13 position is also marked with an asterisk. In RpoS-dependent promoters, the 15-bp-long sequence between the −10 and −35 regions is AT rich. K, G or T; W, A or T; R, A or G.
DISCUSSION
The developmental commitment that E. coli makes when RpoS levels increase is immense; therefore, the cellular content of RpoS is tightly regulated. RpoS is controlled at multiple levels and regulation of its degradation by the ClpXP protease is considered a major level of control (13, 32). Although it has been known for several years that the response regulator SprE orchestrates this degradation of RpoS (22, 25), a critical question remains to be answered: how is SprE's activity regulated? We believe that the knowledge gained from understanding how SprE expression is regulated will help us to answer this question.
The studies presented here were aimed at examining sprE expression. By using rssA null alleles as well as reporter fusions and primer extension analysis, we have demonstrated that rssA and sprE are cotranscribed from a promoter, P1, that is regulated by RpoS. We have also identified an additional promoter, P2, located in the rssA-sprE intergenic region from which sprE is transcribed in an RpoS-dependent fashion. Furthermore, we have shown that under the conditions tested in this report, RssA is not involved in the regulation of either SprE or RpoS.
Interestingly, the P2 promoter has features found in the consensus for promoters recognized by RpoS recently proposed by Becker and Hengge-Aronis (3). Specifically, it lacks a recognizable −35 region but it contains a run of A/T between the −30 and −14 positions. It also contains the GC motif at the −14 and −13 positions and the highly conserved T at position −6, besides a −10 region partially homologous to the proposed TATACT consensus. Although the existence of an RpoS-specific consensus is somewhat controversial (see below), it is important to emphasize that Becker and Hengge-Aronis have shown both allelic suppression between a C at position −13 and residue 173 of RpoS (i.e., proving direct interaction) and the necessity of having either a G or a T at position −14 for maximal expression of RpoS-dependent promoters (3). The sprE P2 promoter fulfills both criteria, having the highly conserved C at the −13 position and a G at the −14 position. More experiments are required, though, to demonstrate the specific role of these positions in sprE expression.
Although many RpoS-dependent promoters have been identified through the years, deriving a consensus from them has not been an easy task. This difficulty arises because RpoS and ς70 are so similar (19). In a recent report, in vitro selection studies searching for the promoter sequences best recognized by RpoS showed that both RpoS and ς70 prefer the same consensus sequences. These studies propose that the specificity of these sigma factors is dictated by how they differ in tolerating binding to nonpreferred sequences (i.e., tolerance to binding to sites with deviations from the well-characterized ς70 consensus sequences) (T. Gaal, W. Ross, S. T. Estrem, L. H. Nguyen, R. R. Burgess, and R. L. Gourse, submitted for publication). Regardless of whether there is a clear RpoS promoter consensus sequence or not, depletion of RpoS significantly decreases transcription of sprE from both P1 and P2 promoters, proving RpoS-dependent transcription of sprE.
Previously, it was reported that although transcription from a fusion containing only the P2 promoter (i.e., it did not include all of rssA) was upregulated in stationary phase, this growth phase regulation was RpoS independent (11). Furthermore, an analogous translational fusion was shown to be RpoS regulated, suggesting translational but not transcriptional control of sprE by RpoS. Additional evidence supporting this RpoS-dependent translational regulation of sprE showed that the levels of SprE present in a strain carrying the sprE19::cam allele were also growth phase regulated (i.e., they increased in stationary phase) (11). Note that the sprE19::cam allele carries a mini-Tncam cassette inserted 27 bp upstream of the translational start of sprE and it is believed to be constitutively transcribed (25).
In contrast, we found that the sprE P2 promoter is RpoS regulated. We have recently isolated a strain that contains a mini-Tncam cassette inserted 95 bp from the translational start of sprE and, in this strain, sprE is constitutively expressed regardless of the growth phase or the presence or absence of RpoS (data not shown), which argues against an RpoS-dependent translational control. To resolve this controversy, we sequenced the region upstream of sprE and found that several strains previously used (11) contain an IS1E element in the rssA-sprE intergenic region which interferes with native sprE expression and regulation.
The paradox of RpoS being necessary for the expression of its negative regulator SprE remains (11). We speculate that by having this feedback loop, the cell ensures a proper amount of RpoS at all times. It is known that under certain conditions, too much RpoS is not beneficial to the cell and can even be fatal. For example, Zambrano et al. showed that during prolonged starvation, cells with an altered form of RpoS, which is less active as sigma factor, results in growth advantage (31). In addition, cells that have exceptionally high levels of RpoS (i.e., sprE null strains) cannot grow in media containing either succinate or acetate as the sole carbon source (24). Thus, coupling the levels of SprE to those of RpoS might serve as a safety mechanism to ensure that the levels of RpoS are appropriate in the cell at all times. Interestingly, it has been reported that in addition to its role in orchestrating RpoS degradation, SprE has anti-sigma factor activity (4, 33). Thus, it is possible that in stationary-phase cells, when SprE-mediated degradation of RpoS does not occur, SprE itself might be acting as an anti-RpoS factor. This could explain, at least in part, why SprE levels need to increase when ClpXP is not degrading RpoS.
In addition, as previously proposed, having SprE already present in stationary phase might be beneficial to cells once they encounter more favorable conditions, thus providing a growth advantage (11). High levels of SprE could ensure rapid degradation of RpoS when nutrients become available. RpoS-dependent transcription would cease, and the cell would then focus its transcriptional and translational machinery on producing proteins that are necessary for rapid growth (ς70-dependent promoters). Reports showing that cells deficient in ClpP suffer a growth disadvantage during competition experiments (i.e., repeated rounds of glucose starvation and recovery) support this idea (8). Alternatively, growth phase regulation of SprE might be necessary if SprE plays a role, as yet to be identified, during stationary phase.
ACKNOWLEDGMENTS
We are grateful to the members of the Silhavy lab for their critical reading of the manuscript. Special thanks to Susan DiRenzo for her assistance in the preparation of this paper. We also thank Weihong Hsing for her gift of pMBPSprE and for her work, in addition to that of Katherine Gibson, in generating the RpoS and SprE antisera. We thank T. Nystrom for his gift of strain AF633. We are also grateful to R. L. Gourse and C. W. Bowers for sharing their unpublished results.
T.J.S. was supported by a grant from the National Institute of General Medical Sciences (GM35791).
REFERENCES
- 1.Andersson R A, Palva E T, Pirhonen M. The response regulator expM is essential for the virulence of Erwinia carotovora subsp. carotovora and acts negatively on the sigma factor RpoS (ςs) Mol Plant Microbe Interact. 1999;12:575–584. doi: 10.1094/MPMI.1999.12.7.575. [DOI] [PubMed] [Google Scholar]
- 2.Bearson S M, Benjamin W H, Jr, Swords W E, Foster J W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J Bacteriol. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker G, Hengge-Aronis R. What makes an Escherichia coli promoter ςs dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of ςs. Mol Microbiol. 2001;39:1153–1165. doi: 10.1111/j.1365-2958.2001.02313.x. [DOI] [PubMed] [Google Scholar]
- 4.Becker G, Klauck E, Hengge-Aronis R. The response regulator RssB, a recognition factor for ςs proteolysis in Escherichia coli, can act like an anti-ςs factor. Mol Microbiol. 2000;35:657–666. doi: 10.1046/j.1365-2958.2000.01736.x. [DOI] [PubMed] [Google Scholar]
- 5.Bosl M, Kersten H. Organization and functions of genes in the upstream region of tyrT of Escherichia coli: phenotypes of mutants with partial deletion of a new gene (tgs) J Bacteriol. 1994;176:221–231. doi: 10.1128/jb.176.1.221-231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouche S, Klauck E, Fischer D, Lucassen M, Jung K, Hengge-Aronis R. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol Microbiol. 1998;27:787–795. doi: 10.1046/j.1365-2958.1998.00725.x. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 8.Damerau K, St. John A C. Role of Clp protease subunits in degradation of carbon starvation proteins in Escherichia coli. J Bacteriol. 1993;175:53–63. doi: 10.1128/jb.175.1.53-63.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farewell A, Kvint K, Nystrom T. uspB, a new ςs-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J Bacteriol. 1998;180:6140–6147. doi: 10.1128/jb.180.23.6140-6147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson K E, Silhavy T J. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator sprE. J Bacteriol. 1999;181:563–571. doi: 10.1128/jb.181.2.563-571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson K E, Silhavy T J. SprE levels are growth phase regulated in a ςs-dependent manner at the level of translation. J Bacteriol. 2000;182:4117–4120. doi: 10.1128/jb.182.14.4117-4120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hengge-Aronis R. The general stress response in Escherichia coli. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 161–178. [Google Scholar]
- 14.Ji Y, McLandsborough L, Kondagunta A, Cleary P P. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect Immun. 1996;64:503–510. doi: 10.1128/iai.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 16.Kretzschmar D, Hasan G, Sharma S, Heisenberg M, Benzer S. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kushner S R. An improved method for transformation of Escherichia coli with ColE1-derived plasmids. In: Boyer H W, Micosia S, editors. Genetic engineering. New York, N.Y: Elsevier/North Holland Biomedical Press; 1978. pp. 17–23. [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lush M J, Li Y, Read D J, Willis A C, Glynn P. Neuropathy target esterase and a homologous Drosophila neurodegeneration-associated mutant protein contain a novel domain conserved from bacteria to man. Biochem J. 1998;332(Pt. 1):1–4. doi: 10.1042/bj3320001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno M, Audia J P, Bearson S M, Webb C, Foster J W. Regulation of ςs degradation in Salmonella enterica var. typhimurium: in vivo interactions between ςs, the response regulator MviA(RssB) and ClpX. J Mol Microbiol Biotechnol. 2000;2:245–254. [PubMed] [Google Scholar]
- 22.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the ςs subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 23.Ostrow K S, Silhavy T J, Garrett S. cis-acting sites required for osmoregulation of ompF expression in Escherichia coli K-12. J Bacteriol. 1986;168:1165–1171. doi: 10.1128/jb.168.3.1165-1171.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pratt L A, Silhavy T J. Crl stimulates RpoS activity during stationary phase. Mol Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- 25.Pratt L A, Silhavy T J. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweder T, Lee K H, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation sigma factor ςs by ClpXP protease. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 28.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 29.Slauch J M, Silhavy T J. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor L A, Rose R E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988;16:358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 32.Zgurskaya H I, Keyhan M, Matin A. The ςs level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol Microbiol. 1997;24:643–651. doi: 10.1046/j.1365-2958.1997.3961742.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y, Gottesman S. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Gottesman S, Hoskins J R, Maurizi M R, Wickner S. The RssB response regulator directly targets ςs for degradation by ClpXP. Genes Dev. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]