Abstract
The type III secretion system encoded by Salmonella pathogenicity island 2 (SPI2) is required for systemic infections and intracellular accumulation of Salmonella enterica. This system is induced by intracellular Salmonella and subsequently transfers effector proteins into the host cell. Growth conditions either inducing expression of the type III secretion system or the secretion of substrate proteins were defined. Here we report the identification of a set of substrate proteins consisting of SseB, SseC, and SseD that are secreted by the SPI2 system in vitro. Secretion was observed if bacterial cells were exposed to acidic pH after growth in minimal medium with limitation of Mg2+ or phosphate. SseB, -C, and -D were isolated in a fraction detached from the bacterial cell surface by mechanical shearing, indicating that these proteins are predominantly assembled into complexes on the bacterial cell surface. The three proteins were required for the translocation of SPI2 effector proteins SspH1 and SspH2 into infected host cells. Thus, SseB, SseC, and SseD function as the translocon for effector proteins by intracellular Salmonella.
The translocation of virulence proteins into the cells of an infected host organism has a central role for the pathogenesis of infections by many gram-negative bacteria. This specific virulence trait is linked to type III secretion systems (TTSS), complex molecular machines that require the function of at least 20 genes. TTSS are involved in very different aspects of pathogenesis, such as invasion of host cells and paralysis of phagocytes as well as establishment of symbiotic relationships (for a review, see reference 14). In most cases, sets of substrate proteins are secreted by the TTSS. However, only a subset of substrate proteins act as translocated effector proteins. Another subset of substrate proteins is required for the translocation of effector proteins and thus acts as a translocator, e.g., by forming pilus-like structures (17, 27) or by pore formation in the host cell membrane (31, 32) through which effector proteins are translocated.
Salmonella enterica is a highly successful pathogen that causes diseases ranging from mild gastrointestinal infections to life-threatening systemic infections such as typhoid fever. A large number of virulence genes is required for S. enterica to be successful as a pathogen in both forms of disease and in a large number of different hosts. S. enterica possesses two TTSS with different functions in pathogenesis. Both TTSS are encoded by large chromosomal gene clusters referred to as Salmonella pathogenicity islands or SPI. SPI1 encodes a TTSS that is required for the interaction of extracellular Salmonella with cells of the gastrointestinal mucosa (for a review, see reference 7). SPI1 effector proteins have multiple functions in triggering invasion of Salmonella into nonphagocytic cells and induction of apoptosis and gastrointestinal symptoms.
The second TTSS encoded by SPI2 has an entirely different role in Salmonella pathogenesis since its function is mainly restricted to intracellular pathogenesis (for a review, see reference 10). Mutant strains defective in SPI2 are highly attenuated in virulence and have reduced rates of intracellular survival and replication (2, 11, 26).
Several substrate proteins of the TTSS of SPI2 in S. enterica serovar Typhimurium have been identified so far. SseB, a protein with sequence similarity to EspA of enteropathogenic Escherichia coli (EPEC), is secreted by S. enterica serovar Typhimurium after induction of the SPI2 and exposure to acidic pH (1). Substrate proteins SseC and SseD have sequence similarity to EPEC proteins EspD and EspA, respectively, and recently secretion of SseC and SseD has been shown (15). EspA, -B, and -D are substrate proteins of the EPEC TTSS and are required for translocation of the EPEC effector Tir. A number of proteins appear to be translocated by the SPI2 TTSS into infected eukaryotic cells. Several of these, including the leucine repeat proteins SspH1 and SspH2 (24), share a conserved N-terminal domain which may function to target these proteins to the secretion apparatus (23).
Here we report the direct identification of a set of secreted substrate proteins of the TTSS of SPI2. We analyzed the functional requirements for the secretion of these proteins and their role in translocation of SPI2 effector proteins into host cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
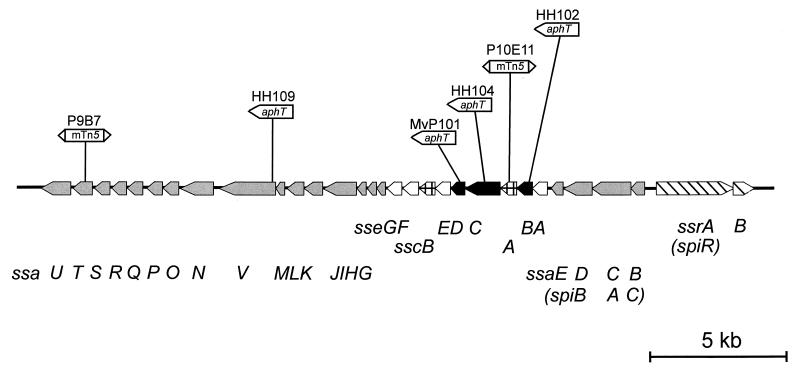
The strains used in this study are listed in Table 1 and the position of mutations in various SPI2 genes are indicated in Fig. 1. S. enterica serovar Typhimurium and E. coli strains were routinely cultivated in Luria broth (LB) containing antibiotics (carbenicillin at 50 μg/ml, kanamycin at 50 μg/ml, and chloramphenicol at 10 to 50 μg/ml) if required to maintain plasmids. The composition of minimal media has been described before (3). Briefly, low-Mg2+ minimal medium containing N-salts [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 100 mM bis-Tris–HCl], 30 μM MgCl2, 38 mM glycerol, and 0.1% Casamino Acids was adjusted to pH 7.0 or 5.0 as indicated. Minimal media containing high (PCN) or low (PCN-P) concentrations of phosphate were used as described before (3). For PCN-P medium at pH 5.8, 80 mM MOPS (morpholinepropanesulfonic acid) was replaced by 80 mM MES (morpholineethanesulfonic acid). Minimal media were prepared with double-distilled water.
TABLE 1.
Bacterial strains and plasmids used in this study
| Designation | Relevant characteristics | Reference or source |
|---|---|---|
| S. enterica serovar Typhimurium strains | ||
| NCTC 12023 | Wild type, identical to ATCC 14028 | ATCCa |
| P10E11 | sscA::mTn5 | 29 |
| P9B7 | ssaT::mTn5 | 29 |
| HH102 | sseB::aphT, Kmr, nonpolar mutation | 11 |
| HH103 | HH102 containing psseB | 11 |
| HH104 | sseC::aphT, Kmr, nonpolar mutation | 11 |
| HH109 | ssaV::aphT, Kmr, nonpolar mutation | 4 |
| MvP101 | sseD::aphT, Kmr, nonpolar mutation | 20 |
| MvP131 | ssaB::luc lacZ | 3 |
| E. coli strain | ||
| M15 | pREP4 | Qiagen |
| Plasmids | ||
| pEM25 | pMJH expressing SspH1-CyaA | 24 |
| pEM30 | pMJH expressing SspH2-CyaA | 24 |
| pQE | His tag expression vectors, Ampr | Qiagen |
| pJD14 | Codons 180 to 484 of sseC in pQE40 | This study |
| pJD15 | Codons 2 to 195 of sseD in pQE30 | This study |
| psseB | sseB in pACYC184, Cmr | 11 |
ATCC, American Type Culture Collection.
FIG. 1.
Genetic organization of SPI2 and position of mutations used in this study. sse genes encoding secreted substrate proteins of the TTSS of SPI2 characterized in this work are indicated by black arrows. Genes encoding the TTSS (ssa), the regulatory system (ssr), and chaperones of substrate proteins (ssc) are represented by shaded, hatched, and cross-hatched symbols, respectively. Open arrows indicate further genes for putative secreted substrate proteins. The alternative designations of SPI2 genes used by Ochman et al. (26) are indicated in parentheses.
Generation of antisera against recombinant SPI2 proteins.
Generation of antisera against recombinant SseB (rSseB) has been described (1) and recombinant SPI2 proteins were expressed and purified essentially as outlined before (3). Briefly, primers were used to amplify sseD and the 3′ portion of sseC and to introduce restriction sites as indicated in Table 2. PCR products were then ligated to vectors of the pQE3x and pQE4x series to generate fusions with an N-terminal tag of six histidines. Fusion proteins were expressed in E. coli M15(pREP) (Qiagen, Hilden, Germany) and purified under denaturing conditions on Hi-trap chelating columns (Pharmacia, Freiburg, Germany) according to the manufacturers' instructions. Recombinant proteins were further purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on preparative gels and electroelution of the relevant protein bands. For the generation of antisera, purified recombinant proteins were emulsified with complete and incomplete Freund's adjuvant for initial and booster immunizations, respectively. Antisera were raised in rabbits following standard procedures (8) and in accordance with the national and institutional guidelines for animal handling.
TABLE 2.
Oligonucleotides used in this study
| Designation | Sequencea |
|---|---|
| sseC-For2-16 | 5′-CTCGGATCCGGAAATCCCGCAGA-3′ |
| sseC-Rev2-2 | 5′-GTACTGCAGATTAAGCGCGATAGC-3′ |
| sseD-For3 | 5′-ATGAGATCTGAAGCGAGTAACGTAGCAC-3′ |
| sseD-Rev2-8 | 5′-TGAAAGCTTACCTCGTTAATGCCC-3′ |
Restriction sites introduced for cloning of PCR products are underlined.
Immunoglobulin G (IgG) of the antiserum against SseB was purified using a protein G-Sepharose column (Pharmacia) according to the manufacturer's instructions. For labeling, the carboxysuccinimidyl ester derivative of fluorescein (NHS-fluorescein; Pierce) was coupled to IgG as described by Hermanson (12). Unbound NHS-fluorescein was removed by gel filtration using a PD-10 column (Pharmacia). The resulting IgG fraction had a fluorescence-to-protein ratio of 0.95.
Preparation of secreted proteins.
For preparation of secreted proteins, bacteria were precultured in LB for 8 h at 37°C. Bacteria were washed once in minimal medium, and equal amounts of bacteria adjusted by optical density at 600 nm (OD600) were used to inoculate cultures in minimal medium at neutral or acidic pH. Cultures (400 ml) in 1-liter glass flasks without baffles were incubated overnight with agitation of 200 rpm at 37°C. Bacteria were pelleted by centrifugation at 6,000 × g for 15 min and resuspended in 20 ml of phosphate-buffered saline (PBS). This suspension was agitated at maximum speed in 50-ml centrifuge tubes (Falcon) on a Vortex mixer (Vortex Genie 2; Scientific Industries) for 60 s. Bacterial cells were pelleted by centrifugation at 10,000 × g for 10 min and the supernatant was passed through a 0.2-μm-pore-size filter to remove residual bacteria. Protein in the supernatant fractions (detached fraction) was recovered by precipitation with trichloroacetic acid (10% [wt/vol] final concentration) on ice for 1 h and centrifugation for 1 h at 10,000 × g. The pellet was washed twice with 15 ml of acetone and recovered by centrifugation at 10,000 × g for 30 min. The final pellet was air dried and resuspended in SDS-PAGE sample buffer (50 mM Tris-HCl [pH 6.8], 4% SDS, 2% β-mercaptoethanol, 12.5% glycerol, and 0.01% bromophenol blue). The final concentration of protein in the detached fraction was adjusted by the cell density of the original culture (OD600 of 100 ml of culture × 100 = x μl of sample buffer).
For the isolation of secreted protein from culture supernatant, bacterial cultures were grown overnight in 40 ml of low-Mg2+ minimal medium, pH 5.0, in flasks without baffles. Bacterial cells were removed by centrifugation (5 min at 4,000 × g) and filtration of the supernatant through a 0.45-μm-pore-size filter. Protein from cell-free culture supernatants was precipitated by addition of trichloroacetic acid (10% [wt/vol] final concentration), recovered by centrifugation for 1 h at 10,000 × g, and washed twice with acetone.
β-Galactosidase activities in various fractions were determined according to a standard procedure (25).
Protein analysis.
Cell-associated and secreted protein profiles of various S. enterica serovar Typhimurium strains were routinely analyzed by SDS-PAGE on Tricine gels (12%) or Tris gels (12%) according to the methods of Schägger and von Jagow (28) and Laemmli (19), respectively. Protein bands were visualized by Coomassie brilliant blue staining or silver staining according to the method of Heukeshoven and Dernick (13). For analysis by Western blotting, proteins were transferred onto nitrocellulose membranes (BA85; Schleicher and Schuell) using the discontinuous semidry blotting procedure (18). Western blots were processed using ECL detection (Amersham-Pharmacia).
For microsequencing, proteins were transferred onto polyvinylidene difluoride membranes (Millipore) and stained with Coomassie brilliant blue. Microsequencing by Edman degradation was performed on an Applied Biosystems A473 sequencer according to the instructions of the manufacturer.
Translocation assay.
Determination of SspH1 and SspH2 translocation by the SPI2 TTSS was performed as previously described (24). Briefly, RAW264.7 murine macrophages were infected with bacteria harboring a plasmid-borne SspH1-CyaA or SspH2-CyaA fusion. For infection, bacteria were grown to stationary phase and were added to macrophages at a multiplicity of infection of 10 for 1 h. Subsequently, medium containing gentamicin was added to kill extracellular bacteria and incubation was performed for seven additional hours to allow for SPI2-dependent translocation by internalized bacteria. Infected macrophages were then lysed in 0.1 N HCl and boiled for 5 min. Cellular cyclic AMP (cAMP) levels were determined with the Direct cAMP Correlate-EIA Kit (Assay Designs, Ann Arbor, Mich.). The cellular cAMP content was then normalized for protein concentration as determined by the Bradford assay and is presented as picomoles of cAMP per microgram of protein. All experiments were performed in triplicate at least twice.
RESULTS
Identification of substrate proteins of the TTSS of SPI2.
Previous work established that SseB, a substrate protein of the TTSS encoded by SPI2, is secreted when bacteria are exposed to media at pH 5.0 or below (1). The majority of SseB, however, was located on the cell envelope rather than being secreted into the media. SseB was recovered from the cell envelope by extraction with organic solvents such as hexadecane. We observed that SseB was also detached from bacterial cells by mechanical shearing. Based on these observations, we set out to identify further substrate proteins secreted by the TTSS of SPI2. Bacteria were grown under conditions inducing the secretion of SPI2 proteins, concentrated, and subjected to vigorous mixing. Protein detached from the bacterial surface by this procedure was concentrated and analyzed (Fig. 2). This fraction of secreted proteins will be referred to as the detached fraction.
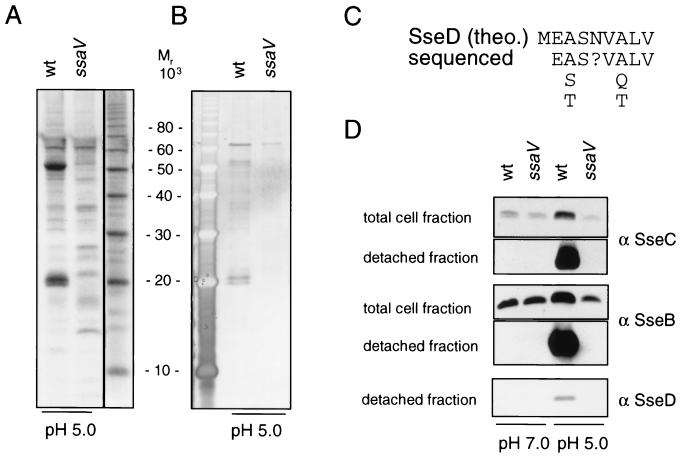
FIG. 2.
Identification of substrate proteins of the TTSS of SPI2. The S. enterica serovar Typhimurium wild-type (wt) and ssaV mutant (HH109 ssaV::aphT) strains were grown overnight in low-Mg2+ minimal medium at pH 7.0 or 5.0. Secreted protein from equal amounts of bacterial cells was recovered by vigorous mixing and concentrated by acetone precipitation as described in Materials and Methods. Protein extracts were subjected to SDS-PAGE on 12% Tricine gels. Subsequently, proteins were visualized by staining with Coomassie brilliant blue (A) or by silver staining (B). The N-terminal animo acid sequence of the protein of about 20 kDa was determined and compared to the predicted gene product of sseD [SseD (theo.)] (C). For Western blot analyses, lysates of equal amounts of bacteria grown in low-Mg2+ minimal medium at pH 7.0 or 5.0 (total cell fraction) or protein recovered by mechanical shearing from equal amounts of bacteria (detached fraction) was separated by SDS-PAGE and transferred onto nitrocellulose membranes. Proteins were detected by Western blotting with antisera raised against rSseC (α SseC), rSseB, or rSseD (D).
To determine whether this procedure affects the integrity of the bacterial cell, we used reporter strain MvP131 (3), which expresses the reporters luciferase and β-galactosidase under the growth conditions used here. Compared to the β-galactosidase activity of the total culture (6,003 Miller units), only very low activities were detected in the culture supernatant (2.1 Miller units) and in the detached fraction (0.3 Miller unit). These data indicate that the mechanical agitation did not result in the release of cytoplasmic protein due to cell lysis.
Analysis by SDS-PAGE revealed that a significantly smaller number of proteins was recovered by mechanical shearing than by extraction with hexadecane (data not shown). The protein pattern obtained by mechanical shearing was compared for wild-type S. enterica serovar Typhimurium and a mutant strain defective in SsaV, a structural component of the SPI2 TTSS (Fig. 2A and B). When bacteria were grown at pH 5.0, proteins with apparent molecular masses of about 20, 21, and 52 kDa were secreted by the wild-type strain but were absent in the detached fraction of the ssaV mutant strain (Fig. 2A and B). Similar protein patterns were observed with strains harboring mutations in ssaC or ssaT, further structural components of the TTSS of SPI2 (data not shown). Therefore, secretion of these three proteins is dependent on the function of the TTSS of SPI2. Another protein of about 63 kDa was present in the detached fraction of the wild-type and ssaV mutant strains. Thus, it is unlikely that this protein is a substrate of the TTSS of SPI2. The N-terminal sequence was determined for the protein of about 20 kDa (Fig. 2C). A BLAST search revealed that this sequence is similar to the predicted N-terminal sequence of SseD. Furthermore, antibodies raised against recombinant SPI2 proteins rSseB, rSseC, and rSseD reacted with the bands at 21, 52, and 20 kDa, respectively (Fig. 2D). Signals for SseB, SseC, and SseD were observed in the detached fraction of the wild-type strain grown at pH 5.0 but not in the detached fraction of bacterial cultures grown at pH 7.0 or in the detached fractions of the ssaV mutant strain (Fig. 2D). A mutation in invG, a structural component of the TTSS encoded by SPI1, had no effect on the secretion of these proteins (data not shown). Therefore, our analyses indicate that SseB, SseC, and SseD are the major secreted proteins of the SPI2 system. We also analyzed proteins secreted into the growth medium under these culture conditions. Western blot analysis indicated low amounts of SseB (data not shown), SseC, and SseD (see Fig. 5) in culture supernatants.
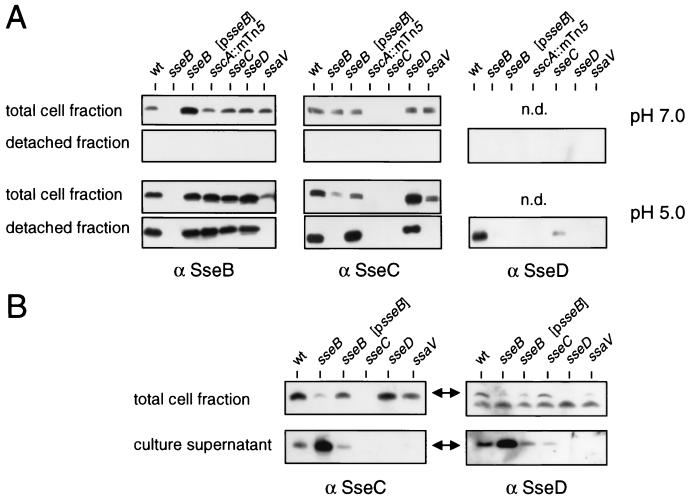
FIG. 5.
Functional requirements for secretion by the TTSS of SPI2. (A) The S. enterica serovar Typhimurium wild type (wt) and various strains harboring mutations in the ssc, sse, or ssa gene were grown in low-Mg2+ minimal medium at pH 7.0 or pH 5.0 to induce expression or expression and secretion, respectively, of SPI2 substrate proteins. Lysates of equal amounts of bacteria were adjusted by OD600 (total cell fraction), or protein recovered from equal amounts of bacteria (detached fraction) was analyzed as described in the legend for Fig. 2. n.d., not done. (B) Role of SseB for secretion of SseC and SseD. Various strains were grown in low-Mg2+ minimal medium at pH 5.0. Protein from culture supernatants was prepared as described in Materials and Methods. Equal amounts of bacterial cells (total cell fraction) and protein prepared from equal amounts of culture supernatants were subjected to Western blot analysis with antibodies raised against rSseC (α SseC) or rSseD.
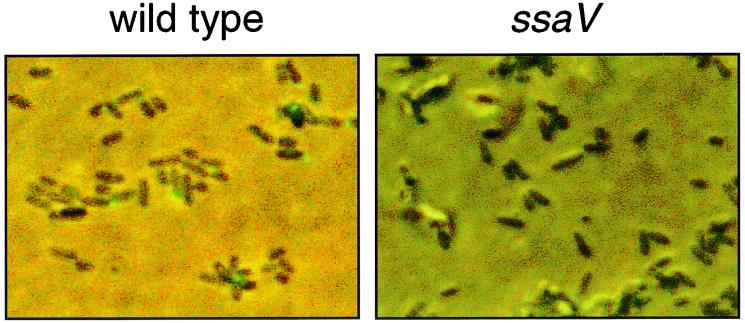
We next used fluorescein isothiocyanate (FITC)-labeled antibodies against SseB to analyze the location of SseB on bacterial cells (Fig. 3). Microscopic analysis revealed that SseB-containing structures were present on the surface on wild-type S. enterica serovar Typhimurium after growth in minimal medium at acidic pH. No staining was detected on a mutant defective in secretion (Fig. 3) or on wild-type S. enterica serovar Typhimurium after growth at neutral pH (data not shown). We observed that SseB-containing structures were not distributed equally on the bacterial surface but were concentrated at one pole of the cell.
FIG. 3.
Detection of surface structures containing SseB. The S. enterica serovar Typhimurium wild-type and ssaV mutant strains were grown overnight in low-phosphate minimal medium at pH 5.8. Bacteria were fixed with 1.0% formaldehyde in PBS for 10 min and washed three times in PBS. Fixed bacteria were incubated with FITC-labeled antiserum against SseB for 1 h, and unbound antibody was removed by washing in PBS. The suspension was then spotted on poly-l-lysine-coated microscope slides (Sigma) and samples were analyzed by epifluorescence microscopy. Phase-contrast images and images of the FITC channel were overlaid using Adobe Photoshop.
Influence of growth conditions on the secretion of SPI2 substrate proteins.
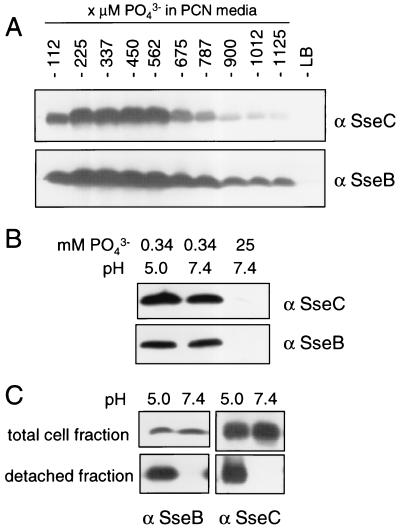
We previously demonstrated that growth in low-Mg2+ minimal medium as well as in low-phosphate minimal medium resulted in the induction of genes encoding the TTSS of SPI2 (3). Levels of substrate proteins SseB and SseC were compared after growth of bacterial cultures in minimal media with different concentrations of phosphate (Fig. 4A and B). A concentration of 337 μM phosphate allowed bacterial growth (OD600 of about 1.0 for overnight cultures) as well as high levels of SseB and SseC synthesis. This phosphate concentration was used for low-phosphate minimal medium in further experiments. Next, we analyzed whether SPI2 substrate proteins were secreted during growth under phosphate limitation at acidic pH. Growth of cultures in minimal medium containing 337 μM phosphate at pH 5.0 did not affect the levels of SseB and SseC synthesis (Fig. 4B). SseB and SseC were detected by antibodies in the detached protein fraction of bacterial cultures grown under phosphate starvation at acidic pH but were absent in the detached fractions of cultures grown at neutral pH (Fig. 4C). High intracellular levels of Sse proteins but no secretion were observed when cultures were grown in these media at neutral pH. Therefore, Mg2+ limitation as well as phosphate starvation results in the synthesis and assembly of a TTSS that is capable of secreting SPI2 substrate proteins under acidic conditions.
FIG. 4.
Effects of phosphate starvation on function of the TTSS of SPI2. The S. enterica serovar Typhimurium wild type was grown in LB or PCN minimal media, pH 7.4, containing various amounts of phosphate as indicated. Equal amounts of bacterial cells were subjected to Western blot analysis with antibodies raised against rSseB (α SseB) or rSseC (A). The amounts of SseB and SseC were analyzed by Western blot assays of total cell fractions of bacteria grown in PCN minimal medium containing high (25 mM) or limiting (0.34 mM) amounts of phosphate at neutral or acidic pH (B). Bacteria were grown in PCN medium containing 0.34 mM phosphate at neutral pH or in PCN medium adjusted to pH 5.0 by the addition of HCl, and protein was prepared from bacterial cells by mechanical shearing as described above. The amounts of SseB and SseC in total cell fractions and in the detached fractions were analyzed by Western blotting (C).
Roles of SseB, SseC, and SseD in secretion.
Sensitivity to mechanical shearing indicates that after secretion by the TTSS, SseB, SseC, and SseD are present mainly on the surface of the bacterial cells, suggesting their association in a surface-located macromolecular structure. An alternative explanation is that substrate proteins secreted into the growth medium associate with the cell surface due to unspecific interactions, e.g., polar interactions between proteins and charged residues in the outer membrane. To distinguish between these possibilities, we analyzed the functional requirements for the secretion of the main substrate proteins of SPI2. Secretion by various strains harboring mutations in the sse, ssc, or ssa gene (see Fig. 1 for positions of mutations) was compared. To exclude possible polar effects of mutations in sse genes on the synthesis of other substrate proteins, the intracellular amounts of SseB and SseC were compared under growth conditions inducing SPI2 expression but not secretion by the TTSS (Fig. 5A, pH 7.0). A nonpolar mutation in sseB (strain HH102) did not affect synthesis of SseC, a mutation in sseC (HH104) had no effect on the synthesis of SseB, and a mutation in sseD (MvP101) did not affect intracellular levels of SseB and SseC. A transposon insertion in sscA encoding a putative chaperone resulted in the absence of SseC but did not affect the amount of SseB. After growth in medium at pH 7.0, SseB, SseC, or SseD was not recovered from the bacterial surface (Fig. 5A).
The amounts of SseB, SseC, and SseD in the total cell fractions and detached fractions were analyzed under growth conditions inducing secretion (Fig. 5A, pH 5.0). The amounts of SseB and SseC in total cell fractions of the ssaV mutant strain were lower than those for the sse and ssc mutant strains, but equal amounts of SseB and SseC were detected in nonsecreting bacteria grown at pH 7.0. This indicates that large amounts of SseB and SseC in bacteria grown at pH 5.0 are secreted and are presumably attached to the cell surface. Mutations in sscA, sseC, or sseD had no effect on the amount of SseB in the detached fraction. In contrast, the sseB mutant strain had no detectable SseC on the cell surface. This defect was complemented by a plasmid-borne allele of sseB. The amount of secreted and surface-located SseD was highly reduced in the sseB mutant background, but no complementation by plasmid-borne sseB was observed. A mutation in sseC resulted in a reduced amount of SseD recovered from the bacterial surface.
We next analyzed if levels of secreted substrate proteins were affected by mutations in sse genes (Fig. 5B). After removal of bacterial cells, protein secreted into the culture supernatant was precipitated and analyzed. Small amounts of SseC and SseD were present in the culture supernatant of the wild-type strain. In contrast, larger amounts of SseC and SseD were present in the culture supernatant of the sseB mutant. SseC was not detected in the supernatant of an sseD mutant strain.
Taken together, these experiments demonstrate that after secretion SseC and SseD are located in the cell-associated fraction if SseB is present but they appear in the culture supernatant if functional SseB is absent.
Kinetics of secretion by the TTSS of SPI2.
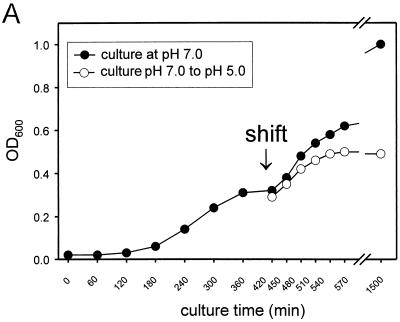
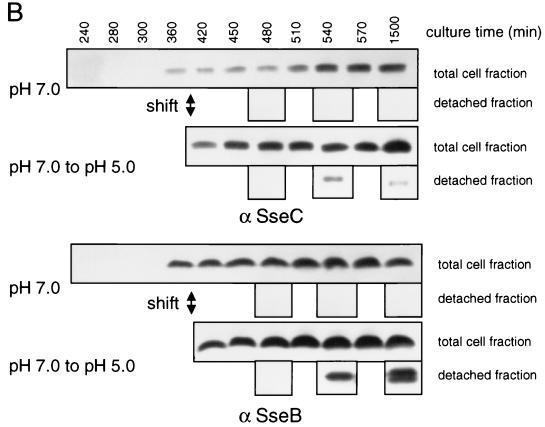
For the kinetic analysis of secretion of SPI2 substrate proteins, the S. enterica serovar Typhimurium wild type and the ssaV mutant strain were grown in minimal medium with Mg2+ limitation at neutral pH. Bacteria were harvested after reaching exponential growth phase and washed, and incubation was continued in minimal medium at neutral or acidic pH (Fig. 6A). Analysis of the detached fraction revealed that SseB and SseC were recovered from the cell surface by mechanical shearing at 2 h after the shift to acidic pH. Neither protein was detected at 1 h after the shift or in the detached fraction of cultures grown at neutral pH (Fig. 6B).
FIG. 6.
Kinetics of SseB secretion. For the analysis of kinetics of secretion by the TTSS of SPI2, the S. enterica serovar Typhimurium wild type was grown in low-Mg2+ minimal medium at pH 7.0. At exponential growth phase (OD600 of about 0.3), bacteria were harvested, washed, and resuspended in fresh medium at pH 7.0 or 5.0 (shift). The incubation was continued and samples of the cultures were taken at various time points after the shift as indicated (A). Total cell fractions and protein detached from bacterial cells as described above were subjected to Western blot analyses (B). α SseC, anti-SseC antibody.
Furthermore, we questioned whether surface-associated Sse proteins are formed by secretion of a presynthesized pool of the substrate proteins or if protein synthesis is required during secretion. Bacteria were grown in low-Mg2+ medium at pH 7.0. At an OD600 of about 0.5, bacteria were washed and shifted to low-Mg2+ medium at pH 5.0, with or without the addition of 50 μg of chloramphenicol/ml to inhibit protein biosynthesis. Secreted proteins in the detached fraction were analyzed as described before. In the presence of chloramphenicol, SseB and SseC were not detected in the detached fraction (data not shown). This result demonstrates that continuous protein biosynthesis is required for the secretion of substrate proteins of SPI2.
SseB, SseC, and SseD are required for the translocation of SPI2 effector proteins.
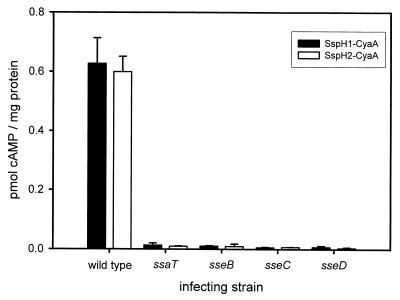
In order to determine if the secreted proteins SseB, SseC, and SseD are required for the translocation of known SPI2 effector proteins, sseB, sseC, or sseD mutations were transduced into S. enterica serovar Typhimurium harboring either the SspH1-CyaA or SspH2-CyaA fusion plasmid. These strains were then used to infect cultured murine macrophages (RAW 264.7 cell line). In addition, the RAW 264.7 cells were infected with the S. enterica serovar Typhimurium wild type and the ssaT mutant strains expressing SspH1-CyaA or SspH2-CyaA as positive and negative controls, respectively. At 8 h after internalization of the bacteria, macrophages were lysed and cellular cAMP levels were determined. Measured cAMP content was then normalized for protein concentration and is expressed as picomoles of cAMP per microgram of protein (Fig. 7). Under these assay conditions, mutations in SPI2 genes did not affect the survival of S. enterica serovar Typhimurium in RAW 264.7 cells (data not shown). Infection of macrophages with wild-type S. enterica serovar Typhimurium resulted in an increase of cAMP levels, indicating that SspH1-CyaA and SspH2-CyaA are translocated; however, no significant increase in cellular cAMP was detected in macrophages infected with the ssaT mutant strain. Likewise, no translocation of either SspH1-CyaA or SspH2-CyaA was detected in macrophages infected with sseB, sseC, or sseD mutant strains.
FIG. 7.
Translocation of SspH1 and SspH2 is dependent upon SseB, SseC, and SseD. Cultured RAW 264.7 murine macrophages were infected with either the wild type or SPI2 mutant (ssaT, sseB, sseC, and sseD) S. enterica serovar Typhimurium harboring CyaA fusions to the SPI2 translocated effector proteins SspH1 and SspH2. The results presented are the means of experiments performed in triplicate, with the error bars representing 1 standard deviation above and below the mean.
DISCUSSION
We performed experimental analyses of the TTSS of SPI2 under in vitro conditions that mimic the phagosomal environment of intracellular S. enterica serovar Typhimurium. Growth of S. enterica serovar Typhimurium with limitation of Mg2+ or starvation of phosphate induces the expression of SPI2 genes (3), and secretion of SPI2 substrate proteins was triggered by media of pH 5.0 or below (1). Under these conditions, three substrate proteins, SseB, SseC, and SseD, are secreted by the TTSS of SPI2 (1, 15; this study). This observation confirms the previous hypothesis about the identity of SPI2 substrate proteins (11). SseC is a member of the YopB family of TTSS substrate proteins, and SseB and SseD are most similar to EspA and EspB of EPEC, respectively (11).
To our knowledge, the SPI2-encoded TTSS is the first example of a TTSS whose expression and secretion can be experimentally controlled by defined growth conditions in synthetic media. Interestingly, both Mg2+ limitation and phosphate starvation result in the synthesis of the TTSS that is capable of secretion of substrate proteins upon induction by acidic environmental conditions.
In a previous study, secretion of SseB was detected within minutes after the shift of SPI2-induced bacteria to acidic pH (1). We observed that incubation of Salmonella at acidic pH for at least 1 h is required before Sse proteins can be detected in the fraction obtained by mechanical shearing or before oligomers of SseB can be detected by cross-linking of surface proteins (J. Deiwick and M. Hensel, unpublished observations). Although secretion of SseB occurs rapidly, the formation of oligomers may require accumulation of SseB over longer periods of secretion by the TTSS of SPI2. Continuous protein biosynthesis is required for this process.
Compared to substrate proteins of S. enterica serovar Typhimurium SPI1 or the Yops of Yersinia spp., only small amounts of the SPI2 proteins SseB, SseC, and SseD are released into the growth medium. SPI2 substrate proteins appear to be predominantly located on the bacterial cell surface. The sensitivity of these surface structures to disruption by mechanical shearing is reminiscent of bacterial flagella and fimbriae, which have been isolated by similar approaches (16, 30). Functional requirements for the secretion of various substrate proteins of SPI2 suggest that these proteins do not associate randomly with the bacterial cell envelope after secretion. In the absence of SseB, substrate proteins SseC and SseD are still secreted but appear in the supernatant rather than in the cell-associated fraction. In contrast, SseC and SseD are not required for the secretion and surface location of SseB. SseC is not required for secretion of SseD and vice versa. Our data demonstrate that SseB has an important role in the assembly of SseC and SseD into a macromolecular structure. Furthermore, polar structures containing secreted SseB were visible by immunofluorescence microscopy.
Surface association has been observed before for substrate proteins of the TTSS of other pathogens, for example, the Ipa proteins of Shigella flexneri (see reference 21 for a review). After secretion, protein-protein interaction was observed for Ipa proteins. In S. flexneri, mutations in IpaB or IpaD result in increased secretion of the remaining Ipa proteins into the culture medium (22). This observation is reminiscent of the effect of the mutation in sseB in S. enterica serovar Typhimurium that resulted in increased amounts of SseC and SseD in the culture supernatant. The organized association of substrate proteins during secretion appears to be essential for the function of the TTSS. Such protein interactions also result in the assembly of macromolecular surface structures, e.g., by the TTSS of Pseudomonas syringae (27) and EPEC (5, 17). SseB, SseC, and SseD are most similar to EspA, EspD, and EspB of EPEC, respectively (11). Secretion of EspA, EspB, and EspD has been demonstrated, and all Esp proteins are required for the attachment and effacement lesions of EPEC-infected epithelial cells (for a review, see reference 6). EspA is the major component of a filamentous surface structure of EPEC. Hartland et al. (9) recently reported that EspB is associated with the EspA filament and that EspA filament formation can occur in the absence of EspB. Our data indicate that surface-associated complexes of SseB were assembled in the absence of SseC or SseD. These findings indicate functional similarities between substrate proteins of EPEC and SPI2 of S. enterica serovar Typhimurium. However, it is not known whether SseB is sufficient to assemble a surface-associated filamentous structure in S. enterica serovar Typhimurium.
Recently, a family of putative translocated proteins of the SPI2 TTSS has been identified (23). These proteins are encoded by genes outside the SPI2 locus and have a conserved N-terminal domain required for translocation. However, this translocation domain is absent in SseB, SseC, SseD, and other proteins encoded by the SPI2 locus, supporting the notion that the function of SseB, SseC, and SseD is distinct from those of the putative effector proteins. There is no evidence that SseB, SseC, or SseD is translocated into the host cell cytoplasm.
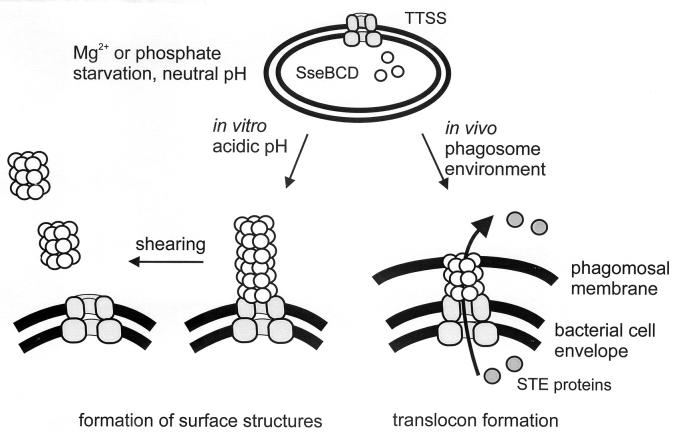
Previous studies revealed that mutations in sseB, sseC, or sseD each result in dramatic attenuation of virulence of S. enterica serovar Typhimurium and in reduced intracellular accumulation (2, 20; unpublished observations). We observed that SseB, SseC, and SseD do not require each other for secretion but that these proteins appear to interact after secretion. Cross-linking experiments support the presence of an oligomeric assembly of SseB subunits after secretion (Deiwick and Hensel, unpublished results). Based on these observations, we propose that after secretion SseB, SseC, and SseD are assembled into a hetero-oligomeric complex (Fig. 8). This complex is formed in vitro after growth of S. enterica serovar Typhimurium in minimal medium at acidic pH. In vivo, SseB, SseC, and SseD are each required for the translocation of effector proteins from intracellular S. enterica serovar Typhimurium into the infected host cell. Translocation of effector proteins is as reduced in sseB, sseC, and sseD mutants as it is in mutants in structural components of the TTSS such as SsaT. Therefore, the secreted proteins SseB, SseC, and SseD act as a translocator for effector proteins in vivo. We hypothesize that in vivo SseB, SseC, SseD, and putative additional substrate proteins form a structure that acts as a translocator to mediate translocation of effector proteins of the TTSS of SPI2 from intraphagosomal S. enterica serovar Typhimurium into the host cell (Fig. 8). Additional analysis by cross-linking experiments may reveal further details of the putative translocator structure.
FIG. 8.
Model for the secretion and function of SseBCD. Expression of the TTSS of SPI2 and SseBCD is induced by growth of S. enterica serovar Typhimurium in minimal media with limiting amounts of phosphate or Mg2+. Growth media with an acidic pH induce secretion of SseBCD by the TTSS of SPI2. Under in vitro conditions, surface structures composed of SseBCD and putative additional proteins are assembled. These surface structures can be detached from the bacterial cell surface by mechanical shearing (this study) or treatment with organic solvents (1). In vivo, SseBCD functions as a translocon for the translocation of STE (Salmonella translocated effector) proteins from intraphagosomal Salmonella over the phagosomal membrane into the host cell.
In conclusion, we have identified that in addition to SseB, substrate proteins SseC and SseD are secreted by the TTSS of SPI2. After secretion, these proteins assemble into a complex exposed on the bacterial cell surface. SseB, -C, and -D are required for translocation of effector proteins into host cells. Further work will reveal whether this structure represents a translocator for SPI2 effector proteins and how such a putative translocator can mediate the transfer of effector proteins by intracellular Salmonella.
ACKNOWLEDGMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft (HE1964/2-3) and the BMBF (01 KI 9606) to M.H. and by grant RO1 AI48683 from the National Institutes of Health to S.I.M. C.R. was supported by a predoctoral fellowship of the Ludwig-Maximilians-Universität.
We are indebted to J. Heesemann for generous support of this work and for stimulating discussions.
REFERENCES
- 1.Beuzon C R, Banks G, Deiwick J, Hensel M, Holden D W. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol. 1999;33:806–816. doi: 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- 2.Cirillo D M, Valdivia R H, Monack D M, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 3.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 4.Deiwick J, Nikolaus T, Shea J E, Gleeson C, Holden D W, Hensel M. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J Bacteriol. 1998;180:4775–4780. doi: 10.1128/jb.180.18.4775-4780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebel F, Podzadel T, Rohde M, Kresse A U, Kramer S, Deibel C, Guzman C A, Chakraborty T. Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol Microbiol. 1998;30:147–161. doi: 10.1046/j.1365-2958.1998.01046.x. [DOI] [PubMed] [Google Scholar]
- 6.Frankel G, Phillips A D, Rosenshine I, Dougan G, Kaper J B, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 7.Galan J E. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr Opin Microbiol. 1999;2:46–50. doi: 10.1016/s1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 8.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 9.Hartland E L, Daniell S J, Delahay R M, Neves B C, Wallis T, Shaw R K, Hale C, Knutton S, Frankel G. The type III protein translocation system of enteropathogenic Escherichia coli involves EspA-EspB protein interactions. Mol Microbiol. 2000;35:1483–1492. doi: 10.1046/j.1365-2958.2000.01814.x. [DOI] [PubMed] [Google Scholar]
- 10.Hensel M. Salmonella pathogenicity island 2. Mol Microbiol. 2000;36:1015–1024. doi: 10.1046/j.1365-2958.2000.01935.x. [DOI] [PubMed] [Google Scholar]
- 11.Hensel M, Shea J E, Waterman S R, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang F, Holden D W. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 12.Hermanson G T. Bioconjugate techniques. San Diego, Calif: Academic Press; 1996. [Google Scholar]
- 13.Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988;9:28–32. doi: 10.1002/elps.1150090106. [DOI] [PubMed] [Google Scholar]
- 14.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein J R, Jones B D. Salmonella pathogenicity island 2-encoded proteins SseC and SseD are essential for virulence and are substrates of the type III secretion system. Infect Immun. 2001;69:737–743. doi: 10.1128/IAI.69.2.737-743.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klemm P, Schembri M, Stentebjerg-Olesen B, Hasman H, Hasty D L. Fimbriae: detection, purification, and characterization. In: Williams P, Ketley J, Salmond G P, editors. Methods in microbiology. San Diego, Calif: Academic Press; 1998. pp. 239–248. [Google Scholar]
- 17.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Medina E, Paglia P, Nikolaus T, Müller A, Hensel M, Guzman C A. Pathogenicity island 2 mutants of Salmonella enterica serovar Typhimurium are efficient carriers for heterologous antigens and enable modulation of immune responses. Infect Immun. 1999;67:1093–1099. doi: 10.1128/iai.67.3.1093-1099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menard R, Dehio C, Sansonetti P J. Bacterial entry into epithelial cells: the paradigm of Shigella. Trends Microbiol. 1996;4:220–226. doi: 10.1016/0966-842X(96)10039-1. [DOI] [PubMed] [Google Scholar]
- 22.Menard R, Sansonetti P, Parsot C. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–5302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miao E A, Miller S I. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao E A, Scherer C A, Tsolis R M, Kingsley R A, Adams L G, Baumler A J, Miller S I. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol. 1999;34:850–864. doi: 10.1046/j.1365-2958.1999.01651.x. [DOI] [PubMed] [Google Scholar]
- 25.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 26.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roine E, Wei W, Yuan J, Nurmiaho Lassila E L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1–100 kDa. Anal Biochem. 1987;266:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 29.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sockett R E. Characterizing flagella and motile behavior. In: Williams P, Ketley J, Salmond G P, editors. Methods in microbiology. San Diego, Calif: Academic Press; 1998. pp. 227–238. [Google Scholar]
- 31.Tardy F, Homble F, Neyt C, Wattiez R, Cornelis G R, Ruysschaert J M, Cabiaux V. Yersinia enterocolitica type III secretion-tanslocation system: channel formation by secreted Yops. EMBO J. 1999;18:6793–6799. doi: 10.1093/emboj/18.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wachter C, Beinke C, Mattes M, Schmidt M A. Insertion of EspD into epithelial target cell membranes by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1999;31:1695–1707. doi: 10.1046/j.1365-2958.1999.01303.x. [DOI] [PubMed] [Google Scholar]