Abstract
A large fragment of the dissimilatory sulfite reductase genes (dsrAB) was PCR amplified and fully sequenced from 30 reference strains representing all recognized lineages of sulfate-reducing bacteria. In addition, the sequence of the dsrAB gene homologs of the sulfite reducer Desulfitobacterium dehalogenans was determined. In contrast to previous reports, comparative analysis of all available DsrAB sequences produced a tree topology partially inconsistent with the corresponding 16S rRNA phylogeny. For example, the DsrAB sequences of several Desulfotomaculum species (low G+C gram-positive division) and two members of the genus Thermodesulfobacterium (a separate bacterial division) were monophyletic with δ-proteobacterial DsrAB sequences. The most parsimonious interpretation of these data is that dsrAB genes from ancestors of as-yet-unrecognized sulfate reducers within the δ-Proteobacteria were laterally transferred across divisions. A number of insertions and deletions in the DsrAB alignment independently support these inferred lateral acquisitions of dsrAB genes. Evidence for a dsrAB lateral gene transfer event also was found within the δ-Proteobacteria, affecting Desulfobacula toluolica. The root of the dsr tree was inferred to be within the Thermodesulfovibrio lineage by paralogous rooting of the alpha and beta subunits. This rooting suggests that the dsrAB genes in Archaeoglobus species also are the result of an ancient lateral transfer from a bacterial donor. Although these findings complicate the use of dsrAB genes to infer phylogenetic relationships among sulfate reducers in molecular diversity studies, they establish a framework to resolve the origins and diversification of this ancient respiratory lifestyle among organisms mediating a key step in the biogeochemical cycling of sulfur.
Siroheme dissimilatory sulfite reductases (EC 1.8.99.3) catalyze the reduction of sulfite to sulfide, an essential step in the anaerobic sulfate-respiration pathway. Consequently, this enzyme has been found in all dissimilatory sulfate-reducing prokaryotes (SRPs) investigated so far. Furthermore, siroheme dissimilatory sulfite reductase-like enzymes have been detected in the hyperthermophilic archaeon Pyrobaculum islandicum capable of using sulfite as terminal electron acceptor (23), the phototrophic bacterium Allochromatium vinosum (10, 12), and the obligate chemolithotrophic species Thiobacillus denitrificans (32). In the latter two organisms the dissimilatory sulfite reductase has a proposed function in sulfide oxidation.
Siroheme sulfite reductases consist of at least two different polypeptides in an α2β2 structure. The genes encoding the two subunits are found adjacent to each other in the respective genomes (see, for example, references 3, 15, 17, 18, and 35) and probably arose from duplication of an ancestral gene (3). Comparative amino acid sequence analysis of the dissimilatory sulfite reductase genes (dsrAB) has recently been used to investigate the evolutionary history of anaerobic sulfate (sulfite) respiration (10, 17, 18, 35). The presence of dsrAB homologs in at least five highly divergent prokaryotic lineages and overall phylogenetic congruence of the dsrAB tree with the 16S rRNA gene tree suggested that the dissimilatory sulfite reductases of extant SRPs evolved vertically from common ancestral protogenotic genes (35). The remarkable degree of conservation of the dsrAB genes also provided a basis for culture-independent molecular diversity studies of natural sulfate-reducing assemblages with the use of PCR primers broadly specific for a large fragment of all known dsrAB genes (1, 22). However, one contradiction between the dsrAB and 16S rRNA phylogenies was recently recognized in that the dsrAB sequences of Desulfotomaculum thermocisternum (17) and Desulfotomaculum ruminis are not monophyletic (18). This finding could indicate that, in addition to vertical transmission, lateral gene transfer is involved in the evolution of SRPs.
In the present study we investigated this question further by phylogenetic analysis of the dsrAB genes from a wide range of cultivated SRPs. We found a clear case for multiple lateral transfer events of the dsrAB genes between major lineages of Bacteria and likely between the domains Bacteria and Archaea, suggesting that genes involved in primary metabolic functions, such as sulfate respiration, may be more prone to lateral transfer than previously thought.
MATERIALS AND METHODS
Bacterial strains.
The investigated reference strains of sulfate- and sulfite-reducing bacteria are listed in Table 1. If necessary, strains were cultured as recommended by the DSMZ type culture collection (Braunschweig, Germany).
TABLE 1.
Physiological and biochemical properties of the sulfite- and sulfate-reducing prokaryotes investigated in this study
| Speciesa | Strain no. | Oxidationb | Topt (°C)c | G+C content (mol%)d
|
Accession no.
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genome′ | dsrAB | Genome′/dsrAB | dsrAB3rd′ | Genome′/dsrAB3rd′ | dsrAB | 16S rDNA | ||||
| Archaea | ||||||||||
| Euryarchaeota | ||||||||||
| Archaeoglobus profundus∗ | DSM5631T | C | 82 | 41 | 47 | 0.87 | 50 | 0.82 | AF071499 | AF297529 |
| Archaeoglobus fulgidus∗ | DSM4304T | C | 83 | 46 | 50 | 0.92 | 59 | 0.78 | M95624 | X05567, Y00275 |
| Bacteria | ||||||||||
| Nitrospira division | ||||||||||
| Thermodesulfovibrio yellowstonii | DSM11347T | I | 65 | 30 | 39 | 0.77 | 28 | 1.07 | U58122/3 | L14619 |
| Thermodesulfovibrio islandicus | DSM12570T | I | 65 | 38 | 39 | 0.97 | 29 | 1.31 | AF334599 | X96726 |
| Thermodesulfobacterium division | ||||||||||
| Thermodesulfobacterium commune∗ | DSM2178T | I | 70 | 34 | 41 | 0.83 | 33 | 1.03 | AF334596 | L10662 |
| Thermodesulfobacterium mobile∗ | DSM1276T | I | 65 | 31 | 41 | 0.76 | 34 | 0.91 | AF334598 | AF334601 |
| Firmicutes: Bacillus/Clostridium group | ||||||||||
| Desulfotomaculum ruminis | DSM2154T | I | 28 | 49 | 46 | 1.07 | 45 | 1.09 | U58118/9 | Y11572 |
| Desulfotomaculum aeronauticum | DSM10349T | I | 37 | 44 | 48 | 0.92 | 52 | 0.85 | AF273033 | X98407 |
| Desulfotomaculum putei | DSM12395T | I | 50–65 | 47 | 52 | 0.90 | 61 | 0.77 | AF273032 | AF053929 |
| Desulfotomaculum geothermicum∗ | DSM3669T | C | 54 | 50 | 53 | 0.94 | 65 | 0.77 | AF273029 | X80789 |
| Desulfotomaculum thermosapovorans∗ | DSM6562T | I | 50 | 51 | 52 | 0.98 | 62 | 0.82 | AF271769 | Y11575 |
| Desulfotomaculum kuznetsovii∗ | DSM6115T | C | 60–65 | 49 | 56 | 0.88 | 74 | 0.66 | AF273031 | Y11569 |
| Desulfotomaculum thermocisternum∗ | DSM10259T | I | 62 | 57 | 55 | 1.04 | 67 | 0.85 | AF074396 | U33455 |
| Desulfotomaculum thermobenzoicum∗ | DSM6193T | C | 62 | 53 | 56 | 0.95 | 74 | 0.72 | AF273030 | L15628 |
| Desulfotomaculum thermoacetoxidans∗ | DSM5813T | C | 55–60 | 50 | 56 | 0.89 | 73 | 0.68 | AF271770 | Y11573 |
| Desulfotomaculum acetoxidans∗ | DSM771T | C | 37 | 38 | 45 | 0.84 | 41 | 0.93 | AF271768 | Y11566 |
| Desulfosporosinus orientis | DSM765T | I | 30 | 45 | 42 | 1.07 | 30 | 1.50 | AF271767 | Y11570 |
| Desulfitobacterium dehalogenans | DSM9161T | I | 37 | 45 | 47 | 0.96 | 46 | 0.98 | AF337903 | L28946 |
| Desulfitobacterium hafniense | DSM10664T | I | 37 | 47 | 48 | 0.98 | 47 | 1.00 | ND | X94975 |
| Proteobacteria delta subdivision | ||||||||||
| Desulfobacter vibrioformis | DSM8776T | C | 33 | 47 | 49 | 0.96 | 55 | 0.85 | AJ250472 | U12254 |
| Desulfobacter latus | DSM3381T | C | 29–32 | 44 | 49 | 0.90 | 54 | 0.81 | U58124/5 | M34414 |
| Desulfobacula toluolica∗ | DSM7467T | C | 28 | 42 | 53 | 0.79 | 66 | 0.64 | AF271773 | X70953 |
| Desulfofaba gelida | DSM12344T | I | 7 | 53 | 52 | 1.02 | 61 | 0.87 | AF334593 | AF099063 |
| Desulfobotulus sapovorans | DSM2055T | I | 34 | 53 | 52 | 1.02 | 59 | 0.90 | U58120/1 | M34402 |
| Desulfosarcina variabilis | DSM2060T | C | 33 | 51 | 55 | 0.93 | 68 | 0.75 | AF191907 | M34407 |
| Desulfococcus multivorans | DSM2059T | C | 35 | 57 | 56 | 1.02 | 72 | 0.79 | U58126/7 | M34405 |
| Desulfovibrio vulgaris | DSM644T | I | 30–36 | 65 | 61 | 1.07 | 82 | 0.79 | U16723 | M34399 |
| Desulfovibrio sp. strain PT-2 | ATCC 49975 | I | 30 | 65 | 62 | 1.05 | 86 | 0.76 | U58114/5 | M98496 |
| Desulfovibrio desulfuricans Essex 6 | DSM642T | I | 30 | 59 | 60 | 0.98 | 79 | 0.75 | AJ249777 | AF192153 |
| Desulfovibrio africanus | DSM2603T | I | 30–36 | 65 | 62 | 1.05 | 86 | 0.76 | AF271772 | X99236 |
| Desulfovibrio desulfuricans El Agheila Z | DSM1926 | NDe | 30 | ND | 56 | ND | 72 | ND | AF334592 | M37316 |
| Desulfoarculus baarsii | DSM2075T | C | 37 | 66 | 62 | 1.06 | 85 | 0.78 | AF334600 | M34403 |
| Desulfomonile tiedjei | DSM6799T | C | 37 | 49 | 53 | 0.92 | 63 | 0.78 | AF334595 | M26635 |
| Desulfobulbus rhabdoformis | DSM8777T | I | 31 | 51 | 52 | 0.98 | 55 | 0.93 | AJ250473 | U12253 |
| Desulfobulbus propionicus | DSM2032T | I | 39 | 60 | 57 | 1.05 | 72 | 0.83 | AF218452 | M34410 |
| Desulfobulbus sp. strain 3pr10 | DSM2058T | I | 29 | ND | 48 | ND | 44 | ND | AF337902 | M34411 |
| Desulforhopalus vacuolatus | DSM9700T | I | 18 | 48 | 47 | 1.02 | 46 | 1.04 | AF334594 | L42613 |
| Desulfovirga adipica | DSM12016T | C | 35 | 60 | 57 | 1.05 | 75 | 0.80 | AF334591 | AJ237605 |
| Desulforhabdus amnigena | DSM10338T | C | 37 | 53 | 52 | 1.02 | 57 | 0.93 | AF337901 | X83274 |
| Thermodesulforhabdus norvegica | DSM9990T | C | 60 | 51 | 53 | 0.96 | 63 | 0.81 | AF334597 | U25627 |
SRPs with a putative xenologous DsrAB are indicated by an asterisk.
C, complete; I, incomplete.
Topt, optimum growth temperature.
Genome/dsrAB and genome/dsrAB 3rd, quotient of genomic and dsrAB G+C content and quotient of genomic and dsrAB third position G+C content, respectively. The accuracy of the genomic G+C values may vary due to the different determination methods used.
ND, no data available.
DNA isolation and PCR amplification.
Genomic DNA of the reference organisms investigated was obtained from logarithmically growing or lyophilized cells by either using the FastPrep FP120 bead beater and the FastDNA Kit MH (Bio-101, Inc., La Jolla, Calif.) or another direct lysis technique (24) modified as described previously (9). An approximately 1.9-kb dsrAB segment was PCR amplified as described previously (35). Since amplification of the dsrAB gene fragment was not possible for all investigated reference strains, additional degeneracies were introduced in the previously published primers DSR1F and DSR4R (DSR1Fdeg, 5′-ACSCAYTGGAARCACG-3′; DSR4Rdeg, 5′-GTGTARCAGTTDCCRCA-3′), making them fully complementary to the respective target sites of recently published dsrAB sequences (17, 18). However, it should be noted that many “non-dsrAB” amplificates of ca. 1.9 kb were obtained using the degenerated primers.
Cloning and sequencing of dsrAB gene fragments.
If not mentioned otherwise dsrAB PCR products of the sulfite- and sulfate-reducing reference strains were ligated into pCR2.1-TOPO or pCR-XL-TOPO vectors (Invitrogen). Clones with approximate 1.9-kb inserts were recovered with the QIAprep spin kit (Qiagen, Hilden, Germany) and sequenced with a 4200L automated Li-Cor Long Reader DNA Sequencer (MWG, Ebersberg, Germany). dsrAB PCR products of the Desulfotomaculum species D. aeronauticum, D. putei, D. geothermicum, D. kuznetsovii, and D. thermobenzoicum were directly sequenced. In addition, dsr sequences of Desulforhabdus amnigena, Desulfobulbus sp., and Desulfitobacterium dehalogenans were determined by direct sequencing as well as sequencing of the cloned PCR product. Previously published (35) partial dsrAB sequences of Desulfotomaculum ruminis, Thermodesulfovibrio yellowstonii, Desulfobacter latus sp., Desulfobotulus sapovorans, Desulfococcus multivorans, and Desulfovibrio sp. strain PT-2 were completed by resequencing of the original clones.
16S rRNA of Thermodesulfobacterium mobile.
The 16S rRNA gene sequence of Thermodesulfobacterium mobile (=T. thermophilum) was obtained as described previously (14).
Phylogeny inference.
Phylogenetic analyses were performed on alignments of the 16S ribosomal DNA (rDNA) nucleotide and the inferred amino acid sequences of the dsrAB genes. Regions of ambiguous positional homology were removed from the 16S rDNA data set using the Lane mask (16) and a DsrAB amino acid alignment mask prepared in ARB (http://www.arb-home.de). A total of 1,335 nucleotides and 543 amino acid positions (alpha subunit, 327; beta subunit, 216) were used in 16S rDNA and DsrAB analyses, respectively. For paralogous rooting DsrA sequences were aligned against DsrB and trees were calculated based on 173 amino acid positions, including positions with insertions and deletions. Phylogenetic analyses were performed with PAUP* version 4.0b2a (D. L. Swofford, Sinauer Associates, Sunderland, Mass.), ARB, or PHYLIP version 3.57c (J. Felsenstein, University of Washington, Seattle). Evolutionary distance (ED) analyses were conducted on the 16S rDNA data set using the Kimura 2 parameter and general time-reversible substitution model corrections with and without rate correction. Rate heterogeneities were corrected using a gamma distribution model (the shape parameter, alpha, was estimated to be 0.52 using a parsimony-based approximation in PAUP*). ED analysis of the DsrAB data set was performed by using a Dayhoff PAM correction and neighbor joining. Maximum parsimony (MP) trees were constructed for both data sets using the default settings in PAUP*. Maximum likelihood (ML) analysis of the 16S rDNA data set was performed in the ARB package using the fastDNAml program (28). Bootstrap resampling of the ED and MP trees was performed for all analyses to provide confidence estimates for the inferred topologies. A total of 1,000 or 2,000 replicates was used in all cases, with the exception of the ED analysis of the DsrAB data set, wherein 100 replicates were calculated.
RESULTS
Dissimilatory sulfite reductase phylogeny.
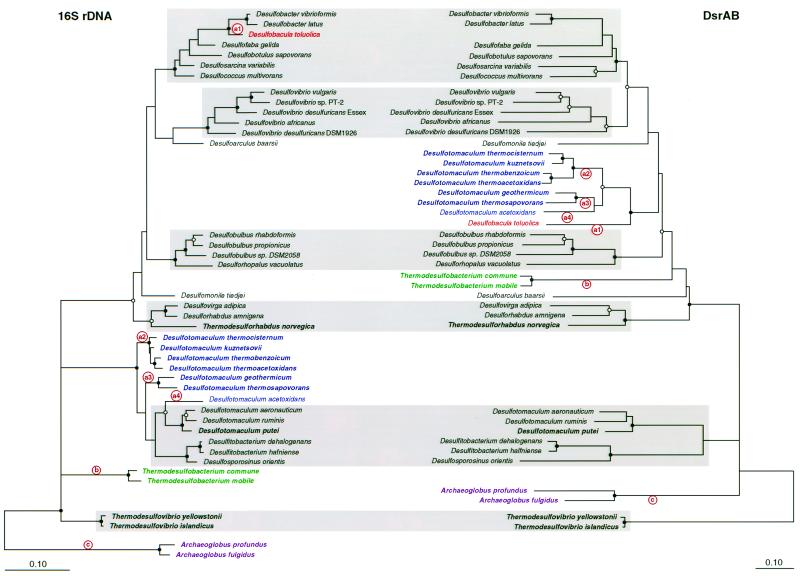
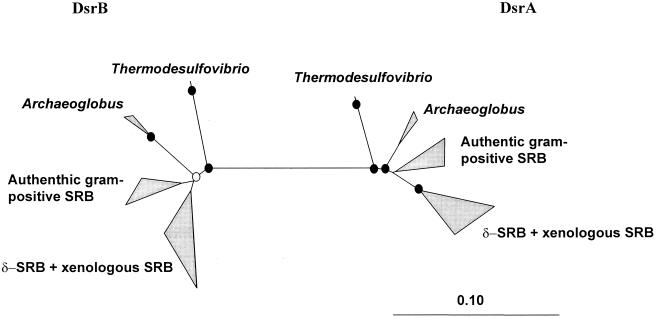
A DNA fragment ca. 1.9 kb in size, encompassing most of the alpha and beta subunit genes of the dissimilatory sulfite reductase, was amplified from 30 sulfite- and sulfate-reducing bacteria (Table 1). Complete sequences of the PCR products were obtained. Compiled sequences were entered into the dsrAB database, translated into amino acids, and manually aligned. Previously published partial length dsr sequences of Desulfovibrio oxyclinae (U58116/7 [35]), Desulfovibrio simplex (U78738 [10]), Desulfovibrio gigas (U80961), Desulfonema limicola (U58128/9 [35]), and Desulfobacterium autotrophicum (Y15478) were not included to avoid resolution loss in phylogenetic analyses. Comparative sequence analyses were performed based on each subunit and both subunits combined. No major differences were noted between the individual and combined subunit tree topologies regardless of the inference method used, indicating a shared evolutionary history for the alpha and beta subunits. Consistent with these findings, the G+C contents of dsrA and dsrB were almost identical for each organism (data not shown). Consequently, detailed phylogenetic analyses were performed on a combined (DsrAB) data set in order to include the maximum number of 543 comparable amino acid positions. For comparison, trees were calculated from the 16S rRNA genes of the identical set of organisms to avoid sampling artifacts (Fig. 1). Since the 16S rDNA sequence of Thermodesulfobacterium mobile was not available, it was determined in this study (1,520 nucleotides). In Fig. 1, the Archaeoglobus sequences were used as the outgroup for the 16S rRNA tree since they are the only representatives of the archaeal domain in an otherwise bacterial tree. In contrast, the Thermodesulfovibrio sequences (bacterial Nitrospira division) were used as the outgroup in the DsrAB analyses since paralogous outgrouping of the alpha and beta subunits suggests that the root of the Dsr tree is along the Thermodesulfovibrio line of descent (Fig. 2). Therefore, it appears likely that the dissimilatory sulfite reductases of the Archaeoglobales have a bacterial origin (see Discussion).
FIG. 1.
Comparison of 16S rRNA (ML) and DsrAB (ED) trees for the sulfate- and sulfite-reducing prokaryotes investigated. Branch points supported by phylogenetic analysis (bootstrap support values of >90% in all ED and MP methods) are indicated by filled circles. Open circles at branch points indicate >75% bootstrap support in most or all analyses, while branch points without circles were not resolved (bootstrap values of <75%) as specific groups in the different analyses. Both trees are collapsed back at the division level. Thermophilic prokaryotes are in boldface. Consistent monophyletic groups between both trees are shaded gray. Microorganisms affected by putative LGT events of the dsrAB genes are color coded. dsrAB recipient or donor lineages are indicated by circled letters (a to c) located above or below the branch, respectively. The bars represent 0.1 changes per nucleotide or amino acid, respectively.
FIG. 2.
Unrooted amino acid tree (ED) based on an alignment of DsrA to DsrB. The dissimilatory sulfite reductases of Allochromatium vinosum and Pyrobaculum islandicum were excluded from the analysis since they likely are members of different enzyme families (9, 22). The bar represents 0.1 changes per amino acid. Bootstrap analyses were performed using the PHYLIP parsimony method with 100 resamplings (Felsenstein, University of Washington). Branch points with parsimony bootstrap support of >85% are indicated by filled circles. Open circles at branch points indicate >50% bootstrap support, while branch points without circles either have parsimony bootstrap values of <50% (authentic gram-positive sulfate-reducing bacteria [SRB] DsrB; δ-SRB + xenologous SRB DsrB) or are not obtained with the parsimony method (Archaeoglobus and authentic gram-positive sulfate-reducing bacteria DsrA sequences form a monophyletic cluster in the parsimony method).
Overall, highly similar orderings of taxa, shaded gray in Fig. 1, were found between the 16S rRNA and DsrAB trees with all treeing methods. However, major incongruencies were found between DsrAB- and 16S rRNA-based analysis for seven members of the genus Desulfotomaculum, for both species of the genus Thermodesulfobacterium, and for the δ-proteobacterium Desulfobacula toluolica (color coded; Fig. 1). In contrast to relationships inferred using the rRNA, the genus Desulfotomaculum, a member of the low G+C gram-positive division (33), is not monophyletic in the DsrAB tree. Desulfotomaculum aeronauticum, D. ruminis, and D. putei form a clearly separated grouping, together with Desulfosporosinus orientis, based on their DsrAB sequences, while the other seven Desulfotomaculum species cluster together with Desulfobacula toluolica within the δ-proteobacterial radiation. Similarly, Thermodesulfobacterium commune and T. mobile comprise a division level lineage by rRNA analysis but branch within the δ-Proteobacteria according to their DsrAB sequences. A final discrepancy recognized is the inconsistent branching point of Desulfobacula toluolica. By 16S rRNA comparison, this species is closely related to Desulfobacter latus and Desulfobacter vibrioformis, while its DsrAB sequence is robustly associated with the Desulfotomaculum group in the δ-Proteobacteria (Fig. 1). The most parsimonious interpretation is that these significant topological conflicts reflect lateral transfer of the DsrAB genes (see Discussion). Points of inferred lateral gene transfer (LGT) are indicated in Fig. 1 by circled letters on the 16S rRNA tree.
Additional evidence for lateral transfer of dissimilatory sulfite reductase.
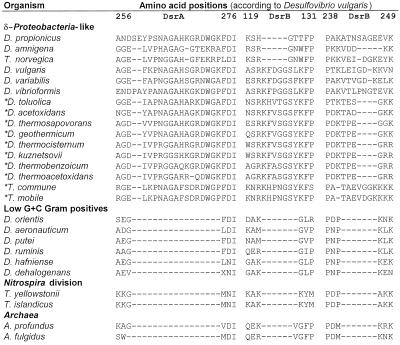
Insertions and deletions within the DsrAB amino acid sequences (excluded in the phylogenetic analyses) were investigated as additional signposts of the deduced evolutionary relationships, particularly with respect to inferred LGT events. In total, three insertions were unique to the δ-Proteobacteria: one in the alpha subunit and two in the beta subunit (Fig. 3). These insertions were also found in the δ-proteobacterium-like DsrAB sequences of the seven Desulfotomaculum species, and two Thermodesulfobacterium species, thus independently supporting the suggested LGT events.
FIG. 3.
Amino acid alignment of DsrA and DsrB showing insertions supporting the δ-proteobacterial origin of the putative laterally transferred sulfite reductases (labeled with an asterisk). It should be noted that the presumably xenologous DsrA and DsrB of Archaeoglobus do not show the typical δ-proteobacterial insertions.
Sizable differences in G+C content of the host genomes and acquired genes have been used to infer recent LGT events (20). A variation of more than 10% between the dsrAB G+C content and the respective genomic G+C content was found in Thermodesulfobacterium mobile, Thermodesulfobacterium commune, Thermodesulfovibrio yellowstonii, Desulfobacula toluolica sp., Desulfotomaculum acetoxidans, Desulfotomaculum kuznetsovii, Desulfotomaculum thermoacetoxidans, and Archaeoglobus profundus (Table 1). In seven of these eight organisms, LGT of dsrAB was predicted by comparison of tree topologies (Fig. 1). We attempted but failed to refine this analysis using the approach of Lawrence and Ochman (20) to identify atypical sequence characteristics (data not shown) since this method produces unreliable estimates for samples containing fewer than 1,500 codons as described previously (20).
Dissimilatory sulfite reductase homolog of Desulfitobacterium dehalogenans
The conserved dsrAB primers also amplified a fragment of the expected length from Desulfitobacterium dehalogenans, a bacterium capable of sulfite reduction but not of sulfate reduction (34). Comparative sequence analysis of the amplicon demonstrated a specific relationship to the dissimilatory sulfite reductase of Desulfosporosinus orientis, a finding consistent with their 16S rRNA-based relationship (Fig. 1). Furthermore, the recently completed genome sequence of Desulfitobacterium hafniense (http://www.jgi.doe.gov) contains a dsrAB sequence highly similar to the one of Desulfitobacterium dehalogenans (97.0% amino acid identity). As expected from the close relationship of both species by 16S rDNA comparison (96.7% similarity), their DsrAB sequences group together independent of the treeing methods applied.
DISCUSSION
In this study we investigated the phylogeny of the dissimilatory sulfite reductase from a study set of reference species encompassing all described lineages of SRPs in order to clarify whether dsr genes, in addition to undergoing vertical transmission, have also been laterally transferred. Using degenerated PCR primers, DNA fragments with strong sequence similarities over their entire length to previously published dsrAB sequences were obtained from all investigated SRPs and from the sulfite reducer Desulfitobacterium dehalogenans.
DSR sequence motifs.
The newly determined dsrAB-like sequences contain the essential cluster-binding residues typical for dissimilatory sulfite reductases. In particular, all alpha-subunit sequences contain the complete (Cys-X5-Cys)-Xn-(Cys-X3-Cys) motif required for coupling of the [Fe4S4]-siroheme cofactor (2). As for other dissimilatory sulfite reductases (10), this Cys motif is truncated in the beta subunit of the newly determined DsrAB sequences. In contrast to the prediction of Dahl et al. (3) the DsrB subunit of Thermodesulfobacterium mobile and Thermodesulfobacterium commune (4, 8) does not contain a complete siroheme-[Fe4S4] binding site that could explain the measured binding of four sirohemes per α2β2 molecule (versus two sirohemes for typical sulfite reductases). Furthermore, all DsrA sequences possess the Cys-Pro and Cys-X2-Cys-X2-Cys motif required for linking [Fe4S4] clusters (3). Since the reverse PCR primers used for amplification target part of the [Fe4S4] cluster binding motif of DsrB only the Cys-Pro signature is present in all deduced DsrB sequences. The absolute conservation of functionally important protein sequences and the absence of frameshift or nonsense mutations suggests that the characterized genes are transcribed and translated and function as dissimilatory sulfite reductases. The sequenced dissimilatory sulfite reductase genes of Thermodesulfobacterium mobile are most likely functionally expressed since the highly variable N-terminal sequence of the beta-subunit is identical to the one determined by Edman degradation (4). Comparison of the 10 N-terminal amino acids of the beta-subunit determined by Edman degradation of the dissimilatory sulfite reductase of Thermodesulfobacterium commune (8) to the sequence deduced in our study revealed a single amino acid difference at position 1 (Thr/Ser predicted by Edman and Gly [GGA codon] found in our study). This inconsistency is either caused by an experimental artifact (mistake in the Edman degradation determination or at least two Taq-induced mutations in the dsrAB clone of Thermodesulfobacterium commune) or by the presence of more than one type of dsrAB genes in this organism. Differences between the deduced N-terminal sequence and that determined by N-terminal polypeptide sequencing were also reported for the DsrB protein of Desulfovibrio desulfuricans (25).
DSR homologs.
Additional homologs to the investigated dsrAB genes may exist in some of the analyzed strains. This is not the case for Desulfitobacterium hafniense and Archaeoglobus fulgidus, since no additional dsrAB homologs are present in their complete genome sequences. Under the assumption that the PCR primers applied would amplify all putative dsr copies, we have indirect evidence that the Desulfotomaculum species, D. aeronauticum, D. putei, D. geothermicum, D. kuznetsovii, and D. thermobenzoicum do not contain multiple dsrAB copies which differ in sequence since the respective dsrAB PCR amplificates could be sequenced directly. For the other analyzed SRPs, knowledge of the copy number of dsrAB-like genes must await an extensive Southern hybridization or complete genome sequence analysis which was beyond the scope of this study.
DSR phylogeny and lateral transfer.
The core of our study was a direct comparison between 16S rRNA and DsrAB trees of the respective SRPs (Fig. 1). In this analysis it is an explicit assumption that the 16S rRNA phylogeny reflects the organismal phylogeny (36), that is, that these highly conserved genes have undergone no lateral transfer in the organisms studied. Based on this supposition, seven Desulfotomaculum species, two Thermodesulfobacterium species, and Desulfobacula toluolica possess nonorthologous dsrAB genes as demonstrated by major inconsistencies between the DsrAB and 16S rRNA trees. These inconsistencies most likely reflect lateral transfer of dsrAB genes rather than the occurrence of dsrAB paralogs which diverged after an initial dsr operon duplication since all nonorthologous dsrAB genes are phylogenetically affiliated with the (presumably orthologous) dsrAB genes of the δ-Proteobacteria (Fig. 1). Furthermore, organisms distantly related by 16S rRNA sequence relationship, such as Desulfobacula toluolica and several Desulfotomaculum species, contain similar nonorthologous dsrAB genes. This close relatedness of dsrAB genes between species belonging to different bacterial divisions is unlikely to be the product of convergent evolution and can more reasonably be explained by multiple lateral acquisitions from a common donor lineage within the δ-Proteobacteria. Consistent with this inference, all putative xenologous dsrAB sequences have insertions typical for the δ-Proteobacteria (Fig. 3).
Five independent LGT events (the red circles in Fig. 1) of dsrAB genes have been postulated to explain the observed discrepancies between the 16S rDNA and DsrAB topologies. It should be noted that for SRPs (i) which do not have close phylogenetic relatives in the current dsrAB data set or (ii) whose positions in the deduced phylogenetic trees vary dependent upon the treeing method used, our analysis cannot rule out that their characterized dsrAB sequences are xenologs. Within the δ-Proteobacteria these limitations apply to Desulfoarculus baarsii and Desulfomonile tiedjei. Furthermore, the characterized DsrAB sequences of Archaeoglobus and Thermodesulfovibrio species and the “authentic” Desulfotomaculum and Desulfitobacterium species possibly could originate from a progenitor of the δ-Proteobacteria or from other as-yet-unidentified SRPs. In fact, it seems likely that the genus Archaeoglobus inherited dsrAB genes from a bacterial donor (i) because the evolutionary distance between Archaeoglobus species and the bacterial sulfate reducers is much shorter in the DsrAB tree than in the 16S rRNA tree and (ii) because the sulfate-reducing phenotype is currently restricted to the genus Archaeoglobus within the archaeal domain. Further support for a lateral transfer of the dsrAB genes to the Archaeoglobales was obtained by a phylogenetic analysis on an alignment of the alpha- against the beta-subunit amino acid sequences. Such analysis can be used to root the Dsr subunit trees (6, 13) (Fig. 2), since the subunits are paralogs arising from an ancestral dsr gene duplication (3). Independent of the treeing method used, the root was consistently indicated between the DsrAB of the Thermodesulfovibrio species and the DsrABs of all other analyzed SRPs, including the Archaeoglobales. This is inconsistent with the 16S rRNA phylogeny and points to a bacterial origin of the Archaeoglobales dsrAB genes ( in the DsrAB tree; Fig. 1). However, the results from the paralogous rooting should not be overemphasized since the alignment of the Dsr subunits against each other (i) is relatively short (173 amino acid positions) and (ii) contains several regions which cannot unambiguously be aligned (caused by the relatively low sequence similarities of the subunits to each other). Furthermore, no evidence for lateral transfer of the Archaeoglobus fulgidus dsrAB genes was indicated by atypical sequence characteristic analysis (27; J. Lawrence, unpublished data), suggesting that, if the genes are xenologs, they have completely ameliorated toward their host genome and were the result of an ancient LGT event.
in the DsrAB tree; Fig. 1). However, the results from the paralogous rooting should not be overemphasized since the alignment of the Dsr subunits against each other (i) is relatively short (173 amino acid positions) and (ii) contains several regions which cannot unambiguously be aligned (caused by the relatively low sequence similarities of the subunits to each other). Furthermore, no evidence for lateral transfer of the Archaeoglobus fulgidus dsrAB genes was indicated by atypical sequence characteristic analysis (27; J. Lawrence, unpublished data), suggesting that, if the genes are xenologs, they have completely ameliorated toward their host genome and were the result of an ancient LGT event.
DSR donor lineages.
The dsrAB gene donors were members of at least two distinct evolutionary lineages within the δ-Proteobacteria ( to
to  and b○ in the DsrAB tree; Fig. 1). Donor lineage a contributed dsrAB genes to two phylogenetically remote groups of bacteria, Desulfobacula toluolica (δ-Proteobacteria) and several Desulfotomaculum species (low G+C gram positives) (
and b○ in the DsrAB tree; Fig. 1). Donor lineage a contributed dsrAB genes to two phylogenetically remote groups of bacteria, Desulfobacula toluolica (δ-Proteobacteria) and several Desulfotomaculum species (low G+C gram positives) (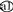 to
to  in the 16S rDNA tree; Fig. 1), suggesting that this lineage is particularly adept at donating dsrAB and possibly other genes. The specific identities of the donor lineages is unknown based on the current data, since no orthologous dsrAB genes were identified within the putative xenolog groups. It is however striking that Desulfobacula toluolica and all but two Desulfotomaculum species which received the xenologous dsrAB are oxidizing their characteristic substrates completely to CO2 while the “authentic” Desulfotomaculum and Desulfitobacterium species are exclusively incomplete oxidizers. One possible explanation for this feature is that the dsrAB donor was a complete oxidizer which bestowed this metabolic capability to the Desulfotomaculum and Desulfobacula species, which subsequently was lost in Desulfotomaculum thermosapovorans and Desulfotomaculum thermocisternum (Table 1). Furthermore, most of the recipients of xenologous dsrAB are thermophilic (Table 1) which could indicate a thermophilic lifestyle of the donor species.
in the 16S rDNA tree; Fig. 1), suggesting that this lineage is particularly adept at donating dsrAB and possibly other genes. The specific identities of the donor lineages is unknown based on the current data, since no orthologous dsrAB genes were identified within the putative xenolog groups. It is however striking that Desulfobacula toluolica and all but two Desulfotomaculum species which received the xenologous dsrAB are oxidizing their characteristic substrates completely to CO2 while the “authentic” Desulfotomaculum and Desulfitobacterium species are exclusively incomplete oxidizers. One possible explanation for this feature is that the dsrAB donor was a complete oxidizer which bestowed this metabolic capability to the Desulfotomaculum and Desulfobacula species, which subsequently was lost in Desulfotomaculum thermosapovorans and Desulfotomaculum thermocisternum (Table 1). Furthermore, most of the recipients of xenologous dsrAB are thermophilic (Table 1) which could indicate a thermophilic lifestyle of the donor species.
DSR recipient lineages.
Desulfobacula toluolica is the recipient of the most recent putative LGT event thus far identified (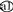 ; Fig. 1) since its close relatives, Desulfobacter latus and Desulfobacter vibrioformis, contain orthologous dsrAB genes. This is also supported by no identifiable amelioration of the third codon position G+C content of the xenologous dsr genes toward the mean G+C content of the host D. toluolica genome (Table 1). The evolution of the genus Desulfotomaculum was also affected by LGT events of dsrAB genes. The number of LGT events within this genus is difficult to predict since the subclustering of its members is not always well supported in the 16S rDNA tree (Fig. 1). On the basis of the presented 16S rDNA tree, it is most parsimonious to postulate at least three LGT events within this genus (
; Fig. 1) since its close relatives, Desulfobacter latus and Desulfobacter vibrioformis, contain orthologous dsrAB genes. This is also supported by no identifiable amelioration of the third codon position G+C content of the xenologous dsr genes toward the mean G+C content of the host D. toluolica genome (Table 1). The evolution of the genus Desulfotomaculum was also affected by LGT events of dsrAB genes. The number of LGT events within this genus is difficult to predict since the subclustering of its members is not always well supported in the 16S rDNA tree (Fig. 1). On the basis of the presented 16S rDNA tree, it is most parsimonious to postulate at least three LGT events within this genus (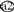 to
to  in the 16S rDNA tree of Fig. 1). Alternatively, one could hypothesize that a single lateral dsrAB gene transfer event occurred to the common ancestor of the genera Desulfotomaculum, Desulfosporosinus, and Desulfitobacterium (which did not displace the orthologous dsr genes), followed by a subsequent xenolog gene loss on at least two independent occasions from the ancestors of the “authentic” Desulfotomaculum and Desulfosporosinus-Desulfitobacterium species, respectively.
in the 16S rDNA tree of Fig. 1). Alternatively, one could hypothesize that a single lateral dsrAB gene transfer event occurred to the common ancestor of the genera Desulfotomaculum, Desulfosporosinus, and Desulfitobacterium (which did not displace the orthologous dsr genes), followed by a subsequent xenolog gene loss on at least two independent occasions from the ancestors of the “authentic” Desulfotomaculum and Desulfosporosinus-Desulfitobacterium species, respectively.
In conclusion, this study demonstrates that the genes encoding the dissimilatory sulfite reductase are subject to frequent LGT events within and across bacterial divisions and possibly even between the bacterial and archaeal domains. This finding was unexpected since the dissimilatory sulfite reductase represents an essential enzyme for anaerobic sulfate and sulfite respiration which acts (at least for the SRPs) in concert with other enzymes. One possible explanation for the observed widespread lateral distribution of dissimilatory sulfite reductases could be that the genes encoding this enzyme are part of a mobilizable metabolic island similar to the genes required for anaerobic nitrate respiration of Thermus thermophilus (30). More generally, our findings add to the accumulating evidence that lateral gene transfer is a potent mechanism shaping the composition of prokaryotic genomes (see, for example, references 5, 7, 19, 20, 21, 26, and 31). On the other hand, our data also demonstrate that the DsrAB phylogeny of most SRPs analyzed is still consistent with the 16S rRNA phylogeny. This observation and the paralogous rooting of the Dsr tree still support an early and thermophilic origin of sulfate respiration.
The use of functional genes, including dsrAB as molecular markers for defined physiological groups of bacteria has become increasingly popular in investigations of complex microbial communities (see, for example, references 1, 22, 29, and 37). If the functional genes are exploited for phylogenetic analysis of the respective bacteria, lateral gene transfer can complicate the interpretation. This has previously been demonstrated for the nifH gene encoding the nitrogenase reductase of nitrogen-fixing bacteria (11) and was shown here to also hold true for the dissimilatory sulfite reductase genes. Therefore, the phylogenetic DsrAB framework established in our study provides an essential basis for better interpreting environmental diversity surveys of SRPs based on comparative DsrAB sequence analysis.
ACKNOWLEDGMENTS
M.K. and H.A. were supported by a grant from the Deutsche Forschungsgemeinschaft to M.W. in the framework of the project “Degradation of marine pollutants by cyanobacterial mats—an interdisciplinary approach.” M.F. was supported by the Max-Planck-Gesellschaft. A.J.R. was funded by NSERC grant 2277085–00. D.A.S. and S.F. were supported by NSF grant DEB-9714303. P.H. is funded by the Cooperative Research Centre for Waste Management and Pollution Control, Ltd., a center established and supported under the Australian Government's Cooperative Research Centres Program.
We thank Bianca Wagner (Marburg, Germany) for excellent technical assistance. We also thank Kathrin Riedel for helpful discussions.
REFERENCES
- 1.Cottrell M T, Cary S C. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl Environ Microbiol. 1999;65:1127–1132. doi: 10.1128/aem.65.3.1127-1132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crane B R, Siegel L M, Getzoff E D. Sulfite reductase structure at 1.6 A: evolution and catalysis for reduction of inorganic anions. Science. 1995;270:59–67. doi: 10.1126/science.270.5233.59. [DOI] [PubMed] [Google Scholar]
- 3.Dahl C, Kredich N M, Deutzmann R, Trüper H G. Dissimilatory sulfite reductase form Archaeoglobus fulgidus: physico-chemical properties of the enzyme and cloning, sequencing and analysis of the reductase gene. J Gen Microbiol. 1993;139:1817–1828. doi: 10.1099/00221287-139-8-1817. [DOI] [PubMed] [Google Scholar]
- 4.Fauque G, Lino A R, Czechowski M, Kang L, DerVartanian D V, Moura J J, LeGall J, Moura I. Purification and characterization of bisulfite reductase (desulfofuscidin) from Desulfovibrio thermophilus and its complexes with exogenous ligands. Biochim Biophys Acta. 1990;1040:112–118. doi: 10.1016/0167-4838(90)90154-8. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gogarten J P, Kibak H, Dittrich P, Taiz L, Bowman E J, Bowman B, Manolson M, Poole R, Date T, Oshima T, Konishi J, Denda K, Yoshida M. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman B S, Kranz R G. Evolution and horizontal transfer of an entire biosynthetic pathway for cytochrome c biogenesis: Helicobacter, Deinococcus, Archae, and more. Mol Microbiol. 1998;27:871–873. doi: 10.1046/j.1365-2958.1998.00708.x. [DOI] [PubMed] [Google Scholar]
- 8.Hatchikian E C, Zeikus J G. Characterization of a new type of dissimilatory sulfite reductase present in Thermodesulfobacterium commune. J Bacteriol. 1983;153:1211–1220. doi: 10.1128/jb.153.3.1211-1220.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hipp W M, Pott A S, Thum-Schmitz N, Faath I, Dahl C, Trüper H G. Towards the phylogeny of APS reductases and siroheme sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology. 1997;143:2891–2902. doi: 10.1099/00221287-143-9-2891. [DOI] [PubMed] [Google Scholar]
- 11.Hurek T, Egener T, Reinhold-Hurek B. Divergence in nitrogenases of Azoarcus spp., Proteobacteria of the beta subclass. J Bacteriol. 1997;179:4172–4178. doi: 10.1128/jb.179.13.4172-4178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imhoff J F, Suling J, Petri R. Phylogenetic relationships among the Chromatiaceae, their taxonomic reclassification and description of the new genera Allochromatium, Halochromatium, Isochromatium, Marichromatium, Thiococcus, Thiohalocapsa, and Thermochromatium. Int J Syst Bacteriol. 1998;48(Pt. 4):1129–1143. doi: 10.1099/00207713-48-4-1129. [DOI] [PubMed] [Google Scholar]
- 13.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juretschko S, Timmermann G, Schmid M, Schleifer K H, Pommerening-Roser A, Koops H P, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karkhoff-Schweizer R R, Huber D P W, Voordouw G. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl Environ Microbiol. 1995;61:290–296. doi: 10.1128/aem.61.1.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 17.Larsen O, Lien T, Birkeland N-K. Characterization of the desulforubidin operons from Desulfbacter vibrioformis and Desulfobulbus rhabdoformis. FEMS Microbiol Lett. 2000;186:41–46. doi: 10.1111/j.1574-6968.2000.tb09079.x. [DOI] [PubMed] [Google Scholar]
- 18.Larsen O, Lien T, Birkeland N-K. Dissimilatory sulfite reductase from Archaeglobus profundus and Desulfotomaculum thermocisternum: phylogenetic and structural implications from gene sequences. Extremophiles. 1999;3:63–70. doi: 10.1007/s007920050100. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence J G. Gene transfer, speciation, and the evolution of bacterial genomes. Curr Opin Microbiol. 1999;2:519–523. doi: 10.1016/s1369-5274(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 20.Lawrence J G, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 21.Logsdon J M, Faguy D M. Thermotoga heats up lateral gene transfer. Curr Biol. 1999;9:R747–R751. doi: 10.1016/s0960-9822(99)80474-6. [DOI] [PubMed] [Google Scholar]
- 22.Minz D, Flax J L, Green S J, Muyzer G, Cohen Y, Wagner M, Rittmann B E, Stahl D A. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory (bi-)sulfite reductase genes. Appl Environ Microbiol. 1999;65:4666–4671. doi: 10.1128/aem.65.10.4666-4671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molitor M, Dahl C, Molitor I, Schäfer U, Speich N, Huber R, Deutzmann R, Trüper H G. A dissimilatory sirohaem-sulfite-reductase-type protein from the hyperthermophilic archaeon Pyrobaculum islandicum. Microbiology. 1998;144:529–541. doi: 10.1099/00221287-144-2-529. [DOI] [PubMed] [Google Scholar]
- 24.More M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morse R, Gibson G R, Collins M D. Secondary structure analysis of the dissimilatory sulphite reductase in Desulfovibrio desulfuricans. Lett Appl Microbiol. 2000;30:375–378. doi: 10.1046/j.1472-765x.2000.00659.x. [DOI] [PubMed] [Google Scholar]
- 26.Nelson K E, Clayton R A, Gill S R, Gwinn M L, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 27.Ochman H, Elwyn S, Moran N A. Calibrating bacterial evolution. Proc Natl Acad Sci USA. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 29.Purkhold U, Pommerening-Roser A, Juretschko S, Schmid M C, Koops H P, Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez-Arcos S, Fernandez-Herrero L A, Marin I, Bergenguer J. Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J Bacteriol. 1998;180:3137–3143. doi: 10.1128/jb.180.12.3137-3143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sassanfar M, Kranz J E, Gallant P, Schimmel P, Shiba K. A eubacterial Mycobacterium tuberculosis tRNA synthetase is eukaryote-like and resistant to a eubacterial-specific antisynthetase drug. Biochemistry. 1996;35:9995–10003. doi: 10.1021/bi9603027. [DOI] [PubMed] [Google Scholar]
- 32.Schedel M, Trüper H G. Purification of Thiobacillus denitrificans siroheme sulfite reductase and investigation of some molecular and catalytic properties. Biochim Biophys Acta. 1979;568:454–466. doi: 10.1016/0005-2744(79)90314-0. [DOI] [PubMed] [Google Scholar]
- 33.Stackebrandt E, Sproer C, Rainey F A, Burghardt J, Päuker O, Hippe H. Phylogenetic analysis of the genus Desulfotomaculum: evidence for misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int J Syst Evol Bacteriol. 1997;47:1134–1139. doi: 10.1099/00207713-47-4-1134. [DOI] [PubMed] [Google Scholar]
- 34.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Evol Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 35.Wagner M, Roger A J, Flax J L, Brusseau G A, Stahl D A. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. Appl Environ Microbiol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zani S, Mellon M T, Collier J L, Zehr J P. Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl Environ Microbiol. 2000;66:3119–3124. doi: 10.1128/aem.66.7.3119-3124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]