Abstract
Sequences of the major outer membrane protein (MOMP) gene (ompA) and the outer membrane complex B protein gene (omcB) from Chlamydia trachomatis, Chlamydia pneumoniae, and Chlamydia psittaci were analyzed for evidence of intragenic recombination and for linkage equilibrium. The Sawyer runs test, compatibility matrices, and index of association analyses provided substantial evidence that there has been a history of intragenic recombination at ompA including one instance of interspecies recombination between the C. trachomatis mouse pneumonitis strain and the C. pneumoniae horse N16 strain. Although none of these methods detected intragenic recombination within omcB, differences in divergence reported in earlier studies suggested that there has been intergenic recombination involving omcB, and the analyses presented in this study are consistent with this. For C. trachomatis, index-of-association analyses suggested a higher degree of recombination for C class than for B class strains and a higher degree of recombination in the downstream half of ompA. In concordance with these findings, many significant breakpoints were found in variable segments 3 and 4 of MOMP for the recombinant strains D/B120, G/UW-57, E/Bour, and LGV-98 identified in this study. We provide examples of how genetic diversity generated by repeated recombination in these regions may be associated with evasion of immune surveillance, serovar-specific differences in tissue tropism, and persistence.
Chlamydiae are gram-negative bacteria responsible for a large variety of diseases in humans, animals, and birds. There are four species currently recognized in the genus: Chlamydia trachomatis, C. pneumoniae, C. psittaci, and C. pecorum. Collectively, they represent a major public health burden. C. trachomatis is the leading cause of preventable blindness and sexually transmitted diseases (STD) in the developing and developed world, respectively (9, 12, 13). C. pneumoniae is an important cause of community-acquired pneumonia (60) and has also been implicated in the etiology of atherosclerosis, stroke, Alzheimer's disease, and multiple sclerosis (6, 59). C. trachomatis and C. pneumoniae are mainly pathogens of humans. However, C. trachomatis can also infect rodents (53, 69) and swine (44) and C. pneumoniae can infect horses and koalas (27, 70). In contrast, C. psittaci mainly infects lower vertebrate mammals and birds; humans are accidental hosts for infection, and to date there has been no evidence suggesting that humans can transmit C. psittaci. Until recently, chlamydial strains of nonhuman origin were uniformly classified as C. psittaci. However, examination of sequence data has revealed that some ruminant strains were not sufficiently similar to any C. psittaci strain, and they have been reclassified as C. pecorum (25).
The chlamydial outer membrane complex is composed primarily of three proteins, specifically the major outer membrane protein (MOMP) and two cysteine-rich proteins, the outer membrane complex B protein (OmcB) and the outer membrane complex A protein (OmcA) (30). Of the three, MOMP is the most extensively studied due to its surface exposure, immunogenicity, and potential function as a cellular adhesin (29). The gene that encodes MOMP, ompA, exhibits extensive DNA sequence variation that is confined mainly to four variable segments or domains (VS or VD 1 to VD 4) (78) that contain subspecies- and serovar-specific antigenic determinants. Important T-cell epitopes are also located in MOMP (1, 39). OmcB, encoded by omcB, does not appear to be surface exposed but is thought to form a supramolecular lattice in the periplasm (51, 62). Another important difference is that OmcB is extremely highly conserved (52). Currently, classification of C. trachomatis is based on the serological differentiation of antigenic epitopes on MOMP into 19 human C. trachomatis serovars (A to K, Ba, Da, Ia, Ja, L1 to L3, and L2a) (72, 73). Based on amino acid similarity, these serovars have been placed into the following serogroup or classes: B class (B, Ba, D, Da, E, L1, L2, and L2a), C class (A, C, H, I, Ia, J, Ja, K, and L3), and intermediate class (F and G) (78).
The inability to serotype several C. trachomatis strains in the last decade prompted investigators to examine the sequence variation of ompA. Some variant strains were observed to have a mosaic structure based on the presence of nucleotide runs with different ancestries. Ia was composed of I/H (46); LGV strains (LGV-98, LGV-224, and LGV-115) were composed of L1 and L2 (34); and 4 to 8% of STD stains were mosaics of C/J and I/H (77) or L1/L2, L2/L1, L3/H, and I/H (7). However, these observed recombination frequencies were dissimilar to those observed in other studies. There were no recombinants among 68 STD ompA strains in San Francisco (15), among 188 trachoma strains in Gambia (32), or among 27 trachoma strains in Tunisia (16). The first computational evidence for recombination in Chlamydia was provided by Fitch, Peterson, and de la Maza (24) based on the analysis of 24 ompA and 10 omcB sequences. They found that phylogenetic reconstructions were not congruent for the C. trachomatis strains and that the genetic distance between L1 and B was 10 times greater for MOMP than for OmcB, despite the fact that the average genetic distance between species was only 25% greater for MOMP than for OmcB. They suggested that the most plausible explanation was genetic exchange among strains with a bias in the direction of recombination. Recently, Jordan et al. (43) identified one apparent gene conversion event between two genes that encode putative outer membrane proteins of C. pneumoniae strain AR39 by comparing complete genome sequences.
While the observational data and computational analyses indicated that recombination had occurred, statistical methods able to detect patterns of apparent recombination among and between chlamydial species in the absence of obvious mosaic structures were not applied to these data. Furthermore, neither the significance of the mosaics nor an accurate identification of breakpoints was assessed. Our approach to providing a comprehensive statistical analysis of recombination at these two loci was twofold. Methods designed to detect unique or rare recombination events as well as those designed to detect repeated recombination were applied to 40 ompA and 19 omcB sequences from C. trachomatis, C. psittaci, and C. pneumoniae. We also evaluated whether any significant intraspecies and/or interspecies gene conversion events had occurred and if there were any barriers to these events. We also analyzed potential mosaics identified by observation to determine their significance and breakpoints. In agreement with Fitch et al. (24), our results revealed an incongruence in the branching orders of phylogenetic trees for C. trachomatis, including one cluster of strains not previously identified. Our data show for the first time statistical evidence to support ompA intragenic recombination for C. trachomatis strains, including a determination of the relative degree of recombination for different regions of ompA and for different classes. Our analyses also reveal ompA intragenic recombination between an equine C. pneumoniae strain and a rodent C. trachomatis strain. We interpret these results in light of their implications for immune evasion, tissue tropism, persistence, and vaccine design.
(This research was presented in part at the Fourth Meeting of the European Society for Chlamydia Research, August, 2000, Helsinki, Finland, abstr. 42.)
MATERIALS AND METHODS
DNA sequences.
Sequence analysis was performed for C. trachomatis, C. pneumoniae, and C. psittaci omcB and ompA sequences. A total of 40 full-length ompA sequences and 12 full-length omcB sequences were from GenBank. Seven omcB sequences were determined by direct sequencing of PCR products using methods described previously (15) except that the following primer pairs were used: omcBF1 (5′-AAAGTTAGTTAATAACAATT-3′) (nucleotides [nt] −71 to −52) plus omcBB1 (5′-CGGATCTCTGGACAAGCGCAT-3′) (nt 632 to 612); omcBF2 (5′-TCCTACTGCTGATGGTAAG-3′) (nt 492 to 510) plus omcBB2 (5′-GCTCCTGCAGCTTCAAGAACT-3′) (nt 1133 to 1113); and omcBF3 (5′-TGTAGAATATGTGATCTCC-3′) (nt 1026 to 1044) plus omcBB3 (5′-AAAGCCGCCCAGGAATCCCT-3′) (nt 1754 to 1733). Using these primers, several fragments of A/Har-13 could not be amplified. The following modified primers were designed and used for this strain; omcBAF1 (5′-AATGTTGAGGGTAAAAGTT-3′) (nt −65 to −47) plus omcBAB1 (5′-ACCTTCTTTAAGAGGTTTTACC-3′) (nt 591 to 570) and omcBAF3 (5′-ACGAGCCTTGCGTACAAGT-3′) (nt 971 to 988) plus omcBAB3 (5′-AAACTCTACAGATTCCTTA-3′) (nt 1566 to 1548). The omcB sequences were truncated to the length of the shortest sequence available (nt 1 to 1566 of the complete 1,676-nt omcB gene). The properties of the chlamydial strains are shown in Table 1. With the exception of strains E-DK20 and D/UW-3, all omcB sequences were derived from the same strains as for ompA.
TABLE 1.
Properties of the chlamydial strains investigated
| Species and serovar/strainb | Yr isolated | Location | Anatomic site/speciesa | Diseaseab | Reference
|
|
|---|---|---|---|---|---|---|
| ompA | omcB | |||||
| C. trachomatis | ||||||
| A/Har-13 | 1958 | Egypt | Co/Hu | Trachoma | 3 | This study |
| A/Sa-1 | 1957 | Saudi Arabia | Co/Hu | Trachoma | 31 | |
| B/Jali20 | 1985 | Gambia | Co/Hu | Trachoma | 33 | 76 |
| B/TW-5 | 1959 | Taiwan | Co/Hu | Trachoma | 14 | |
| Ba/Apache-2 | 1960 | Arizona | Co/Hu | Trachoma | 71 | |
| C/TW-3 | 1959 | Taiwan | Co/Hu | Trachoma | 14 | 18 |
| D/B-120 | 1983? | ? | ? | STD | 64 | |
| D/IC-Cal-8 | 1991? | ? | ? | STD | 64 | |
| D/UW-3 | 1965 | Washington? | Cx?/Hu | Cervicitis? | This study | |
| Da/TW-448 | 1985? | Taiwan | Co/Hu | Trachoma | 64 | |
| E/Bour | 1959 | California | Co/Hu | Trachoma | 54 | 18 |
| E/DK-20 | 1967? | Denmark | Co/Hu | Conjunctivitis | 11 | |
| F/IC-Cal-3 | 1960? | California | Co/Hu | Conjunctivitis | 81 | None |
| G/UW-57 | 1971 | Washington | Cx/Hu | Cervicitis | 71 | This study |
| H/UW-4 | 1965 | Washington | Cx/Hu | Cervicitis | 14 | This study |
| I/UW-12 | 1966 | Washington | Ur/Hu | Urethritis | 71 | This study |
| Ia/IU-4168 | 1987 | Indiana | Ur/Hu | ? | 71 | |
| J/UW-36 | 1971 | Washington | Cx/Hu | Cervicitis | 14 | This study |
| Ja/IU-37538 | 1985 | Indiana | Cx/Hu | ? | 71 | |
| K/UW-31 | 1973? | Washington | Cx/Hu | Cervicitis | 71 | This study |
| L1/440 | 1968 | California | Ln/Hu | LGV | 56 | 10 |
| L2/434 | 1968 | California | Ln/Hu | LGV | 68 | 2 |
| L2a/UW-396 | 1985? | US/Europe | Ln/Hu | LGV? | 14 | |
| L3/404 | 1967 | California | Ln/Hu | LGV | 23 | 18 |
| LGV-98 | 1994? | South Africa | Ln?/Hu | LGV | 34 | |
| LGV-224 | 1994? | South Africa | Ln?/Hu | LGV | 34 | |
| LGV-115 | 1994? | South Africa | Ln?/Hu | LGV | 34 | |
| MoPn/NiggII | 1939 | Minnesota | Lu/Mo | Asymptomatic | 22 | 57 |
| Hamster/SFPD | 1991 | ? | Ileum?/Ha | Ileitis | 79 | |
| C. pneumoniae | ||||||
| /TWAR-IOL-207 | 1967 | Iran | Co/Hu | Trachoma | 8 | 74 |
| /N16 | 1990? | England? | NP/horse | Nasal discharge | 70 | |
| /TWAR-AR-39 | 1983 | Washington | Throat/Hu | Pharyngitis | 48 | |
| /Koala | 1993? | Australia | Co/koala | Conjunctivitis | 27 | |
| /UV-BL | 1996 | Tennessee | CNS/Hu | MS | 66 | |
| C. psittaci | ||||||
| /EAE-A22-M | 1949 | Scotland | Lamb | Abortion | 55 | 75 |
| /EAE-S26-3 | 1984? | Scotland | Lamb | Abortion | 35 | |
| /GPIC1 | 1963? | Massachusetts | Co/GP | Asymptomatic | 80 | |
| /Mn-Cal10 | 1936 | California | Lu/ferret | Asymptomatic | 80 | |
| /6BC | 1941 | California | ?/Parakeet | ? | 19 | 20 |
| AvianC/1 | 1994? | ? | ?/Avian | ? | None | |
| AvianC/2 | 1994? | ? | ?/Avian | ? | None | |
| /N352 | 1982? | England | Cloaca/Duck | Asymptomatic? | 70 | |
| /Fe-pring | 1984 | England? | Co/Cat | Conjunctivitis | 70 | |
Co, conjunctiva; Hu, human; Cx, cervix; Ur, urethra; Ln, lymph node; Lu, lung; RT, respiratory tract; CNS, central nervous system; MS, multiple sclerosis.
LGV, lymphogranuloma venereum; Mo, mouse; GP, guinea pig; Ha, hamster.
Phylogenetic and statistical analyses.
Sequences were aligned manually using the multiple-alignment sequence editor (MASE, version 3.1; Dana-Farber Cancer Institute, Harvard School of Public Health [ftp.ebi.ac.uk/pub/software /unix/]). Neighbor-joining tree topologies (61) were generated by Molecular Evolutionary Genetics Analysis (MEGA, version 1.01; Institute of Molecular Evolutionary Genetics, Pennsylvania State University [http://www.megasoftware.net]), based on distance estimates using a Kimura (45) two-parameter model for substitution events. Bootstrap confidence levels were determined by randomly resampling of the sequence data 1,000 times.
The Sawyer runs test was used to test whether significant intraspecies or interspecies recombination had occurred among the sequences analyzed at the ompA or the omcB locus of Chlamydia. An extension of the nonparametric method of Sawyer (63) that required no phylogenetic inference was performed using GENECONV (version 1.70; Department of Mathematics, Washington University, St. Louis, Mo. [http://www.math.wustl.edu/∼sawyer]). The basic procedure in the method was as follows. Each silent polymorphic codon in the gene was identified. For each sequence pair, the gene was partitioned into fragments. The first fragment was from the beginning of the sequence to the first silent polymorphic codon that differed between the two, the last fragment was from the last difference to the end of the sequence, and fragments in between were bounded by consecutive differences between the two. The fragment length for a given pair was the number of silent polymorphic codons that differed among the other sequences in the data set but were not polymorphic with respect to the given pair. The global fragment score was a linear function of the sum of squares of the fragment lengths over all fragments over all pairs of sequences. We tested the null hypothesis that no significant gene conversion events occurred among the sequences analyzed. The P value was determined empirically.
Sequence data were subdivided to make valid intraspecies and interspecies comparisons. We chose to test these hypotheses for strains that infected humans separately from those that infected lower vertebrate mammals and birds. This was a conservative approach since dramatic differences in coalescence times of the hosts may have influenced the results in ways that were unpredictable. For ompA, we subdivided the sequence data into five groups: human C. trachomatis (all C. trachomatis except MoPn/NiggII and SFPD), lower vertebrate mammal C. trachomatis (MoPn/NiggII and SFPD), human C. pneumoniae (all C. pneumoniae except N16 and Koala), lower vertebrate mammal C. pneumoniae (N16 and Koala), and C. psittaci. For omcB, the data were similarly subdivided into four groups: human C. trachomatis (all except MoPn/NiggII), lower vertebrate mammal C. trachomatis (MoPn/NiggII), human C. pneumoniae (TWAR-IOL-207), and C. psittaci (EAE-A22-M and 6BC).
Intraspecies hypothesis testing involved inspection of the global Sawyer run test score for the appropriate data set. Intraspecies testing was possible for human C. trachomatis and lower vertebrate mammal C. psittaci for ompA and for human C. trachomatis for omcB. Testing was not possible for the other groups because there were either too few sequences or too few polymorphisms to evaluate. Interspecies hypothesis testing was accomplished by combining appropriate data. Evidence was considered significant only if the global score for the combined data set was significant and if there were significant fragments detected between the two species. For the same reasons as noted above, interspecies hypothesis testing for omcB was not possible for lower vertebrate mammal C. trachomatis and C. psittaci. To test hypotheses about barriers to recombination, we compared the global scores of data sets before and after the inclusion of strains from a different species. If the global score decreased from significant to insignificant, a barrier to recombination was considered suggestive for the strains involved.
These hypotheses were further evaluated using compatibility matrices. Compatibility matrices and neighborhood similarity scores were calculated using the program RETICULATE (Human Genetics Group, John Curtin School of Medical Research, Australian National University [http://jcsmr.anu.edu.au/dmm /humgen/ingrid/reticulate.htm]) by the methods of Jakobsen and Easteal (41). In this method, matrices representing the compatibility of all possible pairs of informative sites with a single maximum-parsimony tree were calculated. The neighbor similarity score indicated the degree to which sites were compatible, and its significance was determined empirically. The null hypothesis tested was that sites were randomly distributed with regard to type (incompatible or compatible), where clustering of sites suggested recombination. In this analysis, only parsimoniously informative binary sites (sites with only two different nucleotides present more than once) were included. For these analyses, the ompA and omcB data were subdivided into strains that infect human hosts and those that infect lower vertebrate mammals and birds, for the same reasons as stated above.
The index of association (IA) between codons, according to the methods of Feil et al. (21), was used to test the null hypothesis that ompA polymorphic codons were in complete linkage equilibrium. Briefly, IA was computed by comparing the observed between-strain variance (VO) with the variance that would be expected if polymorphic codons were randomly assorted (VE). Any observed variance that exceeded that expected by chance represented linkage between codons. If the polymorphic codons were randomly assorted and the data were in linkage equilibrium, IA was expected to be zero. The significance of IA was determined empirically. Since VE, and in turn IA, increased with the number of polymorphic codons (r), it was important to compare values only if r was the same and in other case, only to distinguish the IA values that were significantly different from zero from those that were not.
The Recombination Identification Program (RIP, version 1; HIV Database Group, Los Alamos National Laboratories [http://hiv-web.1anl.gov/]) was used as a graphical tool to predict the mosaic structure of a strain. For each of these strains, the Maximum Chi Square program (version 1.0; Molecular Microbiology Group, University of Sussex [http://www.biols.susx.ac.uk/Biochem/Molbiol /maximum-chi-squared.html]) was used according to the methods of Maynard Smith (47) to refine and assess the significance of the structure. In the test, polymorphic sites were defined as sites that varied between the potential recombinant and its possible parental strains. For each of the parental strains, the number of differences between the parental strain and the putative recombinant divided by the total number of differences in the data was calculated before and after a proposed cut. A cut was optimal when the difference in these proportions was maximized. The significance of the division was tested by empirically determining a P value by randomization. An iterative procedure was used to evaluate strains with multiple divisions as described in the method (47).
For all statistical analyses, sites that contained gaps were ignored and analyses were considered significant at P ≤ 0.05 except for the maximum chi-square analysis, which was considered significant at P ≤ 0.001 in order to be conservative.
Nucleotide sequence accession numbers.
The sequences of the omcB genes determined in this study have been submitted to GenBank under accession numbers AF304326-AF304332. The ompA and omcB alignments are available from the authors.
RESULTS
Phylogenetic analysis of ompA and omcB.
In the ompA alignment, the locations of the VS domains as defined by Yuan et al. (78) were as follows: VS1 from 262 to 333, VS2 from 499 to 576, VS3 from 766 to 813, and VS4 from 964 to 1068. At the nucleotide level, there were 691 polymorphic sites within the 1,215 bp of the complete ompA gene and 609 polymorphic sites within the 1,566 bp of the partial omcB gene analyzed. There were 142 polymorphic sites among the B class strains, 100 polymorphic sites among the C class strains, and 36 polymorphic sites among the intermediate class strains.
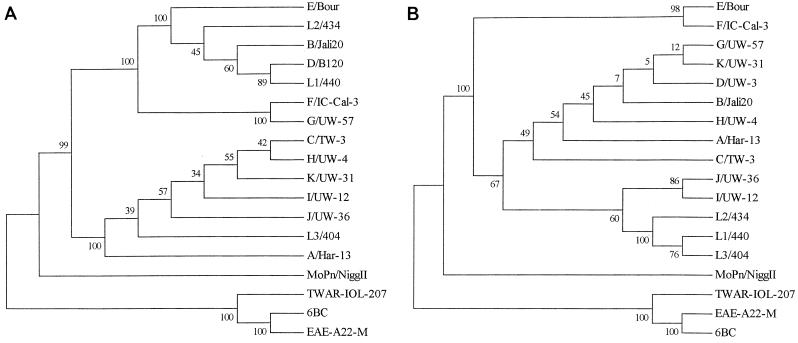
Phylogenetic reconstructions suggest potential differences in the evolutionary histories of ompA and omcB (Fig. 1). Major branching orders of the neighbor-joining ompA trees agreed with those already reported, namely, that the three species were represented by three distinct clades and that the human C. trachomatis strains clustered into B, C, and intermediate classes as described above. However, for omcB, the C. trachomatis strains did not form three classes. The L1, L2, and L3 serovars of the LGV biovar formed a distinct group instead of being split into the B and C classes, consistent with the findings of Fitch et al. (24). In addition, the inclusion of the seven new omcB strains sequenced in this study revealed that F did not cluster with G in the intermediate class but instead grouped with E. These incongruent patterns suggested that recombination had been responsible for driving the divergence of several of the C. trachomatis strains including E, F, G, L1, and L3.
FIG. 1.
Phylogenetic reconstructions for the ompA (A) and omcB (B) gene sequences using the neighbor-joining method. The values at the nodes are the bootstrap confidence levels for the interior branch. The bootstrap confidence level represents the percentage of 1,000 bootstrap resamplings for which the strain to the right was separated from the other strains.
Statistical evidence for or against intraspecies and interspecies recombination in ompA and omcB.
To determine if differences in tree topologies for ompA and omcB could be explained by intragenic recombination, the sequence data were subjected to the Sawyer runs test using GENECONV (63). Table 2 presents the intraspecies and interspecies comparisons for ompA and omcB. Based on this test, there was significant evidence for ompA intraspecies recombination between human C. trachomatis strains (P < 0.0001). Consistent with the phylogenetic data, the pairs of strains E/Bour/G/UW-57 (P = 0.003), B/Jali20/E/Bour (P = 0.007), Ba/Apache-2/E/Bour (P = 0.007), and L1/440/LGV-98 (P = 0.023) had significant ompA gene conversion events. ompA intraspecies recombination between C. psittaci strains was also supported (P = 0.038) with the following pairs of strains identified: N352/EAE-S26-3 (P = 0.038), 6BC/EAE-S26-3 (P = 0.038), AvianC/EAE-S26-3 (P = 0.038), MnCa1-10/EAE-S26-3 (P = 0.038), and EAE-A22-M/EAE-S26-3 (P = 0.038). In terms of ompA interspecies comparisons, there was significant evidence to suggest recombination between the rodent C. trachomatis MoPn/NiggII strain and the C. pneumoniae horse N16 strain (P = 0.033). There was also suggestive evidence for a barrier against recombination between C. trachomatis rodent strains and C. psittaci strains. This latter observation was based on the fact that the inclusion of the rodent strains with the C. psittaci strains weakened the significance of the previous findings from P = 0.038 to P = 0.124. Unlike the results for ompA, there was no statistical evidence to support recombination within omcB. In every analysis, the global score was insignificant.
TABLE 2.
Sawyer runs test results for ompA and omcB for the three species of Chlamydia
| Dataset | Global score | Global P valuea | No. of SDb above simulated mean | SD of simulations |
|---|---|---|---|---|
| Intraspecies comparisons for ompA data | ||||
| Human C. trachomatis | 11.306 | <0.0001 | 6.85 | 1.127 |
| C. psittaci | 4.541 | 0.038 | 2.03 | 1.142 |
| Interspecies comparisons for ompA data | ||||
| Human C. trachomatis and human C. pneumoniae | 8.290 | 0.002 | 4.33 | 1.117 |
| Lower vertebrate animal C. trachomatis and lower vertebrate animal C. pneumoniae | 4.171 | 0.033 | 2.40 | 1.107 |
| Lower vertebrate animal C. trachomatis and C. psittaci | 4.207 | 0.124 | 1.16 | 1.160 |
| Lower vertebrate animal C. pneumoniae and C. psittaci | 5.257 | 0.039 | 2.11 | 1.153 |
| Intraspecies comparisons for omcB data | ||||
| Human C. trachomatis | 0.989 | 0.582 | −0.14 | 0.120 |
| Interspecies comparisons for omcB data | ||||
| Human C. trachomatis and human C. pneumoniae | 0.877 | 0.838 | −0.56 | 0.517 |
After permuting the silent polymorphic sites in the respective data set 10,000 times.
SD, standard deviation.
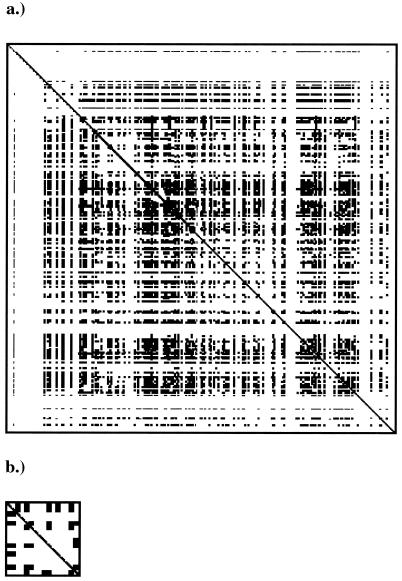
Recombination detection methods are based on the underlying assumption that substitution rates are equal along the gene. Methods based on distributions of polymorphic sites, such as the Sawyer runs test, have a higher false-positive rate due to violations of this assumption than do methods that detect incompatibilities between sites and changes in local estimated phylogenies (C. Wiuf, T. Christensen, and J. Hein, submitted for publication). For this reason, we analyzed the data for compatibility using RETICULATE (41). For ompA, there were large numbers of sites that were incompatible with a single tree, both for all strains that infected humans (neighbor similarity, 0.886) and for all strains that infected lower vertebrate mammals and birds (neighbor similarity, 0.738) (Fig. 2; compatibility matrix not shown for ompA sequences from lower vertebrate mammals and birds). In both cases, the distribution was not random (P = 0.0001). This provides further evidence for recombination within the ompA gene of Chlamydia. In contrast, sites were randomly distributed for omcB sequences from human hosts (P = 0.211), again suggesting no recombination within omcB.
FIG. 2.
Compatibility matrices for the Chlamydia ompA gene from human hosts (a) and the Chlamydia omcB gene from human hosts (b). White spaces represent pairs of informative sites that are compatible with a single maximum-parsimony tree, while black spaces represent pairs that are incompatible with that tree.
The ratio of average synonymous to average nonsynonymous substitutions (dS/dN) was calculated for ompA and omcB by the method of Nei and Gojobori (50). Using the same 17 strains from the three species, the average dS/dN ratio was 6.7:1 for ompA and 5.7:1 for omcB. However, when just the human C. trachomatis strains were examined, the average dS/dN ratio was 6.0:1 for ompA and 1.4:1 for omcB.
Relative degree of recombination in ompA for different regions and classes.
None of the above methods can be used to detect recombination reliably when the number of events is large. In contrast, the IA between codons is a test statistic that can be used to test whether polymorphic codons are in linkage equilibrium. For this reason, we determined the IA between polymorphic codons at the ompA and omcB loci. When all human C. trachomatis sequences were analyzed together, ompA codons were in linkage disequilibrium (P < 0.001). This was also the case for C. psittaci sequences (P < 0.001). Since the degree of recombination tends to increase with increasing similarity between strains, it was possible that human C. trachomatis sequences were in linkage disequilibrium while individual classes were in linkage equilibrium. To test this hypothesis, we subdivided the sequence data into classes and repeated the analysis. For both the B and C classes, polymorphic codons were in linkage disequilibrium when the entire gene was analyzed (P < 0.001). This indicated that, overall, recombination in the ompA gene was not frequent enough to randomize the gene or break up clonal associations between codons, thus indicating a clonal population structure.
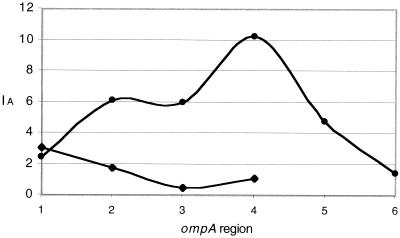
To compare the relative degree of recombination within classes, the entire ompA gene was subdivided into regions, 20 polymorphic codons in length, and the IA test statistic was calculated. For the B class, this resulted in five regions of 20 codons and one region of 12 codons. For the C class, the gene was divided into four regions of 20 codons. For every region except for the first, IA for the B class exceeded that for the C class (Fig. 3). In fact, in region 3, codons for the C class were not in significant linkage disequilibrium (P = 0.103). These results suggest that C class strains experienced gene conversion events to a greater degree than B class strains did. Furthermore, there was a greater degree of recombination in the downstream half of ompA. For omcB, there was no evidence to suggest that polymorphic codons were in linkage equilibrium, indicating that the inability to detect intragenic recombination was not a result of repeated recombination.
FIG. 3.
Linkage between polymorphic codons within successive ompA regions, approximately 20 polymorphisms in length, for C. trachomatis C class strains (⧫) and C. trachomatis B class strains (●).
Identification of breakpoints in ompA mosaics.
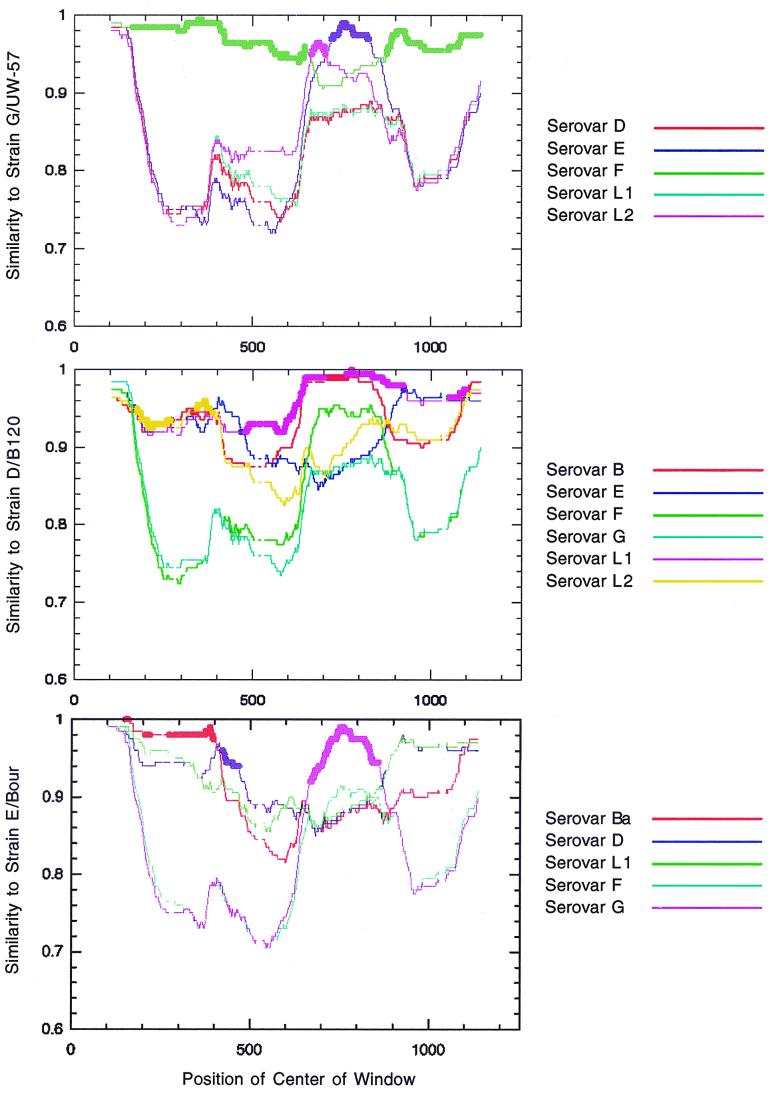
Potential ompA mosaics were identified as the strains detected in the GENECONV analysis, those that clustered differently in tree topologies, and those reported in previous studies. For each strain, the similarity with respect to all other strains was calculated and plotted by nucleotide position using RIP (65). This output was visually inspected to determine potential breakpoints. Figure 4 presents the RIP similarity plots for G/UW-57, D/B-120, and E/Bour (LGV-98 is not shown). These similarity plots provided the initial predictions of the mosaic structure.
FIG. 4.
RIP images depicting the similarity of strain D/B120 (top), strain G/UW-57 (middle), and E/Bour (bottom) against all others with position along the ompA gene. For clarity, only the strains with greatest similarity to the query sequence are shown.
The maximum chi-square test (47) was used to refine the RIP predictions and assess the statistical significance of the refined structures. The G/UW-57, D/B-120, E/Bour, and LGV-98 strains clearly showed mosaic structures, with each cut supported at the P = 0.0001 level (Fig. 5). Table 3 shows the breakpoint analysis of potential ompA mosaics. As suggested in the RIP analysis, G/UW-57 appeared to be a mosaic of strains F/IC-Cal-3 and E/Bour, with two breakpoints at nt 645 and nt 651 and at nt 792 and nt 794. Upstream of the first cut and downstream of the second, G/UW-57 was markedly similar to F/IC-Cal3 and dissimilar to E/Bour. Between the cuts, the opposite was true (Table 3). Strain D/B-120 appeared to be a mosaic of strain L1/440 and a possible divergent segment of E/Bour with a single break point at nt 547 and nt 567. Upstream of the cut, the E/Bour contribution was supported at only the P = 0.01 level. However, downstream of it, D/B-120 was extremely similar to L1/440. E/Bour appeared to be a mosaic of Ba/Apache-2, D/IC-Cal-8, and the aforementioned G/UW-57 with three breakpoints at nt 452 and nt 485, at nt 584 and nt 587, and at nt 851 and nt 867. E/Bour was most similar to Ba/Apache-2 before breakpoint 1, to D/IC-Cal-8 between breakpoints 1 and 2, to G/UW-57 between breakpoints 2 and 3, and to D/IC-Cal-8 again after breakpoint 3. As originally proposed by Hayes et al. (34), LGV-98 was a composite of L1 and L2, with a single breakpoint at nt 567. The upstream portion of LGV-98 most probably originated from L1, while the downstream segment was identical to L2. However, the test predicted two upstream divisions. Due to the small number of polymorphisms, the second division was most probably artifactual. The location of the single division agrees exactly with that proposed by Hayes et al. (34) (nt 537 in the LGV-98 sequence corresponds to nt 567 in this alignment). Ia/IU-4168 was identical to the Ia sequence originally identified to be an I/H mosaic by Lampe et al. (46) over all regions sequenced. Based on the results of the maximum chi-square test, Ia/IU-4168 was not a mosaic of I/H at the P = 0.001 significance level.
FIG. 5.
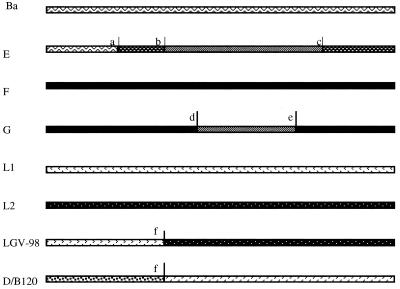
Mosaic structures for strains E/Bour, G/UW-57, LGV-98, and D/B-120 predicted by maximum chi-square analysis. Each line is the ompA gene for the strain given to the left. The shading represents the proposed contributions from parental strains to the genetic composition of the mosaic strain. a to f are the locations of the proposed crossover points at nt 452, 584, 851, 651, 794, and 547, respectively. Symbols: , Ba C. trachomatis -->; , E/G C. trachomatis; , D/IC-Cal-8 C. trachomatis; , F C. trachomatis; , L1 C. trachomatis; , L2 C. trachomatis; , divergent unknown C. trachomatis contribution.
TABLE 3.
Breakpoint analysis of potential ompA mosaics
| ompA mosaic | Parental strain | Location of all cutsa | Proportional differenceb
|
|||
|---|---|---|---|---|---|---|
| Before cut 1 | Between cut 1 and cut 2 | Between cut 2 and cut 3 | After last cut | |||
| G/UW-57 | F/IC-Cal3 | 645, 651 792, 794 | 9/92 (9.8%) | 17/17 (100%) | 9/49 (18%) | |
| E/Bour | 654, 804 | 90/93 (97%) | 2/18 (11%) | 45/47 (96%) | ||
| D/B-120 | L1/440 | 547, 567 | 28/37 (76%) | 9/52 (17%) | ||
| E/Bour | Ba/Apache-2 | 452, 485 584, 587 851, 867 | 4/19 (21%) | 17/20 (85%) | ||
| D/IC-Cal8 | 452, 485 584, 587 851, 867 | 7/40 (18%) | 38/41 (93%) | 11/53 (21%) | ||
| G/UW-57 | 452, 485 584, 587 851, 867 | 14/41 (34%) | 49/53 (93%) | |||
| LGV-98 | L1/440 | 548, 567 | 1/14 (7%) | 47/47 (100%) | ||
| L2/434 | 548, 567 | 13/14 (93%) | 0/47 (0%) | |||
Location of the cut may be represented by two numbers. Only polymorphic sites were analyzed, and thus breakpoint locations are not necessarily consecutive nucleotides.
Proportional difference is number of differences between the parental strain under comparison and the potential recombinant divided by the total number of differences between both parental strains and potential recombinant. The percentage is given in parentheses.
It is interesting that for every significant mosaic strain analyzed, VS3 may have been involved in the gene conversion event. The upstream division of G/UW-57 was just before VS3, and the downstream division cut within VS3. D/B120 and LGV-98 shared a presumed crossover point at nt 547, at the end of VS2. E/Bour had one division at the end of VS3 and another between VS3 and VS4. In fact, E/Bour was the only mosaic with a cut upstream of VS3 (at the beginning of VS2). Identification of most breakpoints just before VS3 is in agreement with the IA results, in which the downstream half of ompA was shown to have a higher degree of recombination than the upstream half. A summary of the test conclusions are presented in Table 4.
TABLE 4.
Summary of recombination test results
| Feature | ompA | omcB |
|---|---|---|
| Intraspecies recombination detected | Human C. trachomatis (P = 0.0001; Sawyer runs test) C. psittaci (P = 0.038; Sawyer runs test) | None |
| Interspecies recombination detected | C. trachomatis MoPn and C. pneumoniae Horse N16 (P = 0.033; Sawyer runs test) | None |
| Barrier to recombination suggested | C. trachomatis MoPn and SFPD and C. psittaci (P = 0.038 0.124: Sawyer runs test) | Not done |
| Linkage equilibrium for entire gene | No | No |
| Linkage equilibrium for regions | C class strains in downstream half (P = 103) | Not done |
| Relative degree of recombination among classes | C class strains higher level of recombination that B class strains | Not done |
| Relative degree of recombination among regions of gene | Downstream half higher level of recombination than upstream half | Not done |
| Significant mosaics found | G/UW-57, D/B-120, E/Bour, LGV-98 (P = 0.001) | Not done |
DISCUSSION
In this study, we provide comprehensive statistical analyses of intraspecies and interspecies recombination for C. trachomatis, C. pneumoniae, and C. psittaci at the ompA and omcB loci. These analyses revealed that intragenic recombination at the ompA locus for C. trachomatis strains is significant and not likely to be due to chance substitution events. The relative degree of recombination for different classes and different ompA regions were consistent with the multiple breakpoints in VS3 and VS4 that we identified for strains D/B120, G/UW-57, E/Bour, and LGV-98. The former three strains were first identified as recombinants in this study. Further, we present the first evidence for intragenic recombination between C. psittaci strains and for interspecies recombination between a rodent C. trachomatis strain and an equine C. pneumoniae strain at the ompA locus.
Three independent statistical methods provided strong and consistent evidence for recombination in ompA but not in omcB. The Sawyer runs test requires no phylogenetic inference and is relatively unbiased by the effects of selection. Monomorphic and amino acid-varying sites that may have clustered under strong selection were excluded. Silent sites should be effectively neutral. In a region that is immunodominant, however, this may not always be the case. A potential shortcoming of the Sawyer runs test is that it can produce false-positive results when mutation rates vary along the gene (Wiuf et al., submitted). However, it is unlikely that variable mutation rates have mimicked recombination in this instance. The incompatibility method we used was less sensitive to this phenomenon (Wiuf et al., submitted), but it also detected recombination in ompA. Further, it should also be noted that recombination in Chlamydia is biologically plausible. Not only has a homolog of RecA been cloned, sequenced, and characterized for C. trachomatis (37), but also homologs of several other enzymes in the RecBCD and RecF recombination pathways have been identified in the complete genome sequence of the C. trachomatis D/UW-3 strain (67).
In contrast to ompA, none of the methods used in this study detected intragenic recombination in omcB. It is unlikely that the failure to detect recombination was due to a high degree of recombination, since the IA test did not provide any evidence for recombination within omcB. To explain the discordant phylogenies for ompA and omcB, there must have been a major breakpoint somewhere in between these two genes. In our analyses, we identified major breakpoints in the downstream half of ompA. These results can be interpreted in at least two ways. One is that only ompA is involved in recombination and that these breakpoints delineate the segments involved. Another is that omcB is involved in recombination in its entirety and that another unidentified breakpoint resides downstream of it. This event cannot be ruled out by our analyses because none of the methods used would have been able to detect recombination without including the intervening sequence that encompassed the major breakpoint(s). However, this latter scenario is consistent with the differences in divergence of the two genes (24). If the difference in divergence was primarily due to differences in selective pressures, there should be the same relative divergence within species as between species, but this was not the case. Further, the ratio of dS/dN for the three species was nearly equal for the two genes. However, when just the human C. trachomatis strains were examined, the average dS/dN was over four times as high for ompA as it was for omcB. This suggests that human C. trachomatis strains evolved more quickly for omcB than for ompA and for strains from other species. Such anomalous patterns of divergence have been seen in other pathogenic bacteria including the Neisseria species, where it was suggested that recombination had obscured the evolutionary history of the organism (21).
Differences in selection pressures may in part explain the relative differences in the degrees to which intragenic and intergenic recombination occurred at the two loci. If omcB provides an essential function to the organism with strong functional and structural constraints, it may be under strong stabilizing selection pressure. In this case, fitness costs attributed to recombination within the reading frame may be too great and may in part prevent it from occurring. On the other hand, conserved regions resulting from strong constraints may have increased the likelihood of intergenic recombination since the degree of homologous recombination increases with increasing similarity between strains. Moreover, these constraints may have provided a selective force leading to the preferential retention of one allele and so to low variation, consistent with the “selective-sweep” model (39). According to this model, large regions of DNA will become fixed in areas of low recombination while only small areas will become fixed in areas of high recombination. For omcB, almost the entire gene has become fixed. This suggests that the degree of recombination must have been low enough to allow a sweep and infers an upper bound for intergenic recombination in omcB. In contrast, ompA is presumably under diversifying immune selection. Recombination within the reading frame of ompA may actually increase fitness and contribute to the organism's success. Since there are few to no regions in ompA that have become fixed, there is no upper bound for the degree of recombination that has occurred. Either recombination was too extensive for a selective sweep, or it was prevented from occurring by diversifying selection pressures. No analyses in this study can distinguish between these possibilities.
We provide evidence for interspecies ompA recombination between C. trachomatis and C. pneumoniae that infect lower vertebrates. Although it has been recognized that interspecies transmission of C. psittaci strains occurs between birds and humans in the form of psittacosis, no other evidence for cross-species transmission exists. Interspecies mosaics have not been identified by sequence observation, and tree topologies over multiple coding regions are congruent with respect to species clustering. Our results are consistent with these findings since these earlier methods were less sensitive than the ones used in this study. Further, there may actually be a barrier to recombination between the C. trachomatis rodent strains and the C. psittaci strains. The nucleotide differences for these species range from 39 to 41%, which is in excess of the presumed upper limit for homologous recombination (58). Thus, recombination between these species may not be feasible for lack of a viable mechanism.
Our analyses of previously identified mosaics and other well-known C. trachomatis strains provided significant evidence that strains D/B120, G/UW-57, E/Bour, and LGV-98 were recombinants. For each of these recombinants, several significant breakpoints just before and within VS3 were identified. These findings were in agreement with the IA analysis, where a higher degree of recombination was found in the downstream half of ompA. Recombination between more distantly related strains drives clonal divergence (28), and diversity in VS3 and VS4 may contribute to the adaptation of the parasite to changing host environments. Specifically, these mechanisms may in part be responsible for differences in immune evasion, persistence, and tissue tropism of the organism.
For C. trachomatis, upstream of and within VS3 are regions that elicit T-helper-cell activity (1, 40). T-cell-dependent antibody production presumably represents an important evolutionary mechanism for protection against microbial pathogens since it is conserved among vertebrate species (38). In turn, pathogens have developed mechanisms to vary T-cell epitopes to evade this host immune response. Altered peptide antagonism is a recognized immune escape mechanism for viruses and malaria where T-cell epitope variants are able to eliminate or downregulate the cell-mediated response to the index peptide (26, 36). In this respect, coevolution of hosts and parasites might be likened to a molecular arms race (26). For Chlamydia, in addition to evidence for the occurrence of point mutations within T-cell epitopes, our results show that a potential mechanism exists via recombination for exchanging T-cell epitopes important for escaping immune surveillance. Immune evasion may result in the failure of the host to clear the organism, resulting in a persistent infection. Further, there is evidence that persistent infections are more commonly associated with C class than B class strains despite the fact that B and intermediate class strains are the most prevalent overall in genital infections (17). Our analyses consistently suggest that the degree of recombination was higher for C class strains than for B class strains. Although the sequences from the above study were not analyzed for mosaic structures and gene conversion events, these data suggest that recombination in ompA may generate the genetic diversity required to evade immune surveillance, ultimately leading to persistence in the host.
In addition to providing the genetic diversity necessary for immune evasion, recombination within VS4 may be important in tissue tropism. Monoclonal antibodies against VS1, VS2, and VS4 neutralize chlamydial infection by inhibiting attachment (49). Differential trypsin inhibition suggests that VS2 and VS4 are critical in this process. Trypsin treatment does not reduce attachment for serovar L2, but it dramatically reduces the attachment for serovar B. This difference may be due to the presence of trypsin-sensitive lysine residues in VS2 and VS4 of serovar B and the absence of these residues in serovar L2 (29). The fact that VS4 is critical for attachment (29) and that there is a high frequency of recombination within VS4 suggests that genetic diversity in this region may contribute to serovar-specific differences in tissue tropism. Our recombination data for serovars D/B-120 and E/Bour are suggestive of this.
Serovar D is the most prevalent serovar in rectal infections except for serovars L1, L2, and L3. This serovar produces mild infections, while serovars L1, L2, and L3 have historically been associated with severe lymphadenitis and proctitis (4). However, it has recently been reported that rectal infections with serovar L1 are milder and similar to those caused by serovar D (5). Our analyses showed that D and L1 were markedly similar within VS3 and VS4 but divergent upstream of these segments, suggesting that D inherited the former VSs via recombination with serovar L1. Thus, recombination with serovar L1 may have allowed serovar D to more effectively invade the rectal mucosa. Additional evidence is provided by clinical data for serovar E. It is the most prevalent serovar in genital infections and outcompetes serovar F for nutrients and resources in tissue culture (41). Our analyses suggest that serovar E is a mosaic of Ba/Apache-2, D/IC-Cal-8, and G/UW-57. E and Ba were markedly similar upstream of VS2. However, Ba is a common serovar in ocular infections and is rarely found in genital infections. E also possesses a long, shared VS4 segment with D, another extremely prevalent serovar in genital infections. It is possible that E was once similar to Ba in its entirety and subsequently acquired DNA from D via recombination in VS4, which contributed to its success in the genital mucosa. This cumulative evidence is suggestive that genetic diversity in VS4 may play a role in determining chlamydial serovar-specific differences in tissue tropism.
The possibility that C. trachomatis C class strains exhibit a panmictic population structure in a region responsible for immune evasion has important implications for the design of a multivalent DNA or protein vaccine. A widely employed approach to the design of a chlamydial vaccine has been to target regions of VSs conserved among strains since VSs are likely to be surface exposed and immunodominant. Our analyses indicate that conserved regions within VS3 and VS4 must be nearly identical such that, within the targeted region, recombination between strains has the effect of homogenization as opposed to diversification. In contrast, targeted regions within VS1 and VS2 may not require the same degree of conservation since recombination occurs to a lesser degree. We can further refine this approach based on the fact that recombination seems to occur primarily between strains of the same class and not between strains of different classes. This is suggested by the fact that the degree of homologous recombination tends to increase with increasing similarity between strains, and in our analyses, an increase in linkage was observed when C class strains were analyzed separately from B class strains. The exception to this is that the intermediate class strains appear to recombine with the B class strains. For this reason, regions within VS3 and VS4 should be nearly identical within each class while some differences between classes may be acceptable where, for the purposes of this discussion, B and intermediate classes are considered together. To be conservative, this rule should be followed for VS1 and VS2 as well.
Following these criteria, we have identified segments in VS3 and VS4 that could be simultaneously targeted for a multistrain vaccine. The segment in VS3 is a hexapeptide located from nt 796 to 813. Among the C class strains the sequence AGTEAA is completely conserved, while among the B and intermediate class strains the sequence AGTDAA is conserved except for A→S in Ba/Apache-2 and AA→GV in L2. The segment in VS4 is a 13-residue peptide located from nt 982 to 1020. Among the C class strains, the sequence DVTTLNPTIAGKG is conserved except for V→T in A/Har-13 and A→T in K/UW-31. Among the B and intermediate class strains, the sequence DVTTLNPTIAGAG is conserved except for V→T in D/B120, E/Bour and L1 and V→I and A→C in both intermediate class strains. Within this 13-residue peptide is a nonapeptide previously identified by Fitch et al. (24) as being an extremely conserved segment that could be targeted for vaccine design. The knowledge that recombination occurs primarily within classes in VS3 and VS4 has provided criteria that has allowed us to take a less conservative approach in identifying conserved segments and thus expand the repertoire of targeted regions with considerable expectation for success.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grant AI39499 (to D. Dean).
We thank Walter Fitch for a thoughtful and critical review of an earlier version of this paper.
REFERENCES
- 1.Allen J E, Locksley R M, Stephens R S. A single peptide from the major outer membrane protein of Chlamydia trachomatis elicits T cell help for the production of antibodies to protective determinants. J Immunol. 1991;147:674–679. [PubMed] [Google Scholar]
- 2.Allen J E, Stephens R S. Identification by sequence analysis of two-site posttranslational processing of the cysteine-rich outer membrane protein 2 of Chlamydia trachomatis serovar L2. J Bacteriol. 1989;171:285–291. doi: 10.1128/jb.171.1.285-291.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baehr W, Zhang Y Z, Joseph T, Su H, Nano F E, Everett K D, Caldwell H D. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci USA. 1988;85:4000–4004. doi: 10.1073/pnas.85.11.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes R C, Rompalo A M, Stamm W E. Comparison of Chlamydia trachomatis serovars causing rectal and cervical infections. J Infect Dis. 1987;156:953–958. doi: 10.1093/infdis/156.6.953. [DOI] [PubMed] [Google Scholar]
- 5.Bauwens J E, Lampe M F, Suchland R J, Wong K, Stamm W E. Infection with Chlamydia trachomatis lymphogranuloma venereum serovar L1 in homosexual men with proctitis: Molecular analysis of an unusual case cluster. Clin Infect Dis. 1995;20:576–581. doi: 10.1093/clinids/20.3.576. [DOI] [PubMed] [Google Scholar]
- 6.Braun J, Laitko S, Treharne J, Eggens U, Wu P, Distler A, Sieper J. Chlamydia pneumoniae—a new causative agent of reactive arthritis and undifferentiated oligoarthritis. Ann Rheum Dis. 1994;53:100–105. doi: 10.1136/ard.53.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunham R, Yang C, Maclean I, Kimani J, Maitha G, Plummer F. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (ompA) gene. J Clin Investig. 1994;94:458–463. doi: 10.1172/JCI117347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter M W, Al-Mahdawi S A H, Giles I G, Treharne J D, Ward M E, Clarke I N. Nucleotide sequence and taxonomic value of the major outer membrane protein gene of Chlamydia pneumoniae IOL-207. J Gen Microbiol. 1991;137:465–475. doi: 10.1099/00221287-137-3-465. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Chlamydia trachomatis genital infections—United States, 1995. Morb Mortal Wkly Rep. 1997;46:193–198. [PubMed] [Google Scholar]
- 10.Clarke I N, Ward M E, Lambden P R. Molecular cloning and sequence analysis of a developmentally regulated cysteine-rich outer membrane protein from Chlamydia trachomatis. Gene. 1988;71:307–314. doi: 10.1016/0378-1119(88)90047-9. [DOI] [PubMed] [Google Scholar]
- 11.Coles A M, Allan I, Pearce J H. The nucleotide and derived amino acid sequence of the omcB gene of Chlamydia trachomatis serovar E. Nucleic Acids Res. 1990;18:6713. doi: 10.1093/nar/18.22.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean D. Chlamydial infections. In: Connor D H, Chandler F W, Schwartz D A, Manz H J, Lack E E, editors. Pathology of infectious diseases. Stamford, Conn: Appleton & Lange; 1999. pp. 473–497. [Google Scholar]
- 13.Dean D. Trachoma. In: Connor D H, Chandler F W, Schwartz D A, Manz H J, Lack E E, editors. Pathology of infectious diseases. Stamford, Conn: Appleton & Lange; 1999. pp. 498–507. [Google Scholar]
- 14.Dean D, Millman K. Molecular and mutation trends analyses of ompA alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J Clin Investig. 1997;99:475–483. doi: 10.1172/JCI119182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean D, Oudens E M, Padian N, Bolan G, Schachter J. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J Infect Dis. 1995;172:1013–1022. doi: 10.1093/infdis/172.4.1013. [DOI] [PubMed] [Google Scholar]
- 16.Dean D, Schachter J, Dawson C, Stephens R S. Comparison of the major outer membrane protein sequence variant regions of B/Ba isolates: a molecular epidemiologic approach to Chlamydia trachomatis infections. J Infect Dis. 1992;166:383–392. doi: 10.1093/infdis/166.2.383. [DOI] [PubMed] [Google Scholar]
- 17.Dean D, Suchland R J, Stamm W E. Evidence for long term cervical persistence of Chlamydia trachomatis by ompl genotyping. J Infect Dis. 2000;182:909–916. doi: 10.1086/315778. [DOI] [PubMed] [Google Scholar]
- 18.de la Maza L M, Fiedler T J, Carlson E J, Markoff B A, Peterson E M. Sequence diversity of the 60-kilodalton protein and of a putative 15-kilodalton protein between the trachoma and lymphogranuloma venereum biovars of Chlamydia trachomatis. Infect Immun. 1991;59:1196–1201. doi: 10.1128/iai.59.3.1196-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett K D, Andersen A A, Plaunt M, Hatch T P. Cloning and sequence analysis of the major outer membrane protein gene of Chlamydia psittaci 6BC. Infect Immun. 1991;59:2853–2855. doi: 10.1128/iai.59.8.2853-2855.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett K D, Hatch T P. Sequence analysis and lipid modification of the cysteine-rich envelope proteins of Chlamydia psittaci 6BC. J Bacteriol. 1991;173:3821–3830. doi: 10.1128/jb.173.12.3821-3830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil E, Zhou J, Maynard Smith J, Spratt B G. A comparison of the nucleotide sequence of the adk and recA genes of pathogenic and commensal Neisseria species: evidence for extensive interspecies recombination within adk. J Mol Evol. 1996;43:631–640. doi: 10.1007/BF02202111. [DOI] [PubMed] [Google Scholar]
- 22.Fielder T J, Pal S, Peterson E M, de la Maza L M. Sequence of the gene encoding the major outer membrane protein of the mouse pneumonitis biovar of Chlamydia trachomatis. Gene. 1991;106:137–138. doi: 10.1016/0378-1119(91)90579-z. [DOI] [PubMed] [Google Scholar]
- 23.Fielder T J, Peterson E M, de la Maza L M. Nucleotide sequence of DNA encoding the major outer membrane protein of Chlamydia trachomatis serovar L3. Gene. 1991;101:159–160. doi: 10.1016/0378-1119(91)90240-c. [DOI] [PubMed] [Google Scholar]
- 24.Fitch W M, Peterson E M, de la Maza L M. Phylogenetic analysis of the outer-membrane-protein genes of Chlamydiae, and its implication for vaccine development. Mol Biol Evol. 1993;10:892–913. doi: 10.1093/oxfordjournals.molbev.a040048. [DOI] [PubMed] [Google Scholar]
- 25.Fukushi H, Hirai K. Proposal of Chlamydia pecorum sp. nov. for Chlamydia strains derived from ruminants. Int J Syst Bacteriol. 1992;42:306–308. doi: 10.1099/00207713-42-2-306. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert S C, Plebanski M, Gupta S, Morris J, Cox M, Aidoo M, Kwiatkowski D, Greenwood B M, Whittle H C, Hill A V. Association of malaria parasite population structure, HLA, and immunological antagonism. Science. 1998;279:1173–1177. doi: 10.1126/science.279.5354.1173. [DOI] [PubMed] [Google Scholar]
- 27.Girjes A, Carrick F, Lavin M. Remarkable sequence relatedness in the DNA encoding the major outer membrane protein of Chlamydia psittaci (koala type I) and Chlamydia pneumoniae. Gene. 1994;138:139–142. doi: 10.1016/0378-1119(94)90796-x. [DOI] [PubMed] [Google Scholar]
- 28.Guttman D S, Dykhuizen D E. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 29.Hackstadt T. Cell biology. In: Stephens R S, editor. Chlamydia: intracellular biology, pathogenesis and immunity. Washington, D.C.: ASM Press; 1999. pp. 101–138. [Google Scholar]
- 30.Hatch T. Developmental biology. In: Stephens R S, editor. Chlamydia: intracellular biology, pathogenesis and immunity. Washington, D.C.: ASM Press; 1999. pp. 29–68. [Google Scholar]
- 31.Hayes L J, Clarke I N. Nucleotide sequence of the major outer membrane protein gene of Chlamydia trachomatis strain A/SAI/OT. Nucleic Acids Res. 1990;18:6136. doi: 10.1093/nar/18.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayes L J, Pecharatana S, Bailey R L, Hampton T J, Pickett M A, Mabey D C, Watt P J, Ward M E. Extent and kinetics of genetic change in the ompl gene of Chlamydia trachomatis in two villages with endemic trachoma. J Infect Dis. 1995;172:268–272. doi: 10.1093/infdis/172.1.268. [DOI] [PubMed] [Google Scholar]
- 33.Hayes L J, Pickett M A, Conlan J W, Ferris S, Everson J S, Ward M E, Clarke I N. The major outer-membrane proteins of Chlamydia trachomatis serovars A and B: intra-serovar amino acid changes do not alter specificities of serovar- and C subspecies-reactive antibody-binding domains. J Gen Microbiol. 1990;136:1559–1566. doi: 10.1099/00221287-136-8-1559. [DOI] [PubMed] [Google Scholar]
- 34.Hayes L J, Yearsley P, Treharne J D, Ballard R A, Fehler G H, Ward M E. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of C. trachomatis. Infect Immun. 1994;62:5659–5663. doi: 10.1128/iai.62.12.5659-5663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herring A J, Tan T W, Baxter S, Inglis N F, Dunbar S. Sequence analysis of the major outer membrane protein gene of an ovine abortion strain of Chlamydia psittaci. FEMS Microbiol Lett. 1989;65:153–158. doi: 10.1016/0378-1097(89)90383-2. [DOI] [PubMed] [Google Scholar]
- 36.Hill A V, Jepson A, Plebanski M, Gilbert S C. Genetic analysis of host-parasite coevolution in human malaria. Philos Trans R Soc London Ser B. 1997;352:1317–1325. doi: 10.1098/rstb.1997.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hintz N J, Ennis D G, Liu W F, Larsen S H. The recA gene of Chlamydia trachomatis: cloning, sequence, and characterization in Escherichia coli. FEMS Microbiol Lett. 1995;127:175–180. doi: 10.1111/j.1574-6968.1995.tb07470.x. [DOI] [PubMed] [Google Scholar]
- 38.Hodgkin P D. An antigen valence theory to explain the evolution and organization of the humoral immune response. Immunol Cell Biol. 1997;75:604–618. doi: 10.1038/icb.1997.95. [DOI] [PubMed] [Google Scholar]
- 39.Hudson R. How can low levels of DNA sequence variation in regions of the Drosophila genome with low recombination rates be explained? Proc Natl Acad Sci USA. 1994;91:6815–6818. doi: 10.1073/pnas.91.15.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishizaki M, Allen J E, Beatty P R, Stephens R S. Immune specificity of murine T-cell lines to the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1992;60:3714–3718. doi: 10.1128/iai.60.9.3714-3718.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jakobsen I B, Easteal S. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. CABIOS. 1996;12:291–295. doi: 10.1093/bioinformatics/12.4.291. [DOI] [PubMed] [Google Scholar]
- 42.Jones R B, Williams J A, van der Pol B. Competitive growth of serovars E and F combined in mixed tissue culture infections. In: Stephens R S, Byrne G I, Christiansen G, Clarke I N, Grayston J T, Rank R G, Ridgway G L, Saikku P, Schachter J, Stamm W E, editors. Chlamydia infections. 1998. pp. 523–526. Proc. Ninth Int. Symp. Hum. Chlamydial Infect. [Google Scholar]
- 43.Jordan I K, Makarova K S, Wolf Y I, Koonin E V. Gene conversions in genes encoding outer-membrane proteins in H. pylori and C. pneumoniae. Trends Genet. 2000;17:7–10. doi: 10.1016/s0168-9525(00)02151-x. [DOI] [PubMed] [Google Scholar]
- 44.Kaltenböck B, Schmeer N, Schneider R. Evidence for numerous ompl alleles of porcine Chlamydia trachomatis and novel chlamydial species obtained by PCR. J Clin Microbiol. 1997;35:1835–1841. doi: 10.1128/jcm.35.7.1835-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 46.Lampe M F, Suchland R J, Stamm W E. Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect Immun. 1993;61:213–219. doi: 10.1128/iai.61.1.213-219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maynard Smith J. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 48.Melgosa M P, Kuo C C, Campbell L A. Sequence analysis of the major outer membrane protein gene of Chlamdia pneumoniae. Infect Immun. 1993;59:2195–2199. doi: 10.1128/iai.59.6.2195-2199.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moulder J W. Interaction of chlamydiae and host cell in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 51.Newhall W J. Biosynthesis and disulfide cross-linking of outer membrane components during the growth cycle of Chlamydia trachomatis. Infect Immun. 1987;55:162–168. doi: 10.1128/iai.55.1.162-168.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newhall W J. Macromolecular and antigenic composition of chlamydiae. In: Barron A L, editor. Microbiology of chlamydiae. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 47–70. [Google Scholar]
- 53.Nigg C. An unidentified virus which produces pneumonia and systemic infection in mice. Science. 1942;95:49–50. doi: 10.1126/science.95.2454.49-a. [DOI] [PubMed] [Google Scholar]
- 54.Peterson E M, Markoff B A, de la Maza L M. The major outer membrane protein nucleotide sequence of Chlamydia trachomatis, serovar E. Nucleic Acids Res. 1990;18:3414. doi: 10.1093/nar/18.11.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pickett M A, Everson J S, Clarke I N. Chlamydia psittaci ewe abortion agent: complete nucleotide sequence of the major outer membrane protein gene. FEMS Microbiol Lett. 1988;55:229–234. [Google Scholar]
- 56.Pickett M A, Ward M E, Clarke I N. Complete nucleotide sequence of the major outer membrane protein gene from Chlamydia trachomatis serovar L1. FEMS Microbiol Lett. 1987;42:185–190. [Google Scholar]
- 57.Read T D, Brunham R C, Shen C, Gill S R, Heidelberg J F, White O, Hickey E K, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg S L, Eisen J, Fraser C M. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roberts M S, Cohan F M. The effect of DNA sequence divergence on sexual isolation in Bacillus. Genetics. 1993;134:401–408. doi: 10.1093/genetics/134.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saikku P. Epidemiology of Chlamydia pneumoniae in atherosclerosis. Am Heart J. 1999;138:S500–S503. doi: 10.1016/s0002-8703(99)70285-1. [DOI] [PubMed] [Google Scholar]
- 60.Saikku P, Wang S P, Kleemola M, Brander E, Rusanen E, Grayston J T. An epidemic of mild pneumonia due to an unusual strain of Chlamydia psittaci. J Infect Dis. 1985;151:832–839. doi: 10.1093/infdis/151.5.832. [DOI] [PubMed] [Google Scholar]
- 61.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 62.Sardinia L M, Segal E, Ganem D. Developmental regulation of the cysteine-rich outer-membrane proteins of murine Chlamydia trachomatis. J Gen Microbiol. 1988;134:997–1004. doi: 10.1099/00221287-134-4-997. [DOI] [PubMed] [Google Scholar]
- 63.Sawyer S A. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- 64.Sayada C, Denamur E, Elion J. Complete sequence of the major outer membrane protein-encoding gene of Chlamydia trachomatis serovar Da. Gene. 1992;120:129–130. doi: 10.1016/0378-1119(92)90022-h. [DOI] [PubMed] [Google Scholar]
- 65.Siepel A C, Halpern A L, Macken C, Korber B T M. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses. 1995;11:1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 66.Sriram S, Mitchell W, Stratton C. Multiple sclerosis associated with Chlamydia pneumoniae infection of the CNS. Neurology. 1998;50:571–572. doi: 10.1212/wnl.50.2.571. [DOI] [PubMed] [Google Scholar]
- 67.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 68.Stephens R S, Mullenbach G, Sanchez-Pescador R, Agabian N. Sequence analysis of the major outer membrane protein gene from Chlamydia trachomatis serovar L2. J Bacteriol. 1986;168:1277–1282. doi: 10.1128/jb.168.3.1277-1282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stills H F, Fox J G, Paster B J, Dewhirst F E. A “new” Chlamydia sp. strain SFPD isolated from transmissable proliferative ileitis in hamsters. Microbiol Ecol Health Dis. 1991;4:S99. [Google Scholar]
- 70.Storey C, Lusher M, Yates P, Richmond S. Evidence for Chlamydia pneumoniae of non-human origin. J Gen Microbiol. 1993;139:2621–2626. doi: 10.1099/00221287-139-11-2621. [DOI] [PubMed] [Google Scholar]
- 71.Stothard D R, Boguslawski G, Jones R B. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potenital pathogenic determinants. Infect Immun. 1998;66:3618–3625. doi: 10.1128/iai.66.8.3618-3625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S P, Grayston J T. Three new serovars of Chlamydia trachomatis: Da, Ia, and L2a. J Infect Dis. 1991;163:403–405. doi: 10.1093/infdis/163.2.403. [DOI] [PubMed] [Google Scholar]
- 73.Wang S P, Kuo C C, Barnes R C, Stephens R S, Grayston S T. Imunotyping of Chlamydia trachomatis with monoclonal antibodies. J Infect Dis. 1985;152:791–800. doi: 10.1093/infdis/152.4.791. [DOI] [PubMed] [Google Scholar]
- 74.Watson M W, al-Mahdawi S, Lamden P R, Clarke I N. The nucleotide sequence of the 60 kDa cysteine rich outer membrane protein of Chlamydia pneumoniae strain IOL-207. Nucleic Acids Res. 1990;17:5299. doi: 10.1093/nar/18.17.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watson M W, Lambden P R, Clarke I N. The nucleotide sequence of the 60 kDa cysteine rich outer membrane protein of Chlamydia psittaci strain EAE/A22/M. Nucleic Acids Res. 1990;18:5300. doi: 10.1093/nar/18.17.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watson M W, Lambden P R, Ward M E, Clarke I N. Chlamydia trachomatis 60 kDa cysteine rich outer membrane protein: sequence homology between trachoma and LGV biovars. FEMS Microbiol Lett. 1989;53:293–297. doi: 10.1016/0378-1097(89)90233-4. [DOI] [PubMed] [Google Scholar]
- 77.Yang C L, Maclean I, Brunham R C. DNA sequence polymorphism of the Chlamydia trachomatis ompA gene. J Infect Dis. 1993;168:1225–1230. doi: 10.1093/infdis/168.5.1225. [DOI] [PubMed] [Google Scholar]
- 78.Yuan Y, Zhang Y X, Watkins N G, Caldwell H D. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect Immun. 1989;57:1040–1049. doi: 10.1128/iai.57.4.1040-1049.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y X, Fox J G, Ho Y, Zhang L, Jr. Stills H F, Smith T F. Comparison of the major outer-membrane protein (MOMP) gene of mouse pneumonitis (MoPn) and hamster SFPD strains of Chlamydia trachomatis with other Chlamydia strains. Mol Biol Evol. 1993;10:1327–1342. doi: 10.1093/oxfordjournals.molbev.a040079. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y X, Morrison S G, Caldwell H D, Baehr W. Cloning and sequence analysis of the major outer membrane protein genes of two Chlamydia psittaci strains. Infect Immun. 1989;57:1621–1625. doi: 10.1128/iai.57.5.1621-1625.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y X, Morrison S G, Caldwell H D. The nucleotide sequence of major outer membrane protein gene of Chlamydia trachomatis serovar F. Nucleic Acids Res. 1990;18:1061. doi: 10.1093/nar/18.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]