Abstract
The emerging Vancomycin-resistant Enterococcus faecium (VRE-fm) is an opportunistic pathogen causing nosocomial infections. The identification of VRE-fm is important for successful prevention and control in healthcare settings. VRE-fm clinical isolates obtained from regional hospitals in northern Taiwan were characterized for antimicrobial susceptibility, virulence genes and biofilm production. Most isolates exhibited multi-drug resistance and carried the virulence genes, esp and hyl. While all isolates produce biofilms, those isolates that carried esp exhibited greater biofilm production. Isolates with different virulence gene carriages were examined for pathogenicity by using a nematode model, Caenorhabditis elegans, for determining microbial-host interactions. The survival assay showed that C. elegans was susceptible to Linezolid-resistant VRE-fm isolates with hyl. Combining the molecular epidemiological profiles regarding pathogenesis in C. elegans can serve as a guide for physicians in limiting opportunistic infections caused by VRE-fm.
Keywords: Vancomycin-resistant Enterococcus faecium, Virulence genes, Biofilm, Caenorhabditis elegans
Introduction
Enterococcus species are facultative anaerobic Gram-positive bacteria and intestinally commensal in humans. Enterococci are well known opportunistic pathogens that cause many infectious diseases, including bacteremia, urinary tract infections (UTI), endocarditis and intra-abdominal or intra-pelvic infections (Miller et al., 2020). Enterococcus faecium (E. faecium) and Enterococcus faecalis (E. faecalis) are the two most prevalent and clinically related pathogens (Michaux et al., 2020). These pathogenic strains are not the same as those colonized in healthy individuals (Lee et al., 2019; Liese et al., 2019). Hence, healthcare settings are important in the prevalence and outbreak of E. faecium infections.
Antimicrobial therapy is the most effective approach to treat bacterial infections. Increasingly, bacterial pathogens have adapted to a variety of antimicrobials. In particular, multi-drug resistant bacteria have become an urgent issue in healthcare systems (van Duin & Paterson, 2020). Compared to E. faecalis, E. faecium is intrinsically tolerant to a spectrum of antibiotics, including aminoglycosides, β-lactams, cephalosporins and sulphonamides. This property enables E. faecium to develop antibiotic resistance in healthcare facilities where antibiotic treatment is common. Since its first appearance in the 1980s, Vancomycin-resistant Enterococcus faecium (VRE-fm) has spread world-wide and has received global awareness. Due to its multi-drug resistance, the World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) have specified VRE-fm as a priority for developing new drugs (Tacconelli et al., 2018).
To establish an infection, Enterococcus spp. carry several virulence factors, including aggregation substance (asa1), gelatinase (gelE), cytolysin (cylA), enterococcal surface protein (esp) and hyaluronidase (hyl) (Gok et al., 2020). A product of asa-1 enhances adherence to cells (Afonina et al., 2018). Gelatinase promotes bacterial colonization and spreading by degrading collagen and gelatin (Pillay, Zishiri & Adeleke, 2018). Cytolysin increases enterococcal virulence and patient mortality (Pillay, Zishiri & Adeleke, 2018). Esp is associated with enhanced virulence, attachment to an abiotic surface, colonization in the urinary tract and biofilm formation (Jovanovic et al., 2018). While esp is encoded on a pathogenicity island in E. faecium and E. faecalis, hyl is specific to E. faecium (Gok et al., 2020). hyl encodes a putative glycosyl hydrolase, yet, its role in virulence is unclear (Panesso et al., 2011).
Biofilms are immobile microbial aggregates that attach to biotic and abiotic surfaces (Yin et al., 2019). The bacteria in biofilms are embedded in the extracellular matrix produced by polysaccharides, nucleic acids, lipids and protein (Powell et al., 2018). Bacteria establish persistent colonization by producing a biofilm as a shield to resist antibiotics and as an escape from the host immune system and harsh environmental factors. Biofilms are associated with 65% of bacterial infections (Rather, Gupta & Mandal, 2021b). In Enterococcus spp., biofilm formation contributes to antimicrobial resistance and virulence (Cepas et al., 2019), results in evasion of the host immune system (Rather, Gupta & Mandal, 2021b) and facilitates the presence of resistant bacteria in health care facilities (Luo et al., 2021). The relationship between biofilm production and enterococci virulence genes remains unclear (Weng, Ramli & Hamat, 2019).
Conventional studies of microbial-host interactions have been carried out in mammalian models. Due to regulatory restrictions, many alternative model organisms are available, including nematodes (Caenorhabditis elegans), fruit flies (Drosophila melanogaster), and zebrafish (Danio rerio) (Kaito et al., 2020). A free-living organism, C. elegans, has been used as an infection model for investigating the virulence of pathogens, including enterococci (Revtovich et al., 2021). In this study, C. elegans was employed for determining the virulence of VRE-fm clinical isolates with different virulence gene carriages. The molecular epidemiological profile of VRE-fm clinical isolates and its pathogenesis in C. elegans can serve as a guide for physicians in limiting opportunistic infections.
Materials and Methods
Collection, identification and antimicrobial susceptibility testing of VRE-fm clinical isolates
Sixty clinical isolates of VRE-fm were collected from urine (46), blood (7), pus (4), body fluid (2) or stool (1) from regional hospitals in northern Taiwan. All clinical isolates were cultured overnight on a blood agar plate (Creative Media Plate, New Taipei City, Taiwan) at 37 °C. A single colony of a clinical isolate was inoculated in tryptic soy broth (TSB) (Neogen, Lansing, MI, USA). The overnight culture was adjusted to McFarland 0.5 by using a densitometer. Identification and antimicrobial susceptibility testing were performed by using the BD Phoenix 100 automatic system (BD, Franklin Laker, NJ, USA). The Taipei City Hospital Institutional Review Board granted Ethical approval to carry out the study within its facilities (TCHIRB-10703123-E, Waiver of Informed Consent).
Multilocus sequence typing (MLST)
MLST is a sequence-based method for establishing the clonal relationship among bacteria (Bello Gonzalez et al., 2017). MLST of the VRE-fm clinical isolates was carried out according to previous reports (Homan et al., 2002; Kariyama et al., 2000). In brief, sequences of seven house-keeping genes specific to E. faecium, including gdh, purK, pstS, atpA, gyd, adk, ddl, were amplified by using PCR followed by electrophoresis analysis with 1.2% Agarose gel (100 V, 30 min). PCR products were purified by a commercial Kit (Favorgen, Ping-Tung, Taiwan) and sent for DNA sequencing (Mission Biotech Co., Taipei, Taiwan). The sequencing results were compared with published alleles, and sequence types (STs) were assigned using the MLST database for E. faecium (https://pubmlst.org/organisms/enterococcus-faecium/).
Virulence gene identification
Genomic DNA extraction of the VRE-fm isolates was performed based on a standard protocol (Kariyama et al., 2000). In brief, DNA was extracted from an overnight bacterial culture by heating at 95 °C for 5 min followed by centrifugation to remove the debris. All samples were subjected to amplification of the virulence genes (asa1, cylA, esp, gelE and hyl) by multiplex PCR and DNA electrophoresis as previously described (Vankerckhoven et al., 2004). The primer sequences were listed in Table S3. In brief, the PCR mixture was prepared in a total volume of 20 μl containing 2 μl of genomic DNA, 0.5 μl of forward (F) and reverse (R) primer (10 μM), 4 μl of 5X PCR Plus Master Mix II solution (Genemark, Taichung, Taiwan), and 9 μl of distilled water. Multiplex PCR was performed in a GeneAmp PCR System 2700 (Perkin-Elmer, Waltham, MA, USA.). The template was initially denatured at 95 °C for 5 min followed by 30 cycles at 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min. Final extension was set at 72 °C for 10 min. The PCR products were analyzed by 1.8% agarose gel electrophoresis and visualized by staining with fluorescent dye.
Biofilm measurements
Biofilm production of the VRE-fm isolates was measured according to a standard protocol with modification (Hashem, Abdelrahman & Aziz, 2021). A single colony of clinical isolates was inoculated in TSB supplemented with 1% sucrose and grown at 37 °C. Staphylococcus epidermidis strain ATCC35984 was used as a positive control (Manandhar et al., 2018). The overnight culture was adjusted to an optical density (OD) at 600 nm with 108 cells/mL in a 96-well culture plate and incubated at 37 °C for 48 h. The culture plate was gently washed thrice with 0.1 ml phosphate-buffered saline (PBS) followed by aspiration and air drying for 30 min (Weng, Ramli & Hamat, 2019). 0.1 ml of 0.1% crystal violet was added to the wells for 15 min, followed by washing thrice with 0.1 ml PBS. 0.2 ml of 95% ethanol was added to the wells for 20 min to dissolve the biofilm-associated dye (Igbinosa & Beshiru, 2019). The optical density of the samples was determined at 570 nm by using a SpectraMax Max ELISA reader (Molecular Devices, San Jose, CA, USA). A negative control was employed to reduce the background absorbance OD values. Since different clinical isolates had varied growth rates, the resulting bacterial cell number would be different after incubation, leading to inaccurate measurement of the biofilm mass. In order to justify the bias due to cell number, the ability to form a biofilm was expressed by using a biofilm formation parameter: (OD570 nm biofilm minus OD570 nm control)/OD600 nm (cells) (Lin, Lin & Lan, 2020). Optical density cut-off (ODc) was defined as three standard deviations above the mean OD of the negative control as described (Stepanovic et al., 2007). Each clinical isolate was classified as follows: no-biofilm detected (ND): OD ≤ ODc; weak biofilm producer: ODc < OD ≤ 2 × ODc; moderate biofilm producer: 2 × ODc < OD ≤ 4 × ODc; and strong biofilm producer: OD > 4 × ODc.
The C. elegans culture and survival assay
Wild-type C. elegans strain, N2, in their larval stage was cultured with non-pathogenic Escherichia coli OP50 as a food source. The C. elegans survival assay was performed based on a previous report (Chen et al., 2020). Briefly, staged young adult worms were transferred to Tryptic Soy Agar (TSA, Neogen, MI, USA) seeded with a bacterial lawn of Linezolid-resistant VRE-fm clinical isolates and incubated at 20 °C. E. coli OP50 were used as a control. Live or dead worms were scored every 24 h. Worms were censored if they crawled off the plate.
Statistical analysis
Statistics of virulence genes, antibiotic susceptibility and biofilm production were performed with a Chi-square test by using SPSS v. 21 software (SPSS Inc., Chicago, IL, USA). The C. elegans lifespan was analyzed by a log-rank test using GraphPad 8 software (GraphPad, San Diego, CA, USA). Statistical significance was indicated by a P value (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant P > 0.05).
Results
Antibiotics susceptibility, molecular typing and virulence gene carriages of VRE-fm clinical isolates
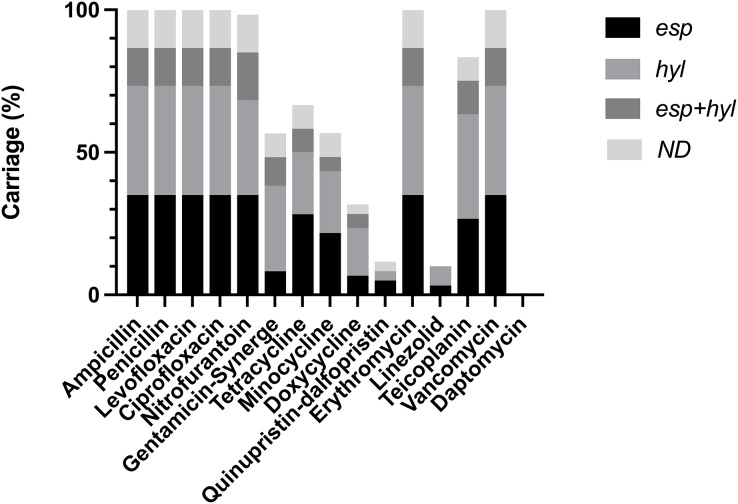
VRE-fm clinical isolates were subjected to analysis of antibiotic susceptibility (Table S1) (Fig. 1). These isolates confer resistance to the most commonly used antibiotics, while being sensitive to a few antibiotics, including Daptomycin, Doxycycline, Linezolid and Quinupristin-dalfopristin. This indicates that a majority of VRE-fm isolates exhibited multi-drug resistance, while a few isolates tolerate Linezolid. Molecular typing based on MLST revealed that three major ST types in clinical isolates, including ST17, ST78 and ST262 (Table 1). The prevalence of ST17 and ST78 is consistent with the findings in a longitudinal study of VRE-fm blood isolates in Taiwan (Kuo et al., 2018).
Figure 1. Distribution of virulence gene carriages in VRE-fm isolates with different antibiotic resistance.
ND: non-detected.
Table 1. Epidemiological profiles and pathogenicity of Vancomycin-resistant Enterococcus faecium clinical isolates in Taiwan analysis of ST types and virulence genes.
| Virulence gene | ST17 (n = 24) | ST78 (n = 17) | ST262 (n = 19) | |||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| esp | 13 | 54.2 | 10 | 58.8 | 6 | 31.6 |
| hyl | 14 | 58.3 | 4 | 23.5 | 13 | 68.4 |
| esp+hyl | 4 | 16.7 | 0 | 0.0 | 4 | 21.1 |
| none | 1 | 4.2 | 3 | 17.6 | 4 | 21.1 |
Five virulence genes were tested, including asa1, gelE, cylA, esp and hyl to determine which enterococcal virulence factor is present in clinical isolates. asa1, gelE and cylA were absent in all isolates, whereas esp and hyl were detected (Table S2). These isolates can be further classified as esp+ (40.0%, n = 24), hyl+ (33.3%, n = 20), esp+/hyl+ (13.3%, n = 8) or esp−/hyl− (13.3%, n = 8) (Table 2). Based on the isolation sites, VRE-fm carrying esp+ and/or hyl+ were isolated mainly from urine (76.6%), followed by blood (11.6%), pus (6.6%), body fluid (3.3%) and stool (1.6%). In urine samples, the percentage of esp+ isolates (39.1%) was higher than that of hyl+ strains (32.6%). The percentage of esp+/hyl+ isolates (15.2%) was higher than esp−/hyl− (13.0%). Nearly all ST17 isolates (95.8%), encoded esp+ or hyl+ (54.2% vs 58.3%). In ST78 isolates, esp+ isolates comprised the major group (58.8%), whereas hyl+ isolates were the major group in ST262 isolates (68.4%) (Table 1).
Table 2. Virulence gene identification of isolated VRE-fm strains.
| Isolated site n (%) | Virulence gene n (%) | ||||
|---|---|---|---|---|---|
| esp | hyl | esp+hyl | None of both | ||
| Urine | 46 (76.6%) | 18 (39.1%) | 15 (32.6%) | 7 (15.2%) | 6 (13.0%) |
| Blood | 7 (11.6%) | 3 (42.8%) | 2 (28.5%) | 1 (14.2%) | 1 (14.2%) |
| Body fluid | 2 (3.3%) | 1 (50.0%) | ND | ND | 1 (50.0%) |
| Stool | 1 (1.6%) | 1 (100.0%) | ND | ND | ND |
| Pus | 4 (6.6%) | 1 (25.0%) | 3 (75.0%) | ND | ND |
Note:
ND: non-detectable.
The relationship between antibiotic resistance/susceptibility and VRE-fm isolates
Isolates carrying esp and/or hyl were resistant to antibiotics, including Ampicillin, Penicillin, Levofloxacin, Ciprofloxacin, Erythromycin, and Vancomycin. Some isolates carrying esp and/or hyl showed either resistance or susceptibility to Doxycyclin, Gentamycin-Synerge, Linezolid, Minocycline, Nitrofurantoin, Quinupristin-dalfopristin, Teicoplanin, and Tetracyclin (Table S2). The esp and/or hyl were predominant in isolates sensitive to Doxycyclin (68.3%), Quinupristin-dalfopristin (88.3%), Linezolid (90.0%) and Daptomycin (100%) (Table S1). These findings indicate that despite the high degree of the virulence gene carriage, there is a lack of a clear association between antibiotic susceptibility and esp/hyl genes in VRE-fm clinical isolates.
Biofilm production in VRE-fm isolates with an esp or hyl carriage
Nearly all isolates displayed moderate to strong biofilm production (96.6%, 58/60), while only two isolates showed weak biofilm production. Among the isolates with moderate to strong biofilm production, a majority of isolates carried either esp (42%) or hyl (42%). While both virulence genes were detected in some isolates (esp/hyl, 16%), the absence of both genes were found in a few isolates (esp−/hyl−, 16%). These results indicate that esp and/or hyl carriage are/is dominant in biofilm producing VRE-fm isolates.
The effect of VRE-fm clinical isolates on C. elegans survival
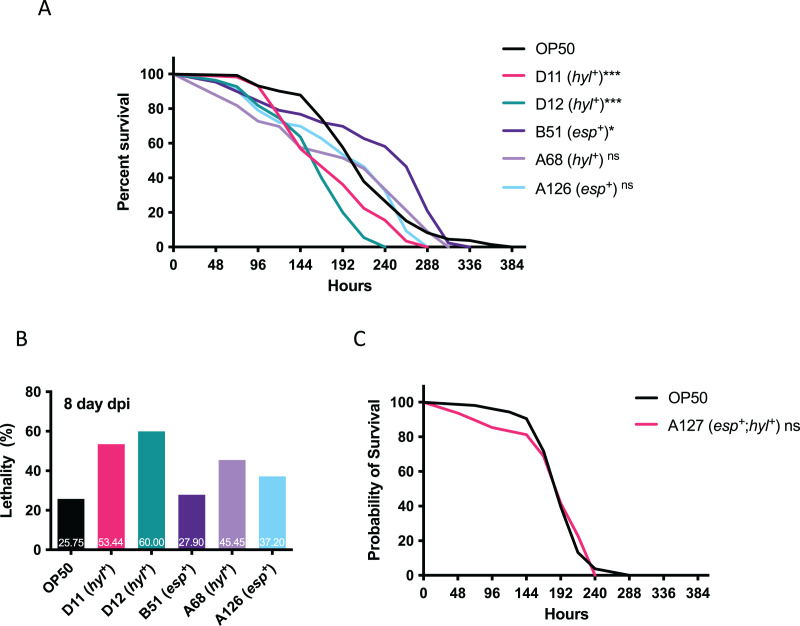
Since multi-drug resistant VRE-fm isolates confer resistance to many clinical antibiotics, this limits treatment options and requires Linezolid as a last resort. The emerging Linezolid-resistant VRE-fm is a pressing issue. To this end, Linezolid-resistant VRE-fm clinical isolates (D11, D12, B51, A68, A126) and a control strain (E. coli OP50) were examined in a C. elegans lifespan assay (Fig. 2). Two hyl+ strains (D11 and D12) reduced C. elegans lifespan (P < 0.001) (Fig. 2A) and survival at the 8th day post infection (Fig. 2B). While the C. elegans lifespan was unaffected by a hyl+ strain (A68) (P = 0.086), this isolate increased C. elegans lethality at the 8th day post infection (Fig. 2B). Two esp+ strains (B51 and A126) were mostly harmless to C. elegans.
Figure 2. Survival analysis of Linezolid-resistant/susceptible VRE-fm in the C. elegans host.
(A) Survival analyses were assayed for the C. elegans challenged by the indicated Linezolid-resistant VRE-fm (n = 132 (OP50), 58 (D11), 55 (D12), 43 (B51), 33 (A68), 46 (A126); *P < 0.05, ***P < 0.001, ns, P > 0.05). (B) Lethality of C. elegans at the 8th day after being infected were derived from (A). Percentage of each condition as indicated. (C) Survival analysis was assayed for C. elegans challenged by the Linezolid-susceptible VRE-fm A127 (n = 48) compared to the E. coli OP50 control (n = 53, P = 0.88).
A esp+/hyl+ strain with Linezolid susceptibility (A127) was used to determine whether or not Linezolid resistance in VRE-fm enhances virulence (Fig. 2C). This isolate did not affect C. elegans survival compared to the control (P = 0.88), suggesting that resistance to certain antibiotics, such as Linezolid, as well as the virulence gene, may be required for VRE-fm pathogenesis.
Discussion
Bacterial virulence factors support pathogenesis by promoting adhesion, host cell lysis, and antibiotic resistance (Leitao, 2020). Pathogenesis of enterococcal infection is achieved partly by the production of virulence factors and resistance to antibiotics. Many enterococcal virulence factors have been identified in E. faecalis and E. faecium. The distribution of putative virulence markers (PVM) has been proposed for studying the diversity of E. faecium. Whether or not these novel putative virulence factors, including Acm (adhesion of collagen from Efm), Scm (second collagen adhesion from Efm), SgrA (serine-glutamine-repeat-containing-protein A) and EcbA (Efm-collagen binding protein A), associate with infection-derived strains is unclear (Freitas et al., 2018). The activity of Esp enhances urinary tract adhesion and biofilm production, while the activity of Hyl increases fatality by promoting colonization in the gastrointestinal tract in a murine peritonitis model (Cho et al., 2018b). The current study revealed that the high frequency (76.7%) of clinical isolates was found in urine samples, suggesting a preference for VRE-fm colonization in the urinary tract. In a similar virulence gene study with 80 VRE-fm isolates, it is shown that the esp carriage (46%) is higher than the hyl carriage (20%) (Arshadi et al., 2018), while a study with 93 VRE-fm isolates, has shown that the esp carriage (60.9%) is higher than the hyl carriage (8.7%) (Say Coskun, 2019). Consistent with the previous reports, the current study revealed that the esp carriage (39.1%) was slightly higher than the hyl carriage (32.6%) in urine. The virulence gene carriage in VRE-fm may be associated with UTIs and warrants that VRE-fm be monitored for potential UTIs.
The emerging Linezolid-resistant VRE-fm is a critical issue. Linezolid is a synthetic antibiotic for effective treatment of VRE-fm infections. Although prevalence is low, Linezolid-resistant VRE-fm has been documented in different countries (Cho et al., 2018a; Wardenburg et al., 2019). Linezolid-resistant VRE-fm may cause higher morbidity and increased medical expenditure than Linezolid-sensitive VRE-fm (Turner et al., 2021), suggesting that multi-drug resistance enhances virulence (Olearo et al., 2021). Even though there are few Linezolid-resistant VRE-fm clinical isolates (5 in 60 isolates) in the current study, whether or not Linezolid-resistance genes are involved in Linezolid resistance/sensitivity of VRE-fm remains unclear and warrants further investigation. Forty six Linezolid-sensitive VREFs contain either the esp or hyl genes, while 6 VREFs contain the esp/hyl genes with Linezolid resistance (Say Coskun, 2019), implying that the relationship between drug resistance and virulence genes remains unclear.
A majority of enterococcal infections are related to biofilm formation, including catheter-related UTI, endocarditis, periodontitis and device-associated infections (Kunz Coyne et al., 2022; Rather et al., 2021a). Biofilms protect enterococci from the host immune response and antibiotics, thus biofilm-producing enterococci pose a greater risk to disease severity. Biofilm formation confers drug resistance and is associated with E. faecium pili (EmpABC) (Almohamad et al., 2014; Fallah et al., 2017). Despite the fact that several virulence genes are associated with biofilm formation, such as esp, it is unclear whether or not esp directly contributes to biofilm formation (Shridhar & Dhanashree, 2019; Tendolkar et al., 2004; Toledo-Arana et al., 2001; Weng, Ramli & Hamat, 2019).
In C. elegans, there are many enterococcal virulence factors reported, including cyl (cytolysin), epaB (Enterococcal polysaccharide antigen), fsrA/B/C (Fsr system), gelE (Gelatinase), lgt (lipoprotein diacylglyceryl transferase), paiA (Transcritional repressor), phrB (Deoxyribodipyrimidine photolyase), recQ (DNA helicase), scrB (Sucrose-6-phosphate hydrolase), and sprE (Serine protease) (Goh et al., 2017). However, the role of esp and hyl has not been studied in the C. elegans model. A high titer of E. faecium can proliferate in the C. elegans intestine but it fails to reduce the lifespan of the host (Revtovich et al., 2021). In contrast, a low inoculum of E. faecalis has caused persistent infection and kills C. elegans adults. The killing of the nematode is ascribed to the presence of virulence factors, such as the quorum-sensing system and a cytolysin (Khan, Jain & Oloketuyi, 2018). C. elegans was mostly affected by the VRE-fm isolates with the hyl gene carriage, but not the esp gene carriage. This indicates that hyl may be associated with virulence during VRE-fm infection in C. elegans. A Linezolid-sensitive VRE-fm isolate carrying both the hyl and esp genes did not reduce the C. elegans lifespan, suggesting that both Linezolid resistance and hyl are important for VRE-fm pathogenesis. The causal relationship between virulence factors and host survival requires further examination with more VRE-fm clinical isolates as well as mutants of the virulence genes. Since Enterococci often cause opportunistic infections, an immunocompromised animal would be a desired host for investigating the exact role of virulence factors in opportunistic infections.
Conclusions
VRE-fm clinical isolates obtained from regional hospitals in northern Taiwan were characterized with the intent of understanding the relationship between antimicrobial susceptibility, virulence genes and biofilm production. Most VRE-fm isolates exhibited multi-drug resistance to commonly used antibiotics and carried the virulence genes esp and hyl. However, there is a lack of association between the specific virulence gene and antibiotic resistance/susceptibility. All VRE-fm isolates were capable of producing biofilms. Isolates carrying esp showed greater biofilm production. The host survival assay indicates that C. elegans is more sensitive to Linezolid-resistant VRE-fm with hyl. This epidemiological information can be beneficial to health care providers in the management of emerging multi-drug resistant microbes. The VRE-fm-C. elegans infection model can possibly facilitate an investigation of the molecular mechanisms of novel virulence factors among the emerging multi-drug resistant VRE-fm.
Supplemental Information
Funding Statement
This work is mainly supported by a grant from the Department of Health, Taipei City Government (11001-62-031 to Pei-Yun Lin). Support was also received from the Ministry of Science and Technology of Taiwan (MOST-109-2320-B-264-001-MY2 to Hung-Chi Yang). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Po-Hsiang Chen, Email: Bob79620@hotmail.com.
Hung-Chi Yang, Email: hcyang@mail.ypu.edu.tw.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Pei-Yun Lin conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Shang-Yih Chan conceived and designed the experiments, authored or reviewed drafts of the article, and approved the final draft.
Arnold Stern performed the experiments, authored or reviewed drafts of the article, and approved the final draft.
Po-Hsiang Chen performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Hung-Chi Yang conceived and designed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
The Taipei City Hospital Institutional Review Board granted Ethical approval to carry out the study within its facilities (Ethical Approval number TCHIRB-10703123-E).
DNA Deposition
Data Availability
The following information was supplied regarding data availability:
The raw data are available in the Supplemental Files.
References
- Afonina et al. (2018).Afonina I, Lim XN, Tan R, Kline KA. Planktonic interference and biofilm alliance between aggregation substance and endocarditis- and biofilm-associated pili in Enterococcus faecalis. Journal of Bacteriology. 2018;200(24):e00361. doi: 10.1128/JB.00361-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almohamad et al. (2014).Almohamad S, Somarajan SR, Singh KV, Nallapareddy SR, Murray BE. Influence of isolate origin and presence of various genes on biofilm formation by Enterococcus faecium. FEMS Microbiology Letters. 2014;353(2):151–156. doi: 10.1111/1574-6968.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshadi et al. (2018).Arshadi M, Mahmoudi M, Motahar MS, Soltani S, Pourmand MR. Virulence determinants and antimicrobial resistance patterns of Vancomycin-resistant Enterococcus faecium isolated from different sources in Southwest Iran. Iranian Journal of Public Health. 2018;47:264–272. [PMC free article] [PubMed] [Google Scholar]
- Bello Gonzalez et al. (2017).Bello Gonzalez TDJ, Pham P, Top J, Willems RJL, van Schaik W, van Passel MWJ, Smidt H. Characterization of Enterococcus isolates colonizing the intestinal tract of intensive care unit patients receiving selective digestive decontamination. Frontiers in Microbiology. 2017;8:1596. doi: 10.3389/fmicb.2017.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepas et al. (2019).Cepas V, Lopez Y, Munoz E, Rolo D, Ardanuy C, Marti S, Xercavins M, Horcajada JP, Bosch J, Soto SM. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microbial Drug Resistance. 2019;25(1):72–79. doi: 10.1089/mdr.2018.0027. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2020).Chen PH, Chen YT, Chu TY, Ma TH, Wu MH, Lin HH, Chang YS, Tan BC, Lo SJ. Nucleolar control by a non-apoptotic p53-caspases-deubiquitinylase axis promotes resistance to bacterial infection. The FASEB Journal. 2020;34(1):1107–1121. doi: 10.1096/fj.201901959R. [DOI] [PubMed] [Google Scholar]
- Cho et al. (2018a).Cho SY, Kim HM, Chung DR, Kim SH, Huh HJ, Kang CI, Peck KR, Lee NY, Song JH. Resistance mechanisms and clinical characteristics of linezolid-resistant Enterococcus faecium isolates: a single-centre study in South Korea. Journal of Global Antimicrobial Resistance. 2018a;12:44–47. doi: 10.1016/j.jgar.2017.09.009. [DOI] [PubMed] [Google Scholar]
- Cho et al. (2018b).Cho SY, Park YJ, Cho H, Park DJ, Yu JK, Oak HC, Lee DG. Comparison of Enterococcus faecium bacteremic isolates from hematologic and non-hematologic patients: differences in antimicrobial resistance and molecular characteristics. Annals of Laboratory Medicine. 2018b;38(3):226–234. doi: 10.3343/alm.2018.38.3.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallah et al. (2017).Fallah F, Yousefi M, Pourmand MR, Hashemi A, Nazari Alam A, Afshar D. Phenotypic and genotypic study of biofilm formation in Enterococci isolated from urinary tract infections. Microbial Pathogenesis. 2017;108(4):85–90. doi: 10.1016/j.micpath.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Freitas et al. (2018).Freitas AR, Tedim AP, Novais C, Coque TM, Peixe L. Distribution of putative virulence markers in Enterococcus faecium: towards a safety profile review. Journal of Antimicrobial Chemotherapy. 2018;73(2):306–319. doi: 10.1093/jac/dkx387. [DOI] [PubMed] [Google Scholar]
- Goh et al. (2017).Goh HMS, Yong MHA, Chong KKL, Kline KA. Model systems for the study of enterococcal colonization and infection. Virulence. 2017;8(8):1525–1562. doi: 10.1080/21505594.2017.1279766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gok et al. (2020).Gok SM, Turk Dagi H, Kara F, Arslan U, Findik D. Investigation of antibiotic resistance and virulence factors of Enterococcus faecium and Enterococcus faecalis strains isolated from clinical samples. Mikrobiyoloji Bulteni. 2020;54(1):26–39. doi: 10.5578/mb.68810. [DOI] [PubMed] [Google Scholar]
- Hashem, Abdelrahman & Aziz (2021).Hashem YA, Abdelrahman KA, Aziz RK. Phenotype-genotype correlations and distribution of key virulence factors in Enterococcus faecalis isolated from patients with urinary tract infections. Infection and Drug Resistance. 2021;14:1713–1723. doi: 10.2147/IDR.S305167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan et al. (2002).Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, Van Embden JD, Willems RJ. Multilocus sequence typing scheme for Enterococcus faecium. Journal of Clinical Microbiology. 2002;40(6):1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbinosa & Beshiru (2019).Igbinosa EO, Beshiru A. Antimicrobial resistance, virulence determinants, and biofilm formation of Enterococcus species from ready-to-eat seafood. Frontiers in Microbiology. 2019;10:728. doi: 10.3389/fmicb.2019.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic et al. (2018).Jovanovic M, Tosic T, Jovanovic S, Stosovic R, Stevanovic G, Velebit B, Zervos MJ. Presence of the esp gene in Enterococcus faecium derived from oropharyngeal microbiota of haematology patients. Archives of Oral Biology. 2018;88(11):54–59. doi: 10.1016/j.archoralbio.2018.01.008. [DOI] [PubMed] [Google Scholar]
- Kaito et al. (2020).Kaito C, Murakami K, Imai L, Furuta K. Animal infection models using non-mammals. Microbiology and Immunology. 2020;64(9):585–592. doi: 10.1111/1348-0421.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariyama et al. (2000).Kariyama R, Mitsuhata R, Chow JW, Clewell DB, Kumon H. Simple and reliable multiplex PCR assay for surveillance isolates of Vancomycin-resistant Enterococci. Journal of Clinical Microbiology. 2000;38(8):3092–3095. doi: 10.1128/JCM.38.8.3092-3095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, Jain & Oloketuyi (2018).Khan F, Jain S, Oloketuyi SF. Bacteria and bacterial products: foe and friends to Caenorhabditis elegans. Microbiological Research. 2018;215(3):102–113. doi: 10.1016/j.micres.2018.06.012. [DOI] [PubMed] [Google Scholar]
- Kunz Coyne et al. (2022).Kunz Coyne AJ, Stamper K, Kebriaei R, Holger DJ, El Ghali A, Morrisette T, Biswas B, Wilson M, Deschenes MV, Canfield GS, Duerkop BA, Arias CA, Rybak MJ. Phage cocktails with daptomycin and ampicillin eradicates biofilm-embedded multidrug-resistant Enterococcus faecium with preserved phage susceptibility. Antibiotics. 2022;11(9):1175. doi: 10.3390/antibiotics11091175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo et al. (2018).Kuo AJ, Shu JC, Liu TP, Lu JJ, Lee MH, Wu TS, Su LH, Wu TL. Vancomycin-resistant Enterococcus faecium at a university hospital in Taiwan, 2002-2015: fluctuation of genetic populations and emergence of a new structure type of the Tn1546-like element. Journal of Microbiology, Immunology and Infection. 2018;51(6):821–828. doi: 10.1016/j.jmii.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2019).Lee T, Pang S, Abraham S, Coombs GW. Antimicrobial-resistant CC17 Enterococcus faecium: the past, the present and the future. Journal of Global Antimicrobial Resistance. 2019;16(Suppl. 1):36–47. doi: 10.1016/j.jgar.2018.08.016. [DOI] [PubMed] [Google Scholar]
- Leitao (2020).Leitao JH. Microbial virulence factors. International Journal of Molecular Sciences. 2020;21(15):5320. doi: 10.3390/ijms21155320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liese et al. (2019).Liese J, Schule L, Oberhettinger P, Tschorner L, Nguyen T, Dorfel D, Vogel W, Marschal M, Autenrieth I, Willmann M, Peter S. Expansion of Vancomycin-resistant Enterococcus faecium in an academic tertiary hospital in Southwest Germany: a large-scale whole-genome-based outbreak investigation. Antimicrobial Agents and Chemotherapy. 2019;63(5):e01978. doi: 10.1128/AAC.01978-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Lin & Lan (2020).Lin MF, Lin YY, Lan CY. Characterization of biofilm production in different strains of Acinetobacter baumannii and the effects of chemical compounds on biofilm formation. PeerJ. 2020;8:e9020. doi: 10.7717/peerj.9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo et al. (2021).Luo Y, Yang Q, Zhang D, Yan W. Mechanisms and control strategies of antibiotic resistance in pathological biofilms. Journal of Microbiology and Biotechnology. 2021;31(1):1–7. doi: 10.4014/jmb.2010.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar et al. (2018).Manandhar S, Singh A, Varma A, Pandey S, Shrivastava N. Evaluation of methods to detect in vitro biofilm formation by staphylococcal clinical isolates. BMC Research Notes. 2018;11(1):714. doi: 10.1186/s13104-018-3820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux et al. (2020).Michaux C, Hansen EE, Jenniches L, Gerovac M, Barquist L, Vogel J. Single-nucleotide RNA maps for the two major nosocomial pathogens Enterococcus faecalis and Enterococcus faecium. Frontiers in Cellular and Infection Microbiology. 2020;10:2246. doi: 10.3389/fcimb.2020.600325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller et al. (2020).Miller WR, Murray BE, Rice LB, Arias CA. Resistance in Vancomycin-resistant Enterococci. Infectious Disease Clinics of North America. 2020;34(4):751–771. doi: 10.1016/j.idc.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olearo et al. (2021).Olearo F, Both A, Belmar Campos C, Hilgarth H, Klupp EM, Hansen JL, Maurer FP, Christner M, Aepfelbacher M, Rohde H. Emergence of linezolid-resistance in Vancomycin-resistant Enterococcus faecium ST117 associated with increased linezolid-consumption. International Journal of Medical Microbiology. 2021;311(2):151477. doi: 10.1016/j.ijmm.2021.151477. [DOI] [PubMed] [Google Scholar]
- Panesso et al. (2011).Panesso D, Montealegre MC, Rincon S, Mojica MF, Rice LB, Singh KV, Murray BE, Arias CA. The hylEfm gene in pHylEfm of Enterococcus faecium is not required in pathogenesis of murine peritonitis. BMC Microbiology. 2011;11(1):20. doi: 10.1186/1471-2180-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay, Zishiri & Adeleke (2018).Pillay S, Zishiri OT, Adeleke MA. Prevalence of virulence genes in Enterococcus species isolated from companion animals and livestock. Onderstepoort Journal of Veterinary Research. 2018;85(1):e1–e8. doi: 10.4102/ojvr.v85i1.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell et al. (2018).Powell LC, Pritchard MF, Ferguson EL, Powell KA, Patel SU, Rye PD, Sakellakou SM, Buurma NJ, Brilliant CD, Copping JM, Menzies GE, Lewis PD, Hill KE, Thomas DW. Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. NPJ Biofilms Microbiomes. 2018;4(1):13. doi: 10.1038/s41522-018-0056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather et al. (2021a).Rather MA, Gupta K, Bardhan P, Borah M, Sarkar A, Eldiehy KSH, Bhuyan S, Mandal M. Microbial biofilm: a matter of grave concern for human health and food industry. Journal of Basic Microbiology. 2021a;61(5):380–395. doi: 10.1002/jobm.202000678. [DOI] [PubMed] [Google Scholar]
- Rather, Gupta & Mandal (2021b).Rather MA, Gupta K, Mandal M. Microbial biofilm: formation, architecture, antibiotic resistance, and control strategies. Brazilian Journal of Microbiology. 2021b;52(4):1701–1718. doi: 10.1007/s42770-021-00624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revtovich et al. (2021).Revtovich AV, Tjahjono E, Singh KV, Hanson BM, Murray BE, Kirienko NV. Development and characterization of high-throughput Caenorhabditis elegans—Enterococcus faecium infection model. Frontiers in Cellular and Infection Microbiology. 2021;11:667327. doi: 10.3389/fcimb.2021.667327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Say Coskun (2019).Say Coskun US. Investigation of the relationship between virulence factors and antibiotic resistance of Enterococci isolates. Cellular and Molecular Biology. 2019;65(2):14–17. doi: 10.14715/cmb/2019.65.2.3. [DOI] [PubMed] [Google Scholar]
- Shridhar & Dhanashree (2019).Shridhar S, Dhanashree B. Antibiotic susceptibility pattern and biofilm formation in clinical isolates of Enterococcus spp. Interdisciplinary Perspectives on Infectious Diseases. 2019;2019(5):7854968. doi: 10.1155/2019/7854968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanovic et al. (2007).Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, Ruzicka F. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. APMIS. 2007;115(8):891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- Tacconelli et al. (2018).Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infectious Diseases. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- Tendolkar et al. (2004).Tendolkar PM, Baghdayan AS, Gilmore MS, Shankar N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infection and Immunity. 2004;72(10):6032–6039. doi: 10.1128/IAI.72.10.6032-6039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana et al. (2001).Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penades JR, Lasa I. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Applied and Environmental Microbiology. 2001;67(10):4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner et al. (2021).Turner AM, Lee JYH, Gorrie CL, Howden BP, Carter GP. Genomic insights into last-line antimicrobial resistance in multidrug-resistant Staphylococcus and Vancomycin-resistant Enterococcus. Frontiers in Microbiology. 2021;12:637656. doi: 10.3389/fmicb.2021.637656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin & Paterson (2020).van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: an update. Infectious Disease Clinics of North America. 2020;34(4):709–722. doi: 10.1016/j.idc.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankerckhoven et al. (2004).Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in Enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. Journal of Clinical Microbiology. 2004;42(10):4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardenburg et al. (2019).Wardenburg KE, Potter RF, D’Souza AW, Hussain T, Wallace MA, Andleeb S, Burnham CD, Dantas G. Phenotypic and genotypic characterization of linezolid-resistant Enterococcus faecium from the USA and Pakistan. Journal of Antimicrobial Chemotherapy. 2019;74(12):3445–3452. doi: 10.1093/jac/dkz367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, Ramli & Hamat (2019).Weng PL, Ramli R, Hamat RA. Antibiotic susceptibility patterns, biofilm formation and esp gene among clinical Enterococci: is there any association? International Journal of Environmental Research and Public Health. 2019;16(18):3439. doi: 10.3390/ijerph16183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin et al. (2019).Yin W, Wang Y, Liu L, He J. Biofilms: the microbial “protective clothing” in extreme environments. International Journal of Molecular Sciences. 2019;20(14):3423. doi: 10.3390/ijms20143423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw data are available in the Supplemental Files.