Abstract
Bioactive small-molecule inhibitors represent a treasure chest for future drugs. In vitro high-throughput screening is a common approach to identify the small-molecule inhibitors that bind tightly to purified targets. Here, we investigate the inhibitor-target binding/unbinding kinetics in E. coli cells using a benzimidazole-derivative DNA inhibitor as a model system. We find that its unbinding rate is not constant but depends on cell growth rate. This dependence is mediated by the cellular activity, forming a feedback loop with the inhibitor’s activity. In accordance with this feedback, we find cell-to-cell heterogeneity in inhibitor-target interaction, leading to co-existence of two distinct subpopulations: actively growing cells that dissociate the inhibitors from the targets and non-growing cells that do not. We find similar heterogeneity for other clinical DNA inhibitors. Our studies reveal a mechanism that couples inhibitor-target kinetics to cell physiology and demonstrate the significant effect of this coupling on drug efficacy.
Subject terms: Antimicrobials, Drug discovery, Biophysics
This study investigates how binding/unbinding kinetics of a benzimidazole-derivative DNA inhibitor in E. coli cells depends on cell growth and physiology and demonstrates the effect of this coupling on drug efficacy.
Introduction
Bioactive small-molecule inhibitors constitute the majority of drugs on the market. However, identifying efficacious lead compounds and developing them as new drugs is a time-consuming and expensive process, which typically requires more than a decade and billions of dollars1. Even these staggering numbers are likely an underestimate, because they do not fully account for numerous failures at early stages2. A major consequence of these inefficiencies is the shortage of new drugs, particularly in the anti-bacterial arsenal. This shortage in light of rising bacterial resistance is a serious medical problem.
In recent years, target-based drug screening has become a central paradigm to streamline the drug discovery process3,4. By screening vast compound libraries against macromolecules essential for cellular function in vitro, target-based screening rapidly identifies a wealth of small molecule inhibitors. Unfortunately, many of these candidates turn out to be therapeutically ineffective and are abandoned after substantial investment5,6. One major issue of this screening is that it is performed with solutions reconstituted with purified targets7–11. In reality, however, the targets reside in/on cells and interact with the inhibitors in highly complex cellular environments. Currently, little is known about how this natural cellular environment affects the inhibitor-target binding/unbinding kinetics. The present study aims to reduce this gap by characterizing the inhibitor-target kinetics in live bacterial cells.
Benzimidazole is one of the most frequent moieties in therapeutic drugs listed by the US Food and Drug Administration12,13. It is being extensively utilized in medicinal chemistry as a promising scaffold for new drug design14–19, because it exhibits high affinity to nucleotides. DNA is a particularly important drug target. DNA-binding inhibitors have been successfully adopted as anticancer drugs20–22 and are being tested to treat other human diseases, including malaria and HIV23–27. Recently, DNA inhibitors have received increasing attention as anti-bacterial drugs due to the rising bacterial resistance28–32.
A benzimidazole derivative known as Hoechst 33342 (HCT)33 binds to DNA to inhibit gene expression34. In this study, HCT was used as a model system because HCT fluoresces only when bound to DNA and loses fluorescence upon unbinding35–38, which allows us to directly monitor the binding and unbinding kinetics. Our data indicate that the rate of unbinding is not constant but is strongly correlated with the rate of cell growth, which was mediated by transcription. This correlation, together with the known mode of action of HCT, suggests a double negative feedback loop between HCT-DNA complex formation and gene expression. In accordance with this loop, we observed a great deal of heterogeneity in the accumulation of HCT on its target, which leads to heterogeneous growth inhibition. These heterogeneous effects were observed for other DNA inhibitors as well.
Results
The rate of HCT unbinding from DNA varies with the HCT concentration
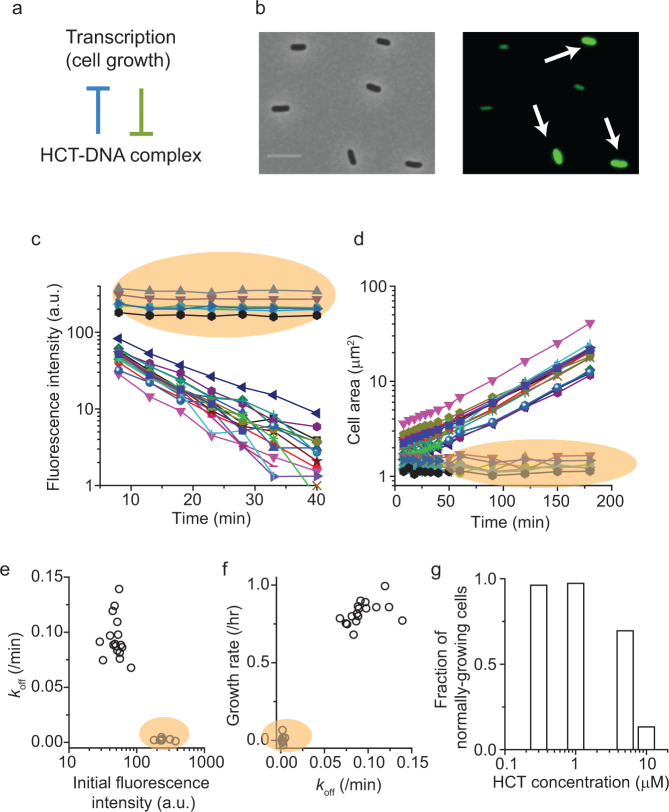
It was previously shown that HCT could fluoresce when embedded in the cell membrane39. However, HCT fluorescence signal arising from the interaction with cell membrane is expected to be weak40. To test this, we co-labeled DNA in live E. coli cells with HCT and mCherry-tagged DNA-associated protein HupA (HupA-mCherry)41. The spatial profiles of their intensities were identical, which demonstrates that the majority of the HCT signal in cells comes from its binding to DNA (Supplementary Fig. 1).
We then characterized the inhibitory effects of HCT on E. coli cells by measuring the growth rate, λ (Fig. 1a, b). λ was unchanged up to 1 μM but decreased sharply at higher HCT concentrations (Fig. 1a, b). The repressive effect of HCT on cell growth is expected based on its ability to bind to DNA, which results in the inhibition of gene expression34. Indeed, when we measured the expression of a reporter gene (lacZ), we found that its expression was inhibited by HCT (Supplementary Fig. 2).
Fig. 1. λ and koff at various HCT concentrations.
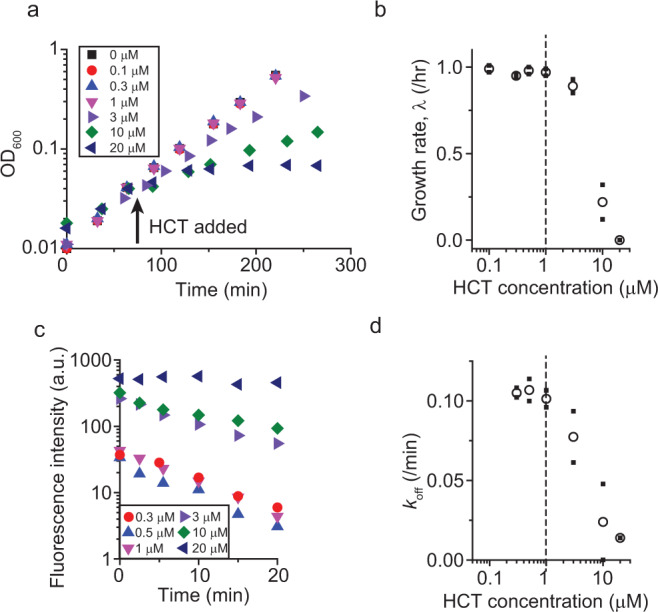
a, b HCT slows down the growth of E. coli cells in bulk culture at concentrations above 1 μM. A t-test (one-tailed) between the growth rates for 1 and 3 μM shows a P-value equal to 0.0325. Therefore, their difference is considered to be statistically significant. This agrees with a noticeable and persistent difference of OD600 between 1 and 3 μM treatment cultures (a). c, d Cells were pre-incubated with HCT and suspended in HCT-free media (which defines time zero). For each data point, the intracellular HCT intensities of ~150 cells were measured and averaged. The slope of the intensity change reflects the koff. The koff decreased at HCT concentrations above 1 μM. a and c show data from a single experiment. A biological replicate was conducted to confirm that the pattern was reproducible. Small solid symbols in b and d indicate the data from two biological repeats, and the open symbols indicate their mean.
We then measured the binding and unbinding kinetic rates kon and koff. To measure the kon, we exposed cells to HCT and monitored an increase in the intracellular HCT fluorescence intensity (Supplementary Fig. 3), as similarly done in previous in vitro studies42,43. The kon was constant over a wide range of HCT concentrations tested (Supplementary Fig. 3). To measure the koff, we pre-incubated cells with HCT, resuspended them in HCT-free media, and measured a decrease in the intercellular HCT intensity (Fig. 1c), the slope of which is equal to the koff. We found that the koff remained constant up to 1 μM HCT at ~0.1/min (Fig. 1c, d). This value is at least one order of magnitude lower than the koff measured in vitro, which is in the range of 1~10/min42,43. The koff in cells was even lower at higher HCT concentrations (Fig. 1c, d).
Correlation between unbinding rate and growth rate
Interestingly, the threshold concentrations above which the koff and λ decrease coincided (1 μM in Fig. 1b and d, dashed line), which suggests that the koff and λ be coupled. This coupling can be more directly visualized by plotting koff as a function of λ, which showed a strong correlation (Fig. 2a). While the HCT dilution due to cell growth, which includes the cell volume expansion and DNA synthesis, could alter koff, its effect (λ ≈ 1/hr = 0.017/min) is an order of magnitude less than a change we observed (Fig. 2a) and thus negligible.
Fig. 2. Cell growth affects HCT unbinding.
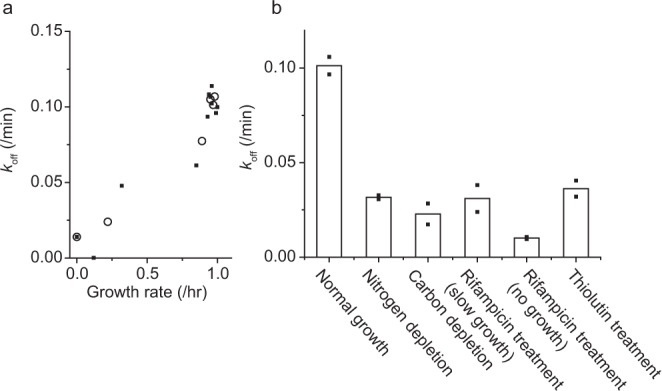
a Data from Fig. 1b and d were put together to find the relationship between koff and λ. b koff under various conditions. Rifampicin and thiolutin were used to repress transcription. The results show an important effect of transcription on koff. See Supplementary Fig. 5 for koff under translation repression by chloramphenicol. See Supplementary Videos 4 and 5 for time-lapse images of HCT intensity under rifampicin treatment and chloramphenicol treatments, respectively. Small solid symbols indicate the data from two biological repeats, and the open symbols and columns indicate their mean.
A recent survey examined the common properties in approved drug molecules and showed that the drug-target unbinding kinetics are a major determinant of drug efficacy44–46. The correlation we observed agrees with this survey result. However, one important distinction should be made. Previously, the koff of inhibitors was measured in test tubes against purified targets and considered as an intrinsic constant value. However, our data show that the koff in cells is not constant but variable.
Motivated by the correlation between λ and koff, we next wondered how repressing cell growth with other means would affect unbinding. We thus quantified the koff in cells whose growth was repressed by nutrient depletion, either nitrogen or carbon. For this experiment, 1 μM HCT was used; note this HCT concentration had little effects on cell growth (Fig. 1b). We found that the growth repression by nutrient depletion led to a decrease in the koff (Fig. 2b).
Next, we repressed cell growth by inhibiting transcription. DNA stores genetic information, which is transcribed by RNA polymerases (RNAPs). RNAPs have high affinities for DNA and are processive47–49. Thus, when they encounter protein roadblocks bound to DNA, they dislodge these proteins and continue their functions. We wondered whether this processivity facilitated unbinding of DNA inhibitors. To test this, we measured the koff in transcription-repressed cells. Rifampicin is known to repress transcription in cells. When cells were treated with rifampicin, it took some time for cell growth to slow down and completely stop (Supplementary Fig. 4). We found that the koff was very low during this slow-down period and even lower when cell growth stopped (Fig. 2b). We repeated the experiment with thiolutin, another transcription repressor. Treatment of thiolutin led to immediate slow-down of cell growth (Supplementary Fig. 4). We again observed that the koff decreased after thiolutin treatment.
This finding with transcription inhibition is consistent with the above measurement with nutrient-deprived cells. As cells are depleted of nutrients, the rate of transcription drops dramatically50–52, which can explain a low koff in nutrient-depleted conditions (Fig. 2b). To further demonstrate the importance of transcription, we measured the koff in cells treated with a translation repressor, chloramphenicol. We found that chloramphenicol treatment reduces the koff only marginally (Supplementary Fig. 5), which further underscores the importance of transcription (but not translation) to the unbinding kinetics.
Heterogeneous effects of DNA inhibitor on cell growth
When HCT (or other drug molecules) binds to DNA, the formation of HCT-DNA complex negatively affects the transcription (Supplementary Fig. 2 and ref. 34). This negative effect is depicted as a blue line in Fig. 3a. Our finding above shows that transcription negatively affects the formation of HCT-DNA by facilitating the unbinding (Fig. 2b); this effect is depicted as a green line in Fig. 3a. As a result, they constitute a double negative feedback loop, which is known to trigger bi-stability in a system53. In the present context, this bi-stability is represented as two subpopulations. In one subpopulation, the inhibitors would unbind from their targets rapidly, leading to low HCT accumulation on the targets. As a result, this subpopulation will exhibit active transcription and fast cell growth. In the other subpopulation, the slow unbinding of inhibitors would lead to higher HCT accumulation on the targets, leading to strong inhibition of transcription and slow (or no) cell growth.
Fig. 3. Heterogeneous inhibitory activities of HCT at single-cell resolution.
a Formation of HCT-DNA complex represses transcription, while transcription represses the complex formation by stimulating unbinding. b Cells were pre-incubated with 5 μM HCT and imaged with a microscope at single-cell resolution. We found two distinct levels of intracellular HCT intensity, indicating that its accumulation on the target (DNA) is heterogeneous across a cell population. Note that HCT emits blue fluorescence but is depicted as green here for the clarity of the images. The scale bar represents 5 μm. c, d Cells pre-incubated with 5 μM HCT were transferred to HCT-free media (which defines time zero), and intracellular HCT intensity and cell area were observed over time using a microscope. Cells exhibiting high HCT intensity (orange shades) exhibited no HCT unbinding and no growth. On the other hand, cells with initially weak HCT intensity exhibited rapid HCT unbinding and active cell growth. e, f Data plotted in c, d were quantitatively analyzed. g The fraction of the subpopulation with rapid HCT unbinding and active cell growth decreases at higher external HCT concentrations. The plotted data were from a single experiment. A biological replicate was conducted to confirm the pattern.
We tested this hypothesis by measuring HCT levels and cell growth with time-lapse single-cell microscopy. In our population study above, we observed a moderate decrease in the koff and λ at 5 μM HCT. Cells were pre-incubated with this concentration and imaged with a microscope. We observed two distinct levels of intracellular HCT accumulation; in Fig. 3b, HCT-bright cells are indicated by white arrows. To determine koff, we resuspended the cells in HCT-free media and measured a decrease in the intercellular HCT intensity. HCT-bright cells maintained their HCT signals, indicating no unbinding (orange shade in Fig. 3c). These HCT-bright cells did not grow (orange shade in Fig. 3d). Longer observation revealed that these cells failed to resume growth and ultimately lysed. On the other hand, cells with weak HCT signals exhibited rapid HCT unbinding and active growth (Fig. 3c, d). While there is intrinsic heterogeneity in cell growth (in the absence of HCT), this heterogeneity is small (<30%)54,55. In fact, we did not observe non-growing cells in the absence of HCT.
When we analyzed these data to quantify the koff and λ of individual cells, we found that the koff and λ were correlated and exhibited a bimodal distribution (Fig. 3e, f). To demonstrate the effects of internal HCT accumulation on gene expression at single-cell resolution, we repeated the experiment with a strain that harbors a fluorescent mCherry gene. We observed that HCT-bright cells did not express the fluorescent protein whereas HCT-weak cells expressed the protein (Supplementary Video 1). This agrees with our previous data that high HCT levels inhibit gene expression (Supplementary Fig. 2), although the latter data were collected at the population level. The fact that some cells were able to keep the internal HCT level low and actively grew reveals that they were resistant to the HCT treatment. When we determined the fraction of this subpopulation, we found that the fraction was high up to 1 μM HCT but decreased at higher HCT concentrations (Fig. 3g).
Our findings collectively demonstrate the co-existence of two subpopulations with distinct unbinding kinetics and growth rates, supporting our hypothesis. We next repeated this single-cell experiment with two commercially available drugs that target DNA and exhibit anti-microbial activities, netropsin and berenil56–58. We again observed two distinct subpopulations (Supplementary Video 2 and 3).
Discussion
Recent survey results show that approved drug molecules generally exhibit slow unbinding44,45. The importance of slow unbinding is logical, because drugs exert their effects only when bound to their targets. Additional beneficial effects of slow unbinding were proposed, e.g., target selectivity59 or enhanced cidality60. This critical effect of koff is well supported by our study as well.
Various intracellular conditions could affect the apparent unbinding rate koff that we reported in this study, e.g., pH, reaction volume, non-specific binding to other intracellular components. In particular, we found transcription has critical effects on koff. As a result, koff is not a fixed value but is variable in cells, which then manifests as heterogeneous activities of the inhibitor at the single-cell level. The unbinding of inhibitors is very slow in some cells, which leads to tight inhibitor-target complex formation, and thus strong inhibition of cell growth (and vice versa in other cells). Interestingly, the heterogeneous effect of antibiotics on cells have been observed for a wide range of antibiotics61–63. Several studies found that this heterogeneous effect could be caused by the variation in the expression of genes that confer resistance to antibiotics64–66. For example, our previous study has shown that as translation inhibitors repress the action of ribosomes at sub-lethal concentrations, cells devote more resources to produce additional ribosomes, leaving little for expressing resistance genes, which then results in heterogenous expression of the resistance genes64. Our finding suggests that the variation in koff could be another mechanism for the heterogenous effect of antibiotics.
Our findings have broad implications to drug discovery. Target-based in vitro assay of small-molecule inhibitors is revolutionizing the drug discovery program3,4. However, the discrepancy between the assay results and therapeutic efficacy has been frequently observed5,6. Precisely understanding what causes this discrepancy is critical to enhancing the predictive power in the drug discovery process. Our study reveals one mechanism that could contribute to this discrepancy, i.e., coupling between unbinding and cell physiology.
Given the important role of koff in drug efficacy and based on our observation that koff is not a fixed value but can vary, is it possible to manipulate koff to improve drug efficacy? We tested this possibility in our proof-of-principle study (Supplementary Fig. 6). We decreased the koff by treating cells with a low dose of a transcription inhibitor, rifampicin. With this treatment, an otherwise growth-permissive HCT concentration became growth-inhibiting, indicating enhanced inhibitory effects of HCT (Supplementary Fig. 6). Therefore, our study opens a door for a novel therapeutic strategy to improve drug efficacy.
Methods
Bacterial strains and culture conditions
E. coli K-12 NCM372267–69 was grown in Neidhart’s MOPS minimal media70 with glucose and NH4Cl as the carbon and nitrogen sources. NCM3722 harboring km::T:ptet:mCherry, sp:Pcon-TetR-LacIq(attB) (NMK104) was used for measurement of mCherry expression under the microscope. NMK80, a NCM3722 harboring Ptet-lacZ (TetR is constitutively expressed in this strain) was used for β-galactosidase assay71. To prepare experimental cultures, cells were taken from −80 °C stocks and streaked on a LB plate. Single colony was inoculated in 2 mL LB medium and grown at 37 °C with constant agitation at 250 rpm in a water bath (New Brunswick Scientific). To monitor growth, the optical density (OD600) of the culture was measured using a Genesys20 spectrophotometer (Thermo-Fisher) with a standard cuvette (16.100-Q-10/Z8.5, Starna Cells Inc). Before cells entered stationary phase, cells were inoculated into 5 mL MOPS minimal medium at very low densities (typically lower than the OD600 of ~0.0001) and cultured them overnight (pre-culture). Next morning, the pre-culture was diluted in pre-warmed, 5 mL MOPS minimal medium (experimental culture) to the OD600 of ~0.01 and allowed to grow exponentially to desired OD600.
Hoechst 33342 unbinding rate measurement
To determine unbinding rate of Hoechst 33342 (HCT), fluorescence intensity of single cells was measured using a microscope. Experimental cultures in MOPS minimal media were prepared as described above. When a culture reached to the OD600 ~ 0.025, HCT was added and incubated for 45 min. For HCT concentrations >5 μM, HCT was added at the OD600 ~0.04. To measure the unbinding rate of HCT under the influence of antibiotics (rifampicin 10 μg/mL, thiolutin 5 μg/mL), drugs were added at same time as HCT was added to the cultures.
After incubation with HCT, 5 μL aliquot of the culture was placed between a no. 1.5 cover glass and 1mm-thick 1.5% agarose pad72 in MOPS minimal media with HCT (with appropriate drugs) and imaged under the microscope. See below for details of microscopy procedures. Fluorescence intensity of these cells was recorded (time 0 min). Then, the culture was centrifuged at 3500 rpm at 37 °C for 5 min and resuspended in fresh media without HCT (with appropriate drugs). Resuspended cells were incubated at 37 °C with agitation in water bath. 5 μL aliquot of the culture was placed between a no. 1.5 cover glass and 1mm-thick 1.5% agarose pad in media without HCT and imaged under the microscope.
HCT unbinding rate was also measured by following individual cells over time (Fig. 3). Cells were prepared as described above with HCT 5 μM. After incubation, cells were placed between agarose pad in media without HCT and a coverslip of 35 mm glass bottom dish (Cellvis) and imaged under the microscope over time. See the Supplementary Table 1 for all the chemicals and concentrations used.
HCT unbinding rate measurement with chloramphenicol
Cells were prepared as described above. At the OD600 ~0.025, 1 μM of HCT and 20 μg/mL of chloramphenicol was added. After 45 min of incubation, 5 μL aliquot of culture was transferred onto a no. 1.5 cover glass, sandwiched by 1mm-thick 1.5% agarose pad72, and imaged with a microscope. For time-lapse microscopy to measure the unbinding rate of HCT in individual cells, 1.5% agarose pad in 35 mm glass bottom dish (Cellvis) was used.
HCT binding rate measurement
Cells were prepared as described above. HCT was added to a culture when OD600 reached to ~0.1. Fluorescence images were acquired periodically by transferring 5 μL aliquot of culture between a no. 1.5 cover glass and 1mm-thick 1.5% agarose pad in media with HCT and using a microscope.
Time-lapse imaging of mCherry expression
NCM3722 harboring km::T:ptet:mCherry, sp:Pcon-TetR-LacIq(attB) (NMK104) was used to visualize the influence of DNA binding drugs on gene expression. Cells were grown as described above. When OD600 reached to ~0.05, 10 μM HCT, 15 μg/mL netropsin, or 30 μg/mL berenil was added to the culture. After incubation with drugs (1 h, 4.5 h, and 4 h for HCT, netropsin, and berenil, respectively), cells were plated between a cover glass and 1.5% agarose in MOPS minimal media with 100 ng/mL aTc on 35 mm glass bottom dish (Cellvis). Time-lapse images were captured using a microscope. mCherry fluorescence was detected using a TRITC filter cube.
β-galactosidase assay
NCM3722 harboring Ptet-lacZ (NMK80, TetR is constitutively expressed in this strain) was used71. For a control (HCT-free), anhydrotetracycline (aTc) was added to a culture at OD600 of ~0.1 to the final concentration of 100 ng/mL. For HCT treatment, HCT was added at OD600 of ~0.05 and then after 3 hr of incubation, aTc was added to the final concentration of 100 ng/mL. Samples were rapidly frozen in ethanol dry ice bath and stored at −80 °C. The β-galactosidase assay was carried out as described73, with some modification. Samples were thawed and kept on ice. Cells were lysed in a mixture of Z-buffer, SDS, and β-mercaptoethanol (final volume of 1 mL). 50 μL of chloroform was added, and solutions were vortexed for 15 sec and incubated at 37 °C in a water bath for 5 min. 200 μL of pre-warmed ONPG (4 mg/mL) was added to the solution. The reaction was stopped by adding 500 μL of 1 M NaCO3 when the color of solution turned to light yellow. 800 μL of reaction was spun down at 10k rpm for 3 min and optical density at 420 nm and 550 nm were measured using a Genesys20 spectrophotometer (Thermo-Fisher) with a standard cuvette (16.100-Q-10/Z8.5, Starna Cells Inc).
Fluorescence microscopy and image analysis
All image acquisition was done using an inverted fluorescence microscope (Olympus IX83) with an oil immersion phase-contrast 60× objective seated inside an incubator chamber (InVivo Scientific) pre-warmed to 37 °C and MetaMorph software (Molecular Devices). HCT fluorescence was measured using a DAPI filter cube. Images were captured using a Neo 5.5 sCMOS camera (Andor).
Fluorescence intensity was obtained with MicrobeJ (5.13 l (4))74, a plug-in for the ImageJ software. MicrobeJ can automatically segment cell boundaries from phase-contrast microscope images and apply the binary masks from the segmentation to measure fluorescence intensities (in mean gray value) inside and outside the cells. The latter (background) is subtracted from the former to determine intracellular fluorescence signals. Autofluorescence of cells was determined by measuring fluorescence intensity in the absence of HCT. Reported intensities are the results of subtracting autofluorescence from intracellular fluorescence signals.
Statistics and reproducibility
At least two biological replicates were conducted for all experiments, and all data were reproducible. For single cell analysis, data points were determined from averaging 100~200 cells. For Figs. 1b, d, 2a, b, and 3g, means and raw data points were plotted. For Fig. 1a, c and 3b–f, data points from a single experiment were plotted to show a typical pattern, and the observed pattern was confirmed in the other biological replicate. For Fig. 1a, one-tailed t-test was conducted to show a statistically significant difference of the growth rates between HCT 1 and 3 μM (P-value = 0.0325).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was funded by Research Corporation for Science Advancement (24097), MP3 Initiative (00097584), and NIH (1U19AI158080).
Author contributions
D.L. and M.K. conceived the study. D.L. and T.A. designed the experiments, analyzed and interpreted the data. D.W. and M.K. secured funding and provided resources. D.L., T.A. and M.K. wrote the manuscript. All authors read and approved the manuscript.
Peer review
Peer review information
Communications Biology thanks Pia Abel zur Wiesch, Ulrich Gerland and Imane El Meouche for their contribution to the peer review of this work. Primary Handling Editors: Wendy Mok and Joao Valente.
Data availability
The datasets are provided as Supplementary Data.
Competing interests
Authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-04604-9.
References
- 1.Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. Br. J. Pharm. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decker, S. & Sausville, E. A. in Principles of Clinical Pharmacology 2nd edn (eds Arthur J. Atkinson et al.) 439–447 (Academic Press, 2007).
- 3.Croston GE. The utility of target-based discovery. Expert Opin. Drug Discov. 2017;12:427–429. doi: 10.1080/17460441.2017.1308351. [DOI] [PubMed] [Google Scholar]
- 4.Alksne LE, Dunman PM. Target-based antimicrobial drug discovery. Methods Mol. Biol. 2008;431:271–283. doi: 10.1007/978-1-60327-032-8_21. [DOI] [PubMed] [Google Scholar]
- 5.Brown D. Unfinished business: target-based drug discovery. Drug Discov. Today. 2007;12:1007–1012. doi: 10.1016/j.drudis.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 6.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discov. Today. 2005;10:139–147. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- 7.WEHRLI W. Kinetic Studies of the Interaction between Rifampicin and DNA-dependent RNA polymerase of Escherichia coli. Eur. J. Biochem. 1977;80:325–330. doi: 10.1111/j.1432-1033.1977.tb11886.x. [DOI] [PubMed] [Google Scholar]
- 8.McPhail D, Cooper A. Thermodynamics and kinetics of dissociation of ligand-induced dimers of vancomycin antibiotics. J. Chem. Soc., Faraday Trans. 1997;93:2283–2289. doi: 10.1039/a701327b. [DOI] [Google Scholar]
- 9.Walkup GK, et al. Translating slow-binding inhibition kinetics into cellular and in vivo effects. Nat. Chem. Biol. 2015;11:416–423. doi: 10.1038/nchembio.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauffer BE, et al. Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J. Biol. Chem. 2013;288:26926–26943. doi: 10.1074/jbc.M113.490706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svetlov MS, Cohen S, Alsuhebany N, Vázquez-Laslop N, Mankin AS. A long-distance rRNA base pair impacts the ability of macrolide antibiotics to kill bacteria. Proc. Natl Acad. Sci. 2020;117:1971–1975. doi: 10.1073/pnas.1918948117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor RD, MacCoss M, Lawson AD. Rings in drugs. J. Med. Chem. 2014;57:5845–5859. doi: 10.1021/jm4017625. [DOI] [PubMed] [Google Scholar]
- 13.Bansal Y, Silakari O. The therapeutic journey of benzimidazoles: a review. Bioorg. Med. Chem. 2012;20:6208–6236. doi: 10.1016/j.bmc.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Göker H, Kuş C, Boykin DW, Yildiz S, Altanlar N. Synthesis of some new 2-substituted-phenyl-1H-benzimidazole-5-carbonitriles and their potent activity against candida species. Bioorg. Med. Chem. 2002;10:2589–2596. doi: 10.1016/S0968-0896(02)00103-7. [DOI] [PubMed] [Google Scholar]
- 15.Klimešová V, et al. Synthesis and preliminary evaluation of benzimidazole derivatives as antimicrobial agents. Eur. J. Med. Chem. 2002;37:409–418. doi: 10.1016/S0223-5234(02)01342-9. [DOI] [PubMed] [Google Scholar]
- 16.Ansari KF, Lal C. Synthesis, physicochemical properties and antimicrobial activity of some new benzimidazole derivatives. Eur. J. Med. Chem. 2009;44:4028–4033. doi: 10.1016/j.ejmech.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 17.Bansal S, et al. Old class but new dimethoxy analogue of benzimidazole: a bacterial topoisomerase I inhibitor. Int. J. Antimicrob. Agents. 2010;35:186–190. doi: 10.1016/j.ijantimicag.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Ranjan N, et al. Selective inhibition of bacterial topoisomerase I by alkynyl-bisbenzimidazoles. Medchemcomm. 2014;5:816–825. doi: 10.1039/C4MD00140K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joubert A, et al. Sequence-selective targeting of long stretches of the DNA minor groove by a novel dimeric bis-benzimidazole. Biochemistry. 2003;42:5984–5992. doi: 10.1021/bi026926w. [DOI] [PubMed] [Google Scholar]
- 20.Cai X, Gray PJ, Von Hoff DD. DNA minor groove binders: back in the groove. Cancer Treat. Rev. 2009;35:437–450. doi: 10.1016/j.ctrv.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 22.Hurley LH, Boyd FL. DNA as a target for drug action. Trends Pharmacol. Sci. 1988;9:402–407. doi: 10.1016/0165-6147(88)90067-3. [DOI] [PubMed] [Google Scholar]
- 23.Scott FJ, Khalaf AI, Duffy S, Avery VM, Suckling CJ. Selective anti-malarial minor groove binders. Bioorg. Med. Chem. Lett. 2016;26:3326–3329. doi: 10.1016/j.bmcl.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal A, et al. Discovery of a selective, safe and novel anti-malarial compound with activity against chloroquine resistant strain of Plasmodium falciparum. Sci. Rep. 2015;5:13838. doi: 10.1038/srep13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayase H, et al. Inhibition of malaria parasite growth by quinomycin A and its derivatives through DNA-intercalating activity. Biosci., Biotechnol., Biochem. 2015;79:633–635. doi: 10.1080/09168451.2014.987205. [DOI] [PubMed] [Google Scholar]
- 26.Collings CK, Little DW, 3rd, Schafer SJ, Anderson JN. HIV chromatin is a preferred target for drugs that bind in the DNA minor groove. PLoS One. 2019;14:e0216515–e0216515. doi: 10.1371/journal.pone.0216515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warowicka A, Nawrot R, Goździcka-Józefiak A. Antiviral activity of berberine. Arch. Virol. 2020;165:1935–1945. doi: 10.1007/s00705-020-04706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hlaka L, et al. Evaluation of minor groove binders (MGBs) as novel anti-mycobacterial agents and the effect of using non-ionic surfactant vesicles as a delivery system to improve their efficacy. J. Antimicrob. Chemother. 2017;72:3334–3341. doi: 10.1093/jac/dkx326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MGB Biopharma Limited. Study to Assess Safety, Tolerability and Efficacy of Incremental Doses of MGB-BP-3 in Patients With CDAD.https://clinicaltrials.gov/ct2/show/NCT03824795 (2020).
- 30.Hartley JA, et al. Pre-clinical pharmacology and mechanism of action of SG3199, the pyrrolobenzodiazepine (PBD) dimer warhead component of antibody-drug conjugate (ADC) payload tesirine. Sci. Rep. 2018;8:10479. doi: 10.1038/s41598-018-28533-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann J, et al. The discovery of a novel antibiotic for the treatment of Clostridium difficile infections: a story of an effective academic–industrial partnership. Medchemcomm. 2015;6:1420–1426. doi: 10.1039/C5MD00238A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization. 2019 Antibacterial Agents in Clinical Development: an Analysis of the Antibacterial Clinical Development Pipeline (World Health Organization, 2020).
- 33.Bucevičius J, Lukinavičius G, Gerasimaitė R. The use of Hoechst dyes for DNA staining and beyond. Chemosensors. 2018;6:18. doi: 10.3390/chemosensors6020018. [DOI] [Google Scholar]
- 34.Baraldi PG, et al. DNA minor groove binders as potential antitumor and antimicrobial agents. Med. Res. Rev. 2004;24:475–528. doi: 10.1002/med.20000. [DOI] [PubMed] [Google Scholar]
- 35.Bontemps J, Houssier C, Fredericq E. Physico-chemical study of the complexes of “33258 Hoechst” with DNA and nucleohistone. Nucleic Acids Res. 1975;2:971–984. doi: 10.1093/nar/2.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisblum B, Haenssler E. Fluorometric properties of the bibenzimidazole derivative hoechst 33258, a fluorescent probe specific for AT concentration in chromosomal DNA. Chromosoma. 1974;46:255–260. doi: 10.1007/BF00284881. [DOI] [PubMed] [Google Scholar]
- 37.Goracci L, Germani R, Savelli G, Bassani DM. Hoechst 33258 as a pH-sensitive probe to study the interaction of Amine oxide surfactants with DNA. ChemBioChem. 2005;6:197–203. doi: 10.1002/cbic.200400196. [DOI] [PubMed] [Google Scholar]
- 38.Downs TR, Wilfinger WW. Fluorometric quantification of DNA in cells and tissue. Anal. Biochem. 1983;131:538–547. doi: 10.1016/0003-2697(83)90212-9. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro AB, Ling V. Extraction of Hoechst 33342 from the cytoplasmic leaflet of the plasma membrane by P-glycoprotein. Eur. J. Biochem. / FEBS. 1997;250:122–129. doi: 10.1111/j.1432-1033.1997.00122.x. [DOI] [PubMed] [Google Scholar]
- 40.Westfall DA, et al. Bifurcation kinetics of drug uptake by Gram-negative bacteria. PloS one. 2017;12:e0184671. doi: 10.1371/journal.pone.0184671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marceau AH, et al. Structure of the SSB–DNA polymerase III interface and its role in DNA replication. EMBO J. 2011;30:4236–4247. doi: 10.1038/emboj.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breusegem, S. Y., Loontiens, F. G., Regenfuss, P. & Clegg, R. M. in Methods in Enzymology Vol 340 (Abelson, J. & Simon, M.) 212–233 (Academic Press, 2001). [DOI] [PubMed]
- 43.Breusegem SY, Clegg RM, Loontiens FG. Base-sequence specificity of Hoechst 33258 and DAPI binding to five (A/T)4 DNA sites with kinetic evidence for more than one high-affinity Hoechst 33258-AATT complex11Edited by I. Tinoco. J. Mol. Biol. 2002;315:1049–1061. doi: 10.1006/jmbi.2001.5301. [DOI] [PubMed] [Google Scholar]
- 44.Copeland RA, Pompliano DL, Meek TD. Drug–target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 45.Swinney DC. The role of binding kinetics in therapeutically useful drug action. Curr. Opin. Drug Disco. Dev. 2009;12:31–39. [PubMed] [Google Scholar]
- 46.Vauquelin G, Charlton SJ. Long-lasting target binding and rebinding as mechanisms to prolong in vivo drug action. Br. J. Pharm. 2010;161:488–508. doi: 10.1111/j.1476-5381.2010.00936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finkelstein IJ, Greene EC. Molecular traffic jams on DNA. Annu. Rev. Biophys. 2013;42:241–263. doi: 10.1146/annurev-biophys-083012-130304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gelles J, Landick R. RNA polymerase as a molecular motor. Cell. 1998;93:13–16. doi: 10.1016/S0092-8674(00)81140-X. [DOI] [PubMed] [Google Scholar]
- 49.Willis IM, Moir RD. Signaling to and from the RNA polymerase III transcription and processing machinery. Annu. Rev. Biochem. 2018;87:75–100. doi: 10.1146/annurev-biochem-062917-012624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lempp M, et al. Systematic identification of metabolites controlling gene expression in E. coli. Nat. Commun. 2019;10:4463. doi: 10.1038/s41467-019-12474-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer S, Park BR, Kim M. Absolute quantitative measurement of transcriptional kinetic parameters in vivo. Nucleic Acids Res. 2016;44:e142. doi: 10.1093/nar/gkw596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schink SJ, Biselli E, Ammar C, Gerland U. Death rate of E. coli during starvation is set by maintenance cost and biomass recycling. Cell Syst. 2019;9:64–73.e63. doi: 10.1016/j.cels.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Mitrophanov AY, Groisman EA. Positive feedback in cellular control systems. BioEssays. 2008;30:542–555. doi: 10.1002/bies.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taheri-Araghi S, et al. Cell-Size control and homeostasis in bacteria. Curr. Biol. 2015;25:385–391. doi: 10.1016/j.cub.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Heerden JH, et al. Statistics and simulation of growth of single bacterial cells: illustrations with B. subtilis and E. coli. Sci. Rep. 2017;7:16094. doi: 10.1038/s41598-017-15895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu SY, Park GY, Kim SH, Hulme J, An SS. Diminazene aceturate: an antibacterial agent for Shiga-toxin-producing Escherichia coli O157:H7. Drug Des., Dev. Ther. 2016;10:3363–3378. doi: 10.2147/DDDT.S114832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khalaf AI, et al. Synthesis and antimicrobial activity of some netropsin analogues. Org. Biomol. Chem. 2004;2:3119–3127. doi: 10.1039/b408386p. [DOI] [PubMed] [Google Scholar]
- 58.Chung J-h, Bhat A, Kim C-J, Yong D, Ryu C-M. Combination therapy with polymyxin B and netropsin against clinical isolates of multidrug-resistant Acinetobacter baumannii. Sci. Rep. 2016;6:28168. doi: 10.1038/srep28168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tonge PJ. Drug–target kinetics in drug discovery. ACS Chem. Neurosci. 2018;9:29–39. doi: 10.1021/acschemneuro.7b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svetlov MS, Vázquez-Laslop N, Mankin AS. Kinetics of drug–ribosome interactions defines the cidality of macrolide antibiotics. Proc. Natl Acad. Sci. USA. 2017;114:13673–13678. doi: 10.1073/pnas.1717168115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coates J, et al. Antibiotic-induced population fluctuations and stochastic clearance of bacteria. eLife. 2018;7:e32976. doi: 10.7554/eLife.32976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.El Meouche I, Siu Y, Dunlop MJ. Stochastic expression of a multiple antibiotic resistance activator confers transient resistance in single cells. Sci. Rep. 2016;6:19538. doi: 10.1038/srep19538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alexander HK, MacLean RC. Stochastic bacterial population dynamics restrict the establishment of antibiotic resistance from single cells. Proc. Natl Acad. Sci. USA. 2020;117:19455–19464. doi: 10.1073/pnas.1919672117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deris B, et al. The innate growth bistability of antibiotic resistant bacteria. Science. 2013;342:1237435–1237431. doi: 10.1126/science.1237435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Angermayr SA, et al. Growth-mediated negative feedback shapes quantitative antibiotic response. Mol. Syst. Biol. 2022;18:e10490. doi: 10.15252/msb.202110490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.El Meouche I, Dunlop MJ. Heterogeneity in efflux pump expression predisposes antibiotic-resistant cells to mutation. Science. 2018;362:686–690. doi: 10.1126/science.aar7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown, S. D. & Jun, S. Complete genome sequence of Escherichia coli NCM3722. Genome Announc.3, e00879–15 (2015). [DOI] [PMC free article] [PubMed]
- 68.Lyons E, Freeling M, Kustu S, Inwood W. Using genomic sequencing for classical genetics in E. coli K12. PloS one. 2011;6:e16717. doi: 10.1371/journal.pone.0016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soupene E, et al. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 2003;185:5611–5626. doi: 10.1128/JB.185.18.5611-5626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iyer S, Le D, Park BR, Kim M. Distinct mechanisms coordinate transcription and translation under carbon and nitrogen starvation in Escherichia coli. Nat. Microbiol. 2018;3:741–748. doi: 10.1038/s41564-018-0161-3. [DOI] [PubMed] [Google Scholar]
- 72.Le D, et al. Active efflux leads to heterogeneous dissipation of proton motive force by protonophores in bacteria. mBio. 2021;12:e00676–00621. doi: 10.1128/mBio.00676-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smale ST. Beta-galactosidase assay. Cold Spring Harb. Protoc. 2010;2010:pdb.prot5423. doi: 10.1101/pdb.prot5423. [DOI] [PubMed] [Google Scholar]
- 74.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The datasets are provided as Supplementary Data.