Abstract
Objective
To investigate the distribution and drug resistance of pathogens among hospitalized patients in the respiratory unit during the COVID-19 pandemic, analyze the risk factors of drug resistance, construct a risk prediction model.
Methods
This study isolated 791 strains from 489 patients admitted to the Affiliated Hospital of Chengdu University, who were retrospectively enrolled between December 2019 and June 2021. The patients were divided into training and validation sets based on a random number table method (8:2). The baseline information, clinical characteristics, and culture results were collected using an electronic database and WHONET 5.6 software and compared between the two groups. A risk prediction model for drug-resistant bacteria was constructed using multi-factor logistic regression.
Results
K. pneumoniae (24.78%), P. aeruginosa (17.19%), A. baumannii (10.37%), and E. coli (10.37%) were the most abundant bacterial isolates. 174 isolates of drug-resistant bacteria were collected, ie, Carbapenem-resistant organism-strains, ESBL-producing strains, methicillin-resistant S. aureus, multi-drug resistance constituting 38.51%, 50.57%, 6.32%, 4.60%, respectively. The nosocomial infection prediction model of drug-resistant bacteria was developed based on the combined use of antimicrobials, pharmacological immunosuppression, PCT>0.5 ng/mL, CKD stage 4–5, indwelling catheter, and age > 60 years. The AUC under the ROC curve of the training and validation sets were 0.768 (95% CI: 0.624–0.817) and 0.753 (95% CI: 0.657–0.785), respectively. Our model revealed an acceptable prediction demonstrated by a non-significant Hosmer-Lemeshow test (training set, p=0.54; validation set, p=0.88).
Conclusion
K. pneumoniae, P. aeruginosa, A. baumannii, and E. coli were the most abundant bacterial isolates. Antimicrobial resistance among the common isolates was high for most routinely used antimicrobials and carbapenems. COVID-19 did not increase the drug resistance pressure of the main strains. The risk prediction model of drug-resistant bacterial infection is expected to improve the prevention and control of antibacterial-resistant bacterial infection in hospital settings.
Keywords: COVID-19, respiratory and critical care medicine, drug-resistant bacteria, risk factors, line chart
Introduction
Antimicrobial resistance (AMR) has become more prevalent in recent years due to the widespread use of antibiotics in clinical practice. Drug-resistant bacterial infection significantly increases the mortality rate, hospitalization time, and treatment cost, which increases with age.1 By 2050, the number of bacterial infections is expected to rise to 10 million, indicating that bacterial resistance is a serious threat to global health.2 Among these, nosocomial infections caused by Gram-negative bacilli (GNB) are the most challenging issue for dealing with AMR.3
Lower respiratory tract infections (LRTIs) are common infections across the globe and a major cause of illness and death targeting people of all ages.4 Antimicrobials are commonly used to treat LRTIs and drug-resistant bacteria are commonly detected in respiratory departments. In line with the standard construction of the national respiratory and critical medicine department in China, large tertiary hospitals are equipped with respiratory and critical medicine departments and independent respiratory intensive care unit (RICU) wards. Patients in RICU usually have severe infections that necessitate the use of high-grade antimicrobials; as a result, the diagnostic and treatment environment for patients has improved due to increased detection of drug-resistant bacteria.5
The Coronavirus Disease 2019 (COVID-19) infection has affected people globally, and the number of cases continues to increase. Studies indicate that the probability of bacterial and fungal infections occurring in patients with COVID-19 infection is extremely low.6–8 Patients hospitalized with COVID-19 typically present with pneumonia with fever, cough, and dyspnoea, similar to atypical bacterial pneumonia.8 Since it is difficult to distinguish them from hospital-acquired pneumonia and ventilator-associated pneumonia, several COVID-19 patients are often administered antimicrobials, which increases the burden of AMR.6,7,9,10 Several studies have shown that the COVID-19 pandemic has further complicated the era of AMR.11,12 During the pandemic, several variants of SARS-CoV-2 with varying transmissibility and severity have emerged. Among these variants, omicron is currently the most prevalent lineage globally, with lower pathogenicity.13 At present, the spread of COVID-19 has peaked in China, and many patients with severe and critical COVID-19 infections are visiting the respiratory general wards or RICU.14 Noteworthy, it is important to appropriately use antimicrobials to prevent aggravating the already unpromising forms of AMR.
This study aims to provide clinical evidence for the management of antibacterial-resistant bacterial infection by analyzing the clinical risk factors of drug-resistant bacteria infection in hospitalized respiratory patients during the COVID-19 pandemic. We will also construct and validate the risk prediction model for drug-resistant bacteria infection.
Materials and Methods
Study Design and Source of Strains
This is a retrospective study conducted between December 2019 and June 2021, at the Affiliated Hospital of Chengdu University in the southwest, China. The hospital is one of the large tertiary general teaching hospitals in southwest China with over 1000 bed capacity. The respiratory medical unit is equipped with over 120 beds. Data on all culture samples were collected from the respiratory and critical care medicine department of the hospital. The samples included valid sputum or tracheal aspirate, urine, blood, pleural fluids, pus swabs, ascitic fluid, feces, and central vein catheter specimens. We implemented the standard operating procedures of sample collection, storage, and transport. Moreover, hospitalized patients above 18 years with complete data were included. Negative and non-bacterial as well as positive cultures but with colonization were excluded. We also excluded patients with hospitalization time<48 hours and incomplete information. Only the first infection was included if the patient had repetitive infections. Eventually, 791 strains from 489 patients were included. General patient information and data on clinical parameters including age, gender, blood count, biochemistry, inflammatory indicators, disease history, types of samples collected, culture results, and antimicrobial sensitivity were collected using the electronic medical record system and WHONET 5.6 software. Subsequently, we analyzed strain susceptibility to antimicrobial agents. The patients were divided into training and validation sets using the random number table method (8:2). A risk prediction model of drug-resistant bacterial infection was constructed using the training set and validated in a validation set. Age, gender, admission to RICU, invasive operation, combined use of antimicrobials, underlying diseases, and outcomes of each group were analyzed. Quality control strains included E. coli ATCC 25922, P. aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 25923. Specimen collection and inspection followed the Clinical Microbiological Specimen Collection and Inspection Guidelines.15
This study was approved by the institutional ethics committee of the Affiliated Hospital of Chengdu University. We confirm that all methods were performed in line with the relevant guidelines and regulations. Since this was a retrospective data review, the need for informed consent from patients (Ethics Committee of Affiliated Hospital of Chengdu University, PJ2020-021-03) was waived.
Sample Processing, Bacterial Identification, and Antimicrobial Susceptibility Testing
The sputum and sterile samples were placed on chocolate, blood, and MacConkey agar plates and incubated in the culture media, in a 5% CO2 at 35°C for 48–72 hours, depending on the specimens. The growth of the medium was manually checked daily. Blood samples were inoculated into blood culture bottles and were incubated in the BD BACTEC FX system (Becton, Dickinson, and Company) at 37°C for 5 days. MicroScan WalkAway 40 automatic microbial identification instrument and drug susceptibility system (Beckman Coulter, Brea, California, USA) were used for simultaneous strain identification and drug susceptibility testing via negative Combo Panel Type 61 and positive Combo Panel Type 11. A microdilution method was used to evaluate the drug sensitivity and establish the minimum inhibitory concentration (MIC). This method uses a small volume of broth to disperse into a round or pointed bottom sterile plastic micro-dilution plate, called micro-dilution. Each hole should contain 0.1mL broth. The bacterial solution was diluted to 0.5 mc, before diluting 1000 times. After sealing, the bacterial solution was placed into a 35 °C ordinary air incubator and incubated for 16~20h, before reading the results. Haemophilus and Streptococcus were detected via incubation for 20–24 hours. Drug sensitivity of Staphylococcus and Enterococcus to oxacillin and vancomycin was evaluated by incubation for 24 hours. The MIC is the minimum drug concentration that can completely suppress bacterial growth in the small hole. It is only meaningful when the bacterial growth test is significant in the positive control hole (without antimicrobial). The maximum drug concentration that inhibits bacterial growth should be recorded when a single hole jumping occurs. The outcomes should not be reported and the test should be repeated in case of multiple hole jumps. The positive and negative drug-sensitive composite plates contain 16 and 20 antibacterial drugs respectively (Reach Surgical (Beijing), Inc.). Results were reported consistent with the Clinical Laboratory Standards Institute Criteria (CLSI) 2021.16
Definitions
Valid sputum and bronchoalveolar lavage fluid: squamous cells < 10/LP and leukocytes > 25/LP were observed microscopically. Extended-Spectrum β-Lactamases (ESBLs), mediated by plasmids, can cause resistance to many β-lactamide antimicrobials. It is an important mechanism of antibiotic resistance in Gram-negative bacteria. ESBL-producing bacteria can develop resistance to various antimicrobials such as penicillins, penicillins, broad spectrum-cephalosporins, and monobactams. The isolates of carbapenem-resistant Enterobacteriaceae (CREs), P. aeruginosa, and A. baumannii were defined as gram-negative bacteria resistant or non-susceptible to one or more carbapenems based on CLSI current breakpoint. Multidrug resistance (MDR) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories (carbapenems, β-lactamase inhibitor combinations, cephalosporins, aminoglycosides, and fluoroquinolones).17 All these mono-microbial infections and polymicrobial infections were included.
Statistical Analysis
Continuous and categorical variables were presented as median (interquartile range (IQR)) and absolute number (percentage), respectively. The risk prediction model of drug-resistant bacterial infection was constructed using a multi-factor logistic regression. A line diagram was determined using the regression coefficient of the model. The model was repeated for 1000 bootstrap self-sampling. Receiver operating characteristic (ROC), area under the curve (AUC), and calibration curve were used to evaluate the discrimination and calibration performance of the model in the training and validation sets. The significance level was set at P < 0.05. The differences between patients with drug-resistant bacteria infections and those without infections were compared using χ2. All statistical analyses were performed using SPSS 24.0. WHONET 5.6 was used to analyze basic information about the strains. The prediction model was constructed and confirmed by R 3.6.3.
Results
Distribution and Constituent Ratio of Pathogens
A total of 5445 specimens were collected, out of which 9 samples were excluded due to incomplete information. The final samples analyzed were 5436; nearly 70% of the patient samples included were male. More than two-thirds (75.26%) of patients aged above 60 years. We excluded negative cultures, non-bacterial cultures, and repeated strains from the same sample of the same patient. Finally, only 791 strains from 489 patients were included in the analysis. The isolation rates of gram-positive and gram-negative bacteria were 12.5% (99/791) and 87.5% (692/791), respectively. K. pneumoniae > P. aeruginosa > A. baumannii = E. coli were the top four isolated strains (isolation rates; 24.78%, 17.19%, 10.37%, and 10.37%, respectively) (Table 1).
Table 1.
Distribution of Pathogens
| Name of Bacteria | Number of Strains | Proportion |
|---|---|---|
| Klebsiella pneumoniae | 196 | 24.78% |
| Pseudomonas aeruginosa | 136 | 17.19% |
| Acinetobacter baumannii | 82 | 10.37% |
| Escherichia coli | 82 | 10.37% |
| Enterococcus faecium | 35 | 4.42% |
| Enterobacter cloacae | 34 | 4.30% |
| Staphylococcus aureus | 31 | 3.92% |
| Stenotrophomonas maltophilia | 25 | 3.16% |
| Enterobacter aerogenes | 24 | 3.03% |
| Serratia marcescens | 22 | 2.78% |
| Proteus mirabilis | 11 | 1.39% |
| Haemophilus influenzae | 11 | 1.39% |
| Klebsiella Oxytoca | 9 | 1.14% |
| Others | 93 | 11.76% |
| Total | 791 | 100% |
Detection of Drug-Resistant Bacteria
A total of 174 isolates of drug-resistant bacteria (174/791, 22.00%) were collected from sputum or tracheal aspirate (n = 136, 78%), urine (n = 25, 14.37%), blood (n = 6, 3.45%), pleural fluids (n = 3, 1.72%), and other specimens (n = 4, 2.30%). The detection rates of carbapenem-resistant A. baumannii, P. aeruginosa, and K. pneumoniae were 43.90%, 16.91%, and 3.57%, respectively. Carbapenem-resistant organism (CRO)-strains, ESBL-producing strains, methicillin-resistant Staphylococcus aureus (MRSA), P. aeruginosa (MDR), A. baumannii (MDR) were 67 (38.51%), 88 (50.57%), 11 (6.32%), 6 (3.45%), and 2 (1.15%) cases, respectively (Table 2). Furthermore, ESBL-producing E. coli (53/174, 30.46%) and K. pneumoniae (28/174, 16.09%), carbapenem-resistant A. baumannii (CR-Ab) (34/179, 19.54%), carbapenem-resistant P. aeruginosa (CR-Pa) (17/174, 9.77%), K. pneumoniae (CR-Kp) (7/174, 4.02%), and MRSA (11/174, 6.32%) were the major isolated drug-resistant bacteria.
Table 2.
Detection of Drug Resistant Bacteria
| Bacterial Species | Number of Strains | Proportion |
|---|---|---|
| Acinetobacter baumannii-CR-Ab | 34 | 19.54% |
| Pseudomonas aeruginosa-CR-Pa | 17 | 9.77% |
| Klebsiella pneumoniae-CR-Kp | 7 | 4.02% |
| Enterobacter aerogenes-CRE | 4 | 2.30% |
| Enterobacter cloacae-CRE | 2 | 1.15% |
| E. coli-CRE | 2 | 1.15% |
| Citrobacter freundii-CRE | 1 | 0.57% |
| Pseudomonas aeruginosa-MDR | 6 | 3.45% |
| Acinetobacter baumannii-MDR | 2 | 1.15% |
| Escherichia coli-ESBL | 53 | 30.46% |
| Klebsiella pneumoniae-ESBL | 28 | 16.09% |
| Proteus mirabilis-ESBL | 7 | 4.02% |
| Staphylococcus aureus-MRSA | 11 | 6.33% |
| Total | 174 | 100% |
Analysis of Drug Resistance of Major Gram-Negative Bacteria
The resistance rates of drug-resistant gram-negative bacteria to tigecycline, cefoxitin, amikacin, ertapenem, meropenem, imipenem, cefepime, ceftazidime, and aztreonam were 0, 23.0%, 39.7%, 24.7%, 54.4%, 41.5%, 87.0%, 88.5%, and 81.6%, respectively. Furthermore, the resistance rate to ampicillin, ceftriaxone, cefotaxime, cefazolin, and cefuroxime was approximately 100%. Major gram-negative bacteria resistant to common major antimicrobials (without removing duplicate strains from similar specimens from the same patients) are shown in Supplementary Material 1.
MRSA Drug Resistance Profile
Although the resistance rate of MRSA to linezolid, vancomycin, daptomycin, quinupristin/dalfopristin, trimethoprim-Sulfa methoxazole, and furantoin was 0, its resistance rate to ampicillin, ceftriaxone, ampicillin-sulbactam, amoxicillin clavulanate potassium was 100%. In contrast, its resistance rate to levofloxacin, moxifloxacin, and ciprofloxacin was more than 50%, and its resistance rate to rifampin was less than 20%.
Patient Profile Analysis
The general information, RICU admission, invasive operations, combined antimicrobial use, underlying disease, albumin level, WBC, and PCT were not significantly different between the two groups (P > 0.05) except for active cancer (P < 0.05, validation set > training set), indicating the suitability of data for training and validation analysis. The training set was divided into drug-resistant and non-drug-resistant groups based on their susceptibility to antimicrobials. Univariate analysis showed that age, admission to RICU, invasive operations (indwelling urinary catheter, tracheal intubation/incision, PICC/CVC), combined antimicrobial use (combined respiratory quinolones, amikacin, antifungal drugs), use of carbapenem antimicrobials, combined chronic underlying diseases (chronic respiratory disease, CKD stage 4–5, chronic cardiac insufficiency, and pharmacological immunosuppression), albumin <30 g/L, leukocytes >10,000/ul, NLR, CRP, and PCT >0.5 ng/mL were significantly different between the two groups (P < 0.05) (Table 3). Notably, admission to RICU was defined as admission to the RICU > within 24 hours.
Table 3.
Clinical Data Analysis of Training Set and Validation Set
| Variant | Training Set, N = 391 | Validation Set, N = 98 | P | Training Set | P | |
|---|---|---|---|---|---|---|
| Sensitive, N = 287 | Drug-Resistance, N = 104 | |||||
| Gender | 0.248 | 0.232 | ||||
| Male | 261 (66.8) | 72 (73.5) | 197 (68.6) | 64 (61.5) | ||
| Female | 130 (33.2) | 26 (26.5) | 90 (31.4) | 40 (38.5) | ||
| Age | 0.556 | 0.001 | ||||
| ≤60 | 94 (24.0) | 27 (27.6) | 82 (28.6) | 12 (11.5) | ||
| >60 | 297 (76.0) | 71 (72.4) | 205 (71.4) | 92 (88.5) | ||
| RICU admission | 0.791 | <0.001 | ||||
| 0 | 312 (79.8) | 76 (77.6) | 246 (85.7) | 66 (63.5) | ||
| Within a week | 18 (4.6) | 4 (4.1) | 12 (4.2) | 6 (5.8) | ||
| >1 week | 61 (15.6) | 18 (18.4) | 29 (10.1) | 32 (30.8) | ||
| Invasive operations | 0.623 | <0.001 | ||||
| No | 279 (71.4) | 73 (74.5) | 225 (78.4) | 54 (51.9) | ||
| Yes | 112 (28.6) | 25 (25.5) | 62 (21.6) | 50 (48.1) | ||
| Retained urinary catheter | 0.627 | <0.001 | ||||
| No | 308 (78.8) | 80 (81.6) | 247 (86.1) | 61 (58.7) | ||
| Yes | 83 (21.2) | 18 (18.4) | 40 (13.9) | 43 (41.3) | ||
| Indwelling drainage tube | 0.809 | 0.387 | ||||
| No | 366 (93.6) | 93 (94.9) | 271 (94.4) | 95 (91.3) | ||
| Yes | 25 (6.4) | 5 (5.1) | 16 (5.6) | 9 (8.7) | ||
| Tracheal intubation/incision | 1 | 0.001 | ||||
| No | 354 (90.5) | 89 (90.8) | 269 (93.7) | 85 (81.7) | ||
| Yes | 37 (9.5) | 9 (9.2) | 18 (6.3) | 19 (18.3) | ||
| PICC/CVC | 0.603 | <0.001 | ||||
| No | 354 (90.5) | 91 (92.9) | 271 (94.4) | 83 (79.8) | ||
| Yes | 37 (9.5) | 7 (7.1) | 16 (5.6) | 21 (20.2) | ||
| Undergoing surgery | 1 | 0.495 | ||||
| No | 382 (97.7) | 96 (98.0) | 279 (97.2) | 103 (99.0) | ||
| Yes | 9 (2.3) | 2 (2.0) | 8 (2.8) | 1 (1.0) | ||
| Antibiotic co-administration | 0.324 | <0.001 | ||||
| No | 274 (70.1) | 63 (64.3) | 222 (77.4) | 52 (50.0) | ||
| Yes | 117 (29.9) | 35 (35.7) | 65 (22.6) | 52 (50.0) | ||
| Amikacin | 0.79 | 0.049 | ||||
| No | 349 (89.3) | 89 (90.8) | 262 (91.3) | 87 (83.7) | ||
| Yes | 42 (10.7) | 9 (9.2) | 25 (8.7) | 17 (16.3) | ||
| Respiratory quinolones | 0.532 | <0.001 | ||||
| No | 357 (91.3) | 92 (93.9) | 273 (95.1) | 84 (80.8) | ||
| Yes | 34 (8.7) | 6 (6.1) | 14 (4.9) | 20 (19.2) | ||
| Combined antifungal | 0.62 | <0.001 | ||||
| No | 356 (91.0) | 87 (88.8) | 272 (94.8) | 84 (80.8) | ||
| Yes | 35 (9.0) | 11 (11.2) | 15 (5.2) | 20 (19.2) | ||
| Tigecycline | 0.945 | <0.001 | ||||
| No | 382 (97.7) | 95 (96.9) | 287 (100.0) | 95 (91.3) | ||
| Yes | 9 (2.3) | 3 (3.1) | 0 (0.0) | 9 (8.7) | ||
| Carbapenems | 0.789 | <0.001 | ||||
| No | 330 (84.4) | 81 (82.7) | 258 (89.9) | 72 (69.2) | ||
| Yes | 61 (15.6) | 17 (17.3) | 29 (10.1) | 32 (30.8) | ||
| Underlying Diseases | 0.243 | 0.013 | ||||
| No | 80 (20.5) | 26 (26.5) | 68 (23.7) | 12 (11.5) | ||
| Yes | 311 (79.5) | 72 (73.5) | 219 (76.3) | 92 (88.5) | ||
| Chronic respiratory diseases | 0.124 | 0.97 | ||||
| No | 183 (46.8) | 55 (56.1) | 135 (47.0) | 48 (46.2) | ||
| Yes | 208 (53.2) | 43 (43.9) | 152 (53.0) | 56 (53.8) | ||
| Dementia | 0.622 | 0.102 | ||||
| No | 333 (85.2) | 86 (87.8) | 250 (87.1) | 83 (79.8) | ||
| Yes | 58 (14.8) | 12 (12.2) | 37 (12.9) | 21 (20.2) | ||
| Pharmacological immunosuppression | 0.137 | 0.002 | ||||
| No | 344 (88.0) | 80 (81.6) | 262 (91.3) | 82 (78.8) | ||
| Yes | 47 (12.0) | 18 (18.4) | 25 (8.7) | 22 (21.2) | ||
| Active cancer | 0.022 | 0.731 | ||||
| No | 322 (82.4) | 70 (71.4) | 238 (82.9) | 84 (80.8) | ||
| Yes | 69 (17.6) | 28 (28.6) | 49 (17.1) | 20 (19.2) | ||
| Chronic cardiac insufficiency | 0.376 | <0.001 | ||||
| No | 309 (79.0) | 82 (83.7) | 248 (86.4) | 61 (58.7) | ||
| Yes | 82 (21.0) | 16 (16.3) | 39 (13.6) | 43 (41.3) | ||
| CKD stage 4–5 | 0.693 | <0.001 | ||||
| No | 373 (95.4) | 95 (96.9) | 283 (98.6) | 90 (86.5) | ||
| Yes | 18 (4.6) | 3 (3.1) | 4 (1.4) | 14 (13.5) | ||
| Diabetes | 0.859 | 0.653 | ||||
| No | 305 (78.0) | 75 (76.5) | 226 (78.7) | 79 (76.0) | ||
| Yes | 86 (22.0) | 23 (23.5) | 61 (21.3) | 25 (24.0) | ||
| WBC | 9.13 ± 5.55 | 9.56 ± 6.25 | 0.509 | 8.72 ± 5.16 | 10.27 ± 6.39 | 0.015 |
| WBC<4000/ul | 1 | 0.764 | ||||
| No | 368 (94.1) | 92 (93.9) | 269 (93.7) | 99 (95.2) | ||
| Yes | 23 (5.9) | 6 (6.1) | 18 (6.3) | 5 (4.8) | ||
| WBC>10000/ul | 0.345 | 0.001 | ||||
| No | 277 (70.8) | 64 (65.3) | 217 (75.6) | 60 (57.7) | ||
| Yes | 114 (29.2) | 34 (34.7) | 70 (24.4) | 44 (42.3) | ||
| N | 7.13 ± 5.44 | 7.56 ± 6.16 | 0.5 | 6.68 ± 5.07 | 8.39 ± 6.23 | 0.006 |
| L | 1.27 ± 0.99 | 1.21 ± 0.74 | 0.565 | 1.31 ± 1.05 | 1.17 ± 0.78 | 0.194 |
| NLR | 10.09 ± 14.77 | 9.28 ± 10.93 | 0.614 | 8.87 ± 13.12 | 13.43 ± 18.25 | 0.007 |
| CRP | 54.14 ± 69.15 | 66.50 ± 74.52 | 0.12 | 47.79 ± 65.89 | 71.66 ± 75.00 | 0.002 |
| PCT | 2.46 ± 8.42 | 4.06 ± 15.71 | 0.17 | 2.07 ± 7.79 | 3.55 ± 9.93 | 0.124 |
| PCT>0.5 ng/mL | 1 | 0.002 | ||||
| No | 243 (62.1) | 61 (62.2) | 192 (66.9) | 51 (49.0) | ||
| Yes | 148 (37.9) | 37 (37.8) | 95 (33.1) | 53 (51.0) | ||
| Serum albumin | 34.32 ± 5.66 | 33.61 ± 5.70 | 0.266 | 35.23 ± 5.57 | 31.81 ± 5.16 | <0.001 |
| Serum albumin<30g/L | 0.659 | <0.001 | ||||
| No | 302 (77.2) | 73 (74.5) | 237 (82.6) | 65 (62.5) | ||
| Yes | 89 (22.8) | 25 (25.5) | 50 (17.4) | 39 (37.5) | ||
| Die | 1 | <0.001 | ||||
| No | 337 (86.2) | 85 (86.7) | 260 (90.6) | 77 (74.0) | ||
| Yes | 54 (13.8) | 13 (13.3) | 27 (9.4) | 27 26.0) | ||
Construction of Predictive Models
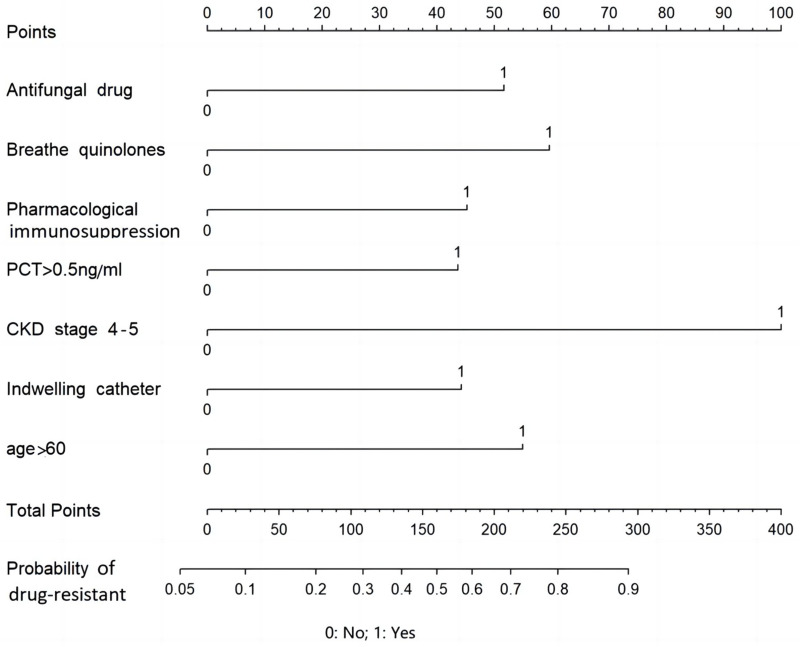
A logistic regression model was constructed based on a drug-resistant organism as the dependent variable (assignment method: no=0, yes=1), the combination of antifungal drugs or a fluoroquinolone, pharmacological immunosuppression, PCT>0.5 ng/mL, CKD stage 4–5, indwelling urinary catheter, and age>60 years as independent variables (assignment method: no=0, yes=1)(Logit (P) = -2.634 + antifungal drugs×0.850 + fluoroquinolone×0.982 + pharmacological immunosuppression×0.746 + PCT>0.5 ng/mL×0.719 + CKD stage 4–5×1.647 + indwelling urinary catheter×0.729 + age>60 years×0.905). The use of combination antifungal drugs or a fluoroquinolone, pharmacological immunosuppression, PCT>0.5 ng/mL, CKD stage 4–5, indwelling urinary catheter, and age>60 years were independent risk factors for hospital-resistant bacterial infections (Table 4). The variance inflation factor (VIF) was less than 2, and no multicollinearity existed among the factors (Table 5). The corresponding predicted probability was defined as the predicted probability of patients with drug-resistant bacteria hospital infection (Figure 1).
Table 4.
Logistic Regression Analysis of Risk of Drug-Resistant Bacterial Infection
| Risk Factors | β | OR | 95% CI | P |
|---|---|---|---|---|
| Combination of antifungal drugs | 0.850 | 2.340 | 1.020~5.366 | 0.043 |
| Combination of respiratory quinolones | 0.982 | 2.670 | 1.191~6.043 | 0.017 |
| Pharmacological immunosuppression | 0.746 | 2.109 | 1.034~4.245 | 0.038 |
| PCT>0.5 ng/mL | 0.719 | 2.052 | 1.233~3.427 | 0.006 |
| CKD stage 4–5 | 1.647 | 5.191 | 1.624~20.026 | 0.009 |
| Indwelling urinary catheter | 0.729 | 2.073 | 1.132~3.763 | 0.017 |
| Age>60 years | 0.905 | 2.472 | 1.253~5.259 | 0.013 |
Table 5.
Multicollinearity Test Result
| Antifungal | Quinolones | Pharmacological Immunosuppression | PCT>0.5 ng/mL | CKD Stage 4–5 | Indwelling Urinary Catheter | Age>60 Years | |
|---|---|---|---|---|---|---|---|
| VIF | 1.086 | 1.102 | 1.047 | 1.037 | 1.025 | 1.168 | 1.020 |
Abbreviation: VIF, variance inflation factor.
Figure 1.
The nomogram for the assessment of the risk of drug-resistant bacteria infection.
Verification of Prediction Model
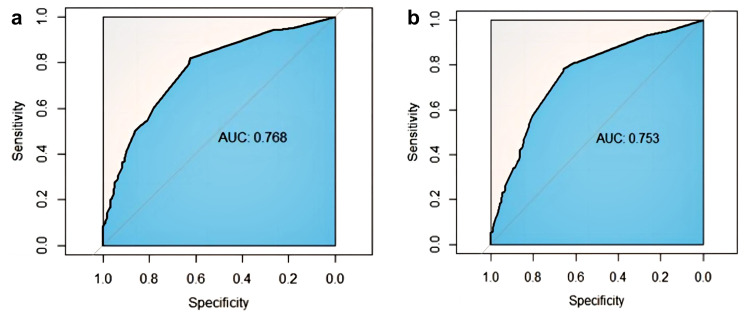
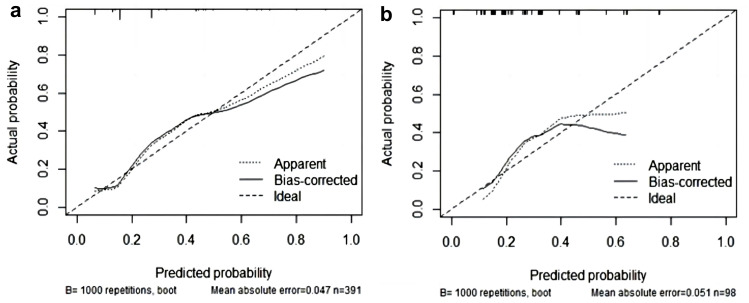
The model was internally and externally verified using the training and validation sets, respectively. All verifications were bootstrap self-sampled 1000 times. The area under the ROC curve of training and validation sets were 0.768 (95% CI: 0.624–0.817) and 0.753 (95% CI: 0.657–0.785), respectively (Figure 2). The predictive performance of the model was evaluated using a calibration curve. The prediction and observation of the model in the training and validation sets showed good consistency. Our model showed acceptable prediction, demonstrated by a non-significant Hosmer-Lemeshow test (training set, p=0.54; validation set, p=0.88) (Figures 2 and 3). In summary, the model displayed good discrimination and consistency.
Figure 2.
ROC curve of the nomogram in the training and validating sets. (a) Training set; (b) Validation set.
Figure 3.
Calibration curve of the nomogram in the training and validating sets. (a) Training set; (b) Validation set.
Discussion
This study found that more than two-thirds (75.26%) of patients were above the age of 60 years, with men accounting for 68.1%. Sputum or tracheal aspirate (78%), urine (14.37%), and blood (3.45%) were the most commonly collected samples. Moreover, the proportion of main sample types was comparable to previous findings reported by China Antimicrobial Surveillance Network (CHINET) 2020 and 2021.18 The proportion of respiratory tract samples was significantly higher than that from CHINET 202019 (38.8%) and 2021 (38.7%), respectively. Out of 5436 specimens during the study period, bacterial growth was only documented in 791 (14.66%) culture samples. The isolation rates of gram-positive and gram-negative bacteria were 12.5% and 87.5%, respectively. This was different from CHINET 2021,18 with gram-negative bacteria accounting for 71.4% and gram-positive bacteria accounting for 28.6%. Here, K. pneumoniae (24.78%), P. aeruginosa (17.19%), A. baumannii (10.37%), and E. coli (10.37%) were the most abundant bacterial isolates from inpatients, and S. aureus (3.92%) ranked 7th. However, our finding was lower than that from CHINET 202118 except E. coli (18.96%). K. pneumoniae (14.12%), P. aeruginosa (7.96%), and A. baumannii (7.28%) were the major clinical isolates in the study hospital. Furthermore, ESBL-producing E. coli (30.46%) and K. pneumoniae (16.09%), CR-Ab (19.54%), CR-Pa (9.77%), CR-Kp (4.02%), and MRSA (6.32%) were the major isolated drug-resistant bacteria. Specifically, (43.90%) of A. baumannii and (16.91%) of P. aeruginosa isolates were carbapenem-resistant, whereas only (3.57%) of K. pneumoniae and (2.43%) of E. coli were carbapenem-resistant. This was different from CHINET 2021. The detection rates of carbapenem-resistant A. baumannii, K. pneumoniae, and P. aeruginosa in Sichuan were 65.40%, 26.70%, and 18.90%, respectively, lower than the detection rates reported in 201918,20 (72.4%, 34%, and 22.4%, respectively) and higher than the detection rates of this study. This suggests that drug resistance rates vary in different regions and years. Similarly, studies in the United States and Japan revealed that carbapenem resistance rates are significantly higher for non-fermenters (>60%) than for fermenters (<10%),21 with ESBL prevalence of 64.6% in E. coli and 14.3% in K. pneumoniae. These findings show that gram-negative drug-resistant bacteria, particularly K. pneumoniae, P. aeruginosa, E. coli, and A. baumannii are the major pathogens among hospital-acquired drug-resistant infections. Several resistance mechanisms exist, including the production of ultra-broad-spectrum β- lactamases, carbapenemases, increased efflux pump activity, alteration of drug binding sites, and alteration of cell membrane permeability.22,23 ESBL production is the primary cause of drug resistance in E. coli and K. pneumonia. The high proportion of carbapenem-resistant gram-negative bacteria indicates that it is difficult to reduce carbapenem-resistant gram-negative bacterial infections. The bacterial isolation rate was different from hospital-acquired infections (HAI) in hospitalized COVID-19 patients.11,24–26 A study of 989 adult hospitalized patients with COVID-19 infections revealed that the most common isolates in COVID-19 patients were S. pneumonia (16.21%), S. aureus (16.21%), P. aeruginosa (13.51%), E. coli (9.46%), and K. pneumoniae (8.11%).24 In a retrospective study in China, the prevalence of carbapenem-resistant Enterobacteriaceae colonization increased from 6.7% in 2019 to 50% in March-April 2020.25 Li et al also discovered A. baumannii (35.8%) and K. pneumoniae (30.8%) as the most common pathogens isolated from COVID-19 inpatients (Wuhan, China), with carbapenem resistance rates of 91.2% and 75.5%, respectively.26 As demonstrated from the above studies, the overall bacterial isolation rate of patients with COVID-19 infections was low.6–8 During the irrational use of antibiotics, the drug resistance rate increased, particularly for carbapenem.6,7,9,10,25,26 The drug resistance rates in our study were generally lower than that in the above studies. Additionally, CHINET 2021 showed that the resistance rates of major strains were lower than that of 2019.18,20 These findings may indicate that COVID-19 did not increase the pressure of drug resistance of key strains in this region.
In this work, the resistance rates of drug-resistant gram-negative bacteria to tigecycline, ertapenem, and cefoxitin were 0, 24.7%, and 23.0%, respectively. Moreover, its resistance rate to carbapenems remained high, however, the resistance rate of ertapenem (24.7%) was lower than that of meropenem (54.4%) and imipenem (41.5%), possibly due to the lower rate of ertapenem use in the hospital. The resistance rates of drug-resistant gram-negative bacteria to ampicillin, ceftriaxone, cefotaxime, cefazolin, and cefuroxime were approximately 100%. Its resistance rate to aztreonam, cefepime, ceftazidime, piperacillin sodium tazobactam, and amikacin was 81.6%, 87.0%, 88.5%, 43.6%, and 39.7%, respectively. This finding might suggest that the combination of amikacin can effectively treat resistant gram-negative bacteria, whereas tigecycline can treat carbapenem-resistant gram-negative bacteria. This finding was also consistent with the other studies.27–29 The current treatments for drug-resistant gram-negative bacteria, particularly CRO and MDR have limitations in clinical settings. Of note, polymyxin, tigecycline, and ceftazidime-avibactam have advantages and disadvantages.30 Therefore, it is important to develop an individualized treatment based on the distribution characteristics of drug-resistant bacteria in the region and the actual situation of patients.3,18,20 Drug resistance varies across countries and regions. As a consequence, an early understanding of the distribution characteristics and drug resistance of pathogenic bacteria in the region, integrated with standardized diagnosis and treatment plan for nosocomial infections, as well as adjusting the type and dose of antibacterial drugs based on the drug sensitivity results are effective strategies for reducing the production of drug-resistant bacteria, while also promoting disease treatment and control.30
Univariate analysis revealed that more than 10 factors, including admission to the RICU, tracheal intubation/incision, PICC/CVC, combined use of fluoroquinolone or antifungal drugs or aminoglycosides, use of carbapenem antimicrobials, combined chronic respiratory disease, chronic cardiac insufficiency, pharmacological immunosuppression, CKD stage 4–5, and albumin<30 g/L were associated with drug-resistant bacterial infections. Similarly, other studies indicate that current or pre-existing RICU admission, invasive operations, use of fluoroquinolone, and use of carbapenems are important risk factors for hospital-acquired drug-resistant bacterial infections.31–34 Moreover, the constructed predictive model had good discrimination, calibration, and clinical utility. Previous studies indicate that elderly patients, specifically those with pharmacological immunosuppression are at a higher risk of opportunistic hospital infections, specifically multi-drug resistant bacterial infections.35 Indwelling urinary catheters also contribute to drug resistance, increasing the risk of bacterial colonization or opportunistic infections, especially in patients in the immunosuppressed state, which was consistent with previous research.36–38 Urinary tract infections (mainly caused by K. pneumoniae) are among the most common hospital-acquired infections (HAI), causing severe bloodstream infections.38,39 The above-mentioned studies are consistent with our findings.31–33,35,39 Most patients in the general respiratory ward, particularly those admitted to the RICU, are often characterized by advanced age, need for mechanical ventilation support, immunocompromised, and combined structural lung disease, and are prone to co-infection with fungal infections.40,41 Excessive antifungal drug exposure may alter the composition of the flora, whereas the combination of antimicrobial drugs may promote resistance gene mutations, thereby causing bacterial resistance.42 Ghannoum et al also supported our research.43 Nonetheless, the specific mechanisms remain unknown and additional studies are necessary for clarification. This study also revealed that combined fluoroquinolone or antifungal drugs are independent risk factors for hospital-acquired drug-resistance infections, consistent with a meta-analysis of HAI in CRKP.44 Another meta-analysis for P. aeruginosa also showed that antimicrobial overuse, particularly quinolones, can significantly cause P. aeruginosa resistance.45 Patients with chronic renal failure often have immune system deficiencies.46 Moreover, patient dialysis often requires frequent infusions, injections, and indwelling catheters, which can easily cause HAI.47 The absorption, distribution, metabolism, and elimination processes of drugs in these patients change.48 Therefore, antimicrobial doses and usage should be adjusted based on the concentration-time curve of drugs because irrational use can result in drug-resistant bacteria.49 PCT has been used as a reference for antimicrobial use in clinical settings.50 Antimicrobials are highly recommended for patients with PCT>0.5 ng/Ml.51 Our study revealed that PCT>0.5ng/mL is also an independent risk factor for drug-resistant bacterial infection. This finding was consistent with research conducted in Italy.52
The COVID-19 pandemic presents new challenges in the operation and management of healthcare systems across the globe. For instance, respiratory clinicians cannot precisely identify viral and bacterial infections, provide timely and adequate empirical treatment for patients at high risk of drug resistance, and safely prevent overuse of antimicrobial overuse in patients without identifiable risk factors.
However, this study has limitations: First, the data were obtained from the Department of Respiratory and Critical Care Medicine of the same tertiary care teaching hospital. Therefore, the clinical applicability of the model should be further verified using data from different multiple centers. Secondly, this is a single retrospective study with limited sample size, potentially resulting in bias.
Conclusion
In conclusion, we analyzed the distribution of bacteria and drug resistance in respiratory inpatients during the COVID-19 pandemic and established a risk prediction model for HAI with drug-resistant bacteria in hospitalized patients. Consequently, we found that K. pneumoniae, P. aeruginosa, A. baumannii, and E. coli were the most abundant bacterial isolates in nosocomial infection. Besides, antimicrobial resistance among common isolates was high for most routinely used antimicrobials and carbapenems. COVID-19 did not increase the drug resistance pressure of the major strains. The combined use of antifungal, fluoroquinolone, indwelling catheter, chronic renal failure, and age > 60 years were the independent risk factors of drug-resistant bacterial infection. The model showed discrimination, consistency, and clinical utility and could be used for hospital-associated infection prevention and control.
Acknowledgments
We acknowledge the contribution of the hospital healthcare workers who participated in this study.
Funding Statement
No specific funding was received for this study.
Abbreviations
RICU, Respiratory intensive care unit; PICC, peripheral inserted central catheter; CVC, Central venous catheter; CKD, Chronic Kidney Disease; WBC, White Blood Cells; N, Neutrophile granulocyte; L, Lymphocyte; NLR, Neutrophil To Lymphocyte Ratio; CRP, C-reactive protein; PCT, Procalcitonin.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
This study has been approved by the ethical committee of Affiliated Hospital of Chengdu University. This clinical study will follow the relevant provisions of the Helsinki Declaration of the World Medical Congress and the Measures for Ethical Review of Biomedical Research Involving Human Beings issued by the National Health and Family Planning Commission of the People’s Republic of China. The research only uses the previous medical record information and has removed the relevant personal information of the subject, which will not cause risks to the subject and will not have adverse effects on the rights and health of the subject, so the application for exemption of informed consent is made. We will make every effort to protect the privacy and personal information of the subject’s personal medical data within the scope permitted by law.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interest.
References
- 1.Cassini A, Hogberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L, Yang Y, Zheng C, et al. Activities of eravacycline, tedizolid, norvancomycin, nemonoxacin, ceftaroline, and comparators against 1871 Staphylococcus and 1068 Enterococcus species isolates from china: updated report of the CHINET study 2019. Microbiol Spectr. 2022;10(6):e0171522. doi: 10.1128/spectrum.01715-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliveira J, Reygaert WC. Gram Negative Bacteria. Treasure Island (FL): StatPearls; 2022. [PubMed] [Google Scholar]
- 4.Troeger C, Forouzanfar M, Rao PC. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the global burden of disease study 2015. Lancet Infect Dis. 2017;17(11):1133–1161. doi: 10.1016/S1473-3099(17)30396-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan N, Du J, Huang C, Li H. Microbial distribution and antibiotic susceptibility of lower respiratory tract infections patients from pediatric ward, adult respiratory ward, and respiratory intensive care unit. Front Microbiol. 2020;11:1480. doi: 10.3389/fmicb.2020.01480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawson TM, Wilson RC, Holmes A. Understanding the role of bacterial and fungal infection in COVID-19. Clin Microbiol Infect. 2021;27(1):9–11. doi: 10.1016/j.cmi.2020.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai CC, Chen SY, Ko WC, Hsueh PR. Increased antimicrobial resistance during the COVID-19 pandemic. Int J Antimicrob Agents. 2021;57(4):106324. doi: 10.1016/j.ijantimicag.2021.106324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak R, Prost K, Yip L, Williams V, Leis JA, Mubareka S. Severity of coronavirus respiratory tract infections in adults admitted to acute care in Toronto, Ontario. J Clin Virol. 2020;126:104338. doi: 10.1016/j.jcv.2020.104338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71(15):769–777. doi: 10.1093/cid/ciaa272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawson TM, Moore LSP, Zhu N, et al. Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;71(9):2459–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nieuwlaat R, Mbuagbaw L, Mertz D, et al. Coronavirus disease 2019 and antimicrobial resistance: parallel and interacting health emergencies. Clin Infect Dis. 2021;72(9):1657–1659. doi: 10.1093/cid/ciaa773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyberg T, Ferguson NM, Nash SG, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022;399(10332):1303–1312. doi: 10.1016/S0140-6736(22)00462-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prevention CCfDCa. The situation of the novel coronavirus infection in China; 2023. Available from: https://www.chinacdc.cn/jkzt/crb/zl/szkb_11803/jszl_13141/202301/t20230115_263381.html. Accessed January 15, 2023.
- 15.Healthcare-associated Infection Control Branch of Chinese Preventive Medicine Association. Guidelines for collection and submission of clinical microbiological specimens. Chin J Nosocomiology. 2018;28(20):3192–3200. [Google Scholar]
- 16.Humphries R, Bobenchik AM, Hindler JA, Schuetz AN, McAdam AJ. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J Clin Microbiol. 2021;59(12):e0021321. doi: 10.1128/JCM.00213-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 18.Network CAS. China antimicrobial surveillance network; 2021. Available from: http://www.chinets.com/Data/AntibioticDrugFast. Accessed February 15, 2023.
- 19.Hu F. CHINET surveillance of bacterial resistance: results of 2020. Chin J Infect Chemother. 2021;21(04):377–387. [Google Scholar]
- 20.Hu F. CHINET surveillance of bacterial resistance across tertiary hospitals in 2019. Chin J Infect Chemother. 2020;20(3):233–243. [Google Scholar]
- 21.Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin Infect Dis. 2019;69(Suppl 7):S521–S528. doi: 10.1093/cid/ciz824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerceo E, Deitelzweig SB, Sherman BM, Amin AN. Multidrug-resistant gram-negative bacterial infections in the hospital setting: overview, implications for clinical practice, and emerging treatment options. Microb Drug Resist. 2016;22(5):412–431. doi: 10.1089/mdr.2015.0220 [DOI] [PubMed] [Google Scholar]
- 23.Morris S, Cerceo E. Trends, epidemiology, and management of multi-drug resistant gram-negative bacterial infections in the hospitalized setting. Antibiotics. 2020;9(4):196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Vidal C, Sanjuan G, Moreno-Garcia E, et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect. 2021;27(1):83–88. doi: 10.1016/j.cmi.2020.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiri B, Sensi E, Marsiliani V, et al. Antimicrobial stewardship program, COVID-19, and infection control: spread of carbapenem-resistant Klebsiella pneumoniae colonization in ICU COVID-19 Patients. What Did Not Work? J Clin Med. 2020;9(9):2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Wang J, Yang Y, et al. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control. 2020;9(1):153. doi: 10.1186/s13756-020-00819-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fritzenwanker M, Imirzalioglu C, Herold S, Wagenlehner FM, Zimmer KP, Chakraborty T. Treatment options for carbapenem- resistant gram-negative infections. Dtsch Arztebl Int. 2018;115(20–21):345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Y, Zhang Q, Wen L, Chen J, Van Tyne D. Combined effect of polymyxin B and tigecycline to overcome heteroresistance in carbapenem-resistant Klebsiella pneumoniae. Microbiol Spectr. 2021;9(2):e0015221. doi: 10.1128/Spectrum.00152-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez MS, Tolmasky ME. Amikacin: uses, resistance, and prospects for inhibition. Molecules. 2017;22(12):2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jean SS, Harnod D, Hsueh PR. Global threat of carbapenem-resistant gram-negative bacteria. Front Cell Infect Microbiol. 2022;12:823684. doi: 10.3389/fcimb.2022.823684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boral B, Unaldi O, Ergin A, Durmaz R, Eser OK, Acinetobacter Study G. A prospective multicenter study on the evaluation of antimicrobial resistance and molecular epidemiology of multidrug-resistant Acinetobacter baumannii infections in intensive care units with clinical and environmental features. Ann Clin Microbiol Antimicrob. 2019;18(1):19. doi: 10.1186/s12941-019-0319-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Qin RR, Huang L, Sun LY. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection and mortality of Klebsiella pneumoniae Infection. Chin Med J. 2018;131(1):56–62. doi: 10.4103/0366-6999.221267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alhussain FA, Yenugadhati N, Al Eidan FA, Al Johani S, Badri M. Risk factors, antimicrobial susceptibility pattern and patient outcomes of Pseudomonas aeruginosa infection: a matched case-control study. J Infect Public Health. 2021;14(1):152–157. doi: 10.1016/j.jiph.2020.11.010 [DOI] [PubMed] [Google Scholar]
- 34.Palavutitotai N, Jitmuang A, Tongsai S, Kiratisin P, Angkasekwinai N, Shafer WM. Epidemiology and risk factors of extensively drug-resistant Pseudomonas aeruginosa infections. PLoS One. 2018;13(2):e0193431. doi: 10.1371/journal.pone.0193431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Guo R, Xie B, et al. Drug resistance of pathogens causing nosocomial infection in orthopedics from 2012 to 2017: a 6-year retrospective study. J Orthop Surg Res. 2021;16(1):100. doi: 10.1186/s13018-021-02234-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foxman B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am. 2014;28(1):1–13. doi: 10.1016/j.idc.2013.09.003 [DOI] [PubMed] [Google Scholar]
- 37.Feifer A, Corcos J. Contemporary role of suprapubic cystostomy in treatment of neuropathic bladder dysfunction in spinal cord injured patients. Neurourol Urodyn. 2008;27(6):475–479. doi: 10.1002/nau.20569 [DOI] [PubMed] [Google Scholar]
- 38.Sedor J, Mulholland SG. Hospital-acquired urinary tract infections associated with the indwelling catheter. Urol Clin North Am. 1999;26(4):821–828. doi: 10.1016/S0094-0143(05)70222-6 [DOI] [PubMed] [Google Scholar]
- 39.Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi: 10.3389/fcimb.2018.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chabi ML, Goracci A, Roche N, Paugam A, Lupo A, Revel MP. Pulmonary aspergillosis. Diagn Interv Imaging. 2015;96(5):435–442. doi: 10.1016/j.diii.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 41.Tiew PY, Thng KX, Chotirmall SH. Clinical Aspergillus signatures in COPD and bronchiectasis. J Fungi. 2022;8(5):480. doi: 10.3390/jof8050480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laws M, Shaaban A, Rahman KM. Antibiotic resistance breakers: current approaches and future directions. FEMS Microbiol Rev. 2019;43(5):490–516. doi: 10.1093/femsre/fuz014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12(4):501–517. doi: 10.1128/CMR.12.4.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu P, Li X, Luo M, et al. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection: a meta-analysis. Microb Drug Resist. 2018;24(2):190–198. doi: 10.1089/mdr.2017.0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raman G, Avendano EE, Chan J, Merchant S, Puzniak L. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2018;7:79. doi: 10.1186/s13756-018-0370-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Betjes MG, Meijers RW, Litjens NH. Loss of renal function causes premature aging of the immune system. Blood Purif. 2013;36(3–4):173–178. doi: 10.1159/000356084 [DOI] [PubMed] [Google Scholar]
- 47.Poinen K, Quinn RR, Clarke A, et al. Complications from tunneled hemodialysis catheters: a Canadian observational cohort study. Am J Kidney Dis. 2019;73(4):467–475. doi: 10.1053/j.ajkd.2018.10.014 [DOI] [PubMed] [Google Scholar]
- 48.Li L, Li X, Xia Y, et al. Recommendation of antimicrobial dosing optimization during continuous renal replacement therapy. Front Pharmacol. 2020;11:786. doi: 10.3389/fphar.2020.00786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eyler RF, Shvets K. Clinical pharmacology of antibiotics. Clin J Am Soc Nephrol. 2019;14(7):1080–1090. doi: 10.2215/CJN.08140718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuetz P, Wirz Y, Sager R, et al. Effect of procalcitonin-guided antibiotic treatment on mortality in acute respiratory infections: a patient level meta-analysis. Lancet Infect Dis. 2018;18(1):95–107. doi: 10.1016/S1473-3099(17)30592-3 [DOI] [PubMed] [Google Scholar]
- 51.Schuetz P, Beishuizen A, Broyles M, et al. Procalcitonin (PCT)-guided antibiotic stewardship: an international experts consensus on optimized clinical use. Clin Chem Lab Med. 2019;57(9):1308–1318. doi: 10.1515/cclm-2018-1181 [DOI] [PubMed] [Google Scholar]
- 52.Huespe I, Prado E, Staneloni I, et al. Cinética de procalcitonina en infecciones causadas por bacterias multirresistentes [Kinetics of procalcitonin in infections caused by multidrug-resistant bacteria]. Medicina. 2020;80(6):599–605. Spanish. [PubMed] [Google Scholar]